Abstract
Estrogens play a pivotal role in breast cancer etiology, and endocrine therapy remains the main first line treatment for estrogen receptor‐alpha (ERα)‐positive breast cancer. ER are transcription factors whose activity is finely regulated by various regulatory complexes, including histone deacetylases (HDACs). Here, we investigated the role of HDAC9 in ERα signaling and response to antiestrogens in breast cancer cells. Various Michigan Cancer Foundation‐7 (MCF7) breast cancer cell lines that overexpress class IIa HDAC9 or that are resistant to the partial antiestrogen 4‐hydroxy‐tamoxifen (OHTam) were used to study phenotypic changes in response to ER ligands by using transcriptomic and gene set enrichment analyses. Kaplan–Meier survival analyses were performed using public transcriptomic datasets from human breast cancer biopsies. In MCF7 breast cancer cells, HDAC9 decreased ERα mRNA and protein expression and inhibited its transcriptional activity. Conversely, HDAC9 mRNA was strongly overexpressed in OHTam‐resistant MCF7 cells and in ERα‐negative breast tumor cell lines. Moreover, HDAC9‐overexpressing cells were less sensitive to OHTam antiproliferative effects compared with parental MCF7 cells. Several genes (including MUC1, SMC3 and S100P) were similarly deregulated in OHTam‐resistant and in HDAC9‐overexpressing MCF7 cells. Finally, HDAC9 expression was positively associated with genes upregulated in endocrine therapy‐resistant breast cancers and high HDAC9 levels were associated with worse prognosis in patients treated with OHTam. These results demonstrate the complex interactions of class IIa HDAC9 with ERα signaling in breast cancer cells and its effect on the response to hormone therapy.
Keywords: antiestrogen resistance, breast cancer, cell proliferation, estrogen receptor, histone deacetylase
Abbreviations
- CGH
comparative genomic hybridization
- ERE
estrogen‐responsive element
- ER
estrogen receptor
- ESR1
estrogen receptor 1
- GO
Gene Ontology
- GSEA
gene set enrichment analysis
- HDAC
histone deacetylase
- HDI
HDAC inhibitor
- IPA
Ingenuity Pathway Analysis
- KO
knockout
- MCF7
Michigan Cancer Foundation‐7
- MTT
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide
- NRIP1
nuclear receptor interacting protein 1
- OHTam
4‐OH‐tamoxifen
- OHTR
OHTam‐resistant
- PR
progesterone receptor
- SD
standard deviation
- TBP
TATA‐binding protein
1. Introduction
Estrogens are important regulators of gene expression in target tissues, such as the mammary gland, and their mitogenic action has a key role in human breast cancer etiology and progression (Henderson et al., 1988). In breast cancer, endocrine therapies based on antiestrogens and aromatase inhibitors are commonly used to counteract estrogen action (O'Regan and Jordan, 2002). However, the initial response often does not last because tumors frequently develop resistance to hormonal manipulation. Therefore, one of the main clinical challenges is to develop new combined targeted therapies for cancers that are insensitive or resistant to endocrine manipulation.
Estrogen activities are mediated via two nuclear estrogen receptors (ERα and ERβ). They belong to a large conserved superfamily of nuclear receptors that function as ligand‐dependent transcription factors (Heldring et al., 2007). Their activities rely on interactions with transcription coregulators that are broadly defined as coactivators or corepressors. Several of these coregulators are involved in chromatin remodeling through multiple enzymatic activities including histone deacetylases (HDACs) (McKenna and O'Malley, 2002).
Human HDACs form a large family of 18 members classified in four groups (I–IV) based on sequence homologies (Haberland et al., 2009). Class I enzymes (HDAC1, 2, 3 and 8) are nuclear proteins with ubiquitous expression. Class II HDACs are divided in two subgroups. Class IIa HDACs (HDAC4, 5, 7 and 9) have a tissue‐specific pattern of expression and can shuttle between nucleus and cytoplasm, depending on their phosphorylation status. These HDACs are mainly involved in tissue differentiation (Parra, 2015). Class IIb includes HDAC10 and HDAC6, which is described as a major cytoplasmic regulator. Class III comprises seven sirtuins with a catalytic site that is structurally different from that of the other three classes. Finally, class IV includes only HDAC11.
It has been shown that HDACs affect estrogen signaling at various levels (for review, see (Linares et al., 2011)). For instance, HDAC1 interacts with ERα in vitro and in vivo and suppresses ERα transactivation (Kawai et al., 2003). Other studies revealed that ERS1 gene transcription is regulated by epigenetic modifications and that HDAC inhibitors (HDI) treatment can induce ER expression in ER‐negative breast cancer cell lines (Thomas and Munster, 2009), although this has been questioned more recently (de Cremoux et al., 2015). The role of epigenetic modifications (including histone acetylation and DNA methylation) in acquired tamoxifen resistance has also been reported in vivo using a preclinical rat model (Hilakivi‐Clarke et al., 2017). Interestingly, combinations of HDIs (vorinostat, panobinostat or entinostat) and hormone therapy, such as tamoxifen, letrozole or exemestane, have been used in metastatic breast cancers leading to an improved response (Munster et al., 2011; Tan et al., 2016; Yeruva et al., 2018).
We recently reported that HDAC9 mRNA expression is markedly increased in the most aggressive breast cancer cell lines, and that HDAC9 expression deregulation (ectopic expression and knockdown) in breast cancer cells significantly alters gene expression, cell proliferation and response to HDIs (Lapierre et al., 2016). Here, we investigated HDAC9 role in ERα signaling in breast cancer cells and its expression in antiestrogen‐resistant breast cancer cell lines. Our results demonstrate that HDAC9 overexpression in Michigan Cancer Foundation‐7 (MCF7) breast cancer cells modulates ERα expression and activity and alters ERα ligand effect on cell proliferation. Moreover, HDAC9 expression was markedly deregulated in antiestrogen‐resistant mammary tumor cell lines. Similarly, in human breast tumor samples, a meta‐analysis revealed that high HDAC9 mRNA levels were associated with decreased overall survival in tamoxifen‐treated patients. Altogether, this work highlights strong interactions between class IIa HDAC9 and ERα signaling, suggesting a key role of HDAC9 in antiestrogen resistance of breast cancers.
2. Materials and methods
2.1. Plasmids and reagents
Estradiol‐17β (E2) was purchased from Sigma‐Aldrich (St Quentin, France). 4‐hydroxy‐tamoxifen (OHTam) and ICI 182 780 (ICI) were kindly donated by Sanofi‐Aventis and AstraZeneca, respectively. Zeocin was from InvivoGen (Toulouse, France). The FLAG‐tagged full length HDAC9 (HDAC9FL) plasmid was a kind gift from A. Zelent (Petrie et al., 2003). For stable transfections, the pcDNA3.1‐HDAC9‐zeo plasmid was obtained by subcloning HDAC9FL in the pcDNA3.1zeo vector (Invitrogen, Cergy Pontoise, France) that confers zeocin resistance. The ERα promoter luciferase reporter plasmid was from B. Westley (Donaghue et al., 1999). The 17EBLuc+ luciferase reporter plasmid contains one copy of the consensus estrogen‐responsive element (ERE). The pRL‐CMV vector (Promega, Charbonnières, France) was used for normalization.
2.2. Cell lines and culture
The MCF7 breast cancer cell line was obtained from the American Type Culture Collection (LGC Standards, Molsheim, France) and was grown using the recommended culture conditions. The OHTam‐resistant MCF7 (MCF7‐OHTR) cell lines have been described previously (Brünner et al., 1993; Badia et al., 2000; Eric Badia et al., 2015) and were authenticated by PCR‐single‐locus technology (Eurofins, Ebersberg, Germany). T47D TamR cells were recently described elsewhere (Mishra et al., 2018). MCF7‐OHTR cells were transitory transfected with small interference RNA directed to HDAC9 (Dharmacon, Lafayette, CO, USA). Control and MCF7 cells that stably overexpress HDAC9 were obtained by transfecting MCF7 cells with pcDNA3.1‐zeo and pcDNA3.1‐HDAC9‐zeo respectively, and were isolated on the basis of their zeocin resistance (0.5% zeocin in Dulbecco's modified Eagle's medium supplemented with 10% FBS). All transfections were carried out using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Cells classified as ERα‐positive (MCF7, T47D) or ERα‐negative (MDA‐MB231, MDA‐MB436) were grown using the recommended culture conditions (Lapierre et al., 2016).
2.3. Cell proliferation
Cells were seeded in triplicate at a density of 2 × 103 cells per well. At the indicated time, 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT; Sigma‐Aldrich®, St Louis, MO, USA) was added at 37 °C for 4 h. Formazan crystals were solubilized in DMSO and absorbance read at 560 nm on a spectrophotometer. The measured absorbance correlates with the number of viable cells. Results were normalized to absorbance at day 0. Stripping of endogenous steroids and incubation with ER ligands were done as previously described (Duong et al., 2008). MCF7 and MCF7‐OHTR cells (transfected or not with siHDAC9) were seeded at a density of 2500 cells per well into E‐Plate 16 (ACEA Biosciences, Inc., San Diego, CA, USA) containing 150 μL of medium supplemented with 10% FBS per well. Dynamic monitoring of cell growth was performed every 24 h during 10 days using the impedance‐based xCELLigence system (ACEA Biosciences, Inc.). The cell index was derived from measurement of cell‐electrode impedance which correlates with the number of viable cells.
2.4. Luciferase reporter assays
Cells were plated in 96‐well plates (105 cells per well) in triplicate 24 h before DNA transfection (250 ng of total DNA) using the Jet PEI (Polyplus, Illkirch, France) method. Firefly luciferase values were measured and normalized to the Renilla luciferase activity [pRL‐CMV‐Renilla plasmid (Ozyme®, Montigny‐le‐Bretonneux, France)]. Values were expressed as the mean ratio of luciferase activities.
2.5. Western blot analysis
Whole cell proteins were extracted as previously described (Duong et al., 2008); 30 μg protein extracts were separated by SDS/PAGE and analyzed by western blotting using primary polyclonal antibodies against ERα (clone sc‐543; Tebu‐bio, Le Perray‐en‐Yvelines, France) or actin (Sigma) and the Chemiluminescence Reagent Plus kit (Perkin‐Elmer Life Science, Villebon S/Yvette, France).
2.6. Immunofluorescence staining
Breast cancer cells were fixed in 4% formaldehyde, permeabilized with 1% Triton X‐100 and saturated with 1% BSA at room temperature for 3 h. Cells were incubated first with an anti‐HDAC9 antibody (Abcam, Cambridge, UK, 18970; 1 : 100 in PBS with 1% BSA) and then with an Alexa‐conjugated secondary antibody (Invitrogen). After washing, cells were counterstained with Hoechst (Sigma‐Aldrich, St Quentin, France) and mounted for fluorescence analysis.
2.7. RNA extraction and real‐time quantitative PCR
Total RNA extraction and reverse transcription were performed as previously described (Duong et al., 2008). Real‐time PCR quantification was done on a LC480 instrument using LightCycler 480 SYBR Green PCR Master (Roche Diagnostics, Meylan, France). For each sample, the expression levels of the target genes were normalized to the mean levels of the housekeeping genes TATA‐binding protein (TBP) and ribosomal protein S9, and expressed relative to a calibrator sample. The primers for the HDAC1 to HDAC11, estrogen receptor 1 (ESR1) and TBP genes have been described elsewhere (Annicotte et al., 2006; Chen et al., 2004; Delfour et al., 2006; Duong et al., 2008). Other primers used in this study are available upon request.
2.8. Breast tumor samples analysis
The datasets used to analyze HDAC9 expression in breast cancers [GSE26459 (Gonzalez‐Malerva et al., 2011), E‐TABM‐158 (Chin et al., 2006), GSE27473 (Al Saleh et al., 2011), GSE47561 (Ur‐Rehman et al., 2013) and the Fan dataset (Fan et al., 2006)] are listed in Table S1. The Oncomine Platform (https://www.oncomine.org) was used to investigate HDAC9 expression in the different breast cancer datasets grouped by ERα status. The BreastMark algorithm (http://glados.ucd.i.e/BreastMark), developed by Molecular Therapeutics for Cancer, Ireland, was used for Kaplan–Meier analysis of several datasets (Madden et al., 2013). The gene set enrichment analysis (GSEA) was implemented with the GSEA Java application (http://software.broadinstitute.org/gsea/index.jsp) (Subramanian et al., 2005). GSEA of the ISDB3008 dataset was performed as previously described (Gao et al., 2014).
2.9. Global gene expression analysis
Global transcriptome analysis of OHTam‐sensitive and OHTR and HDAC9‐overexpressing and control MCF7 cells was performed using human oligonucleotide HG‐U219 microarrays (ThemoFisher Scientific, Waltham, MA, USA) processed at the Microarray Core Facility of the Institute for Regenerative Medicine and Biotherapy (IRMB), CHRU‐INSERM‐UM Montpellier (http://www.chu-montpellier.fr/fr/irmb/). After image processing with the Affymetrix GeneChip command console, the CEL files were analyzed using the affymetrix expression console™ (ThemoFisher Scientific) Software v1.3.1 and normalized with the robust multi‐array average algorithm. Gene annotation was performed using NetAffx (http://www.affymetrix.com; October 2014). The significant analysis of microarrays (Stanford, CA, USA) software with the Wilcoxon test and 300 sample label permutations was used to identify differentially expressed genes relative to control. Hierarchical clustering analyses based on the expression levels of the differentially expressed genes were performed by using the cluster and treeview packages (Eisen et al., 1998). Gene Ontology (GO) biological process enrichment analyses of the differentially expressed genes were generated with the ingenuity pathway analysis (IPA) (Germantown, MD, USA) software. Selected genes were imported in the IPA database and were categorized on the basis of their biological process and molecular functions (www.ingenuity.com).
2.10. Statistical analysis
Results are expressed as the mean ± standard deviation (SD). Data comparison between groups was performed using the Student t‐test or Mann–Whitney U‐test. A probability level of 0.05 was chosen for statistical significance. Statistical analyses were performed using graphpad prism 6, version 6.01 (GraphPad Software, San Diego, CA, USA).
3. Results
3.1. HDAC9 inhibits ERα expression and activity in MCF7 breast cancer cells
A previous work showed that class IIa HDAC9 negatively regulates ERα expression in vivo in mouse heart and in neonatal rat cardiomyocytes (van Rooij et al., 2010). Here, to determine whether HDAC9 also modulates ERα signaling in breast cancer cells, a luciferase reporter assays was used to test the activity of a 4‐kb fragment of the ERα promoter in MCF7 breast cancer cells transfected or not with a plasmid that expresses HDAC9FL. HDAC9 significantly decreased ERα promoter activity in a dose‐dependent manner (Fig. 1A). Similarly, HDAC9 overexpression inhibited ERE‐mediated transcriptional activation in MCF7 cells (Fig. 1B). Altogether, these data suggested that HDAC9 represses estrogen signaling in breast cancer cells.
Figure 1.
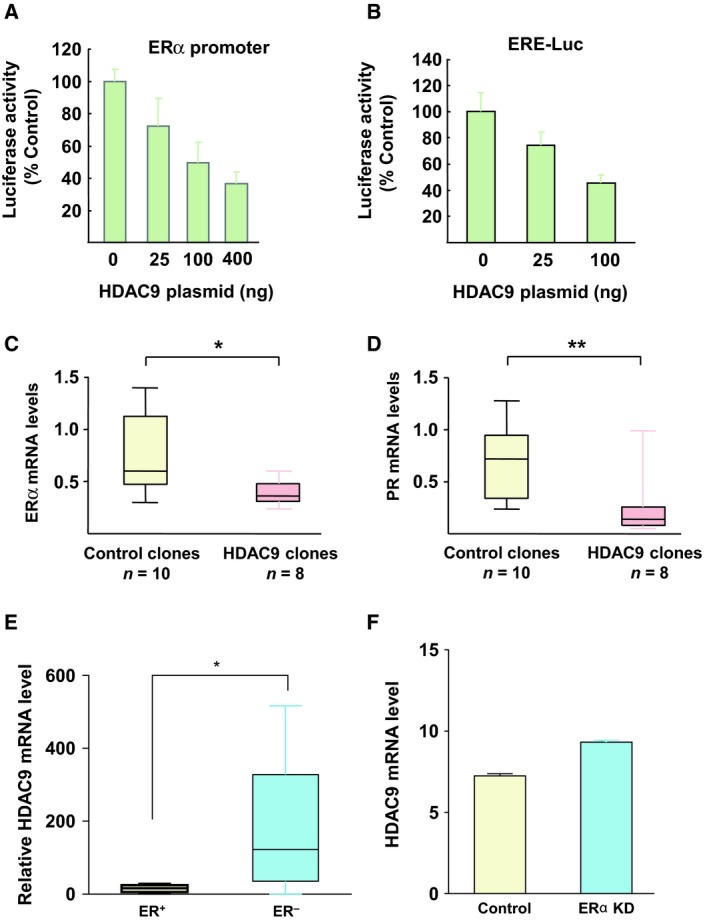
Cross talk between HDAC9 and ERα signaling in breast cancer cells. (A) MCF7 cells were cotransfected with a 4‐kb fragment of the ERα promoter and increasing concentrations of full length HDAC9. Results represent the luciferase activity measured after normalization to Renilla luciferase activity and relative to the values obtained in cells not transfected with the HDAC9 plasmid (control). Data are the mean ± SD of triplicate wells and are representative of two independent experiments. (B) The same as in panel A, but with cells cotransfected with an ERE‐luciferase reporter plasmid and increasing concentrations of full length HDAC9. (C) Total RNA was extracted from control (empty vector; n = 10) and HDAC9‐overexpressing cell clones (n = 8). ERα mRNA levels were quantified using RT‐qPCR. Results represent the mean FC ± SD vs control MCF7 cells; *P < 0.05, **P < 0.01 (Mann–Whitney test). (D) The same as in panel C, but for PGR mRNA. (E) Total RNA was extracted from ERα‐positive or ERα‐negative breast cancer cell lines and ERα mRNA levels were quantified using RT‐qPCR; *P < 0.05. (F) HDAC9 expression levels in parental and MCF7 cells with silenced ERα expression (205659_at probe set) were extracted from the GEO profile GSE27473 and compared (Al Saleh et al., 2011).
3.2. Inhibition of ERα signaling in MCF7 breast cancer cells that stably overexpress HDAC9
To better characterize HDAC9 effect on estrogen signaling in breast cancer cells, two ERα‐positive MCF7 cell clones that stably express HDAC9FL (MCF7‐HDAC9FL, clones 4‐1 and 1‐1) were selected and characterized relative to control clones (P1 and P2; empty vector). HDAC9 mRNA and protein levels were higher in MCF7‐HDAC9FL than in control clones (~ 4.8 and ~ 8.5 times, respectively, for mRNA levels; Fig. S1A,B). Moreover, in agreement with the luciferase reporter assay results, the mRNA levels of endogenous ERα (Fig. 1C) and its target progesterone receptor (PR; Fig. 1D) were significantly decreased in MCF7‐HDAC9FL cells compared with controls (mean ± SD for ERα: 0.38 ± 0.12 and 0.74 ± 0.37 respectively, P = 0.02; and for PR: 0.24 ± 0.32 and 0.71 ± 0.36 respectively, P = 0.007). Western blot analysis confirmed the marked decrease of ERα expression in MCF7‐HDAC9FL cells (Fig. S2A). Finally, GSEA of microarray data obtained from an independent pool of MCF7 cell clones that overexpress or not HDAC9 (Lapierre et al., 2016) confirmed that HDAC9 overexpression led to a significant decrease of genes belonging to the ERα signaling pathway (Fig. S2B). Altogether, these results demonstrated that HDAC9 inhibits ERα expression and activity in human breast cancer cells.
3.3. ERα inhibits HDAC9 expression in human breast cells and tumors
We previously reported that HDAC9 expression was significantly lower in ERα‐positive breast cancer cell lines, such as MCF7 and T47D, compared with ERα‐negative breast cancer cell lines, such as MDA‐MB231 and MDA‐MB436 (Fig. 1E) (Lapierre et al., 2016). Interestingly, the same inverse correlation was found by analyzing several human breast tumor datasets, including the E‐TABM‐158 breast cancer dataset (Chin et al., 2006). In this cohort, HDAC9 mRNA levels were significantly higher in ERα‐negative than in ERα‐positive breast tumors (P < 0.0001; Fig. S2C). Similar findings were obtained by analysis of the NKI (Fan et al., 2006) and the GSE47561 datasets (Ur‐Rehman et al., 2013) (data not shown). Moreover, meta‐analyses performed using the breast cancer gene‐expression miner v3 tool (expression and correlation analysis modules) (Jézéquel et al., 2013) also showed an inverse correlation between ERα and HDAC9 expression (Fig. S2D) as well as between HDAC9 and PR expression in breast tumors (Fig. S2E).
Finally, analysis of the GSE27473 dataset (Al Saleh et al., 2011) indicated that ERα silencing in MCF7 cells resulted in a significant increase of HDAC9 mRNA levels (Fig. 1F). Conversely, ectopic expression of ERα in ER‐negative cells reduced HDAC9 mRNA levels (Fig. S2F). Altogether, these data indicated that ERα loss is involved in the increased HDAC9 gene expression detected in ER‐negative breast cancer cells.
3.4. HDAC9 expression in antiestrogen‐sensitive and antiestrogen‐resistant breast cancer cells
As ERα expression is lost or decreased in OHTR cells (Al Saleh et al., 2011), we next asked whether breast cancer cell resistance to OHTam could be associated with changes in HDAC9 gene expression. Comparison of the HDAC mRNA profile in previously described MCF7 cell lines that are responsive or resistant to OHTam (Badia et al., 2000, 2015) showed that among the 11 HDACs analyzed, only HDAC9 expression was significantly higher (7.6 times) in OHTR than in parental OHTam‐responsive cells (Fig. 2A). Similar results were obtained with OHTR T47D (Mishra et al., 2018) (Fig. 2B) as well as in other MCF7 cell lines (Brünner et al., 1993; Eric Badia et al., 2015; Gonzalez‐Malerva et al., 2011) (Fig. S3A,B). As expected, HDAC9 mRNA overexpression in OHTR cells was associated with decreased ERα levels (Figs 2C,D and S3C). HDAC9 overexpression in OHTR MCF7 cells was confirmed also by immunofluorescence analysis (Fig. 2E).
Figure 2.
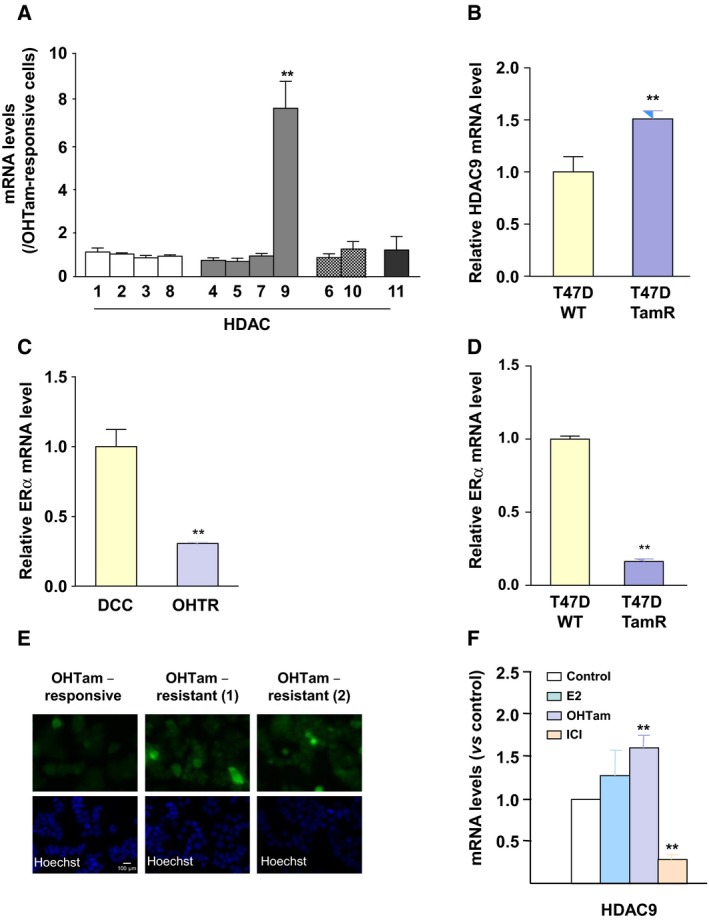
HDAC9 expression in antiestrogen‐resistant breast cancer cells. (A) Total RNA was extracted from OHTam‐sensitive and OHTam‐resistant MCF7 cell lines and HDAC mRNA levels were quantified by RT‐qPCR. Results represent the mean ± SD of three independent cell cultures and are expressed relative to the HDAC mRNA levels of OHTam‐responsive cells, used as reference; **P < 0.01 (Mann–Whitney test compared with OHTam‐responsive cells). (B) HDAC9 mRNA levels were quantified by RT‐qPCR in T47D TamR cells (Mishra et al., 2018). Results are expressed relative to the HDAC mRNA levels in parental cells and represent the mean ± SD of five independent cell cultures; **P < 0.001 (t‐test compared with parental T47D cells). (C) Total RNA was extracted from OHTam‐responsive (DCC) and resistant (OHTR) MCF7 cells and ERα mRNA levels were quantified by RT‐qPCR. Results represent the mean ± SD of three independent cell cultures and are expressed relative to the ERα mRNA levels in DCC cells, used as reference; **P < 0.01 (Mann–Whitney test compared with DCC cells). (D) The same as in panel B for the quantification of ERα mRNA levels by RT‐qPCR in T47D TamR cells. (E) HDAC9 expression was analyzed by immunofluorescence in OHTam‐responsive MCF7 cells and in two sets of OHTam‐resistant MCF7 cells (1 and 2) using an anti‐HDAC9 antibody (top panel); nuclei were stained with Hoechst (bottom panel). Scale bar corresponds to 100 μm. (F) MCF7 cells were incubated with 10−8 M E2, OHTam, ICI or solvent alone (EtOH; Control) for 20 h and then HDAC9 mRNA levels were quantified by RT‐qPCR. Results are expressed relative to the HDAC mRNA levels in control cells and represent the mean ± SD of three independent cell cultures; **P < 0.01 (Mann‐Whitney test compared with control cells).
Then, to determine whether HDAC9 expression could be regulated by short‐term treatments with estrogens and antiestrogens, class I, II and IV HDAC mRNA levels were quantified in ERα‐positive MCF7 cells incubated with E2 or with the antiestrogens OHTam and ICI for 24 h. These ER ligands affected significantly only the expression of several class IIa HDACs, including HDAC9 (Figs 2F and S3D–G). Specifically, the pure antagonist ICI had a strong inhibitory effect on HDAC9 gene expression, while OHTam, and to a lesser extent, E2 had a significant positive effect (Fig. 2F).
3.5. Effect of HDAC9 on ER‐dependent cell proliferation
To investigate whether HDAC9 overexpression modulated ER‐dependent cell proliferation, MCF7‐HDAC9FL (4‐1 and 1‐1) and control clones (P1 and P2) were cultured in the presence of E2 or OHTam. MCF7‐HDAC9FL cells still responded to estrogen stimulation, although to a lesser extent than controls, but remarkably could proliferate also in the presence of the antiestrogen OHTam (Fig. 3A,B). Quantification of the results from independent cell cultures confirmed that MCF7‐HDAC9FL cells were significantly less sensitive to estrogen stimulation and to OHTam inhibition than controls (P < 0.001; Fig. 3C).
Figure 3.
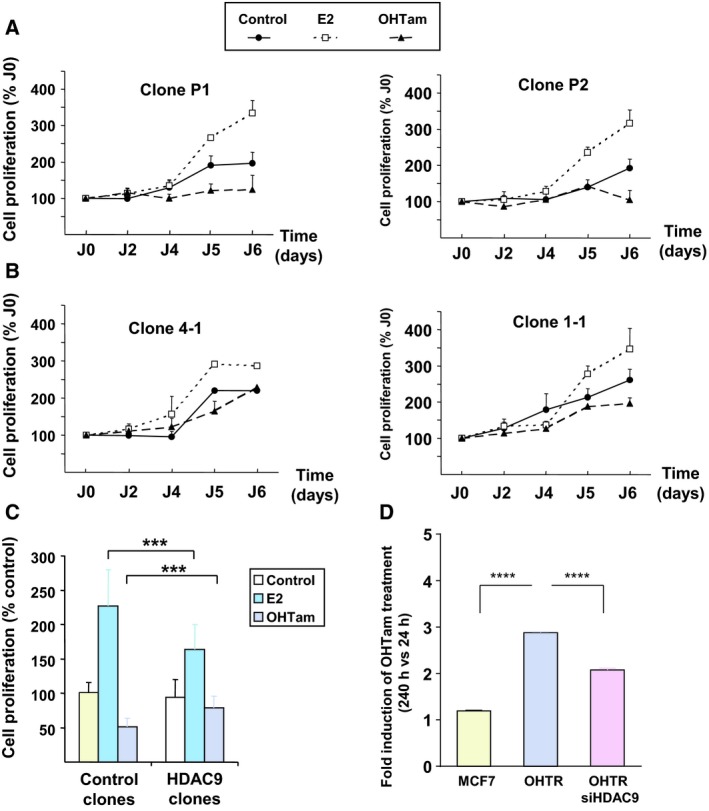
HDAC9 and the regulation of breast cancer cell proliferation by ER ligands. (A) Control MCF7 cell clones (empty vector; P1 and P2) were cultured in DCC medium supplemented with E2, OHTam or solvent alone (Control) for 5 days. Cell proliferation was measured with the MTT assay at days 2, 4, 5 and 6 and expressed relative to the absorbance at day 0. Results are the mean ± SD of three independent experiments, performed in triplicates. (B) The same as in A, but with MCF7‐HDAC9 cell clones (4‐1 and 1‐1). (C) Data obtained for each condition (control and MCFT‐HDAC9 cell clones) were pooled and compared. Data represent the mean ± SD of three independent experiments for each clone; ***P < 0.001 (Mann–Whitney test). (D) The growth of MCF7 and MCF7‐OHTR cells (transfected or not with a siRNA directed against HDAC9) was monitored using the XCELLigence system for a total duration of 10 days. The cell index corresponding to the number of viable cells was determined. The results represent, for each condition, cell proliferation measured after 240 h of culture in the presence of OHTam normalized by the values measured at 24 h (means ± SD; two independent experiments). ****P < 0.0001 (Mann‐Whitney test).
To emphasize these results, we then knocked down HDAC9 expression in MCF7‐OHTR cells and monitored the effect on OHTam resistance. Using a siRNA approach, HDAC9 expression was efficiently silenced in MCF7‐OHTR cells leading to HDAC9 mRNA levels comparable to that of MCF7 cells (Fig. S4). In the presence of OHTam, parental MCF7 cells almost did not grow reflecting their sensitivity to the inhibition of E2 signaling. By contrast, MCF7‐OHTR cell number significantly increased almost by threefold during the same period of time confirming their resistance to OHTam. Interestingly, when treated with the HDAC9 siRNA, MCF7‐OHTR cells grew significantly less in the presence of OHTam (Fig. 3D), strongly suggesting that silencing HDAC9 expression decrease their resistance to OHTam treatment. Altogether, these data clearly supported a link between HDAC9 overexpression and antiestrogen resistance in breast cancer cells.
3.6. Identification of genes coregulated by HDAC9 and resistance to antiestrogens
In order to determine whether HDAC9 overexpression and resistance to OHTam led to the deregulation of common genes, a global transcriptome analysis of MCF7 cells sensitive and resistant to OHTam was performed and compared to that performed in MCF7‐Control and MCF7‐HDAC9FL cells (Lapierre et al., 2016).
Comparison of the expression profiles of OHTam‐sensitive and OHTR MCF7 cells identified 1719 transcripts that were differentially expressed with a fold change (FC) ≥ 2 and a false discovery rate ≤ 0.05 (734 transcripts were downregulated and 985 transcripts upregulated in OHTR cells; see Table S2 for the list of the top up‐ and downregulated genes). Hierarchical clustering of OHTam‐sensitive and OHTR MCF7 cells based on the expression profile of the 1719 transcripts (Fig. 4A) showed that all OHTam‐sensitive MCF7 cell samples clustered in one branch and all OHTR samples in another branch. The GO biological process enrichment analysis of these transcriptomic data with IPA revealed that in resistant cells, the most significantly upregulated biological process was ‘Cell death and survival’, whereas ‘Cellular growth and proliferation’ was the most significantly downregulated process (Table S3).
Figure 4.
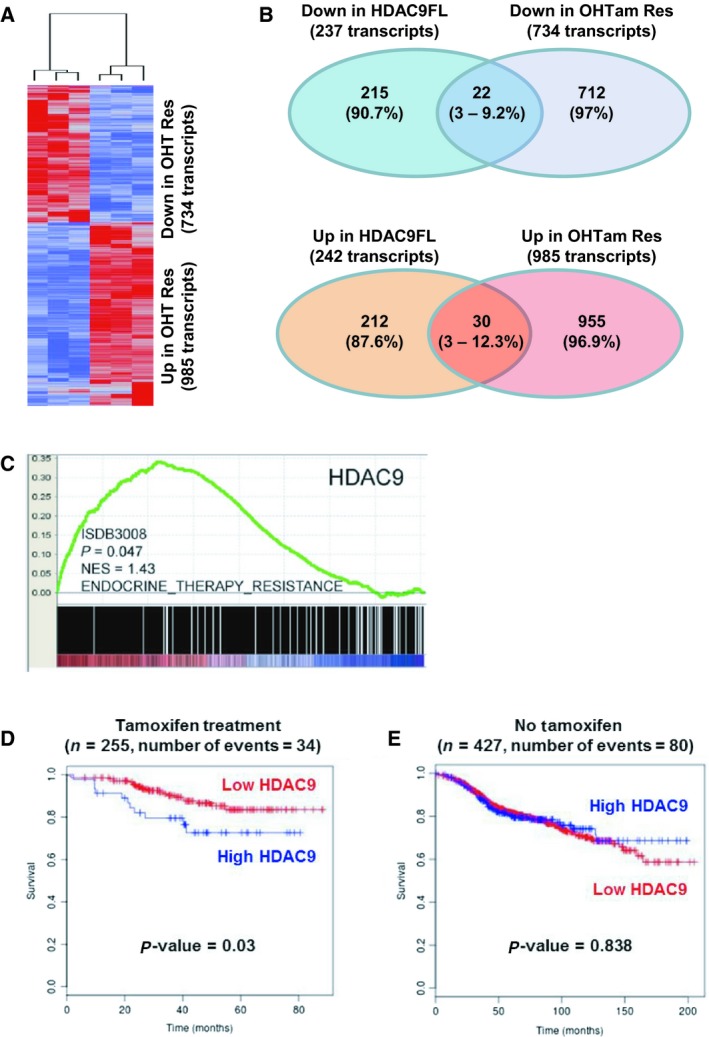
Transcriptomic analyses in MCF7 cells and expression in human breast cancer tumors. (A) The molecular signatures of OHTam‐sensitive and OHTam‐resistant MCF7 cells were visualized by hierarchical clustering of the 1719 genes that are differentially expressed in the two conditions (genes are arranged in rows and samples in columns). For each condition, a tree represents the relationship among samples and the branch lengths reflect the degree of similarity between samples according to the gene expression profile. In red, upregulated genes; in blue, downregulated genes. (B) Schematic representation showing the overlap between downregulated (top drawing) and upregulated (bottom drawing) transcripts in MCF7‐HDAC9FL and OHTam‐resistant MCF7 cells. The number (percentage) of transcripts deregulated in each condition is indicated. (C) GSEA of HDAC9 mRNA expression relative to the endocrine therapy resistance gene set in breast cancer (n = 2795 samples). (D) Kaplan–Meier analysis with the BreastMark algorithm of survival in patients with breast cancer treated with tamoxifen (n = 255 tumor samples) relative to HDAC9 expression using a low cutoff analysis (lower quartile). Disease‐free survival was considered as the survival end point and if not available, distant disease‐free survival or overall survival was used. Significance was calculated using the log rank test. (E) The same as in panel D, but for untreated patients (n = 427 tumor samples).
The transcriptomic analysis of MCF7‐Control and MCF7‐HDAC9FL cells (Lapierre et al., 2016) identified 237 probe sets that corresponded to genes downregulated in MCF7‐HDAC9FL cells (22 also downregulated in OHTR‐MCF7 cells) and 242 probe sets corresponding to genes upregulated in MCF7‐HDAC9FL cells (30 also upregulated in OHTR cells; Fig. 4B). The list of genes similarly regulated by HDAC9 overexpression and OHTam resistance is in Table S4. GFRA1, SGK3, nuclear receptor interacting protein 1 (NRIP1), RAB31 and SLC39A6 were among the most represented downregulated genes, whereas MUC1 and, to a lesser extent, MAGED2 were represented more than once in the list of common upregulated genes. Particularly, ESR1 and some of its target genes, such as NRIP1 (Augereau et al., 2006) and SGK3 (Wang et al., 2011), were downregulated in both conditions, in agreement with the data in Figs 1, 2, 3.
Moreover, GSEA (Subramanian et al., 2005) performed using the transcriptomic data of MCF7‐Control and MCF7‐HDAC9FL cells (Lapierre et al., 2016) showed that gene sets related to tamoxifen resistance, including ‘Endocrine Therapy Resistance’ and ‘Tamoxifen Resistance DN’ were differentially represented (P < 0.05) in MCF7‐HDAC9FL samples compared with MCF7‐Control (Fig. S5). This supports the hypothesis that HDAC9‐regulated genes are involved in antiestrogen resistance.
3.7. Prognostic value of HDAC9 expression in human breast tumors
To investigate the putative role of HDAC9 expression in the patient response to endocrine therapies, GSEA was performed using expression data from the ISDB3008 dataset (2795 breast cancer samples), as already described (Gao et al., 2014). This analysis indicated that HDAC9 expression was positively correlated with genes overexpressed in tumor samples from patients resistant to endocrine therapy (Fig. 4C, P = 0.047), confirming the data shown in Fig. S4.
Finally, HDAC9 expression prognostic value was determined using the BreastMark database (Madden et al., 2013) that allows the reanalysis of 26 different transcriptomic datasets to correlate the clinical outcome with differential gene expression levels and to define the prognostic potential of a given gene. The analysis was restricted to patients with tumors being estrogen and PR‐positive and HER2‐negative. HDAC9 levels were of significant prognostic value, and high levels were correlated with shorter overall survival in tamoxifen‐treated (n = 255; P = 0.03; Fig. 4D), but not in untreated patients (n = 427; P = 0.838; Fig. 4E). Altogether, these results supported our in vitro data and demonstrated the prognostic value of HDAC9 levels in patients with breast cancer who received antiestrogen therapy.
4. Discussion
Previous studies have underlined the potential role of HDACs in breast tumor progression and their cross talk with ER signaling (Linares et al., 2011; Thomas and Munster, 2009). Here, we demonstrated the strong interaction of class IIa HDAC9 with ERα signaling and its implication in OHTam resistance in breast cancer cell lines and tumor samples. These results are schematically summarized in Fig. 5.
Figure 5.
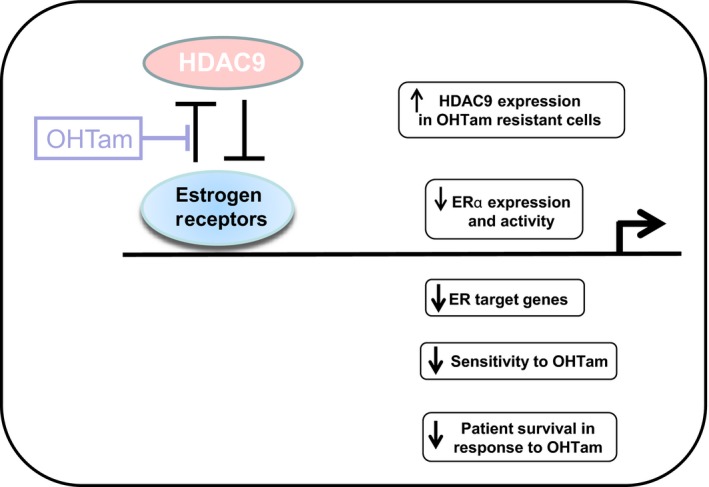
Effect of HDAC9 on estrogen signaling and tamoxifen response in breast cancer cells. Schematic model of the putative role of HDAC9 in estrogen signaling in human breast cancer cells. HDAC9 expression is inversely correlated with ERα expression in breast cancer and is upregulated in OHTam‐resistant breast cancer cells. HDAC9 decreases ERα expression and activity, ER target gene expression, OHTam cytotoxicity and survival of patients treated with tamoxifen.
Since its cloning (Zhou et al., 2001) and thanks to the analysis of knockout (KO) mice, HDAC9 has been involved in various pathophysiological processes including muscle differentiation (Zhang et al., 2001), heart response to stress (Zhang et al., 2002) or immune regulatory T cell function (de Zoeten et al., 2010). HDAC9 plays also an important role in various type of cancers including, among others, oral squamous cell carcinoma (Rastogi et al., 2016), retinoblastoma (Zhang et al., 2016) and breast cancer (Lapierre et al., 2016).
Few years ago, van Rooij et al. (2010) reported that class IIa HDAC9 and HDAC5 reduce ERα expression in rat cardiomyocytes via the control of MEF2 transcription factor activity, which could explain ERα cardioprotective effect in HDAC9 KO female mice. Similarly, our study showed that HDAC9 decreases ERα expression (ERα mRNA and protein levels, ESR1 gene promoter transcription) and transcriptional activity (ERE‐mediated response in luciferase reporter assays and expression of the E2‐regulated PGR gene).
Our work also demonstrated that HDAC9 is overexpressed in different OHTR MCF7 cell lines. The precise mechanisms involved remain to be determined, although loss of ERα expression could contribute to HDAC9 upregulation in antiestrogen‐resistant breast cancer cell lines. Indeed, HDAC9 expression is increased upon ERα silencing in breast cancer cell lines (Al Saleh et al., 2011). Moreover, our analysis of publicly available cDNA array data showed a significant increase in HDAC9 mRNA levels in ERα‐negative tumors compared with ERα‐positive samples, validating the biological relevance of the results obtained in breast cancer cell lines. Other mechanisms could explain HDAC9 gene upregulation in ER‐negative and in antiestrogen‐resistant breast cancer cells. For instance, comparative genomic hybridization (CGH) array analysis from 532 breast tumors indicated a significant copy number gain (23.7% of the cases) for the 7p21‐p15.3 region that encompasses the HDAC9 gene (Mahlknecht et al., 2002; B. Orsetti, data not shown). Similarly, in cervical carcinoma, the HDAC9 gene was identified by CGH array in a region of high copy number gain (Choi et al., 2007) thus suggesting that an increase in gene copy number could also be involved in HDAC9 gene overexpression. More recently, it was reported that HDAC9 is hypomethylated in hepatocellular carcinoma (Archer et al., 2010) suggesting that epigenetic modifications could represent another way to overexpress HDAC9.
We then showed that HDAC9 overexpression decreases the sensitivity of MCF7 breast cancer cells to ER ligands in proliferation assays. Most importantly, we found that silencing HDAC9 expression in MCF7‐OHTR cells reduced their OHTam resistance. The decreased ERα expression in MCF7 cells upon HDAC9 overexpression, even at moderate levels, may be one of the mechanisms by which deregulated HDAC9 expression is linked to OHTam resistance in ERα‐positive breast tumor cells. In line with this hypothesis, we found that ERα expression was decreased in the various OHTR breast tumor cell lines developed by our group. However, other mechanisms have been involved in breast tumor resistance to OHTam, including activation of growth factor pathways (Riggins et al., 2007). The impact of HDAC9 expression on such mechanisms should be further studied to better understand the development of OHTam resistance.
We also observed the association of HDAC9 with OHTam resistance in breast cancer samples. Indeed, HDAC9 expression was positively correlated with genes overexpressed in endocrine therapy‐resistant tumor samples, and had a prognostic significance (shorter overall survival with high HDAC9 expression) only in patients who received OHTam. These data are in accordance with our in vitro observations showing that HDAC9 overexpression reduces OHTam antiproliferative effect in breast cancer cell lines. Moreover, in our global transcriptome analyses, we identified several genes that are similarly deregulated in OHTR cells and HDAC9‐overexpressing cells. For instance, we confirmed the downregulation of the SMC3 gene (Mendes‐Pereira et al., 2012) and the overexpression of the MUC1 (Kharbanda et al., 2013) and S100P (Zhou et al., 2012) genes in OHTR cells and demonstrated, for the first time, their regulation also by HDAC9 overexpression. Altogether, these results argue for an important role of HDAC9 overexpression in the acquisition of tamoxifen resistance by breast cancer cells.
5. Conclusions
Antiestrogen‐based endocrine therapies are commonly used to treat ER‐positive breast cancers, but tumors usually become resistant to such hormonal manipulation. Our study underlines strong interactions between class IIa HDAC9 and ERα signaling (Fig. 5) and undercovers the key role of HDAC9 in antiestrogen resistance of breast cancer cells and tumors. Our data suggest that HDAC9 might represent a new marker of antiestrogen response. Moreover, based on our results, it appears likely that targeting HDAC9 might represent a novel therapeutic target to treat endocrine therapy‐resistant cancers.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
VC and NB designed and supervised the study; AL, ML, ET, CD, CF and AB performed the experiments; SA, GT and JJ performed the bioinformatic analyses; OS and EB provided reagents; AL, ML, NB and VC analyzed the data; ML, NB and VC wrote the manuscript.
Supporting information
Fig. S1. Characterization of HDAC9‐overexpressing MCF7 cell clones.
Fig. S2. Expression of HDAC9 mRNA in breast cancer cells and tissue samples.
Fig. S3. Expression of HDAC9 mRNA in antiestrogen‐resistant cells.
Fig. S4. SiRNA‐mediated knockdown of HDAC9 expression in MCF7‐OHTR cells.
Fig. S5. GSEA of genes that are differentially regulated by HDAC9 in breast cancer cells.
Table S1. List of the different datasets reanalyzed with the corresponding reference and the main observation obtained from their use.
Table S2. List of the most deregulated genes in OHTR vs OHTam‐sensitive MCF7 breast cancer cells.
Table S3. List of the most deregulated GO biological profiles in OHTR vs OHTam‐sensitive MCF7 breast cancer cells.
Table S4. List of the most common deregulated genes in OHTR and HDAC9‐overexpressing MCF7 cells.
Acknowledgements
We thank Caroline Desmetz for technical help and Elisabetta Andermarcher for language editing service. We are also grateful to Stéphan Jalaguier and Catherine Teyssier for critical reading of the manuscript. This work was supported by the INCa (2011‐054), the ‘Prix Ruban Rose’, ‘Institut National de la Santé et de la Recherche Médicale’, the University of Montpellier, the ‘Ligue Nationale contre le Cancer’ and the ‘Association pour la Recherche sur le Cancer’ and the SIRIC (Grant INCa‐DGOS‐Inserm 6045).
2.11. Data accessibility
The datasets used and/or analyzed during the current study are available from the corresponding author.
References
- Al Saleh S, Al Mulla F and Luqmani YA (2011) Estrogen receptor silencing induces epithelial to mesenchymal transition in human breast cancer cells. PLoS ONE 6, e20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annicotte JS, Iankova I, Miard S, Fritz V, Sarruf D, Abella A, Berthe ML, Noël D, Pillon A, Iborra F et al (2006) Peroxisome proliferator‐activated receptor gamma regulates E‐cadherin expression and inhibits growth and invasion of prostate cancer. Mol Cell Biol 26, 7561–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer KJ, Mas VR, Maluf DG and Fisher RA (2010) High‐throughput assessment of CpG site methylation for distinguishing between HCV‐cirrhosis and HCV‐associated hepatocellular carcinoma. Mol Genet Genomics 283, 341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augereau P, Badia E, Fuentes M, Rabenoelina F, Corniou M, Derocq D, Balaguer P and Cavailles V (2006) Transcriptional regulation of the human NRIP1/RIP140 gene by estrogen is modulated by dioxin signalling. Mol Pharmacol 69, 1338–1346. [DOI] [PubMed] [Google Scholar]
- Badia E, Duchesne MJ, Semlali A, Fuentes M, Giamarchi C, Richard‐Foy H, Nicolas JC and Pons M (2000) Long‐term hydroxytamoxifen treatment of an MCF‐7‐derived breast cancer cell line irreversibly inhibits the expression of estrogenic genes through chromatin remodeling. Cancer Res 60, 4130–4138. [PubMed] [Google Scholar]
- Badia E, Morena M, Lauret C, Boulahtouf A, Boulle N, Cavaillès V, Balaguer P and Cristol JP (2015) Effect of tamoxifen and fulvestrant long‐term treatments on ROS production and (pro/anti)‐oxidant enzymes mRNA levels in a MCF‐7‐derived breast cancer cell line. Breast Cancer 23, 692–700. [DOI] [PubMed] [Google Scholar]
- Brünner N, Frandsen TL, Holst‐Hansen C, Bei M, Thompson EW, Wakeling AE, Lippman ME and Clarke R (1993) MCF7/LCC2: a 4‐hydroxytamoxifen resistant human breast cancer variant that retains sensitivity to the steroidal antiestrogen ICI 182,780. Cancer Res 53, 3229–3232. [PubMed] [Google Scholar]
- Chen Y‐X, Fang J‐Y, Zhu H‐Y, Lu R, Cheng Z‐H and Qiu D‐K (2004) Histone acetylation regulates p21WAF1 expression in human colon cancer cell lines. World J Gastroenterol 10, 2643–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T et al (2006) Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10, 529–541. [DOI] [PubMed] [Google Scholar]
- Choi YW, Bae SM, Kim YW, Lee HN, Kim YW, Park TC, Ro DY, Shin JC, Shin SJ, Seo JS et al (2007) Gene expression profiles in squamous cell cervical carcinoma using array‐based comparative genomic hybridization analysis. Int J Gynecol Cancer 17, 687–696. [DOI] [PubMed] [Google Scholar]
- de Cremoux P, Dalvai M, N'Doye O, Moutahir F, Rolland G, Chouchane‐Mlik O, Assayag F, Lehmann‐Che J, Kraus‐Berthie L, Nicolas A et al (2015) HDAC inhibition does not induce estrogen receptor in human triple‐negative breast cancer cell lines and patient‐derived xenografts. Breast Cancer Res Treat 149, 81–89. [DOI] [PubMed] [Google Scholar]
- de Zoeten EF, Wang L, Sai H, Dillmann WH and Hancock WW (2010) Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology 138, 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfour C, Roger P, Bret C, Berthe ML, Rochaix P, Kalfa N, Raynaud P, Bibeau F, Maudelonde T and Boulle N (2006) RCL2, a new fixative, preserves morphology and nucleic acid integrity in paraffin‐embedded breast carcinoma and microdissected breast tumor cells. J Mol Diagn 8, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghue C, Westley BR and May FE (1999) Selective promoter usage of the human estrogen receptor‐alpha gene and its regulation by estrogen. Mol Endocrinol 13, 1934–1950. [DOI] [PubMed] [Google Scholar]
- Duong V, Bret C, Altucci L, Mai A, Duraffourd C, Loubersac J, Harmand PO, Bonnet S, Valente S, Maudelonde T et al (2008) Specific activity of class II histone deacetylases in human breast cancer cells. Mol Cancer Res 6, 1908–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO and Botstein D (1998) Cluster analysis and display of genome‐wide expression patterns. Proc Natl Acad Sci USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van't Veer LJ and Perou CM (2006) Concordance among gene‐expression‐based predictors for breast cancer. N Engl J Med 355, 560–569. [DOI] [PubMed] [Google Scholar]
- Gao T, Han Y, Yu L, Ao S, Li Z and Ji J (2014) CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen resistance. PLoS ONE 9, e91771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Malerva L, Park J, Zou L, Hu Y, Moradpour Z, Pearlberg J, Sawyer J, Stevens H, Harlow E and LaBaer J (2011) High‐throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc Natl Acad Sci USA 108, 2058–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL and Olson EN (2009) The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 10, 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Ström A, Treuter E, Warner M et al (2007) Estrogen receptors: how do they signal and what are their targets. Physiol Rev 87, 905–931. [DOI] [PubMed] [Google Scholar]
- Henderson BE, Ross R and Bernstein L (1988) Estrogens as a cause of human cancer: the Richard and Hinda Rosenthal Foundation award lecture. Cancer Res 48, 246–253. [PubMed] [Google Scholar]
- Hilakivi‐Clarke L, Wärri A, Bouker KB, Zhang X, Cook KL, Jin L, Zwart A, Nguyen N, Hu R, Cruz MI et al (2017) Effects of in utero exposure to ethinyl estradiol on tamoxifen resistance and breast cancer recurrence in a preclinical model. J Natl Cancer Inst 109, djw188–000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jézéquel P, Frénel J‐S, Campion L, Guérin‐Charbonnel C, Gouraud W, Ricolleau G and Campone M (2013) bc‐GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database 2013, bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Li H, Avraham S, Jiang S and Avraham HK (2003) Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int J Cancer 107, 353–358. [DOI] [PubMed] [Google Scholar]
- Kharbanda A, Rajabi H, Jin C, Raina D and Kufe D (2013) Oncogenic MUC1‐C promotes tamoxifen resistance in human breast cancer. Mol Cancer Res 11, 714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre M, Linares A, Dalvai M, Duraffourd C, Bonnet S, Boulahtouf A, Rodriguez C, Jalaguier S, Assou S, Orsetti B et al (2016) Histone deacetylase 9 regulates breast cancer cell proliferation and the response to histone deacetylase inhibitors. Oncotarget 7, 19693–19708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares A, Dalenc F, Balaguer P, Boulle N and Cavailles V (2011) Manipulating protein acetylation in breast cancer: a promising approach in combination with hormonal therapies? J Biomed Biotechnol 2011, 856985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden SF, Clarke C, Gaule P, Aherne ST, O'Donovan N, Clynes M, Crown J and Gallagher WM (2013) BreastMark: an integrated approach to mining publicly available transcriptomic datasets relating to breast cancer outcome. Breast Cancer Res 15, R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlknecht U, Schnittger S, Will J, Cicek N and Hoelzer D (2002) Chromosomal organization and localization of the human histone deacetylase 9 gene (HDAC9). Biochem Biophys Res Commun 293, 182–191. [DOI] [PubMed] [Google Scholar]
- McKenna NJ and O'Malley BW (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108, 465–474. [DOI] [PubMed] [Google Scholar]
- Mendes‐Pereira AM, Sims D, Dexter T, Fenwick K, Assiotis I, Kozarewa I, Mitsopoulos C, Hakas J, Zvelebil M, Lord CJ et al (2012) Genome‐wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci USA 109, 2730–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RR, Belder N, Ansari SA, Kayhan M, Bal H, Raza U, Ersan PG, Tokat ÜM, Eyüpoğlu E, Saatci Ö et al (2018) Reactivation of cAMP pathway by PDE4D inhibition represents a novel druggable axis for overcoming tamoxifen resistance in ER‐positive breast cancer. Clin Cancer Res 24, 1987–2001. [DOI] [PubMed] [Google Scholar]
- Munster PN, Thurn KT, Thomas S, Raha P, Lacevic M, Miller A, Melisko M, Ismail‐Khan R, Rugo H, Moasser M et al (2011) A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy‐resistant breast cancer. Br J Cancer 104, 1828–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Regan RM and Jordan VC (2002) The evolution of tamoxifen therapy in breast cancer: selective oestrogen‐receptor modulators and downregulators. Lancet Oncol 3, 207–214. [DOI] [PubMed] [Google Scholar]
- Parra M (2015) Class IIa HDACs ‐ new insights into their functions in physiology and pathology. FEBS J 282, 1736–1744. [DOI] [PubMed] [Google Scholar]
- Petrie K, Guidez F, Howell L, Healy L, Waxman S, Greaves M and Zelent A (2003) The histone deacetylase 9 gene encodes multiple protein isoforms. J Biol Chem 278, 16059–16072. [DOI] [PubMed] [Google Scholar]
- Rastogi B, Raut SK, Panda NK, Rattan V, Radotra BD and Khullar M (2016) Overexpression of HDAC9 promotes oral squamous cell carcinoma growth, regulates cell cycle progression, and inhibits apoptosis. Mol Cell Biochem 415, 183–196. [DOI] [PubMed] [Google Scholar]
- Riggins RB, Schrecengost RS, Guerrero MS and Bouton AH (2007) Pathways to tamoxifen resistance. Cancer Lett 256, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Fielitz J, Sutherland LB, Thijssen VL, Crijns HJ, Dimaio MJ, Shelton J, De Windt LJ, Hill JA and Olson EN (2010) Myocyte enhancer factor 2 and class II histone deacetylases control a gender‐specific pathway of cardioprotection mediated by the estrogen receptor. Circ Res 106, 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al (2005) Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan WW, Allred JB, Moreno‐Aspitia A, Northfelt DW, Ingle JN, Goetz MP and Perez EA (2016) Phase I study of panobinostat (LBH589) and letrozole in postmenopausal metastatic breast cancer patients. Clin Breast Cancer 16, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S and Munster PN (2009) Histone deacetylase inhibitor induced modulation of anti‐estrogen therapy. Cancer Lett 280, 184–191. [DOI] [PubMed] [Google Scholar]
- Ur‐Rehman S, Gao Q, Mitsopoulos C and Zvelebil M (2013) ROCK: a resource for integrative breast cancer data analysis. Breast Cancer Res Treat 139, 907–921. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhou D, Phung S, Masri S, Smith D and Chen S (2011) SGK3 Is an estrogen‐inducible kinase promoting estrogen‐mediated survival of breast cancer cells. Mol Endocrinol 25, 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeruva SLH, Zhao F, Miller KD, Tevaarwerk AJ, Wagner LI, Gray RJ, Sparano JA and Connolly RM (2018) E2112: randomized phase iii trial of endocrine therapy plus entinostat/placebo in patients with hormone receptor‐positive advanced breast cancer. NPJ Breast Cancer 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA and Olson EN (2002) Class II histone deacetylases act as signal‐responsive repressors of cardiac hypertrophy. Cell 110, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA and Olson EN (2001) The transcriptional corepressor MITR is a signal‐responsive inhibitor of myogenesis. Proc Natl Acad Sci USA 98, 7354–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu D, Xia F, Xian H, Zhu X, Cui H and Huang Z (2016) Downregulation of HDAC9 inhibits cell proliferation and tumor formation by inducing cell cycle arrest in retinoblastoma. Biochem Biophys Res Commun 473, 600–606. [DOI] [PubMed] [Google Scholar]
- Zhou X, Marks PA, Rifkind RA and Richon VM (2001) Cloning and characterization of a histone deacetylase, HDAC9. Proc Natl Acad Sci USA 98, 10572–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Zhong Q, Rhodes LV, Townley I, Bratton MR, Zhang Q, Martin EC, Elliott S, Collins‐Burow BM, Burow ME et al (2012) Proteomic analysis of acquired tamoxifen resistance in MCF‐7 cells reveals expression signatures associated with enhanced migration. Breast Cancer Res 14, R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Characterization of HDAC9‐overexpressing MCF7 cell clones.
Fig. S2. Expression of HDAC9 mRNA in breast cancer cells and tissue samples.
Fig. S3. Expression of HDAC9 mRNA in antiestrogen‐resistant cells.
Fig. S4. SiRNA‐mediated knockdown of HDAC9 expression in MCF7‐OHTR cells.
Fig. S5. GSEA of genes that are differentially regulated by HDAC9 in breast cancer cells.
Table S1. List of the different datasets reanalyzed with the corresponding reference and the main observation obtained from their use.
Table S2. List of the most deregulated genes in OHTR vs OHTam‐sensitive MCF7 breast cancer cells.
Table S3. List of the most deregulated GO biological profiles in OHTR vs OHTam‐sensitive MCF7 breast cancer cells.
Table S4. List of the most common deregulated genes in OHTR and HDAC9‐overexpressing MCF7 cells.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author.


