Abstract
Background
Several neurodegeneration (N) metrics using structural MRI are used for the purpose of Alzheimer's disease (AD)-related staging, including hippocampal volume, global atrophy, and an “AD signature” composite consisting of thickness or volumetric estimates derived from regions impacted early in AD. This study sought to determine if less user-intensive estimates of global atrophy and hippocampal volume were equivalent to a thickness-based AD signature from FreeSurfer for defining N across the AD continuum (i.e., individuals who are amyloid-positive (A+)).
Methods
Cognitively unimpaired (CU) late middle-aged and older adults, as well as A+ mild cognitive impairment (MCI) and A+ AD dementia individuals, with available CSF and structural MRI scan <1.5 years apart, were selected for the study (n = 325, mean age = 62). First, in a subsample of A+ AD dementia and matched biomarker-negative (i.e., A- and tau tangle pathology (T)-) CU controls (n = 40), we examined ROC characteristics and identified N cut-offs using Youden's J for neurofilament light chain protein (NfL) and each of three MRI-based measures: a thickness-based AD signature from FreeSurfer, hippocampal volume (using FIRST), and a simple estimate of global atrophy (the ratio of intracranial CSF segmented volume to brain tissue volume, using SPM12). Based on the results from the ROC analyses, we then examined the concordance between NfL N positivity and N positivity for each MRI-based metric using Cohen's Kappa in the remaining subsample of 285 individuals. Finally, in the full sample (n = 325), we examined the relationship between the four measures of N and group membership across the AD continuum using Kruskal-Wallis tests and Cliff's deltas.
Results
The three MRI-based metrics and CSF NfL similarly discriminated between the A-T- CU (n = 20) and A+ AD (n = 20) groups (AUCs ≥0.885; ps < 0.001). Using the cut-off values derived from the ROCs to define N positivity, there was weak concordance between NfL and all three MRI-derived metrics of N in the subsample of 285 individuals (Cohen's Kappas ≤0.429). Finally, the three MRI-based measures of N and CSF NfL showed similar associations with AD continuum group (i.e., Kruskal-Wallis ps < 0.001), with relatively larger effect sizes noted when comparing the A-T- CU to the A+ MCI (Cliff's deltas ≥0.741) and A+ AD groups (Cliff's deltas ≥0.810) than to the A+T- CU (Cliff's deltas = 0.112–0.298) and A + T+ CU groups (Cliff's deltas = 0.212–0.731).
Conclusions
These findings suggest that the three MRI-based morphometric estimates and CSF NfL similarly differentiate individuals across the AD continuum on N status. In many applications, a simple estimate of global atrophy may be preferred as an MRI marker of N across the AD continuum given its methodological robustness and ease of calculation when compared to hippocampal volume or a cortical thickness AD signature.
Keywords: Neurodegeneration, Hippocampus, Alzheimer's disease, Amyloid, FreeSurfer
Highlights
-
•
Hippocampus, global atrophy and AD signature had strong diagnostic accuracy in AD.
-
•
There was weak concordance between NfL and all three MRI-derived metrics.
-
•
NfL and the three MRI-derived metrics performed similarly across the AD spectrum.
1. Introduction
The preclinical phase of Alzheimer's disease (AD), which is thought to begin a decade or more prior to a clinical diagnosis of dementia due to AD, is characterized by neuropathological changes but intact cognitive and functional abilities (Price et al., 2009; Price and Morris, 1999). Several biomarkers exist for detecting AD-related neuropathology in vivo across the AD continuum. Recently, Jack and colleagues proposed a framework for defining abnormal pathophysiology in AD by dividing these biomarkers into three distinct categories (Jack et al., 2016, Jack et al., 2018) of amyloid burden (A) (e.g., decreased cerebrospinal fluid (CSF) levels of Aβ42 or elevated retention of amyloid PET tracers), tau tangle pathology (T) (e.g., elevations in tau tracer retention on PET or increased CSF levels of phosphorylated tau (ptau181)), and neurodegeneration (N) (e.g., [18F]-fluorodeoxyglucose (FDG)-PET hypometabolism, elevated CSF levels of total tau, or gray matter loss measured using structural MRI). Within the context of this AT(N) framework, individuals can be categorized as A, T, and/or N positive or negative, resulting in eight possible groups.
In order to classify individuals across the AD continuum (i.e., individuals who are A+) using A, T, and N, one must first develop cut-off values for each of these three categories of biomarkers, and such thresholds may be site-specific. Numerous studies demonstrate sensitive and specific cut-off values for both A and T in individuals in the earliest stages of AD (e.g., Clark et al., 2018; Hansson et al., 2006; Mattsson et al., 2014; Sjögren et al., 2001), and the field is improving precision in this regard with new CSF platforms for analysis (Bittner et al., 2016; Vandijck et al., 2017). There is relatively greater heterogeneity in defining N across the AD continuum, particularly with MRI. MRI morphometric approaches for assessing atrophy are abundant, but may vary at several levels including acquisition (field strength and sequence parameters), neuroanatomic operationalization of structures of interest, choice of software, reliability and precision of the measure of interest, computational availability, and end-user expertise and decision rules to evaluate the output. Furthermore, unlike A and T, N is not necessarily specific to AD pathophysiology. For example, a marked increase in CSF total tau (as a surrogate indicator of N) is seen both with acute brain damage, such as ischemic stroke (Hesse et al., 2001), as well as in rapidly progressive neurodegenerative disorders such as Creutzfeldt-Jakob disease (Skillbäck et al., 2014). Nevertheless, prior investigations report that A + T + N+ individuals are more likely to evidence cognitive decline than not only A-T-N- individuals, but also their A + T + N- counterparts (Aschenbrenner et al., 2018; Bouwman et al., 2007; Davatzikos et al., 2011; Jack et al., 2017; Soldan et al., 2019; van Maurik et al., 2017; van Rossum et al., 2012; Vemuri et al., 2009). These findings suggest that N, in conjunction with A and T, may provide important staging information for AD (Jack et al., 2018).
A number of N metrics are currently available for AD-related staging. One of the most readily available methods comes from morphometric estimates obtained using structural MRI. Structural MRI studies examining N in AD have frequently focused on hippocampal volume (Jack et al., 1992; Storandt et al., 2009; van Maurik et al., 2017), global atrophy (Fagan et al., 2009; Fox et al., 1999; Schott et al., 2010; van Maurik et al., 2017), and an “AD signature” consisting of a composite of thickness or volumetric estimates derived from regions impacted early during the course of AD (Dickerson et al., 2011; Dickerson and Wolk, 2012; Schwarz et al., 2016). A previous study, which compared multiple thickness- and volumetric-based methods for obtaining an AD signature, found that thickness-based AD signatures from FreeSurfer and Statistical Parametric Mapping (SPM) + DiReCT (the cortical thickness algorithm from Advanced Normalization Tools (ANTs)) had the strongest correlations with Braak neurofibrillary tangle stage at autopsy (Schwarz et al., 2016). It should be noted that SPM + DiReCT was found to have better test-retest reliability than FreeSurfer, although FreeSurfer performed better in terms of diagnostic separability. As a result, Schwarz et al. recommended either method as appropriate for obtaining an AD signature estimate.
Compared to completely automated methods, FreeSurfer can require iterative manual editing of segmentation output and this practice may vary between laboratories. Further, the automated segmentation component itself can be time consuming, with a recent study suggesting that FreeSurfer 6.0 takes approximately 6–9 h per scan to complete this component (Ellis et al., 2016), with the total amount of time depending upon a number of parameters (e.g., processor speed and workflow type) and control point editing iterations. Given these drawbacks, it may be that less user- and computationally-intensive estimates obtained from structural MRI with quicker quality control procedures, such as hippocampal volume and global atrophy estimates derived from FMRIB Software Library (FSL)-FIRST (a tool designed to segment subcortical structures) and SPM respectively, are sufficient in many settings for defining N in the context of AD.
One way to determine whether hippocampal volume and global atrophy are equivalent to the thickness-based AD signature from FreeSurfer is to follow a similar approach as outlined in Schwarz et al. (2016) by comparing the diagnostic accuracy of these three estimates for separating individuals with AD dementia from cognitively unimpaired (CU) individuals. Alternatively, we could compare the three MRI-based morphometric estimates to a reference standard. Unfortunately, no “gold standard” for defining N currently exists; however, CSF levels of neurofilament light chain protein (NfL) may be a relatively good reference standard in light of prior investigations.
Several previous studies have demonstrated that CSF NfL is a biomarker of axonal injury in multiple neurodegenerative conditions, including AD (Olsson et al., 2016; Petzold, 2005; Zetterberg, 2016; Zetterberg et al., 2016). Furthermore, CSF NfL has been found to be associated with accelerated disease progression and atrophy rates (both hippocampal and whole brain), as well as differentiates individuals with AD from CU individuals and those with mild cognitive impairment (MCI) (Zetterberg et al., 2016). A prior meta-analysis noted that CSF NfL had a large effect size for differentiating CU individuals from those with AD, whereas CSF levels of other N markers (e.g., visinin-like protein 1 and neuron-specific enolase) only demonstrated moderate effect sizes (Olsson et al., 2016). Finally, recent research has found elevated CSF levels of NfL in both symptomatic and asymptomatic carriers of the presenilin-1 or amyloid precursor protein mutation, as well as noted associations between CSF NfL and both cognitive and imaging measures (e.g., global atrophy rate) in this group with familial AD (Weston et al., 2017). Together, these findings suggest that CSF NfL is a useful metric for defining N across the AD continuum, including during the earliest phase of the disease.
In light of the findings reviewed above, the primary goal of the current study was to compare the more computationally expensive and user intensive AD signature to automated, robust, and computationally efficient estimates of hippocampal volume and global atrophy for the purpose of defining N in individuals across the AD continuum (i.e., individuals who are A+). To accomplish this goal, we: 1) examined the diagnostic accuracy of each measure for separating A+ AD individuals from A-T- CU individuals; 2) identified cut-off values for several measures of N; 3) evaluated the association between N defined using CSF NfL and the three MRI-based measures; and 4) examined the ability of these N measures to differentiate A-T- CU individuals from those along the AD continuum (i.e., from A+T- CU to A+ AD individuals).
2. Materials and methods
2.1. Participants
Participants consisted of late middle-aged and older adults (n = 325, mean age = 62; 309 individuals reported their race as White, 1 as Hispanic, 6 as Black or African American, 2 as Asian, and 7 as Native American or Alaskan Native; see Table 1 for sample characteristics) enrolled in longitudinal cohort studies at the Wisconsin Alzheimer's Disease Research Center (WADRC) or the Wisconsin Registry for Alzheimer's Prevention (WRAP; Johnson et al., 2018). To qualify for the current analysis, participants were required to have at least one T1-weighted structural MRI scan (to define N) and one lumbar puncture (with available CSF data for Aβ42, ptau181, total tau, and NfL) within 1.5 years of each other (mean time between lumbar puncture and MRI scan = 0.03 years, standard deviation = 0.14 years, range = 0–1.17 years). Participants were also required to be classified as CU (i.e., no clinical diagnosis of MCI or dementia), MCI due to AD, or dementia due to AD. These classifications were made by a team of clinicians (neuropsychologists, physician dementia specialists, and nurse practitioners) blind to biomarker status, and were based on consensus conference research criteria established by the National Institute on Aging-Alzheimer's Association (Albert et al., 2011; McKhann et al., 2011). Participants were then categorized as biomarker negative (i.e., A-T-) or biomarker positive (A+) based on available CSF data (for more details about cut-off values for A and T, see the section on amyloid and tau status determination below). Participants in the MCI or dementia due to AD groups were required to be A+ and participants in the CU group were required to be either A-T-, A+T-, or A + T+ so as to represent individuals across the AD continuum (Jack et al., 2018). All participants were free from major psychiatric conditions (e.g., schizophrenia or bipolar disorder) and excluded from the current analysis if they failed processing or had an abnormal MRI as described below (see Supplemental Fig. 1 for a flow diagram detailing eligibility for the current study).
Table 1.
Demographics and biomarkers.
| Total sample | ROC sample |
Concordance sample |
|||||
|---|---|---|---|---|---|---|---|
| CU A-T- | AD A+ | CU A-T- | CU A+T- | CU A + T+ | MCI A+ | ||
| N | 325 | 20 | 20 | 229 | 24 | 16 | 16 |
| Sex (male; %)^ | 109 (33.5%) | 10 (50.0%) | 10 (50.0%) | 71 (31.0%) | 4 (16.7%) | 4 (25.0%) | 10 (62.5%) |
| Age (years) (mean (SD))* | 61.9 (8.0) | 66.7 (5.6) | 66.6 (5.6) | 59.7 (7.2) | 63.1 (7.5) | 68.5 (6.9) | 73.0 (8.6) |
| Education (years) (mean (SD))+ | 16.0 (2.5) | 15.2 (2.8) | 15.3 (2.8) | 16.1 (2.4) | 16.0 (2.8) | 15.8 (2.4) | 16.1 (2.8) |
| LN of CSF levels of Aβ42 (pg/mL) (mean (SD)) | 6.4 (0.3) | 6.7 (0.2) | 5.8 (0.2) | 6.6 (0.2) | 6.0 (0.2) | 6.0 (0.1) | 5.9 (0.2) |
| CSF levels of ptau181 (pg/mL) (mean (SD)) | 43.6 (17.9) | 39.5 (8.6) | 70.0 (21.2) | 38.6 (10.0) | 38.5 (13.3) | 69.5 (16.5) | 69.8 (37.0) |
| CSF levels of tau (pg/mL) (mean (SD)) | 339.1 (204.1) | 293.0 (62.9) | 714.3 (262.4) | 268.3 (84.3) | 282.3 (112.5) | 636.0 (178.1) | 728.4 (347.7) |
| CSF levels of NfL (pg/mL) (mean (SD)) | 731.9 (457.0) | 692.9 (160.7) | 1281.1 (717.9) | 622.1 (286.5) | 701.4 (373.8) | 1025.7 (304.2) | 1417.2 (1043.3) |
| AD Signature (z-score) (mean (SD)) | 0.0 (1.0) | 0.2 (0.7) | −1.6 (1.0) | 0.2 (0.9) | −0.1 (0.7) | −0.2 (0.8) | −1.3 (1.0) |
| Hippocampal Volume (z-score) (mean (SD)) | 0.0 (1.0) | −0.2 (0.5) | −1.4 (0.9) | 0.3 (0.9) | −0.2 (0.7) | −0.1 (0.8) | −1.4 (0.7) |
| Global Atrophy (z-score) (mean (SD)) | 0.0 (1.0) | 0.2 (0.8) | 1.8 (0.8) | −0.3 (0.7) | 0.1 (0.9) | 0.3 (1.0) | 1.5 (1.0) |
Notes: ROC = receiver operating characteristic; CU = cognitively unimpaired; AD = Alzheimer's disease; MCI = mild cognitive impairment; LN = natural-log; CSF = cerebrospinal fluid; NfL = neurofilament light chain protein; ^there were significantly more females in the CU group than the MCI group; *the CU group was significantly younger than the AD and MCI groups, but the MCI group was significantly older than the AD group; +one CU A-T- participant in the concordance sample was missing education.
The inclusion of human participants was approved by the University of Wisconsin-Madison Institutional Review Board, and all participants provided informed consent. The current work was carried out in accordance with The Code of Ethics of the World Medical Association.
2.1.1. ROC participants
A+ individuals with a clinical diagnosis of probable AD dementia were matched to biomarker-negative CU individuals (i.e., A-T- individuals based on available CSF) on the following demographic variables: age, sex, and education. Six individuals with dementia due to AD were excluded from the analysis because there was not an age-, sex-, and education-matched biomarker-negative CU individual available. This resulted in 20 A+ individuals with a clinical diagnosis of probable AD and 20 A-T- CU individuals for the ROC analyses. Of the 20 A+ individuals with a clinical diagnosis of probable AD, 18 were A + T+ and 2 were A+T-. The Clinical Dementia Rating Scale (CDR; Morris, 1993) was administered to informants of these 20 participants to determine the severity of dementia (9 were classified as CDR = 0.5 (i.e., very mild dementia) and 11 were classified as CDR = 1.0 (i.e., mild dementia)). For more information on the demographic characteristics of these individuals, please refer to Table 1 (ROC sample).
2.1.2. Participants used for NfL N concordance analysis
A-T- CU individuals who were not included in the ROC analyses, along with CU and MCI individuals who were A+, were included in analyses examining the relationship between N positivity by NfL and the MRI-based measures of N. This resulted in the inclusion of 285 individuals with known biomarker status in these analyses. For information on the demographic characteristics of these individuals, please refer to Table 1 (Concordance Sample).
2.2. Structural MRI
Two identical GE 3.0 Tesla MR750 scanners (Waukesha, WI, USA) with an 8-channel head coil (Excite HD Brain Coil; GE Healthcare) were used to acquire a T1-weighted brain volume in the axial plane, with a 3-D inversion-recovery prepared fast spoiled gradient-echo sequence using the following parameters: inversion time (TI) = 450 ms, repetition time (TR) = 8.1 ms, echo time (TE) = 3.2 ms, flip angle = 12°, acquisition matrix = 256 × 256 × 156 mm, field of view (FOV) = 256 mm, slice thickness = 1.0 mm. Additionally, seven subjects were scanned with a shorter sequence that was less susceptible to motion artifacts, after it was determined their first scan would likely be unusable. The shorter sequence parameters were: TI = 450 ms, TR = 6.044 ms, TE = 2.204 ms, flip angle = 12°, acquisition matrix = 256 × 256 × 130 mm, FOV = 256 mm, slice thickness = 1.2 mm, in-plane resolution 1 mm. Cushions inside the head coil were used to reduce head movement during scanning. All scans were reviewed by a radiologist (H.A.R.) and 14 CU individuals were excluded for abnormalities (e.g., hydrocephalus, prior craniotomy, CHS rating >4; Manolio et al., 1994).
FreeSurfer 6.0 (Fischl, 2012) was used for the purpose of defining an “AD signature,” which consisted of a composite of thickness estimates derived from the following regions of interest (ROIs) based on previous research: entorhinal, inferior temporal, middle temporal, inferior parietal, fusiform, and precuneus (Schwarz et al., 2016). Within FreeSurfer, each gray matter voxel in an MRI image is mapped to a cortical surface mesh and given a neuroanatomical label based on a probabilistic atlas from a manually labeled training set consisting of both healthy younger and older adults as described in the software documentation (Fischl et al., 2002). We followed a quality control process similar to that outlined by Schwarz et al. (2016) in which FreeSurfer segmentation outputs were visually inspected by S.L.A. and R.P.C. for severe errors, which included when FreeSurfer failed to complete topology correction (two participants with dementia due to AD) or a significant portion of the cortical ribbon was placed incorrectly (two MCI and two CU individuals). No manual correction of FreeSurfer was performed and these six participants were excluded from analyses.
Volumetric estimates were derived from the T1-weighted IRSPGR sequence. Tissue segmentation into CSF, gray matter, white matter, and intracranial volume (ICV) was performed using SPM12 (www.fil.ion.ucl.ac.uk/spm), which is an automated and completely reproducible process (Ashburner et al., 2016). Brain (the sum of total gray matter and white matter) and CSF segmentation was the objective of this process and no failures (defined as gross tissue misclassification based on visual inspection of the images by S.L.A) occurred. Hippocampal volume was estimated using the completely automated FSL-FIRST algorithm (Patenaude et al., 2011). Visual inspection of the hippocampal segmentations by S.L.A. revealed that the output misclassified the hippocampus in one dementia due to AD case that had significant structural atrophy, and this participant was excluded from analyses. For quantitative analyses, hippocampal volume was corrected for head size by proportional scaling to total intracranial volume (brain plus CSF volume derived from the SPM12 segmentation output). A simple measure of global atrophy was estimated as the ratio of CSF volume to brain volume (total gray matter volume + total white matter volume). In order to compare the three metrics, they were scaled to have a mean of 0 and a standard deviation of 1.
2.3. Cerebrospinal fluid collection
Twenty-two mL of CSF were removed from the L3-L4 or L4-L5 vertebral interspace. These samples (sent in batches at two time points) were analyzed at the Clinical Neurochemistry Laboratory at the Sahlgrenska Academy of the University of Gothenburg, Sweden using commercially available enzyme-linked immunosorbent assay methods. CSF samples were assayed for Aβ42, ptau181, total tau, and NfL, and corrected for batch when appropriate as described in Clark et al., 2018.
2.4. Amyloid and tau status determination
For the current study, the natural log of CSF Aβ42 < 6.156 was defined as A+, whereas CSF levels of total tau >461.26 or ptau181 > 59.50 were defined as T+. We defined T+ based on either CSF levels of total tau or ptau181 given the high correlation between these two CSF measures in the current study (Spearman rank correlation = 0.916, p < .001). These cut-off values are based on prior ROC analyses for discriminating CU individuals from those with a clinical diagnosis of MCI or dementia due to AD (Clark et al., 2018).
2.5. Statistical analyses
Analyses were conducted in SPSS 23. First, ROC analyses were performed to determine the sensitivity and specificity of hippocampal volume, the AD signature, global atrophy, and CSF NfL for separating A+ individuals with a clinical diagnosis of AD from A-T- CU individuals. The area under the curve (AUC), which may be defined as the accuracy of the diagnostic test (values range from 0 to 1), was computed for each metric. For each of these metrics, the value that maximized Youden's Index (sensitivity + specificity - 1) was chosen as the threshold for N positivity. In the event that different cut-off values evidenced identical Youden's Indices, the cut-off value with the highest sensitivity was selected. Cohen's Kappa was then used to examine the relationship between N positivity defined by NfL and each of the three MRI-based measures of N. Cohen's Kappa was also used to examine the relationships among the three MRI-based measures of N. We reported positive percent agreement, negative percent agreement, and overall percent agreement for each MRI-based measure in relation to NfL and in relation to each other. Separate correlation matrices of all relevant continuous measures of A, T, and N were also computed for the whole sample (n = 325) and for the concordance sample (n = 285) using age-adjusted partial correlations and Spearman rank correlations.
Finally, we wanted to examine the measures of N in the context of staging across the AD continuum. Therefore, we performed Kruskal-Wallis tests (Breslow, 1970) to determine if there was a significant relationship between group membership (i.e., A-T- CU, A+T- CU, A + T+ CU, A+ MCI, and A+ AD individuals) and each measure of N (i.e., CSF NfL, AD signature, hippocampal volume, and global atrophy). Post-hoc analyses comparing the A-T- CU group to each of the four remaining groups were accomplished using Mann-Whitney U tests (Mann and Whitney, 1947). Cliff's delta was used to compute the effect size of each of the three MRI-based measures of N and NfL (Cliff, 1993).
Post-hoc analyses adjusting for age were also performed in the context of staging across the entire AD continuum for Kruskal-Wallis tests, Cliff's deltas, and Mann-Whitney U tests. First, using only the A-T- CU group, separate regressions were performed with each measure of N as an outcome in separate analyses and age as the predictor. The intercept β, age β, and the square root of the mean square error (RMSE) were saved. These parameters were then used for the whole sample (n = 325) to get the predicted N value based on the A-T- CU group age-related change. An individual's unadjusted z-score on each N metric was converted to an age-adjusted z-score as follows:
age-adjusted z-score for the N metric = (observed z-score - predicted z-score)/RMSE.
3. Results
3.1. Demographics
Participants were on average 62 years old, 34% male, and highly educated (see Table 1).
3.2. Determining cut-off values for each neurodegeneration measure1
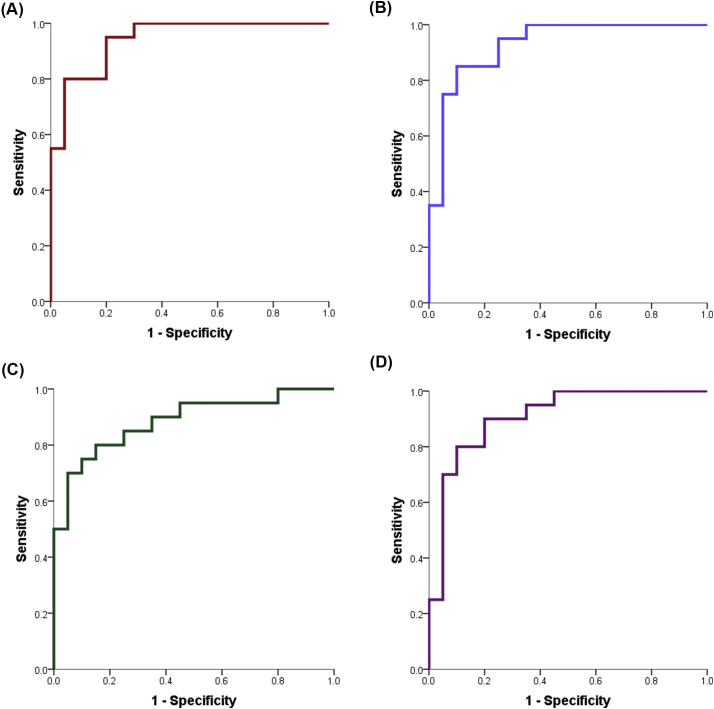
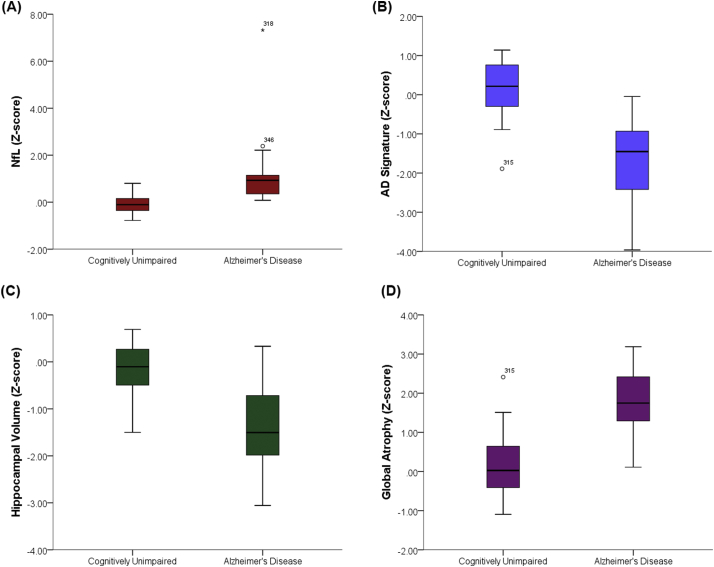
CSF NfL (Fig. 1A), the AD signature (Fig. 1B), hippocampal volume (Fig. 1C), and global atrophy (Fig. 1D) all significantly discriminated between the A-T- CU (n = 20) and A+ AD dementia (n = 20) groups (see Table 2 for each metric's AUC, sensitivity, specificity, p-value, and Youden's Index). Using the selected thresholds, NfL had the highest sensitivity and AUC, whereas the AD signature had the highest specificity. However, examination of the AUC confidence intervals indicated that all confidence intervals overlapped. That is, no one metric was significantly better than any of the other metrics at discriminating between the groups. Please refer to Fig. 2 for boxplots of the neurodegeneration measures comparing A+ AD and A-T- CU individuals.
Fig. 1.
A-D represent ROC curves relating sensitivity and specificity of each measure for differentiating A-T- CU from A+ AD individuals; A) CSF NfL; B) the AD signature; C) hippocampal volume; D) global atrophy.
Table 2.
Diagnostic accuracy.
| AUC (SE) | AUC 95% CI | P-Value | Sensitivity | Specificity | Youden's J | |
|---|---|---|---|---|---|---|
| NfL | 0.943 (0.034) | 0.877–1.000 | <0.001 | 95.0% | 80.0% | 0.750 |
| AD signature | 0.928 (0.041) | 0.847–1.000 | <0.001 | 85.0% | 90.0% | 0.750 |
| Hippocampal volume | 0.885 (0.054) | 0.780–0.990 | <0.001 | 80.0% | 85.0% | 0.650 |
| Global atrophy | 0.908 (0.048) | 0.814–1.000 | <0.001 | 90.0% | 80.0% | 0.700 |
Notes: AUC = area under the curve; SE = standard error; NfL = neurofilament light chain protein;
AD = Alzheimer's disease.
Fig. 2.
A-D represent box plots for each metric comparing A-T- CU to A+ AD individuals; A) CSF NfL; B) the AD signature; C) hippocampal volume; D) global atrophy.
3.3. Relationship between NfL and the MRI-based measures of N
Using the cut-off values derived from the ROCs to define N positivity (both for NfL and the MRI-based metrics), there was a significant relationship between NfL and all three MRI-based metrics of N (i.e., hippocampal volume, the AD signature, and global atrophy), with global atrophy having the highest concordance with NfL. Of note, the agreement among the measures of N was relatively weak. See Table 3A, Table 3B, Table 3C for information regarding Cohen's Kappa, positive percent agreement, negative percent agreement, and overall percent agreement for the measures of N.
Table 3A.
Concordance between the MRI estimates and NfL.
| NfL N+ |
NfL N- |
Positive % agreement | Negative % agreement | Overall % agreement | Kappa (p-value) |
||
|---|---|---|---|---|---|---|---|
| AD signature | N+ | 22 | 32 | 32.35% | 85.25% | 72.63% | 0.189 (p = .001) |
| N- | 46 | 185 | |||||
| Hippocampal volume | N+ | 24 | 33 | 35.29% | 84.79% | 72.98% | 0.213 (p < .001) |
| N- | 44 | 184 | |||||
| Global atrophy | N+ | 30 | 14 | 44.12% | 93.55% | 81.75% | 0.429 (p < .001) |
| N- | 38 | 203 |
Notes: NfL = neurofilament light chain protein; AD = Alzheimer's disease.
Table 3B.
Concordance between AD signature and both hippocampal volume and global atrophy.
| AD signature N+ | AD signature N- | Positive % agreement | Negative % agreement | Overall % agreement | Kappa (p-value) |
||
|---|---|---|---|---|---|---|---|
| Hippocampal volume | N+ | 22 | 35 | 40.74% | 84.85% | 76.49% | 0.251 (p < .001) |
| N- | 32 | 196 | |||||
| Global atrophy | N+ | 21 | 23 | 38.89% | 90.04% | 80.35% | 0.311 (p < .001) |
| N- | 33 | 208 |
Note: AD = Alzheimer's disease.
Table 3C.
Concordance between global atrophy and hippocampal volume.
| Global atrophy N+ | Global atrophy N- | Positive % agreement | Negative % agreement | Overall % agreement | Kappa (p-value) |
||
|---|---|---|---|---|---|---|---|
| Hippocampal volume | N+ | 23 | 34 | 52.27% | 85.89% | 80.70% | 0.341 (p < .001) |
| N- | 21 | 207 |
3.4. Correlations
Using the full sample, the AD signature, hippocampal volume, and global atrophy were all significantly associated with CSF levels of Aβ42, total tau, ptau181, and NfL (ps < 0.05). These associations were similar, but attenuated, in the concordance sample. Of note, there was a strong association between CSF total tau and ptau181 in both samples. See Table 4A, Table 4B, Table 4C, Table 4D for the intercorrelations among the N metrics and CSF biomarkers.
Table 4A.
Age-adjusted partial correlations in the full sample (n = 325).
| AD signature | HC volume | Global atrophy | LN AB42 | Total Tau | Ptau181 | |
|---|---|---|---|---|---|---|
| AD signature | ||||||
| HC volume | 0.252⁎⁎ | |||||
| Global atrophy | −0.431⁎⁎ | −0.433⁎⁎ | ||||
| LN AB42 | 0.358⁎⁎ | 0.317⁎⁎ | −0.329⁎⁎ | |||
| Total Tau | −0.347⁎⁎ | −0.215⁎⁎ | 0.251⁎⁎ | −0.356⁎⁎ | ||
| Ptau181 | −0.231⁎⁎ | −0.130+ | 0.110+ | −0.229⁎⁎ | 0.878⁎⁎ | |
| NfL | −0.185⁎ | −0.148⁎ | 0.263⁎⁎ | −0.134+ | 0.455⁎⁎ | 0.336⁎⁎ |
Notes: AD = Alzheimer's disease; HC = hippocampal; LN = natural log; NfL = neurofilament light chain protein.
p < .001.
p < .01.
p < .05.
Table 4B.
Age-adjusted partial correlations in the concordance sample (n = 285).
| AD signature | HC volume | Global atrophy | LN AB42 | Total Tau | Ptau181 | |
|---|---|---|---|---|---|---|
| AD signature | ||||||
| HC volume | 0.110 | |||||
| Global atrophy | −0.272⁎⁎ | −0.325⁎⁎ | ||||
| LN AB42 | 0.247⁎⁎ | 0.219⁎⁎ | −0.162⁎ | |||
| Total Tau | −0.208⁎⁎ | −0.089 | 0.033 | −0.238⁎⁎ | ||
| Ptau181 | −0.120+ | −0.031 | −0.066 | −0.106 | 0.878⁎⁎ | |
| NfL | −0.116+ | −0.068 | 0.173⁎ | −0.045 | 0.407⁎⁎ | 0.291⁎⁎ |
Notes: AD = Alzheimer's disease; HC = hippocampal; LN = natural log; NfL = neurofilament light chain protein.
p < .001.
p < .01.
p < .05.
Table 4C.
Spearman rank correlations in the full sample (n = 325).
| AD signature | HC volume | Global atrophy | LN AB42 | Total Tau | Ptau181 | |
|---|---|---|---|---|---|---|
| AD signature | ||||||
| HC volume | 0.255⁎⁎ | |||||
| Global atrophy | −0.380⁎⁎ | −0.504⁎⁎ | ||||
| LN AB42 | 0.323⁎⁎ | 0.330⁎⁎ | −0.301⁎⁎ | |||
| Total Tau | −0.253⁎⁎ | −0.258⁎⁎ | 0.338⁎⁎ | −0.106 | ||
| Ptau181 | −0.189⁎ | −0.216⁎⁎ | 0.270⁎⁎ | −0.079 | 0.916⁎⁎ | |
| NfL | −0.303⁎⁎ | −0.348⁎⁎ | 0.568⁎⁎ | −0.156⁎ | 0.653⁎⁎ | 0.573⁎⁎ |
Notes: AD = Alzheimer's disease; HC = hippocampal; LN = natural log; NfL = neurofilament light chain protein.
p < .001.
p < .01.
Table 4D.
Spearman rank correlations in the concordance sample (n = 285).
| AD signature | HC volume | Global atrophy | LN AB42 | Total Tau | Ptau181 | |
|---|---|---|---|---|---|---|
| AD signature | ||||||
| HC volume | 0.159⁎ | |||||
| Global atrophy | −0.296⁎⁎ | −0.444⁎⁎ | ||||
| LN AB42 | 0.240⁎⁎ | 0.257⁎⁎ | −0.211⁎⁎ | |||
| Total Tau | −0.150+ | −0.167⁎ | 0.236⁎⁎ | 0.030 | ||
| Ptau181 | −0.090 | −0.118+ | 0.173⁎ | 0.060 | 0.920⁎⁎ | |
| NfL | −0.218⁎⁎ | −0.279⁎⁎ | 0.508⁎⁎ | −0.044 | 0.618⁎⁎ | 0.532⁎⁎ |
Notes: AD = Alzheimer's disease; HC = hippocampal; LN = natural log; NfL = neurofilament light chain protein.
p < .001.
p < .01.
p < .05.
3.5. Staging across the AD continuum
Kruskal-Wallis tests demonstrated that there was a statistically significant association between the four measures of N (i.e., CSF NfL, AD signature, hippocampal volume, global atrophy) and AD continuum group (ps < 0.001). Findings from Mann-Whitney U tests suggest that the four measures of N performed similarly in terms of differentiating A-T- CU individuals from both A+ MCI and A+ AD individuals (ps < 0.001), with relatively large effect sizes noted (Cliff's deltas ≥0.741).
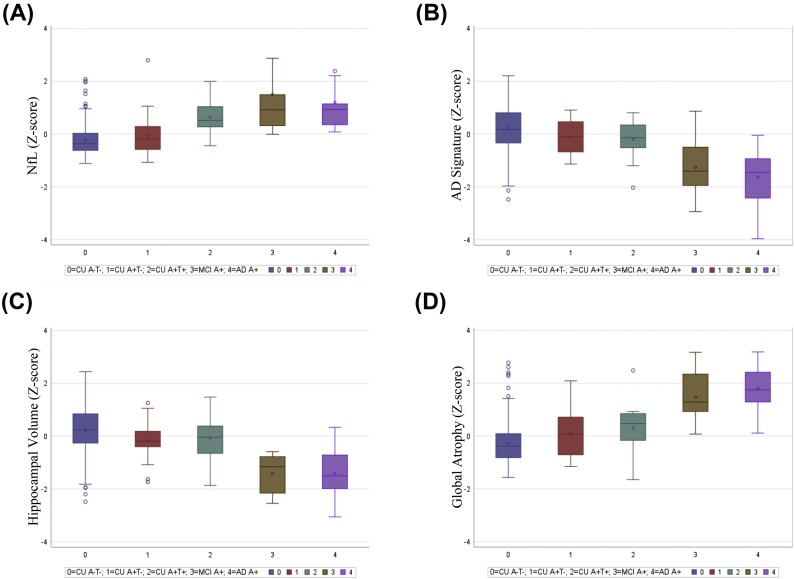
When comparing A-T- CU to A+T- CU individuals, only hippocampal volume was significantly different between these two groups (p = .016, Cliff's delta = 0.298), although there was a non-significant trend for both the AD signature (p = .051, Cliff's delta = 0.242) and global atrophy (p = .088, Cliff's delta = 0.211). Finally, when comparing A-T- CU to A + T+ CU individuals, CSF NfL (p < .001, Cliff's delta = 0.731) and global atrophy (p = .006, Cliff's delta = 0.410) were significantly different between the two groups, with a non-significant trend for the AD signature (p = .058, Cliff's delta = 0.284). It is important to note that examination of the confidence intervals for Cliff's delta suggest that no one metric was significantly better than any of the other metrics at discriminating between the groups, with the exception that CSF NfL had a significantly larger effect size than the hippocampus when discriminating between A-T- CU and A + T+ CU individuals. See Table 5 for information regarding Mann-Whitney U p-values, as well as Cliff's deltas and associated confidence intervals. See Fig. 3 for boxplots of the N measures for each of the five groups (i.e., A-T- CU, A+T- CU, A + T+ CU, A+ MCI, A+ AD).
Table 5.
Comparing measures of neurodegeneration across the AD continuum.
| Statistic | CU A-T- vs. CU A+T- | CU A-T- vs. CU A + T+ | CU A-T- vs. MCI A+ | CU A-T- vs. AD A+ | |
|---|---|---|---|---|---|
| NfL | Mann-Whitney U p-value | 0.364 | <0.001 | <0.001 | <0.001 |
| Cliff's Delta | 0.112 | 0.731 | 0.841 | 0.866 | |
| Confidence interval for Cliff's Delta | −0.152–0.362 | 0.514–0.860 | 0.639–0.935 | 0.712–0.940 | |
| AD signature | Mann-Whitney U p-value | 0.051 | 0.058 | <0.001 | <0.001 |
| Cliff's Delta | 0.242 | 0.284 | 0.741 | 0.859 | |
| Confidence interval for Cliff's Delta | 0.008–0.451 | 0.012–0.516 | 0.469–0.885 | 0.727–0.930 | |
| Hippocampal volume | Mann-Whitney U p-value | 0.016 | 0.156 | <0.001 | <0.001 |
| Cliff's delta | 0.298 | 0.212 | 0.866 | 0.810 | |
| Confidence interval for Cliff's Delta | 0.084–0.485 | −0.078–0.469 | 0.778–0.921 | 0.649–0.902 | |
| Global atrophy | Mann-Whitney U p-value | 0.088 | 0.006 | <0.001 | <0.001 |
| Cliff's Delta | 0.211 | 0.410 | 0.856 | 0.915 | |
| Confidence interval for Cliff's Delta | −0.056–0.450 | 0.051–0.675 | 0.507–0.964 | 0.644–0.982 |
Notes: NfL = neurofilament light chain protein; CU = cognitively unimpaired; MCI = mild cognitive impairment; AD = Alzheimer's disease.
Fig. 3.
A-D represent box plots for each metric comparing the different groups (CU A-T-, CU A+T-, CU A+T+, MCI A+, AD A+); A) CSF NfL; B) the AD signature; C) hippocampal volume; D) global atrophy.
3.6. Post-hoc analyses: staging across the AD continuum, adjusting for age
Results adjusting for age were largely unchanged, with the exception that there was no longer a trend for global atrophy when comparing A-T- CU to A+T- CU individuals (p = .562, Cliff's delta = 0.072). Furthermore, only CSF NfL significantly differed between the A-T- CU and the A + T+ CU groups (p < .001, Cliff's delta = 0.527). See Supplemental Table 1 for information regarding Mann-Whitney U p-values, as well as Cliff's deltas and associated confidence intervals. See Supplemental Fig. 2 for boxplots of the N measures for each of the five groups (i.e., A-T- CU, A+T- CU, A + T+ CU, A+ MCI, A+ AD), adjusting for age.
4. Discussion
The current study sought to compare three commonly used MRI morphometric operationalizations of N: global atrophy, hippocampal volume, and an implementation of the AD signature from FreeSurfer. These three metrics were selected for study because they are commonly used in the field and prior literature has suggested all three are associated with structural changes in AD (Dickerson et al., 2011; Dickerson and Wolk, 2012; Fagan et al., 2009; Fox et al., 1999; Jack et al., 1992; Schott et al., 2010; Schwarz et al., 2016; Storandt et al., 2009). Regarding diagnostic accuracy, hippocampal volume, global atrophy, and the thickness-based AD signature, along with CSF levels of NfL, performed similarly in terms of differentiating A+ AD from A-T- CU individuals. The highest sensitivity and AUC were reported for CSF NfL, whereas the highest specificity was noted for the AD signature; however, the overlapping confidence intervals suggest no strong inferences are warranted. The similar performance of these three metrics may be due to the fact that at the dementia stage of AD, atrophy is present throughout the brain (Scahill et al., 2002; Sluimer et al., 2009). In summary, these findings suggest that for the purpose of identifying individuals with dementia due to AD, N could be defined using any of the three available MRI metrics or CSF NfL.
When applying the N cut-off values derived from the ROC analyses to a larger sample containing A-T- CU, A+T- CU, A + T+ CU, and A+ MCI individuals, global atrophy had a relatively higher concordance with CSF NfL than the other two MRI-based metrics of N; however, it is important to emphasize that all three metrics had weak agreement with CSF NfL and with each other (i.e., Cohen's Kappa values ≤0.429; Fleiss, 1971; Fleiss et al., 2013). Similarly, small to moderate inter-correlations were observed for these four metrics of N. These findings are consistent with prior research (Alexopoulos et al., 2014; Jack et al., 2015; Toledo et al., 2014; Vos et al., 2016), suggesting that these measures appear to provide different information related to N. Despite this low level of agreement, results from Mann-Whitney U tests and Cliff's deltas, unadjusted for age, indicated similar performance in terms of differentiating the A-T- CU group from the AD continuum groups (i.e., A+T- CU, A + T+ CU, A+ MCI, and A+ AD). These findings fit with previous research indicating that different definitions of N produce similar results, including that individuals defined as N+ using a number of different approaches (e.g., hippocampal volume, thickness-based AD signature, FDG-PET) performed more poorly on tests of learning and memory than N- individuals (Jack et al., 2015).
The findings were largely unchanged when adjusting for age, with the exception that there was no longer a trend for global atrophy when comparing A-T- CU to A+T- CU individuals. Furthermore, only CSF NfL significantly differed between the A-T- CU and the A + T+ CU groups. The best approach to account for “normal” (i.e., non-pathologic) age-related change is yet to be determined in the literature for not only structural MRI and CSF NfL, but also for other areas of study such as cognition. Furthermore, previous investigations differ in whether and how they adjust for age when examining N (e.g., Alexopoulos et al., 2014; Jack et al., 2015; Toledo et al., 2014; Vos et al., 2016). In the current study, we tried to estimate age-related changes by examining the relationship between N and age in the A-T- CU group. We then applied the parameters from this group to calculate age-adjusted values of N for individuals across the AD continuum. The findings from the current study do indicate that CSF NfL has the added benefit of similarly distinguishing A-T- CU from A + T+ CU individuals, with and without adjusting for age. Unfortunately, adjusting for age resulted in none of the MRI-based measures differing between the A-T- CU and A + T+ CU groups. Furthermore, this approach did not account for other pathologic changes (e.g., cerebrovascular disease) that may impact estimates of N in the context of aging. The field would benefit from future studies with larger groups across the AD continuum to determine the best approach for adjusting for non-pathologic aging and whether any of the three MRI-based metrics of N is preferable for defining N when accounting for age-associated changes.
Although the present results indicate that any of the three MRI-based measures may be used to define N across the AD continuum, the measure of global atrophy in the current study may be preferable to the other measures for methodological reasons. First, the global atrophy metric is less affected by the limitations of current segmentation techniques. In particular, previous research indicates reduced gray to white contrast with aging, which will affect the estimates of any metric requiring segmentation of gray from white matter structures, and may result in less accurate volumetric and thickness estimates (Fischl et al., 2002). Because the measure of global atrophy was a ratio of CSF volume to white and gray matter volumes (summed), this metric is by design impervious to gray and white matter segmentation uncertainties that were inherent to thickness and hippocampal volumetric estimates. This benefit of the global atrophy estimate is worth emphasizing given existing literature demonstrating that white matter signal intensity declines with age (Salat et al., 2009) and is impacted by disease status, with AD patients evidencing lower signal intensity and gray to white matter contrast than their CU counterparts (Westlye et al., 2009). Together, these findings would suggest that estimates that do not rely on differentiating gray from white matter may be preferable when defining N across the AD continuum.
In contrast to the hippocampal volumetric estimate, both the global atrophy metric and the AD signature had the added benefit of not needing to correct for head size as it was a ratio of CSF to white and gray matter volumes (summed). In the current study, we controlled for head size by dividing hippocampal volume by intracranial volume. A number of other approaches have been used to correct for head size variation (e.g., Buckner et al., 2004; O'Brien et al., 2011; Raz et al., 1997), and currently, there is no universally agreed upon method. Considering that the method employed to account for head size can result in significant variability across studies, a metric like global atrophy, which does not require head size correction, may be preferable when defining N across the AD continuum.
Even though the thickness-based AD signature did not require head size correction, the potential uncertainties related to the accuracy and reliability of FreeSurfer may make this measure less than optimal for defining N across the AD continuum. Specifically, prior research suggests some problems with the accuracy of atlas-based FreeSurfer generated estimates in the temporal lobe (a region included in the current AD signature; Desikan et al., 2006; Oguz et al., 2008). Furthermore, although the AD signature was composed of estimates from a number of regions, a prior investigation indicates lower reliability for regions with smaller surface areas, such as the entorhinal cortex, which was included in the AD signature in this study (Desikan et al., 2006). For these reasons, global atrophy may be preferable to the thickness-based AD signature derived from FreeSurfer for defining N across the AD continuum.
Despite the emphasis of the present study on MRI-derived measures of N and CSF NfL, it is important to highlight the strong association noted between total tau and ptau181 in the current analysis. The AT(N) framework developed by Jack et al. (2018) classifies CSF total tau as a measure of N, whereas ptau181 is considered a metric of T. Despite prior investigations indicating that CSF levels of total tau and ptau181 may diverge in other neurodegenerative conditions, such as Creutzfeldt-Jakob disease (Skillbäck et al., 2014) and certain clinical variants of frontotemporal dementia (Meeter et al., 2018), treating CSF measures of total tau and ptau181 as reflecting N and T, respectively, may be less helpful in terms of staging individuals across the AD continuum given their strong association. Within the context of the current findings, a different measure of N, such as one obtained using structural MRI or CSF NfL, may be preferable to the use of total tau for defining N in individuals across the AD continuum.
The current findings should be considered within the context of several limitations. First, even though the present work suggests similar effect sizes for the four measures when defining N across the AD continuum, this study only examined CSF NfL and three out of a potentially very large number of candidate MRI metrics of atrophy that have been presented in the literature (Atiya et al., 2003; Frisoni et al., 2010). In addition to constraining our examination of N to four measures, each of the four measures was examined independently. There is some research to suggest that a combination of N markers may be the best approach when defining N. For example, relatively recent work suggests that A+ CU individuals who are N+ based on both CSF total tau and hippocampal volume tend to be at greater risk of clinical decline when compared to A+ CU individuals who are N+ on only CSF total tau or hippocampal volume (Vos et al., 2016). Additionally, a recent study found that a combined MRI measure of atrophy and cerebrovascular disease was associated with PET amyloid imaging, CSF biomarkers, and neuropathology at autopsy in individuals across the AD continuum (Brickman et al., 2018). Subsequent work should explore how independent measures compare to a combination of measures (e.g., structural MRI estimates, CSF levels of NfL, FDG-PET, and/or diffusion tensor imaging) for defining N across the AD continuum. Considering that individuals with AD frequently have cerebrovascular disease (Kapasi et al., 2017), future AD-related research could compare different N measures in conjunction with measures of cerebrovascular disease (e.g., cerebral blood flow and pulsatility) (Gu et al., 2005).
Limitations related to the sample in this study should also be acknowledged. Although a strength of the current study is that individuals in the A+ AD group were matched to A-T- CU individuals on age, sex, and education, the sample sizes for the groups were relatively small (i.e., n = 20 in each of the AD and biomarker-negative CU groups). Similar small sample sizes were observed for the other groups across the AD continuum (i.e., n = 20 A+T- CU, n = 16 A + T+ CU, and 16 A+ MCI). Furthermore, the participants were generally well educated with a mean education level of approximately 16 years. These constraints may limit the generalizability of the current findings. Additional research with a larger and more diverse sample is needed. In particular, considering that we constrained our examination to individuals who were along the AD continuum, as well as excluded CU individuals with significant cerebrovascular disease, subsequent research is needed to determine the optimal way to define N in individuals with other neurodegenerative conditions, including those who are A-T+. Finally, future research using the three MRI-based metrics from the current study may also examine the degree to which biomarker-positive CU individuals who are N+ go on to develop dementia due to AD.
A final limitation of the current study is that the cut-off values used to define N were determined based on their ability to differentiate A+ individuals with a clinical diagnosis of probable AD from A-T- CU individuals. These cut-off values were then used in a subsequent analysis to examine the concordance between the three MRI-based metrics and CSF NfL. In an ideal scenario, there would be some “true” cut-off value of NfL, or another marker of N, that defines the presence of neurodegeneration independent from clinical status. We would then use this cut-off value to evaluate our MRI-based metrics. Unfortunately, no agreed upon measure currently exists for NfL or any other measure of neurodegeneration (separate from clinical status) for defining N (Jack et al., 2016, Jack et al., 2018). Future work should reexamine this question once an independent reference standard becomes available.
Within the context of these limitations, the current study suggests that the global atrophy metric may represent a convenient MRI-based measure for defining N across the AD continuum. Specifically, global atrophy was similar to hippocampal volume and the AD signature for differentiating A+ AD dementia individuals from A-T- CU individuals, as well as for differentiating A-T- CU individuals from A+T- CU, A + T+ CU, and A+ MCI. The fact that the global atrophy metric required less human and computational resources and is not affected by methodological issues that may affect complex approaches (e.g., gray from white matter demarcation and atlas based parcellation) also favors the use of global atrophy as a measure of neurodegeneration. The choice of an MRI neurodegeneration metric employed in any particular study will depend on the specific research question and should consider the precision, reliability, and complexity of the approach given the quality of the input image data. Taken together, these findings provide justification for considering global atrophy as a general and robust MRI-based metric for evaluating N across the AD continuum.
The following are the supplementary data related to this article.
Flow diagram depicting eligibility for the current study. Arrows pointing downward indicate number of participants excluded at each decision point. Arrows pointing upward indicate number of participants included at each decision point.
Adjusting for age (see text for more details), A-D represent box plots for each metric comparing the different groups (CU A-T-, CU A+T-, CU A+T+, MCI A+, AD A+); A) CSF NfL; B) the AD signature; C) hippocampal volume; D) global atrophy.
Acknowledgments
Acknowledgments
We would like to thank the researchers and staff at the Wisconsin Registry for Alzheimer's Prevention and Wisconsin Alzheimer's Disease Research Center for assistance in recruitment and data collection. Most importantly, we would like to acknowledge the participants for their dedication, support, and participation in this research.
Funding
The current study was supported by ADRC P50 AG033514 (SA), R01 AG021155 (SCJ), R01 AG027161 (SCJ), and R01 AG037639 (BBB).
Disclosures
-
•
Samantha Allison has no disclosures.
-
•
Paul Cary has no disclosures.
-
•
Erin Jonaitis was employed by Frontier Science from 2015 to 2017 and received funding for travel from Frontier Science and from Actelion Pharmaceuticals during that time. Her spouse is employed by Epic Systems Corporation and owns both stock and stock appreciation rights in the company.
-
•
Rebecca Koscik has no disclosures.
-
•
Howard Rowley is a consultant for GE HealthCare and Bracco.
-
•
Nathaniel Chin has no disclosures.
-
•
Henrik Zetterberg has served at scientific advisory boards for Roche Diagnostics, Wave, CogRx and Samumed and is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg.
-
•
Kai Blennow has served as a consultant or at advisory boards for Alector, Alzheon, CogRx, Biogen, Lilly, Novartis and Roche Diagnostics, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Venture-based platform company at the University of Gothenburg.
-
•
Cynthia Carlsson is a site PI for the NIH/Eli Lilly (A4 Study), and will be receiving a stipend for participating in a one-time Roche Advisory Board meeting.
-
•
Sanjay Asthana has received grants from Eisai Pharmaceutical, Toyoma, Merck Pharmaceutical, and Lundbeck.
-
•
Barbara Bendlin has no disclosures.
-
•
Sterling Johnson has served at scientific advisory boards for Roche Diagnostics.
Footnotes
Results were unchanged when only A + T+ AD individuals and their matched A-T- CU counterparts (n = 36) were included in the analyses.
References
- Albert M.S., DeKosky S.T., Dickson D., Dubois B., Feldman H.H., Fox N.C., Snyder P.J. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulos P., Kriett L., Haller B., Klupp E., Gray K., Grimmer T., Drzezga A. Limited agreement between biomarkers of neuronal injury at different stages of Alzheimer's disease. Alzheimers Dement. 2014;10(6):684–689. doi: 10.1016/j.jalz.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Aschenbrenner A.J., Gordon B.A., Benzinger T.L., Morris J.C., Hassenstab J.J. Influence of tau PET, amyloid PET, and hippocampal volume on cognition in Alzheimer disease. Neurology. 2018;91(9):e859–e866. doi: 10.1212/WNL.0000000000006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J., Barnes G., Chen C., Daunizeau J., Flandin G., Friston K.…Penny W. SPM12 manual. 2016. http://www.fil.ion.ucl.ac.uk/spm/doc/spm12manual.pdf URL.
- Atiya M., Hyman B.T., Albert M.S., Killiany R. Structural magnetic resonance imaging in established and prodromal Alzheimer disease: a review. Alzheimer Dis. Assoc. Disord. 2003;17(3):177–195. doi: 10.1097/00002093-200307000-00010. [DOI] [PubMed] [Google Scholar]
- Bittner T., Zetterberg H., Teunissen C.E., Ostlund R.E., Jr., Militello M., Andreasson U., Eichenlaub U. Technical performance of a novel, fully automated electrochemiluminescence immunoassay for the quantitation of β-amyloid (1–42) in human cerebrospinal fluid. Alzheimers Dement. 2016;12(5):517–526. doi: 10.1016/j.jalz.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Bouwman F.H., Schoonenboom S.N.M., van Der Flier W.M., Van Elk E.J., Kok A., Barkhof F.…Scheltens P. CSF biomarkers and medial temporal lobe atrophy predict dementia in mild cognitive impairment. Neurobiol. Aging. 2007;28(7):1070–1074. doi: 10.1016/j.neurobiolaging.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Breslow N. A generalized Kruskal-Wallis test for comparing K samples subject to unequal patterns of censorship. Biometrika. 1970;57(3):579–594. [Google Scholar]
- Brickman A.M., Tosto G., Gutierrez J., Andrews H., Gu Y., Narkhede A.…Vonsattel J.P. An MRI measure of degenerative and cerebrovascular pathology in Alzheimer disease. Neurology. 2018 doi: 10.1212/WNL.0000000000006310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner R.L., Head D., Parker J., Fotenos A.F., Marcus D., Morris J.C., Snyder A.Z. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. NeuroImage. 2004;23(2):724–738. doi: 10.1016/j.neuroimage.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Clark L.R., Berman S.E., Norton D., Koscik R.L., Jonaitis E., Blennow K., Zetterberg H. Age-accelerated cognitive decline in asymptomatic adults with CSF β-amyloid. Neurology. 2018 doi: 10.1212/WNL.0000000000005291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff N. Dominance statistics: ordinal analyses to answer ordinal questions. Psychol. Bull. 1993;114(3):494. [Google Scholar]
- Davatzikos C., Bhatt P., Shaw L.M., Batmanghelich K.N., Trojanowski J.Q. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol. Aging. 2011;32(12):2322.e19–2322.e27. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R.S., Ségonne F., Fischl B., Quinn B.T., Dickerson B.C., Blacker D., Hyman B.T. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dickerson B.C., Wolk D.A. MRI cortical thickness biomarker predicts AD-like CSF and cognitive decline in normal adults. Neurology. 2012;78(2):84–90. doi: 10.1212/WNL.0b013e31823efc6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson B.C., Stoub T.R., Shah R.C., Sperling R.A., Killiany R.J., Albert M.S., Blacker D. Alzheimer-signature MRI biomarker predicts AD dementia in cognitively normal adults. Neurology. 2011;76(16):1395–1402. doi: 10.1212/WNL.0b013e3182166e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D.G., Kim R.E., Oguz I., Johnson H.J. Wrapping FreeSurfer 6 for use in high-performance computing environments. Gigascience. 2016;5(Suppl_1):16. [Google Scholar]
- Fagan A.M., Head D., Shah A.R., Marcus D., Mintun M., Morris J.C., Holtzman D.M. Decreased cerebrospinal fluid Aβ42 correlates with brain atrophy in cognitively normal elderly. Ann. Neurol. 2009;65(2):176–183. doi: 10.1002/ana.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. NeuroImage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., Busa E., Albert M., Dieterich M., Haselgrove C.…Klaveness S. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fleiss J.L. Measuring nominal scale agreement among many raters. Psychol. Bull. 1971;76(5):378. [Google Scholar]
- Fleiss J.L., Levin B., Paik M.C. John Wiley & Sons; 2013. Statistical Methods for Rates and Proportions. [Google Scholar]
- Fox N.C., Warrington E.K., Rossor M.N. Serial magnetic resonance imaging of cerebral atrophy in preclinical Alzheimer's disease. Lancet. 1999;353(9170):2125. doi: 10.1016/S0140-6736(99)00496-1. [DOI] [PubMed] [Google Scholar]
- Frisoni G.B., Fox N.C., Jack C.R., Scheltens P., Thompson P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010;6(2):67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T., Korosec F.R., Block W.F., Fain S.B., Turk Q., Lum D.…Mistretta C.A. PC VIPR: a high-speed 3D phase-contrast method for flow quantification and high-resolution angiography. Am. J. Neuroradiol. 2005;26(4):743–749. [PMC free article] [PubMed] [Google Scholar]
- Hansson O., Zetterberg H., Buchhave P., Londos E., Blennow K., Minthon L. Association between CSF biomarkers and incipient Alzheimer's disease in patients with mild cognitive impairment: a follow-up study. The Lancet Neurology. 2006;5(3):228–234. doi: 10.1016/S1474-4422(06)70355-6. [DOI] [PubMed] [Google Scholar]
- Hesse C., Rosengren L., Andreasen N., Davidsson P., Vanderstichele H., Vanmechelen E., Blennow K. Transient increase in total tau but not phospho-tau in human cerebrospinal fluid after acute stroke. Neurosci. Lett. 2001;297(3):187–190. doi: 10.1016/s0304-3940(00)01697-9. [DOI] [PubMed] [Google Scholar]
- Jack C.R., Petersen R.C., O'Brien P.C., Tangalos E.G. MR-based hippocampal volumetry in the diagnosis of Alzheimer's disease. Neurology. 1992;42(1):183–188. doi: 10.1212/wnl.42.1.183. [DOI] [PubMed] [Google Scholar]
- Jack C.R., Wiste H.J., Weigand S.D., Knopman D.S., Mielke M.M., Vemuri P., Machulda M.M. Different definitions of neurodegeneration produce similar amyloid/neurodegeneration biomarker group findings. Brain. 2015;138(12):3747–3759. doi: 10.1093/brain/awv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Bennett D.A., Blennow K., Carrillo M.C., Feldman H.H., Frisoni G.B., Knopman D.S. A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology. 2016;87(5):539–547. doi: 10.1212/WNL.0000000000002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Wiste H.J., Weigand S.D., Therneau T.M., Knopman D.S., Lowe V., Senjem M.L. Age-specific and sex-specific prevalence of cerebral β-amyloidosis, tauopathy, and neurodegeneration in cognitively unimpaired individuals aged 50–95 years: a cross-sectional study. The Lancet Neurology. 2017;16(6):435–444. doi: 10.1016/S1474-4422(17)30077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B.…Karlawish J. NIA-AA research framework: toward a biological definition of Alzheimer's disease. Alzheimer's & Dementia. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.C., Koscik R.L., Jonaitis E.M., Clark L.R., Mueller K.D., Berman S.E.…Hogan K.J. The Wisconsin registry for Alzheimer's prevention: a review of findings and current directions. Alzheimer's & Dementia. 2018;10:130–142. doi: 10.1016/j.dadm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapasi A., DeCarli C., Schneider J.A. Impact of multiple pathologies on the threshold for clinically overt dementia. Acta Neuropathol. 2017;134(2):171–186. doi: 10.1007/s00401-017-1717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H.B., Whitney D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947;18(1):50–60. [Google Scholar]
- Manolio T.A., Kronmal R.A., Burke G.L., Poirier V., O'Leary D.H., Gardin J.M., Bryan R.N. Magnetic resonance abnormalities and cardiovascular disease in older adults. The cardiovascular health study. Stroke. 1994;25(2):318–327. doi: 10.1161/01.str.25.2.318. [DOI] [PubMed] [Google Scholar]
- Mattsson N., Insel P.S., Landau S., Jagust W., Donohue M., Shaw L.M., Weiner M. Diagnostic accuracy of CSF Ab42 and florbetapir PET for Alzheimer's disease. Annals of Clinical and Translational Neurology. 2014;1(8):534–543. doi: 10.1002/acn3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H.…Mayeux R. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's & Dementia: The Journal of the Alzheimer's Association. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeter L.H., Vijverberg E.G., Del Campo M., Rozemuller A.J., Kaat L.D., de Jong F.J.…Pijnenburg Y.A. Clinical value of neurofilament and phospho-tau/tau ratio in the frontotemporal dementia spectrum. Neurology. 2018;90(14):e1231–e1239. doi: 10.1212/WNL.0000000000005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.C. The clinical dementia rating (CDR): current version and scoring rules. Neurology. 1993;43(11):2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- O'Brien L.M., Ziegler D.A., Deutsch C.K., Frazier J.A., Herbert M.R., Locascio J.J. Statistical adjustments for brain size in volumetric neuroimaging studies: some practical implications in methods. Psychiatry Res. Neuroimaging. 2011;193(2):113–122. doi: 10.1016/j.pscychresns.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguz I., Cates J., Fletcher T., Whitaker R., Cool D., Aylward S., Styner M. Biomedical Imaging: From Nano to Macro, 2008. ISBI 2008. 5th IEEE International Symposium. vols. 1637-1640. 2008, May. Cortical correspondence using entropy-based particle systems and local features. [Google Scholar]
- Olsson B., Lautner R., Andreasson U., Öhrfelt A., Portelius E., Bjerke M., Strobel G. CSF and blood biomarkers for the diagnosis of Alzheimer's disease: a systematic review and meta-analysis. The Lancet Neurology. 2016;15(7):673–684. doi: 10.1016/S1474-4422(16)00070-3. [DOI] [PubMed] [Google Scholar]
- Patenaude B., Smith S.M., Kennedy D.N., Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold A. Neurofilament phosphoforms: surrogate markers for axonal injury, degeneration and loss. J. Neurol. Sci. 2005;233(1):183–198. doi: 10.1016/j.jns.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Price J.L., Morris J.C. Tangles and plaques in nondemented aging and "preclinical" Alzheimer's disease. Ann. Neurol. 1999;45(3):358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Price J.L., McKeel D.W., Buckles V.D., Roe C.M., Xiong C., Grundman M., Dickson D.W. Neuropathology of nondemented aging: presumptive evidence for preclinical Alzheimer disease. Neurobiol. Aging. 2009;30(7):1026–1036. doi: 10.1016/j.neurobiolaging.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N., Gunning F.M., Head D., Dupuis J.H., McQuain J., Briggs S.D.…Acker J.D. Selective aging of the human cerebral cortex observed in vivo: differential vulnerability of the prefrontal gray matter. Cereb. Cortex. 1997;7(3):268–282. doi: 10.1093/cercor/7.3.268. [DOI] [PubMed] [Google Scholar]
- Salat D.H., Lee S.Y., Van der Kouwe A.J., Greve D.N., Fischl B., Rosas H.D. Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. NeuroImage. 2009;48(1):21–28. doi: 10.1016/j.neuroimage.2009.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scahill R.I., Schott J.M., Stevens J.M., Rossor M.N., Fox N.C. Mapping the evolution of regional atrophy in Alzheimer's disease: unbiased analysis of fluid-registered serial MRI. Proc. Natl. Acad. Sci. 2002;99(7):4703–4707. doi: 10.1073/pnas.052587399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott J.M., Bartlett J.W., Fox N.C., Barnes J. Increased brain atrophy rates in cognitively normal older adults with low cerebrospinal fluid Aβ1-42. Ann. Neurol. 2010;68(6):825–834. doi: 10.1002/ana.22315. [DOI] [PubMed] [Google Scholar]
- Schwarz C.G., Gunter J.L., Wiste H.J., Przybelski S.A., Weigand S.D., Ward C.P., Dickson D.W. A large-scale comparison of cortical thickness and volume methods for measuring Alzheimer's disease severity. NeuroImage. 2016;11:802–812. doi: 10.1016/j.nicl.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren M., Vanderstichele H., Ågren H., Zachrisson O., Edsbagge M., Wikkelsø C.…Marcusson J. Tau and Aβ42 in cerebrospinal fluid from healthy adults 21–93 years of age: establishment of reference values. Clin. Chem. 2001;47(10):1776–1781. [PubMed] [Google Scholar]
- Skillbäck T., Rosén C., Asztely F., Mattsson N., Blennow K., Zetterberg H. Diagnostic performance of cerebrospinal fluid total tau and phosphorylated tau in Creutzfeldt-Jakob disease: results from the Swedish mortality registry. JAMA Neurology. 2014;71(4):476–483. doi: 10.1001/jamaneurol.2013.6455. [DOI] [PubMed] [Google Scholar]
- Sluimer J.D., van der Flier W.M., Karas G.B., van Schijndel R., Barnes J., Boyes R.G., Scheltens P. Accelerating regional atrophy rates in the progression from normal aging to Alzheimer's disease. Eur. Radiol. 2009;19(12):2826. doi: 10.1007/s00330-009-1512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldan A., Pettigrew C., Fagan A.M., Schindler S.E., Moghekar A., Fowler C., Masters C.L. ATN profiles among cognitively normal individuals and longitudinal cognitive outcomes. Neurology. 2019;92(14):e1567–e1579. doi: 10.1212/WNL.0000000000007248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storandt M., Mintun M.A., Head D., Morris J.C. Cognitive decline and brain volume loss as signatures of cerebral amyloid-β peptide deposition identified with Pittsburgh compound B: cognitive decline associated with Aβ deposition. Arch. Neurol. 2009;66(12):1476–1481. doi: 10.1001/archneurol.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo J.B., Weiner M.W., Wolk D.A., Da X., Chen K., Arnold S.E.…Shaw L.M. Neuronal injury biomarkers and prognosis in ADNI subjects with normal cognition. Acta Neuropathologica Communications. 2014;2(1):26. doi: 10.1186/2051-5960-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maurik I.S., Zwan M.D., Tijms B.M., Bouwman F.H., Teunissen C.E., Scheltens P., van der Flier W.M. Interpreting biomarker results in individual patients with mild cognitive impairment in the Alzheimer's biomarkers in daily practice (ABIDE) project. JAMA Neurology. 2017;74(12):1481–1491. doi: 10.1001/jamaneurol.2017.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum I.A., Vos S.J., Burns L., Knol D.L., Scheltens P., Soininen H., Minthon L. Injury markers predict time to dementia in subjects with MCI and amyloid pathology. Neurology. 2012;79(17):1809–1816. doi: 10.1212/WNL.0b013e3182704056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandijck M., Dauwe M., Huyck E., Le Bastard N., Lawson J., Traynham C.…Jannes G. LUMIPULSE® G total tau: key performances of a fully automated chemiluminescent immunoassay. Alzheimer's & Dementia. 2017;13(7) [Google Scholar]
- Vemuri P., Wiste H.J., Weigand S.D., Shaw L.M., Trojanowski J.Q., Weiner M.W.…Initiative A.D.N. MRI and CSF biomarkers in normal, MCI, and AD subjects predicting future clinical change. Neurology. 2009;73(4):294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos S.J., Gordon B.A., Su Y., Visser P.J., Holtzman D.M., Morris J.C., Benzinger T.L. NIA-AA staging of preclinical Alzheimer disease: discordance and concordance of CSF and imaging biomarkers. Neurobiol. Aging. 2016;44:1–8. doi: 10.1016/j.neurobiolaging.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlye L.T., Walhovd K.B., Dale A.M., Espeseth T., Reinvang I., Raz N., Fjell A.M. Increased sensitivity to effects of normal aging and Alzheimer's disease on cortical thickness by adjustment for local variability in gray/white contrast: a multi-sample MRI study. NeuroImage. 2009;47(4):1545–1557. doi: 10.1016/j.neuroimage.2009.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston P.S., Poole T., Ryan N.S., Nair A., Liang Y., Macpherson K., Klimova J. Serum neurofilament light in familial Alzheimer disease: a marker of early neurodegeneration. Neurology. 2017;89(21):2167–2175. doi: 10.1212/WNL.0000000000004667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg H. Neurofilament light: a dynamic cross-disease fluid biomarker for neurodegeneration. Neuron. 2016;91(1):1–3. doi: 10.1016/j.neuron.2016.06.030. [DOI] [PubMed] [Google Scholar]
- Zetterberg H., Skillbäck T., Mattsson N., Trojanowski J.Q., Portelius E., Shaw L.M., Blennow K. Association of cerebrospinal fluid neurofilament light concentration with Alzheimer disease progression. JAMA Neurology. 2016;73(1):60–67. doi: 10.1001/jamaneurol.2015.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow diagram depicting eligibility for the current study. Arrows pointing downward indicate number of participants excluded at each decision point. Arrows pointing upward indicate number of participants included at each decision point.
Adjusting for age (see text for more details), A-D represent box plots for each metric comparing the different groups (CU A-T-, CU A+T-, CU A+T+, MCI A+, AD A+); A) CSF NfL; B) the AD signature; C) hippocampal volume; D) global atrophy.