Abstract
Breast imaging is a multimodal approach that plays an essential role in the diagnosis of breast cancer. Mammography, sonography, magnetic resonance, and image-guided biopsy are imaging techniques used to search for malignant changes in the breast or precursors of malignant changes in, e.g., screening programs or follow-ups after breast cancer treatment. However, these methods still have some disadvantages such as interobserver variability and the mammography sensitivity in women with radiologically dense breasts. In order to overcome these difficulties and decrease the number of false positive findings, improvements in imaging analysis with the help of artificial intelligence are constantly being developed and tested. In addition, the extraction and correlation of imaging features with special tumor characteristics and genetics of the patients in order to get more information about treatment response, prognosis, and also cancer risk are coming more and more in focus. The aim of this review is to address recent developments in digital analysis of images and demonstrate their potential value in multimodal breast imaging.
Keywords: Breast cancer, Big data, Computer analysis, Artificial intelligence
Introduction
In Europe, breast cancer (BC) is the leading cause of death from cancer in women, and it is the most commonly diagnosed cancer worldwide except in Eastern Africa. The prevalence of BC has been increasing due to the possibility of early BC diagnosis, population aging, and changes in risk factors, but independent of further development of new treatment options, early diagnosis is very important as it increases the chances of survival [1].
Breast imaging is a subspecialty in the diagnostic field of radiology, which involves several imaging techniques for the early detection of BC. It also plays a fundamental role in the assessment and follow-up of BC [2, 3, 4, 5]. It requires a multimodal approach that includes mammography (MG) as a gold standard supplemented by sonography (US) and dynamic contrast-enhanced magnetic resonance tomography (DCE-MRI). These techniques are used to search for malignant changes in the breast, e.g., in the screening program, intensified screening, or follow-up monitoring [6, 7].
Although breast imaging is currently the most effective tool for detecting BC, there are still deficits in the detection and interpretation of lesions by the human reader [8]. In addition, the number of dedicated breast imager is limited. For this reason, improvements in breast imaging and image-based features are crucial and currently a very active topic in radiological research. Image analysis with the help of computers has shown that computerized feature extraction and new algorithms classification are great tools to assist physicians in the detection and classification of abnormalities [9]. Moreover, they are playing an increasingly important role in predicting clinical prognosis [10].
This review addresses recent developments in digital analysis and their potential value in the context of multimodal breast imaging.
Mammography
A mammogram is an X-ray method for breast imaging that has been constantly evolving in recent years from a film-based analog MG in the 1960s to full-field digital techniques. While analog MG can lead to an inaccurate diagnosis or false positive results, digital MG is more robust and superior, especially in lesion detection in dense breasts or premenopausal woman [11]. In addition, the radiation exposure of digital MG is around 20–30% lower than screen film for MG [12]. Furthermore, the images can be subsequently altered and easily exchanged electronically.
Since to detect an ion of masses is usually not the biggest challenge for radiologists, if they are not obscured by overlying dense breast tissue, the interpretation is trickier. On the other hand, the detection of small microcalc clusters after a long day or reading a big batch of images in screening can become difficult due to lack of concentration. In order to support the radiologists' detection rate and interpretation of breast imaging, innovative image-based features such as computer-assisted diagnostic (CAD) systems have been developed. CAD systems identify abnormalities on the breast image and can be used after or before the radiological interpretation of the image [13, 14, 15, 16]. Even though these systems can be applied on digitized analog images, the development of digital MG made it much easier to apply these computer tools and will also enable artificial intelligence algorithm in the future.
Magnetic Resonance-Mammography
MRI [17] is usually used as a complement to X-ray MG for the diagnosis of BC. It offers the advantage of providing not only 3-dimensional spatial information but also temporal information. DCE-MRI in breast imaging is considered to be the technique with the highest sensitivity, exceeding 90% [18]. Current data also confirm high accuracies in the detection of ductal carcinomas in situ or the diagnosis of lesions presenting with microcalcifications [19, 20, 21, 22].
MRI allows the acquisition of morphological and functional information of breast tissue and breast lesions and the evaluation of pathological features. Functional information can be obtained using two different techniques: dynamic and morphological enhancement assessment [23] and diffusion-weighted imaging (DWI) [24, 25]. The contrast dynamic technique is based on different enhancement patterns of breast lesions over time after the administration of a contrast agent [26, 27, 28]. DWI, on the other hand, is a technique that measures the diffusion of water molecules in tissue and does not require a contrast medium. Due to the water molecules' mobility, DWI reflects biophysical properties such as cell density, integrity of the membrane, and tissue microstructure [25, 29]. MRI, however, is a method that requires a high level of experience, which may be limited [30, 31]. Besides, image interpretation can show inter- and intraobserver variations [32]. Therefore, CAD programs were also developed for MRI in order to detect and clarify suspicious image features, to assist radiologists in distinguishing lesions, and to save time [33, 34, 35, 36].
Computer Assistance for Image-Based Diagnosis of BC
The long history of CAD systems began between 1960 and 1970, when research studies presented a new automatic computer model that was able to detect and classify suspicious abnormalities [37]. It the 1980s, medical physicists and physicians at the Kurt Rossmann Laboratories (Department of Radiology at the University of Chicago) developed two comprehensive categories of computer models: computer-aided diagnosis (CADx) and computer-assisted detection (CADe) [38].
CAD systems analyze the mammographic images and mark abnormal densities and microcalcifications assisting the radiologist to incorporate the machine results into their decision-making process. The initial systems had a high rate of false positive marks, and not every suspicious finding was safely marked [39]. Therefore, CAD systems should only be used as support for radiologists; the responsibility lies solely with the physician. CAD systems are nowadays widely accepted and used throughout the field of radiology [16, 40]. The image analysis of CAD systems is based on image acquisition, segmentation, processing, detection, and output [15, 16, 41]. CAD can be applied in screen-film MG as well as digital MG. However, conventional images must be digitized first, as CAD systems can only analyze digital datasets [42].
Emerging studies introduce new CAD systems based on neural networks. These innovative systems have the supplementary ability to use already collected information as a pool of data in order to compare with new mammograms [43]. With the new technology, researchers hope that the new CAD method will replace the second imaging reader in the future [44, 45].
The CAD system in MG is used as an assistant to the radiologist by enhancing potential findings that may get overlooked and by reducing the time of image interpretation. CAD in MG is advantageous when there is a high inconsistency among observers, a lack of trained observers, or double reading with two or more radiologists is impractical [46]. Clinical studies have shown that the addition of CAD leads to a sensitivity increase from 20 to 21% in the detection of BC by radiologists [46, 47, 48, 49].
CAD for breast MRI has a different function from its application in MG. It is a component in the daily routine that serves more like a computer-assisted evaluation and visualization tool than only as a lesion detection [33]. Radiologists usually detect the lesions easily, but only the most experienced ones can safely assess the lesion [50]. MRI interpretation requires knowledge of the morphology and kinetics of the lesion as well as its enhancement patterns. Malignant tumors tend to enhance and wash out quickly. An abnormal suspicious morphology of a lesion added to type III kinetics [34] indicates malignancy and increases the likelihood of biopsy.
CAD is a system able to organize this multifaceted image information, reducing the number of false positives and, thereby, unnecessary biopsies [51]. In addition, it has been shown that CAD may also be useful for assessing changes in enhancement patterns of tumors after chemotherapy [52].
Breast Density Assessment
In order to be able to predict the individual risk of women to develop BC, researchers' interest in imaging risk indicators, such as breast density, fibroglandular tissue (FGT), and background parenchymal enhancement (BPE) after approximately 90 s, has increased in recent years. The assessment of the density of the breast using the BI-RADS lexicon quite often shows a significant variability among readers. Therefore, more objective measurement tools would be helpful. Automatic assessment of volumetric percentage density using dedicated software reduces these variabilities. Studies have shown that mammographic percent density, BPE, and FGT are independent imaging biomarkers as they are also risk factors for the development of BC [53, 54, 55].
In addition, several research studies assessed whether a computer-assisted program would also find less BC due to increased tissue density of the breast: Bolivar et al. [56] presented a significant difference in sensitivity between the nondense breasts compared to dense breasts (95 vs. 89%) after using a CAD system. In contrast, Brem et al. [47], Birdwell et al. [57], and Yang et al. [58] showed no impact of breast density on the performance of CAD systems. Engelken et al. [39] showed an influence of the volume of breast tissue but not its percentage on the performance of a CAD system. However, the groups used different CAD systems and partly digital partly screen-film mammograms.
Breast density in MRI is a volumetric assessment of FGT, since fat and FGT are two major components of breast MRI. It is strongly correlated with mammographic percent density [59, 60] and associated with BC risk [55]. BPE in MRI is recognized as enhancement of normal breast tissue to varying degrees after injection of the contrast agent [61, 62]. Breast density, FGT, and BPE may be altered as they represent dynamic physiological processes subject to hormonal changes [63]. In order to assess these imaging biomarkers, radiologists use a 4-point scale that qualifies the breast density according to a visual criterion established by the American College of Radiology: Breast Imaging Reporting and Data System (BI-RADS) criteria. In MRI, BI-RADS assessment should occur on the first image after contrast injection at approximately 90 s. However, the manual assessment of BI-RADS can lead to inconsistencies between inter- and intrareaders and obstructs the prediction of BC [64, 65]. To overcome these limitations, other algorithms are being developed [66, 67].
The initial computer-assisted systems and software algorithms where using more basic techniques and were limited by the performance speed of the available computer technique at that time. But in the last years, more and more sophisticated computer techniques using neuronal networks, deep learning (DL), and artificial intelligence came more and more into focus. These techniques also enable researchers to extract multiple imaging features and correlate them to biological markers to predict, for example, the type and behavior of a tumor. These techniques are described in more detail in the following section.
Radiomics and DL
Radiomics is a technique that converts radiological imaging data, such as CT or MR images, into a high-dimensional feature space, where the term feature refers to a scalar number that encodes a specific property of the tumor. This feature space is hypothesized to contain a link to a clinical or biological endpoint – such as tumor (sub-)type, prognosis, or treatment response. Computational modelling and machine learning (ML) approaches are often used in a hypothesis-generating approach to link the high-dimensional image feature space to a specific endpoint. As such, radiomics can be seen as the radiological interpretation of the systems biology approach [68]: Systems biology is the study of complex biologic systems, which relies on integrating multifaceted data about genes, metabolism, protein expression, and other components. Systems biology approaches encompass data from many scientific disciplines and form an integral part of precision medicine.
Technical Introduction
Radiomics was introduced in a ground-breaking work by Aerts et al. [69]. This work demonstrated in independent CT datasets of patients with lung or head-and-neck cancer that radiomics features, generated by automated image analysis, contain prognostic information. Fundamentally, this work demonstrated that imaging phenotype becomes accessible to computational analysis by the proposed approach termed “radiomics,” which is described in the following subsections.
This initial paper was followed by an abundance of publications which investigated radiomics approaches for the analysis of CT and MR data, with the clinical endpoints of classification, e.g., of tumor types or the presence or absence of genetic subtypes, or progression-free or overall survival [70]. It is noteworthy that the majority of radiomics studies has focused on oncologic questions, although, in principle, the radiomics approach is feasible for all questions that can be formulated as classification, regression, or survival analysis. For the sake of simplicity, the following discussion is limited to radiomics analysis of tumors.
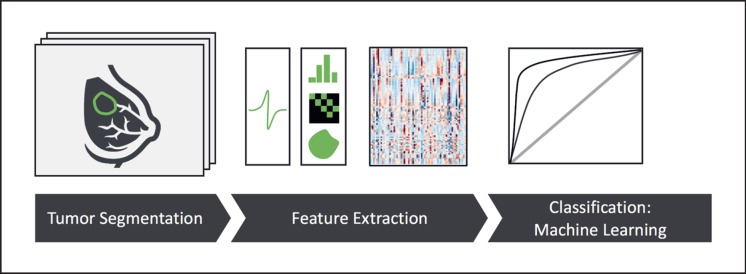
Figure 1 outlines a typical radiomics analysis. Starting with high-quality tomographic images, the tumor is segmented or contoured on each image slice, resulting in a 3-dimensional definition of the tumor shape. In the initial radiomics studies, this tumor segmentation was performed manually by expert readers, typically radiologists with sufficient domain expertise. Depending on image contrast, tumor boundaries, and image resolution, this manual segmentation can be an extremely time-consuming and user-dependent task.
Fig. 1.
Outline of a state-of-the-art radiomics analysis [88]. Briefly, a tumor is outlined or contoured on tomographic images. In the next step, a range of quantitative image features is computed, typically comprising shape features, signal intensity distributions, and image texture. Finally, machine learning is used to establish a link between this high-dimensional data space and the relevant clinical endpoint or outcome.
Once the tumor is segmented, radiomics analysis proceeds by calculating a large number of quantitative image features, which can roughly be categorized into three groups. The first group of features describes tumor shape and encompasses simple and well-understood parameters such as maximum diameter, tumor volume, or tumor surface area. Often, additional parameters such as sphericity, the ratio of surface to volume, or other parameters are calculated. The second group of image features describes the histogram of signal intensities within the tumor. Again, this encompasses simple and straightforward parameters such as mean or median, minimum or maximum signal intensity, but also energy, skewness, and kurtosis of the histogram. With these groups of features, the tumor is already well characterized, but the relation of neighboring pixels is not taken into account. To this end, the third group of features comprises image texture, which essentially captures the spatial relations of different gray levels, by means of gray-level co-occurrence or gray-level run-length matrices. Notably, these texture features are able to capture various aspects of tumor heterogeneity, a measure which often has strong prognostic relevance.
After feature extraction, imaging data are converted into a long vector of image features. These data can be readily enhanced with additional information, such as demographic, clinical, or pathological data, or even additional -omics data, e.g., from genomics. This wealth of data, however, should be interpreted with caution: it may well be that a large number of these features is of little use for endpoint prediction. For example, it may be that some image features are highly sensitive with respect to image segmentation, so that feature changes are dominated by small changes in segmentation, as, for example, caused by interreader variability. Other features may have no association with the outcome at all, and again others may have high intercorrelation and, thus, act as confounders in further analysis. For this reason, the high-dimensional feature space is often narrowed down by means of feature selection, which may employ a large number of feature quality measures to narrow down the number of predictors.
The link between feature space and clinical endpoint then needs to be provided either by statistical modeling or ML, often in a classification or survival setting. In ML terms, this problem can be formulated as “supervised learning:” for each patient, the feature vector and the associated label, i.e., the clinical endpoint, is known; the ML model needs to establish the link between the two entities, which is performed in a so-called training step. After training, the model can then predict an outcome for new hitherto unseen patient data. During training, special care needs to be taken in order to arrive at meaningful predictions – otherwise, a model might simply memorize training data and be unable to predict on new data. A popular strategy for this purpose is to split the data into training and testing data; during training, the model sees only the training data, and after training, the model performance is evaluated on the testing data. A generalization of this approach is cross validation: here, the data is divided into several folds, one fold is held out during training and used only for prediction. This is then permuted, so that predictions for each fold are based on training with all other folds.
Applications in BC
The key application of radiomics in oncological settings is the prediction of a clinical endpoint. In the context of BC, radiomics applications have up to now focused on the characterization of suspicious lesions, on the characterization of tumors, and on prognosis and risk prediction. In the following, we present key studies in these fields. For a more comprehensive overview, please refer, for example, to Pinker et al. [68] or Crivelli et al. [71]. It should be noted, however, that most of these studies have been performed in a retrospective fashion and require validation in prospective studies.
The diagnostic workup of suspicious lesions on conventional MG is a central application of MRI in BC. The additional value of radiomics feature as compared to the maximum linear size was evaluated by Whitney et al. [72], their study demonstrated that radiomics features improved the ability to distinguish benign lesions from luminal A BCs. A distinguishing feature of MRI is its ability to provide information beyond pure morphology, a very useful application is imaging of water diffusion in tissue, based on the notion that densely packed tumor cells provide more diffusion restriction than healthy tissue and, thus, appear dark in so-called apparent diffusion coefficient images. Radiomics analyses of diffusion-weighted MRI in BC have been shown to be superior to mere mean apparent diffusion coefficient values in the discrimination between benign and malignant lesions [73].
In a prospective bi-centric study [74], a specialized DWI protocol was used for imaging patients with suspicious lesions detected on X-ray MG. Here, the radiomics analysis was demonstrated to have higher specificity than conventional image analysis, even with MR scanners from different vendors. Radiomics has also been used to differentiate BC molecular subtypes. Here, it was demonstrated that a quantitative MRI radiomics approach may be very helpful for the molecular classification of invasive BC – e.g., the differentiation of ER+ versus ER– or HER2+ versus HER2– receptor status [75]. In a similar approach, radiomics analysis of DCE-MRI was used to differentiate luminal A, luminal B, HER2, and basal-like subtypes [76].
With respect to prognosis, Li et al. [77] aimed to use MRI features to predict BC recurrence scores from multigene assays. The findings of their study suggested that radiomics features used for image-based phenotyping may be helpful in assessing the risk of cancer recurrence. With respect to the endpoint of disease-free survival, Park et al. [78] demonstrated that a radiomics signature from preoperative MRI, termed “Rad-Score,” has prognostic value. In particular, when combined with clinicopathological and MRI findings, this signature improved the estimation of disease-free survival, suggesting that the radiomics signature provides additional prognostic value. Finally, a recent study investigated the potential of radiomics to predict pathological complete response of patients with locally advanced BC under neoadjuvant chemotherapy [79, 80].
Deep Learning
In contrast to radiomics, where image features are “handcrafted,” DL approaches promise to learn relevant image features on their own. By combining image analysis and ML in one network, convolutional neural networks (CNN) have had a disruptive effect on a large number of ML approaches, in particular with respect to computer vision and image analysis. This has raised considerable interest in the medical imaging community, where such approaches have been shown to provide value in a large number of clinical applications [81]. While DL approaches are useful for a number of tasks in medical imaging, we limit the brief overview below to the application of DL for the classification of conventional MG. For a detailed discussion and a review of recent publications, please refer to Hamidinekoo et al. [82].
On MG images, a tumor may appear as mass, through the presence of microcalcifications or through distortion of tissue. These features can readily be detected through feature extraction by dedicated conventional CAD systems (see above), but these approaches have been challenged through the rise of DL approaches for classification. In a recent study [83], a DL system was trained and evaluated on a large dataset of approx. 45,000 images; the system's performance to detect cancer was compared against a state-of-the-art MG CAD system and against human readers. This study found that the CNN approach outperformed the CAD system at low sensitivity and yielded comparable results at high sensitivity; there was no significant difference in performance between the CNN and human readers.
A critical requirement for training DL algorithms is the availability of sufficient amounts of training data, where the relevant clinical endpoint is available as label to each dataset. This has been a considerable limitation of the application of DL algorithms in medical imaging, which has recently been mitigated through several challenges, where large amounts of labeled data were made available to the public; participants in these challenges trained their algorithms on the public data and submitted them to the competition. For an unbiased evaluation of these trained algorithms, their performance was determined on a held-back validation dataset, which determined the position of the algorithm on the so-called private leaderboard. In the field of MG, an influential challenge was the DREAM challenge which aimed to assess algorithms for risk stratification in screening MG [84]. In this challenge, 86,000 exams were made available, together with a binary label for each image, stating whether BC was diagnosed within 12 months after diagnosis. The winning algorithms achieved AUC values of 0.874 and 0.843, respectively. The second-best algorithm [85] not only predicted the diagnosis of BC on the breast level, but it was also able to localize and detect lesions. As a detector system, this algorithm achieved high sensitivity with very few false positive marks.
Outlook and Perspectives
Imaging methods become more and more complex, promising individualized treatment options arise, and the number of BC patients is increasing – this highlights the crucial role of correct assessment and interpretation of imaging data. At the same time, the workload of experienced breast radiologists increases and the recruitment of new specialists becomes difficult.
Whereas early CAD systems have focused on the mere detection of lesions, recent advances in radiomics and DL promise not only to detect, but also a complete lesion characterization, including tumor type specification and prognosis. Modern CAD systems have reached a reading level that is comparable to human readers, and radiologists should consider options and modalities how these systems can be incorporated into clinical care, especially in a screening setting – for example, CAD systems might replace a second human reader (they reached the same sensitivity as average screening readers in a retrospective study [86]) or preselect normal images to reduce the readers' workload [87].
In addition, much more information about the type of lesions we detect in our patients, the heterogeneity of tumors, and the differences in treatment response and prognosis is inherited in our images and has so far maybe been hidden to the human reader's eyes or is too complex to be completely assessed by the human read-only. In the past, CAD tools helped mainly to detect lesions but did not add much more information for their interpretation. With more advanced diagnostic techniques, like radiomics and DL, a translation from imaging extracted features into tumor-type specifications or prognosis prediction will come more and more into focus.
Computer systems may also be helpful for risk assessment and stratification in high-risk patients, taking into account breast density, tissue structure, background enhancement, or other image features; this is an active area of research. In this sense, the replacement debate loses momentum; instead, radiologists should appreciate and embrace the added value that artificial intelligence systems provide – quite possibly, these systems will relieve radiologists from routine work, will improve the level of patient-centered care, and, maybe most importantly, will enable the radiologist to focus on human interaction and individual treatment.
Disclosure Statement
The authors have nothing to disclose.
Funding Sources
No funding was provided for finalizing this paper.
Author Contributions
All authors of this paper contributed to the structure of the paper, were active in literature review, and careful correction of the draft.
References
- 1.Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018 Nov;103:356–87. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Mann RM, Balleyguier C, Baltzer PA, Bick U, Colin C, Cornford E, et al. European Society of Breast Imaging (EUSOBI), with language review by Europa Donna–The European Breast Cancer Coalition Breast MRI: EUSOBI recommendations for women's information. Eur Radiol. 2015 Dec;25((12)):3669–78. doi: 10.1007/s00330-015-3807-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sardanelli F, Fallenberg EM, Clauser P, Trimboli RM, Camps-Herrero J, Helbich TH, et al. European Society of Breast Imaging (EUSOBI), with language review by Europa Donna–The European Breast Cancer Coalition Mammography: an update of the EUSOBI recommendations on information for women. Insights Imaging. 2017 Feb;8((1)):11–8. doi: 10.1007/s13244-016-0531-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sardanelli F, Aase HS, Alvarez M, Azavedo E, Baarslag HJ, Balleyguier C, et al. Eur Radiol. 27. Germany. 2017. Position paper on screening for breast cancer by the European Society of Breast Imaging (EUSOBI) and 30 national breast radiology bodies from Austria, Belgium, Bosnia and Herzegovina, Bulgaria, Croatia, Czech Republic, Denmark, Estonia, Finland, France, Germany, Greece, Hungary, Iceland, Ireland, Italy, Israel, Lithuania, Moldova, The Netherlands, Norway, Poland, Portugal, Romania, Serbia, Slovakia, Spain, Sweden, Switzerland and Turkey; pp. p. 2737–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans A, Trimboli RM, Athanasiou A, Balleyguier C, Baltzer PA, Bick U, et al. European Society of Breast Imaging (EUSOBI), with language review by Europa Donna–The European Breast Cancer Coalition Breast ultrasound: recommendations for information to women and referring physicians by the European Society of Breast Imaging. Insights Imaging. 2018 Aug;9((4)):449–61. doi: 10.1007/s13244-018-0636-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bick U, Engel C, Krug B, Heindel W, Fallenberg EM, Rhiem K, et al. German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC) High-risk breast cancer surveillance with MRI: 10-year experience from the German consortium for hereditary breast and ovarian cancer. Breast Cancer Res Treat. 2019 May;175((1)):217–28. doi: 10.1007/s10549-019-05152-9. [DOI] [PubMed] [Google Scholar]
- 7.Nadler M, Al-Attar H, Warner E, Martel AL, Balasingham S, Zhang L, et al. MRI surveillance for women with dense breasts and a previous breast cancer and/or high risk lesion. Breast. 2017 Aug;34:77–82. doi: 10.1016/j.breast.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Kolb TM, Lichy J, Newhouse JH. Comparison of the performance of screening mammography, physical examination, and breast US and evaluation of factors that influence them: an analysis of 27,825 patient evaluations. Radiology. 2002 Oct;225((1)):165–75. doi: 10.1148/radiol.2251011667. [DOI] [PubMed] [Google Scholar]
- 9.Pinker K, Shitano F, Sala E, Do RK, Young RJ, Wibmer AG, et al. Background, current role, and potential applications of radiogenomics. J Magn Reson Imaging. 2018 Mar;47((3)):604–20. doi: 10.1002/jmri.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couture HD, Williams LA, Geradts J, Nyante SJ, Butler EN, Marron JS, et al. NPJ Breast Cancer. 4. United States. 2018. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype; p. p. 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pisano ED, Gatsonis C, Hendrick E, Yaffe M, Baum JK, Acharyya S, et al. Digital Mammographic Imaging Screening Trial (DMIST) Investigators Group Diagnostic performance of digital versus film mammography for breast-cancer screening. N Engl J Med. 2005 Oct;353((17)):1773–83. doi: 10.1056/NEJMoa052911. [DOI] [PubMed] [Google Scholar]
- 12.Hendrick RE, Pisano ED, Averbukh A, Moran C, Berns EA, Yaffe MJ, et al. Comparison of acquisition parameters and breast dose in digital mammography and screen-film mammography in the American College of Radiology Imaging Network digital mammographic imaging screening trial. AJR Am J Roentgenol. 2010 Feb;194((2)):362–9. doi: 10.2214/AJR.08.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert FJ, Astley SM, Gillan MG, Agbaje OF, Wallis MG, James J, et al. CADET II Group Single reading with computer-aided detection for screening mammography. N Engl J Med. 2008 Oct;359((16)):1675–84. doi: 10.1056/NEJMoa0803545. [DOI] [PubMed] [Google Scholar]
- 14.Karssemeijer N, Otten JD, Verbeek AL, Groenewoud JH, de Koning HJ, Hendriks JH, et al. Computer-aided detection versus independent double reading of masses on mammograms. Radiology. 2003 Apr;227((1)):192–200. doi: 10.1148/radiol.2271011962. [DOI] [PubMed] [Google Scholar]
- 15.Obenauer S, Hermann KP. CAD in der Mammadiagnostik: Einsatz bei Mammografie und MRT. Radiologie up2date. 2012;12((01)):55–72. [Google Scholar]
- 16.Yassin NI, Omran S, El Houby EM, Allam H. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: A systematic review. Comput Methods Programs Biomed. 2018 Mar;156:25–45. doi: 10.1016/j.cmpb.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Janky R, Verfaillie A, Imrichová H, Van de Sande B, Standaert L, Christiaens V, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLOS Comput Biol. 2014 Jul;10((7)):e1003731. doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters NH, Borel Rinkes IH, Zuithoff NP, Mali WP, Moons KG, Peeters PH. Meta-analysis of MR imaging in the diagnosis of breast lesions. Radiology. 2008 Jan;246((1)):116–24. doi: 10.1148/radiol.2461061298. [DOI] [PubMed] [Google Scholar]
- 19.Kuhl C. The current status of breast MR imaging. Part I. Choice of technique, image interpretation, diagnostic accuracy, and transfer to clinical practice. Radiology. 2007 Aug;244((2)):356–78. doi: 10.1148/radiol.2442051620. [DOI] [PubMed] [Google Scholar]
- 20.Vag T, Baltzer PA, Renz DM, Pfleiderer SO, Gajda M, Camara O, et al. Diagnosis of ductal carcinoma in situ using contrast-enhanced magnetic resonance mammography compared with conventional mammography. Clin Imaging. 2008 Nov-Dec;32((6)):438–42. doi: 10.1016/j.clinimag.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Bennani-Baiti B, Baltzer PA. MR Imaging for Diagnosis of Malignancy in Mammographic Microcalcifications: A Systematic Review and Meta-Analysis. Radiology. 2017 Jun;283((3)):692–701. doi: 10.1148/radiol.2016161106. [DOI] [PubMed] [Google Scholar]
- 22.Baltzer PA, Bennani-Baiti B, Stöttinger A, Bumberger A, Kapetas P, Clauser P. Is breast MRI a helpful additional diagnostic test in suspicious mammographic microcalcifications? Magn Reson Imaging. 2018 Feb;46:70–4. doi: 10.1016/j.mri.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Hylton N. Dynamic contrast-enhanced magnetic resonance imaging as an imaging biomarker. J Clin Oncol. 2006 Jul;24((20)):3293–8. doi: 10.1200/JCO.2006.06.8080. [DOI] [PubMed] [Google Scholar]
- 24.Chopra I, Mattes MD, Findley P, Tan X, Dwibedi N, Sambamoorthi U. Impact of Incident Cancer on Short-Term Coronary Artery Disease-Related Healthcare Expenditures Among Medicare Beneficiaries. J Natl Compr Canc Netw. 2019 Feb;17((2)):149–58. doi: 10.6004/jnccn.2018.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Partridge SC, Nissan N, Rahbar H, Kitsch AE, Sigmund EE. Diffusion-weighted breast MRI: clinical applications and emerging techniques. J Magn Reson Imaging. 2017 Feb;45((2)):337–55. doi: 10.1002/jmri.25479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnall MD, Blume J, Bluemke DA, DeAngelis GA, DeBruhl N, Harms S, et al. Diagnostic architectural and dynamic features at breast MR imaging: multicenter study. Radiology. 2006 Jan;238((1)):42–53. doi: 10.1148/radiol.2381042117. [DOI] [PubMed] [Google Scholar]
- 27.Moon M, Cornfeld D, Weinreb J. Dynamic contrast-enhanced breast MR imaging. Magn Reson Imaging Clin N Am. 2009 May;17((2)):351–62. doi: 10.1016/j.mric.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Yabuuchi H, Matsuo Y, Kamitani T, Setoguchi T, Okafuji T, Soeda H, et al. Non-mass-like enhancement on contrast-enhanced breast MR imaging: lesion characterization using combination of dynamic contrast-enhanced and diffusion-weighted MR images. Eur J Radiol. 2010 Jul;75((1)):e126–32. doi: 10.1016/j.ejrad.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Parsian S, Giannakopoulos NV, Rahbar H, Rendi MH, Chai X, Partridge SC. Diffusion-weighted imaging reflects variable cellularity and stromal density present in breast fibroadenomas. Clin Imaging. 2016 Sep-Oct;40((5)):1047–54. doi: 10.1016/j.clinimag.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhl CK, Braun M. [Magnetic resonance imaging in preoperative staging for breast cancer: pros and contras] Radiologe. 2008 Apr;48((4)):358–66. doi: 10.1007/s00117-008-1665-2. [DOI] [PubMed] [Google Scholar]
- 31.Sardanelli F, Boetes C, Borisch B, Decker T, Federico M, Gilbert FJ, et al. Magnetic resonance imaging of the breast: recommendations from the EUSOMA working group. Eur J Cancer. 2010 May;46((8)):1296–316. doi: 10.1016/j.ejca.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Beresford MJ, Padhani AR, Taylor NJ, Ah-See ML, Stirling JJ, Makris A, et al. Inter- and intraobserver variability in the evaluation of dynamic breast cancer MRI. J Magn Reson Imaging. 2006 Dec;24((6)):1316–25. doi: 10.1002/jmri.20768. [DOI] [PubMed] [Google Scholar]
- 33.Wood C. Computer Aided Detection (CAD) for breast MRI. Technol Cancer Res Treat. 2005 Feb;4((1)):49–53. doi: 10.1177/153303460500400107. [DOI] [PubMed] [Google Scholar]
- 34.Meeuwis C, van de Ven SM, Stapper G, Fernandez Gallardo AM, van den Bosch MA, Mali WP, et al. Computer-aided detection (CAD) for breast MRI: evaluation of efficacy at 3.0 T. Eur Radiol. 2010 Mar;20((3)):522–8. doi: 10.1007/s00330-009-1573-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W, Giger ML, Newstead GM, Bick U, Jansen SA, Li H, et al. Computerized assessment of breast lesion malignancy using DCE-MRI robustness study on two independent clinical datasets from two manufacturers. Acad Radiol. 2010 Jul;17((7)):822–9. doi: 10.1016/j.acra.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renz DM, Durmus T, Böttcher J, Taupitz M, Diekmann F, Huppertz A, et al. Comparison of gadoteric acid and gadobutrol for detection as well as morphologic and dynamic characterization of lesions on breast dynamic contrast-enhanced magnetic resonance imaging. Invest Radiol. 2014 Jul;49((7)):474–84. doi: 10.1097/RLI.0000000000000039. [DOI] [PubMed] [Google Scholar]
- 37.Winsberg F, Elkin M, Macy J, Jr, Bordaz V, Weymouth W. Detection of Radiographic Abnormalities in Mammograms by Means of Optical Scanning and Computer Analysis. Radiology. 1967;89((2)):211–5. [Google Scholar]
- 38.Ayer T, Ayvaci MU, Liu ZX, Alagoz O, Burnside ES. Computer-aided diagnostic models in breast cancer screening. Imaging Med. 2010 Jun;2((3)):313–23. doi: 10.2217/IIM.10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelken F, Bremme R, Bick U, Hammann-Kloss S, Fallenberg EM. Factors affecting the rate of false positive marks in CAD in full-field digital mammography. Eur J Radiol. 2012 Aug;81((8)):e844–8. doi: 10.1016/j.ejrad.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Henriksen EL, Carlsen JF, Vejborg IM, Nielsen MB, Lauridsen CA. The efficacy of using computer-aided detection (CAD) for detection of breast cancer in mammography screening: a systematic review. Acta Radiol. 2019 Jan;60((1)):13–8. doi: 10.1177/0284185118770917. [DOI] [PubMed] [Google Scholar]
- 41.Duffy SW, Gabe R. What should the detection rates of cancers be in breast screening programmes? Br J Cancer. 2005 Feb;92((3)):597–600. doi: 10.1038/sj.bjc.6602345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eadie LH, Taylor P, Gibson AP. A systematic review of computer-assisted diagnosis in diagnostic cancer imaging. Eur J Radiol. 2012 Jan;81((1)):e70–6. doi: 10.1016/j.ejrad.2011.01.098. [DOI] [PubMed] [Google Scholar]
- 43.Elter M, Schulz-Wendtland R, Wittenberg T. The prediction of breast cancer biopsy outcomes using two CAD approaches that both emphasize an intelligible decision process. Med Phys. 2007 Nov;34((11)):4164–72. doi: 10.1118/1.2786864. [DOI] [PubMed] [Google Scholar]
- 44.Azavedo E, Zackrisson S, Mejàre I, Heibert Arnlind M. Is single reading with computer-aided detection (CAD) as good as double reading in mammography screening? A systematic review. BMC Med Imaging. 2012 Jul;12((1)):22. doi: 10.1186/1471-2342-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hadjiiski L, Sahiner B, Chan HP. Advances in computer-aided diagnosis for breast cancer. Curr Opin Obstet Gynecol. 2006 Feb;18((1)):64–70. doi: 10.1097/01.gco.0000192965.29449.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calas MJ, Gutfilen B, Pereira WC. Pereira WCdA. CAD e mamografia: por que usar esta ferramenta? Radiol Bras. 2012;45((1)):46–52. [Google Scholar]
- 47.Brem RF, Baum J, Lechner M, Kaplan S, Souders S, Naul LG, et al. Improvement in sensitivity of screening mammography with computer-aided detection: a multiinstitutional trial. AJR Am J Roentgenol. 2003 Sep;181((3)):687–93. doi: 10.2214/ajr.181.3.1810687. [DOI] [PubMed] [Google Scholar]
- 48.Baker JA, Lo JY, Delong DM, Floyd CE. Computer-aided detection in screening mammography: variability in cues. Radiology. 2004 Nov;233((2)):411–7. doi: 10.1148/radiol.2332031200. [DOI] [PubMed] [Google Scholar]
- 49.Wei J, Sahiner B, Hadjiiski LM, Chan HP, Petrick N, Helvie MA, et al. Computer-aided detection of breast masses on full field digital mammograms. Med Phys. 2005 Sep;32((9)):2827–38. doi: 10.1118/1.1997327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorrius MD, Jansen-van der Weide MC, van Ooijen PM, Pijnappel RM, Oudkerk M. Computer-aided detection in breast MRI: a systematic review and meta-analysis. Eur Radiol. 2011 Aug;21((8)):1600–8. doi: 10.1007/s00330-011-2091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castellino RA. Computer aided detection (CAD): an overview. Cancer Imaging. 2005 Aug;5((1)):17–9. doi: 10.1102/1470-7330.2005.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Demartini WB, Lehman CD, Peacock S, Russell MT. Computer-aided detection applied to breast MRI: assessment of CAD-generated enhancement and tumor sizes in breast cancers before and after neoadjuvant chemotherapy. Acad Radiol. 2005 Jul;12((7)):806–14. doi: 10.1016/j.acra.2005.03.055. [DOI] [PubMed] [Google Scholar]
- 53.Grubstein A, Rapson Y, Benzaquen O, Rozenblatt S, Gadiel I, Atar E, et al. Comparison of background parenchymal enhancement and fibroglandular density at breast magnetic resonance imaging between BRCA gene mutation carriers and non-carriers. Clin Imaging. 2018 Sep-Oct;51:347–51. doi: 10.1016/j.clinimag.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 54.McCormack VA, dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006 Jun;15((6)):1159–69. doi: 10.1158/1055-9965.EPI-06-0034. [DOI] [PubMed] [Google Scholar]
- 55.King V, Jennifer D., Brooks, Jonine L., Bernstein, Anne S., Reiner, Malcolm C., Pike, Elizabeth A. Morris. Background Parenchymal Enhancement at Breast MR Imaging and Breast Cancer Risk. Radiology. 2011 doi: 10.1148/radiol.11102156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolivar AV, Gomez SS, Merino P, Alonso-Bartolomé P, Garcia EO, Cacho PM, et al. Computer-aided detection system applied to full-field digital mammograms. Acta Radiol. 2010 Dec;51((10)):1086–92. doi: 10.3109/02841851.2010.520024. [DOI] [PubMed] [Google Scholar]
- 57.Birdwell RL, Ikeda DM, O'Shaughnessy KF, Sickles EA. Mammographic characteristics of 115 missed cancers later detected with screening mammography and the potential utility of computer-aided detection. Radiology. 2001 Apr;219((1)):192–202. doi: 10.1148/radiology.219.1.r01ap16192. [DOI] [PubMed] [Google Scholar]
- 58.Yang SK, Moon WK, Cho N, Park JS, Cha JH, Kim SM, et al. Screening mammography-detected cancers: sensitivity of a computer-aided detection system applied to full-field digital mammograms. Radiology. 2007 Jul;244((1)):104–11. doi: 10.1148/radiol.2441060756. [DOI] [PubMed] [Google Scholar]
- 59.Klifa C, Carballido-Gamio J, Wilmes L, Laprie A, Shepherd J, Gibbs J, et al. Magnetic resonance imaging for secondary assessment of breast density in a high-risk cohort. Magn Reson Imaging. 2010 Jan;28((1)):8–15. doi: 10.1016/j.mri.2009.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson DJ, Leach MO, Kwan-Lim G, Gayther SA, Ramus SJ, Warsi I, et al. UK study of MRI screening for breast cancer in women at high risk (MARIBS) Assessing the usefulness of a novel MRI-based breast density estimation algorithm in a cohort of women at high genetic risk of breast cancer: the UK MARIBS study. Breast Cancer Res. 2009;11((6)):R80. doi: 10.1186/bcr2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morris EA, Kuhl CK, Lehman CD. Ensuring high-quality breast MR imaging technique and interpretation. Radiology. 2013 Mar;266((3)):996–7. doi: 10.1148/radiol.12121320. [DOI] [PubMed] [Google Scholar]
- 62.Brooks JD, Sung JS, Pike MC, Orlow I, Stanczyk FZ, Bernstein JL, et al. MRI background parenchymal enhancement, breast density and serum hormones in postmenopausal women. Int J Cancer. 2018 Aug;143((4)):823–30. doi: 10.1002/ijc.31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heller SL, Young Lin LL, Melsaether AN, Moy L, Gao Y. Hormonal Effects on Breast Density, Fibroglandular Tissue, and Background Parenchymal Enhancement. Radiographics. 2018 Jul-Aug;38((4)):983–96. doi: 10.1148/rg.2018180035. [DOI] [PubMed] [Google Scholar]
- 64.Ciatto S, Houssami N, Apruzzese A, Bassetti E, Brancato B, Carozzi F, et al. Categorizing breast mammographic density: intra- and interobserver reproducibility of BI-RADS density categories. Breast. 2005 Aug;14((4)):269–75. doi: 10.1016/j.breast.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 65.Spayne MC, Gard CC, Skelly J, Miglioretti DL, Vacek PM, Geller BM. Reproducibility of BI-RADS breast density measures among community radiologists: a prospective cohort study. Breast J. 2012 Jul-Aug;18((4)):326–33. doi: 10.1111/j.1524-4741.2012.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eng A, Gallant Z, Shepherd J, McCormack V, Li J, Dowsett M, et al. Digital mammographic density and breast cancer risk: a case-control study of six alternative density assessment methods. Breast Cancer Res. 2014 Sep;16((5)):439. doi: 10.1186/s13058-014-0439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mohamed AA, Berg WA, Peng H, Luo Y, Jankowitz RC, Wu S. A deep learning method for classifying mammographic breast density categories. Med Phys. 2018 Jan;45((1)):314–21. doi: 10.1002/mp.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinker K, Chin J, Melsaether AN, Morris EA, Moy L. Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment. Radiology. 2018 Jun;287((3)):732–47. doi: 10.1148/radiol.2018172171. [DOI] [PubMed] [Google Scholar]
- 69.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014 Jun;5((1)):4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ingrisch M, Schneider MJ, Nörenberg D, Negrao de Figueiredo G, Maier-Hein K, Suchorska B, et al. Radiomic Analysis Reveals Prognostic Information in T1-Weighted Baseline Magnetic Resonance Imaging in Patients With Glioblastoma. Invest Radiol. 2017 Jun;52((6)):360–6. doi: 10.1097/RLI.0000000000000349. [DOI] [PubMed] [Google Scholar]
- 71.Crivelli P, Ledda RE, Parascandolo N, Fara A, Soro D, Conti M. A New Challenge for Radiologists: Radiomics in Breast Cancer. BioMed Res Int. 2018 Oct;2018:6120703. doi: 10.1155/2018/6120703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whitney HM, Taylor NS, Drukker K, Edwards AV, Papaioannou J, Schacht D, et al. Additive Benefit of Radiomics Over Size Alone in the Distinction Between Benign Lesions and Luminal A Cancers on a Large Clinical Breast MRI Dataset. Acad Radiol. 2019 Feb;26((2)):202–9. doi: 10.1016/j.acra.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bickelhaupt S, Steudle F, Paech D, Mlynarska A, Kuder TA, Lederer W, et al. PLoS One. 12. United States. 2017. On a fractional order calculus model in diffusion weighted breast imaging to differentiate between malignant and benign breast lesions detected on X-ray screening mammography; p. p. e0176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bickelhaupt S, Jaeger PF, Laun FB, Lederer W, Daniel H, Kuder TA, et al. Radiomics Based on Adapted Diffusion Kurtosis Imaging Helps to Clarify Most Mammographic Findings Suspicious for Cancer. Radiology. 2018 Jun;287((3)):761–70. doi: 10.1148/radiol.2017170273. [DOI] [PubMed] [Google Scholar]
- 75.Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2((1)):2. doi: 10.1038/npjbcancer.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan M, Li H, Wang S, Zheng B, Zhang J, Li L. PLoS One. 12. United States. 2017. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer; p. p. e0171683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H, Zhu Y, Burnside ES, Drukker K, Hoadley KA, Fan C, et al. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology. 2016 Nov;281((2)):382–91. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park H, Lim Y, Ko ES, Cho HH, Lee JE, Han BK, et al. Radiomics Signature on Magnetic Resonance Imaging Association with Disease-Free Survival in Patients with Invasive Breast Cancer. Clin Cancer Res. 2018 Oct;24:4705–4714. doi: 10.1158/1078-0432.CCR-17-3783. [DOI] [PubMed] [Google Scholar]
- 79.Braman N, Prasanna P, Whitney J, Singh S, Beig N, Etesami M, et al. Association of Peritumoral Radiomics With Tumor Biology and Pathologic Response to Preoperative Targeted Therapy for HER2 (ERBB2)-Positive Breast Cancer. JAMA Netw Open. 2. United States. 2019:p. e192561. doi: 10.1001/jamanetworkopen.2019.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, et al. Breast Cancer Res. 19. England. 2017. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI; p. p. 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Litjens G, Kooi T, Bejnordi BE, Setio AA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017 Dec;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 82.Hamidinekoo A, Denton E, Rampun A, Honnor K, Zwiggelaar R. Deep learning in mammography and breast histology, an overview and future trends. Med Image Anal. 2018 Jul;47:45–67. doi: 10.1016/j.media.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 83.Kooi T, Litjens G, van Ginneken B, Gubern-Mérida A, Sánchez CI, Mann R, et al. Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal. 2017 Jan;35:303–12. doi: 10.1016/j.media.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 84. https://www.synapse.org/#!Synapse:syn4224222/wiki/401743. [Google Scholar]
- 85.Ribli D, Horvath A, Unger Z, Pollner P, Csabai I. Sci Rep. 8. England. 2018. Detecting and classifying lesions in mammograms with Deep Learning; p. p. 4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodriguez-Ruiz A, Lång K, Gubern-Merida A, Broeders M, Gennaro G, Clauser P, et al. Stand-Alone Artificial Intelligence for Breast Cancer Detection in Mammography: Comparison With 101 Radiologists. J Natl Cancer Inst. 2019 Mar;:djy222. doi: 10.1093/jnci/djy222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodriguez-Ruiz A, Lang K, Gubern-Merida A, Teuwen J, Broeders M, Gennaro G, et al. A feasibility study. Germany: Eur Radiol; 2019. Can we reduce the workload of mammographic screening by automatic identification of normal exams with artificial intelligence? [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016 Feb;278((2)):563–77. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]