ABSTRACT
Background
Controlled glycemic concentrations are associated with a lower risk of conditions such as cardiovascular disease and diabetes. Models commonly used to guide interventions to control the glycemic response to food have low efficacy, with recent clinical guidelines arguing for the use of personalized approaches.
Objective
We tested the efficacy of a predictive model of personalized postprandial glycemic response to foods that was developed with an Israeli cohort and that takes into consideration food components and specific features, including the microbiome, when applied to individuals from the Midwestern US.
Design
We recruited 327 individuals for this study. Participants provided information regarding lifestyle, dietary habits, and health, as well as a stool sample for characterization of their gut microbiome. Participants were connected to continuous glucose monitors for 6 d, and the glycemic response to meals logged during this time was computed. The ability of a model trained using meals logged by the Israeli cohort to correctly predict glycemic responses in the Midwestern cohort was assessed and compared with that of a model trained using meals logged by both cohorts.
Results
When trained on the Israeli cohort meals only, model performance for predicting responses of individuals in the Midwestern cohort was better (R = 0.596) than that observed for models taking into consideration the carbohydrate (R = 0.395) or calorie content of the meals alone (R = 0.336). Performance increased (R = 0.618) when the model was trained on meals from both cohorts, likely because of the observed differences in age distribution, diet, and microbiome.
Conclusions
We show that the modeling framework described in Zeevi et al. for an Israeli cohort is applicable to a Midwestern population, and outperforms commonly used approaches for the control of blood glucose responses. The adaptation of the model to the Midwestern cohort further enhances performance and is a promising means for designing effective nutritional interventions to control glycemic responses to foods. This trial was registered at clinicaltrials.gov as NCT02945514.
Keywords: glycemic response, personalized nutrition, diabetes, continuous glucose monitors, carbohydrate content, microbiome
Introduction
Glycemic concentrations are associated with the risk of diseases such as obesity, cardiovascular disease, metabolic syndrome, and type II diabetes mellitus (1–4). Diet directly affects glycemic concentrations, and dietary interventions aimed at controlling these concentrations are commonly prescribed (5).
Common dietary approaches for preventing frequent high blood glucose include calorie or carbohydrate counting, and approaches based on the glycemic index (6–8); however, their efficacy varies across individuals (5). Besides characteristics of the food consumed (9), such variability may stem from, among other things, lifestyle, genetics, degree of insulin sensitivity, and exocrine pancreatic and glucose transporter activity concentrations (10), as well as intestinal transit time (11). There is no evidence supporting the view that one nutritional strategy is consistently superior to others, with researchers suggesting that the most successful strategy for controlling blood glucose concentrations is dependent on the characteristics of each particular individual (12).
A previous study (13) showed that the microbiome of an individual was also one of the parameters associated with postprandial glucose responses (PPGRs) to food, in addition to other physiological and clinical parameters. Using a wide set of features, the team were able to construct a predictive model of greater accuracy than state-of-the-art models for PPGRs, and show that this model could be used to construct personalized diets that induce a low glycemic response in tested participants.
One caveat of the study by Zeevi et al. (13) was the fact that it focused on a specific population, raising questions regarding the generality of the model used to predict PPGR. The microbiome, for example, has been shown to vary across populations as a consequence of economic and geographic factors (14), and diet composition (15). Dietary habits, clinical features, and other personal characteristics may also be significantly different between populations. So, it is possible that observations, or results of a study involving one population, may not translate to other populations (16).
Here, we tested the ability of a model trained on the Israeli population to predict the PPGRs of a cohort of participants mostly based in the Midwestern region of the US. We replicated the approach in the original study as closely as possible to reduce the number of potential confounding variables affecting the results, and used the data collected to improve the predictive model.
Methods
The central objective of this study was to measure the accuracy of a personalized model for predicting PPGRs in individuals without diabetes in a Midwestern US cohort.
Study participants
A total of 327 participants were recruited from October 2016 to December 2017, in Olmsted and Hennepin counties in Minnesota, and Duval County in Florida, US, under the Mayo Clinic Institutional Review Board protocol number 16–0,05208. It is also registered at clinicaltrials.gov as NCT02945514. Inclusion criteria included men and women > 18 y, with access to a mobile device and web browser. Individuals were excluded if they were < 18 y, were prediagnosed with type I or type II diabetes mellitus, used antibiotics in the 3 mo prior to study participation, were pregnant, were substance abusers, had a chronic medical condition, treatment, or medication known to affect glucose metabolism, such as fertility treatment (e.g., clomiphene, gonadotropins) or metformin-based medications, had undergone bariatric weight loss surgery, had undergone further fertility treatments in the 3 mo prior to study participation, had undergone chemotherapy or radiation treatment of cancer within the last 2 y or had active cancer, had a chronic gastrointestinal disorder, had chronic anemia, or were unable to safely perform finger pricking.
Study design
This study follows, as closely as possible, the methodology described in Zeevi et al. (13).
The study subjects were recruited in Olmsted and Hennepin counties, Minnesota (n = 318), and Duval County, Florida (n = 9), US. The subjects are henceforth designated the Midwestern cohort. After screening regarding eligibility, study subjects were asked to provide written consent to participate in the study. Participants were asked to answer a series of questions regarding their health, lifestyle, activities, and diet preferences. In the 2 d prior to the beginning of the study week, participants were asked to provide a stool sample using an OMNIgene-Gut stool collection kit (DNA Genotek). Samples were shipped at room temperature to the DayTwo processing facility in Israel.
Participants were also asked to attend a connection meeting at the beginning of the study week. During this meeting, study staff provided a review of the study purpose and requirements and offered an opportunity to answer questions. Study staff also measured height, weight, waist, and hip circumference. They took blood pressure and pulse, drew blood for estimation of HbA1c% concentrations (for which fasting was not required), attached the continuous glucose monitor (CGM) and provided standardized meals. Study participants were also instructed on the use of the food and activity logging mobile application to be used throughout the week and the manual blood monitor (Bayer Contour Next Link Glucometer). During the study week, study subjects were asked to wear the CGM, complete manual glucose monitoring ≥ 4 times a day for added accuracy, log food intake (including meal content, duration, and time), activity (including intensity, duration, and time), anthropometric characteristics, medications, and sleep. They were asked to maintain their normal eating habits for the week, except for four breakfasts, which were composed of defined food items and provided by the study team.
Just like the study by Zeevi et al. (13), measures of anthropometric and physiological parameters were conducted by trained technicians. The participants also wore a CGM for the study week and logged their meal information exclusively in a mobile device application similar to the one used for the Israeli cohort, bypassing the risk of introducing methodological variability observed when different food logging methods are used.
During the week, study personnel, including a registered dietician, were available to assist study subjects with the logging of food and activity items.
Focused set of the Zeevi et al. cohort
To eliminate potential biases, when comparing the Midwestern cohort to the Israeli cohort, we subset the cohort described in Zeevi et al. (13). First, only the 413 subjects from the original cohort whose stool samples were collected using the same collection method were used in all comparative analyses (Genotek kits; see Stool Sample Processing). To account for different sequencing depths and for paired compared with single end sequencing differences, we used only the forward reads from the Israeli cohort samples and downsampled all samples to an equal sequencing depth of 5 Million reads (the same depth used for the Midwestern cohort). Second, since diabetic subjects were excluded from the Midwestern cohort, we further excluded 16 diabetic subjects from the Israeli cohort (Figure 1). The resulting subset consisted of 397 participants from the original study, representing the Israeli cohort used in all further analyses. Both the Midwestern and Israeli cohorts were subjected to the same bioinformatics analysis pipeline.
FIGURE 1.
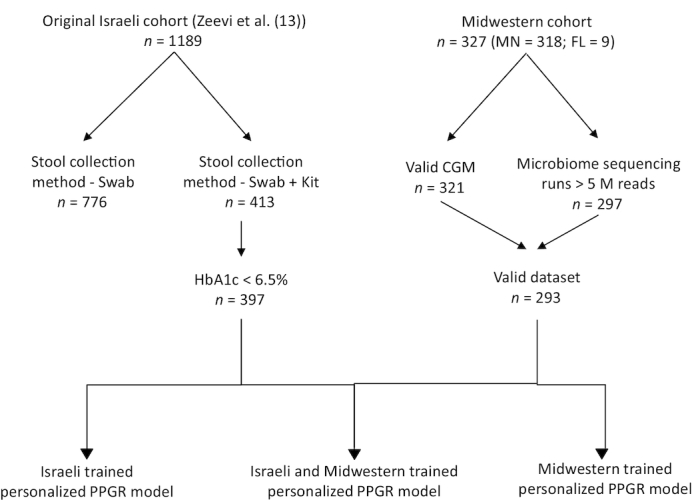
Flow diagram of the subjects included in the study. Israeli cohort used in this study represents a subset of the full cohort in Zeevi et al. (13). CGM, continuous glucose monitor; MN, Minnesota; FL, Florida.
Stool sample processing
Participants sampled their stool following detailed printed and online video clip instructions. Sampling was done using the OMNIgeneGUT (OMR-200; DNA Genotek) stool collection kit. Samples were stored at room temperature, as per the manufacturer's instructions, until delivered to the processing facilities.
Genomic DNA extraction and filtering
Genomic DNA was purified using the PowerMag Soil DNA isolation kit (MoBio) optimized for the Tecan automated platform. Illumina compatible libraries were prepared as described (17), and sequenced on an Illumina Nextera 500 (75 base pair, single end).
Microbial and metagenomic analysis
Reads were processed with Trimmomatic (18) (version 0.32, parameters used: -phred33 ILLUMINACLIP: < adapter file > :2:30:10 LEADING:25 TRAILING:25 MINLEN:50) to remove reads containing Illumina adapters, filter low-quality reads, and trim low-quality read edges. Reads mapping to host DNA were detected by mapping with GEM (19; parameters used: -q offset-33 –gem-quality-threshold 26 -e 0.1 –min-matched-bases 0.8 –max-big-indel-length 15 -s 3 -d 200 -D 1 -v -m 0.05) and removed from downstream analysis. All samples were subsequently downsampled to a depth of 5 M reads. Samples with fewer reads were removed from further analysis, leaving us with samples from 297 Midwestern cohort participants that were used for downstream analyses. Relative abundance for members of the microbial community were obtained with MetaPhlAn2 (20; default parameters).
CGM
Glucose was measured using the iPro2 CGM (Medtronic), which measures interstitial glucose concentrations every 5 min using the subcutaneous Enlite sensors. CareLink online software (Medtronic) was used to perform calibration for CGM measurements, as directed by the iPro2 manual. Subjects were connected to the CGM for 6 d [compared with 7 d for the Israeli cohort (13)].
Real-time meal logging and meal data preprocessing
Meals were logged in real time by the participants using a mobile device application available for both iOS and Android devices (“DayTwo Food & Activity Logger”). Participants were asked to choose the food items consumed from the MyNetDiary database with over 400,000 items for which curated nutrient content information is available and to log the amount consumed, and the time and duration of the meal. Reported meal times were rounded to the closest 5-min interval. Meals logged < 30 min apart were merged. We also removed meals with very large (>1 kg and >20 kcal) components, meals with incomplete logging, and meals with unreasonable nutritional values (defined as meeting one or more of these conditions: >5000 kcal, >500 g sugar, >1000 g carbohydrate, >1000 g protein, >500 g fat, >5000 mg sodium, >600 mg caffeine, >300 g dietary fibers), as they were likely to be the result of logging errors. Meals logged within 90 min of other meals and meals consumed at the first and last 12 h of the connection week were not used for algorithm training, to avoid inaccuracies in PPGR measurements. Lastly, meals with >40 g carbohydrate and PPGR values <5 mg/dl·h were excluded from the analysis as manual inspection confirmed that in many cases, such meals showed faulty CGM measurements or misreporting of meal times.
Standardized meals
The four standardized meals used with the Israeli cohort (bread, bread + butter, glucose, fructose) were described in Zeevi et al. (13). For the American cohort, participants were supplied with four standardized meals representing typical Midwestern breakfast foods that were to be eaten alone, as the first meal of the day; two of them consisted of one plain bagel with or without cream cheese; the other two consisted of cereals (participants chose one of three brands) with or without a cup of milk (milk was not supplied and participants were instructed to use either soy milk or 2% milk). Each meal was provided in duplicate to allow estimation of the degree of replicability of the glycemic measures by analyzing the intra-individual variability in PPGRs for the standardized meals. In practice, many participants reported varying amounts and numerous types of milk, and some participants reported eating standardized foods along with other items and/or at different times of day. As a result of this misreporting, combined with a relatively small cohort, only the bagel with cream cheese meals had sufficient replicates from a large enough group of participants to allow calculation of overall intra-individual variability in our cohort (n = 111). Since the standardized meals were not chosen by, and may not represent the eating habits of, particular participants, they were not used when analyzing macronutrient consumption differences between cohorts.
Computing PPGRs
Five subjects from the Midwestern cohort had corrupted CGM files that could not be processed. These were thus excluded from the PPGR predictor construction and analyses. A total of 293 participants that had both valid CGM measurements and over 5 M reads depth in the microbiome samples were used for training the model (see below).
PPGRs were computed as described in Zeevi et al. (13): logged meal times and CGM measurements were used to calculate the incremental area under the curve (iAUC) in the 2 h following a meal as previously described (21). To reduce noise, the median of all glucose values from the 30-min period prior to the meal was taken as the initial glucose concentration, above which the incremental area was calculated. Missing values in ≤ 25 consecutive minutes were interpolated. Following this interpolation, any meals that still had incomplete glucose measurements in the time window of 30 min before and 2 h after the logged meal time were filtered out. The resulting quantity is referred to as the “measured PPGR” throughout the manuscript.
PPGR predictor
As in Zeevi et al. (13), we predicted PPGRs based on stochastic gradient boosting regression. Using this type of model, PPGRs are predicted as the sum of predictions from thousands of decision trees. The trees were learned iteratively, where each tree tried to minimize the residuals in predictions of all previous trees. Here, the model was implemented using the XGBoost package (version 0.6, https://xgboost.readthedocs.io/en/latest/, 22), XGBRegressor class. The model used 2,000 tree estimators with a maximal depth of 9 and with a learning rate of 0.002, with the objective of minimizing squared errors in prediction. At each iteration half of the samples were used to prevent overfitting.
Features used in each tree estimator were chosen using an inference process from a pool of 72 features highly similar to the ones used in the Zeevi et al. (13) original study, with the exception of some blood test results, bacterial growth dynamics, and KEGG-based (Kyoto Encyclopedia of Genes and Genomes) bacterial functional features, that were excluded from our analysis (see Online Supporting Information (OSM), Supplemental Table 1, for a full list of the features used). The features included: 1) meal features, 2) meal context features, 3) blood tests, 4) personal features, 5) features derived from CGM measurements, and 6) microbiome features. For microbiome features we used the relative abundances for taxa used by Zeevi et al. (13) with the addition of the abundances of Prevotella and Bacteroides genera. Prevotella and Bacteroides features were added to the model based on results below, showing that the abundances of these genera varied significantly between cohorts. Microbiome features whose median abundance was <10−4 in the focused Israeli cohort and the Midwestern cohort combined were excluded.
Model performance was evaluated using 10-fold cross validation, where the data is split into 10 groups (making sure that all meals reported by a single client are grouped in the same fold). The model is trained on 9 of the 10 folds and its performance is assessed by its ability to accurately predict the PPGR for the held out fold (1/10 of the data) that was not used for training, which is repeated for all folds.
Statistical tests for significance
The chi-square statistic was used for testing the significance of gender distribution differences between cohorts. Welch's t-test was used for all other tests of significance (personal features, macronutrient consumption, phyla/genera abundance distributions, etc.). When multiple hypotheses were tested, P values were corrected using the Bonferonni correction (23).
Partial dependence plots
Although the significance of each feature of the model is hard to assess in models created through the use of gradient boosting, their individual effects can be analyzed using partial dependent plots. For each feature f (e.g., carbohydrate grams in meal), the average predicted PPGR is computed at each feature value (e.g., at x = 1…80 g of carbohydrate per meal) by setting the feature value to x (e.g., carbohydrates = 20 g) across the entire set of reported meals, and keeping all other feature values as originally reported. The vector of average predicted PPGR is then centered to yield the partial dependence values observed in the partial dependence plots.
Principal coordinate analysis
To determine whether cohorts were different in terms of their gut microbial profile, we performed an unsupervised analysis using principal coordinate analysis on dissimilarities between all species. Analysis was performed using the PC-ord tool v7 (https://www.pcord.com) using Bray–Curtis distances.
Results
We focused our work on testing and improving the performance of a model for the personalized prediction of the glycemic response to foods created for an Israeli cohort on a population of the Midwest of the US. For this, we recruited individuals and collected personal information, a variety of anthropometric and physiologic parameters, stool samples, and continuously monitored their blood glucose concentrations and dietary habits in real time for 6 consecutive days, using a food logging mobile application and following a previously described methodology (13). A companion report with a more focused clinical analysis of the Midwestern cohort is currently in press (Mendes-Soares et al., 24)
Midwestern and Israeli cohorts differ in physiological parameters
General attributes of the cohorts are shown in Table 1, and provide a basic overview of the differences between the study populations. The Midwestern cohort that was part of the analysis was composed of 297 nondiabetic individuals (77% females, 23% males), aged 45.1 ± 12.3 y, with a BMI of 27.9 ± 6.0 kg/m2 and HbA1c of 5.2% ± 0.3%. Although both cohorts are skewed in favor of females, there is an even stronger gender bias in the Midwestern cohort relative to the Israeli cohort (Table 1). The total percentage of overweight or obese individuals was significantly higher in the Midwestern cohort compared with the Israeli cohort (64% and 55%, respectively), but these values are in accordance with national and regional statistics [63% overweight reported in the state of Minnesota in 2015 (25), 49.8% overweight reported in Israel in 2011 (26)]. The percentage of prediabetics (defined as 5.7% < = HbA1c% <6.5%) in the Midwestern cohort was lower than expected based on national values [8.4% in our cohort compared with 38% in the US (27)].
TABLE 1.
Demographic and physiologic comparison of the Midwestern and Israeli cohorts
| Midwestern cohort | Israeli cohort | t-test P value | |
|---|---|---|---|
| # Participants | 297 | 397 | |
| Gender, % females | 77% | 60% | 3e-61 |
| Age, y mean ± SD | 45.1 ± 12.3 | 42.0 ± 12.4 | 0.0012 |
| BMI, kg/m2 mean ± SD | 27.9 ± 6.0 | 26.6 ± 5.0 | 0.0022 |
| BMI ≥ 25 | 64% (189) | 55% (217) | |
| BMI ≥ 30 | 27% (81) | 21% (84) | |
| HbA1c% mean ± SD | 5.2 ± 0.3 | 5.4 ± 0.4 | 8.5e-102 |
| HbA1c% ≥ 5.7% | 8.4% (25) | 21% (85) |
1Chi-square test of independence in a contingency table.
2Two-sided t-test for samples with unequal variances (Welch's t-test).
Carbohydrate response sensitivity to diverse foods varies across individuals
Participants were asked to wear a CGM for the duration of the study, allowing a complete description of their daily glucose concentrations. Of the participants from the Midwestern cohort, 93.5% exhibited a positive correlation between the amount of carbohydrates consumed in a meal and the PPGR to that meal compared with 98% of the participants in the Israeli cohort (Figure 2A). However, the extent of the correlation varied significantly, with some people showing high sensitivity (Figure 2B) to the amount of carbohydrates consumed, and others showing limited sensitivity (Figure 2C). This supports the view that the glycemic response is not easily predicted from simple meal features and varies across individuals.
FIGURE 2.
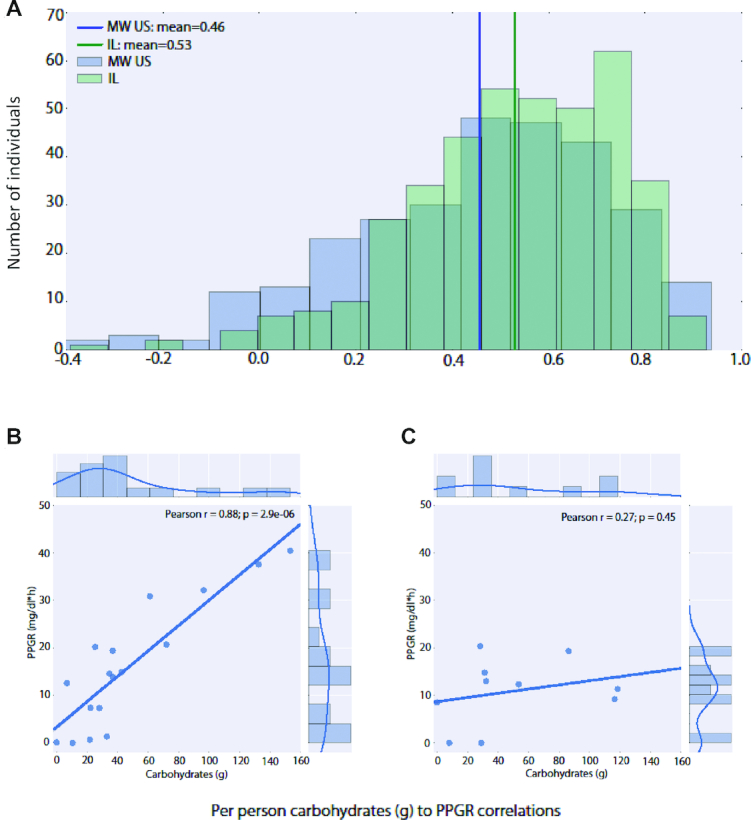
Carbohydrate sensitivity in individuals of the cohorts of the study. (A) Histogram of per person Pearson correlation R values between carbohydrates and postprandial glycemic response (PPGR) for individuals in the Israeli (IL, green; mean 0.53, std 0.22) and Midwestern (MW, blue; mean 0.46, std 0.26) cohorts. Note the high variability across individuals in sensitivity to carbohydrates. (B) An example for an individual with a high carbohydrate to PPGR correlation (left) and an individual with a low carbohydrate to PPGR correlation (right). For each reported meal, the carbohydrate values (x-axis) and the PPGR values (y-axis) are shown.
PPGRs to identical meals show reproducibility in individuals and high variability across the study population
During the study, participants were asked to maintain their dietary habits, except for the consumption of identical standardized meals provided in duplicate by the study team that would allow the comparison of PPGRs within and across individuals. Two types of meals were provided to participants, but only one was reported by enough participants to allow analysis.
To understand the reproducibility of the glycemic response to foods for each individual, we calculated the correlation between the PPGRs to the same meal for each individual participant. Each participant was asked to consume two identical meals of bagel and cream cheese provided by the study team. The PPGRs to the two meals were highly correlated (n = 111, Pearson's correlation R = 0.66, Figure 3), although these replicates show a somewhat lower reproducibility than that of the four standardized meals (bread, bread and butter, glucose, fructose) described in Zeevi et al. (13; R = 0.71–0.77).
FIGURE 3.
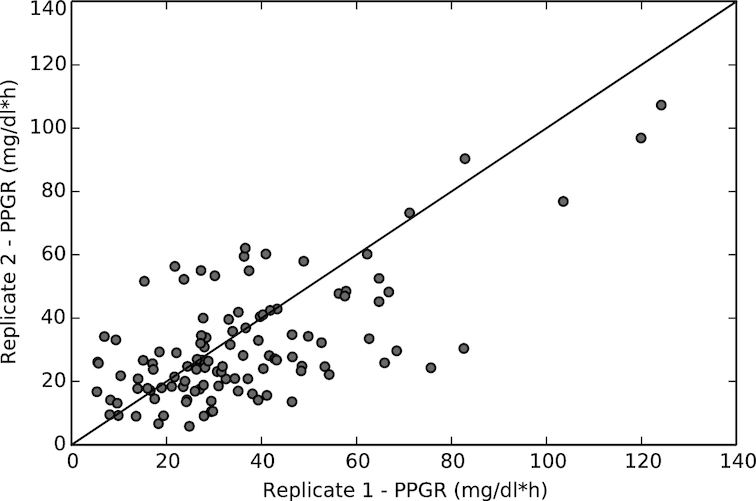
Reproducibility of postprandial glycemic responses to a standardized meal, as given by the correlation between the glycemic responses to replicate bagel and cream cheese meals consumed by individuals in the Midwestern cohort (n = 111; R = 0.66; see Methods).
Similarly to what has been observed in previous studies (13, 28), there was a significant spread of PPGR values across individuals. However, interpersonal differences were much larger than intrapersonal differences, with glycemic responses to the same bagel and cream cheese meal across individuals spanning almost the entire range of measured responses (the lowest 10% of responses to this meal had PPGR values <14 mg/dl·h, the highest 10% spanned PPGR values between 59 and 124 mg/dl·h).
Glycemic response in the Midwestern cohort can be predicted by a model trained with the Israeli cohort
Next, we tested whether a model trained on information from a subset of the Israeli cohort from Zeevi et al. (13) that used the same stool collection methods as our cohort could be used to predict glycemic responses for individuals in the Midwestern cohort. The model predicted glycemic responses based on a pool of 72 features from the data collected (OSM, Supplemental Table 1), including meal nutrients, personal questionnaires, daily activities, CGM measurement-derived features, blood tests, and microbiome composition. Just as described in Zeevi et al. (13), to account for nonlinear interactions between the features, the model was based on gradient boosting regression (29) in which PPGRs are predicted as the sum of predictions from thousands of decision trees. Each decision tree is a weak predictor that uses a subset of features. The trees are built iteratively, with each new tree trained on the residuals of all previous trees, so that prediction errors decrease with each iteration.
When trained on data from 397 Israeli participants alone, performance on held out data from the Israeli cohort (using 10-fold cross validation) was similar but slightly inferior to that described previously [R = 0.671 compared with R = 0.68 in Zeevi et al. (13)], likely stemming from a reduced data set and some discrepancies in feature sets used.
The performance of this model on the Midwestern cohort was R = 0.596 (Figure 4). This level of performance was better than that achieved by linear models that rely on meal carbohydrate or caloric content alone (R = 0.395, R = 0.336, respectively). Given the importance of clinical and physiological parameters of the individuals, diet, and microbiome in the performance of the predictive model, this result may be indicative of differences between the two cohorts in regards to these features.
FIGURE 4.
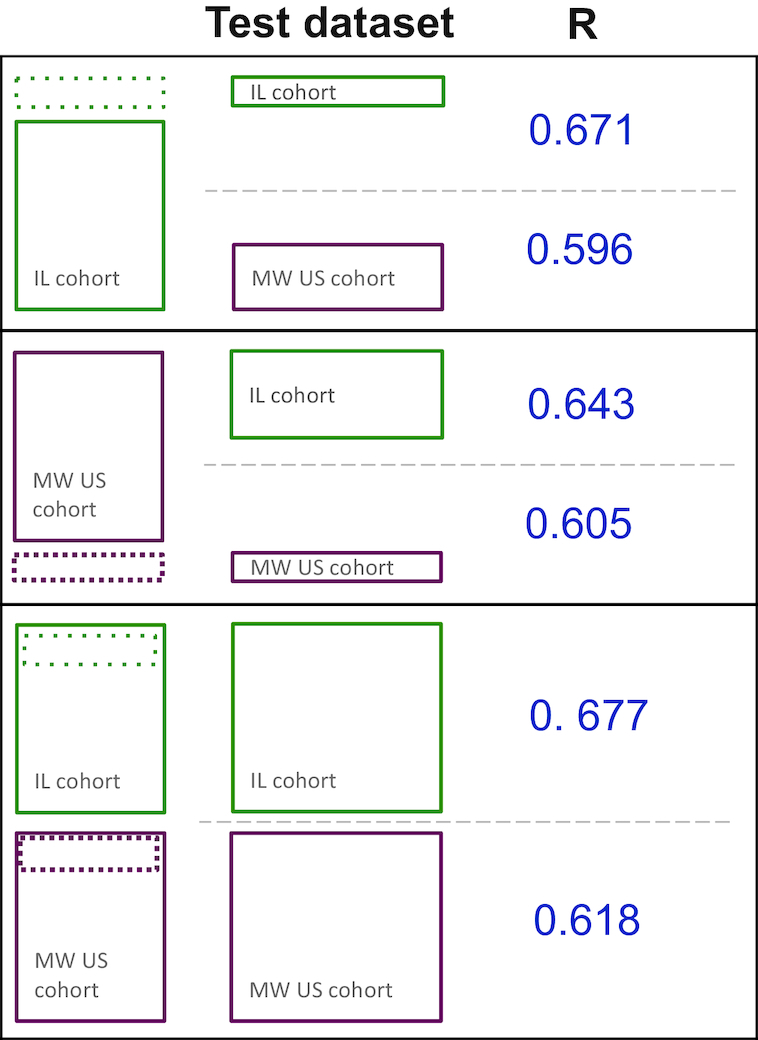
Schematic of the performance of the models trained with data exclusively from the Israeli (IL) cohort, exclusively from the Midwestern (MW) cohort, or both.
Macronutrient and fiber intake differ between cohorts
Participants were asked to log their eating patterns with a mobile application available for the most common operating systems. In total, 2094 d with meals defined as being valid for our analysis and a total of ∼3.2 M kcal were logged for the Midwestern cohort, and 3006 d and a total of 4.6 M kcal for the Israeli cohort. The observed differences result from a slightly shorter connection week for the Midwestern cohort.
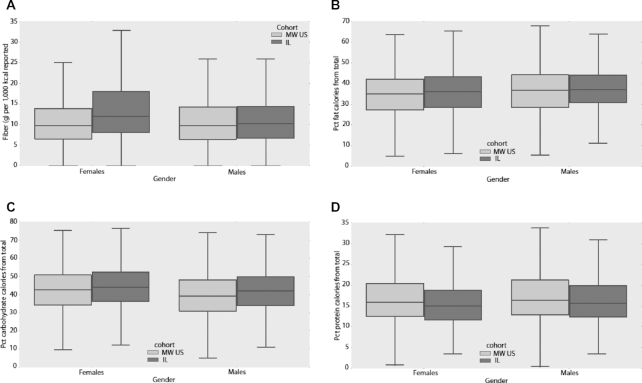
Comparison of macronutrient intake between cohorts revealed several significant differences, mostly in the female population, which is likely due to the relatively small number of male participants. The total reported caloric intake was somewhat higher in the Midwestern cohort compared with that in the Israeli cohort, but differences were significant only for females (1467 ± 755 compared with 1368 ± 712 mean daily kcal of reported consumption, P value = 9x10-5). Daily reported fiber consumption was lower in the Midwestern cohort compared with the Israeli one, but only significantly lower in females (11.1 compared with 14.2 dietary fiber g per 1000 kcal reported, P value = 8e-24; Figure 5A). In females, the percentage of calories from fat was slightly but significantly lower in the Midwestern cohort (34.3% compared with 35.8%, P value = 0.0005; Figure 5B). The percentage of calories from carbohydrates was slightly lower for the Midwestern cohort than for the Israeli cohort for males (39.5% compared with 41.7%, P value = 0.008) and females (43.1% compared with 44.7%, P value = 0.0007; Figure 5C). In contrast, the percentage of calories from protein was slightly larger in both males and females from the Midwestern relative to the Israeli cohort (males: 16.7% compared with 18.0%, P value = 0.004; females: 15.7% compared with 17.1%, P value = 8x10-10; Figure 5D). Sodium consumption was lower in Midwestern compared with Israeli males (2550 ± 1635 compared with 2850 ± 1612 mg sodium on average per day, P value = 0.001). Overall, these findings align well with the different diets adopted preferentially by these populations, with a lower prevalence of a Mediterranean diet, rich in raw vegetables and lower in animal products, in the Midwestern compared with the Israeli study subjects.
FIGURE 5.
The Israeli and Midwestern cohorts showed substantial differences in terms of their nutritional behavior. (A), (B), (C), and (D) show the amounts of each nutrient consumed by the female and male participants of the Midwestern (MW) and Israeli (IL) cohorts [(A) fiber, (B) fat, (C) carbohydrates, (D) protein]. Note significant differences in dietary fiber and fat consumption in females (P values = 8e-24 and 0.0005, respectively), as well as significant differences in carbohydrate and protein consumption for both genders (males – P values = 0.008 and 0.004, respectively; females – P values = 0.0007 and 8e-10, respectively). All P values were computed using Welch's t-test. Boxes extend from the lower to upper quartile of the data, with a line at the median. Whiskers show the range of the data and flier points [>Q3 + 1.5(Q3–Q1) or <Q1 – 1.5(Q3–Q1)] are considered outliers. Pct stands for percentage.
Microbiome analysis reveals lower diversity, increased Firmicutes/Bacteroidetes and decreased Prevotella/Bacteroides ratios in the Midwestern cohort
The microbiome of the participants was studied through analysis of stool samples collected by the participants themselves. Whole genome shotgun sequencing of fecal DNA was performed. As described in the Methods section, to eliminate biases in the comparison of microbial community analyses between the Midwestern and Israeli cohort, in this study we analyzed only a subset of 397 nondiabetic participants from the Zeevi et al. study (13) whose samples were collected with the method used by the Midwestern cohort.
Due to the slight differences between the two cohorts, in terms of population characteristics and dietary habits, particularly fiber consumption, we expected their microbiomes to show differences in terms of diversity and microbial composition.
α diversity in the Midwestern cohort is lower than in the Israeli cohort
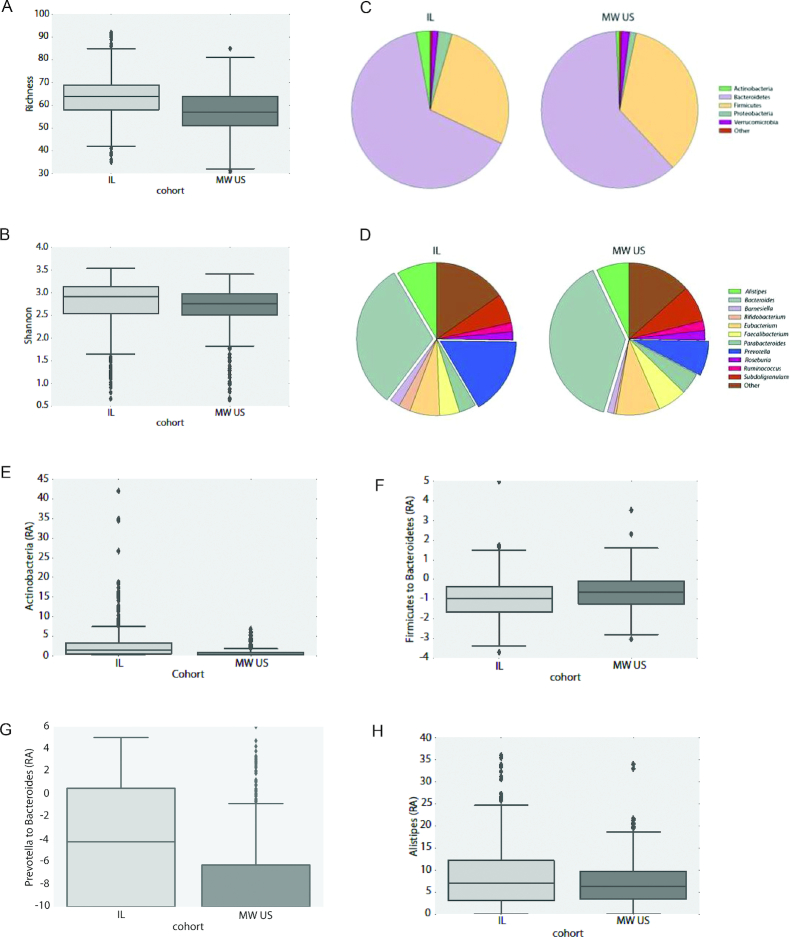
We compared average Species Richness (57 ± 10 Midwestern compared with 64 ± 9 Israeli, Figure 6A) and Shannon diversity index (2.65 ± 0.5 Midwestern compared with 2.75 ± 0.6 Israeli, Figure 6B) between cohorts. In both measures, the average diversity of the Midwestern cohort was significantly lower than the Israeli cohort (P value < 0.05). This is in accordance with observations in populations with a lower consumption of dietary fiber as is the case for the Midwestern cohort (Figure 5A, 30).
FIGURE 6.
Box plots showing Species Richness (A) and Shannon's species diversity index (B) for the Midwestern (MW) and Israeli (IL) cohorts. Note the significantly lower diversity and richness of the Midwestern cohort (P value <0.05 for both measures). (C) Average phyla abundances in the different cohorts. Abundances for phyla with abundance <1% were aggregated and labeled as “other” for visualization purposes. (D) same as (C) but for genera. (E) Box plots showing the relative abundance (RA) of Actinobacteria in the different cohorts (P value = 1e-17). (F) Box plots showing the Firmicutes to Bacteroidetes log-abundance in the different cohorts (P value = 7e-6). Abundances were capped from below by 10−6% and log abundances were clipped to the range of [-5, 5]. (G) Same as (F) but for the Prevotella to Bacteroides genera ratio (P value = 1e-18). In this case, due to the different distribution of abundances, log-abundance values were clipped to the range of [-10, 6]. (H) Same as (F) but for Alistipes genus abundance (P value = 0.002). Boxes extend from the lower to upper quartile of the data, with a line at the median. Whiskers show the range of the data and flier points [>Q3 + 1.5(Q3–Q1) or <Q1–.5(Q3–Q1)] are considered outliers. All P values were computed using Welch's t-test.
To test whether differences in diversity seen between cohorts can be explained by differences in gender, HbA1c% concentrations, or BMI in the different cohorts, we computed diversity values also based on these parameters, regardless of the participant's cohort. Although gender and HbA1c concentrations could not account for observed differences (data not shown), comparisons between overweight and obese (BMI > 25) and healthy individuals (BMI < 25) showed significant differences. Lower diversity in overweight and obese subjects was observed using both diversity measures (Richness 59.95 ± 10.4 in overweight, 61.64 ± 9.56 in healthy; Shannon index 2.66 ± 0.57 in overweight, 2.77 ± 0.53 in healthy; P value < 0.05). This is consistent with previous observations (31). When restricted only to overweight individuals (189 subjects in the Midwestern cohort, 217 in the Israeli cohort), comparison of diversity indices for the Midwestern cohort were still lower, but the differences were no longer statistically significant. Thus, it seems like higher BMI is likely partly accountable for the lower diversity observed in the Midwestern cohort.
Microbial composition comparison
Figure 6C, D shows the average distribution of phyla and genera in the two cohorts, respectively. At the phylum level, the decreased abundance of Actinobacteria (Figure 6E; P value = 1e-17) and an increased Firmicutes/Bacteroidetes ratio (Figure 6F; P value = 7e-6) in the Midwestern cohort stand out as the major differences between the Midwestern and Israeli cohorts. The finding of decreased Actinobacteria is intriguing, as high levels of Actinobacteria have been previously associated with low-fiber diets (32). High Firmicutes/Bacteroidetes ratios have previously been positively associated with higher BMI and reduced glycemic control (33), though these associations are controversial (34, 35).
At the genus level, a significantly decreased Prevotella/Bacteroides ratio (Figure 6G; P value = 1e-18), was observed in the Midwestern cohort, in line with decreased fiber consumption (36). In addition, decreased numbers of Alistipes (Figure 6H; P value = 0.002), as well as decreased numbers of the genera Bifidobacterium (mean relative abundance: 0.51 ± 0.97% in the Midwestern cohort, 2.48 ± 4.35% in the Israeli cohort, P value = 6e-17) and Lactobacillus (mean relative abundance: 0.02 ± 0.19% in the Midwestern cohort, 0.21 ± 0.84% in the Israeli cohort, P value = 1e-5), were observed in the Midwestern cohort. The observed differences in the ratio of Prevotella/Bacteroides were further supported by unsupervised methods of data analysis (Supplemental Figure 1A–C). For differences at the species level, see Supplemental Figure 1D.
Inclusion of data from the Midwestern cohort improves accuracy of model predictions
Given the observed differences between the cohorts, we decided to examine the effect of incorporating data from the Midwestern cohort on prediction accuracy. A model trained solely with data from the Midwestern cohort reduced prediction accuracy on the Israeli cohort, but improved accuracy for prediction for the Midwestern cohort (Figure 4). However, a model re-trained using data from both cohorts, and again validated using a 10-fold cross validation, showed improved accuracy in both cohorts. As expected, introduction of the data from the Midwestern cohort allowed the model to better predict the glycemic response to meals for individuals in the Midwestern cohort, and also allowed for a minor improvement of prediction accuracy on Israeli participants (Midwestern: R = 0.618, Israeli: R = 0.677, Figure 4). Treating the reproducibility of glucose responses to standardized foods as an upper bound for explainable variance (R2 = 0.662 = 0.44 for the Midwestern cohort, R2 = 0.712 = 0.50 to R2 = 0.772 = 0.59 for the Israeli cohort, depending on food), performance in the two cohorts was rather similar, with 88% of explainable variance explained for the Midwestern American cohort, and 78–91% of explainable variance explained for the Israeli cohort.
Features contributing to model predictions are in high agreement with those reported by Zeevi et al.
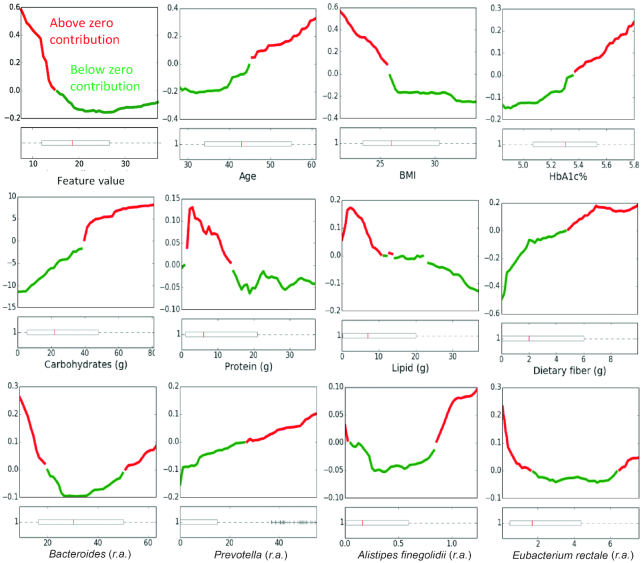
To understand how the different features of the model may affect the predictions, we constructed partial dependence plots (PDPs). These show the marginal effects of each feature on predictions after averaging the contributions from all other features. By and large, the effects of the features of our model agree with the ones presented previously (13) (Figure 7 and OSM Supplemental Figure 2). As expected, on average, increased carbohydrate intake leads to high predicted PPGR values.
FIGURE 7.
Partial dependence plots showing the marginal contribution of various features (x-axis) to the predicted postprandial glycemic response (y-axis). Red and green indicate above and below zero contributions, respectively. Boxplots (bottom) indicate the feature distribution across the cohort. Boxes extend from the lower to upper quartile of the data, with a line at the median. Whiskers show the range of the data and flier points [>Q3 + 1.5(Q3–Q1) or < Q1–1.5(Q3–Q1)] are considered outliers. Partial dependence plots for all features can be seen in Online Supporting Material, Supplemental Figure 2. r.a., relative abundance.
Furthermore, in agreement with previous work, the inverse relations observed between fat content and total dietary fiber consumed in the 24 h prior to each meal with the predicted PPGR values and the direct relation between dietary fiber in each meal and predicted PPGR values. In addition, we observed that lower BMIs show a direct relation with the predicted PPGR values, which although surprising, has been previously reported (37–40).
As for microbiome-related features, here too we report high agreement with the findings in Zeevi et al. (13). For example, similar to their report, the relative abundances of Actinobacteria, Eubacterium eligens, Alistipes putredinis, and Subdoligranulum showed mostly a direct relation, however, those of Eubacterium rectale showed mostly an inverse relation with predicted PPGR values (Figure 7). In addition to the findings of Zeevi et al., we observed a direct relation for the abundance of organisms from the Prevotella genus. Literature describing the effects of Prevotella on glycemic control are conflicting and may be strain specific (41, 42). Stratifying participants based on the occurrence of Prevotella (>0 relative abundance) or a high Prevotella/Bacteroides log ratio (>−4 log ratio of relative abundance) revealed slightly lower average PPGR for high Prevotella (18.06 ± 8.53 compared with 18.9 ± 7.99) or high Prevotella/Bacteroides ratio (17.56 ± 8.84 compared with 19.22 ± 8.27) participants, but these differences were not significant (Welch's t-test P value >0.1). Together with the observed PDP plots, these results may indicate a complex relation between these species and glycemic control that may be reflective of contrasting effects caused by different Prevotella strains.
Finally, in contrast to the report of Zeevi et al. (13), the relative abundance of Bacteroides dorei shows a direct relation with predicted PPGR values but Alistipes finegoldii shows mixed effects, depending on relative abundance. For both of these species, the average relative abundance in the Midwestern and Israeli cohorts were significantly different (P value <0.0005), potentially explaining the differences seen in PDPs.
Discussion
Balanced glycemic concentrations are associated with an improvement in a variety of health markers (6, 7), and although a variety of nutrition interventions exist which aim to reduce the glycemic response to food, all have shown limited efficacy and/or compliance. In this study, we successfully applied the methodologies of the work performed by Zeevi at al. (13) to show that the model trained with an Israeli cohort predicts PPGRs for a cohort of Midwestern individuals despite differences between the two populations, and outperforms common approaches used to inform dietary interventions to regulate glycemic control. Re-training the model on data collected for the Midwestern population further improved its performance, though the extent of improvement was mild. This may be due to the integration of some cohort-specific characteristics, such as different average age, nutrient consumption, and composition of the microbiome.
Diverse nutritional strategies aimed at reducing PPGRs to food exist, but they tend to ignore the characteristics of the individuals (8), and their variability. The interindividual variability commonly observed for the glycemic response to the consumption of foods used as standards for glycemic response (43) was also seen in the Midwestern cohort studied here, as was the variable degrees of correlation between carbohydrate content and PPGR (Figure 2).
In contrast, the model presented in this study considers, in addition to meal features, a variety of personal characteristics, such as age, BMI, and the microbiome. Through the integration of individual traits in addition to food characteristics, it performed significantly better than models based on the calorie or carbohydrate content of meals. Although the latter may prove useful for guiding food intake in some individuals, current guidelines suggest the use of personalized dietary interventions that take into consideration characteristics of individuals (5). The results by Zeevi et al. (13) and this study, strongly support this suggestion, by showing the utility of integrating information about the microbiome, as well as physiological factors and meal features, for the construction of accurate individualized predictions of PPGRs.
Food consumption varies significantly across countries, and besides varying depending on age and socio-economic status (44), the way populations eat foods may also be a consequence of cultural and ethnic heritage, food availability, and the ability to purchase and prepare them, etc. (45). It was therefore important that the methods for collection of data regarding the nutritional behavior of our participants was consistent across the two populations. In general, we have tried to replicate, as much as possible, the framework described by Zeevi et al. (13). The logging app used closely matched the one described by Zeevi et al. (13); however, some modifications were nevertheless inevitable. Standardized meals were chosen as ones more appropriate for a Midwestern cohort (e.g., bagel and cream cheese), and an appropriate food database containing relevant food items was used for each population (see Methods). To reduce noise in microbiome data, only participants whose stools were collected using the same collection methods and subjected to the same downstream analyses were used to construct the model. Some features used in the original study that were not available in the current study (including some blood tests, inferred replication rates, and functions of bacterial species) were removed in all models considered (Supplemental Table 1), to facilitate comparison of the models across different populations.
The correlation between the predicted and observed values of glycemic response to meals in the Midwestern cohort, although still shy of the value of 0.7 observed for the Israeli population, is close to the degree of reproducibility observed in this cohort using standardized meals. This means that the predicted glycemic responses for each individual may in fact be close to the best possible predictive performance in this study setting. The glycemic responses to specific foods have been shown to vary significantly even in the same individual both in this study and in published literature (44). Given that the experimental setting was highly similar between the two studies, these differences may stem from issues of compliance and inaccurate reporting (for example, of portions actually eaten or additional items consumed with standardized meals) that are more pronounced in smaller cohorts, like the Midwestern cohort, but may also partly stem from the difference in the composition of the standardized meal. This is supported by the varying reproducibility of different standardized meals described in Zeevi et al. (R = 0.77 for glucose, whereas R = 0.71 for bread).
Our work demonstrates the potential of the use of personalized predictive models to guide interventions to improve health. However, this type of model could further benefit from adaptation for specific populations. For instance, still in the sphere of glycemic control, further work integrating physical activity information at finer resolution could allow better control of glycemic concentrations through exercise, in addition to dietary interventions. Knowing how exercise affects the rate of glucose decrease in the blood would help individuals regulate the lower range of their glycemic responses and potentially avoid hypoglycemia. In addition, models targeted for specific populations of interest, such as patients of diabetes or other metabolic diseases, can be developed by collecting data in these populations, and can potentially help to provide more accurate nutritional guides in these cases.
Diabetes and cardiovascular disease are two conditions that have increased dramatically in the last 30 y, and are projected to continue to rise unless better control of glycemic concentrations is achieved (10). We believe that the type of modeling framework developed by Zeevi et al. (13). and its adaptations to target populations of interest, as was done here, will significantly help nutrition experts delineate personalized nutrition plans to guide nutritional behaviors of individuals that aim to control their glycemic response to foods, and thus significantly reduce the risk of developing these conditions.
Supplementary Material
Acknowledgments
The authors’ contributions were as follows—DayTwo is a company providing personalized nutrition recommendations for normalizing blood sugar concentrations. HM-S: designed research, conducted research, wrote the article, and had primary responsibility for final content; TR-S: analyzed data and performed statistical analysis, wrote the article, and had primary responsibility for final content; SA, YB-S, and YC: analyzed data and performed statistical analysis; TO, DB, and JS: designed research; LS: designed research, provided essential materials; PK and HN: designed research, provided essential materials, and the wrote article, and all authors: read and approved the final manuscript.
HM-S, KE, DC, PK, and HN disclose that Mayo Clinic received support for this study from DayTwo and has a financial interest in this company. LS is a co-founder of DayTwo. TRS, SA, YBS, YC, TO, DB, JS, and LS are DayTwo employees. They report investments from Angels Hi-tech Investments, Mayo Foundation for Medical Education And Research (Mayo Clinic), Johnson & Johnson Innovation - JJDC, Inc, HFL SCA, HFL ALPHA, and I.B.I. Trust Management Ltd; in addition, DayTwo has two pending patents US2016/0232311A1 and WO 2015/166489 A2.
References
- 1. Brand-Miller JC. Glycemic load and chronic disease. Nutrition Reviews. 2003;61(5):S49–55. [DOI] [PubMed] [Google Scholar]
- 2. de Vegt F, Dekker JM, Ruhe HG, Stehouwer CD, Nijpels G, Bouter LM, Heine RJ. Hyperglycaemia is associated with all-cause and cardiovascular mortality in the Hoorn population: the Hoorn study. Diabetologia. 1999;42:926–31. [DOI] [PubMed] [Google Scholar]
- 3. Glucose tolerance and mortality: comparison of WHO and American Diabetic Association diagnostic criteria. The Lancet. 1999;354(9179):617–21. [PubMed] [Google Scholar]
- 4. American Diabetes Association. Postprandial blood glucose. Diabet Care. 2001;24(4):775–7. [DOI] [PubMed] [Google Scholar]
- 5. Bantle JP, Wylie-Rosett J, Albright AL, Apovian CM, Clark NG, Franz MJ, Hoogwerf BJ, Lichtenstein AH, Mayer-Davis E, Mooradian ADAmerican Diabetes Association, et al., American Diabetes Association Nutrition recommendations and interventions for diabetes: a position statement of the American Diabetes Association. Diabetes Care. 2008;31(Suppl 1):S61–78. [DOI] [PubMed] [Google Scholar]
- 6. Ajala O, English P, Pinkney J. Systematic review and meta-analysis of different dietary approaches to the management of type 2 diabetes. Am J Clin Nutr. 2013;97(3):505–16. [DOI] [PubMed] [Google Scholar]
- 7. Livesey G, Taylor R, Hulshof T, Howlett J. Glycemic response and health–a systematic review and meta-analysis: relations between dietary glycemic properties and health outcomes. Am J Clin Nutr. 2008;87(suppl):258S–68S. [DOI] [PubMed] [Google Scholar]
- 8. Evert AB, Boucher JL, Cypress M, Dunbar SA, Franz MJ, Mayer-Davis EJ, Neumiller JJ, Nwankwo R, Verdi CL, Urbanski P et al.. Nutrition therapy recommendations for the management of adults with diabetes. Diabetes Care. 2013;36(11):3821–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Englyst KN, Englyst HN, Hudson GJ, Cole TJ, Cummings JH. Rapidly available glucose in foods: an in vitro measurement that reflects the glycemic response. Am J Clin Nutr. 1999;69:448–54. [DOI] [PubMed] [Google Scholar]
- 10. Dodds RF. Understanding Diabetes: A Biochemical Perspective. Hoboken, NJ: John Willey & Sons; 2013. [Google Scholar]
- 11. Rayner CK, Samson M, Jones KL, Horowitz M. Relationships of upper gastrointestinal motor and sensory function with glycemic control. Diabetes Care. 2001;24:371–81. [DOI] [PubMed] [Google Scholar]
- 12. Davis N, Forbes B, Wylie-Rosett J. Nutritional strategies in type 2 diabetes mellitus. Mt Sinai J Med. 2009;76(3):257–68. [DOI] [PubMed] [Google Scholar]
- 13. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M et al.. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–94. [DOI] [PubMed] [Google Scholar]
- 14. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP et al.. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA et al.. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, Israeli D, Zmora N, Gilad S, Weinberger A et al.. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514(7521):181–6. [DOI] [PubMed] [Google Scholar]
- 18. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marco-Sola S, Sammeth M, Guigo R, Ribeca P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods. 2012;9(12):1185–8. [DOI] [PubMed] [Google Scholar]
- 20. Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12(10):902–3. [DOI] [PubMed] [Google Scholar]
- 21. Wolever TMS, Jenkins DJ. The use of the glycemic index in predicting the blood glucose response to mixed meals. Am J Clin Nutr. 1986;43:167–72. [DOI] [PubMed] [Google Scholar]
- 22. Chen T, Guestrin C.. XGBoost: a scalable tree boosting system. arXiv 2016;1603.02754v3. [Google Scholar]
- 23. Goeman JJ, Solari A. Multiple hypothesis testing in genomics. Statistics in Medicine. 2014; 33:1946–78. [DOI] [PubMed] [Google Scholar]
- 24. Mendes-Soares H, Raveh-Sadka T, Azulay S, Edens K, Ben-Shlomo Y, Cohen Y, Ofek T, Bachrach D, Stevens J, Colibaseanu D et al.. Assessment of a personalized approach to predicting postprandial glycemic responses to food among individuals without diabetes, JAMA Network Open, accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Health MDo. Adult obesity: facts and figures. [Internet]. Available from: https://apps.health.state.mn.us/mndata/obesity_basic. [Google Scholar]
- 26. Health SoIMo. Health 2013. 2013. [Google Scholar]
- 27. Menke A, Casagrande S, Geiss L, Cowie CC. Prevalence of and trends in diabetes among adults in the United States, 1988–2012. JAMA. 2015;314(10):1021–9. [DOI] [PubMed] [Google Scholar]
- 28. Vrolix R, Mensink RP. Variability of the glycemic response to single food products in healthy subjects. Contemp Clin Trials. 2010;31(1):5–11. [DOI] [PubMed] [Google Scholar]
- 29. Friedman JH. Greedy function approximation: a gradient boosting machine. The Annals of Statistics. 2001;29(5):1189–232. [Google Scholar]
- 30. Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, Ahn J. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One. 2015;10(4):e0124599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen J, Ryu E, Hathcock M, Ballman K, Chia N, Olson JE, Nelson H. Impact of demographics on human gut microbial diversity in a US Midwest population. Peer J. 2016;4:e1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu GD, Chen J, Hogffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R et al.. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koliada A, Syzenko G, Moseiko V, Budovska L, Puchkov K, Perederiy V, Gavalko Y, Dorofeyev A, Romanenko M, Tkach S et al.. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017;17(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chakraborti CK. New-found link between microbiota and obesity. World J Gastrointest Pathophysiol. 2015;6(4):110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fernandes J, Su W, Rahat-Rozenbloom S, Wolever TM, Comelli EM. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr Diabetes. 2014;4:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gorvitovskaia A, Holmes SP, Huse SM. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. 2016;4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vazquez LA, Rodriguez A, Salvador J, Ascaso JF, Petto H, Reviriego J. Relationships between obesity, glycemic control, and cardiovascular risk factors: a pooled analysis of cross-sectional data from Spanish patients with type 2 diabetes in the preinsulin stage. BMC Cardiovascular Disorders. 2014;14:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang J, Yan R, Wen J, Kong X, Li H, Zhou P, Zhu H, Su X, Ma J. Association of lower body mass index with increased glycemic variability in patients with newly diagnosed type 2 diabetes: a cross-sectional study in China. Oncotarget. 2017;8(42):73133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jauch-Chara K, Schmoller A, Oltmanns KM. Impaired glucose tolerance in healthy men with low body weight. Nutr J. 2011;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee EY, Lee YH, Jin SM, Yang HK, Jung CH, Park CY, Cho JH, Lee WJ, Lee BW, Kim JH. Differential association of body mass index on glycemic control in type 1 diabetes. Diabetes Metab Res Rev. 2017;33(1):e2815. [DOI] [PubMed] [Google Scholar]
- 41. Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, Forslund K, Hildebrand F, Prifti E, Falony G et al.. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–81. [DOI] [PubMed] [Google Scholar]
- 42. Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Bjork I, Backhed F. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metabolism. 2015;22:971–82. [DOI] [PubMed] [Google Scholar]
- 43. Vega-Lopez S, Ausman LM, Griffith JL, Lichtenstein AH. Interindividual variability and intra-individual reproducibility of glycemic index values for commercial white bread. Diabetes Care. 2007;30(6):1412–7. [DOI] [PubMed] [Google Scholar]
- 44. Imamura F, Micha R, Khatibzadeh S, Fahimi S, Shi P, Powles J, Mozaffarian D; Global Burden of Diseases Nutrition and Chronic Diseases Expert Group (NutriCoDE). Dietary quality among men and women in 187 countries in 1990 and 2010: a systematic assessment. The Lancet Global Health. 2015;3(3):e132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Auestad N, Fulgoni VL 3rd. What current literature tells us about sustainable diets: emerging research linking dietary patterns, environmental sustainability, and economics. Adv Nutr. 2015;6(1):19–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.