Abstract
Targeted metabolomics and biochemical studies complement the ongoing investigations led by the Human Proteome Organization (HUPO) Biology/Disease-driven-Human Proteome Project (B/D-HPP). However, it is challenging to identify and prioritize metabolite and chemical targets. Literature mining-based approaches have been proposed for target proteomics studies, but text mining methods for metabolite and chemical prioritization is hindered by a large number of synonyms and non-standardized names of each entity. In this study, we developed a cloud-based literature mining and summarization platform that maps metabolites and chemicals in the literature to unique identifiers and summarizes the co-publication trends of metabolite/chemicals and B/D-HPP topics using the Protein Universal Reference Publication-Originated Search Engine (PURPOSE) scores. We successfully prioritized metabolites and chemicals associated with the B/D-HPP targeted fields, with the results validated by checking against expert-curated associations and enrichment analyses. Comparing with existing algorithms, our system achieved better precision and recall in retrieving chemicals related to B/D-HPP focused area. Our cloud-based platform enables queries on all biological terms in multiple species, which will contribute to B/D-HPP and targeted metabolomics/chemical studies.
Keywords: Metabolomics, Chemicals, Biology/Disease-Driven Human Proteome Project, Literature mining, PURPOSE (Protein Universal Reference Publication-Originated Search Engine), FACTA+ (Finding Associated Concepts with Text Analysis), BEST (Biomedical Entity Search Tool)
INTRODUCTION
The Human Proteome Organization (HUPO) Biology/Disease-driven-Human Proteome Project (B/D-HPP) is a coordinated comprehensive proteomics profiling effort that focuses on human biology and diseases1–3. Investigations of metabolites and chemicals associated with human biology and diseases can enhance and complement the ongoing studies on B/D-HPP1. With the advancement of targeted assays, researchers can quantify hundreds of metabolites or chemical compounds simultaneously4. These high-throughput approaches have the potential to characterize the chemical landscape of human biology in various organs and identify the metabolomics disturbance under disease conditions5, 6, which will contribute to a holistic understanding of biology and diseases.
Similar to proteomics studies, target prioritization is crucial for targeted metabolomics and chemical investigations7. There are more than tens of thousands of metabolites and hundreds of thousands of exogenous and endogenous chemicals8; however, many modern targeted assays can only handle hundreds to thousands of targets at a time9. In order to maximize the utility of the targeted approaches, it is crucial to prioritize the metabolites and chemicals relevant to the study. Previously, researchers have proposed computational approaches to prioritize proteins using literature mining algorithms10–12. Nevertheless, due to the plethora of metabolites and chemicals, a comprehensive tool for their prioritization is lacking. In addition, many metabolites and chemicals have a great number of synonyms and non-standardized names13, 14, which hindered the development of automated approaches for their identification15.
Recent studies present efficient algorithms that summarize the strength and specificity of protein-topic co-publication patterns in the PubMed literature11, 12. Such methods prioritized the associations between any topic and any protein in the PubMed abstract. With the ongoing curation efforts of the Human Metabolome Database (HMDB)8, Chemical Entities of Biological Interest (ChEBI)16, and updates in the Medical Subject Headings (MeSH)17, there is an opportunity to extend the literature mining algorithms to characterize the relations between metabolites/chemicals and any search topic systematically.
In this study, we implemented a cloud-based system for prioritizing metabolites and chemicals for the B/D-HPP targeted fields and any custom search terms. Our system employs the state-of-the-art approach of bio-entity tagging and PubMed literature mining18, searches the PubMed database in real time, compiles the results automatically, and ranks the retrieved metabolites and chemicals within a few seconds using an efficient co-publication summarization algorithm12. Our system will enable the comprehensive investigations of metabolites and chemicals in all targeted areas of B/D-HPP, complementing the ongoing efforts on proteomic profiling in these areas of interest.
METHODS
Data Retrieval for Literature Mining
The targeted areas of B/D-HPP are retrieved from the B/D-HPP website19. The identified B/D-HPP topics are brain, cancers, cardiovascular, diabetes, extreme conditions, eyeOme, food and nutrition, glycoproteomics, immune-peptidome, infectious diseases, kidney and urine, liver, mitochondria, model organisms, musculoskeletal, PediOme, plasma, protein aggregation, and rheumatic disorders. Table S-1 shows the PubMed search terms for the B/D-HPP targeted fields.
To systematically identify metabolites and chemicals from the PubMed literature, the chemical and species tags from PubTator was obtained for each PubMed article18. The retrieved tags were intersected with the Medical Subject Headings (MeSH) subtrees17 of known chemicals. Through obtaining the PubTator taggings and filtering them by the MeSH ontology tree, the unique identifier of each chemical was identified. This approach effectively mapped the synonyms of chemicals to unique identifiers. To ensure that the most updated metabolite, chemical, and species tags were retrieved, an automated downloader was implemented to retrieve PubTator data files from its File Transfer Protocol (FTP) site periodically. To enable metabolite prioritization, the list of human metabolites was retrieved from the HMDB8. The chemicals included in the HMDB list were employed in the metabolite prioritization tasks.
For each PubMed article with relevant tags, the NLM Entrez Programming Utilities (E-utilities)20 were used to obtain the title, authors, journal, year of publication, and the number of citations.
Metabolite and Chemical Prioritization through PURPOSE score
The Protein Universal Reference Publication-Originated Search Engine (PURPOSE) score was used to prioritize metabolites and chemicals for each of the B/D-HPP targeted area12. The score is defined as:
where nTC denote the number of papers associated with both the topic and the chemical/metabolite (TC), Sum(Cit/Yr) is the sum of yearly citation numbers of TC, nU is the number of PubMed publications, nT is the number of publications related to the topic, and nC is the number of publications associated with the chemical/metabolite. This scoring scheme accounts for the strength and the specificity of topic-chemical associations. In particular, the first parenthesis of the formula summarized the frequency of the topic-chemical co-publication, and the number of annualized citations was included in the algorithm to put higher weights to seminal papers and landmark studies12. The second parenthesis of the formula takes into account the overall popularity of the queried topic and the chemicals. This scoring formula is related to the term frequency-inverse document frequency statistic21, and a similar approach achieved superior performance in proteomics literature mining12.
Enrichment Analyses and Pathway Visualization
In order to identify the biological pathways associated with the retrieved chemicals and metabolites, the Search Tool for Interactions of Chemicals (STITCH) tool was employed to identify the known associations among chemicals, metabolites, genes, and proteins22. The STITCH tool conducts enrichment analysis on an open-sourced database containing 500,000 chemicals, 9.6 million proteins, and 1.6 billion interactions22. The database is maintained by the European Molecular Biology Laboratory, the Swiss Institute of Bioinformatics, and the Center for Protein Research22. Gene Ontology enrichment analyses, KEGG pathway analyses, and network analyses were performed by the STITCH tool22. Network statistics of the gene-metabolite and gene-chemical interaction networks, including centralization, Krackhardt efficiency, transitivity, and connectedness scores, were computed by the R ‘sna’ package23. The centralization of a network was evaluated by Freeman’s centrality score24; the Krackhardt efficiency score computed the proportion of necessary edges that could not be removed without disconnecting the nodes in the network; the transitivity score assessed the proportion of connections where transitivity holds (whether node A is directly connected to node C when node A is connected to node B and node B is connected to node C); and the connectedness score identified the proportion of connected node pairs in the networks23. The Metscape app25–27 in Cytoscape28 was used to visualize the interactions among metabolites, genes, and enzymes. All analyses were conducted on May 20, 2018.
Metabolites/Chemicals-B/D-HPP Linkage Visualization
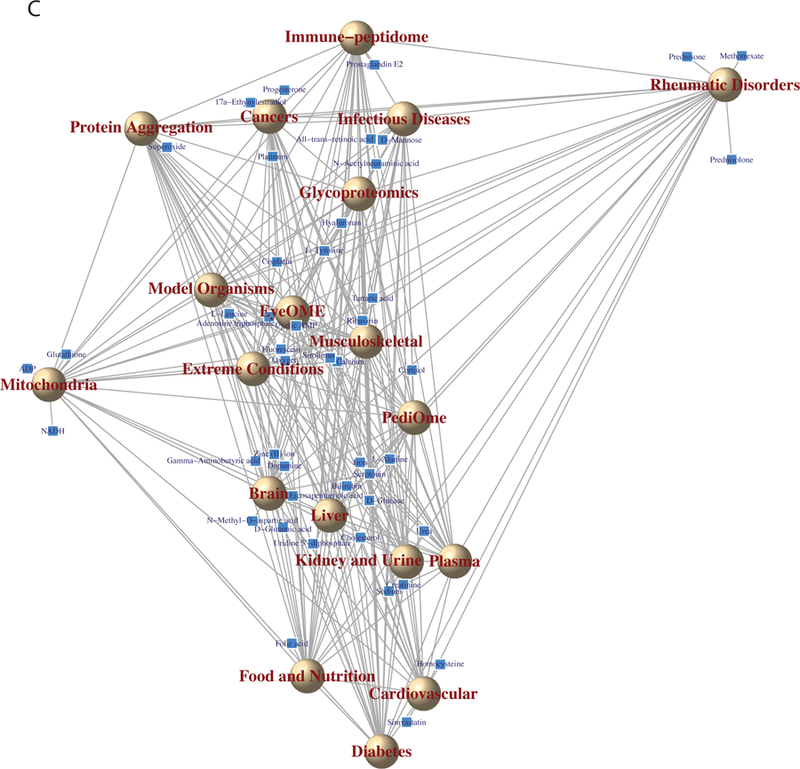
To summarize the linkages among B/D-HPPs and metabolites/chemicals, the correlations among B/D-HPP targeted fields and the associations between the most prominent metabolites/chemicals and the related B/D-HPP areas were visualized. For each pair of B/D-HPP targeted areas, pairwise Spearman’s correlation coefficient was computed for the associated metabolites’ or chemicals’ PURPOSE scores, and one minus the Spearman’s correlation coefficients were defined as the distance between the B/D-HPP fields. Multidimensional scaling (MDS)29 was employed to map the distances among B/D-HPP fields into a two-dimensional graph. The most prominent metabolites and chemicals were added to the resulting graph. The pairwise distances among the B/D-HPP areas reflected their correlations in the PURPOSE scores, and the connections between metabolites/chemicals and B/D-HPP areas visualized the most prominent linkages (metabolites and chemicals were shown in the graphs if their PURPOSE scores in the respective B/D-HPP area were in the top 2.5 percentile and the scores were greater than 20). For metabolites/chemicals strongly associated with only one B/D-HPP, the distances between the metabolites/chemicals and the B/D-HPP areas were inversely proportional to their PURPOSE scores. For metabolites/chemicals strongly correlated with two or more B/D-HPP areas, the distances between the metabolites/chemicals and the associated B/D-HPP areas reflected both their PURPOSE scores in the associated B/D-HPP areas as well as the general correlations among the associated B/D-HPP areas. The figures were generated by R version 3.3 on the Extreme Science and Engineering Discovery Environment (XSEDE) platform30.
Evaluation of the Prioritization Results
Curated chemical-topic associations in the Comparative Toxicogenomics Database (CTD) database31 were employed as the ground truth for evaluating chemical prioritization results. The precision, recall, and the F1 measure (the harmonic mean of precision and recall) of the PURPOSE algorithm and those of the Finding Associated Concepts with Text Analysis (FACTA+) tool32, 33 and the Biomedical Entity Search Tool (BEST)34 were compared. MeSH terms were used to aggregate the synonyms of a chemical. B/D-HPP areas of cancers, diabetes, rheumatic, and liver were selected as the topics for evaluation, due to the availability of the curated annotations and their clean MeSH organization.
Cloud-based User Interface
To facilitate user interaction, a user interface is built with the “shiny” package in R. The system is deployed to a cloud server, allowing researchers to access the system with ease. All statistical analysis was conducted using R version 3.3. The source codes of the cloud-based system, the literature mining backend, and the automated updater for PubTator data files, are available at http://rebrand.ly/metapurposesourcecode.
RESULTS
Summary of Metabolites and Chemicals Published in the PubMed Literature
At the time of evaluation, there were 27 million PubMed articles. PubTator tagged 79,948 chemicals in 9.04 million PubMed articles. 7,508 chemicals (9.39%) are labeled as human metabolites by the HMDB and are mentioned in 7.29 million articles in PubMed. The publication trend of all PubMed articles and articles associated with at least one chemical or metabolite since 1950 is shown in Figure 1A. The number of publications per year on human, chemicals related to human, and human metabolites increased steadily since 1950. The annualized number of publications on human is strongly correlated with the annualized number of papers describing human metabolites (Spearman’s correlation coefficient = 0.998) and the annualized number of publications mentioning chemicals related to human (Spearman’s correlation coefficient = 0.996).
Figure 1.
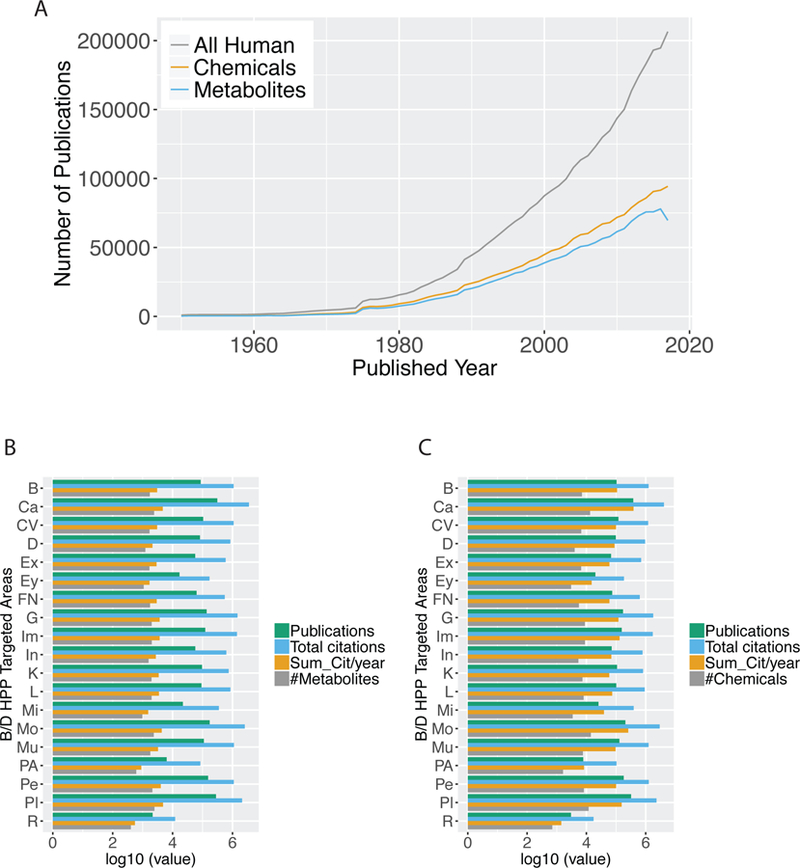
Summary of metabolites and chemical publication patterns in the B/D-HPP targeted areas. (A) The number of all PubMed publications on human, publications associated with any chemical, and publications associated with any metabolite since 1950. The number of PubMed publications increased exponentially since 1975. (B) The number of publications, total citations, citations per year (Sum_Cit/year), and the number of associated metabolites in the B/D-HPP fields. Note that the X-axis is log10-transformed. (C) The number of publications, total citations, citations per year (Sum_Cit/year), and the number of associated chemicals in the B/D-HPP areas. Note that the X-axis is log10-transformed. Abbreviations: B: brain; Ca: cancers; CV: cardiovascular; D: diabetes; Ex: extreme conditions; Ey: EyeOME; FN: food and nutrition; G: glycoproteins; Im: immune-peptidome; In: infectious diseases; K: kidney and urine; L: liver; Mi: mitochondria; Mo: model organisms; Mu: musculoskeletal; Pe: PediOme; Pl: plasma; PA: protein aggregation; R: rheumatic disorders.
Publication Patterns of Metabolites and Chemicals Related to the B/D-HPP Targeted Areas
To prioritize the metabolites and chemicals associated with the B/D-HPP targeted area through literature mining, we implemented the PURPOSE algorithm to summarize the topic- metabolite/chemical co-publication strength in the PubMed literature. For each targeted area of the B/D-HPP, the numbers of associated metabolite/chemical, publications, total citations, and citations per year are summarized in Figures 1B and 1C. The total number of metabolites associated with each B/D-HPP areas is between 405 (rheumatic) and 2,483 (plasma), whereas that of chemicals is between 705 (rheumatic) and 14,070 (model organisms). Across the B/D-HPP topics, the Spearman’s correlation coefficient between the number of identified chemical and that of metabolites is 0.98. The targeted areas with the greatest number of metabolite-related publication are cancers (310,537 publications), plasma (281,874), model organisms (172,860), PediOme (153,053), and glycoproteomics (137,234). The areas with the most chemical-related publications are also cancers (378,929 publications), plasma (321,201), model organisms (205,747), PediOme (181,353), and glycoproteomics (172,064). For the 19 B/D-HPP topics, the Spearman’s correlation coefficient between the number of publications associated with metabolites and the number of publications associated with chemicals is 0.998, and the correlation coefficient between the annualized citation numbers associated with metabolites and that of chemicals is 0.875. All B/D-HPP topics have at least 2,150 publications associated with any metabolites or chemicals, indicating the rich information in the published literature.
Prioritizing Metabolites in the B/D-HPP Targeted Fields
To prioritize metabolites related to the B/D-HPP targeted areas, a list of human metabolites were identified from the HMDB8, where a number of drugs, drug metabolites, and chemical compounds were annotated as metabolites. The metabolites associated with each B/D-HPP were ranked by the PURPOSE score, which balanced the strength (quantified by the number of co-publications in PubMed and the citation numbers of the papers per year) and the specificity (accounted for by the number of publications associated with the topics and that of the proteins in general) of the associations12. As an illustration, L-tyrosine (PURPOSE score = 43.44), sirolimus (43.13), 17a-ethynylestradiol (41.67), docetaxel (41.62), and progesterone (41.32) were the metabolites with the highest PURPOSE score in cancers (Figure 2A). These metabolites were enriched in the epidermal growth factor receptor signaling pathway, protein autophosphorylation, and Fc receptor signaling pathway (Figure 2B). Metscape revealed that these metabolites participated in the metabolism of phosphatidylinositol phosphate and purine (Figure S-1). For diabetes, metabolites D-glucose (44.95), 1,1-dimethylbiguanide (39.80), cholesterol (37.59), adenosine monophosphate (36.64), and creatinine (36.43) had the highest scores (Figure 2A). These metabolites and chemicals were enriched in the PPAR signaling pathway and a number of biological processes including the regulation of cellular ketone metabolic process (Figure 2B). Metscape showed that the prioritized metabolites were involved in glycolysis, gluconeogenesis, cholesterol biosynthesis, and de novo fatty acid biosynthesis pathways (Figure S-1). Metabolites L-tyrosine (39.49), adenosine triphosphate (39.08), D-glucose (37.05), hyaluronan (36.47), and N-acetylneuraminic acid (36.37) attained the highest scores in glycoproteomics (Figure 2A). These metabolites participated in the aminosugars metabolism, fructose and mannose metabolism, and glycerophospholipid metabolism pathways (Figures 2B and S-1). Calcium (39.97), L-tyrosine (38.05), tartaric acid (37.06), adenosine triphosphate (36.74), and D-glucose (36.63) were the metabolites most relevant to the musculoskeletal system (Figure 2A). Pathway analysis revealed that these metabolites were associated with the metabolism pathways of carbohydrates (including fructose, mannose, and galactose) and amino acids (such as tyrosine, arginine, proline, glutamate, aspartate, and asparagine) (Figures 2B and S-1). The results indicated that our methods successfully retrieved many known associations between metabolites and the B/D-HPP areas. Network analysis across the four areas revealed that these gene-metabolite interaction networks (Figure 2B) were highly connected (connectedness scores (the proportion of connected node pairs in the network) > 0.93) and moderately centralized (centralization scores 0.40–0.51). In addition, there were multiple interactions connecting the nodes, resulting in moderate connectivity efficiency (efficiency scores (the proportion of edges that could not be removed without disconnecting the nodes) 0.54–0.72) and transitivity (transitivity scores (when node A is connected to node B and node B is connected to node C, the probability that nodes A and C are directly connected in the network) 0.53–0.65; Figure S-2A). We further computed the scores of all related metabolites for each of the B/D-HPP targeted areas, with the results summarized in Data S-1. Figure 2C shows the correlations among the B/D-HPP targeted areas and highlights metabolites strongly associated with each B/D-HPP. Biologically-related concepts, such as cardiovascular, diabetes, as well as food and nutrition, formed a cluster in the figure.
Figure 2.
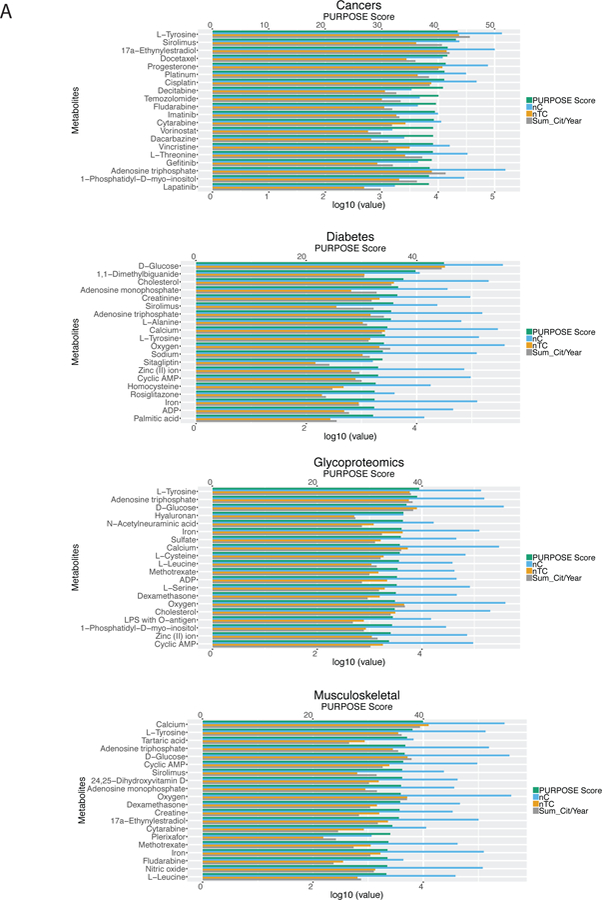
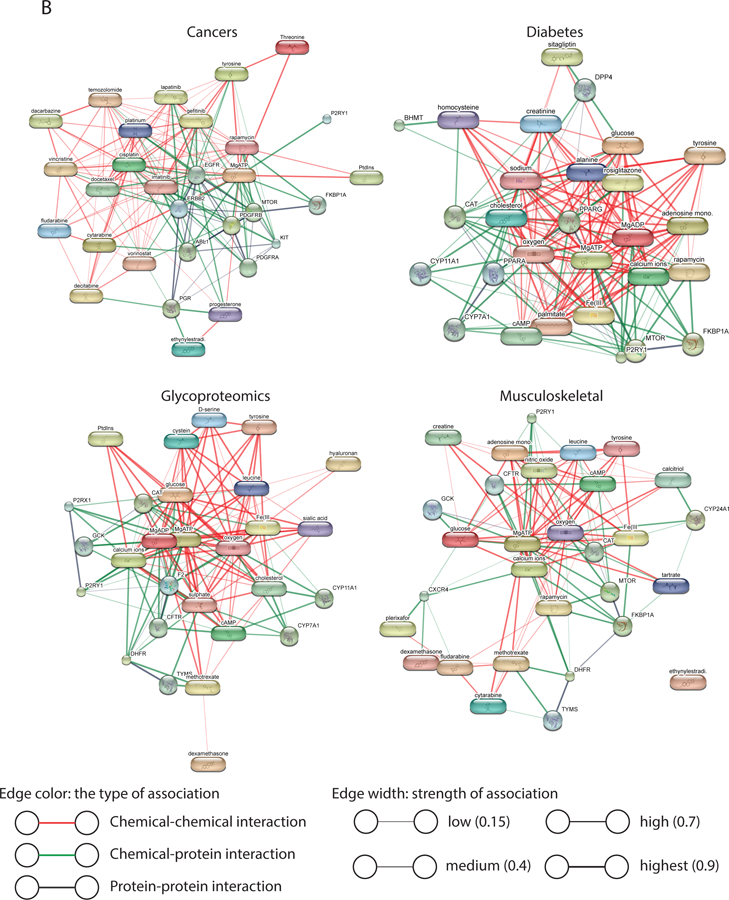
Metabolite prioritization in the selected B/D-HPP targeted areas. (A) Distribution of the Protein Universal Reference Publication-Originated Search Engine (PURPOSE) score of the top metabolites associated with cancers, diabetes, glycoproteomics, and the musculoskeletal system. X-axis at the top: PURPOSE score; X-axis at the bottom: log10(value) of nC (the number of publications associated with the metabolite), nTC (the number of papers associated with both the topic and the metabolite (TC)), and Sum_Cit/Year (citations per year of TC). (B) Network analysis results using the Search Tool for Interactions of Chemicals (STITCH) tool. Metabolites with the highest PURPOSE scores as well as their interacting proteins were shown. (C) Multidimensional scaling (MDS) visualization of the connections among B/D-HPP targeted fields and their associated metabolites. B/D-HPP fields with higher correlation in their associated metabolites’ PURPOSE scores have shorter distances on the graph.
Prioritizing Chemicals in the B/D-HPP Targeted Fields
In addition to metabolites, our algorithm successfully prioritized chemicals associated with the B/D-HPP targeted fields (Data S-2). The identified chemicals ranged from endogenous chemicals (including hormones and neurotransmitters), drugs, drug metabolites, ions, and environmental pollutants, as defined by the PubTator tool18. Based on the DrugBank definition, 1,630 chemicals tagged by PubTator are drugs. Drugs tended to have more PubMed publications (median number of publications = 1195.5) than non-drug chemicals (median number of publications = 4).
Using the PURPOSE score, we identified chemicals implicated with each of the B/D-HPP focused area. For instance, gefitinib (PURPOSE score = 44.35), vemurafenib (42.50), decitabine (42.38), temozolomide (41.79), and lapatinib (41.73) scored the highest among all chemicals in cancers (Figure 3A). These chemicals were enriched in protein autophosphorylation and transmembrane receptor protein tyrosine kinase signaling pathways (Figure 3B). For diabetes, insulin (44.00), C-peptide (42.61), and blood glucose (41.90) had the highest scores (Figure 3A). These chemicals are involved in the PPAR signaling pathway and carbohydrate metabolism mechanisms (Figure 3B). Chemicals proteoglycans (39.15), alpha 1-antitrypsin (38.62), hyaluronic acid (37.35), and glycosylphosphatidylinositols (37.17) scored the highest in glycoproteomics (Figure 3A). The chemicals were highly enriched in the cholesterol metabolic process, carbohydrate derivative binding, and ATP binding functions (Figure 3B). For the musculoskeletal system, parathyroid hormone (39.77), collagen type I trimeric cross-linked peptide (37.85), 1,25-dihydroxyvitamin D (35.96), and zoledronic acid (35.82), and diphosphonates (35.74) were the highest-scoring chemicals (Figure 3A). Pathway analysis revealed that these chemicals were associated with positive regulation of vitamin D 24-hydroxylase activity and the vitamin D catabolic process (Figure 3B). Quantitative analyses on the gene-chemical interaction networks (Figure 3B) showed that these networks are less well-connected than the gene-metabolite interaction networks (Figure 2B) of the same query topic (connectedness scores 0.54–0.87), with low to moderate centralization scores (0.33–0.41). In these B/D-HPP targeted fields, many drugs had high PURPOSE scores, which is consistent with the fact that there were more publications associated with drugs than non-drugs in general. Compared with the gene-metabolite interaction networks, the gene-chemical interaction networks had relatively sparse edges connections, resulting in higher connectivity efficiency scores in general (efficiency score 0.58–0.90) and variable transitivity scores (0.20–0.66; Figure S-2B). Figure S-3 visualized the connections among the B/D-HPP targeted areas and illustrates chemicals strongly associated with each B/D-HPP.
Figure 3.
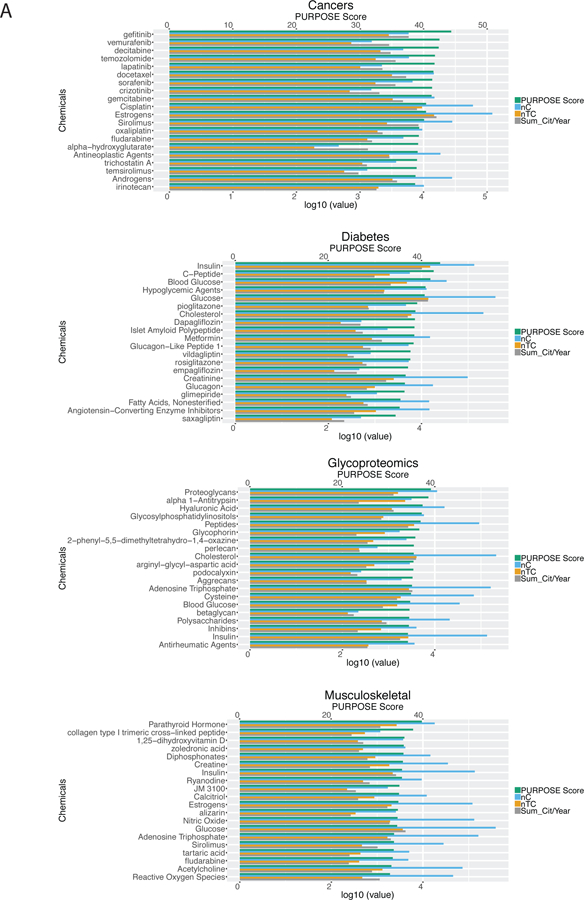
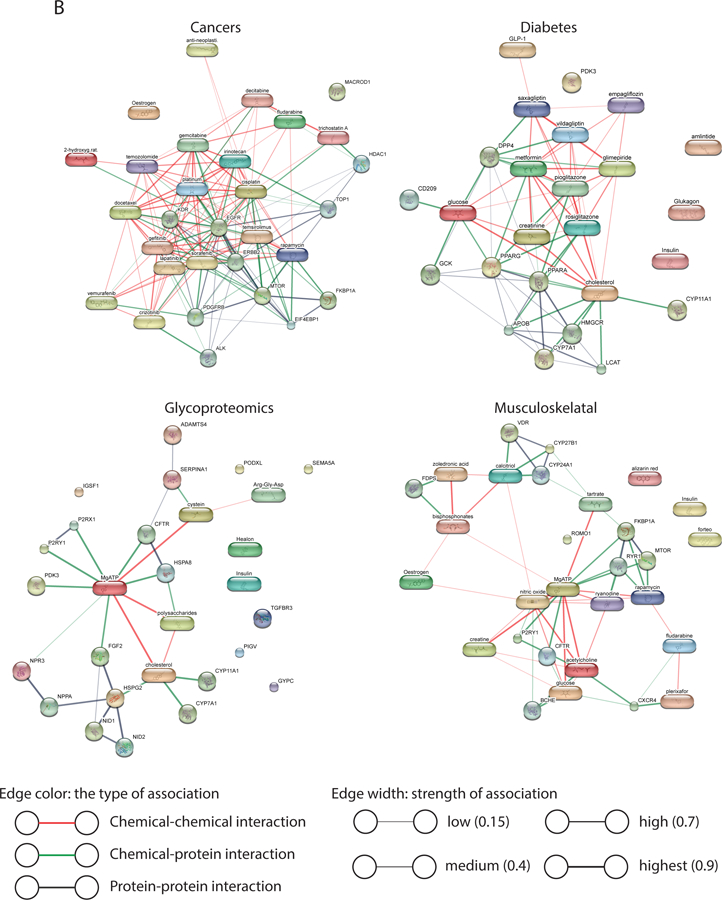
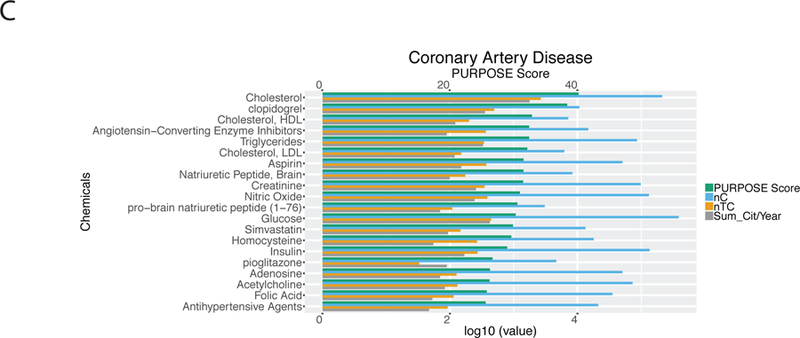
Chemical prioritization in the selected B/D-HPP targeted areas. (A) Distribution of the Protein Universal Reference Publication-Originated Search Engine (PURPOSE) score in the top chemicals associated with cancers, diabetes, glycoproteomics, and the musculoskeletal system. X-axis at the top: PURPOSE score; X-axis at the bottom: log10(value) of nC (the number of publications associated with the chemical), nTC (the number of papers associated with both the topic and the chemical (TC)), and Sum_Cit/Year (citations per year of TC). (B) Network analysis results using the STITCH tool. Chemicals with the highest PURPOSE scores and their interacting proteins were shown. (C) PURPOSE scores of the top chemicals associated with coronary artery disease.
Our algorithm can also identify the chemicals associated with specific biological or medical conditions. As an illustration, when querying “coronary artery disease” in human, our method retrieved many well-known chemicals associated with the disease (Figure 3C), such as cholesterol (PURPOSE score 40.18), HDL (32.87), triglycerides (32.42), LDL (32.13), brain natriuretic peptide (31.54), and homocysteine (29.62). In addition, many drugs related to the treatment of coronary artery disease and related comorbidities were identified by our system. For instance, clopidogrel (38.39) and aspirin (31.55) ranked among the top ten chemicals in this query. These results suggested the extensibility of the PURPOSE algorithm to specific biomedical conditions of clinical importance.
Evaluation of the Prioritization Results
Comparing with the curated topic-chemical relations obtained from the CTD31, our tool successfully retrieved relevant chemicals from the literature. The precision and recall of our tool were better than those of the FACTA+32 and BEST34 systems in most B/D-HPP fields with CTD annotations (Figure 4). Among the top 500 retrieved chemicals associated with cancers, diabetes, or liver, our tool achieved a 5.2–11.4% improvement in precision and a 2.0–5.7% improvement in recall compared with FACTA+, and 5.2–16.8% improvement in precision and 1.6–3.2% improvement in recall compared with BEST. FACTA+ performed better than BEST in cancers but had worse performance in liver, and the two systems had similar performance in diabetes. For rheumatic diseases, which had the least number of PubMed publications, the first 390 chemicals retrieved by PURPOSE attained the highest precision and recall among all three tools, but the precision gradually decreased when we went further down the retrieved list to include chemicals with lower PURPOSE scores, indicating that the PURPOSE algorithm worked better in well-published fields and for well-studied chemicals. These results validated the relevance of the PURPOSE algorithm in chemical prioritization tasks.
Figure 4.
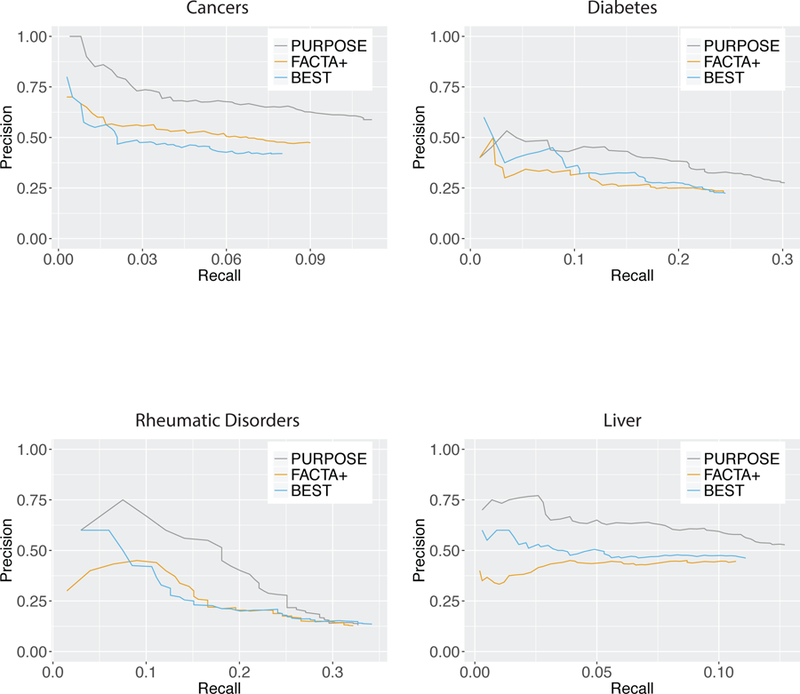
Performance comparison among Protein Universal Reference Publication-Originated Search Engine (PURPOSE), Finding Associated Concepts with Text Analysis (FACTA+), and Biomedical Entity Search Tool (BEST) tools. Precision-recall curves for chemical prioritization for cancers, diabetes, rheumatic diseases, and liver were shown. Biologists-curated topic-chemical relations from the Comparative Toxicogenomics Database (CTD) was used as the ground truth. PURPOSE achieved the best precision and recall in cancers, diabetes, and liver, and have similar performance in rheumatic diseases comparing with FACTA+ and BEST. BEST performed better than FACTA+ in liver but has worse performance in cancers, and the two systems had similar performance in diabetes and rheumatic diseases.
Cloud-Based System Deployment
To facilitate real-time metabolite and chemical prioritization, a cloud-based system was deployed. In addition to the B/D-HPP targeted areas, our system allows users to input any search term of interest and retrieves the results in a few seconds. Modules for enrichment analyses, visualization of PURPOSE score distributions, and summarization of highly-cited publications are available in the browser-based user interface. Our system is freely-accessible at http://rebrand.ly/metapurpose.
DISCUSSION
We have presented a novel general-purpose tool for metabolite and chemical prioritization, with direct applications to the ongoing B/D-HPP investigations1–3. Our cloud-based system automatically obtained the most updated PubMed literature and bio-entity taggings, employed the state-of-the-art literature mining approach to prioritizing metabolites and chemicals, and successfully validated the results in the curated CTD database31. Our approach will facilitate targeted metabolomics and chemical analyses, which is expected to expedite multi-omics integration for investigations on human biology and disease states5, 35, 36.
As many metabolites and chemicals possess a number of evolving synonyms13, it was difficult to track their publication trends, and there was no available tool that prioritizes metabolites for targeted investigations. To address this challenge, our system employed the tagged entities from PubTator18, identified tags for chemicals using the MeSH ontological structure17, and filtered known human metabolites using the curated information from the HMDB8. In addition, we demonstrated the extensibility of the PURPOSE algorithm12, which achieved improved precision and recall compared with the previously-proposed literature mining methods32, 34. Our system allows users to input any search term of interest, queries the most updated PubMed database, retrieves and prioritizes the metabolites and chemicals in real time, and summarizes the results to the users. Our cloud-based system enables enrichment analyses on the retrieved results14, provides external links to curated databases8, and shows the landmark publications describing the relations between the queried topic and the prioritized metabolites and chemicals.
Our results demonstrated that there are a great number of publications describing metabolites and chemicals associated with each of the B/D-HPP targeted fields, indicating the feasibility of building literature mining systems for prioritizing metabolites and chemical targets. The number of publications on human metabolites and chemicals increased steadily since 1950. In recent years, there are more than 70,000 new publications on human metabolites and chemicals (including more than 50,000 papers mentioning drugs) being added to the literature each year. The amount of information posed a challenge to manual literature curation but a unique opportunity for text-mining algorithms in retrieving and aggregating the most updated and relevant information from the literature37. Our system showcased a novel way of utilizing such information, and the prioritized metabolites and chemicals can guide targeted analysis as well as serve as dynamic summaries of the publication trends in the queried fields.
One limitation of our approach is that some newly synthesized chemicals may not have a MeSH term or identifier. Such new chemicals could be missed by PubTator tagging and hence not prioritized by our system. To address this challenge, we implemented an automated updater to obtaining the most recent MeSH entries and PubTator taggings regularly. In addition, like all literature mining tools, the undiscovered topic-chemical associations would not receive a high PURPOSE score. The ongoing efforts on high-throughput metabolomics and chemical profiling could mitigate this issue38.
In summary, our system successfully identified the relevant metabolites and chemicals associated with each of the B/D-HPP focused fields. Together with the previously described protein prioritization framework12, our tools can compile lists of proteins, metabolites, and chemicals related to the B/D-HPP targeted areas and other human organ-systems or disease states, which will facilitate the design of targeted proteomic, metabolomic, and biochemical profiling methods, and expedite integrative multi-omic analyses. The cloud-based metabolites and chemicals prioritization platform can accommodate any custom search term, enabling scientific investigations of any diseases or organs of interest, and contribute to the development of precision medicine.
Supplementary Material
ACKNOWLEDGMENT
The authors express their appreciation to Professor Griffin Weber for his insight on citation counts, Dr. Stephen Bach and Mr. Chen-Ruei Liu for their valuable advice on literature mining and suggestions on the manuscript, Dr. Mu-Hung Tsai for pointing out the literature mining resources, and Ms. Samantha Lemos for her administrative support. The authors thank the AWS Cloud Credits for Research, Microsoft Azure Research Award, and the NVIDIA Corporation for their supports on the computational infrastructure. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) Bridges Pylon at the Pittsburgh Supercomputing Center through allocation TG-BCS180016, which is supported by National Science Foundation grant number ACI-1548562.
Funding Sources
K.-H. Y. is a Harvard Data Science Fellow. This work was supported in part by grants from National Human Genome Research Institute, National Institutes of Health, grant number 5P50HG007735, National Cancer Institute, National Institutes of Health, grant number 5U24CA160036, the Defense Advanced Research Projects Agency (DARPA) Simplifying Complexity in Scientific Discovery (SIMPLEX) grant number N66001–15-C-4043 and the Data-Driven Discovery of Models contract number FA8750–17-2–0095, and the Ministry of Science and Technology Research Grant, Taiwan, grant number MOST 103–2221-E-006–254-MY2.
ABBREVIATIONS
- HUPO
Human Proteome Organization
- B/D-HPP
biology/disease-driven Human Proteome Project
- MeSH
Medical Subject Headings
- GO
Gene Ontology
- STITCH
Search Tool for Interactions of Chemicals
- PURPOSE
Protein Universal Reference Publication-Originated Search Engine
- FACTA+
Finding Associated Concepts with Text Analysis
- BEST
Biomedical Entity Search Tool
Footnotes
Supporting Information.
The following files are available as supporting information.
Figure S-1. Metscape visualized the pathways involved with the prioritized metabolites.
Figure S-2. The quantitative network statistics of the gene-metabolite interaction and gene-chemical interaction networks.
Figure S-3. Multidimensional scaling (MDS) visualization of the connections among B/D-HPP targeted fields and their associated chemicals.
Table S-1. The PubMed search terms for the B/D-HPP targeted fields.
Data S-1. (XLSX) The metabolite prioritization results for B/D-HPP.
Data S-2. (XLSX) The chemical prioritization results for B/D-HPP.
REFERENCES
- 1.Aebersold R; Bader GD; Edwards AM; van Eyk JE; Kussmann M; Qin J; Omenn GS, The biology/disease-driven human proteome project (B/D-HPP): enabling protein research for the life sciences community. J Proteome Res 2013, 12, (1), 23–7. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R; Bader GD; Edwards AM; van Eyk J; Kussman M; Qin J; Omenn GS, Highlights of B/D-HPP and HPP Resource Pillar Workshops at 12th Annual HUPO World Congress of Proteomics: September 14–18, 2013, Yokohama, Japan. Proteomics 2014, 14, (9), 975–88. [DOI] [PubMed] [Google Scholar]
- 3.Van Eyk JE; Corrales FJ; Aebersold R; Cerciello F; Deutsch EW; Roncada P; Sanchez JC; Yamamoto T; Yang P; Zhang H; Omenn GS, Highlights of the Biology and Disease-driven Human Proteome Project, 2015–2016. J Proteome Res 2016, 15, (11), 3979–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei R; Li G; Seymour AB, High-throughput and multiplexed LC/MS/MRM method for targeted metabolomics. Anal Chem 2010, 82, (13), 5527–33. [DOI] [PubMed] [Google Scholar]
- 5.Chen R; Mias GI; Li-Pook-Than J; Jiang L; Lam HY; Chen R; Miriami E; Karczewski KJ; Hariharan M; Dewey FE; Cheng Y; Clark MJ; Im H; Habegger L; Balasubramanian S; O’Huallachain M; Dudley JT; Hillenmeyer S; Haraksingh R; Sharon D; Euskirchen G; Lacroute P; Bettinger K; Boyle AP; Kasowski M; Grubert F; Seki S; Garcia M; Whirl-Carrillo M; Gallardo M; Blasco MA; Greenberg PL; Snyder P; Klein TE; Altman RB; Butte AJ; Ashley EA; Gerstein M; Nadeau KC; Tang H; Snyder M, Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012, 148, (6), 1293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu KH; Snyder M, Omics Profiling in Precision Oncology. Mol Cell Proteomics 2016, 15, (8), 2525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borras E; Sabido E, What is targeted proteomics? A concise revision of targeted acquisition and targeted data analysis in mass spectrometry. Proteomics 2017, 17, (17–18). [DOI] [PubMed] [Google Scholar]
- 8.Wishart DS; Feunang YD; Marcu A; Guo AC; Liang K; Vazquez-Fresno R; Sajed T; Johnson D; Li C; Karu N; Sayeeda Z; Lo E; Assempour N; Berjanskii M; Singhal S; Arndt D; Liang Y; Badran H; Grant J; Serra-Cayuela A; Liu Y; Mandal R; Neveu V; Pon A; Knox C; Wilson M; Manach C; Scalbert A, HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018, 46, (D1), D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusebauch U; Campbell DS; Deutsch EW; Chu CS; Spicer DA; Brusniak MY; Slagel J; Sun Z; Stevens J; Grimes B; Shteynberg D; Hoopmann MR; Blattmann P; Ratushny AV; Rinner O; Picotti P; Carapito C; Huang CY; Kapousouz M; Lam H; Tran T; Demir E; Aitchison JD; Sander C; Hood L; Aebersold R; Moritz RL, Human SRMAtlas: A Resource of Targeted Assays to Quantify the Complete Human Proteome. Cell 2016, 166, (3), 766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam MP; Venkatraman V; Cao Q; Wang D; Dincer TU; Lau E; Su AI; Xing Y; Ge J; Ping P; Van Eyk JE, Prioritizing Proteomics Assay Development for Clinical Translation. J Am Coll Cardiol 2015, 66, (2), 202–4. [DOI] [PubMed] [Google Scholar]
- 11.Lam MP; Venkatraman V; Xing Y; Lau E; Cao Q; Ng DC; Su AI; Ge J; Van Eyk JE; Ping P, Data-Driven Approach To Determine Popular Proteins for Targeted Proteomics Translation of Six Organ Systems. J Proteome Res 2016, 15, (11), 4126–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu KH; Lee TM; Wang CS; Chen YJ; Re C; Kou SC; Chiang JH; Kohane IS; Snyder M, Systematic Protein Prioritization for Targeted Proteomics Studies through Literature Mining. J Proteome Res 2018, 17, (4), 1383–1396. [DOI] [PubMed] [Google Scholar]
- 13.Mattingly CJ; Rosenstein MC; Davis AP; Colby GT; Forrest JN Jr.; Boyer JL, The comparative toxicogenomics database: a cross-species resource for building chemical-gene interaction networks. Toxicol Sci 2006, 92, (2), 587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn M; von Mering C; Campillos M; Jensen LJ; Bork P, STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res 2008, 36, (Database issue), D684–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shatkay H; Feldman R, Mining the biomedical literature in the genomic era: an overview. J Comput Biol 2003, 10, (6), 821–55. [DOI] [PubMed] [Google Scholar]
- 16.Hastings J; Owen G; Dekker A; Ennis M; Kale N; Muthukrishnan V; Turner S; Swainston N; Mendes P; Steinbeck C, ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res 2016, 44, (D1), D1214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipscomb CE, Medical Subject Headings (MeSH). Bull Med Libr Assoc 2000, 88, (3), 265–6. [PMC free article] [PubMed] [Google Scholar]
- 18.Wei CH; Kao HY; Lu Z, PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res 2013, 41, (Web Server issue), W518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Human Proteome Organization Biology/Disease-driven HPP. https://www.hupo.org/B/D-HPP (July 25),
- 20.Sayers E. Entrez programming utilities help. http://www.ncbi.nlm.nih.gov/books/NBK25499 (July 25),
- 21.Leskovec J; Rajaraman A; Ullman JD, Mining of massive datasets Cambridge university press: 2014; p 1–15. [Google Scholar]
- 22.Szklarczyk D; Santos A; von Mering C; Jensen LJ; Bork P; Kuhn M, STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res 2016, 44, (D1), D380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butts CT, sna: Tools for Social Network Analysis. R package version 2.2–0 In 2010. [Google Scholar]
- 24.Freeman LC, Centrality in social networks conceptual clarification. Social networks 1978, 1, (3), 215–239. [Google Scholar]
- 25.Basu S; Duren W; Evans CR; Burant CF; Michailidis G; Karnovsky A, Sparse network modeling and metscape-based visualization methods for the analysis of large-scale metabolomics data. Bioinformatics 2017, 33, (10), 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karnovsky A; Weymouth T; Hull T; Tarcea VG; Scardoni G; Laudanna C; Sartor MA; Stringer KA; Jagadish HV; Burant C; Athey B; Omenn GS, Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2012, 28, (3), 373–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J; Tarcea VG; Karnovsky A; Mirel BR; Weymouth TE; Beecher CW; Cavalcoli JD; Athey BD; Omenn GS; Burant CF; Jagadish HV, Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics 2010, 26, (7), 971–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P; Markiel A; Ozier O; Baliga NS; Wang JT; Ramage D; Amin N; Schwikowski B; Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003, 13, (11), 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox TF; Cox MA, Multidimensional scaling Chapman and hall/CRC: 2000; p 5–8. [Google Scholar]
- 30.Towns J; Cockerill T; Dahan M; Foster I; Gaither K; Grimshaw A; Hazlewood V; Lathrop S; Lifka D; Peterson GD, XSEDE: accelerating scientific discovery. Computing in Science & Engineering 2014, 16, (5), 62–74. [Google Scholar]
- 31.Davis AP; Grondin CJ; Johnson RJ; Sciaky D; King BL; McMorran R; Wiegers J; Wiegers TC; Mattingly CJ, The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res 2017, 45, (D1), D972–D978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuruoka Y; Tsujii J; Ananiadou S, FACTA: a text search engine for finding associated biomedical concepts. Bioinformatics 2008, 24, (21), 2559–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuruoka Y; Miwa M; Hamamoto K; Tsujii J; Ananiadou S, Discovering and visualizing indirect associations between biomedical concepts. Bioinformatics 2011, 27, (13), i111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S; Kim D; Lee K; Choi J; Kim S; Jeon M; Lim S; Choi D; Kim S; Tan AC; Kang J, BEST: Next-Generation Biomedical Entity Search Tool for Knowledge Discovery from Biomedical Literature. PLoS One 2016, 11, (10), e0164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu KH; Berry GJ; Rubin DL; Re C; Altman RB; Snyder M, Association of Omics Features with Histopathology Patterns in Lung Adenocarcinoma. Cell Syst 2017, 5, (6), 620–627 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu KH; Fitzpatrick MR; Pappas L; Chan W; Kung J; Snyder M, Omics AnalySIs System for PRecision Oncology (OASISPRO): A Web-based Omics Analysis Tool for Clinical Phenotype Prediction. Bioinformatics 2018, 34, (2), 319–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu F; Patumcharoenpol P; Zhang C; Yang Y; Chan J; Meechai A; Vongsangnak W; Shen B, Biomedical text mining and its applications in cancer research. J Biomed Inform 2013, 46, (2), 200–11. [DOI] [PubMed] [Google Scholar]
- 38.Wishart DS, Proteomics and the human metabolome project. Expert Rev Proteomics 2007, 4, (3), 333–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


