Abstract
There is a need for new therapeutic targets with which to prevent Alzheimer’s disease (AD), a major contributor to aging-related cognitive decline. Here, we report the construction and validation of a molecular network of the aging human frontal cortex. Using RNA sequence data from 478 individuals, we first build a molecular network using modules of coexpressed genes and then relate these modules to AD and its neuropathologic and cognitive endophenotypes. We confirm these associations in two independent AD datasets as well as in epigenomic data. We also illustrate the use of the network in prioritizing amyloid-associated genes for in vitro validation in human neurons and astrocytes. These analyses based on unique cohorts enable us to resolve the role of distinct cortical modules that have a direct effect on the accumulation of AD pathology from those that have a direct effect on cognitive decline, exemplifying a network approach to complex diseases.
One sentence summary:
Systems biology analysis of RNA sequencing data from the aging human cortex identifies a molecular network which prioritizes groups of genes that influence cognitive decline or neuropathology in Alzheimer’s disease.
Introduction
The incidence of late-onset Alzheimer’s disease (AD) is expected to triple by 20501, yet no therapies are available to treat or prevent the disease2. Possible reasons for the continued failure of AD trials include the biological complexity of the disease and its phenotypic heterogeneity3. Recent genome-wide association studies (GWAS) have identified new potential therapeutic targets involved in endocytosis, metabolism, and inflammation4. However, it has been difficult to transition from mostly non-coding susceptibility variants to molecular mechanisms that lead to the characteristic accumulation of β-amyloid and helical filament tau (PHFtau) pathology as well as the subsequent cognitive decline of AD.
Here, we describe an analysis of participants from two large, longitudinal cohort studies of aging (total n=478), as well as a validation set (n=82), which have careful assessments of both antemortem cognitive function and postmortem neuropathologic burden. We hypothesized that using RNA-sequencing (RNA-Seq) data from the dorsal lateral frontal cortex (DLPFC) would allow us to identify coherent intermediate cellular mechanisms that are associated with cognitive decline and/or neuropathological changes. Noting that dynamic genomic measurements such as RNA-Seq reflect the collective effect of upstream, downstream, and disease-correlated processes, we use a series of network-based approaches that account for known and hidden confounding factors in order to enrich our results for likely upstream associations.
A network-based perspective provides a more nuanced molecular definition of complex disease than do traditional single-gene associations because it provides a natural framework with which to assemble disparate single gene findings into disease mechanisms5–8. Transcriptomic data can be used to identify groups of coexpressed genes (or modules) that represent cellular processes and can be related to phenotypes of interest7,9–11. This approach offers an unsupervised and tissue-specific perspective that identifies transcriptional programs related to disease phenotypes, independent of historical bias arising from research on particular genes and pathways10.
Coexpression approaches have identified genes associated with a syndromic diagnosis of AD dementia12,13. However, these approaches have never been applied to large cohorts with quantitative measurements of both AD neuropathology and cognitive decline. Because neuropathologic burden and cognition show important divergence in AD14,15, jointly modeling both aspects of the disease may better capture the relationship between molecular events and different stages of the disease process. Further, previous efforts did not distinguish between transcriptomic patterns that are indirectly associated with AD phenotypes, via a chain of intermediaries from those that are directly associated12. Here, we build on prior work to address these limitations. Our approach, called gene module trait network analysis (MTN) (Figure 1), constructs gene expression modules and identifies those that are directly associated with cognitive decline, conditioned on neuropathology and other large-scale transcriptomic changes in the aging brain. We confirm the biological plausibility of this systems biology analysis in five other types of independent datasets. Finally, we test the identified associations in a relevant human model system to functionally characterize the role of selected genes with regards to amyloid-β levels (a measurable endophenotype in an experimental system) and to prioritize candidates for further evaluation as possible therapeutic targets.
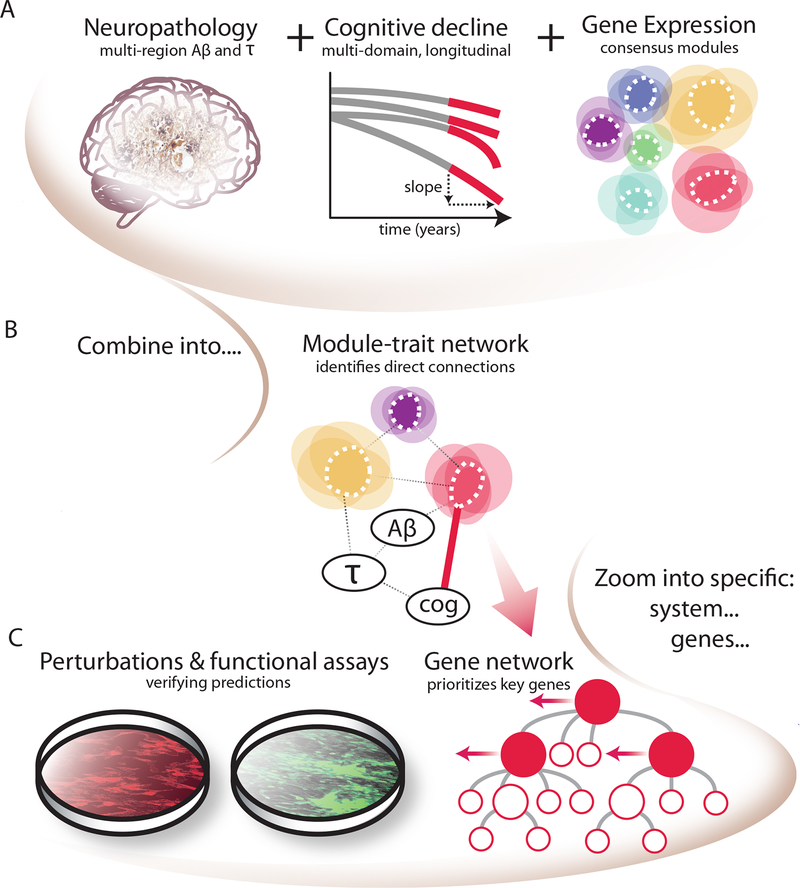
Figure 1. Schematic of the implementation of the module-trait network (MTN) method to prioritize modules and genes directly related to AD-related traits in our study.
(A) Inputs to the MTN method are: 1) AD pathological traits of amyloid and tau measurements, which are aggregated over multiple brain regions; 2) slope of cognitive decline prior to death aggregated over multiple neuropsychologic tests; and 3) average expression of coexpressed gene sets (modules), detected in the same subjects using consensus clustering. (B) These three inputs are combined using conditional independence relationships (via Bayesian networks) to identify direct relationships among coexpression modules, AD traits and cognitive decline. (C) The disease relevance of top predicted genes were tested experimentally in an astrocyte and iPSC-induced neuron in vitro system.
Results
Data origin and phenotypes
Data were derived from subjects enrolled in the Religious Orders Study (ROS) or the Rush Memory and Aging Project (MAP), two prospective clinical-pathologic cohort studies of aging and dementia. All participants are non-demented at enrolment, have annual clinical evaluations and have agreed to brain donation. At death, each brain undergoes a structured, quantitative neuropathologic assessment (see Supplementary methods). The two studies (collectively referred to as ROSMAP) share clinical and neuropathological standards, allowing for joint analyses of the data. Individual trajectories of cognitive decline are calculated from longitudinal cognitive measures that include up to 20 yearly evaluations16,17. For this study, we used data from 478 participants, with a mean age at death was 88.7 years. Over the course of the study, some subjects experience cognitive decline, and, at the time of death, 32% remain cognitively non-impaired, 27% have mild cognitive impairment (MCI), 39% have a diagnosis of AD dementia, and 2% have another form of dementia. Further, 58% of these subjects received a diagnosis of pathologic AD. However, 46% of these individuals with pathologic AD are clinically non-demented, illustrating the well-described divergence of pathologic and clinical diagnoses of AD dementia (Table S1).
Our analysis includes five phenotypic traits related to AD: two of these traits are clinical measures - a clinical diagnosis of AD dementia proximate to death (ClinAD), and a continuous measure of cognitive decline over time quantified as a per-subject slope of the cognitive decline trajectory from a linear mixed-effects model18 (“cognitive decline” abbreviated as “Cog Dec”). The three pathology variables include continuous measures of PHFtau tangle density and β-amyloid burden (both averaged over multiple regions) and a binary diagnosis of pathologic AD (PathoAD) (See the Supplementary methods for details).
In these subjects, we see the expected strong association of APOE ε4 with ClinAD (p=5.55×10−16; used logistic regression to model the number of ε4 while accounting for age and sex), but even this unique genetic risk factor explains little of the variance in ClinAD (2.2% variance explained) or cognitive decline (5.1% variance explained). Subsequent inclusion of the other 21 AD susceptibility variants to represent the known genetic architecture of AD only explains 2.1% of ClinAD and 7.6% of cognitive decline. Thus, while these robustly validated susceptibility variants provide important insights into risk factors that contribute to AD, they capture only a small fraction of the biology of the disease, much of which may be influenced by non-genetic risk factors. The transcriptome, with its dynamic nature that is molded by environmental exposures and life experiences, provides a complementary approach to therapeutic target discovery in AD.
RNA-Seq gene expression data and standard association analysis
After rigorous quality control evaluations, we retained RNA-Seq data from the dorsolateral prefrontal cortex (DLPFC) of 478 individuals for downstream analyses; an average of 95 million (median 90 million) paired-end reads was available for each subject. RNA-Seq data were then normalized to account for the effect of a large number of known biological and technical confounding factors (Supplementary Methods; Figure S1). Genes with low expression were removed to reduce the influence of technical noise, resulting in quantified expression for 13,484 unique genes (25,400 transcripts) (Supplementary methods).
We first performed a standard transcriptome-wide association study (TWAS) to identify genes whose expression levels associate with AD-related phenotypes. We found that the expression levels of thousands of genes associate (FDR<0.05) with these traits (Table S2; Figure 2A). The combination of cognitive and pathological phenotypes allows us to compare their respective effects on gene expression, and we observed that cognitive decline was associated with the largest number of genes (3,025 genes at FDR<0.05), compared to the other AD traits. Accordingly, the key clinical variable of cognitive decline may implicate additional molecular mechanisms in AD, beyond those found by neuropathological markers of AD. Indeed a majority of age-related cognitive decline cannot be accounted for by current measures of AD or other age-related pathologies19. Overall, using the π1 statistic20, we estimated that 55–90% of the associated genes are shared amongst these correlated AD-related (Figure S2). The large number of associated genes highlights the need to assemble these findings into coherent biological processes that are directly associated with specific disease-relevant endpoints.
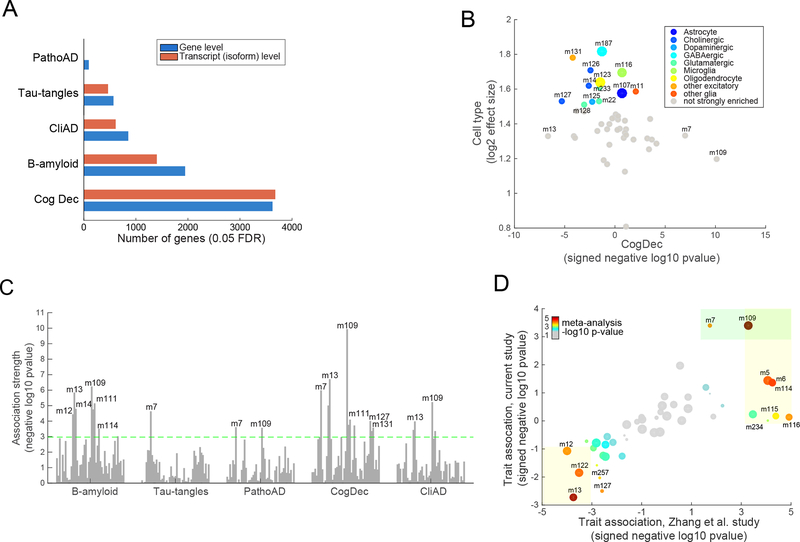
Figure 2. Characterization of human cortical RNA-Seq data and their relation with AD traits and cellular processes.
(A) Figure shows the number of genes whose gene expression levels significantly associate with each of the five tested AD-related traits (0.05 FDR). Results are shown for association testing at the “gene level” and “transcript level” (isoform level). (B) This figure shows the enrichment of each of the 47 modules for cell-specific signature genes defined in mice (y-axis) (see Supplementary methods). The X-axis shows the signed association strength (signed -log10 pvalue) between each module and cognitive decline. Larger sized points (modules) are those that we deem to most strongly represent each of the four major brain cell types: neurons (m187), microglia (m116), oligodendrocytes (123), and astrocytes (m107) (see Supplementary methods for details). (C) Figure shows the association strength (quantified as negative log10 pvalue) between each of the 47 modules of coexpressed genes (visualized as vertical bars) and each of the five tested AD-related trait (x axis). The dashed line depicts the Bonferroni-adjusted significance threshold at the module-level (p<0.001). Only some of the modules that pass Bonferroni threshold are labeled, for visualization ease. (D) This figure shows the strength and direction of each module’s association (signed negative log10 pvalue) for association with a binary diagnosis of pathological AD (PathoAD) in our ROSMAP study on the y-axis. The x-axis shows the signed association strengths between each module and pathological AD in an existing microarray dataset (Zhang et al. dataset)22. Specifically, the modules are defined using the ROSMAP samples, and their definitions are projected onto the Zhang et al. study. The size of each point represents the size of each module (i.e., number of assigned genes). The color of the point is proportional to the significance of the association in a meta-analysis of the ROSMAP and Zhang modules (using the gradient shown in the upper left portion of the graph). The light orange boxes highlight those modules that are significantly associated with PathoAD diagnosis in the microarray (“Zhang”) dataset. The green box highlights the modules that are significantly associated with a PathoAD diagnosis in the ROSMAP data.
Constructing the nodes of our module-trait network
In order to identify coherent cellular processes that impact AD phenotypes, we utilize a module-trait network (MTN) approach. The goal of this approach is to go beyond single gene level associations in defining robust molecular mechanisms while avoiding the limitations of pathways derived from ontology databases. MTN summarizes large-scale transcriptome changes into gene modules and prioritizes specific genes within a module for additional experiments. MTN consists of three steps. As described in detail below, we first identify groups of coexpressed genes or “modules”, which we validate in other datasets. Coexpressed gene sets represent the outcome of transcriptional regulatory mechanisms that include transcription factors, chromatin conformation and latent factors that generate correlations, such as the proportion of different cell types present in the sampled tissue10. In the second step, we identify which modules have direct relationships with cognitive decline and other AD traits using Bayesian networks to prune correlations between modules and AD-related traits that are indirect. In the final analytical step, we select a top-scoring module and prioritize genes that are uniquely influential within that module for validation in our in vitro model systems.
In the first step of MTN, we applied the SpeakEasy consensus clustering algorithm21 to our RNA-Seq data and identified 47 mutually exclusive modules ranging in size between 20 and 556 gene members (Table S3; Figure S3; Supplementary methods). We note that the clustering assignment identified by the SpeakEasy algorithm overlapped significantly with those proposed by the frequently used WGCNA algorithm22 (Supplementary methods; Table S4); however, we chose to use SpeakEasy because of its state-of-the-art performance on benchmark and real-world datasets21.
We validated the biological coherence of the 47 modules from 5 perspectives: (1) Gene Ontology (GO) functional enrichment analysis, (2) module coherence (“preservation”) in a separately processed set of ROSMAP samples and an entirely independent cohort, (3) concordance with co-regulation observed in epigenomic data generated from the same ROSMAP brains, (4) concordance with brain gene expression data from multiple AD mouse models, and (5) cell type-specific expression. In the first validation, we find that 29 (62%) of the modules were enriched for at least one GO functional category (Bonferroni p<0.01, Table S5), which is in the range of enrichments seen in other studies10. Second, we assessed whether the 47 modules are preserved in: (a) a previously published DLPFC microarray dataset of 229 persons with pathologic and clinical AD12 and (b) RNA-Seq data from an independent set of 82 ROSMAP subjects. Using the Z-summary statistic that summarizes multiple measures of module preservation23, we observed significant evidence for preservation in 45 of the 47 modules in the independent microarray dataset and all 47 modules in the separately-processed ROSMAP subjects (Figure S4A). Third, we assessed the robustness of these modules from a transcriptional regulatory perspective with H3K9Ac ChIP-Seq data generated from the same DLPFC samples. The H3K9Ac histone mark is found near active transcription start sites and enhancers24, and so coacetylation would be expected for genes that are coexpressed at the RNA level. In our data, we detected coacetylation for 32 of the 47 RNA-Seq-defined modules, indicating that the modules reflect, in part, the underlying epigenomic architecture (Figure S4A). Fourth, we also found that 31% of the modules are significantly preserved (Bonferroni adjustment) in gene expression data derived from the cortex of several pathology-based mouse models of AD25 (Figure S4A), which may help to prioritize those results that can be pursued further in mouse models. Fifth, we find that 13 modules are strongly enriched for cell-type specific genes, including those from microglia, astrocytes, oligodendrocytes and two neuronal subtypes (GABAergic and pyramidal neurons) (Figure 2B, Table S6) based on purified neuronal and non-neuronal cell populations, derived from the mouse brain26 (Supplementary methods). Thus, some of the modules capture previously identified cell-specific gene sets. Each of these 5 validations in parallel and orthogonal datasets indicates that gene membership of these modules is reproducible and biologically meaningful.
We next assessed the association between each module and each phenotypic trait, (Supplementary methods). Overall, 11 modules were associated (Bonferroni adjusted at module level, p<10−3) with cognitive decline, AD dementia, or AD pathology traits (Figure 2C, Table S7). Consistent with the gene-level results, cognitive decline was associated with the greatest number of modules. In general, modules enriched for GO categories related to immunity, mitochondria, cell cycle and transcriptional regulation show a positive correlation with β-amyloid, AD and cognitive decline, while modules for neuronal or synaptic function showed a negative correlation. To replicate the phenotypic associations of these modules, we projected them onto the largest previous AD brain microarray study12 (the “Zhang study”) (Supplementary methods). Only a pathologically confirmed diagnosis of AD dementia is available in the Zhang study as an outcome, and we observed a strong concordance in both the strength and direction of the module-pathologic AD associations that we defined in ROSMAP subjects (Figure 2D). Notably, we observed that the module-level trait associations were more similar between these two datasets than were the univariate gene-level trait associations, further reinforcing the utility of the module-level approach (Figure S4B).
Because variation in cell type proportions across individuals can drive coexpression patterns between genes, some module-trait associations may be due to changes in cell type proportions that occur over the course of the disease (Figure S5A–B). Importantly, as we describe next, the identification of “cell type-specific modules” enabled us to guide functional validation efforts and to comprehensively model and account for changes in cell type composition that may drive large-scale transcriptomic changes in AD at the bulk tissue level.
Identifying modules associated with disease in networks of the aging human brain
To separate a small number of direct module-trait associations from the larger number of indirect module-trait correlations, we used Bayesian network (BN) inference27. A BN models the joint probabilities of a set of random variables as a directed acyclic graph. Here, the random variables represent module expression levels and trait values across individuals. Edges in a BN represent direct conditional dependencies between two variables: an arrow from X and Y in a BN indicates that a value taken by variable Y depends on the value taken by variable X, conditioned on all the other variables in the BN (see Supplementary methods).
To limit the network size for more accurate inference, only modules associated with any of the three main AD-related traits (β-amyloid load, tau tangle density, and cognitive decline) were included. We also included four modules representing the proportions of four major brain cell types - microglia, astrocytes, oligodendrocytes, and neurons – to account for the effects of changes in cell population frequencies (Supplementary methods). In summary, the resulting BN consisted of 18 nodes: 11 nodes representing trait-associated modules, 3 trait nodes, and 4 “cell type modules” (Figure 3A).
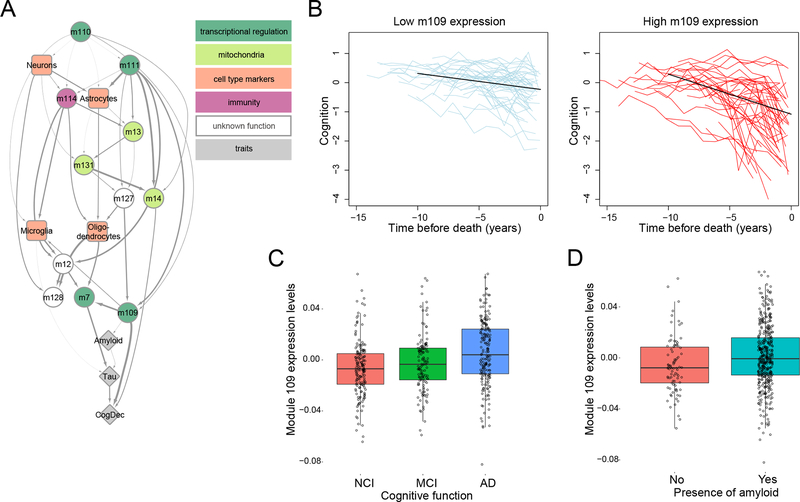
Figure 3. The AD network model prioritizes module 109 as being directly associated with cognitive decline.
(A) A directed acyclic graph (DAG), learned using Bayesian network structure learning, represents the relationships between modules (circles), cell type markers (specific modules, squares), and three relevant AD traits (B-amyloid, Tau-tangles, and CogDec)(diamonds). The thickness of the arrow is proportional to the number of times that a connection was detected (Supplementary methods). The color of the modules relates to annotations using the Gene Ontology database (see Table S5 for details); a color key is found at the upper right aspect of the image. (B) This figure shows the relation of high m109 expression level with more rapid cognitive decline. The two panels present trajectories of cognitive decline for people with low (left panel) or high (right panel) levels of m109 expression. On the left, the segmented light blue lines (“spaghetti plots”) show the annual global cognition scores for 50 randomly selected participants with low m109 expression level (1st quartile) to illustrate the nature the of the longitudinal cognitive data; the solid black line reports the trajectory of cognitive decline for a typical female participant with 85 years of age at the time of death, 15 years of education and mean expression level for the group (approximate to the 10th percentile of m109 expression). On the right panel, the red spaghetti plots show the annual global cognition scores for 50 randomly selected participants with high expression level (4th quartile), and superimposed in black is the model derived cognitive trajectory for a typical female participant with 85 years of age at the time of death, 15 years of education and mean expression level for the group (approximate to the 90th percentile). (C) This boxplot shows expression levels of module 109 (mean expression of genes assigned to m109) for individuals who have no cognitive impairment (NCI)(red), mild cognitive impairment (MCI)(green), or an AD diagnosis (AD)(blue). Each point represents one individual. (D) This boxplot shows expression level of module 109 for individuals without (red box) and with (turquoise box) amyloid deposition at autopsy. Each point represents one individual.
As shown in Figure 3A, module 109 (m109) is the module most strongly associated with cognitive decline, conditioned on all other correlated modules and modules that represent cell type proportions. It consists of 390 genes with diverse functions. Prominent functions that are enriched in this module relate to the regulation of the cell cycle and chromatin modification (Table S5). The association of m109 with cognitive decline replicates in the independently processed set of 82 ROSMAP subjects (p=0.006).
Relation of RNA derived modules to the genetic architecture of AD
The APOE ε4 haplotype has a unique role in the disease given its large effect size (odds ratio > 3 for one copy of the APOE ε4 haplotype) and substantial frequency in human populations28. Therefore, we evaluated its effect on m109 and found that APOE ε4 has a modest association with higher m109 expression (nominal p=0.03, Wilcoxon test, Figure S6), consistent with this haplotype’s known associations with accumulation of β-amyloid pathology and cognitive decline29. In a conditional association analysis (Supplementary methods), after accounting for the APOE ε4 haplotype, m109 remains strongly associated with both pathological AD (effect magnitude reduced by 13%, adjusted p=0.0027) and cognitive decline (effect magnitude reduced by 7.4%, adjusted p=5.03×10−10), implying that the effect of m109 is largely independent of this susceptibility haplotype. We extended our genetic analysis of m109 to other common AD susceptibility variants4 which, in earlier studies, displayed very little or no association with neuropathologic features of AD30,31. The 21 common AD variants did not associate with m109 (i.e., trans-eQTL analysis), consistent with the observed sparsity of trans-associations (trans-eQTLs) reported in studies with larger sample sizes32.
We also assessed whether any of the modules are enriched for genes found in the vicinity of AD variants (nearest genes, as previously reported4) and found one enriched module, m116, (p=0.0018 using the INRICH algorithm33) which mostly contains microglial genes (Table S6). However, m116 is not directly associated with cognitive decline, AD pathology, or AD dementia in this dataset (Table S7). But it is associated with age (p=0.003), highlighting the important role of advancing age in AD susceptibility and the fact that AD GWAS studies use a clinical diagnosis of AD and often younger control subjects to achieve very large sample sizes.
Finally, for completeness, we tested the association between the 21 common AD susceptibility alleles4 and the 47 modules (i.e., a module-QTL analysis) and found no significant associations after multiple testing correction, consistent with the observation noted earlier that, while these susceptibility alleles are robust risk factors, they capture only a small fraction of the variance in these AD-related traits.
Prioritizing genes in module 109 and testing their effect on extracellular β-amyloid levels
Because of m109’s strong direct association with cognitive decline (p<10−9) (Figure 3B and 3C), we elected to focus our functional evaluations on this module. In addition to cognitive decline, m109 is associated with β-amyloid pathology (p<0.0001) (Figure 3A and D, Table S7), but we note that a conditional correlation analysis indicated that the effect of m109 on cognitive decline is not fully mediated by the accumulation of β-amyloid. That is, m109 influences cognitive decline through both β-amyloid and non-β-amyloid processes. Due to the absence of a cellular model of cognitive decline, we focused our validation effort on m109’s association with β-amyloid load.
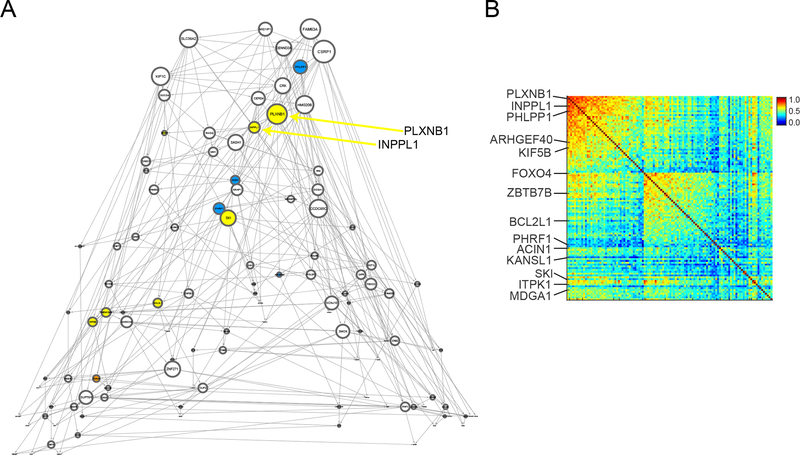
To prioritize genes in m109 for functional validation, we used several criteria: gene network connectivity (Figure 4A and B, Table S8), sufficient expression levels in cultured human astrocytes and/or induced human neurons (our two experimental systems), the magnitude of the gene-level association with our three AD-related phenotypes, and existing knowledge about gene function. We identified 21 genes within m109 that satisfied these criteria and were selected for experimental perturbation (see Supplementary methods).
Figure 4. Identifying specific genes within m109 for experimental follow-up.
(A) This figure shows the estimated gene regulatory network (Bayesian Network) for 112 selected genes in module 109. Each gene is a node (circle) in the displayed graph. Colored nodes are those that are tested in our experimental systems (yellow: tested in both Astrocytes and iPSC-derived neurons (iN), blue: only tested in Astrocytes, orange: only tested in iNs). The size of each node is proportional to its node degree (total number of ingoing and outgoing edges per node). (B) This figure shows the coexpression values for the 112 genes shown in Figure 3A, highlighting the fact that there is sub-structure within the coexpression pattern of m109. Genes that are tested in our experimental systems are highlighted and are found in each subset of the correlation matrix.
For the 21 selected genes, an average of 5 shRNA constructs targeting each selected gene were tested for their knock down of gene expression (Table S9). To be included in our functional screen, genes had to have at least two shRNA constructs meeting our knockdown efficacy criteria of >50% reduction in expression in the target cell type. 12 genes (with 37 shRNA constructs) met this criterion in neurons and 14 genes (with 41 shRNA constructs) met this criterion in astrocytes, 11 genes were tested in both cell types (Table S10).
We performed 78 shRNA experiments to knock down (KD) the selected 14 genes in astrocytes and 12 genes in neurons. With these KD experiments, we measured extracellular levels of the pathogenic β-amyloid Aβ42 peptide, that can be readily assayed in vitro and is related to the defining pathologic lesion in AD: we measured it in conditioned media from astrocyte cultures as well as iPSC-derived neuronal cultures following gene perturbation (see Supplementary methods). These experiments included three types of negative controls, including empty vectors and vectors that only contained GFP (see Supplementary methods). Since m109 is positively correlated with β-amyloid burden (Table S7), the MTN approach predicts that a knockdown of expression would result in reduced β-amyloid levels.
Using an ANOVA model that accounted for testing multiple constructs per gene (Supplementary methods), we evaluated the effect of each shRNA construct on extracellular Aβ42 levels in contrast to the negative controls. In neurons, this outcome measure was not altered. By contrast, we identified two shRNA constructs targeting different genes, INPPL1 and PLXNB1, that exceeded the Bonferroni threshold of significance (p< 0.0012) in astrocytes (Figure 5A; Table S11) in our discovery screen. Two additional constructs for these genes were found to meet a suggestive threshold (p<0.024, defined as one over the number of tests). Notably, these two genes are predicted to be upstream of the other tested genes in our BN model (Figure 4A), are directly connected to one another and are two of the major hubs in the m109 coexpression network (Figure 4B).
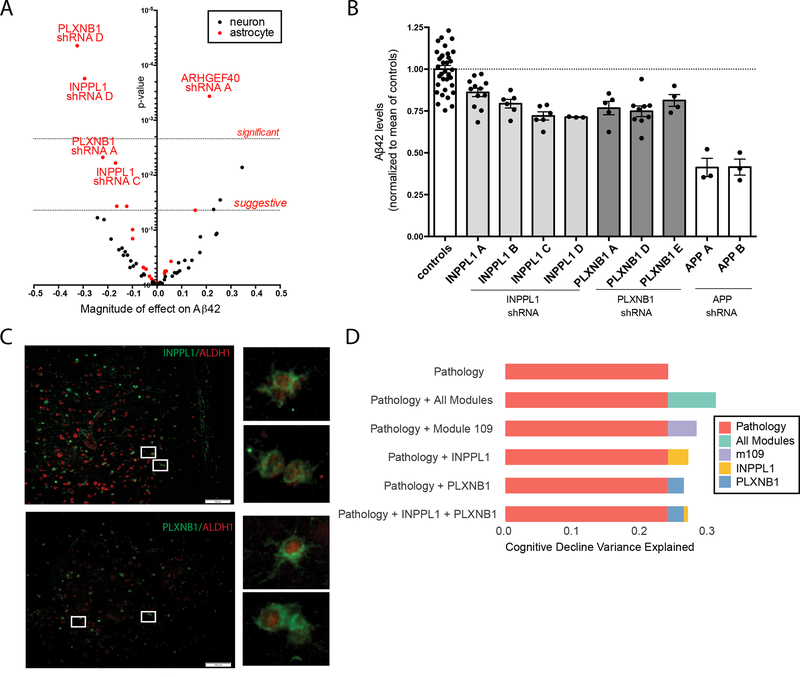
Figure 5. INPPL1 and PLXNB1 knock down in human astrocytes significantly lowers Aβ42 levels.
Human primary astrocytes or iPSC-derived neurons (iNs) were transduced with a lentivirus encoding an shRNA construct targeting one of the selected m109 genes. Aβ42 levels were measured by ELISA in the conditioned media (CM) from successfully targeted cultures. The volcano plot in (A) summarizes the results of the discovery screen in which knockdown experiments were performed in both astrocytes (red dots) and iNs (black dots). Each dot is one construct, with the magnitude of its effect on Aβ42 secretion reported on the x-axis and the statistical significance of the construct’s effect on the y-axis. Dotted lines mark cut-offs for significant (after Bonferroni correction) and suggestive (defined as accepting 1 false positive in the experiment) results for the astrocyte experiments. (B) Replication study: results of INPPL1 and PLXNB1 knock down on Aβ42 secretion were measured in additional experiments using multiple shRNA constructs targeting each of these genes. Knockdown of amyloid precursor protein (APP), which yields the Aβ42 peptide, was performed in parallel for comparison. Each dot represents an individual experiment for a given construct, and the mean level of Aβ42 secretion for all of the instances of a given construct’s evaluation is presented as a solid bar with the 95% confidence interval. The dotted line denotes the mean level of secretion in all of the control experiments. (C) INPPL1 and PLXNB1 immunostaining was performed on a section of DLPFC from post-mortem human brain of a subject with AD. Co-labelling with ALDH1 (a marker specific for cortical astrocytes) was observed in a subset of cells that are labeled with either INPPL1 or PLXNB1. (D) Figure summarizes the proportion of variance in cognitive decline that is explained by different factors. First row (orange) shows the proportion of variance explained (PVE) by measures of amyloid and tau pathology that are captured by the structured post-mortem examination of each subject. The second row shows PVE where we use all 47 modules in addition to pathology (amyloid and tau): RNA data explains more of the variance in cognitive decline than is captured by the two key measures of AD pathology. The third row shows PVE by the m109 module alone (with amyloid and tau). Finally, the next three rows show PVE by INPPL1 and PLXNB1, alone and together. As shown, PLXNB1 and INPPL1 capture much but not all of the effect of m109, and they are largely redundant with each other.
To replicate these findings, we repeated the perturbation experiments, this time including 4 shRNA constructs for INPPL1 and 3 shRNA constructs for PLXNB1, as well as several negative and positive controls (see Supplementary methods). Specifically, as positive control we included shRNA constructs targeting APP, the parent protein of the Aβ42 peptide. In these experiments, knockdown of both genes led to reduced extracellular levels of Aβ42 in astrocyte cultures (pINPPL1 = 2×10−4 and pPLXNB1 = 9×10−6) (Figure 5B). Overall, these results are consistent with the direction of m109 association where higher m109 expression is seen with a greater β-amyloid load. We also immunostained frontal cortex from subjects with pathologic AD and showed that both INPPL1 and PLXNB1 were expressed at the protein level in astrocytes (Figure 5C), confirming that these two genes were expressed in vivo in the human cell type used in the validation experiments. The astrocytes expressing these two genes are found in the vicinity of β-amyloid deposits (data not shown). These genes also are expressed in other cell types, such as neurons, and we cannot rule out that they may be implicated in AD in more than one cell type.
Having functionally confirmed a role in modulating extracellular levels of pathogenic Aβ42 for these two m109 genes in vitro, we returned to our transcriptomic data to evaluate the magnitude of the effect of INPPL1 and PLXNB1 on β-amyloid load. Separately, they each account for a small proportion of the variance in β-amyloid load - 2.8% for INPPL1 and 3.1% for PLXNB1 – and, as is anticipated from the network model, are largely redundant as, together, they explain 3.1% (adjusted R2) of the variance in β-amyloid load (Figure 5D). This compares to <1% variance of this trait explained by validated AD SNPs other than APOE. For example, the CR1 susceptibility allele explains 0.39% of variance in β-amyloid load in our data34. The effect of INPPL1 and PLXNB1 is somewhat stronger on cognitive decline: 5.5% of variance is explained by INPPL1, 4.4% for PLXNB1, and 5.4% (adjusted R2) for both. More broadly, we assessed the degree to which these two genes capture the effect of the entire module. The m109 meta-feature explains 4.3% variance in β-amyloid burden and 8.5% variance in cognitive decline, and, after accounting for INPPL1 and PLXNB1, we see that some of the effect of m109 remains for β-amyloid (0.95% variance, p=0.029), and more of the effect on cognitive decline persists (3.5% variance, p=2.7×10−5). Thus, while INPPL1 and PLXNB1 play an important role in m109, they do not appear to account for the effects of the entire module, suggesting that additional validation work will be needed to identify additional driver genes for m109 and that they are likely driven by non-β-amyloid processes.
Discussion
We deployed MTN, a network-based approach, to identify coherent biological processes and specific genes associated with multiple AD-relevant traits. A key feature of our approach is the identification of unique, direct molecular-pathological-clinical relationships, which should reduce efforts spent on spurious disease associations and on indirect associations. Further, we apply MTN to a cohort with measures of change in cognition over time, which is the most relevant clinical outcome measure of AD clinical trials for both prodromal AD as well as MCI due to AD and AD dementia. This framework and our data allowed us to identify cellular processes in the human cortex that directly relate to cognitive decline, separate from those genes that directly influence the accumulation of AD pathology. These processes are different and complementary to those identified by GWAS-derived genes, as the effect of GWAS-derived SNPs on gene expression in this case is limited.
The central finding of this project is the existence of a robust set of coexpressed genes, supported by multiple other datasets, which is related to both β-amyloid burden and to the slope of cognitive decline in older individuals. Since modeling cognitive decline in vitro is challenging, we explored the relationship of a subset of genes that were predicted to be influential genes in relation to β-amyloid biology within m109. Two of these genes INPPL1 and PLXNB1 showed relationships to extracellular β-amyloid levels in astrocyte cultures, an intriguing preliminary result that now requires further investigation. We note that the overall conclusions of this study are strongest at the systems level and that more in vitro experiments and careful selection of the model system will be needed to test the regulatory structure of this system as a whole.
PLXNB1 is a member of a family of proteins that mediate Semaphorin signalling, which plays a role in a number of neuronal processes including neurite outgrowth, remodelling and synaptic plasticity35. However, little is known about its potential contribution to cognitive decline and AD. INPPL1, also called SHIP2, is a lipid phosphatase that regulates the levels of the important second messenger PIP3. Levels of PIP3 in turn regulate downstream AKT and GSK3β signalling pathways, and AKT is also a member of the m109 module. INPPL1 plays an important role in insulin signalling, and polymorphisms in the INPPL1 gene are associated with type 2 diabetes mellitus (T2D)36. Transgenic mice overexpressing INPPL1 show a disruption in insulin/IGF1 signalling mediated via AKT in cultured neurons, and these mice show impaired memory37. Further, administration of an INPPL1 inhibitor to a mouse model of diabetes was able to rescue the synaptic plasticity and memory defects observed in this model. While these published studies are centered on neurons and not the astrocytes that our shRNA study implicate, they elaborate the picture of how INPPL1 may play a multifaceted role in AD.
In prior work, we found evidence of decreased response to insulin growth factor 1 (IGF1) in brains from persons with mild cognitive impairment and AD dementia relative to controls, regardless of diabetes status.38 Specifically, levels of postranslationally modified forms of the IGF1 receptor were positively related to β-amyloid and negatively related to episodic and working memory, after adjusting for AD pathology. In line with these findings, our study provides evidence that the INPPL1 gene plays a role in cognitive decline and β-amyloid accumulation, and it opens new avenues for investigating the relation of phosphoinositides, insulin signalling, and AD.
It is also important to note the limitations of our approach. First, assessing the false positive rate of MTN or other similar methods would require experiments on a large set of predicted null targets, which is currently cost prohibitive. Also, the MTN framework acts to increase network accuracy by modeling networks at two resolutions (a zoomed-out module/trait network and a zoomed-in gene network within selected modules), but increased accuracy comes at the price of including only a subset of modules in the inference process, resulting in potential loss of information. Additionally, an important “biological” limitation is functionally screening m109 genes for effects on in vitro β-amyloid extracellular levels, when their strongest effects were on cognitive function. This study thus interrogates only part of m109’s function.
In summary, we completed the initial arc of molecular network discovery and validation, and we have illustrated the use of the network in prioritizing a module, m109, and a small subset of genes within it for functional assessment. INPPL1 and PLXNB1 are intriguing candidates that we have connected to amyloid biology in vitro and now require further evaluation to assess their potential as AD therapeutic targets. This report is therefore an initial blueprint for a quantitative systems approach to investigating the pathobiology and function of the aging human brain.
Supplementary Material
Acknowledgements
We thank the participants of ROS and MAP for their essential contributions and the gift of their brain to these projects. All subjects gave informed consent. This work has been supported by many different NIH grants: U01AG046152, R01AG036836, P30AG10161, R01AG015819, R01AG017917, R01AG036547. This work was done as part of the National Institute of Aging’s Accelerating Medicines Partnership for AD (AMP-AD).
Footnotes
Data Availability and Accession Code
All molecular and phenotypic data are available through the Sage Synapse AMP-AD resource (https://www.synapse.org/#!Synapse:syn2580853/wiki/409844) and the RUSH Alzheimer Disease Center Resource Sharing hub (https://www.radc.rush.edu). Synapse codes for each set of data are available in the online methods section.
Competing Financial Interest Statement
There are no competing financial interests.
Supplementary Material
1. Supplementary methods
2. Supplementary Figures S1–S6
3. Supplementary Table S1–S11 as sheets for one excel file
References
- 1.Hebert LE, Weuve J, Scherr PA & Evans DA Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology 80, 1778–1783, doi: 10.1212/WNL.0b013e31828726f5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummings JL, Morstorf T & Zhong K Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res Ther 6, 37, doi: 10.1186/alzrt269 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider JA, Arvanitakis Z, Leurgans SE & Bennett DA The neuropathology of probable Alzheimer disease and mild cognitive impairment. Annals of neurology 66, 200–208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert J-C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature genetics 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaiteri C, Mostafavi S, Honey CJ, De Jager PL & Bennett DA Genetic variants in Alzheimer disease - molecular and brain network approaches. Nat Rev Neurol 12, 413–427, doi: 10.1038/nrneurol.2016.84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitra K, Carvunis AR, Ramesh SK & Ideker T Integrative approaches for finding modular structure in biological networks. Nat Rev Genet 14, 719–732, doi: 10.1038/nrg3552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parikshak NN, Gandal MJ & Geschwind DH Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet 16, 441–458, doi: 10.1038/nrg3934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gustafsson M et al. Modules, networks and systems medicine for understanding disease and aiding diagnosis. Genome Med 6, 82, doi: 10.1186/s13073-014-0082-6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geschwind DH & Konopka G Neuroscience in the era of functional genomics and systems biology. Nature 461, 908–915, doi: 10.1038/nature08537 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaiteri C, Ding Y, French B, Tseng GC & Sibille E Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav 13, 13–24, doi: 10.1111/gbb.12106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B & Horvath S A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4, Article17, doi: 10.2202/1544-6115.1128 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Zhang B et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720, doi: 10.1016/j.cell.2013.03.030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller JA, Woltjer RL, Goodenbour JM, Horvath S & Geschwind DH Genes and pathways underlying regional and cell type changes in Alzheimer’s disease. Genome Med 5, 48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyle PA et al. Much of late life cognitive decline is not due to common neurodegenerative pathologies. Annals of neurology 74, 478–489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack CR et al. Age-specific population frequencies of cerebral β-amyloidosis and neurodegeneration among people with normal cognitive function aged 50–89 years: a cross-sectional study. The Lancet Neurology 13, 997–1005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett DA, Schneider JA, Arvanitakis Z & Wilson RS Overview and findings from the religious orders study. Curr Alzheimer Res 9, 628–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett DA et al. Overview and findings from the rush Memory and Aging Project. Curr Alzheimer Res 9, 646–663 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson RS et al. Conscientiousness, dementia related pathology, and trajectories of cognitive aging. Psychology and aging 30, 74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyle PA et al. Much of late life cognitive decline is not due to common neurodegenerative pathologies. Ann Neurol 74, 478–489, doi: 10.1002/ana.23964 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storey JD & Tibshirani R Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America 100, 9440–9445, doi: 10.1073/pnas.1530509100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaiteri C et al. Identifying robust communities and multi-community nodes by combining top-down and bottom-up approaches to clustering. Sci Rep 5, 16361, doi: 10.1038/srep16361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P, Luo R, Oldham MC & Horvath S Is my network module preserved and reproducible? PLoS Comput Biol 7, e1001057, doi: 10.1371/journal.pcbi.1001057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan C & Boyd DD Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Molecular and cellular biology 26, 6357–6371 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matarin M et al. A genome-wide gene-expression analysis and database in transgenic mice during development of amyloid or tau pathology. Cell reports 10, 633–644 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Mancarci BO et al. Cross-Laboratory Analysis of Brain Cell Type Transcriptomes with Applications to Interpretation of Bulk Tissue Data. eNeuro 4, doi: 10.1523/ENEURO.0212-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koller D & Friedman N Probabilistic graphical models: principles and techniques. (MIT press, 2009). [Google Scholar]
- 28.Raber J, Huang Y & Ashford JW ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiology of aging 25, 641–650 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Yu L, Boyle PA, Leurgans S, Schneider JA & Bennett DA Disentangling the effects of age and APOE on neuropathology and late life cognitive decline. Neurobiology of aging 35, 819–826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beecham GW et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet 10, e1004606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farfel JM et al. Relation of genomic variants for Alzheimer disease dementia to common neuropathologies. Neurology, 10.1212/WNL. 0000000000002909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battle A et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res 24, 14–24, doi: 10.1101/gr.155192.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee PH, O’Dushlaine C, Thomas B & Purcell SM INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics 28, 1797–1799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chibnik LB et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Annals of neurology 69, 560–569 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perälä N, Sariola H & Immonen T More than nervous: the emerging roles of plexins. Differentiation 83, 77–91 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Suwa A, Kurama T & Shimokawa T SHIP2 and its involvement in various diseases. Expert opinion on therapeutic targets 14, 727–737 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Soeda Y et al. The inositol phosphatase SHIP2 negatively regulates insulin/IGF-I actions implicated in neuroprotection and memory function in mouse brain. Molecular Endocrinology 24, 1965–1977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talbot K et al. Brain insulin resistance demonstrated in Alzheimer’s disease is closely associated with elevated IRS-1 pS and cognitive deficits. Alzheimer’s & Dementia 7, S683 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.