Abstract
Background:
The presence of allergic sensitization has a major influence on the development and course of common childhood conditions such as asthma and rhinitis. The etiology of allergic sensitization is poorly understood, and its underlying biological mechanisms are not well established. Several studies showed that DNA methylation (DNAm) at some CpGs is associated with allergic sensitization. However, no studies have focused on the critical adolescence period.
Methods:
We assessed the association of pre- and postadolescence genome-wide DNAm with allergic sensitization against indoor, outdoor and food allergens, using linear mixed models. We hypothesized that DNAm is associated with sensitization in general, and with poly-sensitization status, and these associations are age- and gender-specific. We tested these hypotheses in the IoW cohort (n = 376) and examined the findings in the BAMSE cohort (n = 267).
Results:
Via linear mixed models, we identified 35 CpGs in IoW associated with allergic sensitization (at false discovery rate of 0.05), of which 33 were available in BAMSE and replicated with respect to the direction of associations with allergic sensitization. At the 35 CpGs except for cg19210306 on C13orf27, a reduction in methylation among atopic subjects was observed, most notably for cg21220721 and cg11699125 (ACOT7). DNAm at cg10159529 was strongly correlated with expression of IL5RA in peripheral blood (P-value = 6.76 × 10−20). Three CpGs (cg14121142, cg23842695, and cg26496795) were identified in IoW with age-specific association between DNAm and allergic sensitization.
Conclusion:
In adolescence, the status of allergic sensitization was associated with DNAm differentiation and at some CpGs the association is likely to be age-specific.
Keywords: adolescence, allergic sensitization, DNA methylation, longitudinal, skin prick test
Graphical Abstract
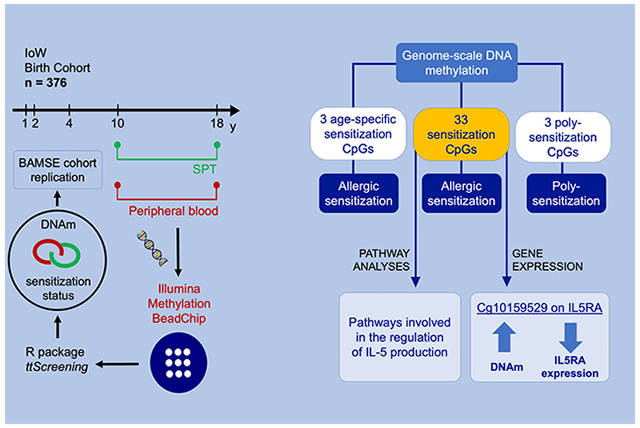
DNA methylation is associated with allergic sensitizations, findings from a genome-scale longitudinal study during adolescence.
1 |. INTRODUCTION
The presence of allergic sensitization has a major influence on the development and course of common childhood conditions such as asthma and rhinitis.1,2 In clinical practice, allergic sensitization is defined as the presence of sensitization to allergen by skin prick testing (SPT),3,4 or by the presence of serum specific immunoglobulin E (IgE).5 The etiology of allergic sensitization is poorly understood, and its underlying biological mechanisms are not well established, although it was suggested that gene-environment interactions in early life may play a role in atopy and atopic-disease susceptibility.6
A covalent addition of a methyl group to the DNA at a cytosine residue that is followed by a guanine (CpG site), denoted in our study as DNA methylation (DNAm) at CpG sites, is an important epigenetic mechanism that regulates gene transcription.7 Several studies have shown that epigenetic changes, in particular methylation of CpGs, are associated with allergic sensitization. Most studies in humans have focused on methylation measured in DNA from peripheral blood cells. Studying DNAm of Alu and LINE-1 repetitive elements (as a proxy for global methylation levels) in older adults (mean age 73 years) from the Normative Ageing Study cohort, Sor-dillo et al. showed that sensitization to aeroallergens was associated with increased Alu methylation.8 White et al. examined the association between atopic status and epigenetic changes in peripheral blood among children (6 years or younger) and adults in the promoter region of IFN-γ gene.9 They found that methylation was reduced in naive CD8 + T cells among atopic children, implying that differential regulation of IFN-γ promoter methylation in T cells may contribute to development of allergic sensitization. However, this difference was not observed in cord blood CD8+ T cells between infants who subsequently went on to be sensitized to ≥ 1 allergen by age 2 years compared to those remained nonatopic.
The studies described above have focused on candidate genes. In one cross-sectional study, the association of DNAm with allergic sensitization was examined at a genome scale and 13 CpGs were identified.10 Participants in this study were young females aged 18 years, while participants in the aforementioned studies were either in early childhood (6 years or younger), adults, or older men. To our knowledge, no studies have focused on the critical period of adolescence. Adolescence is accompanied by significant changes, for example, puberty, rapid growth, and often body mass index (BMI) increase, which may be associated with changes in sensitization and DNAm.5,11,12 We also detected changes in DNAm in genes in the Th2 pathways between pre- and postadolescence and their association with risk of asthma and with changes in asthma status between pre- and postadolescence.13 These findings led to the present study to examine whether similar association patterns between allergic sensitization and DNAm would be observed in childhood and young adults, and if not, then to what extent they differ. To this end, we assessed the association of pre- and postadolescence genome-scale DNAm with any sensitizations (yes vs no) against 11 allergens, including indoor, outdoor, and food allergens, using linear mixed models. We hypothesized that DNAm is associated with allergic sensitization and the associations are gender-specific and are different between pre- and postadolescence. We tested these hypotheses in subsamples from two independent cohorts.
2 |. METHODS
2.1 |. Participants and allergic sensitization assessments
Data analyzed in this study were from a birth cohort of children born between January 1, 1989, and February 28, 1990, on the Isle of Wight (IoW), United Kingdom.14 Of the 1536 children born and recruited in this period, 1456 were available for further follow-up with data collected at ages 1, 2, 4, 10, and 18 years. Questionnaires that included the questions of the International Study of Asthma and Allergy in Childhood (ISAAC) were administered at each follow-up. Information such as disease status, age of disease onset, and tobacco smoke exposure was collected based on responses to the questionnaires. The Local Research Ethics Committee approved this study (06/Q1701/34). The informed consent was written for in-person visits.
Skin prick test (SPT) assessment was conducted at ages 4, 10, and 18 years for 11 common allergens (house dust mite, cat dander, dog dander, grass pollen mix, tree pollen mix, Alternaria alternata, Cladosporium herbarium, cow’s milk, hen’s egg, peanut, and cod). Standard lancets (ALK, Horsholm, Denmark) were used. Participants were asked to avoid taking oral antihistamines for 72 hours. The average of the longest diameter and the diameter perpendicular to it was assessed at 15 minutes. Results were discarded if the wheal size of positive control was smaller than 3 mm in diameter. Allergic sensitization to an allergen was defined as a mean wheal size of 3 mm greater than the negative control. Specific IgEs were measured at ages 10 and 18 using the FX5 test for a mixture of five common food allergens (milk, egg, cod, peanut, and soy), and the Phadiatop test for mix inhalant allergens (grasses, trees, weeds, cat, dog, mites, and mold). Positivity was defined as serum IgE > 0.35 kU/L.
2.2 |. DNA methylation
Peripheral blood samples were collected in IoW at ages 10 and 18 years. DNA was extracted using a standard salting out procedure.15 The methylation level for each CpG was assessed using one of two platforms, the Illumina Infinium HumanMethylation450 Bead-Chip or the MethylationEPIC BeadChip (Illumina, Inc., San Diego, CA). DNA concentration was estimated by fluorometric quantitation. For each sample, one microgram DNA was bisulfite-treated for cytosine to thymine conversion using the EZ 96-DNA Methylation Kit (Zymo Research, Irvine, CA), following the manufacturer’s protocol. HumanMethylation450 BeadChip interrogates > 484 000 CpGs and MethylationEPIC BeadChip > 850 000 CpGs associated with over 24 000 genes. Arrays were processed using a standard protocol6 with multiple identical control samples assigned to each bisulfite conversion batch to assess assay variability. DNAm was measured by a ratio of intensities, denoted as β values for each CpG site. It is a ratio of methylated (M) over the sum of methylated and unmethylated (U) probes (β = M/[c+M+U]), where c is a constant to prevent zero in the denominator. In total, 376 subjects with DNAm at both ages and allergic sensitization status on 11 allergens available at one or both ages were included in the study.
2.3 |. Preprocessing
Probes not reaching a detection P-value of 10−16 in at least 95% of samples were excluded. CpGs on sex chromsomes were also excluded to avoid bias. DNAm were then preprocessed using the CPACOR pipeline for data from both platforms (HumanMethylation450 and MethylationEPIC).16 Specifically, the data were quantile-normalized using the R package, minfi.17 Next, principal components (PCs) inferred based on control probes were used to represent latent chip-to-chip and technical variations. Since DNAm data were from two different platforms, we determined the PCs based on DNAm at shared control probes. In total, 195 control probes were shared between the two arrays and used to calculate the control probe PCs with the top 15 to represent latent batch factors.16
After preprocessing, a total of 439 635 CpGs overlapped between the two platforms were included in subsequent analyses. Probes that contained single nucleotide polymorphisms (SNPs) within 10 base pairs of a targeted CpG site with minor allele frequency in at least 0.7% subjects (corresponding to at least 10 subjects in IoW with n = 1456) were excluded due to their influence to DNAm.
2.4 |. Statistical methods
2.4.1 |. Comparing with the complete cohort
To compare the data used in our study and in the complete cohort at ages 10 and 18 years, respectively, we tested proportions of subjects being sensitized at each age and different demographic variables such as gender and socio-economic status.
2.4.2 |. Screening CpGs
We excluded CpGs potentially not associated with allergic sensitization and CpGs showing no difference between mono- and poly-sensitization in DNAm. An R package ttScreening18,19 was implemented for this purpose. The method in ttScreening utilizes training and testing data in robust linear regressions with surrogate variables included to adjust for unknown effects (Appendix S1). We applied the package to assess the association of genome-scale DNAm with the status of sensitization (yes/no) and with the status of poly-sensitization status (poly-vs mono-sensitization). The screening was performed for each gender at ages 10 and 18 years, separately. Subjects with DNAm and allergic sensitization data at one or both ages were included in the screening. CpGs that passed the screening at either age for one or both genders were included in subsequent analyses.
2.4.3 |. Detecting CpGs associated with status of sensitization or status of poly-sensitization
CpGs that passed the screening were further examined. We implemented two linear mixed models to assess the associations with DNAm, one model focusing on the effect of sensitization and the other on the effect of poly-sensitization. The dependent variable was DNAm of a CpG at each age (10 and 18 years). To remove effects of batches and technical variations on DNAm measurements, we first regressed logit-transformed DNAm at each CpG on the aforementioned 15 PCs and an indicator for the two platforms (450 and EPIC). The residuals were treated as batch-effect-adjusted DNAm, and included in the models as dependent variables. The main independent variable was the atopic status at each age or the status of mono-/poly-sensitization. Gender and age were included in the model as covariates.20–23 DNAm is influenced by heterogeneity of cell compositions in peripheral blood. Thus, cell type proportions at each age were inferred using the minfi R package24 and included in the model.25 Finally, to test the moderating effects of gender and age on the association of DNAm with sensitizations, we considered additional linear mixed models with two-way interactions, one for moderating effect of gender and the other for age. Multiple testing was corrected by controlling for a false discovery rate (FDR) of 0.05.
2.5 |. Replication cohort—the BAMSE cohort
CpGs shown to be associated with allergic sensitization or poly-sensitization in IoW were further tested in an independent cohort, the BAMSE study (sensitization defined as specific IgE > 0.35 kU/L to cat, dog, grass, house dust mite, egg, or peanut).26 As in IoW, DNA was extracted from peripheral whole blood. DNAm was analyzed for 267 subjects at 16 years of age using the Illumina HumanMethylation450 array, and quality control details have been presented elsewhere.27 Robust linear regressions were implemented with similar covariates, gender, and cell type proportions, included in the analyses. Genome-scale gene expressions in peripheral white blood cells of the 267 participants aged 16 years were used to assess biological relevance for the identified CpGs.28
2.6 |. Bioinformatic assessment
Genes annotated to the CpGs replicated in BAMSE were identified based on Illumina’s manifestation file. Bioinformatic assessment of the genes was conducted using ToppFun, available in the ToppGene Suite.29 ToppFun is for functional enrichment analyses based on a list of genes in Gene Ontology (GO) for molecular functions and biological processes. Multiple testing was adjusted by controlling FDR of 0.05.
3 |. RESULTS
The subsamples included in the study represented the whole cohort with respect to demographics related to allergy, except for maternal smoking during pregnancy (P-value = 0.04, Table 1a). Proportions of allergic sensitization to any allergen between the subsample and the whole cohort were also compared. No statistically significant difference was observed at the age of 10 years, but at age 18 years, the proportion of being sensitized was higher in the subsamples (P-value = 0.01, Table 1b).
TABLE 1.
Comparing the subsample included in the current study with the complete Isle of Wight cohort
| (a) on demographics via proportion tests | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Complete cohort | Subsample | P-value | |||||||
| Gender (female) | 786; 51.2% (Na = 1536) | 193; 51.3% (N = 376) | 0.99 | |||||||
| Season of birth | ||||||||||
| Winter | 499; 32.5% | 116; 30.9% | 0.78 | |||||||
| Spring | 364; 23.7% | 86; 22.9% | ||||||||
| Summer | 353; 23.0% (N = 1536) | 87; 23.1% (N = 376) | ||||||||
| Low birthweight (yes) | 66; 4.4% (N = 1497) | 17; 4.7% (N = 360) | 0.80 | |||||||
| Socio-economic status | ||||||||||
| Low | 209; 15.4% | 51; 13.8% | 0.73 | |||||||
| Medium | 1037; 76.4% (N = 1357) | 287; 77.6% (N = 370) | ||||||||
| Maternal smoking during pregnancy (yes) | 384; 25.3% (N = 1521) | 75; 20.1% (N = 374) | 0.04 | |||||||
| Parental allergy (yes) | 756; 50.0% (N = 1514) | 206; 55.4% (N = 372) | 0.06 | |||||||
| (b) on allergic sensitizations via proportion tests | ||||||||||
| 10 y (n$; %) |
18 y (n; %) |
|||||||||
| Complete cohort | Subsample | P-value | Complete cohort | Subsample | P-value | |||||
| Allergic sensitization | 279; 26.9% (Nb=1036) | 108; 29.8% (N = 363) | 0.26 | 353; 41.4% (N = 853) | 176; 48.2% (N = 365) | 0.01 | ||||
| Mono-sensitizationc | Female (N = 520) | Male (N = 516) | Female (N = 168) | Male (N = 195) | Female (N = 446) | Male (N = 407) | Female (N = 172) | Male (N = 193) | ||
| Alternaria | 1; 0.19% | 3; 0.58% | 1; 0.60% | 3; 1.54% | 6; 1.35% | 3; 0.74% | 4; 2.33% | 2; 1.04% | ||
| Cladosporium | 1; 0.19% | 1; 0.19% | — | 1; 0.51% | 1; 0.22% | — | — | — | ||
| HDM | 38; 7.31% | 55; 10.66% | 15; 8.93% | 18; 9.23% | 38; 8.52% | 48; 11.79% | 14; 8.14% | 22; 11.40% | ||
| Cat dander | 4; 0.77% | 5; 0.97% | — | 4; 2.05% | 2; 0.45% | 1; 0.25% | — | — | ||
| Dog dander | 2; 0.38% | — | — | — | 1; 0.22% | 1; 0.25% | 1; 0.58% | 1; 0.52% | ||
| Grass | 13; 2.50% | 17; 3.29% | 6; 3.57% | 7; 3.59% | 25; 5.61% | 7; 1.72% | 15; 8.82% | 4; 2.07% | ||
| Cod | — | 1; 0.19% | — | 1; 0.51% | — | — | — | — | ||
| Peanut | — | 1; 0.19% | — | 1; 0.51% | 1; 0.22% | 1; 0.25% | 1; 0.58% | 1; 0.52% | ||
| Totalc | 59; 11.35% | 83; 16.09% | 22; 13.10% | 34; 17.44% | 74; 16.59% | 61; 14.99% | 35; 20.34% | 30; 15.54% | ||
| Poly-sensitizationc | Female (N = 520) | Male (N = 516) | Female (N = 195) | Male (N = 168) | Female (N = 446) | Male (N = 407) | Female (N = 172) | Male (N = 193) | ||
| 59; 11.35% | 77; 14.92% | 20; 11.90% | 32; 16.41% | 83; 18.61% | 129; 31.70% | 42; 24.42% | 68; 35.23% | |||
All Ns denote the number of subjects with no missing values for each variable. In total, N = 376 subjects were included in the analyses.
All Ns denote the number of subjects with no missing values for each variable. In total, N = 376 subjects were included in the analyses.
Number and percentage of subjects mono-sensitized against one allergen for each gender at each age (10 and 18 y), and “Total” refers to the number and percentage of subjects mono-sensitized against one of the 11 allergens, stratified by gender and age. For mono-sensitizations, only allergens with at least one subject with positive SPT at ages 10 or 18 are included in the table.
All n’s denote the number of subjects in each sensitization category, and %’s are calculated by n/N × 100%.
In total, 1437 CpGs passed screening at either age for one or both genders for their potential association with allergic sensitization against one or more allergens, and these CpGs were analyzed further using linear mixed models. Of the 1437 CpGs, DNAm at 35 CpGs was shown to be associated with the status of allergic sensitization (FDR = 0.05), controlling for the effects of age, gender, cell type compositions, and random subject effects and technical variation (Table 2). Consistent findings were observed when sensitization were defined using specific IgE (Table S1). Out of the 35 CpGs, four (14%) were on CpG islands, and 17 CpGs (48.6%) were not close to any islands. Except for cg19210306 (5’ UTR of gene C13orf27, alias for TEX30, coefficient = 0.22, P-value = 4.63 × 10−4), being atopic was associated with lower DNAm at other 34 CpGs (Table 2, Figure 1). Three CpGs are on one gene, ACOT7 (cg09249800, cg11699125, and cg21220721), and two of these three CpGs, cg21220721 and cg11699125, exhibited the largest DNAm reduction (coefficients = −0.20 with P-values = 1.12 × 10−4 and 4.34 × 10−5, respectively).
TABLE 2.
Effects of any allergic sensitization (based on SPT) on DNAm via linear mixed models in the loW cohort and in the replication cohort BAMSE on each of the 35 identified CpGs
| Name | Relation to CpG islands | Gene name | Location | Chr | loW cohort | BAMSE | |||
|---|---|---|---|---|---|---|---|---|---|
| Est. | Raw P-value | FDR P-value | Est. | Raw P-value | |||||
| cg01888561 | Island | SEC14L1 | TSS1500 | 17 | −0.070 | 1.36 × 10−4 | 0.050 | −0.036 | 0.13 |
| cg02245534 | N_Shore | METRNL | Body | 17 | −0.061 | 4.41 × 10−5 | 0.016 | −0.060 | 0.14 |
| cg02475695 | S_Shore | NHLRC4 | TSS1500 | 16 | −0.077 | 1.22 × 10−4 | 0.045 | −0.111 | 0.0026 |
| cg02803925 | — | PCYT1A | Body | 3 | −0.178 | 1.14 × 10−4 | 0.042 | −0.136 | 0.016 |
| cg03493123 | N_Shore | B4GALT7 | Body | 5 | −0.062 | 6.17 × 10−5 | 0.023 | −0.012 | 0.69 |
| cg05072552 | N_Shore | CFL1 | Body | 11 | −0.053 | 1.26 × 10−4 | 0.046 | NA | NA |
| cg05390183 | N_Shelf | Intergenic | 1 | −0.058 | 5.57 × 10−5 | 0.020 | −0.053 | 0.010 | |
| cg06070625 | — | MITF | Body; TSS200 | 3 | −0.075 | 8.71 × 10−5 | 0.032 | −0.086 | 0.0065 |
| cg06099697 | Island | ZFPM1 | Body | 16 | −0.067 | 8.79 × 10−5 | 0.032 | −0.024 | 0.44 |
| cg07124719 | — | GDEP | TSS200 | 4 | −0.075 | 7.85 × 10−5 | 0.029 | −0.064 | 0.031 |
| cg07721901 | — | FAM81B | TSS1500 | 5 | −0.073 | 3.32 × 10−5 | 0.012 | −0.042 | 0.11 |
| cg09249800 | Island | ACOT7 | Body | 1 | −0.160 | 1.17 × 10−4 | 0.043 | NA | NA |
| cg09705784 | — | DNAH17 | Body | 17 | −0.105 | 8.74 × 10−5 | 0.032 | −0.145 | 0.00066 |
| cg10159529 | — | IL5RA | TSS1500 | 3 | −0.085 | 5.49 × 10−5 | 0.020 | −0.113 | 0.00026 |
| cg11699125 | Island | ACOT7 | Body | 1 | −0.198 | 4.34 × 10−5 | 0.016 | −0.264 | 0.00011 |
| cg11988722 | S_Shelf | Intergenic | 20 | −0.067 | 9.04 × 10−5 | 0.033 | −0.086 | 0.0029 | |
| cg12077460 | — | MFHAS1 | Body | 8 | −0.077 | 2.98 × 10−5 | 0.011 | −0.133 | 7.03 × 10−8 |
| cg12105691 | — | C3orf50 | Body | 3 | −0.100 | 2.25 × 10−5 | 0.008 | −0.133 | 8.74 × 10−5 |
| cg14011077 | — | Intergenic | 9 | −0.183 | 2.83 × 10−5 | 0.010 | −0.235 | 1.16 × 10−4 | |
| cg14436861 | — | WEE1 | 3’UTR | 11 | −0.089 | 6.74 × 10−5 | 0.025 | −0.108 | 4.26 × 10−4 |
| cg15482717 | S_Shelf | FADD | 3’UTR | 11 | −0.063 | 1.32 × 10−4 | 0.048 | −0.054 | 0.020 |
| cg15710961 | — | DST | Body | 6 | −0.113 | 1.15 × 10−5 | 0.004 | −0.097 | 0.0018 |
| cg17203290 | — | C8orf47 | 3’UTR | 8 | −0.086 | 4.38 × 10−5 | 0.016 | −0.101 | 0.0085 |
| cg17429587 | S_Shelf | NCOR2 | Body | 12 | −0.086 | 2.69 × 10−5 | 0.010 | −0.117 | 1.83 × 10−4 |
| cg17933300 | S_Shelf | SCOC | TSS200 | 4 | −0.079 | 1.03 × 10−5 | 0.037 | −0.061 | 0.028 |
| cg17971251 | — | SEC16B | Body | 1 | −0.089 | 1.46 × 10−5 | 0.005 | −0.159 | 3.38 × 10−7 |
| cg18666454 | N_Shore | KCNH2 | Body | 7 | −0.135 | 1.11 × 10−4 | 0.040 | −0.089 | 0.018 |
| cg19210306 | Island | C13orf27 | 5’UTR | 13 | 0.217 | 4.63 × 10−5 | 0.017 | 0.014 | 0.72 |
| cg20315954 | — | PMP22 | Body | 17 | −0.087 | 7.91 × 10−5 | 0.029 | −0.097 | 0.0091 |
| cg21220721 | Island | ACOT7 | Body | 1 | −0.202 | 1.12 × 10−4 | 0.041 | −0.294 | 7.07 × 10−4 |
| cg25087851 | S_Shelf | GPR44 | TSS1500 | 11 | −0.069 | 1.37 × 10−4 | 0.050 | −0.082 | 0.011 |
| cg25479097 | S_Shelf | C13orf35 | 5’UTR | 13 | −0.092 | 4.36 × 10−5 | 0.016 | −0.093 | 0.0066 |
| cg25854298 | — | ASCC1 | Body | 10 | −0.102 | 6.28 × 10−5 | 0.023 | −0.131 | 6.38 × 10−4 |
| cg26508444 | — | FAM53B | Body | 10 | −0.054 | 1.37 × 10−4 | 0.050 | −0.047 | 0.0087 |
| cg27469152 | — | EPX | 3’UTR | 17 | −0.070 | 1.62 × 10−5 | 0.006 | −0.071 | 0.016 |
Chr., chromosome; Est., estimation.
P-values that are statistically significant at the 0.05 level in the BAMSE are in bold.
The “—” in the second column represents CpGs not identified as being related to CpG islands. Following Illumina’s definition, a CpG island is defined as regions > 500 bp with more than 55% GC and expected/observed CpG ratio of more than 0.65. Around 40% of gene promoters contain islands. CpG shelves are ~4 kbp from islands, and CpG shores are ~2 kbp from islands.47
FIGURE 1.
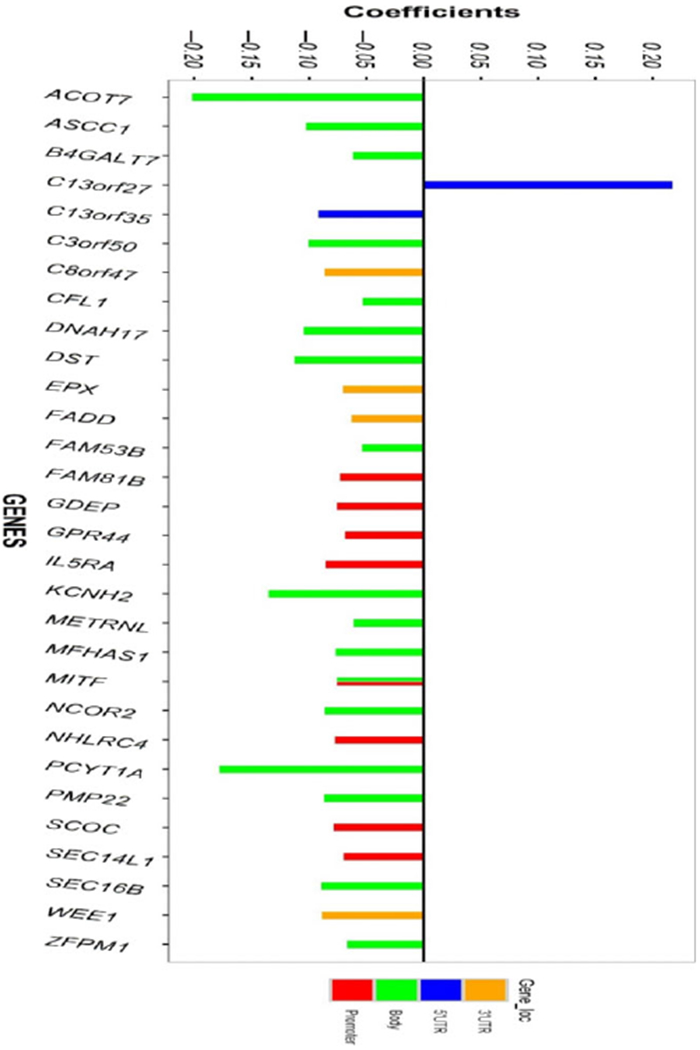
Bar plots of the regression coefficients and the location of CpGs on their corresponding genes. The coefficients represent changes in DNA methylation associated with allergic sensitizations. Negative coefficients reflect decrease in DNA methylation in sensitized patients
We also assessed whether the association of DNAm at each of the 1437 CpGs with allergic sensitization was gender-specific. No statistically significant interaction effects were identified at FDR = 0.05. In addition, stability of DNAm at all these 35 identified CpGs was suggested by results from linear mixed models.23 Stability in DNAm was defined as statistically insignificant changes in DNAm between ages 10 and 18 years.
Replication analyses were carried out in BAMSE. Of the 35 CpGs, DNAm on 33 CpGs was available in BAMSE (cg05072552 and cg09249800 were not available). Altogether, 27 of the 33 CpGs had a nominal P-value < 0.05 in BAMSE. Notably, at all the 33 CpGs, the pattern of DNAm differentiation due to atopy was replicated (Table 2). We also assessed the association of methylation with expression of the genes in Table 2 (Tables S2a,b). The most significant association was observed at cg10159529 with expression of IL5RA (coefficient = −12.58, P-value = 6.76 × 10−20). CpG cg10159529 was located in the TSS1500 of IL5RA.
We further conducted bioinformatic functional analyses on the 33 CpGs using ToppFun, based on 29 genes that these 33 CpGs mapped to. We identified seven enriched GO molecular functions (Table 3). Five of the 29 genes are involved in these seven enrichment terms, B4GALT7, SEC14L1, GPR44, PCYT1A, and IL5RA. A further 15 pathways enriched in GO biological processes are also identified (Table 3). The findings were not statistically significant at FDR = 0.05.
TABLE 3.
Enriched pathways in GO molecular function and biological processes identified using genes from the longitudinal assessment of SPT on DNAm. For molecular functions, all detected pathways are with P-value of 0.091, and for biological processes, the P-values are 0.099
| Pathway ID | Name | Raw P-value | FDR P-value | Genes from our study involved in each term |
|---|---|---|---|---|
| GO molecular function | ||||
| GO:0046525 | Xylosylprotein 4-beta-galactosyltransferase activity | 0.0014 | B4GALT7 | |
| GO:0039552 | RIG-I binding | 0.0014 | SEC14L1 | |
| GO:0004956 | Prostaglandin D receptor activity | 0.0028 | GPR44 | |
| GO:0001785 | Prostaglandin J receptor activity | 0.0028 | 0.091 | GPR44 |
| GO:0004105 | Choline-phosphate cytidylyltransferase activity | 0.0028 | PCYT1A | |
| GO:0004958 | Prostaglandin F receptor activity | 0.0028 | GPR44 | |
| GO:0004914 | Interleukin-5 receptor activity | 0.0028 | IL5RA | |
| GO biological process | ||||
| GO:0032674 | Regulation of interleukin-5 production | 0.0004 | 0.099 | EPX, IL5RA |
| GO:0032634 | Interleukin-5 production | 0.0004 | EPX, IL5RA | |
| GO:0045598 | Regulation of fat cell differentiation | 0.0006 | NCOR2, ZFPM1, METRNL | |
| GO:0002761 | Regulation of myeloid leukocyte differentiation | 0.0007 | FADD, ZFPM1, MITF | |
| GO:0032673 | Regulation of interleukin-4 production | 0.001 | ZFPM1, EPX | |
| GO:0045403 | Negative regulation of interleukin-4 biosynthetic process | 0.0014 | ZFPM1 | |
| GO:0036116 | Long-chain fatty-acyl-CoA catabolic process | 0.0014 | ACOT7 | |
| GO:2000454 | Positive regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation | 0.0014 | FADD | |
| GO:0036114 | Medium-chain fatty-acyl-CoA catabolic process | 0.0014 | ACOT7 | |
| GO:0072365 | Regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter | 0.0014 | NCOR2 | |
| GO:0060377 | Negative regulation of mast cell differentiation | 0.0014 | ZFPM1 | |
| GO:1902303 | Negative regulation of potassium ion export | 0.0014 | KCNH2 | |
| GO:1900533 | Palmitic acid metabolic process | 0.0014 | ACOT7 | |
| GO:1900535 | Palmitic acid biosynthetic process | 0.0014 | ACOT7 | |
| GO:0032633 | Interleukin-4 production | 0.0015 | ZFPM1, EPX |
For the 33 CpGs tested in BAMSE, 13 were on a list of 190 672 potentially spurious CpGs.30 These sites may be more likely to contain outliers that influence results. To examine the possibility of this concern, we tested for multimodality using the R package diptests31 and visually inspected the density plots of DNAm beta values. None of the multimodality tests were statistically significant at the 0.05 significance level, and the sample density plots of all the 13 CpGs did not support multimodality either (Table S3 and Figure S1).
Of the 1437 CpGs, we identified three CpGs (cg14121142 on DDN, cg23842695 on PRKD2, and cg26496795 on ANKRD20A8P) showing an interaction effect between any allergic sensitization and age (FDR = 0.05). At these three CpGs, the effect of sensitization on DNAm was strengthened at age 18 years compared to 10 years (Table 4). Associations at these CpGs were consistent with those when specific IgEs were applied to determine allergic sensitization status (Table S4). In addition, DNAm at these three CpGs was shown to be stable between ages 10 and 18 years.23 A ToppFun analysis indicated that PRKD2 is possibly involved in one enriched GO pathway of molecular functions (protein kinase C activity, P-value = 0.077) and in five GO biological processes (Table 5, P-value = 0.022).
TABLE 4.
Identified three CpGs such that age moderates the association of DNAm with SPT (genome-scale study)
| CpG name | Gene name | Location | Chr. | Main effects (SPT) | Interaction effects (SPT × age) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Est. | Raw P-value | FDR P-value | Est. | Raw P-value | FDR P-value | ||||
| cg14121142 | DDN | Body | 12 | −0.073 | 0.0003 | 0.090 | 0.95 | 4.7 × 10−6 | 0.0058 |
| cg23842695 | PRKD2 | Body | 19 | 0.012 | 0.0005 | 0.11 | −0.24 | 1.2 × 10−5 | 0.015 |
| cg26496795 | ANKRD20B | Body | 2 | 0.050 | 0.94 | 0.99 | −0.10 | 1.9 × 10−5 | 0.023 |
Chr., chromosome; Est., estimation.
TABLE 5.
Enriched pathways identified based on the three CpGs in GO biological process with gene PRKD2 included. All the detected pathways are with FDR-adjusted P-value of 0.022
| Pathway ID | Name | Raw P-value | FDR 2-value | Genes from our study included in the pathways |
|---|---|---|---|---|
| GO:0089700 | Protein kinase D signaling | 0.00032 | 0.022 | PRKD2 |
| GO:1901727 | Positive regulation of histone deacetylase activity | 0.00054 | ||
| GO:0038033 | Positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway | 0.00064 | ||
| GO:0038089 | Positive regulation of cell migration by vascular endothelial growth factor signaling pathway | 0.00075 | ||
| GO:1901725 | Regulation of histone deacetylase activity | 0.00075 |
Turning to mono-sensitization vs poly-sensitizations, 48 CpGs passed screening based on comparison in DNAm between the two groups at each age for each gender (Table S5). Via linear mixed models, DNAm at three of the 48 CpGs was shown to be significantly different between mono- and poly-sensitized subjects, adjusting for the same confounders and covariates as in the analyses for status of sensitization to any allergens. At cg11371879 (in the body of DENND3 gene) and cg08762603 (intergenic), DNAm was lower among subjects with poly-sensitization (cg11371879, coefficient = −0.31, P-value 5.03 × 10−4; and cg08762603, −0.32 and 8.46 × 10−4, respectively). At cg01381613 (TSS200 of PRM3 gene), DNAm was higher for poly-sensitized subjects (coefficient = 0.18, P-value 2.25 × 10−4). However, when testing these three CpGs in BAMSE, none were statistically significant at 0.05 level.
For the testing of moderating effects of gender or age in IoW on the association of mono- vs poly-sensitization with DNAm, no statistically significant interactions were identified at FDR = 0.05.
4 |. DISCUSSION
We examined the association of allergic sensitization with DNAm at the genome scale. In the IoW, 35 CpGs showed an association with the status of sensitization, of which 33 were replicated in BAMSE. The consistent findings between the two independent cohorts suggested informativeness of the detected CpGs. In IoW, DNAm at three CpGs was shown to be different between subjects allergic to one allergen and subjects allergic to more than one allergens, but the findings were not replicated in BAMSE.
Based on the 33 CpGs, genes were identified with biological functions relevant to allergic sensitization. The top enriched pathways in GO biological processes were involved in the regulation of IL-5 production, and these processes included IL5RA and EPX. Furthermore, when assessing the association of DNAm with gene expression, the strongest association was found between cg10159529 (in the promoter region of IL5RA) and expression of IL5RA. The alpha subunit of the IL-5 receptor complex is expressed on eosinophils and B cells and plays an important role in allergic inflammation through the regulation of expression of genes involved in proliferation, cell survival, and maturation and effector functions of B cells and eosinophils. Genetic variation in IL5RA has been found to be associated with allergic sensitization,32 asthma,33–35 and atopic dermatitis.36,37 Likewise, EPX encodes eosinophil peroxidase, a component of the matrix of the cytoplasmic granules of eosinophils. It is likely that both these signals arise from the higher numbers of blood eosinophils present in atopic individuals.38
The ACOT7 gene was found enriched in four biological processes. Effect sizes of three CpGs (cg09249800, cg11699125, and cg21220721) in this gene were among the largest. These three CpGs were also detected in a genome-scale epigenetic meta-analysis of serum total IgE in Hispanic children by Chen et al.,39 and in an epigenome-scale association study of asthma and wheeze in a European cohort.33 In a recent association study of allergen specific IgE,40 ACOT7 was identified as well, but it was not associated with self-reported allergies. The previous findings and results in the present study indicate a potentially critical role of CpGs on the ACOT7 gene in allergic sensitizations and related conditions. It was observed that in the assessment of association of DNAm with expression of ACOT7, DNAm at two CpGs available in BAMSE (cg11699125 and cg21220721 located in gene body) did not show any statistically significant association.
Among all the detected CpGs, decreased DNAm was observed in subjects allergic to any allergen, except for cg19210306 located on the 5’ UTR region of the TEX30 gene. DNAm reduction on most CpGs has also been observed in a recent epigenome-scale study on asthma.27 The TEX30 gene has been shown to be mutated in patients with allergic eczema and their fathers.41 A further investigation is warranted to examine the role of cg19210306 in the development of allergic sensitization leading to atopic eczema.
At three CpGs (cg14121142, cg23842695, and cg26496795) in IoW, the period of adolescence transition seemed to strengthen the association of overall sensitization with DNAm. However, insignificant moderating effects of age were suggested on the association of mono- vs poly-sensitizations with DNAm. It is likely that the moderating effects of age only occurred to atopic status and are irrelevant to the number of allergic sensitizations.
It is not clear whether there is a fundamental difference in IgE responses in mono- and poly-sensitized subjects or poly-sensitized subjects are merely “more reactive” than mono-sensitized subjects; a quantitative rather than qualitative difference.42 In this study, a statistically significant difference in DNAm between mono- and poly-sensitized subjects was observed on three CpGs in IoW, but the findings were not replicated in BAMSE. Immune responses in poly-sensitized subjects are likely not to be “qualitatively” different. Pepys detailed atopic status in quantity terms based on the number of positive SPT results, and classified patients to 0, 1, 2, or 3 or more groups.43 Although disagreement between IgE and SPT has been discussed,42 there is evidence that compared to mono-sensitized patients, poly-sensitized patients tend to have a higher serum total and specific IgE levels.44–46 Our findings with specific screening IgE also demonstrated the general agreement between IgE and SPT with respect to their association with DNAm. It is possible that the sample sizes in both cohorts were insufficient to identify CpGs showing differentiation in DNAm between mono- and poly-sensitized patients and further studies in larger cohorts are warranted.
The findings of this study were based on data such that allergic sensitization status was assessed at the same age when blood samples collected, and that only pre- and postadolescence in IoW were considered. We thus were not able to assess the trend of DNAm changes and differentiate whether DNAm changes were casual or a consequence of allergic sensitization. It would be desirable to have allergic sensitization assessment at denser time points (starting from nonsensitization) during adolescence along with DNAm at those time points or at lagged time points. In addition, BAMSE did not have paired pre-adolescence DNAm available in sufficient numbers and we thus did not replicate age-specific associations with allergic sensitization. Finally, it would be necessary to include information on genetic polymorphisms that may modify susceptibility of allergic sensitization with similar allergen exposure.
Supplementary Material
ACKNOWLEDGMENT
The authors are thankful to Ramya Velampati for the help in the analyses, and the nurses and staff at the David Hide Asthma & Allergy Research Centre, Isle of Wight, UK, for their help in recruitment and sample collections. Our special thanks also go to the High Performance Computing Facility provided by the University of Memphis. The BAMSE team would like to thank Professors Jean Bousquet and Josep Maria Antó for coordinating the MeDALL project and Professor Charles Auffray and his team for generating the transcriptomics data in BAMSE.
Funding information
This study was supported by the National Institutes of Health research funds R21AI099367 (PI: H Zhang), R01AI121226 (MPI: H Zhang and JW Holloway), and R01AI091905 (PI, W Karmaus). BAMSE was supported by the Swedish Heart-Lung Foundation, the Swedish Research Council, Stockholm County Council (ALF), the Swedish Research Council Formas and the Mechanisms of the Development of ALLergy (MeDALL) project, European Framework Programme 7, Project No: 261357. Erik Melen was supported by grants from the Strategic Research Programme (SFO) in Epidemiology at Karolinska Institutet and the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (grant agreement n° 757919, TRIBAL).
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Sly PD, Boner AL, Björksten B, et al. Early identification of atopy in the prediction of persistent asthma in children. Lancet. 2008;372 (9643):1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arshad SH, Kurukulaaratchy RJ, Fenn M, Matthews S. Early life risk factors for current wheeze, asthma, and bronchial hyperresponsiveness at 10 years of age. CHEST J. 2005;127(2):502–508. [DOI] [PubMed] [Google Scholar]
- 3.Heinzerling L, Frew AJ, Bindslev-Jensen C, et al. Standard skin prick testing and sensitization to inhalant allergens across Europe - a survey from the GA2LEN network. Allergy. 2005;60(10):1287–1300. [DOI] [PubMed] [Google Scholar]
- 4.Food Allergy Research and Education. Skin Prick Tests. McLean, VA: Food Allergy Research and Education; 2016:1. [Google Scholar]
- 5.Roberts G, Zhang H, Karmaus W, et al. Trends in cutaneous sensitization in the first 18 years of life: results from the 1989 Isle of Wight birth cohort study. Clin Exp Allergy. 2012;42(10): 1501–1509. [DOI] [PubMed] [Google Scholar]
- 6.Lockett GA, Huoman J, Holloway JW. Does allergy begin in utero? Pediatr Allergy Immunol. 2015;26(5):394–402. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484. [DOI] [PubMed] [Google Scholar]
- 8.Sordillo JE, Lange NE, Tarantini L, et al. Allergen sensitization is associated with increased DNA methylation in older men. Int Arch Allergy Immunol. 2012;161(1):37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White GP, Hollams EM, Yerkovich ST, et al. CpG methylation patterns in the IFNgamma promoter in naive T cells: variations during Th1 and Th2 differentiation and between atopics and non-atopics. Pediatr Allergy Immunol. 2006;17(8):557–564. [DOI] [PubMed] [Google Scholar]
- 10.Everson TM, Lyons G, Zhang H, et al. DNA methylation loci associated with atopy and high serum IgE: a genome-wide application of recursive Random Forest feature selection. Genome Med. 2015;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guthikonda K, Zhang H, Nolan VG, et al. Oral contraceptives modify the effect of GATA3 polymorphisms on the risk of asthma at the age of 18 years via DNA methylation. Clin Epigenetics. 2014;6(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yousefi M, Karmaus W, Zhang H, Ewart S, Arshad H, Holloway JW. The methylation of the LEPR/LEPROT genotype at the promoter and body regions influence concentrations of leptin in girls and BMI at age 18 years if their mother smoked during pregnancy. Int J Mol Epidemiol Genet. 2013;4(2):86. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Tong X, Holloway JW, et al. The interplay of DNA methylation over time with Th2 pathway genetic variants on asthma risk and temporal asthma transition. Clin Epigenetics. 2014;6(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arshad SH, Holloway JW, Karmaus W, et al. Cohort profile: the Isle of Wight Whole Population Birth Cohort (IOWBC). Int J Epidemiol. 2018;47(4):1043–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehne B, Drong AW, Loh M, et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray MX, Tong X, Zhang H. ttScreening: Genome-wide DNA methylation sites screening by use of training and testing samples (R Package); 2014. [Google Scholar]
- 19.Ray MA, Tong X, Lockett GA, Zhang H, Karmaus WJ. An efficient approach to screening epigenome-wide data. Biomed Res Int. 2016;2016:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang FF, Cardarelli R, Carroll J, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6(5):623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Maarri O, Becker T, Junen J, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122(5):505–514. [DOI] [PubMed] [Google Scholar]
- 22.Xu C-J, Bonder MJ, Söderhäll C, et al. The emerging landscape of dynamic DNA methylation in early childhood. BMC Genom. 2017;18 (1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han L, Zhang H, Kaushal A, et al. Stability of DNA methylation pre- and post-adolescence and related risk factors; 2018. Division of Epidemiology, Biostatistics, and Environmental Health, School of Public Health, University of Memphis, Memphis, TN. [Google Scholar]
- 24.Hansen KD, Aryee M, Irizarry RA, et al. minfi: Analyze Illumina Infinium DNA Methylation Arrays in R Package; 2018. [Google Scholar]
- 25.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickman M, Asarnoj A, Tillander H, et al. Childhood-to-adolescence evolution of IgE antibodies to pollens and plant foods in the BAMSE cohort. J Allergy Clin Immunol. 2014;133(2):580–582. [DOI] [PubMed] [Google Scholar]
- 27.Xu CJ, Söderhäll C, Bustamante M, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med. 2018;6(5):379–388. [DOI] [PubMed] [Google Scholar]
- 28.Gref A, Merid SK, Gruzieva O, et al. Genome-wide interaction analysis of air pollution exposure and childhood asthma with functional follow-up. Am J Respir Crit Care Med. 2017;195(10):1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8(1):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naeem H, Wong NC, Chatterton Z, et al. Reducing the risk of false discovery enabling identification of biologically significant genomewide methylation status using the HumanMethylation450 array. BMC Genom. 2014;15:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maechler M Diptest: Hartigan’s dip test statistic for unimodality—corrected. R package version 0.75–7; 2015. See https://CRAN.R-project.org/package=diptest [Google Scholar]
- 32.Tripathi P, Hong X, Caruso D, Gao P, Wang X. Genetic determinants in the development of sensitization to environmental allergens in early childhood. Immun Inflamm Dis. 2014;2(3):193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arathimos R, Suderman M, Sharp GC, et al. Epigenome-wide association study of asthma and wheeze in childhood and adolescence. Clin Epigenetics. 2017;9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forno E, Wang T, Yan Q, et al. A multiomics approach to identify genes associated with childhood asthma risk and morbidity. Am J Respir Cell Mol Biol. 2017;57(4):439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheong HS, Kim LH, Park BL, et al. Association analysis of interleukin 5 receptor alpha subunit (IL5RA) polymorphisms and asthma. J Hum Genet. 2005;50(12):628–634. [DOI] [PubMed] [Google Scholar]
- 36.Namkung JH, Lee JE, Kim E, et al. IL-5 and IL-5 receptor alpha polymorphisms are associated with atopic dermatitis in Koreans. Allergy. 2007;62(8):934–942. [DOI] [PubMed] [Google Scholar]
- 37.Kotsimbos AT, Hamid Q. IL-5 and IL-5 receptor in asthma. Mem Inst Oswaldo Cruz. 1997;92(suppl 2):75–91. [DOI] [PubMed] [Google Scholar]
- 38.Lewis SA, Pavord ID, Stringer JR, Knox AJ, Weiss ST, Britton JR. The relation between peripheral blood leukocyte counts and respiratory symptoms, atopy, lung function, and airway responsiveness in adults. Chest. 2001;119(1):105–114. [DOI] [PubMed] [Google Scholar]
- 39.Chen W, Wang T, Pino-Yanes M, et al. An epigenome-wide association study of total serum IgE in Hispanic children. J Allergy Clin Immunol. 2017;140(2):571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ek WE, Ahsan M, Rask-Andersen M, et al. Epigenome-wide DNA methylation study of IgE concentration in relation to self-reported allergies. Epigenomics. 2017;9(4):407–418. [DOI] [PubMed] [Google Scholar]
- 41.Luo W, Xu W, Xia L, et al. Family-based whole exome sequencing of atopic dermatitis complicated with cataracts. Oncotarget. 2017;8 (35):59446–59454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bousquet J, Anto JM, Wickman M, et al. Are allergic multimorbidities and IgE polysensitization associated with the persistence or reoccurrence of foetal type 2 signalling? The MeDALL hypothesis Allergy. 2015;70(9):1062–1078. [DOI] [PubMed] [Google Scholar]
- 43.Pepys J “Atopy”: a study in definition. Allergy. 1994;49(6):397–399. [DOI] [PubMed] [Google Scholar]
- 44.Lee JH, Noh G. Clinical significance and evaluation of polysensitisation using skin sensitisation profiling in atopic dermatitis. Inflamm Allergy Drug Targets. 2013;12(3):212–220. [DOI] [PubMed] [Google Scholar]
- 45.Rudin A, Macaubas C, Wee C, Holt BJ, Slya PD, Holt PG. “Bystander” amplification of PBMC cytokine responses to seasonal allergen in polysensitized atopic children. Allergy. 2001;56(11):1042–1048. [DOI] [PubMed] [Google Scholar]
- 46.Bousquet J, Knani J, Hejjaoui A, et al. Heterogeneity of atopy I Clinical and immunologic characteristics of patients allergic to cypress pollen. Allergy. 1993;48(3):183–188. [DOI] [PubMed] [Google Scholar]
- 47.Illumina, Field Guide to Methylation Methods Illumina Sequencing Methods; 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


