Abstract
Acetaminophen (APAP) is the most commonly used analgesic and antipyretic drug in the world. Yet, it poses a major risk of liver injury when taken in excess of the therapeutic dose. Current clinical markers do not detect the early onset of liver injury associated with excess APAP—information that is vital to reverse injury progression through available therapeutic interventions. Hence, several studies have used transcriptomics, proteomics, and metabolomics technologies, both independently and in combination, in an attempt to discover potential early markers of liver injury. However, the casual relationship between these observations and their relation to the APAP mechanism of liver toxicity are not clearly understood. Here, we used Sprague-Dawley rats orally gavaged with a single dose of 2 g/kg of APAP to collect tissue samples from the liver and kidney for transcriptomic analysis and plasma and urine samples for metabolomic analysis. We developed and used a multi-tissue, metabolism-based modeling approach to integrate these data, characterize the effect of excess APAP levels on liver metabolism, and identify a panel of plasma and urine metabolites that are associated with APAP- induced liver toxicity. Our analyses, which indicated that pathways involved in nucleotide-, lipid-, and amino acid-related metabolism in the liver were most strongly affected within 10 h following APAP treatment, identified a list of potential metabolites in these pathways that could serve as plausible markers of APAP-induced liver injury. Our approach identifies toxicant- induced changes in endogenous metabolism, is applicable to other toxicants based on transcriptomic data, and provides a mechanistic framework for interpreting metabolite alterations.
Keywords: Acetaminophen, genome-scale model, glycine limitation, blood and urine metabolites, glutathione
Introduction
Acetaminophen (N-acetyl-p-aminophenol, APAP) is a commonly used analgesic and antipyretic drug, which is safe and effective when administered appropriately at therapeutic doses. However, an acute overdose causes liver damage by inducing centrilobular cell death (Boyd and Bereczky 1966; Prescott 1980a). APAP toxicity is the most frequent cause of acute liver failure in the U.S. and several other countries (Bunchorntavakul and Reddy 2013; Lee 2008), with unintentional overdoses often being more common than intentional ones (Lee 2017; Reuben et al. 2016). N- acetylcysteine (NAC), which is currently the only clinically approved antidote against APAP- induced liver damage, is most effective when administered immediately after exposure (Larson 2007). Therefore, identification of early markers indicative of APAP overdose that can lead to liver damage are of importance for effective therapeutic interventions.
The liver metabolizes APAP, and under normal conditions predominantly converts it into nontoxic glucuronide and sulfate conjugates, which are then safely eliminated by the kidneys in the urine (McGill and Jaeschke 2013; Watari et al. 1983). However, depending on the dose, a fraction of APAP is converted into a highly reactive intermediate metabolite, N-acetyl-p- benzoquinone imine (NAPQI), by cytochrome P450 (CYP) enzymes (Patten et al. 1993). Glutathione (GSH) effectively eliminates substantial amounts of NAPQI by conjugating it into APAP-cysteine and APAP-mercapturate (Prescott 1980b). However, during an overdose of APAP, glucuronide and sulfate conjugation becomes saturated and the buildup of NAPQI depletes GSH, which causes further accumulation of NAPQI (Xie et al. 2015). This leads to excess binding of NAPQI with proteins and subcellular structures and induces rapid cell death (necrosis), which, in turn, can lead to acute liver failure (Bunchorntavakul and Reddy 2018; James et al. 2003; Kon et al. 2004).
APAP-induced liver toxicity is diagnosed by evaluating liver function through serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST). Unfortunately, neither enzyme is an ideal marker because each lacks specificity. In addition, these enzymes cannot predict the onset or severity of liver injury because they are elevated only when pronounced liver injury has already occurred (Kaplowitz 2005; Korones et al. 2001; Watkins 2009). Advances in high- throughput technologies have paved the way for studying toxicant-induced cellular perturbations at the molecular level (transcriptomic and metabolomic changes) to identify toxicant-specific alterations of compound-specific modes of action, which may precede traditional toxicological endpoints (Kroeger 2006). Indeed, several studies evaluated changes in liver gene expression induced by APAP in rats under various doses and exposure durations and have provided potential indicators of adverse effects, such as mitochondrial damage and oxidative stress associated with APAP toxicity (Eakins et al. 2015; Minami et al. 2005; Morishita et al. 2006; Powell et al. 2006). Similarly, various studies have identified several endogenous metabolites in biofluids as potential markers that predict APAP-induced hepatotoxicity earlier than do clinical chemistry markers (Bhushan et al. 2013; Kumar et al. 2012; Luo et al. 2014; Sun et al. 2008; Sun et al. 2014; Yamazaki et al. 2013).
These high-throughput platforms, which generate large volumes of data, are increasingly being used to search for novel markers and gain a deeper understanding of the toxic effects of drugs. The power of these platforms can be further harnessed by concurrently measuring data from multiple sources and analyzing them together, providing detailed information about correlations among them and thereby allowing for further identification of targeted organs, time courses of injury, and markers. However, it remains a daunting task to integrate these data into a comprehensive, mechanistic framework, which clarifies how best to characterize the state of a cell, tissue, or organism in terms of the underlying biological processes. To this end, genome- scale metabolic network reconstructions (GENREs) have proven to be a good platform for integrating high-throughput data to elucidate genotype-phenotype relationships and identify various biological processes associated with disease states (Blais et al. 2017; Duarte et al. 2007; Mardinoglu et al. 2014; Pannala et al. 2018; Shlomi et al. 2009). Moreover, for most toxicants, the metabolites they generate and the toxicity mechanisms they engage are unknown; hence, a metabolic network-based approach that does not rely on knowledge of how CYP450 enzymes metabolize a toxicant can offer an alternative approach by capturing changes in the endogenous metabolism. Metabolic network models, which account for gene-protein-reaction relationships (GPR), offer a way to mechanistically incorporate transcriptomic data and thereby predict changes in endogenous metabolites and their subsequent secretion into plasma and urine, where they can be detected. Hence, these models provide an opportunity to distinguish metabolites in biofluids that are strongly associated with tissue-specific gene expression changes among all other possibilities through which metabolite alterations occur.
The present study employed an approach that integrated transcriptomic and metabolomic data to examine and improve our understanding of early metabolite changes in plasma and urine collected from rats exposed to APAP. Building on a rat genome-scale model (Blais et al. 2017; Pannala et al. 2018), we developed a new rat multi-tissue metabolic model comprising liver and kidney compartments connected via exchange of metabolites with blood and urine. We integrated high-throughput data into the multi-tissue model and investigated the metabolic differences in blood and urine between control and APAP-treated conditions. Integration of transcriptomic data obtained concurrently from the liver and kidney exposed to APAP suggested that at least 66% of the significantly changed metabolites in the blood were strongly correlated with the gene expression changes in the liver. Furthermore, using the metabolic network model together with pathway enrichment analysis, we observed that most of the genes involved in glycine, serine, and threonine metabolism were upregulated, suggesting the use of compensatory pathways for glycine production to keep up with the synthesis of glutathione that was depleted, owing to the high dose of APAP. Similarly, many genes involved in pyrimidine metabolism were downregulated, suggesting that APAP-mediated redox imbalance leads to altered nucleotide metabolism. Finally, using our computational approach, we identified a potential list of metabolites in the blood and urine that were strongly associated with gene expression changes, and which could be proposed for further targeted and clinical assessment for their potential as early indicators of liver damage. As such, our approach can be used to analyze large-scale high- throughput data obtained from rats exposed to other toxicants [e.g., gene expression data in the Open TG-Gates database (Igarashi et al. 2015), obtained from rats exposed to various drugs and chemicals], and provide an initial, hypothesis-based identification of metabolites to be tested in further targeted studies.
Materials and methods
Animal handling and treatment
Male Sprague-Dawley rats (10 weeks of age) purchased from Charles River Laboratories (Wilmington, MA) were fed with Formulab Diet 5001 (Purina LabDiet; Purina Miles, Richmond, IN) and given water ad libitum in an environmentally controlled room, set at 23 °C and on a 12:12-h light-dark cycle. All experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the U.S. Department of Agriculture, using protocols approved by the Vanderbilt University Institutional Animal Care and Use Committee, and the U.S. Army Medical Research and Materiel Command Animal Care and Use Review Office.
Rats were anesthetized with isoflurane and surgery was performed seven days before each experiment to implant the catheters for sample drawing (Shiota 2012). After surgery, the rats were housed individually. Two days before each study, the rats were moved from their regular housing cages to metabolic cages (Harvard Apparatus, Holliston, MA). To determine the appropriate APAP dose and time duration after exposure, rats were treated with either vehicle (6 ml/kg of 50% polyethylene glycol, n = 6) or either 1 g/kg (n = 6) or 2 g/kg (n = 7) of APAP at 7 a.m. by gavage. Blood and accumulated urine were collected at 7 a.m. and 5 p.m. daily for 3 days to measure serum ALT and AST levels for the liver and urine creatine levels for the kidney, to examine their injury status. Recently, we showed that ALT and AST levels increase markedly 24 h after APAP treatment for the 2 g/kg dose, and subsequent histological analysis at 58 h after treatment indicates extensive liver injury (Pannala et al. 2018).
Based on the results of these studies, we chose 2 g/kg as the appropriate APAP dose and two time points, 5 h and 10 h after treatment (n = 8 each), to obtain transcriptomic and metabolomic data. Following blood collection, animals were given either vehicle (6 ml/kg of 50% polyethylene glycol) or APAP (2 g/kg) by gavage at 7 a.m. and moved to new housing cages, where they could access water ad libitum but not food. At 12 p.m. (the 5-h group) or 5 p.m. (the 10-h group), after blood was collected from each group, animals were anesthetized by an intravenous injection of sodium pentobarbital through the jugular vein catheter and a laparotomy was performed immediately. Urine samples were collected directly from the bladder at the two time points. The liver and kidney were first dissected, and then frozen using Wollenberger tongs precooled in liquid nitrogen. The collected blood, urine, and organ samples were kept in a −80 °C freezer prior to analyses.
RNA isolation and sequencing
Because kidney tissue is histologically heterogeneous, the frozen whole kidney samples were powdered in liquid nitrogen. Total RNA was isolated from the liver or powered kidney, using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA) and the direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA). The isolated RNA samples were then submitted to the Vanderbilt University Medical Center VANTAGE Core (Nashville, TN) for RNA quality determination and sequencing. Total RNA quality was assessed using a 2100 Bioanalyzer (Agilent, Santa Clara, CA). At least 200 ng of DNase-treated total RNA with high RNA integrity was used to generate poly-A-enriched mRNA libraries, using KAPA Stranded mRNA sample kits with indexed adaptors (Roche, Indianapolis, IN). Library quality was assessed using the 2100 Bioanalyzer (Agilent), and libraries were quantitated using KAPA library Quantification kits (Roche). Pooled libraries were subjected to 75-bp single-end sequencing according to the manufacturer’s protocol (Illumina HiSeq3000, San Diego, CA). Bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed Fastq files.
Analysis of RNA-sequencing data
The analysis of RNA-seq data involved two stages: 1) determination of transcript abundance and 2) determination of differentially expressed genes (DEGs). To determine transcript abundance levels from Fastq files, which consist of raw sequence reads, we used Kallisto, a recently published software tool (Bray et al. 2016). Using this tool, we first generated a reference transcriptome index from cDNA files based on the rat genome assembly Rnor6.0, published on ENSEMBL Release 92 (Zerbino et al. 2018). We then determined transcript abundance levels by pseudoaligning raw sequence reads (single) with the reference transcriptome index. We used the analysis tool Sleuth (Pimentel et al. 2017) to investigate differential expression of genes between the two time points in each study from the transcript abundance levels thus determined.
To determine statistically significant changes in gene expression between the control and treatment groups, we used a likelihood ratio test. We followed this with a Wald test to determine the effect sizes (analogous to fold-change values) for all genes, and thus obtained effect sizes for genes determined by the likelihood ratio test to have changed significantly. Genes that changed significantly, and for which the absolute effect size fell within the top 10th percentile, were deemed biologically significant.
To understand the biological significance of each such gene, we identified the DEGs derived from Kallisto-Sleuth analyses that mapped to the rat GENRE, and used KEGG pathways to identify molecular pathways that were significantly enriched. We used the online tool Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Huang da et al. 2009) to perform this task.
Metabolomic analysis
Sample preparation was carried out at Metabolon Inc. (Durham, NC), in a manner similar to a previous study (Hatano et al. 2016). Briefly, individual samples were subjected to methanol extraction and then split into aliquots for analysis by ultrahigh performance liquid chromatography/MS (UHPLC/MS). The global biochemical profiling analysis comprised four unique arms, consisting of reverse-phase chromatography positive ionization methods optimized for hydrophilic compounds (LC/MS Pos Polar) and hydrophobic compounds (LC/MS Pos Lipid), reverse-phase chromatography with negative ionization conditions (LC/MS Neg), as well as a hydrophilic interaction liquid chromatography (HILIC) method coupled to negative ionization (LC/MS Polar) (Evans et al. 2014). All methods alternated between full scan MS and data- dependent MSn scans. The scan range varied slightly between methods but generally covered 70–1000 m/z.
Metabolites were identified by automated comparison of the ion features in the experimental samples to a reference library of chemical standard entries. These included retention time, m/z, preferred adducts, and in-source fragments, as well as associated MS spectra, and curated by visual inspection for quality control using software developed at Metabolon. Identification of known chemical entities was based on comparisons with metabolomic library entries of purified standards (Dehaven et al. 2010).
Two types of statistical analyses were performed: 1) significance tests and 2) classification analysis. Standard statistical analyses were performed in ArrayStudio on log- transformed data. The R program (http://cran.r-project.org) was used for non-standard analyses. Following log transformation and imputation of missing values, if any, with the minimum observed value for each compound, Welch’s two-sample t-test was used to identify biochemicals that differed significantly (p < 0.05) between experimental groups. An estimate of the false discovery rate (FDR, or q- value) was calculated to take into account the multiple comparisons that normally occur in metabolomics-based studies.
To understand the biological significance of metabolites, we identified those whose levels were altered by APAP exposure and that mapped to the rat GENRE, and used KEGG pathways to identify molecular pathways that were significantly enriched. We used the online tool MetaboAnalyst 4.0 (Chong et al. 2018) to perform this task.
Development of a multi-tissue rat metabolic model
The original rat genome-scale model was developed to provide a global description of rat metabolism, without considering tissue-specific information. In general, because particular cell types in each tissue do not use the functional capabilities of all metabolic enzymes encoded in the genome, procedures have been developed to contextualize the genome-scale models into tissue- or cell-specific models (Jerby et al. 2010; Pannala et al. 2018; Shlomi et al. 2008). Furthermore, to understand the importance of the tissue specificity of disease processes, several tissue-specific metabolic reconstructions have been developed by reducing the genome-scale models, using different algorithmic approaches based on genomic and proteomic data from the target tissues (Chang et al. 2010; Gille et al. 2010; Mardinoglu et al. 2013; Opdam et al. 2017; Schultz and Qutub 2016; Wang et al. 2012). However, tissues in multi-cellular organisms are highly interconnected, and in several disease processes, multiple tissues (e.g., skeletal muscle, liver, and adipose tissues in insulin resistance leading to type 2 diabetes mellitus) regulate secretion of metabolites in the blood. Similarly, metabolite secretions in the blood and urine due to exposure to a toxicant can be a consequence of alterations in several tissues. Hence, we need multi-tissue metabolic network models to interpret biofluid metabolite changes stemming from both normal and disease-induced perturbations arising from multiple organs.
Using the updated rat genome-scale model (Pannala et al. 2018), and physiological uptake and secretion data available through published literature (Banta et al. 2007; Banta et al. 2005; Izamis et al. 2011), we developed metabolic reconstructions for the rat liver and kidney. To determine the reactions specific to each tissue, we obtained information on tissue specificity from the gene expression data by evaluating whether the gene is present or absent in the data based on the TPM (transcripts per million) values (Roux et al. 2017) in our experiments. We also used baseline expression data on the rat RNA-seq transcriptomic body map (Yu et al. 2014), downloaded from Expression Atlas (Petryszak et al. 2016). Specifically, we used gene expression data from the liver and kidney of adult rats, with expression cutoff values of TPM greater than one to indicate the presence of the gene in the tissue of concern.
We used the mCADRE algorithm (Wang et al. 2012) to reduce the rat genome-scale model into tissue-specific models of the liver and kidney. Using the gene expression data, mCADRE defined a set of core reactions and pruned all other reactions based on their connectivity to the core reactions using a confidence score. The algorithm then removed reactions that were not necessary to support the core reaction set and defined metabolic functionalities of the specific tissue under reconstruction. We set the algorithm parameters such that no core reaction was allowed to be removed unless it was accompanied by removal of a large number of reactions with zero-expression evidence. We then manually curated the developed reconstructions and tested them on the metabolic functional tasks of the liver (Blais et al. 2017) and kidney (Supplementary Table S1), using the flux balance approach (Orth et al. 2010).
Subsequently, we integrated the tissue-specific reconstructions for the liver and kidney into a multi-tissue model (Fig. 1a). First, we renamed the reactions and metabolites in each reconstruction for proper compartmentalization. Then, we constructed new blood and urine compartments that connected the liver to the kidney as well as the kidney to the urine compartment. We removed the exchange reactions in the tissue-specific reconstructions, allowing only gene-associated transporters and free diffusion for intracellular metabolite transport. We then added exchange reactions to allow the blood compartment to exchange metabolites with the external system (i.e., to represent the exchange of blood metabolites with other organs of the body). We constructed the kidney exchange reactions with the urine compartment based on the metabolites identified in the global metabolomics data obtained as part of this study (Supplementary Table S2). We allowed metabolite exchange (i.e., both uptake and secretion) between the blood compartment and the external system, but only metabolite secretion from the urine compartment to the external system. Over all, the resulting multi-tissue model contained 1,982 unique genes and 12,049 reactions, connected by 7,042 metabolites. We provide the complete model in SBML format in the Supplementary Information (iRnoMTmodel).
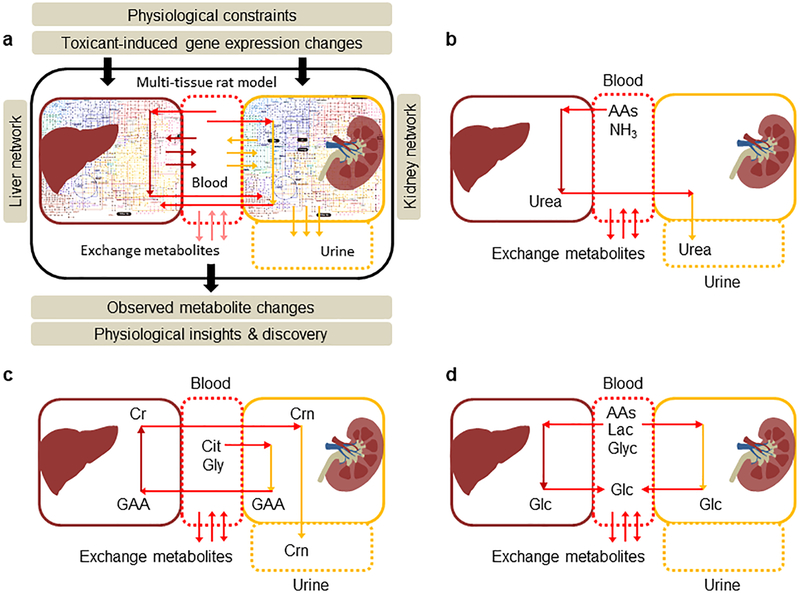
Figure 1:
A rat multi-tissue metabolic model that captures systems-level functionalities. a) Overall summary of a rat multi-tissue model comprised of liver and kidney tissues connected via a blood compartment, with the kidney connected to a urine compartment. Physiological uptake and secretion rates and gene expression changes are supplied as constraints to the model for simulating metabolite alterations in blood and urine. b) Inter-organ connectivity of the excretion of urea in urine. Schematic representation of urea synthesis in the liver from amino acids (AAs) and ammonia (NH3), followed by its excretion in urine by the kidneys. c) Inter-organ connectivity of the excretion of creatinine (Crn) in urine. Schematic representation of creatine (Cr) synthesis in the liver from guanidinoacetic acid (GAA), produced in the kidney by citrulline (Cit) and glycine (Gly). The liver transports Cr back into the blood, where it continuously degrades to Crn, which is subsequently excreted by the kidneys in urine. d) Inter-organ connectivity of glucose homeostasis under the fasting state. Schematic representation of glucose production in the liver and kidney from AAs, lactate (Lac), and glycerol (Glyc) under fasting conditions.
To evaluate the consistency of the multi-tissue model, we tested it on three functional tasks at the systems level: excretion of urea and creatinine in the urine, and production of glucose in the blood. We tested the capability of the liver to detoxify ammonia (NH3), and to transport the resulting urea to the blood compartment and then into the kidney for excretion (Fig. 1b). We achieved this by first blocking the reactions in the kidney compartment (L-arginine aminohydrolase, EC: 3.5.3.1) that produce urea, and blood exchange of ammonia from the external system that can lead to urea production. Similarly, for creatinine excretion in the urine, we blocked the production of guanidinoacetate in the liver (guanidinoacetate methyltransferase, EC: 2.1.1.2), because it is produced only in the kidney (Fig. 1c). We then evaluated the model’s ability to produce urea and its ability to produce creatinine in the urine compartment as the objective functions, given any amino acid exchange reaction as the input in the absence of all other carbon sources. Finally, we assessed the ability of the model to maintain blood glucose by constraining the respective glucose transport reactions for the liver and kidney with the glucose exchange reaction in the blood compartment as the objective function (Fig. 1d).
Algorithm for high-throughput data integration and metabolite predictions
We used the transcriptionally inferred metabolic biomarker response (TIMBR) algorithm (Blais et al. 2017) to integrate the observed changes in liver and kidney gene expression into the multi- tissue model and make predictions for metabolite alterations in the blood and urine. Briefly, using the GPR relations in the model, the TIMBR algorithm converts the log2 fold changes of all liver- and kidney-specific alterations in gene expression into reaction weights. It then calculates the global network demand required for producing a metabolite in the blood and urine. The objective function minimizes the weighted sum of fluxes across all reactions for each condition and metabolite, so as to satisfy the associated mass balance and an optimal fraction of the maximum network capability to produce a metabolite. Based on values reported in the literature, we used appropriate uptake and secretion rates for the exchange reactions of the liver and kidney under short-term fasting conditions (Supplementary Table S3).
Results
Rat multi-tissue model captures systems-level functionalities
Here, we developed metabolic network reconstructions for two major tissue types in the rat: the liver and kidney. We manually curated and tested the quality of these reconstructions using well- defined tissue-specific metabolic tasks. The liver reconstruction satisfied all 327 metabolic tasks identified as part of the original rat genome-scale model (Blais et al. 2017). For the kidney, we assembled a comprehensive collection of 155 metabolic tasks, each representing a known biological process (Supplementary Table S1). The kidney reconstruction successfully satisfied all 155 functionalities, thereby validating the quality of the developed tissue-specific metabolic reconstructions that we used to generate the multi-tissue network model (see Methods for a detailed description of the network reconstruction process).
We assessed the functional capability of the model by testing three physiologically important metabolic states of rat metabolism that encompass both the liver and kidney compartments: excretion of urea in urine, excretion of creatinine in urine, and production of glucose in the blood. In the process of urea excretion, excess amino acids in the body undergo deamination in the liver to be broken down into keto acids, leaving ammonia as the by-product. While keto acids are used to produce energy and as substrates for gluconeogenesis, the liver converts relatively toxic ammonia into relatively less toxic urea, which it releases into the blood. The urea is then removed from the blood as it passes through the kidneys. To reproduce this scenario in the multi- tissue model, we used urea production in the urine as the objective function, while AAs or NH3 from blood as input to the model (Fig. 1b). Our model simulations showed that the reactions responsible for urea synthesis in the liver and subsequent exchange reactions to remove it in the urine were actively carrying flux indicating the model capability in capturing the functional connectivity between the liver and kidney.
Similarly, creatine is an organic nitrogenous compound that plays an important role in cellular energy metabolism. However, once creatine is synthesized in the liver and released into the blood, a portion of it continuously degrades into creatinine, which is eliminated in urine by the kidneys (Fig. 1c). The multi-tissue model successfully produced flux distributions satisfying all mass-balanced steady-state assumptions under these conditions when simulated with the appropriate boundary conditions. Finally, under fasting conditions, one of the major functions of the liver and kidneys is to maintain blood glucose concentrations within a narrow range (Fig. 1d). Although both the liver and kidney contain gluconeogenesis pathways for this purpose, the contributions from the kidney are smaller under short-term fasting and increase as the fasting period increases (Gerich 2010). The multi-tissue model simulated glucose from both the liver and kidney tissues, given the appropriate boundary conditions, (e.g., imposing a lower bound constraint on the transport reactions responsible for glucose secretion into the blood from both tissues) depicting the functional integrity of the developed systems-level model.
Metabolic changes induced by APAP in the rat liver and kidney
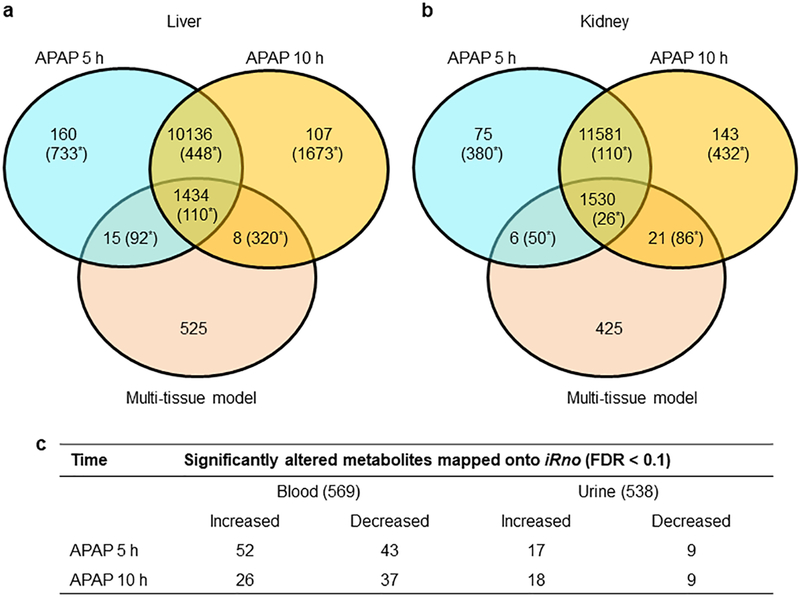
Figure 2 shows a summary of the APAP-induced changes at the levels of genes and metabolites. To obtain the gene-level changes in the liver and kidney, we analyzed the raw RNA-sequencing data collected from rats exposed to a single dose of APAP (2 g/kg) and observed for 5 h or 10 h after treatment. Overall, we identified roughly equal numbers of global changes (Fig. 2, values not in parentheses) at both time points in the liver (Fig. 2a) and kidney (Fig. 2b), although the total number of identified genes was slightly higher for the kidney compared to the liver (see details in Supplementary Table S4). Furthermore, to differentiate the genes that drive metabolic reactions from all other genes, we mapped all of the genes identified in the RNA-sequencing analysis onto the multi-tissue model. APAP exposure showed a clear demarcation between the expression patterns in the liver and kidney when we accounted for the differentially expressed genes (DEGs, FDR < 0.1; Fig. 2, values in parentheses), where the number of changes in the liver was significantly higher (1,383 at 5 h) than that in the kidney (566 at 5 h). This effect was even more pronounced at 10 h, with far more changes in the liver (2,551) than in the kidney (654)—a result clearly indicating the well-known hepatotoxicity of APAP. The effect was similar when we differentiated the metabolic genes from non-metabolic genes in the liver and kidney, by mapping them onto the multi-tissue model. Interestingly, changes in liver gene expression induced as early as 5 h after a single dose of APAP, many of which were apparent even after 10 h, indicated that persistent perturbations in liver metabolism could be major contributors to the metabolite alterations in biofluids.
Figure 2:
Summary of APAP-induced changes in the metabolism of the liver and kidney of a rat. Venn-diagrams representing the total number of global gene-expression changes observed in the liver (a) and kidney (b) at 5 h and 10 h following APAP treatment, and their mapping onto the multi-tissue metabolic model. Values in parentheses show the respective numbers of differentially expressed genes (FDR < 0.1) for each case. c) Summary of the total number of metabolites detected in the plasma (569) and urine (538) metabolic profiling studies, and the number of significantly altered metabolites (FDR < 0.1) that were mapped onto the multi-tissue model, along with their direction of change.
Global metabolic profiling of plasma and urine samples revealed significant changes in a number of metabolites at 5 h and 10 h after APAP administration (Supplementary Tables S2 and S5). Overall, we identified 569 metabolites in the plasma and 538 in urine. Principal component analysis performed on both samples showed a clear separation between the control and APAP- treatment groups (results not shown). Furthermore, to differentiate the metabolites and determine the causality of these changes in relation to gene-level changes, we mapped all of the metabolites identified by the metabolic analysis onto the multi-tissue model. Of the total metabolites detected in the plasma and urine, we mapped 226 and 197, respectively, onto the model using KEGG ID annotation and biochemical names of the metabolites. Figure 2c shows a summary of the significantly altered blood and urine metabolites (FDR < 0.1) mapped onto the model, along with their direction of change. These numbers show that APAP clearly induced more perturbations in the plasma than in urine.
Gene expression-induced metabolite changes predicted by a multi-tissue model
Using the boundary constraints on the uptake and secretion of exchange metabolites for the liver and kidney under fasting conditions (Supplementary Table S3), we integrated the concomitantly measured APAP-induced gene expression changes in the liver and kidney into the multi-tissue model with the TIMBR algorithm. TIMBR calculates the relative propensity score for the production of a metabolite to be elevated or reduced in the blood and urine, based on the gene expression changes, for all exchangeable metabolites between control and APAP-treatment conditions (see Methods). We then compared the log2 fold changes of the metabolites identified from the global metabolic profiling analysis (Fig. 1c and Supplementary Tables S2 and S5) with the model predictions, and assessed how accurate the model was in predicting the direction of APAP-induced changes in metabolite level. When we used only the liver compartment by blocking all reactions in the kidney, the model predicted only 41% and 57% of the plasma metabolite changes in the correct direction at 5 h and 10 h following APAP treatment, respectively. However, when we incorporated both the liver and kidney compartments, the model predictions increased to 50% and 66% at the corresponding time points (Supplementary Tables S5 and S7). The multi-tissue model also predicted 57% and 48% of the altered metabolites in the urine compartment accurately at 5 h and 10 h following APAP treatment. These predictions clearly indicate the importance of multi-tissue models, which show greater concordance with the data and provide better constraints by capturing the interconnectivity between the organs.
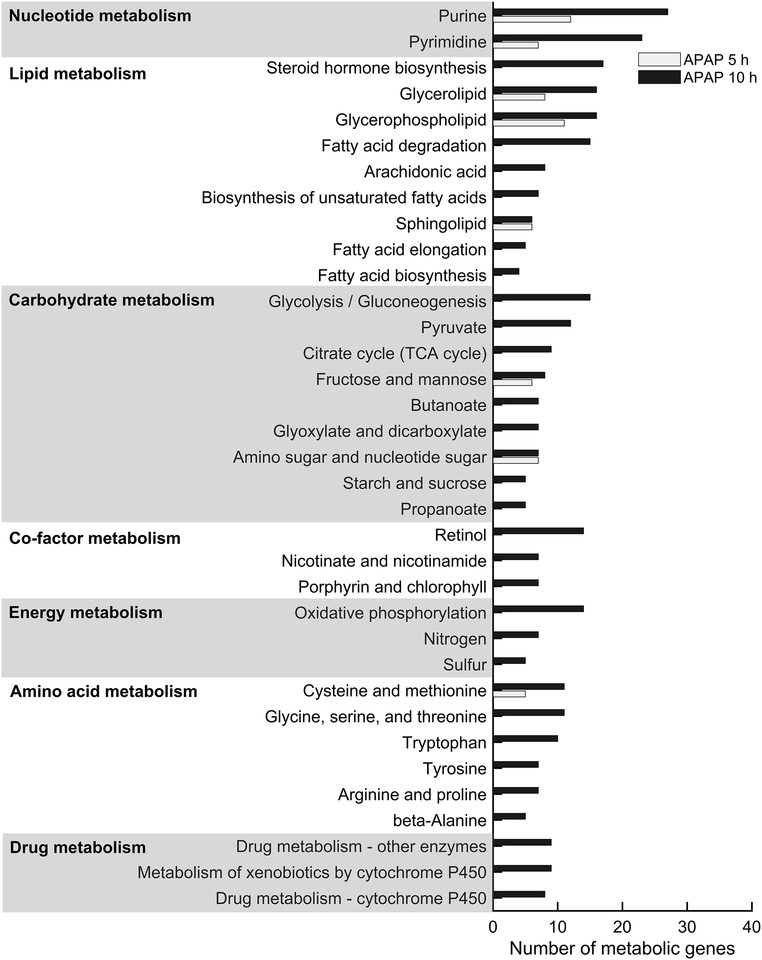
To further ascertain whether the gene expression changes were causally related to the metabolite alterations in the plasma and urine, we used the significantly altered genes and metabolites mapped onto the model to perform pathway enrichment analysis. Specifically, we used the significantly altered genes (FDR < 0.1) from the liver (Fig. 2a) and kidney tissues (Fig. 2b) and performed gene enrichment analysis using the DAVID web tool for functional annotation. Our analysis showed significant perturbations induced by APAP (Benjamini p < 0.1) in many metabolism-related pathways in the liver at 10 h (Fig. 3). Genes within the nucleotide, carbohydrate, and lipid metabolism pathways were significantly altered at both time points, indicating that APAP exposure induced persistent modifications in these pathways. In contrast, most genes in the drug and amino acid metabolism pathways were altered only at 10 h, indicating that APAP metabolism caused excess accumulation of toxic intermediates in the APAP detoxification process leading to progressive liver injury. A similar analysis using significantly altered genes in kidney tissue showed that APAP altered only arginine and proline metabolism, and had a less severe effect on glycerolipid metabolism (data not shown).
Figure 3:
Pathway enrichment analysis of significantly altered metabolic genes in the liver. We used the most significantly differentially expressed metabolic genes (FDR < 0.1) represented in the network model of the liver as input for the functional annotation with DAVID. Significantly enriched pathways are marked as bar graphs for the 5 h (white) and 10 h (black) time points, as appropriately.
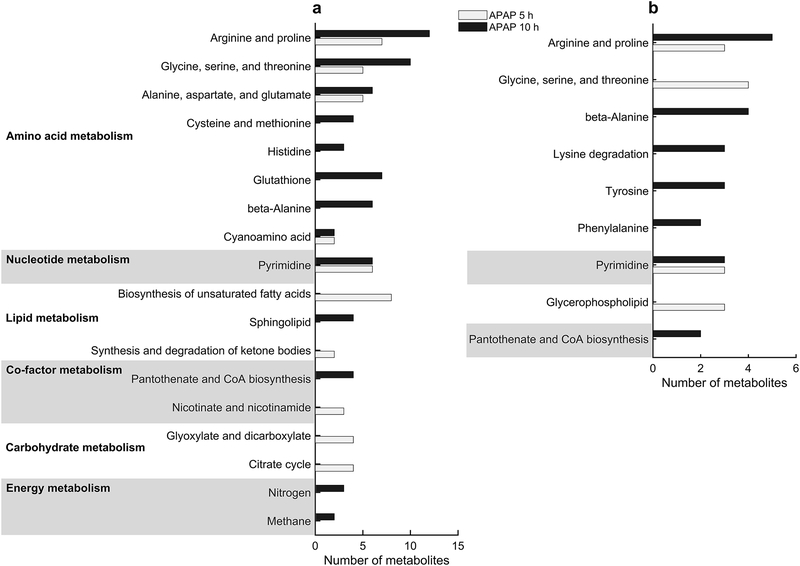
After mapping the significantly altered plasma and urine metabolites onto the multi-tissue model, we also performed pathway enrichment analysis, using the MetaboAnalyst platform. This analysis showed good agreement with the gene enrichment analysis, in that metabolites in the amino acid, nucleotide, and lipid metabolism pathways were significantly altered in the plasma and urine (Fig. 4). Specifically, metabolites in the arginine and proline; glycine, serine, and threonine; and pyrimidine metabolism pathways were significantly altered at both time points in the plasma (Fig. 4a) and urine (Fig. 4b).
Figure 4:
Pathway enrichment analysis of significantly altered metabolites in the plasma and urine. We used the most significantly altered metabolites (FDR < 0.1) represented in the network model as input to the MetaboAnalyist web tool for pathway enrichment with the rat pathway library, and over-representation and pathway topology analyses using hypergeometric and relative-betweenness centrality tests, respectively. The most significantly enriched pathways are marked as bar graphs for the 5 h (white) and 10 h (black) time points, for metabolites detected in the plasma (a) and urine (b).
APAP overdose upregulates glycine, serine, and threonine metabolism and downregulates pyrimidine metabolism
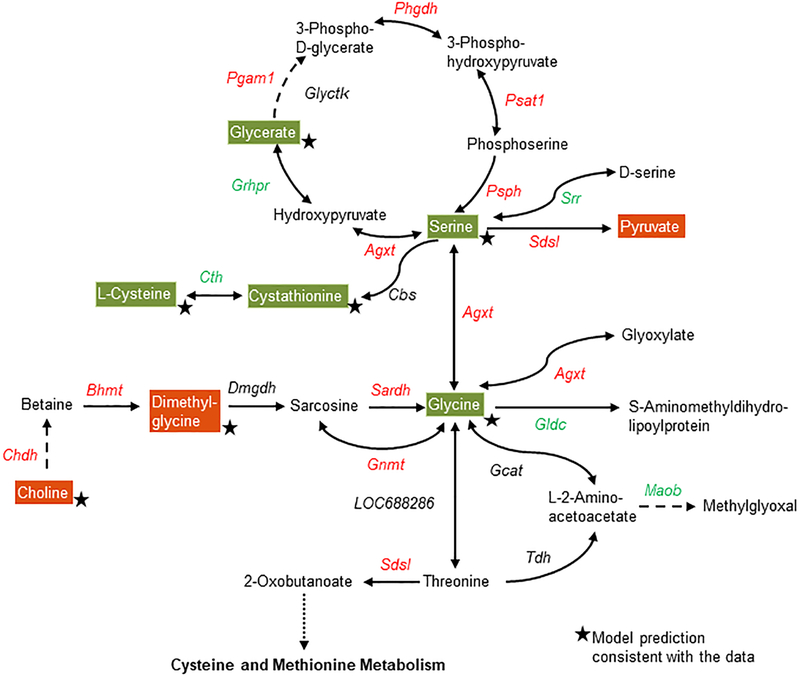
Using the GPR relationships in the metabolic network model together with pathway enrichment analysis, we further investigated the alterations in genes and metabolites at the pathway level in some highly enriched pathways. Ten hours after APAP treatment, many genes and metabolites in the glycine, serine, and threonine metabolism pathway were significantly altered relative to the control treatment (Fig. 5). Most genes responsible for serine and glycine production (shown in red) were upregulated, indicating an increased demand for serine and glycine after APAP overdose. The cause for such upregulation can be readily observed in the reduced levels of serine and glycine (Fig. 5, green) in the plasma, together with the increase in the metabolite precursors of choline and dimethylglycine (Fig. 5, red), which are required for glycine production. Interestingly, the multi-tissue model simulations, which account for the entire network demand to calculate the propensity of a metabolite to be secreted based on gene expression, accurately predicted most of these metabolite changes (Fig. 5, stars).
Figure 5:
Summary of APAP-induced gene and metabolite perturbations in the glycine, serine, and threonine metabolism pathway. Each arrow indicates the direction of a reaction converting a substrate into a product, with the name of the gene indicated next to the arrow. Upregulated and downregulated genes are shown in red and green, respectively. Increased and decreased metabolites in the plasma are shown in white text with red and green backgrounds, respectively. Stars indicate model predictions consistent with the data. Dashed arrows indicate multiple steps involved in a reaction; the dotted line indicates metabolite precursors involved in other pathways.
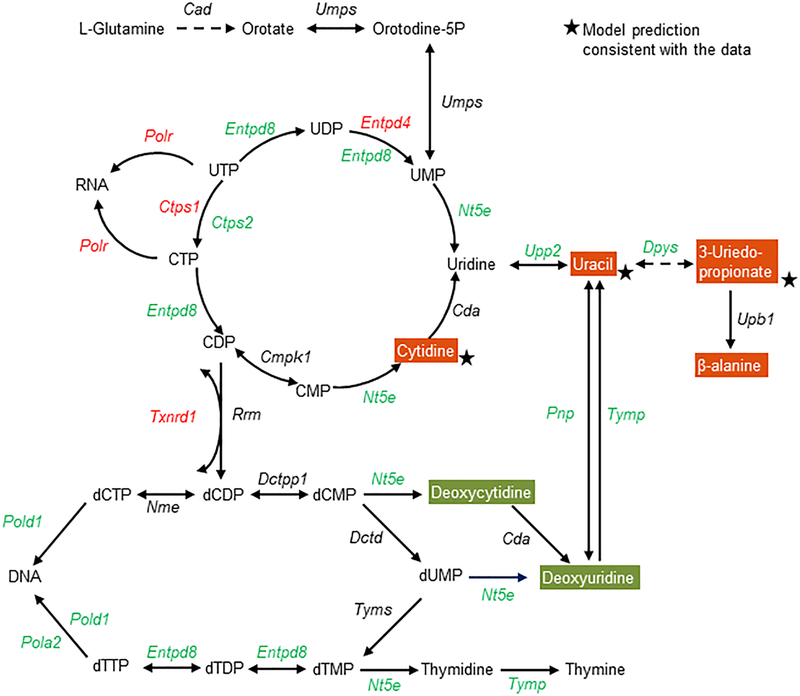
Similarly, we analyzed the genes and metabolites that were mapped onto pyrimidine metabolism (Fig. 6). Many genes that regulate the reactions driving uridine production were significantly downregulated (Fig. 6, green), together with upregulation of some genes for RNA synthesis. However, most genes responsible for DNA synthesis were downregulated, indicating reduced capability after an overdose of APAP. The multi-tissue model accurately predicted some metabolites (cytidine, uracil, and 3-ureidopropionate) that increased significantly in the plasma. We also analyzed glycerophospholipid metabolism (Fig. S1), as well as arginine and proline metabolism (Fig. S2), to identify individual changes in the reactions of these pathways, based on a combination of gene expression and metabolite alterations. Our model accurately predicted most metabolites in these pathways, indicating that the metabolites detected in highly enriched pathways correlate better with the model predictions and, hence, may serve as potential plasma metabolite markers.
Figure 6:
Summary of the APAP-induced gene and metabolite perturbations in the pyrimidine metabolism pathway at 10 h following APAP treatment. Each arrow indicates the direction of a reaction converting a substrate into a product, with the name of the gene indicated next to the arrow. Upregulated and downregulated genes are shown in red and green, respectively. Increased and decreased metabolites in the plasma are shown in white text with red and green backgrounds, respectively. Stars indicate model predictions consistent with the data. Dashed arrows indicate multiple steps involved in a reaction.
Metabolites in highly enriched pathways as markers of APAP-induced liver toxicity
Using our multi-tissue model together with pathway enrichment analysis, we determined highly enriched pathways and compiled a plausible panel of significantly altered plasma and urine metabolites that were strongly correlated with gene expression changes (Supplementary Table S6). Our multi-tissue model accurately predicted several metabolites in these pathways that showed a strong correlation with the gene expression changes. Table 1 shows a potential list of these metabolites, correctly predicted by the model in the highly enriched pathways and arranged according to their main and subordinate pathways. Specifically, we identified several consistently increased metabolites in the nucleotide- and lipid-metabolism pathways (Table 1a), and consistently decreased metabolites in the amino acid- and carbohydrate-metabolism pathways (Table 1b). Our computational model predicted most of these metabolites as early as 5 h post exposure, suggesting that they could serve as early markers of APAP-induced liver damage.
Table 1a:
List of significantly altered metabolites in the nucleotide and lipid metabolism based on the model predictions for 5 and 10 h post APAP exposure.
| Main pathway | Subordinate pathway | Metabolite name | Log2(Fold change) | |||
|---|---|---|---|---|---|---|
| Blood | Urine | |||||
| 5h | 10 h | 5h | 10 h | |||
| Nucleotide metabolism | Pyrimidine | Cytidine | 1.56 | 0.99 | 4.16 | 2.18 |
| Deoxyuridine | 1.52 | |||||
| 3-Ureidopropionate | 0.69 | 1.10 | 0.72 | |||
| Thymidine | 0.90 | 3.11 | ||||
| Uracil | 0.72 | 0.51 | ||||
| Uridine | 0.66 | |||||
| Purine | Adenosine | 2.21 | ||||
| Urate | 0.73 | 0.99 | ||||
| Lipid metabolism | Glycerolipid & phospholipid | Choline | 0.30 | 0.29 | ||
| Glycerol 3-phosphate | −0.47 | −0.60 | ||||
| Acetylcholine | 1.60 | |||||
| Phosphocholine | 1.56 | |||||
| Sphingolipid | Sphingosine 1-phosphate | 0.37 | ||||
| Sphingosine | 0.62 | |||||
| Stearoyl sphingomyelin | 0.32 | 0.83 | ||||
| Fatty acid | Margaric acid | 1.48 | ||||
| Oleate | 0.86 | |||||
| 10-Heptadecenoate | 0.83 | |||||
| Stearate | 0.71 | |||||
| Mead acid | 1.20 | |||||
| Pentadecanoate | 0.55 | |||||
| Butanoylcarnitine | −0.69 | −0.60 | ||||
| Propanoylcarnitine | −0.69 | −0.68 | ||||
| Methylmalonate | −1.06 | |||||
| Bile acids | Tauro-a-muricholate | 1.16 | ||||
| Taurocholate | 1.97 | |||||
| Tauro-P-muricholate | 1.95 | |||||
| taurochenodeoxycholate | 1.77 | |||||
| Chenodeoxycholic acid | −1.10 | −1.64 | ||||
| Glycoursodeoxycholate | −0.62 | |||||
| Cholate | −3.84 | |||||
| β-muricholic acid | −1.84 | |||||
| Glycocholate | −2.25 | |||||
| Hyodeoxycholate | −2.94 | |||||
| Glycodeoxycholate | −3.47 | |||||
| Hyocholate | −3.64 | |||||
Table 1b:
List of significantly altered metabolites in the amino acid and carbohydrate metabolism based on the model predictions for 5 and 10 h post APAP exposure.
| Log2(Fold change) | ||||||
|---|---|---|---|---|---|---|
| Main pathway | Subordinate pathway | Metabolite name | Blood | Urine | ||
| 5h | 10 h | 5h | 10 h | |||
| Amino acid metabolism | Arginine and proline | Proline | −0.20 | −0.34 | ||
| Citrulline | −0.20 | |||||
| Argininosuccinate | −0.36 | |||||
| Ornithine | −0.64 | |||||
| trans-4-Hydroxyproline | −0.36 | −0.71 | ||||
| Creatine | 0.43 | |||||
| Guanidinoacetate | −0.80 | |||||
| Spermidine | −0.81 | 1.10 | 2.93 | |||
| 4-Acetamidobutanoate | −2.12 | −1.15 | ||||
| Agmatine | −1.51 | |||||
| Glycine, serine, and threonine | Glycine | −0.25 | −0.36 | |||
| Dimethylglycine | 0.36 | 0.79 | 0.54 | |||
| Serine | −0.22 | −0.42 | ||||
| Glycerate | −0.22 | −0.30 | ||||
| Sarcosine | −0.30 | 1.50 | ||||
| Cysteine and methionine | N-acetylmethionine | −0.27 | ||||
| Cystathionine | −0.67 | |||||
| Cystein | −0.56 | |||||
| Alanine, aspartate, and glutamate | Alanine | −0.14 | −0.14 | |||
| Aspartate | −0.38 | |||||
| Glutamate | −0.43 | |||||
| l-Pyrroline-5-carboxylate | −0.67 | −0.64 | ||||
| N-acetylaspartate | −0.43 | |||||
| Asparagine | −0.36 | −0.40 | ||||
| Glutathione | Oxidized glutathione | −1.15 | ||||
| 5-Oxoproline | −1.15 | −1.40 | ||||
| Histidine | Methylimidazoleacetic acid | −1.10 | −1.03 | −0.56 | ||
| Carbohydrate & energy metabolism | Fructose and mannose | Fructose | 0.53 | |||
| Mannose | 0.37 | 0.21 | 1.95 | |||
| Pentose | Arabitol | −0.43 | ||||
| Xylitol | −0.51 | |||||
| Ribose | −0.81 | |||||
| RibM | −0.34 | |||||
| Energy | Malate | −0.45 | ||||
| Fumarate | −0.51 | |||||
Discussion
Multiple studies have investigated the potential of several markers to identify APAP-induced liver injury. For example, biomarkers such as APAP-protein adducts can assist in diagnosing the likely cause of liver injury and severity of overdose several days after exposure (Davern et al. 2006). Others, such as, miR-122, full-length cytokeratin-18, and high mobility group box 1 (HMGB1) protein are generally considered indicators of necrotic cell death (Antoine et al. 2009; McGill and Jaeschke 2014; Wang et al. 2009; Weemhoff et al. 2017). In addition to the above markers, previous studies have also analyzed mtDNA, glutamate dehydrogenase, and nuclear DNA fragments as mechanistic biomarkers of mitochondrial damage due to APAP overdose (McGill et al. 2014; Shi et al. 2015). However, most of these biomarkers depend on cell lysis to be detectable in plasma.
Global metabolic profiling has proven to be highly reliable and reproducible in detecting molecular perturbations at the cellular level before increases in AST and ALT are first detected.
In numerous studies, small changes in the physiological processes of cellular metabolism, as observed in accessible biofluids (e.g., blood and urine), have been assigned to a specific toxicological mode of action. In particular, it has been shown that plasma metabolite changes in amino acids, lipids, and bile acids, which may be indicative of liver toxicity induced by various drugs in animal studies, have greater potential for human translation (Beger et al. 2015; Iruzubieta et al. 2015; Mesnage et al. 2018; Sun et al. 2014; van Ravenzwaay et al. 2007). Several studies have reported that elevated levels of fatty acids, acylcarnitines, and medium chain fatty acids in the serum could be potential markers of liver injury induced by APAP, although they have yet to be demonstrated as satisfactory indicators of injury progression (Chen et al. 2009; Coen et al. 2003; Kumar et al. 2012; Luo et al. 2014; Yamazaki et al. 2013). Similarly, studies employing transcriptomics to analyze the liver toxicity of APAP (Beyer et al. 2007; Heinloth et al. 2007; Heinloth et al. 2004; Kikkawa et al. 2006) have indicated that the gene expression changes could discriminate between sub-toxic and toxic doses of APAP examined at different time points. Furthermore, these gene expression changes occurred earlier than clinical chemistry markers and were closely correlated with these markers at later time points.
Most of the studies described above identified APAP-induced perturbations in gene expression at the tissue level and metabolite alterations in biofluids. However, the extent to which changes in the former influence those in the latter has been unclear. In a previous study using a rat genome- scale metabolic model, we evaluated the plasma metabolite changes induced by liver gene expression for APAP (Pannala et al. 2018). Our results indicated that the model satisfactorily predicted plasma metabolite changes and suggested that gene perturbations in the liver alone could explain a considerable portion of the metabolite changes and that unexplained changes could be associated with contributions from other organs. Furthermore, our analysis suggested that plasma predictions could be improved by better physiological bounds, such as the addition of central carbon fluxes as constraints on the model.
Although APAP is generally known as a liver toxicant, the intermediate free radical molecules, which are generated after APAP is metabolized in the liver and transported into the blood circulation, could affect several other organs (Craig et al. 2011; Mirochnitchenko et al. 1999). Hence, the APAP-induced metabolite changes observed in biofluids may not reflect changes specific to liver tissues alone or at least have contributions from the kidney, the organ involved in APAP elimination in the urine. Furthermore, they are difficult to distinguish from other pathological factors. Few, if any studies, however, have considered contributions from multiple sources. We tackled this issue here by using healthy rats under fasting conditions with corresponding control groups exposed to a single high dose of APAP (2 g/kg), and simultaneously capturing changes in gene expression in the liver and kidney, as well as changes in blood and urine metabolites. Furthermore, use of our model-based approaches to integrate the metabolic data with gene expression changes provides added confidence that the identified candidate markers are specific to APAP-induced changes and may be detected in, and not influenced by, other pathological conditions. Building on our metabolic network model of the liver described above, we developed a multi-tissue model that also includes the kidney and captures the inter-organ connectivity, to understand the contributions of each organ to gene expression-induced metabolite changes in biofluids. The extended model successfully accounted for both tissue-level metabolic functions (Supplementary Table S1) and various systems-level tasks (Fig. 1) (e.g., urea/creatinine excretion in urine, glucose production in the blood) encompassing both liver and kidney tissues. It also predicted metabolite alterations in the blood more accurately than did the liver-only metabolic network model. These results suggest that the multi-tissue model provides the ability to predict altered metabolite levels in both the blood and urine, given gene expression changes in the liver only, the kidney only, or both organs.
Consistent with the notion that APAP induces liver toxicity, the metabolic changes in the liver were more pronounced than those in the kidney (Fig. 2). Approximately 10% and 20% of the genes were significantly and differentially expressed in the liver at 5 h and 10 h, respectively, compared to ~5% at both time points in the kidney. In line with the gene expression changes, global metabolic analysis revealed significant changes in many metabolites in the plasma and urine. However, we could not attribute any of these changes to the gene perturbations in the liver. Thus, we used the multi-tissue model and simultaneously integrated the gene expression data from the liver and the kidney, and predicted the associated changes in plasma and urine metabolites. Interestingly, a comparison of model predictions with the significantly changed metabolites (Fig. 2c) in the plasma indicated that the multi-tissue model provided better predictions at both time points (50% and 65% at 5 h and 10 h, respectively) than did the liver- only model. These results suggest that the multi-tissue model, which captures the inter-organ connectivity between the liver and kidney, is physiologically well constrained. Furthermore, the model allowed us to predict APAP-induced metabolite changes in the urine, which was not possible using the liver-only model. The level of agreement between the model predictions and data is notable, despite the fact that the multi-tissue network model 1) did not capture the kinetics or gene regulatory and signaling pathways, 2) did not consider the concentration of enzymes, which might affect the flux bounds, and 3) lacked precise information on the physiological uptake and secretion fluxes under which the tissues operate.
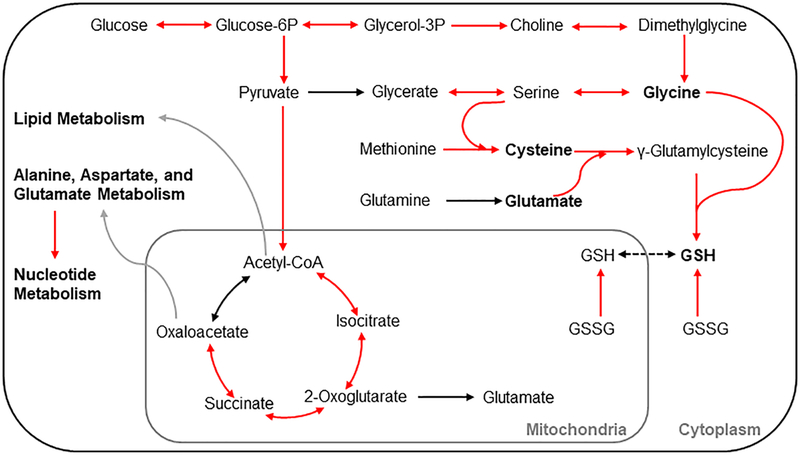
The inherently complex relationships between genes, proteins, and metabolites have heretofore made it difficult to track the factors and detailed mechanisms that contribute to metabolite changes in the blood and urine. Our approach here, in which we utilized the metabolic genes and metabolites mapped onto the network model and performed pathway-level perturbations, made it possible to address this challenge. The gene expression changes in several pathways in the liver were consistent with a widely accepted mechanism of APAP-induced liver toxicity, in which the excess APAP following an overdose is biotransformed into a reactive intermediate molecule (NAPQI) that overwhelms the detoxification mechanism by GSH-conjugation. Depletion of GSH leads to excess NAPQI in cells causing mitochondrial damage, which in turn can causes further damage by releasing reactive oxygen species (ROS) (Du et al. 2016). GSH also plays a major role in the detoxification of ROS (Mailloux et al. 2013) and its depletion by NAPQI leads to further damage by inefficient ROS removal. Genes in amino acid metabolism pathways, which are prerequisites for GSH synthesis, were significantly upregulated (Fig. 3), indicating that the high dose of APAP resulted in depletion of GSH and that cells utilized a compensatory mechanism to replenish GSH. Specifically, our integrated analysis revealed that the metabolism of glycine, one of the three amino acids required in the synthesis of the tripeptide GSH (Fig. 7), was significantly upregulated at 10 h post-APAP treatment, suggesting that glycine is another potential limiting substrate under stress conditions, a result not readily apparent in the literature for acetaminophen toxicity (Mardinoglu et al. 2017). In support of this view, we also observed significant reductions in the levels of glycine and serine in the plasma, along with the well- known limiting factor cysteine (Tateishi et al. 1974). Furthermore, consistent with APAP- mediated perturbations in GSH, we also observed significant downregulation of several genes in nucleotide metabolism pathways (Fig. 6), suggesting that APAP-mediated redox imbalance leads to reduced DNA synthesis.
Figure 7:
Summary of APAP-induced gene perturbations in various metabolic pathways at 10 h following APAP treatment. A schematic representation of the most significantly differentially expressed genes in the pyruvate; TCA-cycle; glycine, serine, and threonine; and cysteine and methionine metabolism pathways leading to glutathione (GSH) synthesis. Each arrow indicates the direction of a reaction converting a substrate into a product, with upregulation and no change indicated by red and black, respectively. Grey arrows indicate metabolite precursors involved in other pathways. Metabolites shown in bold indicate the amino acids involved in GSH synthesis.
The network model integration and pathway enrichment analysis revealed a set of metabolites in the plasma and urine, whose alterations strongly correlated with the changes in gene expression induced by APAP (Table 1). Of the metabolites that decreased, we consistently detected reductions in arginine, proline, argininosuccinate, trans-4-hydroxy-proline, glycine, and serine in the plasma at 5 h and 10 h post-APAP treatment, which were also well correlated with the gene expression changes. Similarly, we consistently detected reductions in 4-acetamidobutanoate and 5-oxoproline in urine (Fig. 5 and Table 1). Of the metabolites that increased, we detected elevated levels of choline, uracil, and 3-ureidopropionate in the plasma at both time points, consistent with model predictions. Specifically, cytidine levels significantly increased in the plasma and urine at both time points—an effect strongly correlated with gene expression changes as predicted by the multi-tissue model.
Overall, our model predictions indicate that the metabolites in the nucleotide- and lipid- metabolism pathways are early indicators of APAP exposure, whereas those in the amino acid- and carbohydrate-metabolism pathways are late indicators. We provide a complete list of significantly altered metabolites, re-grouped by pathway enrichment analysis and the agreement of their changes with the alterations predicted by the metabolic network model (Table 1). These metabolites can be further assessed in targeted analyses to identify their potential as early markers of APAP-induced liver toxicity.
In summary, we have developed a new multi-tissue metabolic network model, which considers all liver- and kidney-specific metabolic functional tasks, while capturing the overall physiological systems-level metabolism in the rat. The use of a multi-tissue model to integrate concomitantly measured gene expression changes in the liver and kidney facilitated a better understanding of the mechanisms underlying the metabolite alterations in the plasma and urine compartments than did the use of a liver-only model. Our integrated metabolic network and pathway enrichment analyses suggest that APAP overdose invokes a compensatory amino acid- related pathway, namely the metabolic pathway for glycine, serine, and threonine—the precursors for GSH production and significant alterations in the nucleotide and lipid metabolism. Using our model predictions, we provide a complete set of metabolite markers (Table 1) that could facilitate early diagnosis of APAP-induced liver toxicity. The developed computational approach has the potential to be broadly applied to other toxicant studies to analyze omics data and identify metabolites as markers of organ injuries.
Supplementary Material
Figure S1: Summary of APAP-induced gene and metabolite perturbations in the glycerophospholipid metabolism pathway.
Figure S2: Summary of APAP-induced gene and metabolite perturbations in the arginine and proline metabolism pathway.
Supplementary Table S1: Compilation of kidney metabolic functional tasks
Supplementary Table S2: Global metabolomic profiling analysis of urine
Supplementary Table S3: Compilation of literature data on the uptake/secretion rates for the liver and kidney
Supplementary Table S4: Results of RNA-sequencing analysis for the liver and kidney
Supplementary Table S5: Global metabolomic profiling analysis of blood
Supplementary Table S6: Model comparison and list of potential metabolites in the highly enriched pathways
Supplementary Table S7: Multi-tissue model predictions for blood and urine metabolite changes
iRnoMTmodel: SBML version of the rat multi-tissue model
Highlights:
Toxic levels of acetaminophen induce rapid metabolic changes in rat-liver
A genome-scale network model to analyze high-throughput data from multiple tissues
Metabolomic and transcriptomic data indicate altered liver and kidney metabolism
Coupled liver/kidney metabolic network model predicts circulating metabolites
Changes in injury-specific pathways can serve as early indicators of liver damage
Acknowledgements
The authors gratefully acknowledge the assistance of Drs. Francisco Vital-Lopez and Shivendra G. Tewari for their valuable suggestions in developing the model and methodology. The authors also gratefully acknowledge the assistance of Dr. Tatsuya Oyama in editing the manuscript. The Vanderbilt University Medical Center VANTAGE Core provided the genome-wide RNA sequencing data; Metabolon Inc. provided the global metabolic profiling data and some technical assistance for this work. The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Army or of the U.S. Department of Defense, or the Henry M. Jackson Foundation for Advancement of Military Medicine, Inc. This paper has been approved for public release with unlimited distribution. The authors were supported by the U.S. Army Medical Research and Materiel Command, Ft. Detrick, MD, as part of the U.S. Army’s Network Science Initiative and under Contract No. W81XWH-14-C-0058 (to JDY). VANTAGE is supported in part by a Clinical and Translational Science Awards (CSTA) Grant (5UL1 RR024975–03), the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Vision Center (P30 EY08126), and NIH/NCRR (G20 RR030956).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors declare no competing interests.
References
- Antoine DJ, Williams DP, Kipar A, et al. (2009) High-mobility group box-1 protein and keratin-18, circulating serum proteins informative of acetaminophen-induced necrosis and apoptosis in vivo. Toxicol Sci 112(2):521–31 doi: 10.1093/toxsci/kfp235 [DOI] [PubMed] [Google Scholar]
- Banta S, Vemula M, Yokoyama T, Jayaraman A, Berthiaume F, Yarmush ML (2007) Contribution of gene expression to metabolic fluxes in hypermetabolic livers induced through burn injury and cecal ligation and puncture in rats. Biotechnol Bioeng 97(1):118–37 doi: 10.1002/bit.21200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta S, Yokoyama T, Berthiaume F, Yarmush ML (2005) Effects of dehydroepiandrosterone administration on rat hepatic metabolism following thermal injury. J Surg Res 127(2):93–105 doi: 10.1016/j.jss.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Beger RD, Bhattacharyya S, Yang X, et al. (2015) Translational biomarkers of acetaminophen-induced acute liver injury. Arch Toxicol 89(9):1497–522 doi: 10.1007/s00204-015-1519-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer RP, Fry RC, Lasarev MR, et al. (2007) Multicenter study of acetaminophen hepatotoxicity reveals the importance of biological endpoints in genomic analyses. Toxicol Sci 99(1):326–37 doi: 10.1093/toxsci/kfm150 [DOI] [PubMed] [Google Scholar]
- Bhushan B, Borude P, Edwards G, et al. (2013) Role of bile acids in liver injury and regeneration following acetaminophen overdose. Am J Pathol 183(5):1518–26 doi: 10.1016/j.ajpath.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais EM, Rawls KD, Dougherty BV, et al. (2017) Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions. Nat Commun 8:14250 doi: 10.1038/ncomms14250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EM, Bereczky GM (1966) Liver necrosis from paracetamol. Br J Pharmacol Chemother 26(3):606–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34(5):525–7 doi: 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Bunchorntavakul C, Reddy KR (2013) Acetaminophen-related hepatotoxicity. Clin Liver Dis 17(4):587–607, viii doi: 10.1016/j.cld.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Bunchorntavakul C, Reddy KR (2018) Acetaminophen (APAP or N-Acetyl-p-Aminophenol) and Acute Liver Failure. Clin Liver Dis 22(2):325–346 doi: 10.1016/j.cld.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Chang RL, Xie L, Xie L, Bourne PE, Palsson BO (2010) Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLOS Comput Biol 6(9):e1000938 doi: 10.1371/journal.pcbi.1000938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Krausz KW, Shah YM, Idle JR, Gonzalez FJ (2009) Serum metabolomics reveals irreversible inhibition of fatty acid beta-oxidation through the suppression of PPARalpha activation as a contributing mechanism of acetaminophen-induced hepatotoxicity. Chem Res Toxicol 22(4):699–707 doi: 10.1021/tx800464q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Soufan O, Li C, et al. (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46(W1):W486–W494 doi: 10.1093/nar/gky310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen M, Lenz EM, Nicholson JK, Wilson ID, Pognan F, Lindon JC (2003) An integrated metabonomic investigation of acetaminophen toxicity in the mouse using NMR spectroscopy. Chem Res Toxicol 16(3):295–303 doi: 10.1021/tx0256127 [DOI] [PubMed] [Google Scholar]
- Craig DG, Bates CM, Davidson JS, Martin KG, Hayes PC, Simpson KJ (2011) Overdose pattern and outcome in paracetamol-induced acute severe hepatotoxicity. Br J Clin Pharmacol 71(2):273–82 doi: 10.1111/j.1365-2125.2010.03819.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davern TJ 2nd, James LP, Hinson JA, et al. (2006) Measurement of serum acetaminophen-protein adducts in patients with acute liver failure. Gastroenterology 130(3):687–94 doi: 10.1053/j.gastro.2006.01.033 [DOI] [PubMed] [Google Scholar]
- Dehaven CD, Evans AM, Dai H, Lawton KA (2010) Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform 2(1):9 doi: 10.1186/1758-2946-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K, Ramachandran A, Jaeschke H (2016) Oxidative stress during acetaminophen hepatotoxicity: Sources, pathophysiological role and therapeutic potential. Redox Biol 10:148–56 doi: 10.1016/j.redox.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, et al. (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A 104(6):1777–82 doi: 10.1073/pnas.0610772104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakins R, Walsh J, Randle L, et al. (2015) Adaptation to acetaminophen exposure elicits major changes in expression and distribution of the hepatic proteome. Sci Rep 5:16423 doi: 10.1038/srep16423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A, Bridgewater B, Liu Q, et al. (2014) High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 4(2):1 doi: 10.4172/2153-0769.1000132 [DOI] [Google Scholar]
- Gerich JE (2010) Role of the kidney in normal glucose homeostasis and in the hyperglycaemia of diabetes mellitus: therapeutic implications. Diabet Med 27(2):136–42 doi: 10.1111/j.1464-5491.2009.02894.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille C, Bolling C, Hoppe A, et al. (2010) HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol Syst Biol 6:411 doi: 10.1038/msb.2010.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T, Saiki S, Okuzumi A, Mohney RP, Hattori N (2016) Identification of novel biomarkers for Parkinson’s disease by metabolomic technologies. J Neurol Neurosurg Psychiatry 87(3):295–301 doi: 10.1136/jnnp-2014-309676 [DOI] [PubMed] [Google Scholar]
- Heinloth AN, Boorman GA, Foley JF, Flagler ND, Paules RS (2007) Gene expression analysis offers unique advantages to histopathology in liver biopsy evaluations. Toxicol Pathol 35(2):276–83 doi: 10.1080/01926230601178207 [DOI] [PubMed] [Google Scholar]
- Heinloth AN, Irwin RD, Boorman GA, et al. (2004) Gene expression profiling of rat livers reveals indicators of potential adverse effects. Toxicol Sci 80(1):193–202 doi: 10.1093/toxsci/kfh145 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57 doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Igarashi Y, Nakatsu N, Yamashita T, et al. (2015) Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res 43(Database issue):D921–7 doi: 10.1093/nar/gku955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iruzubieta P, Arias-Loste MT, Barbier-Torres L, Martinez-Chantar ML, Crespo J (2015) The Need for Biomarkers in Diagnosis and Prognosis of Drug-Induced Liver Disease: Does Metabolomics Have Any Role? Biomed Res Int 2015:386186 doi: 10.1155/2015/386186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izamis ML, Sharma NS, Uygun B, et al. (2011) In situ metabolic flux analysis to quantify the liver metabolic response to experimental burn injury. Biotechnol Bioeng 108(4):839–52 doi: 10.1002/bit.22998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LP, Mayeux PR, Hinson JA (2003) Acetaminophen-induced hepatotoxicity. Drug Metab Dispos 31(12):1499–506 doi: 10.1124/dmd.31.12.1499 [DOI] [PubMed] [Google Scholar]
- Jerby L, Shlomi T, Ruppin E (2010) Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol 6:401 doi: 10.1038/msb.2010.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplowitz N (2005) Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov 4(6):489–99 doi: 10.1038/nrd1750 [DOI] [PubMed] [Google Scholar]
- Kikkawa R, Fujikawa M, Yamamoto T, Hamada Y, Yamada H, Horii I (2006) In vivo hepatotoxicity study of rats in comparison with in vitro hepatotoxicity screening system. J Toxicol Sci 31(1):23–34 doi: 10.2131/jts.31.23 [DOI] [PubMed] [Google Scholar]
- Kon K, Kim JS, Jaeschke H, Lemasters JJ (2004) Mitochondrial permeability transition in acetaminophen-induced necrosis and apoptosis of cultured mouse hepatocytes. Hepatology 40(5):1170–9 doi: 10.1002/hep.20437 [DOI] [PubMed] [Google Scholar]
- Korones DN, Brown MR, Palis J (2001) “Liver function tests” are not always tests of liver function. Am J Hematol 66(1):46–8 doi: 10.1002/1096-8652 (200101) [DOI] [PubMed] [Google Scholar]
- Kroeger M (2006) How omics technologies can contribute to the ‘3R’ principles by introducing new strategies in animal testing. Trends Biotechnol 24(8):343–6 doi: 10.1016/j.tibtech.2006.06.003 [DOI] [PubMed] [Google Scholar]
- Kumar BS, Chung BC, Kwon OS, Jung BH (2012) Discovery of common urinary biomarkers for hepatotoxicity induced by carbon tetrachloride, acetaminophen and methotrexate by mass spectrometry-based metabolomics. J Appl Toxicol 32(7):505–20 doi: 10.1002/jat.1746 [DOI] [PubMed] [Google Scholar]
- Larson AM (2007) Acetaminophen hepatotoxicity. Clin Liver Dis 11(3):525–48 doi: 10.1016/j.cld.2007.06.006 [DOI] [PubMed] [Google Scholar]
- Lee WM (2008) Acetaminophen-related acute liver failure in the United States. Hepatol Res 38 Suppl 1:S3–8 doi: 10.1111/j.1872-034X.2008.00419.x [DOI] [PubMed] [Google Scholar]
- Lee WM (2017) Acetaminophen (APAP) hepatotoxicity-Isn’t it time for APAP to go away? J Hepatol 67(6):1324–31 doi: 10.1016/j.jhep.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Schomaker S, Houle C, Aubrecht J, Colangelo JL (2014) Evaluation of serum bile acid profiles as biomarkers of liver injury in rodents. Toxicol Sci 137(1):12–25 doi: 10.1093/toxsci/kft221 [DOI] [PubMed] [Google Scholar]
- Mailloux RJ, McBride SL, Harper ME (2013) Unearthing the secrets of mitochondrial ROS and glutathione in bioenergetics. Trends Biochem Sci 38(12):592–602 doi: 10.1016/j.tibs.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Mardinoglu A, Agren R, Kampf C, et al. (2013) Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol Syst Biol 9:649 doi: 10.1038/msb.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinoglu A, Agren R, Kampf C, Asplund A, Uhlen M, Nielsen J (2014) Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat Commun 5:3083 doi: 10.1038/ncomms4083 [DOI] [PubMed] [Google Scholar]
- Mardinoglu A, Bjornson E, Zhang C, et al. (2017) Personal model-assisted identification of NAD(+) and glutathione metabolism as intervention target in NAFLD. Mol Syst Biol 13(3):916 doi: 10.15252/msb.20167422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MR, Jaeschke H (2013) Metabolism and disposition of acetaminophen: recent advances in relation to hepatotoxicity and diagnosis. Pharm Res 30(9):2174–87 doi: 10.1007/s11095-013-1007-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MR, Jaeschke H (2014) Mechanistic biomarkers in acetaminophen-induced hepatotoxicity and acute liver failure: from preclinical models to patients. Expert Opin Drug Metab Toxicol 10(7):1005–17 doi: 10.1517/17425255.2014.920823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MR, Staggs VS, Sharpe MR, Lee WM, Jaeschke H, Acute Liver Failure Study G (2014) Serum mitochondrial biomarkers and damage-associated molecular patterns are higher in acetaminophen overdose patients with poor outcome. Hepatology 60(4):1336–45 doi: 10.1002/hep.27265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R, Biserni M, Balu S, et al. (2018) Integrated transcriptomics and metabolomics reveal signatures of lipid metabolism dysregulation in HepaRG liver cells exposed to PCB 126. Arch Toxicol 92(8):2533–47 doi: 10.1007/s00204-018-2235-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami K, Saito T, Narahara M, et al. (2005) Relationship between hepatic gene expression profiles and hepatotoxicity in five typical hepatotoxicant-administered rats. Toxicol Sci 87(1):296–305 doi: 10.1093/toxsci/kfi235 [DOI] [PubMed] [Google Scholar]
- Mirochnitchenko O, Weisbrot-Lefkowitz M, Reuhl K, Chen L, Yang C, Inouye M (1999) Acetaminophen toxicity. Opposite effects of two forms of glutathione peroxidase. J Biol Chem 274(15):10349–55 doi: 10.1074/jbc.274.15.10349 [DOI] [PubMed] [Google Scholar]
- Morishita K, Mizukawa Y, Kasahara T, et al. (2006) Gene expression profile in liver of differing ages of rats after single oral administration of acetaminophen. J Toxicol Sci 31(5):491–507 doi: 10.2131/jts.31.491 [DOI] [PubMed] [Google Scholar]
- Opdam S, Richelle A, Kellman B, Li S, Zielinski DC, Lewis NE (2017) A Systematic Evaluation of Methods for Tailoring Genome-Scale Metabolic Models. Cell Syst 4(3):318–29 doi: 10.1016/j.cels.2017.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BO (2010) What is flux balance analysis? Nat Biotechnol 28(3):245–8 doi: 10.1038/nbt.1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannala VR, Wall ML, Estes SK, et al. (2018) Metabolic network-based predictions of toxicant-induced metabolite changes in the laboratory rat. Sci Rep 8(1):11678 doi: 10.1038/s41598-018-30149-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten CJ, Thomas PE, Guy RL, et al. (1993) Cytochrome P450 enzymes involved in acetaminophen activation by rat and human liver microsomes and their kinetics. Chem Res Toxicol 6(4):511–8 doi: 10.1021/tx00034a019 [DOI] [PubMed] [Google Scholar]
- Petryszak R, Keays M, Tang YA, et al. (2016) Expression Atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res 44(D1):D746–52 doi: 10.1093/nar/gkv1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L (2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14(7):687–90 doi: 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Powell CL, Kosyk O, Ross PK, et al. (2006) Phenotypic anchoring of acetaminophen-induced oxidative stress with gene expression profiles in rat liver. Toxicol Sci 93(1):213–22 doi: 10.1093/toxsci/kfl030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott LF (1980a) Hepatotoxicity of mild analgesics. Br J Clin Pharmacol 10 Suppl 2:373S–379S doi:0306-5251/80/140373-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott LF (1980b) Kinetics and metabolism of paracetamol and phenacetin. Br J Clin Pharmacol 10 Suppl 2:291S–298S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben A, Tillman H, Fontana RJ, et al. (2016) Outcomes in Adults With Acute Liver Failure Between 1998 and 2013: An Observational Cohort Study. Ann Intern Med 164(11):724–32 doi: 10.7326/M15-2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux J, Liu J, Robinson-Rechavi M (2017) Selective Constraints on Coding Sequences of Nervous System Genes Are a Major Determinant of Duplicate Gene Retention in Vertebrates. Mol Biol Evol 34(11):2773–91 doi: 10.1093/molbev/msx199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A, Qutub AA (2016) Reconstruction of Tissue-Specific Metabolic Networks Using CORDA. PLOS Comput Biol 12(3):e1004808 doi: 10.1371/journal.pcbi.1004808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Yang X, Mattes WB, Mendrick DL, Harrill AH, Beger RD (2015) Circulating mitochondrial biomarkers for drug-induced liver injury. Biomark Med 9(11):1215–23 doi: 10.2217/bmm.15.59 [DOI] [PubMed] [Google Scholar]
- Shiota M (2012) Measurement of glucose homeostasis in vivo: combination of tracers and techniques. Methods Mol Biol 933:229–53 doi: 10.1007/978-1-62703-068-7_15 [DOI] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E (2008) Network-based prediction human tissue-specific metabolism. Nat Biotechnol 26(9):1003–10 doi: 10.1038/nbt.1487 [DOI] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Ruppin E (2009) Predicting metabolic biomarkers of human inborn of metabolism. Mol Syst Biol 5:263 doi: 10.1038/msb.2009.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Schnackenberg LK, Holland RD, et al. (2008) Metabonomics evaluation of urine from given acute and chronic doses of acetaminophen using NMR and UPLC/MS. J Chromatogr B Analyt Technol Biomed Life Sci 871(2):328–40 doi: 10.1016/j.jchromb.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Sun J, Slavov S, Schnackenberg LK, et al. (2014) Identification of a metabolic biomarker panel in rats for prediction of acute and idiosyncratic hepatotoxicity. Comput Struct Biotechno J 10(17):78–89 doi: 10.1016/j.csbj.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi N, Higashi T, Shinya S, Naruse A, Sakamoto Y (1974) Studies on the regulation of glutathione level in rat liver. J Biochem 75(1):93–103 [DOI] [PubMed] [Google Scholar]
- van Ravenzwaay B, Cunha GC, Leibold E, et al. (2007) The use of metabolomics for the discovery of new biomarkers of effect. Toxicol Lett 172(1–2):21–8 doi: 10.1016/j.toxlet.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang S, Marzolf B, et al. (2009) Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A 106(11):4402–7 doi: 10.1073/pnas.0813371106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Eddy JA, Price ND (2012) Reconstruction of genome-scale metabolic models for 126 human tissues using mCADRE. BMC Syst Biol 6:153 doi: 10.1186/1752-0509-6-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari N, Iwai M, Kaneniwa N (1983) Pharmacokinetic study of the fate of acetaminophen and its conjugates in rats. J Pharmacokinet Biopharm 11(3):245–72 [DOI] [PubMed] [Google Scholar]
- Watkins PB (2009) Biomarkers for the diagnosis and management of drug-induced liver injury. Semin Liver Dis 29(4):393–9 doi: 10.1055/s-0029-1240008 [DOI] [PubMed] [Google Scholar]
- Weemhoff JL, Woolbright BL, Jenkins RE, et al. (2017) Plasma biomarkers to study mechanisms of liver injury in patients with hypoxic hepatitis. Liver Int 37(3):377–384 doi: 10.1111/liv.13202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, McGill MR, Cook SF, et al. (2015) Time course of acetaminophen-protein adducts and acetaminophen metabolites in circulation of overdose patients and in HepaRG cells. Xenobiotica 45(10):921–9 doi: 10.3109/00498254.2015.1026426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Miyake M, Sato H, et al. (2013) Perturbation of bile acid homeostasis is an early pathogenesis event of drug induced liver injury in rats. Toxicol Appl Pharmacol 268(1):79–89 doi: 10.1016/j.taap.2013.01.018 [DOI] [PubMed] [Google Scholar]
- Yu Y, Fuscoe JC, Zhao C, et al. (2014) A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun 5:3230 doi: 10.1038/ncomms4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, et al. (2018) Ensembl 2018. Nucleic Acids Res 46(D1):D754–D761 doi: 10.1093/nar/gkx1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Summary of APAP-induced gene and metabolite perturbations in the glycerophospholipid metabolism pathway.
Figure S2: Summary of APAP-induced gene and metabolite perturbations in the arginine and proline metabolism pathway.
Supplementary Table S1: Compilation of kidney metabolic functional tasks
Supplementary Table S2: Global metabolomic profiling analysis of urine
Supplementary Table S3: Compilation of literature data on the uptake/secretion rates for the liver and kidney
Supplementary Table S4: Results of RNA-sequencing analysis for the liver and kidney
Supplementary Table S5: Global metabolomic profiling analysis of blood
Supplementary Table S6: Model comparison and list of potential metabolites in the highly enriched pathways
Supplementary Table S7: Multi-tissue model predictions for blood and urine metabolite changes
iRnoMTmodel: SBML version of the rat multi-tissue model