Abstract
1,1-Dimethyl-3-phenylurea (known as fenuron) which is a phenyl urea-based widely used herbicide exhibits interesting structural and conformational properties and a notable biological activity. A detailed analysis on the vibrational, molecular and electronic characteristics of fenuron has been carried out. Potential energy scans (PESs) performed at the B3LYP/6-311++G(d,p) level of theory predicted two possible minima corresponding to the optimized anti and synforms resulting from the internal rotation about the N-C bond. The presence of an auxochrome together with the interaction with DMSO solvent exhibited a blue shift corresponding to the C=O orbitals. Delocalization of HOMO and LUMO orbital facilitated the charge transfer effect in the molecule. The calculated HOMO-LUMO energies, chemical potential, energy gap and global hardness suggested a low softness value for the compound while its biological activity was described by the value of electrophilicity. Chlorine substitution in the phenyl ring influenced the orbital delocalization for ortho and para substitutions but that of meta remained unaffected. NLO properties were noticed to increase due to chlorine substitution in the parent molecule. The docking results suggested that the compound exhibits an inhibitory activity against mitochondrial ubiquinol-cytochrome-c reductase and can be developed as a potential anticancer agent.
Keywords: Organic chemistry, Pharmaceutical chemistry, Theoretical chemistry, DFT, Fenuron, Molecular docking, NBO, NLO
1. Introduction
Fenuron is phenyl urea-based herbicide used in agriculture for weeds' control [1]. It functions by inhibiting photosynthetic process of weeds [1, 2]. Fenuron is among the organic compounds that are not easily degradable by microbes and as such it is relatively persistent in the environment and gets its way through runoff to both surface and ground water as a contaminant [2]. A prominent method being employed in treating fenuron for its removal from environment and other phenyl urea herbicides is oxidative degradation. Ozonization using O3 and O3/H2O2 had been employed to degrade phenyl urea herbicides in water but with the formation of by-products [3]. Furthermore, an improved method known as electro-Fenton process was developed and applied for the degradation of herbicides in water [2, 4]. In this method, O3/H2O2 was replaced by Fenton's reagent (Fe2+/H2O2 and Fe3+/H2O2) which electrochemically produced highly reactive hydroxyl radicals [5]. Another method is hydrothermal oxidation usually carried out between 200 and 540 °C [6]. On the other hand, the method involving the use of direct solar light for degradation of phenyl urea was found to be slow [7], while radiolysis technique has been proven to be efficient for the degradation of fenuron in water [8] because the hydroxyl radicals used to degrade fenuron are being efficiently produced during the process. Considering the different degradation approaches applied on fenuron and in order to provide a satisfactory valuation of such techniques at the atomistic level, a thorough investigation of the electronic and molecular properties of fenuron will be helpful to study the stability and degradation properties of the compound. Therefore, in this work we have explored the electronic, conformational and spectroscopic characteristics for the title compound, and the effect of chlorine substitution in the phenyl ring on its biological activity has been studied.
2. Experimental
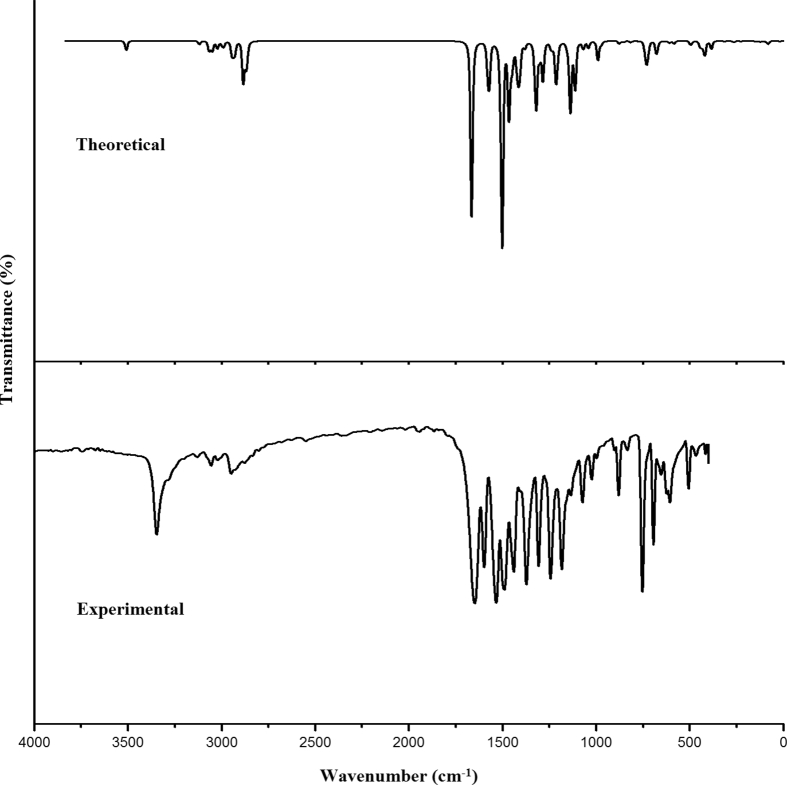
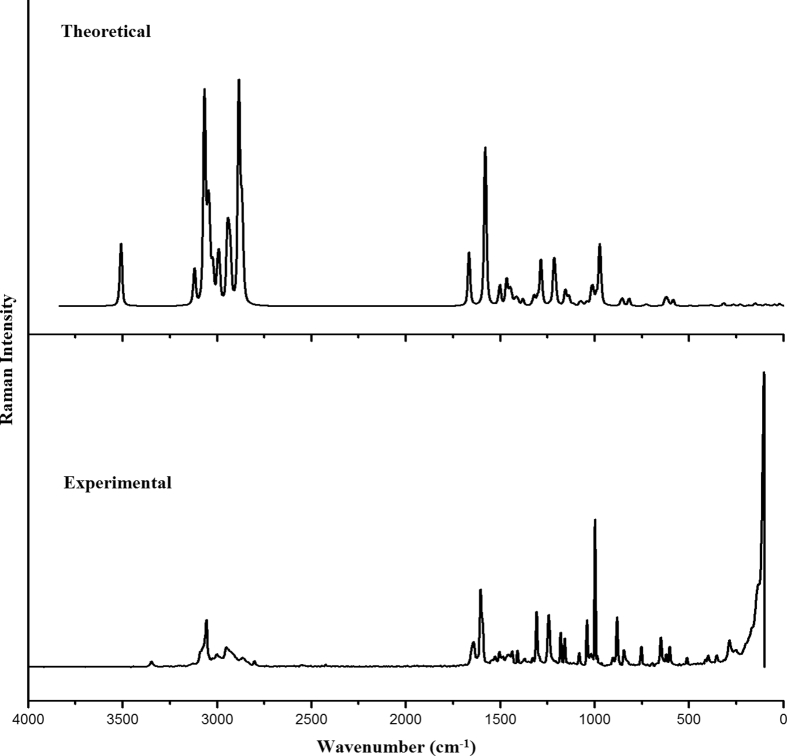
The FT-IR spectrum of solid fenuron sample (Fig. 1) was recorded using a Nicolet 6700 FT-IR spectrometer equipped with a global source, a KBr beam splitter and a DTGS KBr detector in the range of 4000–400 cm−1 at a resolution of 4 cm−1. A Nicolet NXR FT-Raman module equipped with CaF2 beam splitter, a Helium–Neon detector was used to collect the Raman spectrum (Fig. 2) in the range of 4000–100 cm−1 at a resolution of 4 cm−1. The TGA analysis was carried out using an SDT Q600 V20.9 Build 20, Module DSC-TGA standard instrument. Measurements were carried out under nitrogen atmosphere, at a temperature range of 25–800 °C/min. Firstly, the TGA inbuilt balance which contains two pans of alumina, one for sample and the second for reference was tared. 5–10 mg samples of fenuron were loaded in the alumina pans and ramped at 10 °C/min from 25 °C to 800 °C in the dry at a flow rate of 50 ml min. The UV–Visible spectrum of fenuron solution was recorded using a GENESYS10S UV–Visible spectrophotometer (Thermo Fisher Scientific) at room temperature in a range of 200–800 nm. The fenuron solution was prepared by dissolving a 10mg of the solid fenuron in 10 ml DMSO.
Fig. 1.
FT-IR spectrum of fenuron.
Fig. 2.
FT-Raman spectrum of fenuron.
3. Calculation
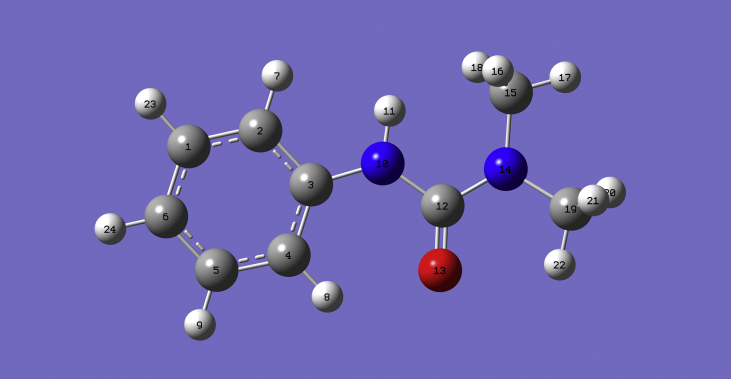
The Gaussian09 software program [9] using the DFT-B3LYP with the 6-311++G(d,p)(5D,7F) basis set was used to predict the structure (Fig. 3) and vibrational wavenumbers. The theoretically obtained wavenumbers were visualized and analyzed with Gaussview 5 [10]and assigned by potential energy distribution method [11]. To obtain a better harmony with experimental results, a scaling factor of 0.9613 was used to scale the theoretical wavenumbers [12]. The hydrogen atoms, H7 (ortho), H23 (meta) and H24 (para) of the title compound were replaced by chlorine atom to find the drug activity, binding affinities, global chemical descriptors, NLO behavior and other parameters.
Fig. 3.
Optimized geometry of fenuron.
4. Results and discussion
4.1. Conformational analysis
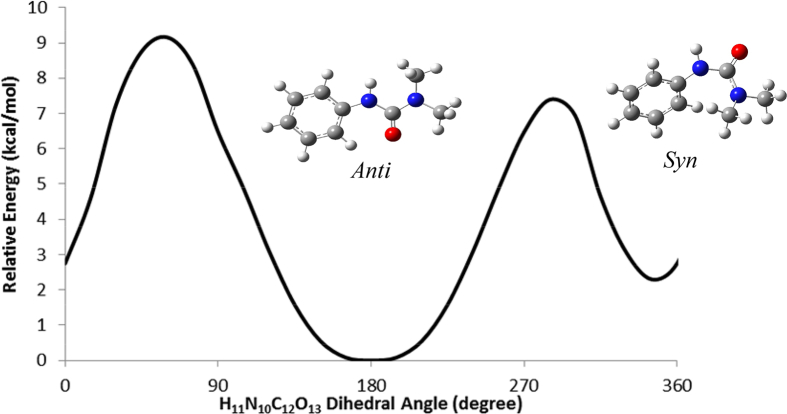
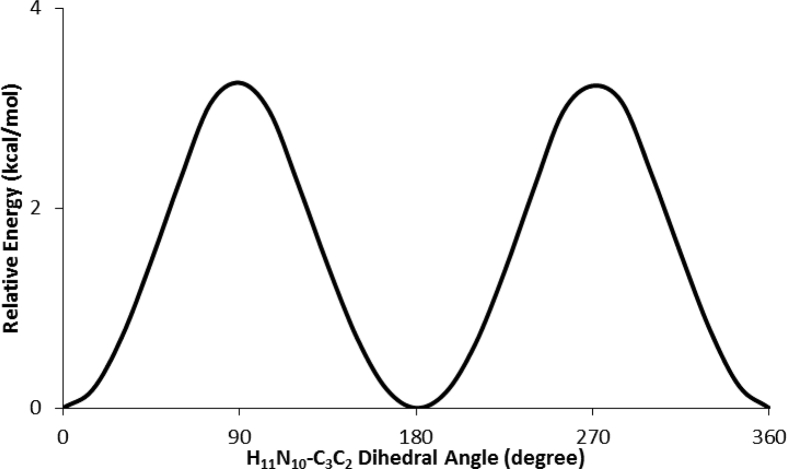
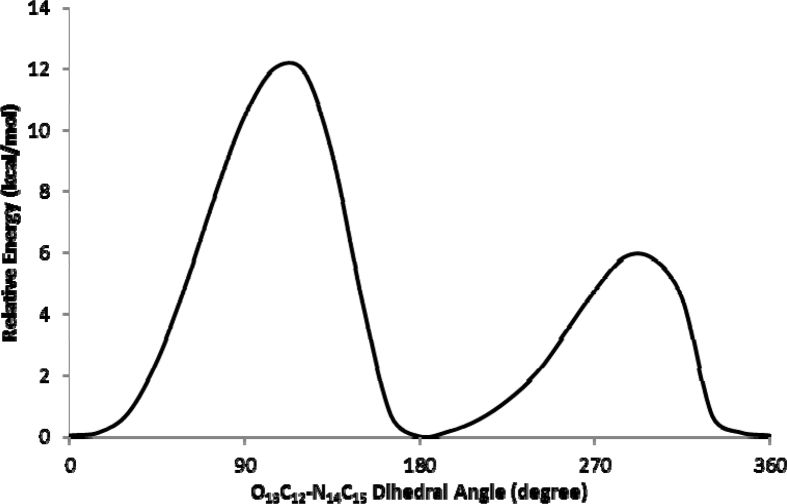
In the most stable form of fenuron (Fig. 3), the oxygen atom was predicted to be at an anti position with respect to the N-H bond, forming an estimated N10-C12-O13 angle of 118°. Conformational study was carried out to understand the role of various intramolecular interactions in the compound, and potential energy scans (PESs) were carried out about the N10-C12 (Fig. 4), C3-N10 (Fig. 5) and the C12-N14 (Fig. 6) bonds. Fig. 4 shows two possible minimas with their corresponding optimized structural anti and synforms, that resulted from the internal rotation about the N10-C12 bond. The anti form was predicted to be about 2 kcal/mol more stable than the syn form (Table 1). This is in consistence with previously reported conformational analysis for urea and dimethylurea at the DFT and MP2 levels of theories [13, 14], for urea and its dimer at the AM1, PM3 and MP2 levels of theory [15], and for thiourea at the MP2 level of theory [16]. The anti and syn conformations have also been reported as the two possible minima stable for urea, methylurea, ethylurea, isopropylurea, tert-butylurea and phenylurea at different levels of theory [17]. The anti form was predicted to be more stable for the urea and all the urea derivatives. This can also be due to the stabilization via intra molecular hydrogen bonding between the functional groups. The relative stabilities between the anti and the syn forms were reported to be 1.02 kcal/mol for urea, 0.93 kcal/mol for methylurea, 1.65 kcal/mol for ethylurea, 1.67 kcal/mol for isopropylurea, 2.41 kcal/mol for tert-butylurea and 0.16 kcal/mol for phenylurea using the DFT/B3LYP approach [17]. The high energy conformational interchange barrier of about 7 kcal/mol for fenuron (Fig. 4) is comparable with those reported for urea [17, 18, 19, 20]. Such a high barrier is a result of the inversion at N10 atom [15] as well as the repulsive interaction between the N10 lone pair of electrons and the methyl hydrogen atoms. Moreover, Fig. 5 shows the potential scan of the rotation of the phenyl group of the anti form. Such an internal rotation leads to break-up of the conjugation, and hence two equivalent minimum forms could be obtained. The relatively low energy barrier of about 3 kcal/mol is attributed to the modest electron delocalization across the aliphatic chain of the molecule. On the other hand, the energy scan carried out for the dimethyl urea moiety about the amide linkage (Fig. 6) exhibits the effect of the electron lone pair of the N14 on the overall stability of the molecule. The rotation of the -N(CH3)2 group yields to identical conformations with dihedral angles (175o, 9o) that are in good agreement with the OCNCH3 dihedral angle (117o and 9o) at the B3LYP level of theory and (177o and 14o) at the MP2 level of theory reported for dimethylurea [13]. This indicates that the methyl substitution for hydrogen atom in fenuron has no significant effect on the urea dihedral angle. Previous studies showed that the methyl group rotation in methylurea, ethyl group rotation of ethylurea, isopropyl group rotation of isopropylurea, and tert-butyl group rotation of tert-butylurea resulted with two identical forms in the PES of the molecules [17], similar to the case of fenuron (Fig. 6). In the low-energy form, the lone pair of electrons of N14 tends to defuse causing the amide and dimethyl urea bonds being coplanar due to a predominant hyper-conjugation effect. The rotation of the N(CH3)2 moiety across the amide bond resulted with more pronounced sp3 character on urea nitrogen, and two non-equivalent transition states would develop. The first transition structure is characterized by a strong repulsive interaction between N14 lone pair of electrons and the carbonyl group, while the other transition structure resulted from the repulsion of the carbonyl and methyl groups. The latter transition structure was predicted to be more feasible as its activation energy is approximately half of that of the high-energy one.
Fig. 4.
Potential energy scan resulting from the rotation of H11N10-C12O13 dihedral angle.
Fig. 5.
Potential energy scan resulting from the rotation of H11N10-C3C2 dihedral angle.
Fig. 6.
Potential energy scan resulting from the rotation of O13C12-N14C15 dihedral angle.
Table 1.
Relative stability and calculated H11N10—C12O11 dihedral angle for anti and syn forms.
| Conformer | Relative Energy (kcal/mol) | H11N10-C12O13 Dihedral Angle (degree) |
|---|---|---|
| Anti | 0.00 | 176.9 |
| Syn | 2.29 | -12.5 (347.5) |
4.2. IR, Raman and VCD spectra
The vibrational assignments of the anti form of the title molecule with observed and calculated wavenumbers are given in Table 2.
Table 2.
Calculated scaled wavenumbers, observed IR, Raman bands and assignments.
| B3LYP/6-311++G(d,p) (5D, 7F) |
IR |
Raman |
Assignmentsa |
||
|---|---|---|---|---|---|
| υ(cm−1) | IRI | RA | υ(cm−1) | υ(cm−1) | - |
| 3509 | 19.83 | 100.98 | 3345 | 3348 | υNH(100) |
| 3120 | 7.10 | 52.99 | 3124 | 3123 | υCH(100) |
| 3067 | 17.85 | 266.59 | - | 3075 | υCH(95) |
| 3051 | 23.65 | 84.76 | 3053 | 3054 | υCH(95) |
| 3043 | 0.12 | 94.48 | - | - | υCH(90) |
| 3039 | 1.14 | 3.33 | - | - | υCH3(92) |
| 3021 | 16.10 | 53.25 | - | 3024 | υCH(94) |
| 2992 | 19.01 | 108.35 | 3005 | 2995 | υCH3(96) |
| 2942 | 38.45 | 138.57 | 2950 | 2945 | υCH3(99) |
| 2931 | 29.72 | 79.21 | 2929 | - | υCH3(91) |
| 2883 | 91.58 | 316.63 | - | - | υCH3(100) |
| 2866 | 52.21 | 117.32 | 2870 | 2863 | υCH3(93) |
| 1664 | 339.35 | 49.45 | 1656 | 1648 | υC = O(73) |
| 1579 | 44.32 | 141.52 | 1585 | - | υPh(57), δNH(18) |
| 1570 | 72.37 | 5.69 | - | - | υPh(44), δNH(29) |
| 1503 | 475.79 | 21.34 | - | 1505 | δNH(45), δCH3(17) |
| 1469 | 6.64 | 4.15 | 1475 | - | δCH3(22), υPh(53) |
| 1465 | 132.11 | 191.2 | - | 1461 | δCH3(60), υPh(11) |
| 1452 | 58.24 | 6.86 | - | - | δCH3(47), υPh(14) |
| 1446 | 3.89 | 9.35 | - | - | δCH3(51), υPh(20) |
| 1443 | 13.09 | 7.52 | 1440 | 1439 | δCH3(22), υPh(53) |
| 1418 | 93.01 | 7.83 | - | - | δCH3(55), υPh(20) |
| 1409 | 73.48 | 6.64 | 1407 | 1407 | δCH3(58), υPh(20) |
| 1381 | 11.09 | 5.91 | 1377 | 1378 | δCH3(60), υPh(18) |
| 1321 | 169.65 | 10.75 | - | 1325 | δCH3(56), υPh(21) |
| 1301 | 29.38 | 6.59 | 1304 | 1303 | δCH(64), υPh(11) |
| 1283 | 75.34 | 39.64 | - | 1285 | υPh(57), δCH(15) |
| 1237 | 22.24 | 1.58 | 1240 | 1240 | υCN(37), δNH(42) |
| 1212 | 119.83 | 58.39 | - | 1208 | υCN(41), δNH(23) |
| 1155 | 6.60 | 12.71 | - | 1156 | δCH(46), δNH(18) |
| 1138 | 109.94 | 2.57 | 1137 | - | υCN(38), δCH(22) |
| 1135 | 43.14 | 6.99 | - | - | δCH(55), υCN(17) |
| 1112 | 92.65 | 1.29 | - | 1110 | δCH3(49), δCH(32) |
| 1078 | 1.76 | 2.65 | - | 1080 | δCH3(54), δCH(23) |
| 1067 | 13.66 | 2.13 | 1069 | - | δCH(47), δCH3(17) |
| 1040 | 15.17 | 3.27 | - | 1041 | δCH3(45), δCH(22) |
| 1012 | 0.28 | 25.85 | 1020 | 1015 | δCH(52), δCH3(23) |
| 991 | 36.91 | 6.94 | 993 | 993 | δCH3(39), υCN(35) |
| 971 | 6.94 | 53.76 | - | 977 | υPh(44), δCH3(15), υCN(12) |
| 961 | 0.83 | 0.21 | 961 | 962 | γCH(65), υPh(12) |
| 935 | 0.22 | 0.11 | - | - | γCH(57), υPh(22) |
| 876 | 6.67 | 0.05 | 880 | 880 | γCH(71) |
| 855 | 3.11 | 8.80 | - | 849 | υCN(38), γCH(19) |
| 817 | 2.73 | 5.49 | 825 | - | γCH(63), τPh(15) |
| 805 | 0.12 | 0.09 | - | - | γCH(44), τPh(23) |
| 734 | 56.84 | 1.14 | 736 | - | γCH(57), γC = O(12) |
| 723 | 16.61 | 0.74 | - | 722 | γC = O(33), γCH(18), δPh(21) |
| 676 | 32.63 | 0.01 | 685 | - | γNH (41), γC = O(20) |
| 622 | 0.34 | 10.00 | 623 | 620 | δPh(35), δC = O(34) |
| 610 | 3.81 | 2.04 | 608 | 607 | δPh(29), δCN(23) |
| 585 | 6.04 | 4.66 | - | - | δPh(22), δCN(27) |
| 494 | 11.14 | 0.29 | 501 | - | τPh(40), γCN(22) |
| 441 | 19.90 | 0.41 | - | - | γNH(28), τPh(22), γCN(15) |
| 422 | 35.89 | 0.45 | 419 | 419 | γNH(19), δCH3(28), τPh(16) |
| 400 | 1.00 | 0.06 | - | 399 | τPh(33), δCH3(13), γNH(15) |
| 385 | 14.86 | 0.69 | - | - | τCH3(36), τPh(28), γCN(23) |
| 317 | 1.09 | 2.49 | - | - | δPh(24), γCN(27), δCN(22) |
| 267 | 4.39 | 1.42 | - | 260 | τCH3(29), δPh(32), γCN(12) |
| 230 | 1.19 | 1.30 | - | - | τPh(41), τCH3(18), δCN(10) |
| 187 | 0.65 | 0.43 | - | 193 | τCH3(33), τPh(17), τCN(12) |
| 149 | 2.09 | 2.13 | - | 139 | τCH3(25), τPh(18), τCN(12) |
| 124 | 2.12 | 0.59 | - | - | δPh(32), τC = O(20) |
| 95 | 2.31 | 1.34 | - | - | τCH3(34), τNH(18) |
| 82 | 6.00 | 0.36 | - | - | τPh(26), τNH(15) |
| 49 | 0.64 | 1.03 | - | - | τPh(31), τCN(27) |
| 20 | 1.16 | 1.57 | - | - | τPh(29), τCH3(32) |
υ-stretching; δ-in-plane deformation; γ-out-of-plane deformation; τ-torsion; Ph-Phenyl ring; IRI-IR intensity(KM/Mole); RA-Raman activity(Ǻ4/amu).
For the title compound, υN-H are assigned at 3345 cm−1 (IR), 3348 cm−1 (Raman) and at 3509 cm−1 theoretically, and bending modes of N-H are assigned at 1503, 1237, 676 cm−1 theoretically while the experimentally modes are seen at 1505, 1240 cm−1 (Raman) and 1240, 685 cm−1 (IR) [21, 22]. The reported values of NH modes are at 3462 cm−1 (IR), 3450 cm−1 (Raman), 3400 cm−1 (DFT) (stretching), 1508, 1219, 655 cm−1 (DFT) (deformation) [23] and 1587, 1250, 650 cm−1 (IR), 1580, 1227, 652 cm−1 (DFT) (deformation modes) [24]. In the present case the NH stretching mode in the IR spectrum is downshifted by 164 cm−1 from the computed value, which is due to the strong hyper-conjugative interaction as given by NBO analysis (n1(O13) to σ*(N10-C12)) as well as the high rotational barriers (6–12 kcal/mol).
For fenuron, the CN stretching modes are assigned at 991 and 855 cm−1 theoretically and other CN stretching modes are assigned at 1237, 1212, 1138 cm−1 theoretically, while bands are observed at 1240, 1137, 993 cm−1 (IR) and at 1240, 1208, 993, 849 cm−1 (Raman) [25]. For nitrogen bonded methyl molecules, the CN stretching mode are reported at 1141, 1009 cm−1 and 992 cm−1 [26] and at 1005, 855 cm−1 (IR), and at 1008, 850 cm−1 (DFT) [27] and at 996, 954 cm−1 (DFT) by Mary et al. [28]. For fenuron, the C=O stretch is observed at 1656 cm−1 (IR), 1648 cm−1 (Raman), 1664 cm−1 theoretically [29] and reported values are at 1694 cm−1 (IR), 1696 cm−1 (Raman) and at 1699 cm−1 theoretically [23].
The CH3 stretching modes of fenuron are observed at 3005, 2950, 2929, 2870 cm−1 (IR) and at 2995, 2945, 2863 cm−1 (Raman) [21, 25]. The CH3 bending vibrations are observed at 1407, 1377, 993 cm−1 (IR) and at 1461, 1407, 1378, 1110, 1080, 993 cm−1 (Raman). From the DFT analysis, these modes are in the ranges 3039-2866 cm−1 and 1465-991 cm−1 [21, 25].
The stretching of phenyl C-H are observed at 3124, 3053 cm−1 (IR) and at 3123, 3075, 3054, 3024 cm−1 (Raman) [25]. DFT results give these modes in the range 3120–3021 cm−1 [25]. The υPh modes are in the range 1579–1283 cm−1 while bands are observed at 1585, 1475, 1440 cm−1 (IR) and at 1439, 1285 cm−1 (Raman) [25]. For fenuron, the ring breathing mode of phenyl ring is assigned at 971 cm−1 theoretically as expected [30]. The phenyl CH deformation modes are observed at 1304, 1069, 1020 cm−1 (IR), 1303, 1156, 1015 cm−1 (Raman) (in-plane deformation) and at 961, 880, 825, 736 cm−1 (IR), 962, 880 cm−1 (Raman) (out-of-plane deformation) as expected [25]. The RMS error between theoretical wavenumbers and observed IR and Raman bands are 4.84 and 4.59, respectively.
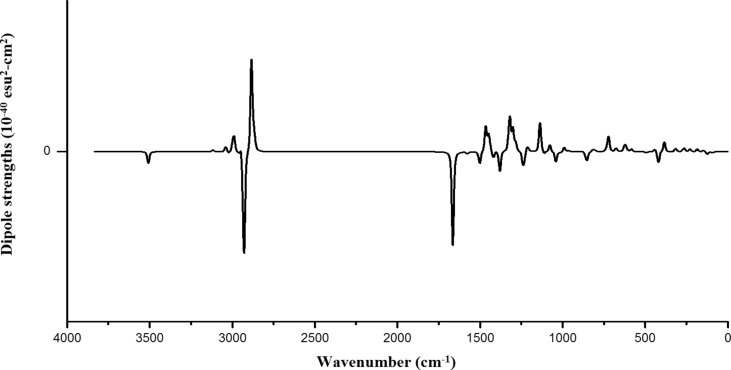
The various stretching modes produce VCD signals that help in configuration identification of molecular systems [31]. The VCD bands (Fig. 7) at 2883, 1465, 1321 cm−1 corresponding to CH3 groups are good markers of pattern showing left polarization while the band at 2931 cm−1 show right polarization and are good markers of the configuration. The VCD spectrum shows right polarization at 3508 cm−1 for the NH stretching and at 1664 cm−1 for C=O stretching.
Fig. 7.
VCD spectrum of fenuron.
4.3. TG/DTG and UV and natural bond orbital analysis
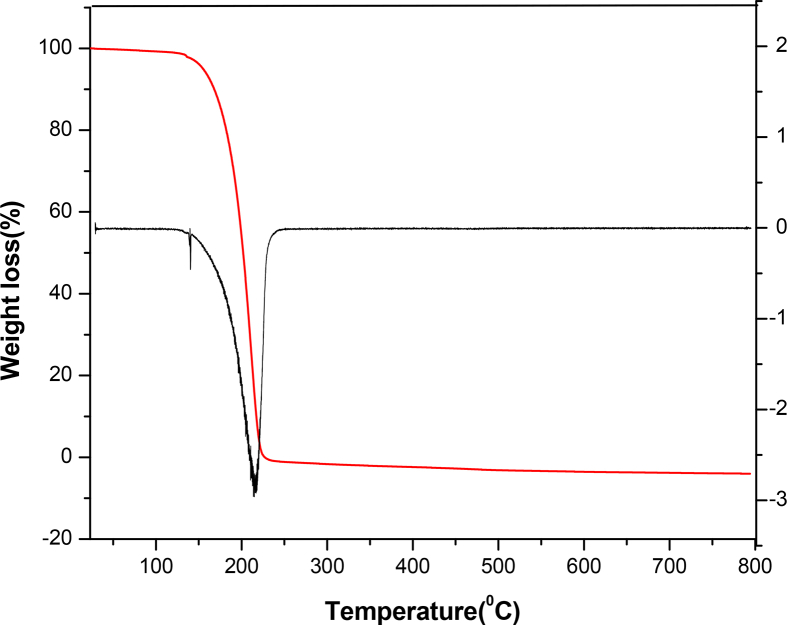
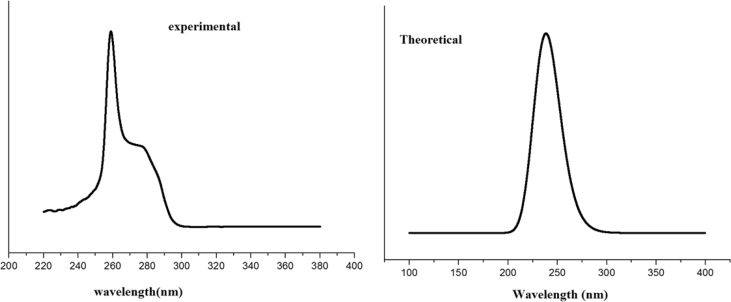
The TG curve (Fig. 8) of fenuron shows that the sample is stable up to 130 °C and shows only stage decomposition. The decomposition starts at 130 °C and completed at 248 °C. This is denoted by a DTG peak at 213 °C. Fenuron is derived from urea and as a result, its chromophore property might be attributed to C=O functional group. However, its C=O is bonded to N(CH3)2 and NHC6H5 that are basic auxochromes significantly altering chromophore absorption λmax. As presented in Fig. 9, maximum absorption was recorded for fenuron at 259 nm which quite agreed with calculated λmax (243 nm). The result suggests that the absorption might have resulted from n-π* transition, which is the only electronic transition for C=O. In addition, the blue shift observed in wavelength with respect to the absorption λmax (270 nm) (Table 3) known for C=O could be associated with the presence of auxochrome together with the interaction with chloroform solvent. The natural bond orbital (NBO) analysis was performed using NBO 3.1 program [32] as implemented in the Gaussian09 package at the DFT/B3LYP level. The main interaction are: n1(N10)→π*(C3-C4), n1(N10)→ σ*(C12-O13) and n1(N14)→σ*(C12-O13) with energies 33.63, 23.86 and 23.73 kcal/mol. Also there is an interaction from n1(O13) to σ*(N10-C12) and σ*(C12-N14) with energies 25.85 and 25.11 kcal/mol. Nearly 100% p-character observed in n1N10, n2O13 and N14 atoms of the title compound. Important results are tabulated in Tables 4 and 5.
Fig. 8.
TG/DTG spectrum of Fenuron.
Fig. 9.
UV spectrum of Fenuron.
Table 3.
Calculated electronic absorption spectrum.
| Excitation |
CI expansion |
Energy |
Wavelength |
Oscillator |
|---|---|---|---|---|
| - | coefficient | (eV) | calc. (nm) | strength(f) |
| Excited State 1 | ||||
| 44→45 | 0.69008 | 4.7415 | 261.49 | 0.0036 |
| Excited state 2 | ||||
| 44→46 | 0.55533 | 4.8660 | 254.79 | 0.0253 |
| Excited state 3 | ||||
| 44→48 | 0.14074 | 5.2152 | 237.74 | 0.3564 |
Table 4.
Second-order perturbation theory analysis of Fock matrix in NBO basis corresponding to the intramolecular bonds of neuron.
| Donor(i) | Type | ED/e | Acceptor(j) | Type | ED/e | E(2)a | E(j)-E(i)b | F(ij)c |
|---|---|---|---|---|---|---|---|---|
| LPN10 | σ | 1.70801 | C3-C4 | π* | 0.38222 | 33.63 | 0.30 | 0.092 |
| - | - | C12-O13 | σ* | 0.27259 | 23.86 | 0.47 | 0.095 | |
| - | - | C12-O13 | π* | 0.14268 | 6.55 | 0.67 | 0.061 | |
| LPO13 | σ | 1.97277 | N10-C12 | σ* | 0.07558 | 2.17 | 1.11 | 0.044 |
| - | π | 1.84383 | N10-C12 | σ* | 0.07558 | 25.85 | 0.68 | 0.120 |
| - | - | C12-N14 | σ* | 0.08185 | 25.11 | 0.69 | 0.119 | |
| LPN14 | σ | 1.73229 | C12-O13 | σ* | 0.27259 | 23.73 | 0.46 | 0.094 |
| - | - | C12-O13 | π* | 0.14268 | 7.78 | 0.66 | 0.066 |
E(2) means energy of hyper-conjugative interactions (stabilization energy in kcal/mol).
Energy difference (a.u) between donor and acceptor i and j NBO orbitals.
F(i,j) is the Fock matrix elements (a.u) between i and j NBO orbitals.
Table 5.
NBO results showing the formation of Lewis and non-Lewis orbitals.
| Bond(A-B) | ED/ea | NBO | s% | p% |
|---|---|---|---|---|
| n1N10 | 1.70801 | sp1.00 | 0.00 | 100 |
| - | -0.25334 | |||
| n1O13 | 1.97277 | sp0.68 | 59.54 | 40.45 |
| - | -0.66004 | |||
| n2O13 | 1.84383 | sp1.00 | 0.00 | 100 |
| - | -0.22719 | |||
| n1N14 | 1.73229 | sp85.71 | 1.15 | 98.85 |
| - | -0.24343 |
ED/e is expressed in a.u.
4.4. Nonlinear optical properties
The computed values of dipole moment, polarizability, first order hyperpolarizability and second order hyperpolarizability values are: 4.3301 Debye, 1.919 × 10−23esu, 2.869 × 10−30esu, -5.948 × 10−37esu. The energy gap for the title molecule is 3.558 eV less than that of urea (6.706eV) and the first hyperpolariazability is 22.07 times that of urea [33, 34]. Ortho and meta substitution of chlorine atom reduces the dipole moment while for para substitution dipole moment increases from that of parent molecule (Tables 6.1 and 6.2). All the polarizability values increases due to chlorine substitution and second hyperpolarizability shows an increase from that of parent atom, which means NLO property increases due to halogen substitution and it can be attributed to the electronegativity of the group, which decreases electron density from the ring and enhances nonlinear properties.
Table 6.1.
Chemical descriptors.
| HOMO | LUMO | I = -EHOMO | A = -ELUMO | Gap | η=(I-A)/2 | μ = -(I + A)/2 | ω = μ2/2η | |
|---|---|---|---|---|---|---|---|---|
| fenuron | -8.099 | -4.541 | 8.099 | 4.541 | 3.558 | 1.779 | -6.320 | 11.23 |
| Ortho | -7.744 | -4.233 | 7.744 | 4.233 | 3.511 | 1.756 | -5.989 | 10.21 |
| Meta | -8.124 | -4.320 | 8.124 | 4.320 | 3.804 | 1.902 | -6.222 | 10.18 |
| Para | -7.605 | -4.528 | 7.605 | 4.528 | 3.077 | 1.539 | -6.067 | 11.96 |
Table 6.2.
NLO properties.
| μ | α ×10−23 | β ×10−30 | γ ×10−37 | MR = 1.333παN | |
|---|---|---|---|---|---|
| fenuron | 4.3301 | 1.919 | 2.869 | -5.948 | 109.16 |
| Ortho | 2.7936 | 2.095 | 3.688 | -7.006 | 70.27 |
| Meta | 4.2488 | 2.119 | 3.081 | -8.577 | 107.11 |
| Para | 5.9146 | 2.135 | 3.679 | -9.218 | 149.11 |
4.5. Molecular reactivity
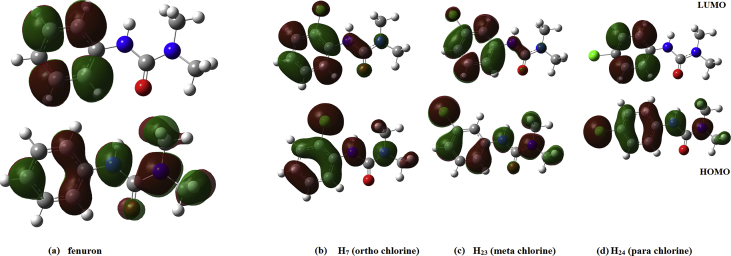
A number of fundamental electronic parameters were calculated to provide insights on the molecular reactivity of fenuron and its chloro-substituted derivatives. The HOMO-LUMO plots are presented in Fig. 10 and HOMO is delocalized over the whole molecule while LUMO is only delocalized over the phenyl ring. HOMO plots of chlorine substitution changes for ortho and para substitution, while for the meta there is no change in the delocalization. For LUMO, para delocalization is predicted to be similar to that of the parent molecule, yet the delocalization changes for ortho and meta substitutions. From the calculated HOMO and LUMO energies, the ionization potential (obtained as I = -EHOMO), the electron affinity (obtained as A = -ELUMO) [35] and the HOMO-LUMO energy gap were computed, respectively, as 8.099, 4.541 and 3.558eV; which indicates that the third compound is relatively more reactive due to its low energy gap. Moreover, the chemical descriptors are given by hardness η= (I-A)/2 = 1.779; chemical potential μ = -(I + A)/2 = -6.320 and electrophilicity index ω = μ2/2η = 11.23 [36]. There was no significant change noticed in the chemical descriptors due to halogen substitutions in the parent molecule (Tables 6.1 and 6.2). Molar refractivity (MR) is a term used in structure property activity and is given as MR = 1.333παN, where α is the polarizability and N is the Avogadro number [37]. For the title compound, MR value is 109.16, which gives the binding nature of fenuron and can be used for the cure of different diseases [38].
Fig. 10.
HOMO-LUMO plots of (a) fenuron (b) ortho chlorine (c) meta chlorine (d) para chlorine.
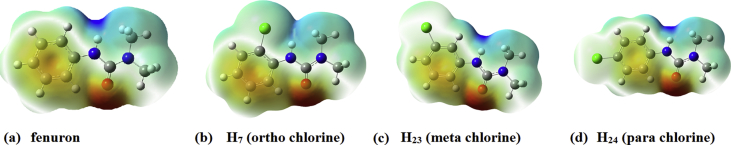
Additionally, the probable reactive sites of electrophilic and nucleophilic attacks in fenuron were predicted with the help of the molecular electrostatic potential (MEP) map calculated at the DFT level [39]. The negative (red and yellow) regions in the MEP plot were related to electrophilic reactivity and positive, (blue region) to nucleophilic reactivity. From the MEP plot (Fig. 11) it is clear that the carbonyl oxygen atom and phenyl ring are the most negative regions and the NH moiety is the most positive region. The electrophilic and nucleophilic sites have no change in chlorine substituted parent molecule.
Fig. 11.
MEP plots of (a) fenuron (b) ortho chlorine (c) meta chlorine (d) para chlorine.
4.6. Homology and molecular docking
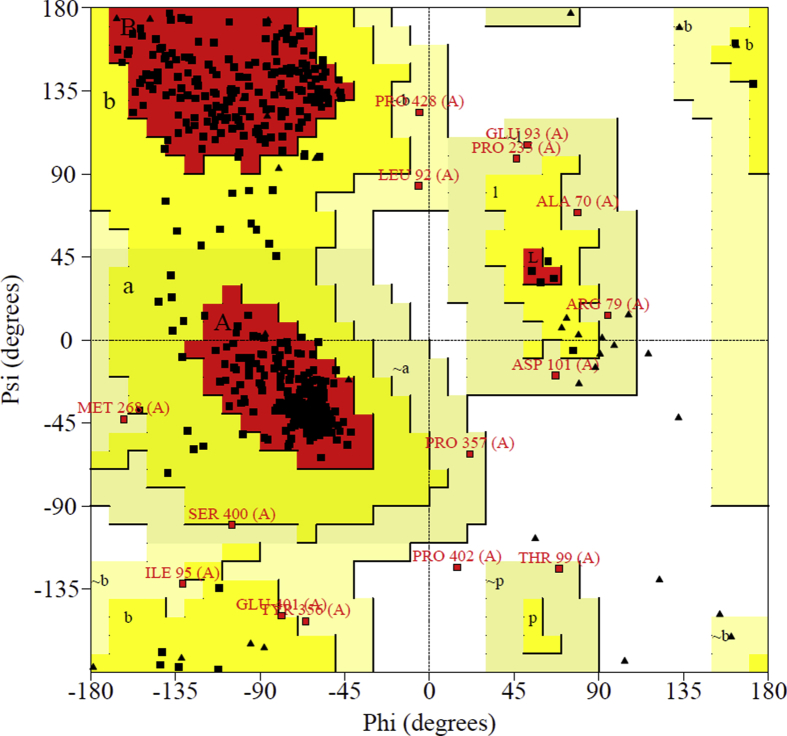
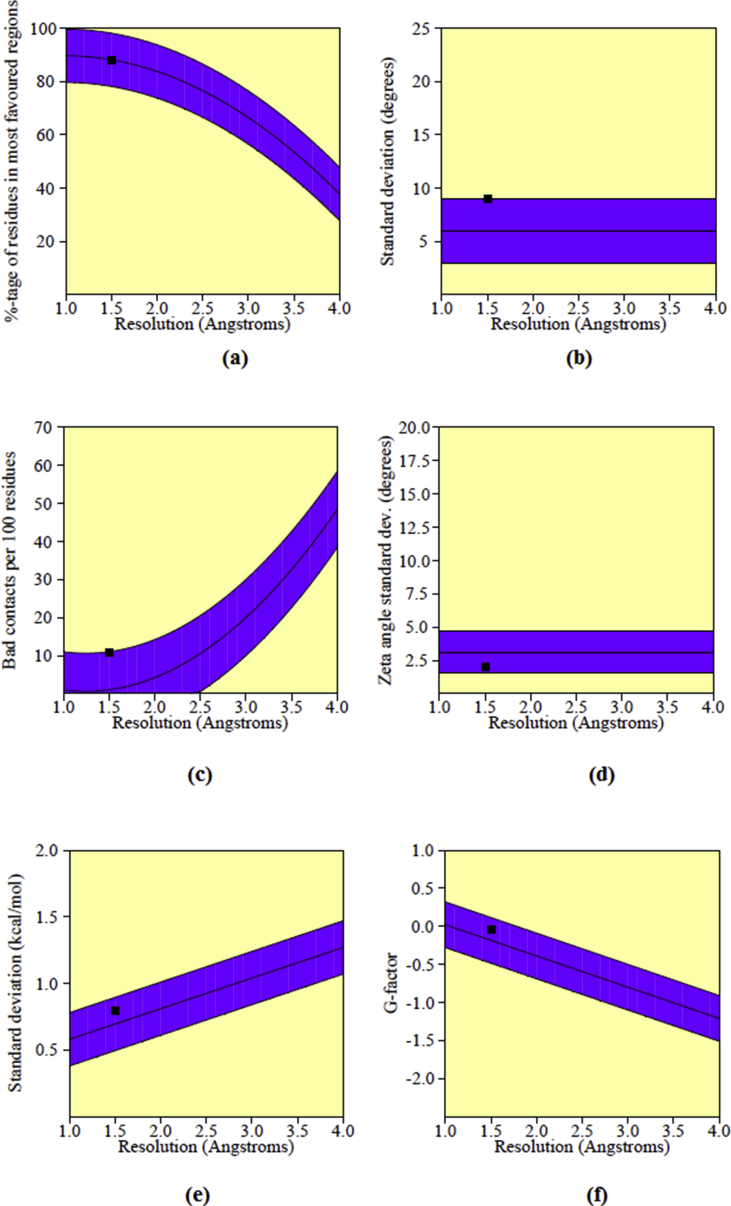
PASS analysis (Prediction of Activity Spectra) [40] of the title compound gives activities given in the Table 7, Ubiquinol-cytochrome-c reductase inhibitor activity with probability to be active (Pa) value of 0.950. High resolution crystal structure of mitochondrial ubiquinol-cytochrome-c reductase was downloaded from the protein data bank website with PDB ID: 3I73. Mitochondria came into the focus of research of cancer biologists. The importance of mitochondria as potential targets in cancer cells stems from the fact that they are a pool of proteins that promote the apoptotic death when mobilized into the cytosol [41, 42]. Mitochondrial biogenesis (MtBIO) is to be involved in chemo resistance, the foremost obstacle in the treatment of patients with ovarian cancer [43]. Thus mitochondrial ubiquinol-cytochrome-c reductase is chosen as target for docking study. Utilizing PROCHECK server, the homology property of the selected protein, 3I73 was verified since validation is used to find any deviations or errors from the normal protein and before docking study. From the Ramachandran plot, it is clear that the most of the amino acid residues were in red color core regions with 88.1% (over 90% represent the ideal value), 9.4% and 2.5% of residues were in allowed yellow color regions and 0.0% were in generously allowed light yellow color regions (Fig. 12) for 1V93 and 2Y71, respectively. Also a very low percentage 0.0% of residues was seen in the disallowed white color regions (due to large distance from the enzyme's active site). In the present case, the G-factors are found above the unusual values, which give information about the quality of dihedral, overall bond angles and covalency of residues [44].
Table 7.
PASS prediction for the activity spectrum of fenuron Pa represents probability to be active and Pi represents probability to be inactive.
| Pa | Pi | Activity |
|---|---|---|
| 0.950 | 0.003 | Ubiquinol-cytochrome-c reductase inhibitor |
| 0.901 | 0.002 | Phospholipid-translocating ATPase inhibitor |
| 0.881 | 0.002 | N-acylmannosamine kinase inhibitor |
| 0.834 | 0.022 | Phobic disorders treatment |
| 0.828 | 0.030 | Membrane integrity agonist |
| 0.805 | 0.011 | NADPH peroxidase inhibitor |
| 0.801 | 0.013 | Glycosylphosphatidylinositol phospholipase D inhibitor |
| 0.801 | 0.018 | Mucomembranous protector |
| 0.751 | 0.021 | Taurine dehydrogenase inhibitor |
| 0.695 | 0.005 | N-methylhydantoinase (ATP-hydrolysing) inhibitor |
| 0.734 | 0.044 | Testosterone 17beta-dehydrogenase (NADP+) inhibitor |
| 0.696 | 0.009 | Insulysin inhibitor |
| 0.712 | 0.031 | Nicotinic alpha6beta3beta4alpha5 receptor antagonist |
| 0.686 | 0.009 | Trimethylamine-oxide aldolase inhibitor |
| 0.702 | 0.026 | Nicotinic alpha2beta2 receptor antagonist |
| 0.681 | 0.007 | Eye irritation. inactive |
| 0.683 | 0.016 | Dimethylargininase inhibitor |
| 0.709 | 0.042 | Acrocylindropepsin inhibitor |
| 0.709 | 0.042 | Chymosin inhibitor |
| 0.709 | 0.042 | Saccharopepsin inhibitor |
| 0.671 | 0.008 | Cytochrome P450 stimulan |
Fig. 12.
Ramachandran plot PDB ID 3I73.
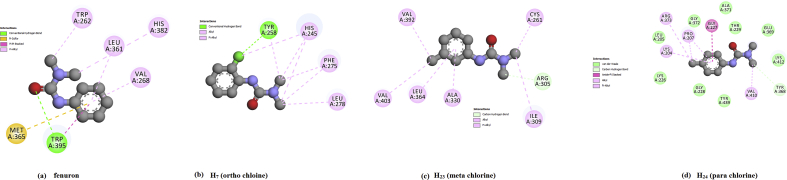
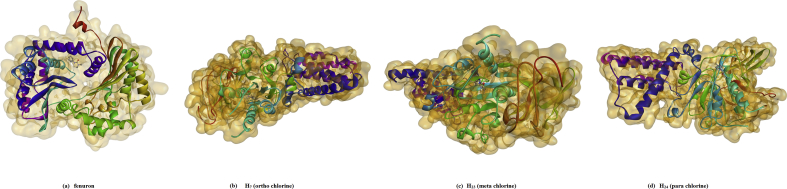
For the plot of the six properties shown in Fig. 13, the measured parameters were all inside the dark band in each graph and this indicates that the present models are well within the limits of reliable structures. From the validation results, 3I73 was reasonably good in geometry and stereochemistry and are appropriate for the ligand-protein docking studies of the title compounds. Molecular docking analysis was done on Auto Dock-Vina software and as in literature [45, 46, 47, 48, 49]. The ligand binds at the active site of the substrate by weak non-covalent interactions as detailed in 2D plot (Fig. 14). The docked ligands form a stable complex with mitochondrial ubiquinol-cytochrome-c reductase as shown in Fig. 15. The binding affinity value of original compound is -6.6 kcal/mol (Tables 8.1, 8.2, 8.3, and 8.4) while ortho, meta and para substitution the binding affinity values lowered as 5.8, 5.5, 5.3 kcal/mol respectively. These preliminary results suggest that the compounds might exhibit inhibitory activity against mitochondrial ubiquinol-cytochrome-c reductase and the compounds can be developed as a new anticancer agent. The improvement in sequencing and bioinformatics technology lead to a realization that the improbable tasks of finding a good cancer cure that would target only a single gene or a single signaling pathway.
Table 8.1.
The binding affinity values of different poses of the fenuron compound predicted by Autodock Vina.
| Mode |
Affinity (kcal/mol) |
Distance from best mode (Å) |
|
|---|---|---|---|
| - | - | RMSD l.b. | RMSD u.b. |
| 1 | -6.6 | 0.000 | 0.000 |
| 2 | -6.4 | 1.298 | 2.034 |
| 3 | -6.2 | 2.072 | 2.345 |
| 4 | -5.1 | 19.094 | 19.535 |
| 5 | -5.1 | 15.838 | 17.979 |
| 6 | -5.1 | 8.495 | 11.459 |
| 7 | -5.0 | 7.151 | 8.665 |
| 8 | -5.0 | 16.062 | 17.364 |
| 9 | -5.0 | 15.230 | 17.809 |
Table 8.2.
The binding affinity values of different poses of the ortho chlorine predicted by AutodockVina.
| Mode |
Affinity (kcal/mol) |
Distance from best mode (Å) |
|
|---|---|---|---|
| - | - | RMSD l.b. | RMSD u.b. |
| 1 | -5.8 | 0.000 | 0.000 |
| 2 | -5.5 | 16.238 | 17.533 |
| 3 | -5.3 | 16.392 | 17.677 |
| 4 | -5.1 | 16.920 | 18.045 |
| 5 | -5.0 | 14.841 | 15.463 |
| 6 | -5.0 | 2.915 | 3.768 |
| 7 | -5.0 | 17.723 | 18.657 |
| 8 | -4.9 | 2.040 | 5.153 |
| 9 | -4.9 | 19.116 | 20.076 |
Table 8.3.
The binding affinity values of different poses of the meta chlorine predicted by AutodockVina.
| Mode |
Affinity (kcal/mol) |
Distance from best mode (Å) |
|
|---|---|---|---|
| - | - | RMSD l.b. | RMSD u.b. |
| 1 | -5.5 | 0.000 | 0.000 |
| 2 | -5.3 | 18.858 | 19.966 |
| 3 | -5.2 | 2.926 | 5.426 |
| 4 | -5.2 | 11.689 | 13.120 |
| 5 | -5.2 | 1.918 | 2.473 |
| 6 | -5.1 | 19.399 | 20.417 |
| 7 | -5.0 | 3.548 | 3.968 |
| 8 | -4.8 | 3.124 | 4.799 |
| 9 | -4.8 | 12.944 | 14.272 |
Fig. 13.
Main chain parameters of PDB ID 3I73 (a) Ramanchandran plot quality assessment (b) peptide bond planarity-omega sd (c) Measure of bad non-bonded interactions (d) Alpha carbon tetrahedral distortion (e) Hydrogen bond energies (f) Overall G-factor.
Fig. 14.
2D interactive plot of ligand with the residues of themitochondrialubiquinol-cytochrome-c reductase of (a) fenuron (b) ortho chlorine (c) meta chlorine (d) para chlorine.
Fig. 15.
The docked ligand at the active site of mitochondrial ubiquinol-cytochrome-c reductase of (a) fenuron (b) ortho chlorine (c) meta chlorine (d) para chlorine.
Table 8.4.
The binding affinity values of different poses of the para chlorine predicted by AutodockVina.
| Mode |
Affinity (kcal/mol) |
Distance from best mode (Å) |
|
|---|---|---|---|
| - | - | RMSD l.b. | RMSD u.b. |
| 1 | -5.3 | 0.000 | 0.000 |
| 2 | -5.2 | 20.843 | 21.905 |
| 3 | -5.2 | 17.760 | 18.632 |
| 4 | -5.1 | 17.742 | 18.571 |
| 5 | -5.1 | 20.571 | 22.534 |
| 6 | -5.1 | 17.673 | 18.453 |
| 7 | -5.1 | 20.491 | 21.385 |
| 8 | -5.0 | 15.811 | 17.352 |
| 9 | -5.0 | 17.978 | 19.874 |
5. Conclusion
Detailed spectroscopic analysis together with DFT calculations was performed. Significant LUMO and HOMO energy difference is an evidence for the charge transfer within the molecule. PES shows two possible minima with their corresponding optimized structural form (anti and sym forms), that result from the internal rotation about the N10-C12 bond and the anti conformer was predicted to be more stable. The first hyperpolariazability of fenuron is 22.07 times that of urea and chlorine substitution increases the NLO properties. From the MEP plot gives carbonyl oxygen atom and phenyl ring as the most negative regions and the NH moiety as the most positive region. PASS analysis predicts activity, Ubiquinol-cytochrome-c reductase inhibitor activity and the docked compound has a binding affinity of-6.6 kcal/mol while ortho, meta and para substitution the binding affinity values are lowered. The title compound and halogenated substituents can be developed as a new anticancer agent.
Declarations
Author contribution statement
Y.Sheena Mary, Kabiru Haruna, Saheed Popoola, Renjith Thomas, Roxy M S, Abdulaziz Al-Saadi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Experimental and computational resources provided by King Fahd University of Petroleum and Minerals (KFUPM) to support this work are appreciated.
Contributor Information
Y. Sheena Mary, Email: marysheena2018@rediffmail.com.
A.A. Al-Saadi, Email: asaadi@kfupm.edu.sa.
References
- 1.Gangwar S.K., Rafiquee M.Z.A. Kinetics of the alkaline hydrolysis of fenuron in aqueous and micellar media. Int. J. Chem. Kinet. 2007;39:638–644. [Google Scholar]
- 2.Oturan M.A., Edelahi M.C., Oturan N., El K., Aaron J. Environmental kinetics of oxidative degradation/mineralization pathways of the phenylurea herbicides diuron, monuron and fenuron in water during the application of the electro-Fenton process. Appl. Catal. B Environ. 2010;97:82–89. [Google Scholar]
- 3.Tahmasseb L.A., Nelieu S., Kerhoas L., Einhorn J. Ozonation of chlorophenylurea pesticides in water, reaction monitoring and degradation pathways. Sci. Total Environ. 2002;291:33–44. doi: 10.1016/s0048-9697(01)01090-7. [DOI] [PubMed] [Google Scholar]
- 4.Gallard H., De Laat J. Kinetics of oxidation of chlorobenzenes and phenylureas by Fe(II)/H2O2 and Fe(III)H2O2, evidence of reduction and oxidation reactions of intermediates by Fe(II) or Fe(III) Chemosphere. 2001;42:405–413. doi: 10.1016/s0045-6535(00)00076-x. [DOI] [PubMed] [Google Scholar]
- 5.Mazellier D.L.J., Busset P., Delmont A. A comparison of fenuron degradation by hydroxyl and carbonate radicals in aqueous solution. Water Res. 2007;41:4585–4594. doi: 10.1016/j.watres.2007.06.066. [DOI] [PubMed] [Google Scholar]
- 6.Aymonier C., Beslin P., Jolivalt C. Hydrothermal oxidation of a nitrogen containing compound, the fenuron. J. Supercrit. Fluids. 2000;17:45–54. [Google Scholar]
- 7.Brahmia O., Boulkamh A., Sehili T., Aguer J.P., Richard C. Kinetics of photocatalytic decomposition of fenuron over TiO2 in aqueous solution. Int. J. Photoenergy. 2002;4:85–89. [Google Scholar]
- 8.Kovacs K., Mile V., Csay T. Hydroxyl radical induced degradation of fenuron in pulse and gamma radiolysis, kinetics and product analysis. Environ. Sci. Pollut. Res. 2014;21 doi: 10.1007/s11356-014-3197-9. 0 12693-12700. [DOI] [PubMed] [Google Scholar]
- 9.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., Nakatsuji H., Caricato M., Li X., Hratchian H.P., Izmaylov A.F., Bloino J., Zheng G., Sonnenberg J.L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J.A., Jr., Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Keith T., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Rega N., Millam J.M., Klene M., Knox J.E., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Martin R.L., Morokuma K., Zakrzewski V.G., Voth G.A., Salvador P., Dannenberg J.J., Dapprich S., Daniels A.D., Farkas O., Foresman J.B., Ortiz J.V., Cioslowski J., Fox D.J. Gaussian, Inc.; Wallingford CT: 2013. Gaussian 09, Revision D.01. [Google Scholar]
- 10.Dennington R., Keith T., Millam J. Semichem. Inc.; Shawnee Mission, KS: 2009. Gaussview, Version 5. [Google Scholar]
- 11.Martin J.M.L., Van Alsenoy C. University of Antwerp; Belgium: 2007. GAR2PED, a Program to Obtain a Potential Energy Distribution from a Gaussian Archive Record. [Google Scholar]
- 12.Foresman J.B., Pittsburg P.A. In: Exploring Chemistry with Electronic Structure Methods: a Guide to Using Gaussian. Frisch E., editor. 1996. [Google Scholar]
- 13.Lecomte F., Lucas B., Gregoire G., Schermann J.P., Desfrancois C. Urea and methyurea dipole-bound anions. Phys. Chem. Chem. Phys. 2003;5:3120–3125. [Google Scholar]
- 14.Skurski P., Simons J. An excess electron bound to urea. I. Canonical and zwitterionictautomers. J. Chem. Phys. 2001;115:8373. [Google Scholar]
- 15.Masunov A., Dannenberg J.J. Theoretical study of urea. I. Monomers and Dimers. J. Phys. Chem. A. 1999;103:178–184. [Google Scholar]
- 16.Lesarri A., Mata S., Blanco S., López J.C., Alonso José. L. A rotational study of laser ablated thiourea. J. Chem. Phys. 2004;120:6191. doi: 10.1063/1.1642585. [DOI] [PubMed] [Google Scholar]
- 17.Bryantsev V.S., Firman T.K., Hay B.P. Conformational analysis and rotational barriers of alklyl- and phenyl-substituted urea derivatives. J. Phys. Chem. A. 2005;109:832–842. doi: 10.1021/jp0457287. [DOI] [PubMed] [Google Scholar]
- 18.Kontoyianni M., Hoffman A.J., Bowen J.P. Ab initio and molecular mechanical calculations on imine derivatives: a study of the rotational barriers and the development of MM2 parameters. J. Comput. Chem. 1992;13:57–65. [Google Scholar]
- 19.Gobbi A., Frenking G. Y-conjugated compounds: the equilibrium geometries and electronic structures of guanidine, guanidiniumcation, urea, and 1,2-diaminoethylene. J. Am. Chem. Soc. 1993;115:2362–2372. [Google Scholar]
- 20.Dixon D.A., Matsuzawa N. Density functional study of the structures and nonlinear optical properties of urea. J. Phys. Chem. 1994;98:3967–3977. [Google Scholar]
- 21.Colthup N.B., Daly L.H., Wiberly S.E. Academic Press; New York: 1975. Introduction of Infrared and Raman Spectroscopy. [Google Scholar]
- 22.Bellamy L.J. third ed. Chapman and Halls; London: 1975. The Infrared Spectrum of Complex Molecules. [Google Scholar]
- 23.Sebastian S.H.R., Al-Alshaikh M.A., El-Emam A.A., Panicker C.Y., Zitko J., Dolezal M., Van Alsenoy C. Spectroscopic, quantum chemical sttudies, Fukui functions, in vitro antiviral activity and molecular docking of 5-chloro-N-(3-nitrophenyl)pyrazine-2-carboxamide. J. Mol. Struct. 2016;1119:188–199. [Google Scholar]
- 24.Mary Y.S., Varghese H.T., Panicker C.Y., Dolezal M. Vibrational spectroscopic studies andab initio calculations of a substituted amide of pyrazine-2carboxylic acid – C12H10ClN3O. Spectrochim. Acta. 2008;71:725–730. doi: 10.1016/j.saa.2007.12.055. [DOI] [PubMed] [Google Scholar]
- 25.Roeges N.P.G. John Wiley and Sons; New York: 1994. A Guide to the Complete Interpretation of Infrared Spectra of Organic Structures. [Google Scholar]
- 26.Mary Y.S., Panicker C.Y., Yamuna T.S., Siddegowda M.S., Yathirajan H.S., Al- Saadi A.A., VanAlsenoy C. Theoretical investigations on the molecular structure, vibrational spectral, HOMO-LUMO and NBO analysis of 9-[3-(dimethylamino)propyl]-2-trifluoro-methyl-9H-thioxanthen-9-ol. Spectrochim. Acta. 2014;132:491–501. doi: 10.1016/j.saa.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Resmi K.S., Haruna K., Mary Y.S., Panicker C.Y., Saleh T.A., Al-Saadi A.A., Van Alsenoy C. Conformational, NBO, NLO, HOMO-LUMO, NMR, electronic spectral study and molecular docking study of N,N-dimethy,-3-(10H-phenothiazin-10yl)-1-propanamine. J. Mol. Sruct. 2016;1122:268–279. [Google Scholar]
- 28.Mary Y.S., Jojo P.J., Van Alsenoy C., Kaur M., Siddegowda M.S., Yathirajan H.S., Nogueira H.I.S., Cruz S.M.A. Vibrational spectroscopic (FT-IR, FT-Raman, SERS) and quantum chemical calculations of 3-(10,10-dimethyl-anthracen-9-ylidene)-N,N,N-trimethylpropanaminiium chloride (Melitracenium chloride) Spectrochim. Acta. 2014;120:370–380. doi: 10.1016/j.saa.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 29.Socrates G. John Wiley and Sons; New York: 1981. Infrared Characteristic Group Frequencies. [Google Scholar]
- 30.Varsanyi G. Wiley; New York: 1974. Assignments of Vibrational Spectra of Seven Hundred Benzene Derivatives. [Google Scholar]
- 31.Keiderling T.A., Baianu I.C., Pessen H., Kumosinski T. Van Norstrand Reinhold; New York: 1993. Physical Chemistry of Food Processes, Advanced Techniques, Structures and Applications; pp. 307–337. [Google Scholar]
- 32.Glendening E.D., Reed A.E., Carpenter J.E., Weinhold F. University of Wisconsin; Madison: 1998. NBO Version 3.1, TCI. [Google Scholar]
- 33.Raja M., Muhamed R.R., Muthu S., Suresh M. Synthesis, spectroscopic (FT-IR, FT- Raman, NMR, UV-Visible), first order hyperpolarizability, NBO and molecular docking study of (E)-1-(4-bromobenzylidene)semicarbazide. J. Mol. Struct. 2017;1128:481–492. [Google Scholar]
- 34.Kuruvilla T.K., Prasana J.C., Muthu S., George J., Mathew S.A. Quantum mechanical and spectroscopic (FT-IR, FT-Raman) study, NBO analysis, HOMO-LUMO, first order hyperpolarizability and molecular docking study of methy[(3R)-3-(2-methylphenoxy)-3-phenylpropyl]amine by density functional method. Spectrochim. Acta. 2018;188:382–393. doi: 10.1016/j.saa.2017.07.029. [DOI] [PubMed] [Google Scholar]
- 35.Mary Y.S., Varghese H.T., Panicker C.Y., Girisha M., Sagar B.K., Yathirajan H.S., Al-Saadi A.A., Van Alsenoy C. Vibrational spectra, HOMO, LUMO, NBO, MEP analysis and molecular docking study of 2,2-diphenyl-4-(piperidin-1-yl)butanamide. Spectrochim. Acta. 2015;150:543–556. doi: 10.1016/j.saa.2015.05.090. [DOI] [PubMed] [Google Scholar]
- 36.Parr R.G., Pearson R.G. Absolute hardness: companion parameter to absolute electronegativity. J. Am. Chem. Soc. 1983;105:7512–7516. [Google Scholar]
- 37.Sureshkumar B., Mary Y.S., Panicker C.Y., Resmi K.S., Suma S., Armakovic S., Armakovic S.J., Van Alsenoy C. Spectroscopic analysis of 8-hydroxyquinoline-5-sulphonic acid and investigation of its reactive properties by DFT and molecular dynamics simulations. J. Mol. Struct. 2017;1150:540–552. [Google Scholar]
- 38.Verma R.P., Hansch C. A comparison between two polarizability parameters in chemical biological interactions. Bioorg. Med. Chem. 2005;13:2355–2372. doi: 10.1016/j.bmc.2005.01.051. [DOI] [PubMed] [Google Scholar]
- 39.Abraham C.S., Prasana J.C., Muthu S. Quantum mechanical, spectroscopic and docking studies of 2-amino-3-bromo-5-nitropyridine by density functional method, Spectrochim. Acta. 2017;181:153–163. doi: 10.1016/j.saa.2017.03.045. [DOI] [PubMed] [Google Scholar]
- 40.Lagunin A., Stepanchikova A., Filimonov D., Poroikov V. PASS: prediction of activity spectra for biologically active substances. Bioinformatics. 2000;16:747–748. doi: 10.1093/bioinformatics/16.8.747. [DOI] [PubMed] [Google Scholar]
- 41.Green D.R., Kroemer G. vol. 305. 2004. pp. 626–629. (The Pathophysiology of Mitochondrial Cell Death Science). [DOI] [PubMed] [Google Scholar]
- 42.Kluckova K., Bezawork-Geleta A., Rohlena J., Dong L., Neuzil J. Mitochondrial complex II, a novel target for anti-cancer agents. Biochim. Biophys. Acta Bioenerg. 2013;1827:552–564. doi: 10.1016/j.bbabio.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 43.Uddin M.H., Kim B., Such D.H., Song Y.S. Anticancer strategy targeting mitochondrial biogenesis in ovarian cancer. J. Cancer Sci. Ther. 2014;6:422–428. [Google Scholar]
- 44.Abrigach F., Rokni Y., Takfaoui A., Khoutoul M., Doucet H., Aseshraou A., Touzani R. In vitro screening, homology modeling and molecular docking studies of some pyrazole and imidazole derivatives. Biomed. Pharmacother. 2018;103:653–661. doi: 10.1016/j.biopha.2018.04.061. [DOI] [PubMed] [Google Scholar]
- 45.Trott O., Olson A.J. AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kramer B., Rarey M., Lengauer T. Evaluation of the FlexX incremental construction algorithm for protein ligand docking. Proteins Struct. Funct. Genet. 1999;37:228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 47.Thomas R., Hossain M., Mary Y.S., Resmi K.S., Armaković S., Armaković S.J., Nanda A.K., Ranjan V.K., Vijayakumar G., Van Alsenoy C. Spectroscopic analysis and molecular docking of imidazole derivatives and investigation of its reactive properties by DFT and molecular dynamics simulations. J. Mol. Struct. 2018;1156:336–347. [Google Scholar]
- 48.Hossain M., Thomas R., Mary Y.S., Resmi K.S., Armaković S., Armaković S.J., Nanda A.K., Vijayakumar G., Alsenoy C. Van. Understanding reactivity of two newly synthetized imidazole derivatives by spectroscopic characterization and computational study. J. Mol. Struct. 2018;1158:176–196. [Google Scholar]
- 49.Mary Y.S., Aswathy V.V., Panicker C.Y., Bielenica A., Brzoka P., Savczenko O., Armakovic S., Armakovic S.J., Van Alsenoy C. Spectroscopic, single crystal XRD structure, DFT and molecular dynamics investigation of 1-(3-chloro-4-fluorophenyl)-3-[3-(trifluoromethyl)phenyl]thiourea. RSC Adv. 2016;113:111997–112015. [Google Scholar]