Abstract
Enterococcus faecalis are a major cause of nosocomial infection worldwide, and the spread of vancomycin resistant strains (VRE) limits treatment options. Tigecycline-resistant VRE began to be isolated from inpatients at a Brazilian hospital within months following the addition of tigecycline to the hospital formulary. This was found to be the result of a spread of an ST103 E. faecalis clone. Our objective was to identify the basis for tigecycline resistance in this lineage. The genomes of two closely related tigecycline-susceptible (MIC = 0.06 mg/L), and three representative tigecycline-resistant (MIC = 1 mg/L) ST103 isolates were sequenced and compared. Further, efforts were undertaken to recapitulate the emergence of resistant strains in vitro. The specific mutations identified in clinical isolates in several cases were within the same genes identified in laboratory-evolved strains. The contribution of various polymorphisms to the resistance phenotype was assessed by trans-complementation of the wild type or mutant alleles, by testing for differences in mRNA abundance, and/or by examining the phenotype of transposon insertion mutants. Among tigecycline-resistant clinical isolates, five genes contained non-synonymous mutations, including two genes known to be related to enterococcal tigecycline resistance (tetM and rpsJ). Finally, within the in vitro-selected resistant variants, mutation in the gene for a MarR-family response regulator was associated with tigecycline resistance. This study shows that E. faecalis mutates to attain tigecycline resistance through the complex interplay of multiple mechanisms, along multiple evolutionary trajectories.
Keywords: Vancomycin-resistant enterococci, Tigecycline resistance, tetM, rpsJ
1. Introduction
Enterococci are opportunistic pathogens and one of the major causes of nosocomial infection worldwide. The emergence of vancomycin-resistant enterococci (VRE) has significantly reduced available treatment options (Cetinkaya et al., 2000). Tigecycline is an antibiotic of last resort for infections caused by a number of multidrug-resistant organisms, including VRE (Cunha et al., 2017). This glycylcycline antibiotic, synthetic derivate from tetracycline, was approved for the treatment of complicated skin and soft-structure infections, complicated intra-abdominal infections, and community-acquired bacterial pneumonia in United States (Food and Drug Administration, 2005) and Brazil (Agência Nacional de Vigilância Sanitária, n.d.) in 2005, and in Europe (European Medicines Agencies, n.d.) in 2006.
Drug efflux is the most frequently reported mechanism of resistance to tigecycline (Zhong et al., 2014; Peleg et al., 2007; Ruzin et al., 2007). In Staphylococcus aureus, tigecycline resistance has been ascribed to mutations in the mepRAB efflux pump (McAleese et al., 2005; Dabul et al., 2017) and response regulator (Dabul et al., 2017). Monooxygenases capable of modifying the tigecycline molecule have been identified in Acinetobacter baumannii (Costello et al., 2016) and Bacillus subtilis (Bartha et al., 2011). Tigecycline resistance in multiple organisms has also been associated with mutations in the gene encoding the ribosomal protein S10 (Beabout et al., 2015). Tigecycline-resistant isolates of E. faecalis have been reported previously (Tyson et al., 2018; Werner et al., 2008), but resistance mechanisms have not been fully explored in this pathogen.
In the process of infection control surveillance at Risoleta Tolentino Neves Hospital in Belo Horizonte, Brazil in 2009, 63 VRE were isolated from sites of infection and colonization (Merlo et al., 2015). Of these VRE, 14 strains were identified as E. faecalis (22.2%) and none exhibited resistance to tigecycline (Merlo et al., 2015). In 2011, tigecycline was introduced in this hospital for treatment of soft tissue infections caused by multidrug-resistant Acinetobacter sp., and also for abdominal infections caused by VRE. Subsequent surveillance from March to June 2011 showed an increase in the proportion of E. faecalis to 29 out of 47 VRE (61.7%). Further, ten of these vancomycin-resistantE. faecalis strains were found to be resistant to tigecycline. Because resistance to tigecycline in vancomycin-resistant E. faecalis is uncommon (Kuch et al., 2012; Cordina et al., 2012), it was important to determine the genetic background and features of these tigecycline-resistant VRE isolates, and to identify mutations as either markers for or contributing to tigecycline resistance.
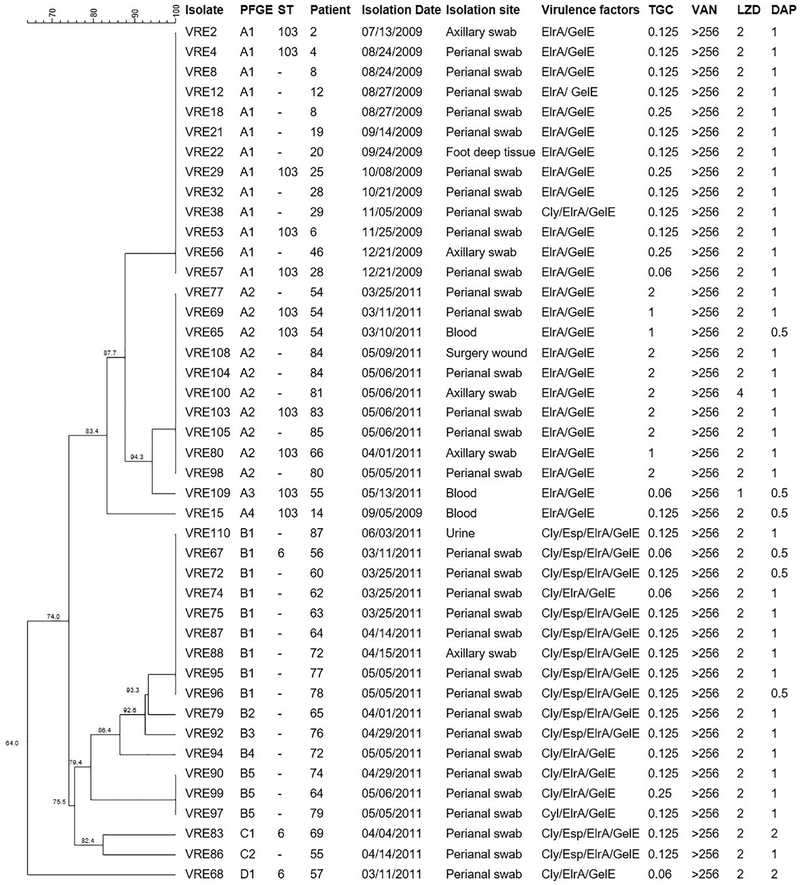
Prior PFGE analysis revealed that E. faecalis VRE57 (Merlo et al., 2015), from 2009, and E. faecalis VRE109, from 2011, were the most closely related tigecycline-susceptible isolates to the tigecycline-resistant ST103 strains isolated in 2011 (Fig. 1). Thus, our approach to identify candidate genes or mutations that could contribute to tigecycline resistance was to first compare the genomes of the tigecycline-resistant strains VRE65, VRE69 and VRE80, with the tigecycline-susceptible strain VRE109 and VRE57, and then perform an adaptation experiment in vitro with VRE109 in increasing concentrations of tigecycline, following genome analysis of the adapted strains.
Fig. 1.
Dendrogram of vancomycin-resistant Enterococcus faecalis isolated from Risoleta Tolentino Neves Hospital in 2009 and 2011. Legend: PFGE – Pulsotype; ST – Sequence Type; TGC – Tigecycline; VAN – Vancomycin; LZD – Linezolid; DAP – Daptomycin. Breakpoints: TGC – S ≤ 0. 25 mg/L/R > 0.5 mg/L; VAN – S ≤ 4 mg/L/ I = 8–16 mg/L/R ≥ 32 mg/L; LZD – S ≤ 2 mg/L/I = 4 mg/L/R ≥ 8 mg/L; DAP – S ≤ 4 mg/L.
2. Materials and methods
2.1. Source of strains
During routine infection control surveillance between March and June 2011 at Risoleta Tolentino Neves, 29 VRE of the species E. faecalis were isolated from patients. The hospital sees an average of 280 patients per day in its emergency room, contains 345 beds, and is located in Belo Horizonte, the capital of the state of Minas Gerais and the sixth most populous city in Brazil.
2.2. Antimicrobial susceptibility
Minimum inhibitory concentrations (MICs) for vancomycin, line-zolid, daptomycin and tigecycline were determined by broth micro-dilution following Clinical and Laboratory Standards Institute (CLSI) reccomendations (Clinical and Laboratory Standards Institute, 2018). The tigecycline breakpoint used to define resistance was 0.25 mg/L, as defined by the European Committee on Antimicrobial Susceptibility Testing (European Committee on Antimicrobial Susceptibility Testing, 2018).
2.3. Molecular characterization
PCR was used to detect the presence of vanA (Woodford et al., 1993) and to assess strains for virulence genes elrA, cylLL, esp and gelE, essentially as described (Camargo et al., 2006; Leavis et al., 2007; Brinster et al., 2007). Extended PCR was used to characterize Tn1546 (Woodford et al., 1997). Molecular typing of strains was performed by pulsed-field gel electrophoresis (PFGE) after macrorestriction of genomic DNA with SmaI (Tenover et al., 1995), and by Multi-Locus Sequence Typing (MLST) (Ruiz-Garbajosa et al., 2006). E. faecalis strains from the same hospital, isolated during the earlier 2009 surveillance and described previously by our group (Merlo et al., 2015) were included for comparison. Plasmid rep types present in all isolates were determined essentially as described (Jensen et al., 2010).
2.4. In vitro-selection of tigecycline-resistant variants
Tigecycline-susceptible isolate E. faecalis VRE109 was selected as the parent strain for attempting to select tigecycline-resistant mutations in vitro. The experiment was performed in triplicate as previously described (Dabul et al., 2017), and conducted over 42 days. Briefly, an overnight culture of each of three VRE109 colonies was grown in Brain Heart Infusion (BHI) broth, then diluted to an OD600 = 0.1. From this, 30 μL (approximately 3 × 106 CFU) were inoculated into tubes containing 3 mL of BHI and varying concentrations of tigecycline: a) 1/2 MIC (0.03 mg/L); b) 1× MIC (0.06 mg/L); c) 2× MIC (0.125 mg/L); and d) 4× MIC (0.25 mg/L). All tubes were protected from light and grown at 37 °C overnight without shaking. The next day, the tube of each replicate with the highest tigecycline concentration showing visible growth was used to inoculate fresh tubes with that and increasing drug concentrations.
2.5. Genome sequencing, assembly and comparative analysis
Three naturally occurring tigecycline-resistant E. faecalis strains, VRE65, VRE69 and VRE80, isolated in March and April 2011 were selected for genome sequencing because they were representative of the first tigecycline-resistant E. faecalis to be isolated in the hospital. For comparison, tigecycline susceptible VRE109, and in vitro-selected tigecycline resistant variants A1, A11, A20, A42, B1, B21, B42, C1, C6, and C42 were also sequenced (letter designations correspond to the experimental replicate, and numbers correspond to the day of the experiment from which the variant was isolated). Additionally, two tigecycline-susceptible VRE clinical strains, one from 2009 and one from 2011, were selected for sequencing and comparison based on PFGE similarity to the tigecycline-resistant E. faecalis strains.
Total genomic DNA was extracted from cultures of each strain using the DNeasy Blood & Tissue kit (QIAGEN, Valencia, USA) following the manufacturer’s recommendations for Gram-positive organisms. Sequencing libraries were prepared using the Illumina Nextera XT DNA sample preparation kit (Illumina, San Diego, USA), with recommended modifications for 2 × 250 bp paired-end sequencing. One nanogram of DNA was used for library preparation. Libraries were multiplexed and sequenced on an Illumina MiSeq at the Ocular Genomics Institute in the Massachusetts Eye and Ear Infirmary.
CLC Genomics Workbench v.7.0.4 (Qiagen, Aarhus, Denmark) was used for genome assembly using default parameters. Genomes were annotated through the NCBI Prokaryotic Annotation Pipeline. Variants were called using CLC Genomics Workbench by comparing the genome sequences of the resistant strains to the annotated sequences of tigecycline-susceptible isolates VRE57 and VRE109. Altered nucleotides and corresponding amino acid changes were further analyzed using BlastN and BlastX from the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences of genes possessing polymorphisms potentially associated with tigecycline resistance were confirmed by Sanger sequencing in all ten tigecycline-resistant clinical isolates.
All genomes have been deposited at DDBJ/EMBL/GenBank under the accessions: JTEX00000000 (VRE57), JTEY00000000 (VRE65), JTEZ00000000 (VRE69), JTFA00000000 (VRE80), JTFB00000000 (VRE109), LGCB00000000 (A1), LGCA00000000 (A11), LGBZ00000000 (A20), LGBY00000000 (A42), LGBX00000000 (B1), LGBW00000000 (B21), LGBV00000000 (B42), LGBU00000000 (C1), LGBT00000000 (C6), and LGBS00000000 (C42). The versions described in this article are versions JTEX01000000 (VRE57), JTEY01000000 (VRE65), JTEZ01000000 (VRE69), JTFA01000000 (VRE80), JTFB01000000 (VRE109), LGCB01000000 (A1), LGCA01000000 (A11), LGBZ01000000 (A20), LGBY01000000 (A42), LGBX01000000 (B1), LGBW01000000 (B21), LGBV01000000 (B42), LGBU01000000 (C1), LGBT01000000 (C6), and LGBS01000000 (C42).
2.6. Construction of overexpressing strains
Genes of interest from genome sequence comparisons were amplified using primers designed to add BamHI and XbaI restriction sites (Table 1). Genes rpsJ (QP83_07505), tetM (QP83_12955), a hypothetical protein (QP83_08660), lepA (QP83_09250), and the MarR-family transcriptional regulator (QP83_14295) were each amplified from resistant/mutated isolates. Amplicons included the inferred promoter region for each gene, and were digested and ligated into the pAT28 shuttle vector (Trieu-Cuot et al., 1990). Transformants were selected on 100 mg/L spectinomycin for E. coli, or 500 mg/L for E. faecalis. Since no promoter is predicted immediately adjacent to the gene encoding the multidrug ABC transporter ATP-binding protein (QP83_01175), the reading frame was amplified and inserted into the pMSP3535 expression vector (Bryan et al., 2000). When cloning into pMSP3535, transformants were selected on 150 mg/L erythromycin for E. coli, and 10 mg/L for E. faecalis.
Table 1.
Primers used for cloning genes of interest into pAT28 or pMSP3535.
| Gene | Plasmid | Primers F/R (5′→3′) | Amplicon size (bp) |
|---|---|---|---|
| rpsJ | pAT28 | F: CGCGGATCCCGCAGAAAATCCTTG | 544 |
| R: CGCTCTAGAGAGTACACCTCCATCTTAATT | |||
| tetM | pAT28 | F: CGCGGATCCTTTGATAAAAAATTG | 2290 |
| R: CGCTCTAGATTTATATAACAACATAAAATACA | |||
| QP83_08660 | pAT28 | F: CGCGGATCCCTGGTTTCCGCCAGCTG | 839 |
| R: CGCTCTAGACATTTAAGCATCAATAATCAAGG | |||
| lepA | pAT28 | F: CGCGGATCCTTTTCCATCTCCAATAAATTTA | 2103 |
| R: CGCTCTAGATTATGATTTTTTCTGATCATCTTCA | |||
| QP83_14295 | pAT28 | F: CGCGGATCCCCTTTTTCTGTGAAAATGGT | 672 |
| R: CGCTCTAGATCCTCACTTTTTATTTGTATTCTTGC | |||
| QP83_01175 | pMSP3535 | F: CGCGGATCCAAAATTGAAGGGAGTGA | 1853 |
| R: CGCTCTAGAGGTTATTTAACTCTTTCTATTGTTACG |
Following initial propagation and isolation from E. coli DH5-α, pAT28-based constructs were transformed into E. faecalis VRE80 and VRE109 by electroporation. For the pMSP3535-cloned ABC transporter QP83_01175, following amplification in E. coli, it was electroporated into E. faecalis OG1RF (selected because VRE80 and VRE109 are resistant to erythromycin).
2.7. Efflux pump inhibitor assays
The effect of efflux pump inhibitors verapamil, reserpine and carbonyl cyanide 3-chlorophenylhydrazone (CCCP) on tigecycline MIC was determined. First, efflux pump inhibitor concentration was titrated to identify the maximum levels usable without themselves inhibiting growth. Pilot experiments identified 800 mg/L for verapamil, 20 mg/L for reserpine, and 1 mg/L for CCCP, as the highest concentrations that did not inhibit E. faecalis growth.
2.8. Analysis of transposon insertion mutants in genes of interest
Individual, presumed loss-of-function E. faecalis OG1RF mariner transposon insertion mutants (Dale et al., 2018) in lepA, and genes encoding the multidrug ABC transporter ATP-binding protein and the hypothetical protein (kindly provided by Gary Dunny), were tested for their tigecycline sensitivities in a broth microdilution format as follows: Two-fold serial dilutions of tigecycline in Mueller Hinton II were made in a 96-well plate, and wells were inoculated with 100 μL of a 1:5000 dilution of culture normalized to OD600 = 0.5, for a total volume of 200 μL and final bacterial dilution of 1:10,000. Plates were incubated at 37 °C for 24 h and OD600 measured using a Synergy2 Biotek plate reader (Winooski, VT) with Gen5 software.
2.9. Relative gene expression
To determine whether any of the observed resistance-associated mutations resulted in changes in gene expression, clinical isolates VRE57, VRE65, VRE69, VRE80 and VRE109 were cultured in BHI media overnight at 37 °C with shaking. The next day, all cultures were diluted 1:25 into fresh BHI, and grown to OD600 = 0.6–1.0. Cells were collected by centrifugation for 2 min at 14,000 x g at 4 °C and lysed by the addition of 3 mg/mL of lysozyme. RNA was then extracted using the SV Total RNA Isolation System (Promega Corporation, Madison, USA), according to manufacturer’s recommendations. The concentration of RNA was determined by determining the 260 nm/280 nm absorbance ratio using a NanoDrop 2000c Spectrophotometer (Thermo Scientific, Waltham, USA). All RNA preparations yielded absorbance ratios > 1.8.
Expression of rpsJ, tetM, and the genes encoding multidrug ABC transporter ATP-binding protein, hypothetical protein and MarR-family transcriptional regulator, and reference genes for normalization of expression data (Table 2) was quantified by qPCR. For reverse transcription, a SuperScript™ III First-Strand kit (Invitrogen, Carlsbad, USA), with 50 ng of random hexamers and 1 μg of RNA was used according to manufacturer’s recommendations to generate ~ 1 μg of cDNA. The quality of cDNA was assessed by PCR amplification, and verified by gel electrophoresis, the presence of one amplicon of the correct size was observed, the cDNA was considered a good and specific. A cDNA pool, prepared with a mixture of all enterococcal cDNA samples, was used for optimization of annealing temperatures. Additionally, the presence of single peaks in melting curve analyses demonstrated that amplifications by all pairs of primers were target-specific. Then, the cDNA pool was also used to generate a standard curve for primer efficiency, which was used to correct expression values (Bustin et al., 2009), and to evaluate the expression stability of the reference genes gdh, pyrC and gyrA with RefFinder (Xie et al., 2012). All three genes were considered stable for normalization of relative expression.
Table 2.
List of genes and primers used for qPCR analysis.
| Gene | Gene type | Primers | Tm (°C) | Amplicon size |
|---|---|---|---|---|
| gdh | Reference | F: 5′- TGTCGGTGAGCTGCCTAAAG −3′ | 60.0 °C 59.8°C | 120 pb |
| R: 5′- AAAGCGGTTTCAGCTAACGC −3′ | ||||
| pyrC | Reference | F: 5′- TCGGTAGTGAAACGGCCTTC −3′ | 60.0 °C 59.4°C | 106 pb |
| R: 5′- ATTTCTGCCGGTTTCACTGC −3′ | ||||
| gyrA | Reference | F: 5′- AGGTGTTCGTGGAATCCGTC −3′ | 60.0 °C 60.6°C | 148 pb |
| R: 5′- ACCGCCACGTCCTTTAACTG −3′ | ||||
| rpsJ | Target | F: 5′- AAGAACTGGAGCTGACGTATC −3′ | 57.5 °C 58.7°C | 80 pb |
| R: 5′- TATGAGTCGCACGAACAACTG −3′ | ||||
| tetM | Target | F: 5′- TGTATCACCGCTTCCGTTGG −3′ | 60.4 °C 60.0°C | 114 pb |
| R: 5′- TCGCAACCATAGCGTATCCC −3′ | ||||
| QP83_08660 | Target | F: 5′- AAAACGGTCAGCAAAGAGGC −3′ | 59.3°C 60.1°C | 121 pb |
| R: 5′- GGTACTCCCCGTTTTCTCCG −3′ | ||||
| QP83_01175 | Target | F: 5′- CGTTTCGACGCATGTTTTAC −3′ R: 5′- GCATTGTAGCAGTTTTAGGC −3′ | 56.4 °C 56.8°C | 126 pb |
| QP83_14295 | Target | F: 5′- CCGTCGGAACACTTACTGTAG-3′ | 56.0 °C 59.6°C | 98 pb |
| R: 5′- GCCAAGCTTTACTACACGTCG −3′ |
For qPCR amplification, the fluorescent DNA-binding dye PowerUP™ SYBR® Green PCR Master Mix (Thermo Fisher Scientific, Waltham, USA) was used according to manufacturer’s recommendations, with 0.5 ng cDNA added per reaction. A CFX96 Touch™ Real Time PCR Detection System (Bio-Rad, Hercules, USA) was used, with cycling conditions: Initial equilibration at 50 °C for 2 min, followed by denaturation at 95 °C for 2 min, then 40 cycles of 15 s at 95 °C and 1 min at 60 °C. A quantitative melting curve was performed on each product (using a temperature ramp of 0.5 °C per second, from 65 to 95 °C), to verify amplicon identity in each reaction. Each qPCR reaction was performed in triplicate. Relative expression was calculated by normalization to levels of expression for each of the three reference genes, by comparing ΔΔCT values with PCR efficiency correction (Vandesompele et al., 2002; Livak and Schmittgen, 2001).
3. Results and discussion
3.1. Molecular characterization
The emergence of tigecycline resistance among vancomycin-resistant E. faecalis was noticed within months of its introduction into use at a Brazilian hospital. It was therefore of considerable interest to understand the nature and basis for this phenotype.
As shown in Fig. 1, 43 VRE E. faecalis strains were studied. These included 29 strains collected in 2011 (ten of which were tigecycline-resistant), and 14 additional VRE strains collected in 2009, prior to the spread of tigecycline resistance at this site (Merlo et al., 2015). All isolates contained the vanA resistance operon carried by an intact Tn1546 transposon, and exhibited MICs for vancomycin > 256 mg/L. All isolates were susceptible to daptomycin, and one isolate had line-zolid MIC = 4 mg/L, which is the clinical susceptibility breakpoint. A plasmid of the rep9 family was detected in every 2011 isolate, which was not surprising given the widespread distribution of rep9-family plasmids among E. faecalis (Wardal et al., 2013; Song et al., 2013). All 2009 isolates were positive for rep1 and seven for rep2, in addition to rep9.
PFGE analysis revealed that most 2009 isolates clustered in a single pulsotype (designated pulsotype A), as did 11 out of 29 isolates from 2011, with 87.7% DNA fragmentation similarity (Fig. 1). The virulence factor profile for the pulsotype A strains was elrA+gelE+. All subtype A2 samples (n = 10) were resistant to tigecycline, and one of them also showed intermediate resistance to linezolid (VRE100). These isolates were collected from seven different patients, indicating dissemination of this resistant clone in the hospital during the 2011 sampling period. VRE109 was the only isolate of subtype A3, and it was susceptible to tigecycline. The 18 remaining vancomycin-resistant E. faecalis strains from 2011 belonged to pulsotypes B-D and were all susceptible to tigecycline. Three representative isolates from these pulsotypes were typed by MLST and identified as ST6. Their virulence factor profile was either cyl+esp+elrA+gelE+ or cyl+elrA+gelE+ (Fig. 1).
Six isolates from pulsotype A (including one from subtype A2) were typed by MLST and found to belong to ST103 (Fig. 1), a sequence type previously detected in this hospital (Merlo et al., 2015), but distinct from a tigecycline-resistant ST6 E. faecalis lineage encountered in Germany (Werner et al., 2008). Tigecycline resistance among E. faecalis isolated in Brazil is a rare event, as highlighted by data from the Tigecycline Evaluation and Surveillance Trial between 2004 and 2015, which showed that only 14.3% of vancomycin-resistant E. faecalis in Brazil were resistant to tigecycline (Vega and Dowzicky, 2017), and also by results from the SENTRY Antimicrobial Surveillance Program, which revealed 100% susceptibility to tigecycline among E. faecalis isolated in Latin America between 2011 and 2014 (Sader et al., 2016), as did a study of isolates from 2016 (Pfaller et al., 2018).
3.2. Analysis of the genome sequence of the ST103 tigecycline-resistant lineage
Virulence determinants and resistant genes are frequently carried by plasmids and disseminated among bacterial cells by conjugation. The fact that no other plasmid family and no other virulence genes were introduced in the tigecycline-resistant isolates could suggest the accessory genome does not play a role in tigecycline resistance, so our deeper analysis focused in the core genome.
PFGE analysis revealed that E. faecalis VRE57 (isolated in December 2009) (Merlo et al., 2015) and E. faecalis VRE109 (isolated in May 2011) were the most closely related tigecycline-susceptible isolates to the tigecycline-resistant ST103 strains isolated in 2011 (Fig. 1). In order to identify candidate genes or mutations that could contribute to tigecycline resistance, we first compared the genomes of the tigecycline-resistant strains VRE65, VRE69 and VRE80, with the tigecycline-susceptible strain VRE109. Results of that comparison were then filtered to remove polymorphisms that were not shared upon comparison with VRE57, the tigecycline-susceptible isolate from 2009. From this analysis, five genetic changes were found to be common to all three tigecycline-resistant clinical isolates, and absent from the two tigecycline-susceptible comparators (Table 3). Further, these five differences were confirmed to occur in the remaining tigecycline-resistant strains VRE77, VRE98, VRE100, VRE103, VRE104, VRE105 and VRE108. These mutations do not occur in publicly accessible sequences in the NCBI database, suggesting that they are unique to this lineage. The wild type alleles for each gene, however, were identified in the database. The nature of the changes in each of the five genes is discussed in greater detail below.
Table 3.
Sequence changes in the tigecycline-resistant clinical isolates VRE65, VRE69, and VRE80 compared to tigecycline-susceptible isolates VRE57 and VRE109.
| Genea | Product | Coding region change | Amino acid change |
|---|---|---|---|
| QP83_07505 | S10 protein of the 30S ribosomal subunit | 165_176del | Δ56_59HKYK |
| QP83_01175 | Multidrug ABC transporter ATP-binding protein | T735TA | D245E |
| QP83_12955 | Tetracycline resistance protein TetM | T1417A | F473I |
| QP83_08660 | Hypothetical protein | G410A | G137E |
| QP83_09250 | GTP-binding protein LepA | A47T | K16I |
Names are from the annotated sequence of the VRE109 isolate.
3.2.1. rpsJ (QP83_07505)
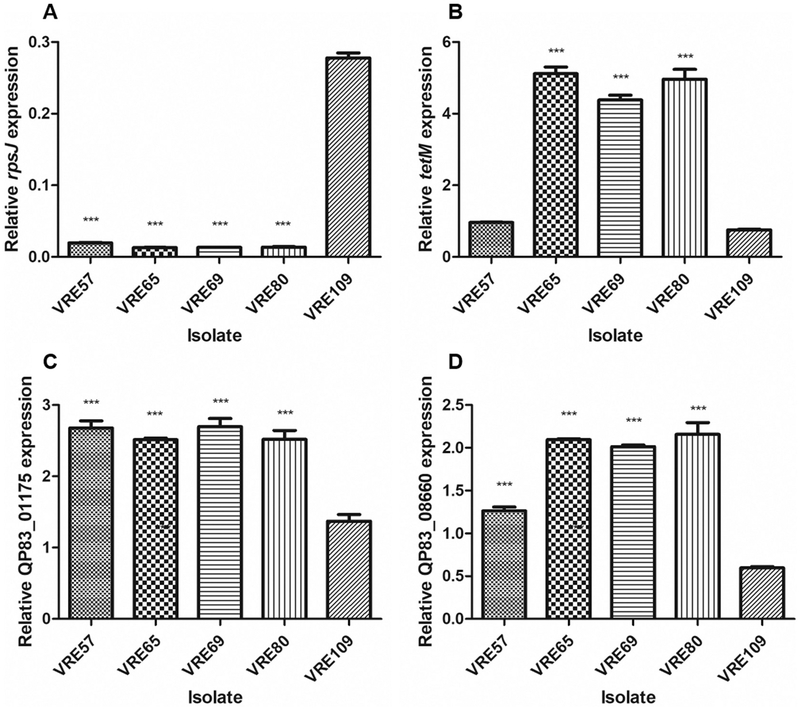
A 12 bp deletion was identified in the rpsJ gene (QP79_02985), encoding the S10 protein of the 30S ribosomal subunit, in each of the tigecycline-resistant isolates. The deletion begins at position 165 of the gene, and results in removal of amino acids HKYK from the protein at positions 56–59 (Table 3). In Enterococcus faecium, an Asp60Tyr substitution in the same gene was found to be responsible for the reduced susceptibility to tigecycline in three mutant strains generated in vitro (Cattoir et al., 2015). Additionally, the same study characterized a clinical isolate of E. faecium with reduced tigecycline susceptibility and found that the isolate had a Lys57Glu mutation in rpsJ (Cattoir et al., 2015). Codon 57 is located at the vertex of a well conserved loop in close proximity to the tigecycline target site in the 30S ribosomal subunit (Villa et al., 2014). Mutations in this region have also been observed in other tigecycline-resistant E. faecium clinical isolates, as well as in K. pneumoniae(Villa et al., 2014; Niebel et al., 2015). Of particular interest, Beabout et al. experimentally proved that a modification (R53Q-Δ54–57ATHK) that occurred in tigecycline-resistant E. faecalis strain selected in vitro resulted in a 4-fold increase in tigecycline resistance (Beabout et al., 2015). Here, we observed a 12 bp deletion at the same region of the S10 protein in clinical E. faecalis isolates, highlighting the relevance of alteration in this specific region of the rpsJ gene for tigecycline resistance in E. faecalis. We attempted to express the mutant rpsJ allele in trans in the tigecycline-susceptible VRE109 strain, but we did not observe a change in tigecycline MIC. This likely stems from the fact that the wild type gene is highly expressed in E. faecalis (Table 4) on a polycistronic message, and the ribosome is assembled co-translationally (Shieh et al., 2015). Allelic replacement at this locus in enterococci has not yet been successful. (Beabout et al., 2015; Cattoir et al., 2015) The polymorphism in rpsJ occurring in tigecycline-resistant strains VRE65, VRE69, and VRE80 was not associated with significantly altered levels of mRNA abundance in these strains (Fig. 2). Nevertheless, the data collectively support the deduction that the deletion in rpsJ observed in these tigecycline-resistant clinical isolates of E. faecalis contributes to the resistant phenotype.
Table 4.
Tigecycline Minimal Inhibitory Concentrations (MICs) in the absence and presence of efflux pump inhibitors.
| Strain | TGC | TGC + CCCP | TGC + RES | TGC + VER |
|---|---|---|---|---|
| (mg/L) | ||||
| VRE57 | 0.060 | 0.060 | 0.060 | 0.030 |
| VRE65 | 1.000 | 1.000 | 1.000 | 0.250 |
| VRE69 | 1.000 | 1.000 | 1.000 | 0.250 |
| VRE80 | 1.000 | 1.000 | 1.000 | 0.250 |
| VRE109 | 0.060 | 0.030 | 0.060 | 0.030 |
| A1 | 0.060 | 0.030 | 0.060 | 0.030 |
| A11 | 0.125 | 0.125 | 0.125 | 0.030 |
| A20 | 0.500 | 0.500 | 0.500 | 0.060 |
| A42 | 1.000 | 1.000 | 1.000 | 0.125 |
| B1 | 0.060 | 0.060 | 0.060 | 0.030 |
| B21 | 0.250 | 0.250 | 0.250 | 0.030 |
| B42 | 0.500 | 0.500 | 0.500 | 0.125 |
| C1 | 0.060 | 0.030 | 0.060 | 0.030 |
| C6 | 0.125 | 0.125 | 0.125 | 0.06 |
| C42 | 1.000 | 1.000 | 0.500 | 0.125 |
| VRE109(pAT28) | 0.060 | N/A | N/A | N/A |
| VRE109(pAT28-hypVRE80) | 0.125 | N/A | N/A | N/A |
| VRE109(pAT28-rpsJVRE80) | 0.060 | N/A | N/A | N/A |
| OG1RF(pMSP3535) | 0.060 | N/A | N/A | N/A |
| OG1RF(pMSP3535-abcVRE80) | 0.060 | N/A | N/A | N/A |
| VRE109(pAT28-marR VRE109) | 0.060 | N/A | N/A | N/A |
| VRE109(pAT28-marRB42) | 0.125 | N/A | N/A | N/A |
| B42(pAT28) | 1.000 | N/A | N/A | N/A |
| B42(pAT28-marRVRE109) | 1.000 | N/A | N/A | N/A |
| B42(pAT28-marRB42) | 2.000 | N/A | N/A | N/A |
TGC: tigecycline; CCCP: carbonyl cyanide 3-chlorophenylhydrazone; RES: reserpine; VER: verapamil; N/A: not applicable.
Fig. 2.
Relative mRNA abundance levels in tigecycline-susceptible and resistant E. faecalis clinical isolates. Mean levels of rpsJ, tetM, Multidrug ABC transporter ATP-binding protein QP83_01175 and Hypothetical protein QP83_08660 mRNA in tigecycline-susceptible VRE57 and VRE109, and tigecycline-resistant VRE65, VRE69, and VRE80 strains normalized to reference genes gdh, pyrC and gyrA (p > .05, as determined by ANOVA). Error bars represent standard deviations of at least three experimental replicates, and statistically significant differences are marked with ***.
3.2.2. tetM (QP83_12955)
Comparison of genomes identified a Phe473Ile substitution in the gene encoding the tetracycline resistance protein TetM. However this polymorphism was located far away from codons for amino acids responsible for excluding tetracycline from the ribosome by altering nucleotide conformation of the 16S rRNA (Donhofer et al., 2012). Linkevicius et al. (2016) reported several tetM mutations involved with increased tigecycline resistance in E. coli, and the one with the greatest capacity to increase tigecycline MIC was a L505 deletion, resulting in a shortened III loop in TetM (Linkevicius et al., 2016). TetM-mediated tigecycline resistance has also been attributed to an overlap of the 9-tbutylglycylamido moiety of tigecycline and the domain IV loop of TetM, but it seems unlikely that the Phe473Ile substitution we identified would alter this overlap, since amino acid 473 does not interact directly with the tigecycline molecule (Jenner et al., 2013). Recently, Fiedler et al. (2016) reported that high-level expression of tetM due to its presence on a high copy-number plasmid was able to confer tigecycline resistance in enterococcal clinical isolates (Fiedler et al., 2016). We therefore examined tetM expression in our tigecycline-susceptible and resistant strains. We found that the abundance of tetM message was in fact higher in VRE65, VRE69 and VRE80 compared to VRE57 and VRE109 strains (Fig. 2). We currently do not know whether the polymorphism we observed in the tigecycline-resistant strains might stabilize the mRNA resulting in greater abundance, or whether increased abundance is due to higher levels of transcription of this gene in these strains. No polymorphisms were detected in the region upstream of the tetM ORF, which is known to regulate expression via transcriptional attenuation (Su et al., 1992).
3.2.3. Multidrug ABC transporter ATP-binding protein (QP83_01175)
Another polymorphism that was common to tigecycline-resistant strains resided in a predicted multidrug ABC transporter ATP-binding protein. The base change causes an Asp245Glu substitution in the amino acid sequence of the protein. This protein has a primary sequence similar to the Gram-negative lipid A export permease/ATP-binding protein MsbA (Doerrler and Raetz, 2002), and to the ABC-type bacteriocin and lantibiotic exporters belonging to the SunT superfamily (Paik et al., 1998). Because this protein appears to be involved in transmembrane transport, we hypothesized that it could play a role in tigecycline efflux. We found that the efflux pump inhibitor verapamil was able to produce a 4-fold decrease in the tigecycline MIC for VRE65, VRE69 and VRE80, but only a 2-fold decrease in the tigecycline MIC for VRE57 and VRE109 (Table 4). On the other hand, reserpine and CCCP were not able to produce any changes in the tigecycline MIC. According to DeMarco et al. (2007), an efflux pump inhibitor-associated 4-fold decrease in MIC is indicative of efflux (Demarco et al., 2007). The observed synergy with verapamil suggests that efflux may contribute to the resistance phenotypes of VRE65, VRE69 and VRE80.
To test whether this multidrug ABC transporter ATP-binding protein could efflux tigecycline, we tested a presumed loss of function transposon insertion mutant that was generated in E. faecalis strain OG1RF. In this strain, a mariner insertion occurs at base 418 of the gene and would be predicted to permit only the first 139 amino acids of 594 in the full-length protein to be made, resulting in truncation to only ~20% of the full-length protein and notably removing the ATP-binding domain. However, no difference in tigecycline MIC was observed between wild type OG1RF and the transposon insertion mutant (Table 4). We also observed no change in tigecycline MIC when the mutant allele was overexpressed in the VRE109 background (Table 4). Finally, no differences in mRNA abundance for this gene were observed when comparing the sensitive and resistant strains (Fig. 2). Thus, it seems that the polymorphism that occurs in the multidrug ABC transporter ATP-binding protein is not involved in the resistance of E. faecalis to tigecycline.
3.2.4. Hypothetical protein (QP83_08660)
We identified a Gly137Glu substitution in the sequence of a gene encoding a hypothetical protein that could be involved in translation, based on the occurrence of conserved domains. Because tigecycline inhibits protein translation by blocking the entry of aminoacyl-tRNA into the A site of the ribosome (Jenner et al., 2013), we could not rule out the possibility that mutating this hypothetical protein was somehow important for tigecycline resistance. Expression of the mutant allele of the hypothetical protein in trans in the VRE109 background did not increase the tigecycline MIC, and a transposon mutant that truncates 96% of the protein did not affect the tigecycline MIC of OG1RF (Table 4). No differences in mRNA abundance for this hypothetical protein were observed when comparing the sensitive and resistant strains to one another (Fig. 2). Thus, it seems that hypothetical protein (QP83_08660) is not involved in tigecycline resistance in these strains.
3.2.5. lepA (QP83_09250)
Another polymorphism that plausibly could be related to tigecycline resistance in E. faecalis occurred in lepA, which encodes a GTP-binding protein (Gibbs et al., 2017). A K16I substitution in tigecycline-resistant isolates distinguished them from sensitive isolates and others in the NCBI database. An OG1RF transposon insertion mutant missing ~70% if the LepA protein did not exhibit an altered tigecycline MIC (Table 4). Thus, we conclude that lepA is unlikely to be involved in the observed tigecycline resistance in E. faecalis.
3.3. Polymorphisms in tigecycline-resistant variants of VRE109 selected in vitro
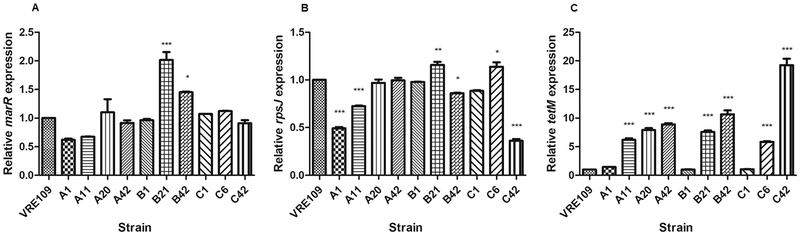
To explore evolutionary trajectories leading to tigecycline resistance in the ST103 lineage, and to attempt to recapitulate some the most relevant polymorphisms, we conducted independent in vitro resistance evolution experiments in triplicate using VRE109 as the tigecycline-sensitive parent strain. Comparison of genome sequences of resistant variants selected in vitro to the VRE109 parent revealed no consistent differences occurring exclusively in reading frames. However, examining of non-coding sequences revealed multiple changes upstream of a gene coding for a MarR-family transcriptional regulator. Mutations occurred in all three evolution experiments in either its predicted promoter region, or within the coding sequence. Tigecycline resistant mutants A11, A20 and A42 had an A → G substitution at position −41 (relative to the marR start codon); mutants B21 and B42 had a G400A transversion in the marR coding sequence resulting in an Ala to Thr change (Ala134Thr), and mutant C42 had a C → T substitution at position −10.
We wondered if the effect of the upstream mutations was to alter marR mRNA abundance in the tigecycline-resistant strains. Counterintuitively, we found a modest but significant increase in marR expression in strains B21 and B42 only (Fig. 3). We constructed several marR trans complementation strains (Table 4), but none affected tigecycline MIC. This mutation may have been selected for simply for enhanced ability to rapidly divide in vitro over the extended course of the experiment.
Fig. 3.
Relative mRNA levels for MarR-family transcriptional regulator QP83_14295 (A), rpsJ (B) and tetM (C) for E. faecalis VRE109 and tigecycline-resistant variants selected in vitro. Mean expression is plotted relative to the reference genes gdh, pyrC and gyrA. Relative QP83_14295, rpsJ and tetM expression were normalized to VRE109 (p > .05, as determined by ANOVA). Error bars represent the standard deviations, and statistically significant differences are marked with *, ** or *** indicating the intensity of the difference, being *** the more intense.
Other resistance-associated mutations that we identified occurred uniquely in each of the triplicate experiments, suggesting that multiple evolutionary trajectories lead to tigecycline resistance (Table 5). Variants in the S10 protein-encoding gene rpsJ as well as tetM arose in resistant strains from experiments A and C, respectively, recapitulating the importance of these genes in the clinical isolates, although we did not observe any relationship between the rpsJ mutations and expression levels (Fig. 3). Additionally, variants in several other ribosomal proteins arose in experiments A and B.
Table 5.
Mutations in coding regions detected among in vitro tigecycline-resistant strains.
| Strain | Tigecycline MIC (mg/L) | Product | Coding region change | Amino acid change |
|---|---|---|---|---|
| A1 | 0.06 | - | - | - |
| A11 | 0.125 | - | - | - |
| A20 | 0.5 | iron ABC transporter substrate-binding protein | G943 T | G315* |
| 30S ribosomal protein S10 | C166T | H56Y | ||
| A42 | 1.0 | iron ABC transporter substrate-binding protein | G943 T | G315* |
| 30S ribosomal protein S10 | C166T | H56Y | ||
| translation initiation factor IF-2 | G1514A | G505D | ||
| 50S ribosomal protein S3 | 493_494insGTTCAG | S164E165insGS | ||
| B1 | 0.06 | - | - | - |
| B21 | 0.25 | MarR-family transcriptional regulator | G400A | A134T |
| B42 | 0.5 | MarR-family transcriptional regulator | G400A | A134T |
| PhoB-family transcriptional regulator | A320G | E107G | ||
| 50S ribosomal protein L11 | C76T | P26Ser | ||
| Integrase | T707A | F239L | ||
| 50S ribosomal protein L6 | G196A | G66Arg | ||
| Membrane protein | A1029T | L343F | ||
| C1 | 0.06 | - | - | - |
| C6 | 0.125 | - | - | - |
| C42 | 1.0 | TetM | G1826T | G609V |
Quantitative PCR showed that tetM expression increased proportionally to changes in tigecycline MIC in each individual experiment (Fig. 3). TetM is a ribosomal protection protein able to bind to the ribosome and chase tetracyclines from their binding sites (Arenz et al., 2015). Interestingly, the tigecycline-resistant strain C42 possesses a 125 bp deletion upstream of tetM, which includes the region that encodes its leader peptide (Roberts and Mullany, 2009), as well as a polymorphism in the tetM structural gene itself. This variant also exhibited the greatest levels of tetM mRNA as well as the highest tigecycline MIC. Since tetM transcription is regulated via attenuation (Su et al., 1992), this could explain why tetM mRNA levels are so much higher in C42 compared to other strains. Similar mutations upstream of tetM were not observed in tigecycline-resistant variants with elevated mRNA levels deriving from the other two in vitro selection experiments. Whether other factors affect tetM regulation in those strains, or mRNA turnover, remains unknown.
4. Conclusions
The introduction of tigecycline into the formulary at Risoleta Tolentino Neves Hospital in early 2011 appears to have selected for the emergence and proliferation of an endemic, highly transmissible ST103 VRE E. faecalis lineage. The Hospital Infection Control Committee was notified so that measures could be taken to limit its spread. Our results suggest that resistance in the ST103 lineage stems from polymorphisms in rpsJ, which encodes the S10 protein of the 30S ribosomal subunit, and polymorphisms leading to increased tetM expression. Selecting for tigecycline-resistant variants in vitro generated similar types of mutations, with the most consistently observed effect being the emergence of strains with altered ribosomal proteins and elevated levels of tetM expression. In one case, this could be directly attributed to loss of the transcription attenuator that limits tetM expression. The fact that resistance to tigecycline can arise from endogenous genetic traits already present in many strains of E. faecalis indicates that to maximize itsutility it should be reserved for treating E. faecalis infections where other options have been exhausted, or where the course of therapy is likely to be short.
Our results suggest that rpsJ and tetM play a major role in the development of resistance to tigecycline in E. faecalis. When it comes to the in vitro settings, the tigecycline resistance phenotype of E. faecalis ST103 strains seems to be a multifactorial event, with contributions of many ribosomal proteins, but increased expression of TetM is a common theme.
Acknowledgements
We thank Edna Leite, Hoberdan Pereira and Hyllo Marcello Jr. from RTNH for providing the isolates, Lars Bogø Jensen for providing the rep-family plasmids controls, and Ana Lúcia Darini for providing controls for species identification, Van element typing and virulence factor detection.
Funding
This work was supported by National Council for Scientific and Technological Development [grant number 472907/2010–7], São Paulo Research Foundation [grant numbers 2011/14592–1 to T. P. M. and 2013/24952–0 to A. N. G. D.], Coordination for the Improvement of Higher Education Personnel88881.030524/2013–01 to I. L. B. C. C., 88887.114824/2015–00 to J. S. A. C. and 88887.063842/2014–00 to R.N. B.). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) -Finance Code 001. Sequencing, cloning, and comparative genome analysis studies conducted in Boston were supported by the Harvard-wide Program on Antibiotic Resistance, AI083214, and AI072360. D.V.T. was supported by National Institutes of Health award EY028222.
Footnotes
Transparency declarations
We declare no conflicts of interest.
References
- Agência Nacional de Vigilância Sanitária Registro ANVISA n° 1211002630013 - TYGACIL- VÁLIDO. https://www.smerp.com.br/anvisa/?ac=prodDetail&anvisaId=1211002630013>.
- Arenz S, Nguyen F, Beckmann R, Wilson DN, 2015. Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9-a resolution. Proc. Natl. Acad. Sci. U. S. A 112, 5401–5406. 10.1073/pnas.1501775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartha NA, Soki J, Urban E, Nagy E, 2011. Investigation of the prevalence of tetQ, tetX and tetX1 genes in Bacteroides strains with elevated tigecycline minimum inhibitory concentrations. Int. J. Antimicrob. Agents 38, 522–525. 10.1016/j.ijantimicag.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Beabout K, et al. , 2015. The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother 59, 5561–5566. 10.1128/aac.00547-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster S, et al. , 2007. Enterococcal leucine-rich repeat-containing protein involved in virulence and host inflammatory response. Infect. Immun 75, 4463–4471. 10.1128/iai.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan EM, Bae T, Kleerebezem M, Dunny GM, 2000. Improved Vectors for Nisin-Controlled Expression in Gram-Positive Bacteria. Plasmid 44, 183–190. 10.1006/plas.2000.1484. [DOI] [PubMed] [Google Scholar]
- Bustin SA, et al. , 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 55, 611–622. 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Camargo IL, Gilmore MS, Darini AL, 2006. Multilocus sequence typing and analysis of putative virulence factors in vancomycin-resistant and vancomycin-sensitive Enterococcus faecium isolates from Brazil. Clin. Microbiol. Infect 12, 1123–1130. 10.1111/j.1469-0691.2006.01496.x. [DOI] [PubMed] [Google Scholar]
- Cattoir V, et al. , 2015. Genomic analysis of reduced susceptibility to tigecycline inEnterococcus faecium. Antimicrob. Agents Chemother 59, 239–244. 10.1128/aac.04174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetinkaya Y, Falk P, Mayhall CG, 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13, 686–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. (CLSI, Wayne, PA, USA, 2018). [Google Scholar]
- Cordina C, Hill R, Deshpande A, Hood J, Inkster T, 2012. Tigecycline-resistant Enterococcus faecalis associated with omeprazole use in a surgical patient. J. Antimicrob. Chemother 67, 1806–1807. 10.1093/jac/dks122. [DOI] [PubMed] [Google Scholar]
- Costello SE, Gales AC, Morfin-Otero R, Jones RN, Castanheira M, 2016. Mechanisms of resistance, clonal expansion, and increasing prevalence of Acinetobacter baumannii strains displaying elevated tigecycline MIC values in Latin America. Microb. Drug Resist 22, 253–258. 10.1089/mdr.2015.0168. [DOI] [PubMed] [Google Scholar]
- Cunha BA, Baron J, Cunha CB, 2017. Once daily high dose tigecycline - pharmacokinetic/pharmacodynamic based dosing for optimal clinical effectiveness: dosing matters, revisited. Expert Rev. Anti-Infect. Ther 15, 257–267. 10.1080/14787210.2017.1268529. [DOI] [PubMed] [Google Scholar]
- Dabul ANG, Avaca-Crusca JS, Van Tyne D, Gilmore MS, Camargo ILBC, 2017. Resistance in In Vitro selected tigecycline-resistant methicillin-resistant Staphylococcus aureus sequence Type 5 is driven by mutations in mepR and mepA genes. Microb. Drug Resist 10.1089/mdr.2017.0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JL, et al. , 2018. Comprehensive Functional Analysis of the Enterococcus faecalisCore Genome Using an Ordered, Sequence-Defined Collection of Insertional Mutations in Strain OG1RF. mSystems 3 10.1128/mSystems.00062-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarco CE, et al. , 2007. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob. Agents Chemother 51, 3235–3239. 10.1128/aac.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerrler WT, Raetz CR, 2002. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem 277, 36697–36705. 10.1074/jbc.M205857200. [DOI] [PubMed] [Google Scholar]
- Donhofer A, et al. , 2012. Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. U. S. A 109, 16900–16905. 10.1073/pnas.1208037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing, 2018. Version 8.1 Breakpoint tables for interpretation of MICs and zone diameters. Vol. 2018.
- European Medicines Agencies Tygacil (tigecycline). http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/human/medicines/000644/human_med_001118.jsp&mid=WC0b01ac058001d124>.
- Fiedler S, et al. , 2016. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J. Antimicrob. Chemother 71, 871–881. 10.1093/jac/dkv420. [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration, 2005. Full Prescribing Information - Tygacil. https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/021821s026s031lbl.pdf>.
- Gibbs MR, et al. , 2017. Conserved GTPase LepA (Elongation factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome. Proc. Natl. Acad. Sci. U. S. A 114, 980–985. 10.1073/pnas.1613665114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, et al. , 2013. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc. Natl. Acad. Sci. U. S. A 110, 3812–3816. 10.1073/pnas.1216691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LB, et al. , 2010. A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods 80, 25–43. 10.1016/j.mimet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Kuch A, et al. , 2012. Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. J. Antimicrob. Chemother 67, 551–558. 10.1093/jac/dkr544. [DOI] [PubMed] [Google Scholar]
- Leavis HL, et al. , 2007. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog 3 (e7). 10.1371/journal.ppat.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkevicius M, Sandegren L, Andersson DI, 2016. Potential of Tetracycline ResistanceProteins to Evolve Tigecycline Resistance. Antimicrob. Agents Chemother 60, 789–796. 10.1128/aac.02465-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McAleese F, et al. , 2005. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob. Agents Chemother 49, 1865–1871. 10.1128/aac.49.5.1865-1871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo TP, Dabul AN, Camargo IL, 2015. Different VanA elements in E. faecalis and in E. faecium suggest at least two origins of Tn1546 among VRE in a Brazilian Hospital. Microb. Drug Resist 21, 320–328. 10.1089/mdr.2014.0077. [DOI] [PubMed] [Google Scholar]
- Niebel M, et al. , 2015. Deletions in a ribosomal protein-coding gene are associated with tigecycline resistance in Enterococcus faecium. Int. J. Antimicrob. Agents 46, 572–575. 10.1016/j.ijantimicag.2015.07.009. [DOI] [PubMed] [Google Scholar]
- Paik SH, Chakicherla A, Hansen JN, 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem 273, 23134–23142. [DOI] [PubMed] [Google Scholar]
- Peleg AY, Adams J, Paterson DL, 2007. Tigecycline Efflux as a Mechanism for Nonsusceptibility in Acinetobacter baumannii. Antimicrob. Agents Chemother 51, 2065–2069. 10.1128/aac.01198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Huband MD, Streit JM, Flamm RK, Sader HS, 2018. Surveillance of tigecycline activity tested against clinical isolates from a global (North America, Europe, Latin America and Asia-Pacific) collection (2016). Int. J. Antimicrob. Agents 10.1016/j.ijantimicag.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Roberts AP, Mullany P, 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17, 251–258. 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Ruiz-Garbajosa P, et al. , 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol 44, 2220–2228. 10.1128/jcm.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Keeney D, Bradford PA, 2007. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J. Antimicrob. Chemother 59, 1001–1004. 10.1093/jac/dkm058. [DOI] [PubMed] [Google Scholar]
- Sader HS, et al. , 2016. Tigecycline antimicrobial activity tested against clinical bacteria from Latin American medical centres: results from SENTRY Antimicrobial Surveillance Program (2011–2014). Int. J. Antimicrob. Agents 48, 144–150. 10.1016/j.ijantimicag.2016.04.021. [DOI] [PubMed] [Google Scholar]
- Shieh Y-W, et al. , 2015. Operon Structure and Cotranslational Subunit Association Direct Protein Assembly in Bacteria. 10.1126/science.aac8171. [DOI] [PubMed]
- Song X, Sun J, Mikalsen T, Roberts AP, Sundsfjord A, 2013. Characterisation of the plasmidome within Enterococcus faecalis isolated from marginal periodontitis patients in Norway. PLoS One 8, e62248 10.1371/journal.pone.0062248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YA, He P, Clewell DB, 1992. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother 36, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, et al. , 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol 33, 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P, 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18, 4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GH, Sabo JL, Rice-Trujillo C, Hernandez J, McDermott PF, 2018. Whole-genome sequencing based characterization of antimicrobial resistance in Enterococcus. Pathog Dis 76 10.1093/femspd/fty018. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, et al. , 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega S, Dowzicky MJ, 2017. Antimicrobial susceptibility among Gram-positive and Gram-negative organisms collected from the Latin American region between 2004 and 2015 as part of the Tigecycline Evaluation and Surveillance Trial. Ann. Clin. Microbiol. Antimicrob 16, 50 10.1186/s12941-017-0222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L, Feudi C, Fortini D, Garcia-Fernandez A, Carattoli A, 2014. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob. Agents Chemother 58, 1707–1712. 10.1128/aac.01803-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardal E, Gawryszewska I, Hryniewicz W, Sadowy E, 2013. Abundance and diversity of plasmid-associated genes among clinical isolates of Enterococcus faecalis. Plasmid 70, 329–342. 10.1016/j.plasmid.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Werner G, Gfrorer S, Fleige C, Witte W, Klare I, 2008. Tigecycline-resistant Enterococcus faecalis strain isolated from a German intensive care unit patient. J. Antimicrob. Chemother 61, 1182–1183. 10.1093/jac/dkn065. [DOI] [PubMed] [Google Scholar]
- Woodford N, et al. , 1993. Application of DNA probes for rRNA and vanA genes to investigation of a nosocomial cluster of vancomycin-resistant enterococci. J. Clin. Microbiol 31, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N, Watson AP, Chadwick PR, 1997. Investigation by long PCR of the genetic elements mediating VanA glycopeptide resistance in enterococci from un-cooked meat in South Manchester. Adv. Exp. Med. Biol 418, 409–412. [DOI] [PubMed] [Google Scholar]
- Xie F, Xiao P, Chen D, Xu L, Zhang B, 2012. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- Zhong X, et al. , 2014. First emergence of acrAB and oqxAB mediated tigecycline resistance in clinical isolates of Klebsiella pneumoniae pre-dating the use of tigecycline in a Chinese hospital. PLoS One 9, e115185 10.1371/journal.pone.0115185. [DOI] [PMC free article] [PubMed] [Google Scholar]