Abstract
Influenza databases now contain over 100,000 worldwide sequence records for strains influenza A(H3N2) and A(H1N1). Although these data facilitate global research efforts and vaccine development practices, they also represent a stumbling block for researchers because of their confusing and heterogeneous annotation. Unclear passaging annotations are particularly concerning given the recent work highlighting the presence and risk of false adaptation signals introduced by cell passaging of viral isolates. With this in mind, we aim to provide a concise outline of why viruses are passaged, a clear overview of passaging annotation nomenclature currently in use, and suggestions for a standardized nomenclature going forward. Our hope is that this summary will empower researchers and clinicians alike to more easily understand a virus sample’s passage history when analyzing influenza sequences.
Keywords: influenza virus, passaging adaptations, H1N1, H3N2
1. Introduction
Thousands of influenza viruses are sequenced annually in a global public health endeavor aimed at understanding and combating seasonal epidemics. The constant, steady proliferation of sequenced viruses from year to year has both led to more information available for vaccine development (Ampofo et al. 2013) and allowed researchers to create progressively more detailed viral phylogenies in an effort to identify regions under selection (Anderson et al. 2016; Timofeeva et al. 2017). Increasingly sophisticated analyses using sequences from various collaborative influenza databases, such as the Influenza Research Database Flu database (IRD) (Zhang et al. 2017) and the Global initiative on sharing all influenza data Epiflu (GISAID) database (https://www.gisaid.org), have helped identify long-term evolutionary trends in influenza viruses (Belanov et al. 2015; Du et al. 2017; Moncla, Florek, and Friedrich 2017). Although these efforts have greatly expanded our understanding of influenza virus evolution and have led to more informed vaccine development, they have also highlighted a major stumbling block in influenza research as a whole: spurious adaptation signals introduced by cell passaging (Gatherer 2010; Lee et al. 2013; Chen et al. 2016; McWhite, Meyer, and Wilke 2016).
Although it has long been known that high levels of passaging and cultivation in certain cell types can alter influenza strain phenotype and sequence (Wyde et al. 1977; Robertson et al. 1993; Bush et al. 2000), recently it has been shown that even low levels of passaging in a wide range of cell types can introduce false adaptation signals. Spurious adaptation signals were first identified in egg-passaged influenza sequences some 25 years ago (Robertson et al. 1993). Since then, similar signals have been shown to originate in samples cultivated in a myriad of cell types derived from diverse species and tissues, including canine (Li et al. 2009; McWhite, Meyer, and Wilke 2016; Lin et al. 2017), monkey (McWhite, Meyer, and Wilke 2016) and hamster (Govorkova et al. 1999) cell lines.
The recent identification of a spurious Zanamivir (influenza neuraminidase inhibitor) resistant mutation in MDCK (Mardin Darby Canine Kidney) passaged sequences (Little et al. 2015) highlights that such false signals represent more than just a theoretical concern for the influenza research and the larger medical communities. Although the Zanamivir example is concerning, the impact of such erroneous information on seasonal vaccine development is of a potential greater medical threat. False signals complicate downstream analysis and can lead to poorly inferred evolutionary trends, which may ultimately result in improper strain selection and the development of less effective vaccines. Indeed, recent sub-optimal vaccine strains may have passaged isolates to blame, as highlighted by structural and biochemical analyses linking the poor performance of vaccines developed from egg-passaged sequences directly to mutations caused by passaging (Wu et al. 2017; Zost et al. 2017; Chen et al. 2018).
Although research efforts into other human viruses may also encounter false adaptation signals related to cell passaging, the issue is particularly pronounced in influenza virus because of the diversity of cell lines used to culture the virus and the seasonal vaccine development efforts. In terms of cell line diversity, most other human viruses are solely cultured in primate cell lines (e.g. Zika virus or Ebola virus) or in human cell lines (HIV) (Krowicka et al. 2008; Broadhurst, Brooks, and Pollock 2016; Himmelsbach and Hildt 2018), and these cell lines are likely to produce less significant adaptation signals than the broad collection of cell lines in which influenza samples are cultivated. Influenza is also unique in that global influenza surveillance efforts are aimed at producing yearly vaccines that are likely more influenced by false adaptation signals compared with vaccines or treatments developed over longer periods for slower evolving and non-seasonal disease agents.
With growing focus on the effects of cell passaging on influenza sequencing data, it is becoming increasingly important for researchers to clearly understand the nomenclature used to annotate passaged sequences. To facilitate this understanding, we provide below a clear outline of existing annotation strategies for common sequences in the IRD, GISAID, and the Swiss Institute of Bioinformatics (SIB) OpenFluDB (OpenFlu, http://OpenFlu.vital-it.ch/) databases, and we propose a standardized approach to annotating isolates. We hope that this perspective will catalyze a more systematic approach to creating, storing, and analyzing passaging information in the influenza research community, and that this effort will ultimately strengthen research efforts that lead to refinements in the seasonal vaccine strain selection process.
2. Passaging: we do it because we have to
Given the growing body of research illuminating issues with cell-passaged influenza sequences and the increasing availability of sequences from original clinical specimens, one has to ask why passage influenza samples at all? Most influenza research, whether the investigation involves vaccine development or not, needs to propagate viruses in vitro in order to analyze host characteristics. In an ideal world, clinical specimens would be sequenced directly, analyzed, and used to create an accurate model of influenza adaption that would inform strain inclusion for vaccine development with little to no bias. This best-case scenario, however, is impeded by the techniques needed to characterize viruses in vitro.
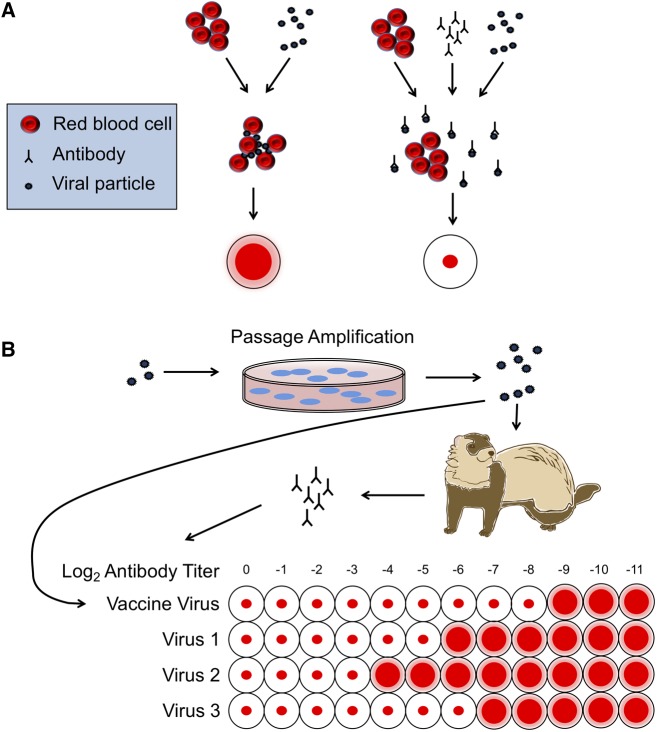
Although it is true that many influenza clinical specimens are directly sequenced without passaging, clinical specimens do not typically provide the amount of virologic material necessary to perform the standard antigenic assays: the hemagglutination-inhibition (HI) test, which is essential in strain selection during vaccine development (Fig. 1) or animal experiments (Krauss, Walker, and Webster 2012; Eisfeld, Neumann, and Kawaoka 2014). Indeed, the HI assay requires a minimum of approximately seven logs of virus per 50 μL (Hierholzer and Killington 1996) (eight hemagglutination units/50 μl; Pedersen 2014), which is acquired through one or more rounds of passaging. Additionally, two types of vaccines require strains which must be passaged either in eggs or in a qualified MDCK cell line (Weir and Gruber 2016; Grohskopf et al. 2018). Thus, while the use of sequences derived from original clinical material represents a research ideal, the current reality is that passaged isolates are a necessary step in obtaining sufficient antigenic and genetic information for vaccine development; therefore, it is important to have a clear understanding of passaging and its effects on viral sequences.
Figure 1.
HI assay for vaccine development. (A) Overview of the HI assay. Hemagglutinin on the surface of viral particles binds red blood cells, creating a lattice of blood cells that show up as a diffuse layer at the top of a microtiter plate well. When enough antibodies with strong affinity for the viral hemagglutinin are present, viral particles are bound and the red blood cells sink, forming a small dot at the bottom of the microtiter plate well. (B) Journey of a viral particle, from isolation to HI assay. Viral particles are isolated, cell passaged (often multiple times), and then either tested for hemagglutination activity or used to produce antibodies via infection of animals with naïve immune systems.
3. Passaging nomenclature
Currently, the vast majority of influenza sequences are passaged isolates from a menagerie of various cell types. This passaging information is indicated via a patchwork of non-standard nomenclature methods that vary wildly across and even within databases. Indeed, the passaging information associated with A (H3N2) samples collected between 2005 and 2018 and stored in OpenFlu, IRD, and GISAID illustrates the haphazard naming and numbering strategies for various cell types used to passage isolates (Table 1). In the GISAID database alone, MDCK passaged samples are indicated with at least fifteen variable naming schemes from different institutions. The absence of clear labeling patterns combined with the extreme variability in naming conventions across cell types and databases create a Gordian knot for researchers seeking to disentangle the effects of passaging on influenza sequences. Despite ongoing work to develop tools to parse passage history abbreviations any such tool will require constant manual updates to keep pace with novel abbreviations introduced by new entries.
Table 1.
Common influenza A(H3N2) passaging annotation patterns across three databases.
| Base pattern | No. cell lines | Example |
|---|---|---|
| Clinical specimen | 0 | Clinical specimen |
| Direct | 0 | Direct |
| OR | 0 | OR |
| Original | 0 | Original |
| Original sample | 0 | Original sample |
| Original specimen | 0 | Original specimen |
| PI | 1 | PI |
| T | 1 | MDCK |
| T# | 1 | MDCK1 |
| T cells | 1 | MDCK Cells |
| P-# | 1 | P-1 |
| P# | 1 | P1 |
| Passage details: T# | 1 | Passage details: PMK01 |
| Passage details: T | 1 | Passage details: MDCK |
| T# (MM/DD/YYYY) | 1 | S1 (9/30/2008) |
| T# (YYYY-MM-DD) | 1 | S2 (2008-9-30) |
| T # +# | 1 | MDCK 1 +1 |
| TT# | 1 | MDCKMDCK1 |
| TT# | 2 | HEPGMDCK1 |
| T/T# | 2 | X/C1 |
| T # +T# | 2 | MDCK 2 +SIAT1 |
| T#/T# | 2 | C1/C2 |
| T# T# | 2 | MDCK2 Siat1 |
| T#/T# (MM/DD/YYYY) | 2 | C1/S1 (01/04/2015) |
| No information | ||
| −N/A- | No information | −N/A- |
T represents type of cell; # represents a single digit number.
Notwithstanding substantial heterogeneity in approach, all passaging annotations aim to provide similar relevant information about the history of the cultivation of an influenza isolate sequence: typically, the type of cell(s) used in passaging, number of passages, and cell handling data (movement between laboratories and/or change in substrate). This information is then used both to identify factors responsible for false adaptation signals and to help distinguish which sequences should be excluded from downstream analyses.
3.1 Cell type
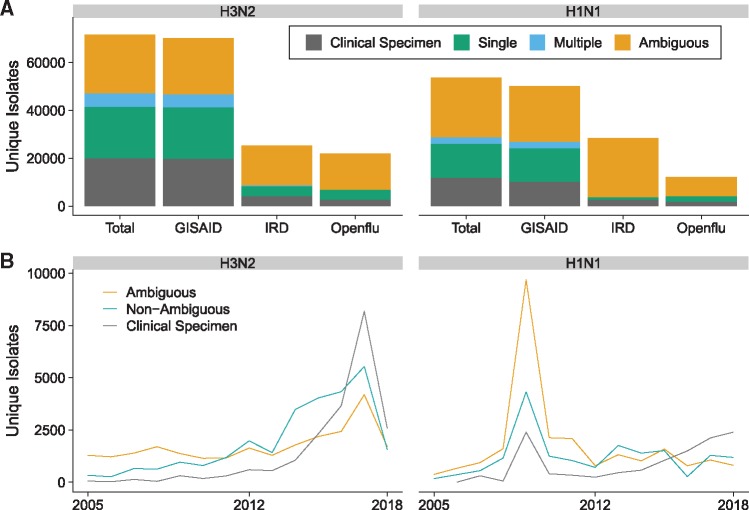
Annotated influenza samples are typically passaged in one or more of only a handful of cell types. Annotations generally begin with an indicator of the cell line used, such as SIAT for MDCK SIAT cells, E for egg, and PMK for primary monkey kidney (Table 2). These indicators are in no way standardized and substantial variation exists for each cell type. Figure 2 illustrates the frequency of unique labels used to identify sequences passaged at least once in a monkey cell line. Although these labels are likely similar enough to allow a researcher to manually distinguish samples that have been passaged in monkey cell lines from those that have not, they are dissimilar enough to make it difficult for an automated script to efficiently do the same. In addition, many indicators are vague or ambiguous (i.e. X1 or C1 for a sample that has been passaged once in ‘an unspecified cell line’), and a significant portion of influenza A(H3N2) and A(H1N1) samples lack any cell type indicator or passaging annotation whatsoever (Fig. 3A). Although the proportion of such unclearly annotated isolates has decreased in recent years, they still accounted for roughly one-third of all recorded A(H3N2) isolates in 2017 (Fig. 3B).
Table 2.
Existing naming conventions and suggested standardized names for common cell lines used to passage influenza viruses.
| Cell type | Common existing annotations | Suggested name |
|---|---|---|
| Egg | E# | Egg# | Embryonated Eggs | AM | EGG |
| Madin-Darby Canine Kidney | MDCK# | M# | MDCK CELLS | MDCK |
| Rhesus Monkey Kidney | RMK# | RHMK# | RII | PMK# | PRHMK# | RhMK |
| Madin-Darby Canine Kidney—SIAT | MDCK-SIAT# | S# | SIATMDCK# | SIAT# | SIAT |
| Unpassaged | Original | OR | Clinical Specimen | No Passage | Primary | Direct | Nasal Swab | CS | Original Specimen |
| Unknown | None | | -N/A- | N/A |
| Unknown Cell | C# | P# | X# | Unknown Cell |
The symbol ‘#’ represents a single digit number other than 0. Note, this table does not attempt to provide a complete list of all possible cell lines but rather focuses on the most common cell types across three databases.
Figure 2.
Word cloud of influenza A(H3N2) sequence annotations indicating passaging in one or more cell lines where at least one is a PMK cell line. Word height corresponds to number of sequences exhibiting a given pattern.
Figure 3.
Influenza A(H3N2) and A(H1N1) sequence isolate counts across three databases, 2005–18. (A) Aggregate isolates data by type. GISAID accounts for the majority of unique isolates in both strains, about half of which either lack clear passage information or have been passaged in multiple cell lines. Isolate types are defined as follows: ‘clinical specimen’: any unpassaged direct clinical specimen; ‘single’: passaged in single identified cell line; ‘multiple’: passaged in multiple identified cell lines; ‘ambiguous’: passaging information unclear (may be single unidentified cell line or multiple cell lines with at least one line unclear). (B) Yearly isolate data by type. In both analyzed strains there is an increase in direct clinical specimen sequences relative to other samples in more recent years. Includes unique records across all three databases (GISAID, IRD, Openflu). Isolate types are as under (A) except ‘non-ambiguous’ which refers to isolates passaged in one or multiple identified cell lines.
Even though many influenza virus isolates have missing passaging annotations or are ambiguously labeled, several studies have successfully identified the impact of passaging on sequence fidelity in the most commonly used cell types (Govorkova et al. 1999; Li et al. 2009; McWhite, Meyer, and Wilke 2016; Lin et al. 2017). For influenza A(H3N2) viruses, these studies have shown that MDCK cells expressing human SIAT1 produce sequences that differ the least from sequences derived from original clinical samples and are now (from 2015 onwards) the predominate cell type used in North America to passage isolates as per GISAID records (Li et al. 2009; Chen et al. 2016; McWhite, Meyer, and Wilke 2016).
3.2 Number of passages
Passage number, the most uniformly recorded annotation aspect, is consistently indicated by a number succeeding a cell type indicator with or without a space between (e.g. MDCK2 or MDCK 2). This convention allows researchers to easily parse annotations for passage number information, although some confusion arises when cell line names include numbers (e.g. MV1LU cells) and when passaging annotations lack clear indicators for all cell types (e.g. C3 + 1). As with cell type, many samples exclude information about passage number. This lack of information is represented either explicitly with an X following a cell type indicator or implicitly with lack of a number indicating no information.
Due to the reasonably clear and consistent nomenclature currently in use, passage number is perhaps the easiest factor to study when focusing on influenza adaptation to cell culture. As such, several groups have been able to show that each additional passage has a consistent, additive impact on the presence of false adaptation signals in sequenced samples. Across the most commonly used cell types, sequences diverge more from original clinical specimens as passage number increases (Wyde et al. 1977; McWhite, Meyer, and Wilke 2016). Since most annotated sequences plainly indicate the number of passages it is also easy to consider this trend when inferring influenza virus phylogenies or selecting vaccine strains. Researchers can simply favor sequences which have been passaged less, although the additive effect of passaging makes it difficult to provide an absolute limit on the number of passages acceptable for any given cell type.
3.3 Heterogeneous passaging and cell handling data
Samples are also often passaged in multiple cell lines and/or cell types. Such passaging is commonly indicated by a wide array of symbols, including ‘and’, blank spaces, ‘,’, ‘−’, ‘_’, ‘;’, and ‘+’. Cell lines can also be listed with no separation, for example, M3C3 to indicate three passages in MDCK and three in some other cell line, and certain symbols are used by some researchers to indicate more information than just passaging, such as ‘/’ indicating the transfer of a strain between labs or institutions. This diversity adds another layer of difficulty to parsing sample annotations, and it has made it particularly difficult to investigate the impact of heterogeneous passaging on sequence fidelity. Consequently, most studies lump such samples together and either analyze them as a heterogeneous group (Chen et al. 2016) or exclude them altogether (McWhite, Meyer, and Wilke 2016). Such lumped analyses mean that little can be determined about the effects of heterogeneous passaging and freeze–thaw cycles in specific cell lines.
3.4 Database differences
In an effort to strengthen influenza surveillance efforts, several databases of influenza sequences are maintained, the three largest of which are GISAID, IRD, and OpenFlu. Each of these databases collects sequences from different sources, although there is a fair amount of overlap (see Fig. 3A) as all include publicly available samples from the International Nucleotide Sequence Database Collaboration (INSDC, http://www.insdc.org), stored in the National Center for Biotechnology Information’s GenBank repository (Benson et al. 2018). Although each database supplements these INSDC samples with user-uploaded sequences, IRD and OpenFlu have far fewer user-uploaded data than GISAID, which includes nearly all available influenza A(H3N2) and (H1N1) data (Fig. 3A). Despite the large amount of shared isolate data across databases, each database parses annotations into differently named fields. For example, the sequence and metadata for the influenza A(H3N2) isolate A/Zhuhai/964/2008 was uploaded to Genbank on 24 July 2016 with passage indicated as ‘MDCK’ under the ‘lab_host’ field of the structured comments. In the corresponding GISAID record the passage information is listed correctly under ‘passage details/history’ as ‘MDCK’ while in IRD and OpenFlu the same sample is listed as ‘N/A’ and ‘no information’ under the fields ‘Passage History’ and ‘passage’, respectively. The correct annotation is present in IRD under ‘lab host’, a field that cannot be included when downloading sample information. The correct annotation is wholly absent from the OpenFlu record, even though other records from the same submission did properly import the passaging information from GenBank to OpenFlu under the ‘passage’ field. Consequently, each database will not only contain some degree of unique samples but also divergent nomenclature standards and metadata that can produce conflicting information for the same sample. These variations make it difficult for researchers to easily integrate and investigate sequences from multiple databases, and they may bring a well annotated isolate’s passaging status into question.
3.5 Towards a standard nomenclature
Because of the great diversity in annotation strategies, it is difficult to effectively establish exclusion criteria for passaged influenza sequences. Until the effects of cell passaging are better understood it will remain unclear which, if any, cell types produce influenza sequences that are truly free from false adaptation signals. We therefore propose the use of new standard names for common cell lines (Table 2) and a new universal passage annotation convention for all influenza samples (Table 3). These new standards use elements from common existing annotations. They were selected for ease of both human and machine parsability, while staying as close as possible to existing annotation practices, to minimize potential confusion and cost of switching over. The vast majority of existing isolates are passaged in one of the cell lines indicated in Table 2, but adding additional names for uncommon or novel cell lines should prove relatively straightforward once initial guidelines are established.
Table 3.
Suggested passaging annotation scheme for influenza isolates.
| Suggested annotation changes | Example |
|---|---|
| One standardized name per cell line | MDCK |
| Cell line names should not end in numbers or X | SIAT |
| Passage number indicated via a number immediately following the cell line | SIAT1 |
| Unknown passage number indicated with an X | SIATX |
| Intra-lab passaging in multiple cell lines is denoted with + | SIAT1+EGG1 |
| Passaging in multiple cell lines with transfer between labs is denoted with / | SIAT1/EGG1 |
Cell lines should be represented by a standardized name not ending in X or a number. This includes SIAT cells which previously were also designated as SIAT1 and should be referenced only as SIAT for consistency with the new scheme. Multiple passages in the same cell line should be represented by a number (if number is known) or an X (if number is not known) immediately following the cell line name. Passages in different cell lines should be separated by a plus (+) to indicate intra-lab passaging or a slash (/) to indicate transfer between labs.
Additionally, we strongly suggest that influenza databases incorporate changes to encourage accurate passaging annotation and facilitate passage-focused research. These include requiring a passage history field for all sequence submissions, validating that passage history entries either match existing standards (for common cell lines) or are further explained in another field (uncommon cell lines), and making passage history searchable by discrete categories such as Egg, Cell, and Original Clinical Specimen.
4. Conclusion
Making sense of influenza passaging annotations is a daunting task. However, it is becoming increasingly important for epidemiologists and vaccine developers to consider passage history of isolates when selecting sequences for inclusion in phylodynamic analyses or in vaccines. Although a clear and definitive understanding of the effects of viral passaging in all cell types is a distant end point of this research efforts, awareness within the influenza community of the negative impact of cell passaging on sequence fidelity is easily and currently attainable and can only improve epidemiological and clinical research efforts. This highlights the need for a more standard approach to passage nomenclature across influenza researchers and producers of sequence data. As the influenza community is looked upon as a model of data sharing for other epidemic viruses (Shu and McCauley 2017), we encourage this highly collaborative community to work together to enact a new global naming convention that further evinces the power and effectiveness of open research.
5. Methods
Annotations were obtained for all (i.e. global) unique, non-laboratory influenza A(H3N2) and A(H1N1) isolates collected from humans between 1 January 2005 and 8 November 2018 and uploaded to GISAID, OpenFlu, or IRD by 8 November 2018 for A(H3N2) isolates and 12 November 2018 for A(H1N1) isolates. All annotations were first converted to uppercase characters and then occurrences of each unique isolate and annotation were counted and manually sorted by cell type and passage number. These data were used to generate Figures 2 and 3 and Tables 1–3.
Funding
This work was supported by NIH (grant R01 GM088344) to C.O.W. and S.M.S. was supported by the A*STAR HEIDI program (grant number H1699f0013). The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
Conflict of interest: None declared.
References
- Ampofo W. K. et al. (2013) ‘Improving Influenza Vaccine Virus Selection: Report of a WHO Informal Consultation Held at WHO Headquarters, Geneva, Switzerland, 14-16 June 2010’, Influenza and Other Respiratory Viruses, 7: 52–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T. K. et al. (2016) ‘A Phylogeny-Based Global Nomenclature System and Automated Annotation Tool for H1 Hemagglutinin Genes from Swine Influenza a Viruses’, mSphere, 1: e00275–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanov S. S. et al. (2015) ‘Genome-Wide Analysis of Evolutionary Markers of Human Influenza A(H1N1)pdm09 and A(H3N2) Viruses May Guide Selection of Vaccine Strain Candidates’, Genome Biology and Evolution, 7: 3472–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A. et al. (2018) ‘GenBank’, Nucleic Acids Research, 46: D41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst M. J., Brooks T. J. G., Pollock N. R. (2016) ‘Diagnosis of Ebola Virus Disease: Past, Present, and Future’, Clinical Microbiology Reviews, 29: 773–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush R. M. et al. (2000) ‘Effects of Passage History and Sampling Bias on Phylogenetic Reconstruction of Human Influenza A Evolution’, Proceedings of the National Academy of Sciences of the United States of America, 97: 6974–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. (2018) ‘Passage Adaptation Correlates with the Reduced Efficacy of the Influenza Vaccine’, Clinical Infectious Diseases. DOI: 10.1093/cid/ciy1065. [DOI] [PubMed] [Google Scholar]
- Chen H. et al. (2016) ‘Dynamic Convergent Evolution Drives the Passage Adaptation across 48 Years’ History of H3N2 Influenza Evolution’, Molecular Biology and Evolution, 33: 3133–43. [DOI] [PubMed] [Google Scholar]
- Du X. et al. (2017) ‘Evolution-Informed Forecasting of Seasonal Influenza A (H3N2)’, Science Translational Medicine, 9: eaan5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld A. J., Neumann G., Kawaoka Y. (2014) ‘Influenza a Virus Isolation, Culture and Identification’, Nature Protocols, 9: 2663–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatherer D. (2010) ‘Passage in Egg Culture Is a Major Cause of Apparent Positive Selection in Influenza B Hemagglutinin’, Journal of Medical Virology, 82: 123–7. [DOI] [PubMed] [Google Scholar]
- Govorkova E. A. et al. (1999) ‘Selection of Receptor-Binding Variants of Human Influenza a and B Viruses in Baby Hamster Kidney Cells’, Virology, 38: 31–8. [DOI] [PubMed] [Google Scholar]
- Grohskopf L. A. et al. (2018) ‘Prevention and Control of Seasonal Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices—United States, 2018–19 Influenza Season’, MMWR Recommendations and Reports, 67: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer J. C., Killington R. A. (1996) ‘Virus Isolation and Quantitation’, in B.W.J., Mahy and H.O., Kangro (eds) Virology Methods Manual, pp. 25–46. Cambridge, MA: Academic Press. [Google Scholar]
- Himmelsbach K., Hildt E. (2018) ‘Identification of Various Cell Culture Models for the Study of Zika Virus’, World Journal of Virology, 7: 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S., Walker D., Webster R. G. (2012). ‘Influenza Virus Isolation’, inKawaoka Y., Neumann G. (eds) Influenza Virus: Methods and Protocols, pp. 11–24. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Krowicka H. et al. (2008) ‘Use of Tissue Culture Cell Lines to Evaluate HIV Antiviral Resistance’, AIDS Research and Human Retroviruses, 24: 957–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. K. et al. (2013) ‘Comparison of Mutation Patterns in Full-Genome a/H3N2 Influenza Sequences Obtained Directly from Clinical Samples and the Same Samples After a Single MDCK Passage’, PLoS One, 8: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. et al. (2009) ‘In Vivo and in Vitro Alterations in Influenza a/H3N2 Virus M2 and Hemagglutinin Genes : Effect of Passage in MDCK-SIAT1 Cells and Conventional MDCK Cells’, Journal of Clinical Microbiology, 47: 466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. et al. (2017) ‘The Characteristics and Antigenic Properties of Recently Emerged Subclade 3C. 3a and 3C. 2a Human Influenza A (H3N2) Viruses Passaged in MDCK Cells’, Influenza and Other Respiratory Viruses, 11: 263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little K. et al. (2015) ‘Zanamivir-Resistant Influenza Viruses with Q136K or Q136R Neuraminidase Residue Mutations Can Arise during MDCK Cell Culture Creating Challenges for Antiviral Susceptibility Monitoring’, Euro Surveillance, 20: pii=30060. DOI: 10.2807/1560-7917.ES.2015.20.45.30060. [DOI] [PubMed] [Google Scholar]
- McWhite C. D., Meyer A. G., Wilke C. O. (2016) ‘Sequence Amplification via Cell Passaging Creates Spurious Signals of Positive Adaptation in Influenza Virus H3N2 Hemagglutinin’, Virus Evolution, 2: doi: 10.1093/ve/vew026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncla L. H., Florek N. W., Friedrich T. C. (2017) ‘Influenza Evolution: New Insights into an Old Foe’, Trends in Microbiology, 25: 432–4. [DOI] [PubMed] [Google Scholar]
- Pedersen J. C. (2014) ‘Hemagglutination-Inhibition Assay for Influenza Virus Subtype Identification and the Detection and Quantitation of Serum Antibodies to Influenza Virus’, Methods in Molecular Biology, 1161: 11–25. [DOI] [PubMed] [Google Scholar]
- Robertson J. S. et al. (1993) ‘The Role of Amniotic Passage in the Egg-Adaptation of Human Influenza Virus Is Revealed by Haemagglutinin Sequence Analyses’, Journal of General Virology, 74: 2047–51. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. (2017) ‘GISAID: Global Initiative on Sharing All Influenza Data - From Vision to Reality’, Euro Surveillance, 22: 30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva T. A. et al. (2017) ‘Predicting the Evolutionary Variability of the Influenza A Virus’, Acta Naturae, 9: 48–54. [PMC free article] [PubMed] [Google Scholar]
- Weir J. P., Gruber F. (2016). ‘An overview of the regulation of influenza vaccines in the United States’, Influenza and Other Respiratory Viruses, 10: 354–60. [DOI] [PMC free article] [PubMed]
- Wu N. C. et al. (2017) ‘A Structural Explanation for the Low Effectiveness of the Seasonal Influenza H3N2 Vaccine’, PLoS Pathogens, 13: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyde P. R. et al. (1977) ‘Effects of Low- and High-Passage Influenza Virus Infection in Normal and Nude Mice’, Infection Immunity, 15: 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. (2017) ‘Influenza Research Database: An Integrated Bioinformatics Resource for Influenza Virus Research’, Nucleic Acids Research, 45: D466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S. J. et al. (2017) ‘Contemporary H3N2 Influenza Viruses Have a Glycosylation Site That Alters Binding of Antibodies Elicited by Egg-Adapted Vaccine Strains’, Proceedings of the National Academy of Sciences of the United States of America, 114: 12578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]