Abstract
Objective
In type 2 diabetes (T2D), oxidative stress contributes to the dysfunction and loss of pancreatic β cells. A highly conserved feature of the cellular response to stress is the regulation of mRNA translation; however, the genes regulated at the level of translation are often overlooked due to the convenience of RNA sequencing technologies. Our goal is to investigate translational regulation in β cells as a means to uncover novel factors and pathways pertinent to cellular adaptation and survival during T2D-associated conditions.
Methods
Translating ribosome affinity purification (TRAP) followed by RNA-seq or RT-qPCR was used to identify changes in the ribosome occupancy of mRNAs in Min6 cells. Gene depletion studies used lentiviral delivery of shRNAs to primary mouse islets or CRISPR-Cas9 to Min6 cells. Oxidative stress and apoptosis were measured in primary islets using cell-permeable dyes with fluorescence readouts of oxidation and activated cleaved caspase-3 and-7, respectively. Gene expression was assessed by RNA-seq, RT-qPCR, and western blot. ChIP-qPCR was used to determine chromatin enrichment.
Results
TRAP-seq in a PDX1-deficiency model of β cell dysfunction uncovered a cohort of genes regulated at the level of mRNA translation, including the transcription factor JUND. Using a panel of diabetes-associated stressors, JUND was found to be upregulated in mouse islets cultured with high concentrations of glucose and free fatty acid, but not after treatment with hydrogen peroxide or thapsigargin. This induction of JUND could be attributed to increased mRNA translation. JUND was also upregulated in islets from diabetic db/db mice and in human islets treated with high glucose and free fatty acid. Depletion of JUND in primary islets reduced oxidative stress and apoptosis in β cells during metabolic stress. Transcriptome assessment identified a cohort of genes, including pro-oxidant and pro-inflammatory genes, regulated by JUND that are commonly dysregulated in models of β cell dysfunction, consistent with a maladaptive role for JUND in islets.
Conclusions
A translation-centric approach uncovered JUND as a stress-responsive factor in β cells that contributes to redox imbalance and apoptosis during pathophysiologically relevant stress.
Keywords: Translational regulation, Oxidative stress, Apoptosis, β cell
Abbreviations: TRAP, Translating Ribosome Affinity Purification; GLT, Glucolipotoxicity
Highlights
-
•
TRAP-seq uncovers genes with altered ribosome occupancy during PDX1 deficiency.
-
•
JUND is post-transcriptionally upregulated in β cells during metabolic stress.
-
•
Depletion of JUND protects β cells from oxidative stress and apoptosis.
-
•
JUND regulates a set of pro-oxidant and pro-inflammatory genes in β cells.
-
•
JUND target genes are commonly dysregulated in models of β cell dysfunction.
1. Introduction
Insufficient secretion of insulin from pancreatic β cells underlies all forms of diabetes. In type 2 diabetes (T2D), β cell failure is caused at least in part by prolonged oxidative stress, which leads to impaired insulin secretion, apoptosis, and loss of cell identity [1], [2]. Thus, elucidating the molecular mechanisms comprising the β cell stress response is central to the development of novel therapeutic strategies. Restricting these searches to the transcriptome of β cells, however, risks missing key players in disease processes that undergo post-transcriptional regulation. Specifically, regulation of mRNA translation is a critical and highly conserved component of the cellular response to stress [3].
β cells are particularly dependent on translational controls as evidenced by reduced β cell mass and permanent neonatal diabetes in patients with homozygous loss of the gene encoding EIF2AK3, a kinase that reduces translation rates during stress [4]. Global translation rates in β cells are responsive to nutrient excess, such as high free fatty acid levels, and excess translation can lead to β cell dysfunction [5]. Furthermore, the acute adaptation of β cells to high levels of glucose includes increased translation of the mRNA encoding proinsulin [6], highlighting the importance of transcript-specific translational controls.
Translational regulation is often overlooked because rapid advances in the depth and accuracy of RNA sequencing technologies have made the measurement of mRNA abundance a convenient readout of gene expression. Certain methodologies, including polysome profiling, Translating Ribosome Affinity Purification (TRAP), RiboTag, and ribosome profiling, leverage the power of RNA-seq to study translation by determining the relative density of ribosomes binding to mRNA, which we will refer to as ribosome occupancy, on a genome-wide scale [7]. Many studies have highlighted the use of these approaches to study translational regulation under stress or disease conditions, leading to the identification of novel stress-responsive factors and providing new insights into mechanisms of disease [8], [9], [10], [11], [12], [13].
For example, TRAP performed in INS-1 cells demonstrated a rapid increase in ribosomes binding to the mRNA encoding thioredoxin-interacting protein (TXNIP) during ER stress, which emphasized the importance of this factor in modulating β cell apoptosis under stress conditions [9]. Similarly, TRAP has been used to broadly characterize changes in gene expression in endothelial cells and cardiomyocytes in heart disease models [10].
In this study, we focus on translational regulation as a means to identify novel stress-responsive factors influencing β cell adaptation to diabetes-associated conditions. In particular, we perform TRAP followed by RNA-seq in a PDX1-deficiency model to screen for translationally regulated genes in β cells. This led us to examine the translational regulation of JUND, a transcription factor previously linked to regulating oxidative stress in other cell types [14], [15], but the role of which in β cells is unknown. In fibroblasts and endothelial cells, loss of JUND increases reactive oxygen species (ROS) levels; however, this effect has been attributed to the dysregulation of distinct target genes [14], [15], consistent with this factor playing cell-type-specific roles.
JUND is a member of the Jun family of transcription factors, which also includes C-JUN and JUNB. These factors form either homo- or hetero-dimers among themselves or with factors from other transcription factor families, including Fos, Atf, and Maf. Importantly, the composition of these dimers dictates the DNA binding preference and transcriptional effect of JUND [16]. Jund −/− mice have a shortened lifespan and hyperinsulinemic hypoglycemia, possibly due to hypervascularization of pancreatic islets [17]. However, whether loss of JUND has cell-autonomous effects in β cells and how this factor may influence adaptation to stress conditions are unknown. Here, we uncover JUND as a novel stress-responsive factor that is translationally upregulated in β cells during metabolic stress. Assessments of oxidative stress and apoptosis in primary islets as well as transcriptome analyses indicate that JUND promotes a maladaptive response in β cells during conditions associated with T2D.
2. Material and methods
2.1. Animals
Animal studies were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Wild type CD1 males were purchased from Jackson Laboratory. Male db/db mice (C57BLKS/J Leprdb/db) or db/+ mice (C57BLKS/J Leprdb/+) were purchased from Jackson Laboratory. Blood glucose levels were determined by handheld glucometers (One Touch). Serum NEFA levels were determined by fluorometric assay (Abcam). Mice were housed in a 12hr light/dark cycle and had ad libitum access to food.
2.2. Lentivirus production
293T cells were transfected for 8 h in OptiMEM using Lipofectamine 2000 (Invitrogen), after which the media was changed to standard high glucose DMEM. psPAX2 and pMD2.G were used for packaging and envelope vectors. These plasmids were a gift from Didier Trono (Addgene plasmid #12260 and # 12259). Media containing virus was collected 2 and 3 days post-transfection. Ultracentrifugation of collected media (19,000rpm for 1.5 h at 4 °C) was used to concentrate virus. Lentivirus was titered by RT-PCR [18].
2.3. Cell line culture
Min6 mouse insulinoma cells passage 20–30 were cultured in high glucose DMEM as described [19], unless otherwise noted. For siRNA-mediated depletion of Pdx1, cells were nucleofected by AMAXA with siRNA for Pdx1 (Dharmacon L-040402-01) or non-targeting control (Dharmacon D-001810-10) and collected 72hrs post-transfection. For lentiviral infections, Min6 cells were transduced for 6 h with virus and polybrene (Sigma) at 8ug/mL. Cells were allowed to recover for 4–5 days before collection or stress treatments. HEK293T cells were cultured in DMEM containing 25 mM glucose.
2.4. GFP-RPL10A Min6 stable cell line
The GFP-RPL10A transgene was generated by cloning PCR amplified fragments for GFP-RPL10A or GFP into the pBABE-puro retroviral vector [20] digested with SalI. Retrovirus was produced in HEK293T cells and added to Min6 cells, followed by two rounds of puromycin selection (5 days, 2ug/mL).
2.5. Islet isolation and culture
Mouse islets were isolated from 6 to 12 week old CD1 male mice unless otherwise noted. Briefly, ductal inflation of the pancreas was performed followed by collagenase digestion (Roche 11213873001). Islets were enriched by density gradient centrifugation with Ficoll–Paque (GE 45-001-751). After handpicking 3–4 times, islets were collected for RNA/protein isolation or cultured overnight for recovery from isolation and stress treatments were started the next day.
Human islets were obtained through the NIH-supported Human Pancreas Analysis Program via the University of Pennsylvania Islet Core facility. The islets were harvested from non-diabetic deceased donors without any identifying information at NIH-approved centers with informed consent and IRB approval at the islet isolation centers. Human islet donor characteristics are provided in Supplementary Table 1.
The culture media used for mouse and human islets was RPMI 1640 (11 mM glucose) supplemented with 10% FBS, 2 mM glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 1% antibiotic antimycotic (Thermo 15240096), and pH was adjusted to 7.3–7.4.
2.6. Islet transductions
Lentiviral infection of mouse islets was performed as described [21]. 100–200 islets were cultured overnight in serum-free islet media containing lentivirus at an MOI of 20.
2.7. Palmitate preparation and glucolipotoxicity conditions
Palmitate (Sigma–Aldrich P9767) was dissolved in 50% ethanol at 65 °C and diluted in 10% fatty acid free BSA (Sigma–Aldrich A7030) to a concentration of 7 mM. The mixture was incubated at 37 °C for 1 h to allow for conjugation before diluting in culturing media to a final concentration of 0.5 mM. Control media was made by performing the same procedure with 50% ethanol and no palmitate.
For islets, control media (described above) contained 11 mM glucose with no added palmitate. Media for glucolipotoxic conditions contained 25 mM glucose with 500uM palmitate.
For Min6, control media (DMEM) contained 5.6 mM glucose with no added palmitate while glucolipotoxic conditions had 25 mM glucose and 500uM palmitate.
2.8. RNA isolation and RT-qPCR
For Min6, cells were washed 2X with cold PBS before addition of TRIZOL (Invitrogen) and RNA was extracted according to manufacturer's instructions. RNA was reverse transcribed with random hexamers using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For islets, handpicked islets were washed 2X with cold PBS and RNA was extracted using RNeasy Mini Kit (Qiagen). RNA was reverse transcribed using oligo(dT) and Superscript III (Invitrogen). Quantitative PCR (BioRad CFX384) was used to measure transcript abundance and normalized to HPRT. See Supplementary Table 2 for a list of primer sequences used for these analyses.
2.9. Western blot
Proteins were separated by SDS-PAGE and immunoblotted with the following antibodies: rabbit anti-JUND (Santa Cruz, sc-74), mouse anti-ATF4 (Santa Cruz, sc-390063), rabbit anti-cleaved caspase-3 (Cell Signaling, 9664S), mouse anti-Tubulin (Sigma, T9026), mouse anti-Ran (B.D. 610340), goat anti-GFP (Abcam, 6673), mouse anti-RPL10A (Novus, 3G2), rabbit anti-RPL7 (Novus, NB100-2269), and rabbit anti-RPS6 (Abcam, ab40820).
2.10. TRAP
TRAP was performed as described [22], with minor modifications. Briefly, after stress treatments of GFP-RPL10A Min6 cells, cycloheximide (Sigma) was added to the culture media at 100ug/mL for 10 min prior to washing 2X with cold PBS. Cells were lysed and protein concentration was measured by BCA (Thermo). Total RNA was isolated (1–5% input) with the RNeasy Mini Kit (Qiagen). For IP RNA, cell lysates encompassing 200ug of protein were added to Protein G Dynabeads bound to GFP antibodies (19C8 and 19F7, Memorial Sloan-Kettering Monoclonal Antibody Facility) and incubated overnight at 4 °C. The next day, the beads were washed 4X with high salt buffer, as described [22]. IP RNA was eluted from beads in RLT buffer and extracted using the RNeasy Mini Kit. Ribosome occupancy was determined by dividing transcript abundance for each gene in the IP RNA fraction by its level in the Total RNA fraction. For TRAP followed by RT-qPCR, RNA was reverse transcribed with random hexamers and Superscript III (Invitrogen) and transcript abundance was first normalized to HPRT for each fraction before determining ribosome occupancy.
2.11. Cycloheximide chase assay
Min6 cells were cultured in control or glucolipotoxic conditions for 30hrs prior to treatment with cycloheximide at a concentration of 200ug/mL. Cells here harvested at 0hr, 3hr, 6hr, and 9hr time points post-cycloheximide treatment and analyzed by western blot.
2.12. shRNA design and cloning
The lentiviral backbone was generated by cloning the rat insulin II promoter (−405 to +7 relative to the TSS), GFP, and the UltramiR mir-30 scaffold [23] into pLenti CMV puro [24] by PCR and Gibson Assembly (replaced CMV promoter with rat insulin promoter). shRNA sequences were designed using the shERWOOD algorithm [23] (shJunD: AGCAGCTCAAACAGAAAGTCC, shNT: GCGCGATAGCGCTAATAATTT).
2.13. CRISPR design and cloning
CRISPR gRNAs were designed using http://crispr.mit.edu/to minimize off-target binding (ROSA26: AAGATGGGCGGGAGTCTTCT, JUND: CAGCTTGCGCTTGCGGCATT). gRNAs were cloned into lentiCRISPR v2, as described [25].
2.14. Immunofluorescence staining of isolated islets
Transduced islets were collected, dispersed to single cell suspension, washed in PBS, and fixed for 10 min at room temperature in 4% PFA. Fixed cells were attached to slides using cytospin. Following permeabilization (0.1% Triton x-100 for 10 min at room temperature), immunofluorescence staining was performed using the following antibodies: guinea pig anti-insulin (Dako, A0564) and goat anti-GFP (Abcam, 6673). Images were taken on a Keyence BZ-X700 microscope and images were analyzed using the BZ-X Advanced Analysis Software.
2.15. Oxidative stress measurement
For mouse islets, following stress treatment, 50–100 intact islets were transferred to a polysterene round bottom tube and CellROX Deep Red Reagent (Invitrogen) was added at 1:500 dilution to culture media followed by incubation at 37 °C for 45 min. Islets were washed 2X with PBS, dispersed to single cell suspension, attached to slides using cytospin, and imaged using fluorescence microscopy (Keyence BZ-X700 microscope). GFP signal was used to identify transduced cells and the CellROX signal from each cell was quantified and normalized by cell area using BZ-X Advanced Analysis Software. Signal from at least 60 GFP positive cells was assessed for each condition per experiment.
For Min6 cells, following stress treatment, CellROX Deep Red Reagent (Invitrogen) was added at 1:500 dilution directly to culture media followed by incubation at 37 °C for 30 min. Cells were washed 2X with PBS and imaged using fluorescence microscopy (Keyence BZ-X700 microscope). Bright field images were used to determine cell areas and CellROX signal was quantified from at least 6 imaging fields for each group using BZ-X Advanced Analysis Software.
2.16. Caspase-3 and-7 activation assay
Following stress treatment, 50–100 intact islets were transferred to a polysterene round bottom tube and fluorescent inhibitor of caspases (FLICA) reagent (Image-iT LIVE Red Caspase-3 and -7 Detect kit, Invitrogen) was added at 1:150 dilution followed by incubation at 37 °C for 1 h. Islets were washed 2X with wash buffer (provided by manufacturer), dispersed to single cell suspension, attached to slides using cytospin, and imaged using fluorescence microscopy (Keyence BZ-X700 microscope). Double positive cells were determined using BZ-X Advanced Analysis Software, and at least 200 GFP positive cells were counted for each condition per experiment.
2.17. RNA-seq and analysis
RNA was isolated using the RNeasy Mini Kit (Qiagen). Libraries were prepared using NEB Next Ultra RNA Library Prep Kit according to manufacturer's instructions with polyA enrichment followed by paired-end sequencing of 150bp using HiSeq (Illumina). Reads were mapped to mm10 using TopHat2 [26] and read counts per gene were determined using featureCounts [27]. For TRAP-seq analysis, ribosome occupancy was calculated by dividing normalized gene counts in the IP RNA samples by that in the total RNA samples. Differential expression analysis was performed using edgeR [28] with significant genes called using fold-change cutoffs of greater than 1.5 or less than −1.5 and FDR less than 0.05. Gene ontology analysis was performed using DAVID [29]. The overlap between RNA-seq data sets was determined using hypergeometric tests.
2.18. Chromatin immunoprecipitation followed by qPCR (ChIP-qPCR)
ChIP was performed as previously described [30], with the following modifications. For each immunoprecipitation, 40ug of lysate was used to prepare cross-linked chromatin and was immunoprecipitated using 6ug of JUND antibody (Santa Cruz, sc-74) or normal rabbit IgG (Santa Cruz). Peak locations and primer sequences to detect JUND occupancy are listed in Supplementary Table 3.
2.19. Statistics
Data are presented as mean ± standard error of the mean unless otherwise noted in figure legends. Statistical analyses were performed using GraphPad PRISM 7 software. Statistical tests used are noted in figure legends and include unpaired two-tailed Student's t-test, one-way analysis of variance (ANOVA), two-way ANOVA, and hypergeometric test.
2.20. Data availability
RNA-seq data that support the findings of this study have been deposited in NCBI's GEO under accession codes GSE115219 and GSE115239. Previously published sequencing data that were re-analyzed here are available under accession codes GSE107489 and GSE53949. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
3. Results
3.1. Genome-wide identification of genes with differential ribosome occupancy
In order to detect changes in mRNA translation on a genome-wide scale, we have employed the Translating Ribosome Affinity Purification (TRAP) methodology in which expression of a tagged ribosomal subunit (GFP-RPL10A) is used to immunoprecipitate ribosomes and their associated mRNA [22]. TRAP can be used to determine the ribosome occupancy, or the density of ribosome binding, for a particular gene by dividing transcript levels in the ribosome-associated RNA fraction (GFP immunoprecipitation) by that in the total RNA fraction (input). This calculation serves as a proxy for the efficiency of mRNA translation [10]. Stable expression of the GFP-RPL10A transgene in the mouse insulinoma Min6 cell line allowed for the isolation of intact ribosomes and their associated mRNAs (Supplementary Figure 1). To confirm that this approach can detect dynamic shifts in ribosome occupancy, TRAP was performed in GFP-RPL10A Min6 cells treated with thapsigargin to induce ER stress, and ribosome occupancy was determined by normalizing transcript abundance in the ribosome pull-down fraction to that in total RNA. This gave a near doubling of ribosome occupancy for the stress-responsive factor ATF4 during thapsigargin treatment (Figure 1A), consistent with previous findings using polysome profiling [5], [31].
Figure 1.
TRAP identifies genes with altered ribosome occupancy during PDX1 deficiency. (A) TRAP in GFP-RPL10a Min6 cells was used to determine ribosome occupancy (IP RNA/Input RNA) after 3hr treatment with thapsigargin (Tg, 1uM) or vehicle (DMSO), followed by RT-qPCR of IP RNA and Total RNA (n = 3). (B) Volcano plot depicting changes in ribosome occupancy with Pdx1 depletion as determined by TRAP-seq in GFP-RPL10a Min6 cells transfected with siRNA targeting Pdx1 or non-targeting (NT) control and harvested 72 h post-transfection (n = 3). Significant changes in ribosome occupancy shown in red (upregulated) or blue (downregulated). (C) Results from TRAP-seq with Pdx1 depletion plotted as a comparison of the change in transcript abundance with Pdx1 depletion in the IP RNA fraction to that in the Total RNA fraction. (D) Heatmap showing genes identified by TRAP-seq as having significant changes in ribosome occupancy but no significant change in total RNA levels after Pdx1 depletion. P values were calculated by unpaired two-tailed Student's t-test. * = p < 0.05.
To screen for translationally regulated genes in β cells, we used deficiency of the transcription factor PDX1 as a means to disrupt normal β cell homeostasis. PDX1 is a human diabetes gene [32], and reduced PDX1 levels cause β cell dysfunction and an impaired stress response [33], [34]. Given these connections to human disease and β cell stress, we reasoned that PDX1 deficiency was a promising model to uncover functionally important translational controls in β cells. Transfection of siRNA in GFP-RPL10A Min6 cells provided a 60% reduction of PDX1 transcript levels, and TRAP-seq identified 53 genes with an increase in ribosome occupancy after PDX1 depletion while 57 genes had a reduction (Figure 1B,C). A subset of these genes demonstrated no change in total mRNA abundance but a significant change in ribosome association (Figure 1D), indicating that the dominant regulatory mechanism for these genes was post-transcriptional. This list included several genes with known importance in β cells such as the transcription factor NKX2-2, a critical regulator of β cell identity and function [35], [36]. Further, several of these genes have potential relevance for the β cell stress response. Of particular interest was the transcription factor JUND, which had previously been linked to oxidative stress levels in other cell types [14], [15], but whose role in β cells during conditions associated with T2D was unknown.
3.2. Post-transcriptional upregulation of JUND in islets during metabolic stress
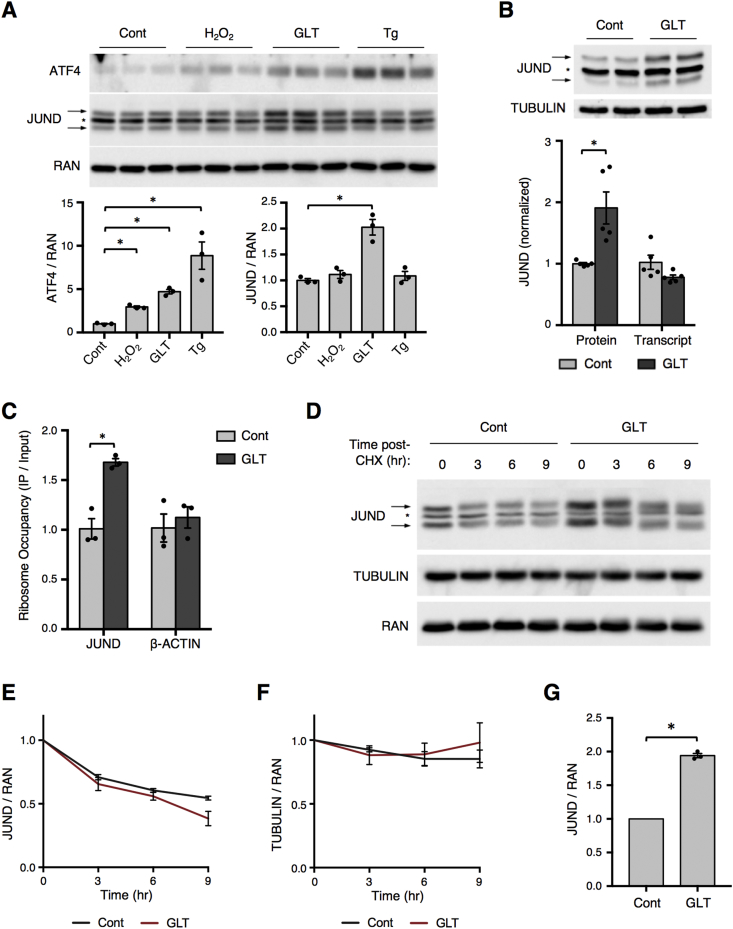
We next investigated whether the translational upregulation of JUND during PDX1 deficiency was generalizable to other models of β cell stress that are associated with T2D pathogenesis. To this end, we exposed mouse islets to a panel of stressors, including hydrogen peroxide (oxidative stress), high levels of glucose and the free fatty acid palmitate, termed glucolipotoxicity (metabolic stress), and thapsigargin (ER stress). While ATF4 was upregulated in all of these stress models, JUND was only induced by metabolic stress (Figure 2A). Despite the increase in JUND protein levels during glucolipotoxicity, there was no change in the abundance of JUND mRNA (Figure 2B). This discordance between steady state protein and mRNA levels could be caused by either an increase in the translation of JUND mRNA or an increase in the stability of JUND protein. In support of a translational mechanism, a TRAP assay performed in GFP-RPL10A Min6 cells showed a significant increase in the density of ribosomes binding to the JUND mRNA during metabolic stress (Figure 2C). We next performed a cycloheximide chase assay in Min6 cells and found no effect of glucolipotoxicity on JUND protein degradation rates compared to the loading controls RAN and TUBULIN (Figure 2D–F) despite the expected increase in JUND levels prior to cycloheximide treatment (Figure 2G). Together, these data support increased mRNA translation as the mechanism controlling JUND levels during metabolic stress.
Figure 2.
Post-transcriptional upregulation of JUND during glucolipotoxicity. (A) Western blot depicting ATF4 and JUND levels in mouse islets treated with hydrogen peroxide (H2O2, 200uM) for 1 h, glucolipotoxicity (GLT, 25 mM glucose, 500uM palmitate) for 2 days, or thapsigargin (Tg, 1uM) for 3 h (n = 3). (B) Increased protein levels of JUND as determined by Western blot, but not transcript levels by RT-qPCR, in mouse islets treated with glucolipotoxicity for 2 days (n = 5). (C) Increased ribosome occupancy of JUND in GFP-RPL10a Min6 cells after 30 h of glucolipotoxic conditions, as determined by TRAP followed by RT-qPCR (n = 3). (D) Representative western blot depicting reduction in JUND protein levels at indicated times after addition of cycloheximide (CHX) to Min6 cells. CHX added after culturing for 30hrs in control or glucolipotoxic (GLT) culturing conditions (n = 3). (E,F) Quantification of western blot signal for JUND or TUBULIN normalized to RAN. Each group normalized to value at 0hr time point to depict reduction in protein over time. (G) Quantification of JUND western blot for CHX chase assay at 0hr time points with normalization to the control group to depict increased JUND levels prior to CHX addition. P values were calculated by unpaired two-tailed Student's t-test, except in (E,F) in which a 2-way ANOVA was used. For western blot images of JUND, arrows denote two bands for JUND and * denotes a non-specific band. Otherwise, * = p < 0.05. Cont denotes control culturing conditions and GLT denotes glucolipotoxic culturing conditions.
To determine if this induction of JUND also occurs in β cells under metabolic stress in vivo, we used 12-week-old db/db mice, which are obese with elevated serum glucose and free fatty acid levels (Supplementary Figure 2). Indeed, compared to non-diabetic db/+ mice, islets from db/db mice had a significant increase in JUND protein but not mRNA (Figure 3A). To extend these findings to a human model, we assessed the induction of JUND during metabolic stress by culturing human islets with high levels of glucose and palmitate. Consistent with our findings in mouse islets, this excess of metabolic fuel caused an increase in JUND protein levels in human islets, indicating that JUND induction by metabolic stress is evolutionarily conserved (Figure 3B). Thus, we have uncovered the post-transcriptional upregulation of JUND as a novel component of the β cell response to metabolic stress.
Figure 3.
Upregulation of JUND in islets from diabetic db/db mice and in human islets treated with glucolipotoxicity. (A) Western blot showing increased JUND levels in islets isolated from db/db mice compared to db/+ at 12 wks of age. No change in JUND transcript as determined by RT-qPCR (n = 3). (B) Representative western blot showing increased JUND levels in human islets treated with glucolipotoxic (GLT) conditions for 2 days. Quantification of results from four human donors are shown. P values were calculated by unpaired two-tailed Student's t-test. For the western blot images of JUND, arrows denote two bands for JUND and * denotes a non-specific band.
3.3. Depletion of JUND blocks the increase in oxidative stress and apoptosis in β cells during glucolipotoxicity
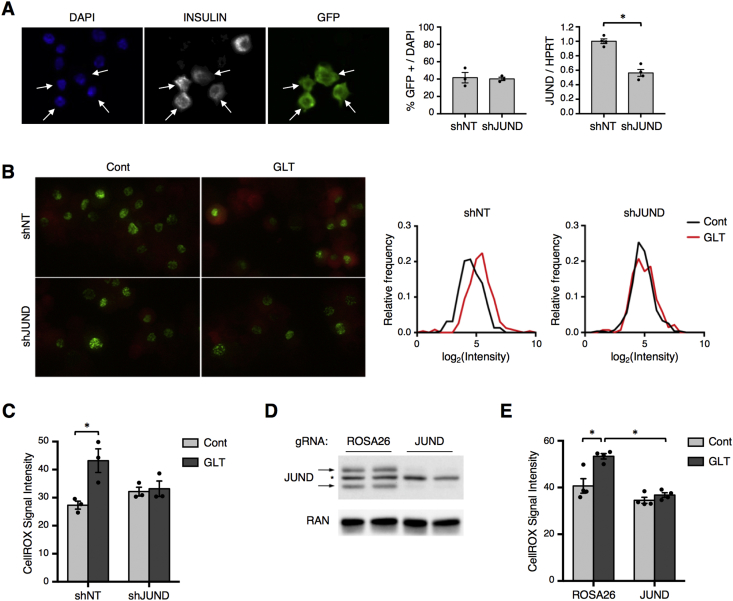
Chronically elevated glucose and free fatty acid levels cause β cell dysfunction and apoptosis at least partly due to increased oxidative stress [37], [38]. Given its connection to oxidative stress in other cell types [14], [15], we hypothesized that JUND influences β cell redox homeostasis during metabolic stress. To avoid cell non-autonomous effects in the Jund −/− model, we developed a system for β cell-specific depletion of JUND in primary islets by generating a lentiviral vector to co-express GFP and an shRNA from the rat insulin promoter (Supplementary Figure 3A). We first confirmed that this shRNA system efficiently targeted JUND in Min6 cells (Supplementary Figure 3B). To assess transduction efficiency in primary mouse islets, transduced islets were allowed to recover in culture media for two days and were subsequently dispersed to single cells for immunofluorescence staining. Nearly half of all islet cells were GFP positive, and all GFP positive cells stained positive for insulin, indicating efficient and β cell-specific transgene expression (Figure 4A). Further, the shRNA targeting JUND provided a 50% reduction in its transcript level in transduced islets (Figure 4A), similar to the rate of transduction and suggesting an effective depletion of JUND in those cells expressing the transgene.
Figure 4.
β cell-specific depletion of JUND in mouse islets prevents the increase in oxidative stress during glucolipotoxicity. (A) Immunofluorescence images to identify GFP and insulin positive cells show β cell-specific expression of GFP in half of mouse islet cells after lentiviral transduction of intact islets. Islets were dispersed to single cell suspensions and attached to slides using cytospin for analysis. Arrows denote β cells staining positive for GFP. Quantification of JUND transcript levels in intact islets by RT-qPCR shows that delivery of shRNA targeting JUND provides a significant reduction in JUND levels compared to a non-targeting (NT) control (n = 3–4). (B) Assessment of oxidative stress in β cells transduced with shRNA vectors targeting JUND or NT control by fluorescence imaging of CellROX Deep Red reagent. Intact mouse islets cultured for 2 days in control or glucolipotoxic conditions prior to oxidative stress assessment. Islets were dispersed to single cell suspensions and attached to slides using cytospin immediately prior to analysis. Representative fluorescence image for GFP and CellROX in islet cells after cytospin are shown, along with representative frequency distributions of CellROX intensity in GFP positive cells. (C) Quantifications of CellROX signal in GFP positive cells, which are averages of 3 independent experiments. (D) Western blot showing depletion of JUND in Min6 cells using lentiviral delivery of CRISPR-Cas9. A gRNA targeting the ROSA26 locus is used as a negative control. (E) Quantification of CellROX signal in Min6 cells with CRISPR-mediated depletion of JUND after culturing in glucolipotoxic conditions for 30hrs (n = 4). P values were calculated by two-way ANOVA. For Western blot images of JUND, arrows denote two bands for JUND and * denotes a non-specific band. Otherwise, * = p < 0.05. Cont denotes control culturing conditions and GLT denotes glucolipotoxic culturing conditions.
To test whether loss of JUND impacted oxidative stress levels in β cells, transduced islets were incubated with a cell-permeable dye that has increased fluorescence signal upon oxidation, and fluorescence intensity was quantified from GFP positive β cells. As expected, β cells exposed to glucolipotoxic conditions showed elevated oxidative stress as seen by increased fluorescence from the oxidation-sensitive dye (Figure 4B,C). Surprisingly, however, depletion of JUND in β cells blocked this increase in oxidative stress caused by high levels of glucose and palmitate (Figure 4B,C). This prevention of redox imbalance during metabolic stress was also observed in Min6 cells with CRISPR-mediated depletion of JUND compared to a control group with a gRNA targeting the ROSA26 locus (Figure 4D,E). These results indicate that JUND promotes oxidative stress in β cells during metabolic stress, which is in contrast to findings in fibroblasts and endothelial cells in which JUND ameliorates oxidative stress [14], [15], highlighting the cell-type-specificity of this factor.
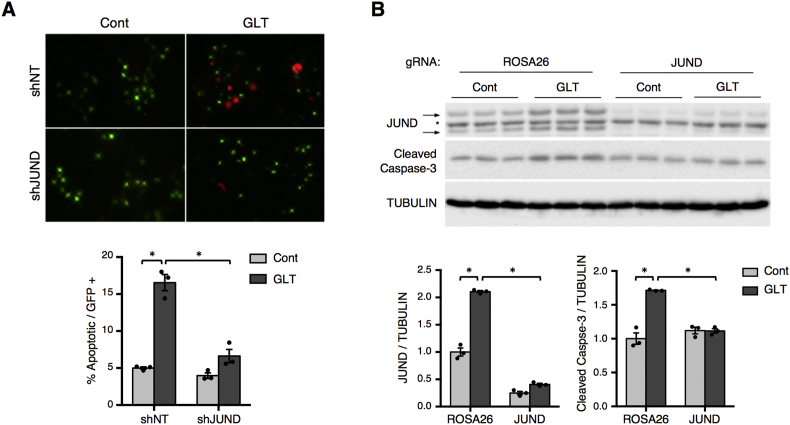
β cells are thought to be particularly sensitive to oxidative stress due to low expression of antioxidant genes [39]. Thus, we investigated whether the impact of JUND on redox homeostasis would also affect β cell apoptosis during metabolic stress. Using a fluorescence readout of caspase-3/7 activation, there was a 3-fold increase in the number of apoptotic β cells after culturing islets with high levels of glucose and palmitate for 3 days (Figure 5A). Consistent with its impact on redox imbalance, depletion of JUND significantly reduced this induction of apoptosis during metabolic stress (Figure 5A). This improvement in cell survival was confirmed in Min6 cells as CRISPR-mediated depletion of JUND completely abrogated the increase in cleaved caspase-3 levels caused by metabolic stress (Figure 5B).
Figure 5.
β cell-specific depletion of JUND in mouse islets reduces apoptosis during glucolipotoxicity. (A) Assessment of apoptosis in transduced β cells by fluorescence imaging of FLICA reagent, which marks apoptotic cells as red, after 3 days of culturing intact mouse islets in control or glucolipotoxic conditions. Islets were dispersed to single cell suspensions and attached to slides using cytospin immediately prior to analysis. Representative images for FLICA and GFP signals in cells after cytospin are shown. The percentage of GFP positive cells that were apoptotic were averaged for 3 independent experiments. (B) Western blot of cleaved caspase-3 used to assess apoptosis in Min6 cells with CRISPR-mediated depletion of JUND and cultured in glucolipotoxic conditions for 30hrs (n = 3). P values were calculated by two-way ANOVA. For western blot images of JUND, arrows denote two bands for JUND and * denotes a non-specific band. Otherwise, * = p < 0.05. Cont denotes control culturing conditions and GLT denotes glucolipotoxic culturing conditions.
3.4. JUND regulates a cohort of genes that are commonly dysregulated in models of β cell dysfunction
In accordance with the distinct pro-oxidant role of JUND in β cells, none of the antioxidant genes reported to be targets of JUND in other cell types were dysregulated in β cells (Supplementary Figure 3C). This indicates that both the transcriptional and phenotypic effects of JUND are cell-type-specific. To investigate how JUND impacts redox homeostasis and cell survival in β cells, we assessed the transcriptome of Min6 cells cultured with high levels of glucose and palmitate after CRISPR-mediated depletion of JUND, leading to the identification of 27 downregulated genes and 10 upregulated genes (Figure 6A). Gene ontology analysis on the downregulated genes demonstrated a significant enrichment in processes including stress response, reactive oxygen species (ROS) metabolism, and inflammation (Figure 6B). Interestingly, several of these genes, including Nos2, Ptgs2, and Steap4, encode proteins with enzymatic activity leading to ROS generation [40], [41], [42], [43], [44], consistent with our observed phenotype that loss of JUND dampens oxidative stress in β cells.
Figure 6.
JUND regulates pro-oxidant and pro-inflammatory genes associated with β cell stress and dysfunction. (A) Volcano plot depicting changes in transcript levels after CRISPR-mediated JUND depletion in Min6 cells cultured in glucolipotoxic conditions for 30hrs, as determined by RNA-seq (n = 3). Significant changes shown in red (upregulated) or blue (downregulated). (B) Gene ontology analysis for downregulated genes. (C) RT-qPCR panel of select JUND target genes in Min6 cells with CRISPR-mediated depletion of JUND and cultured in control or glucolipotoxic (GLT) conditions for 30hrs (n = 3). (D) ChIP-qPCR for specified genomic regions performed in Min6 cells cultured in control or GLT conditions for 30hrs. Values shown as enrichment over IgG control for each treatment group (n = 3). In (C), P values were calculated by two-way ANOVA and significance of p < 0.05 denoted by * for ROSA26 + Cont vs ROSA26 + GLT, # for ROSA26 + GLT vs JUND + GLT, and $ for JUND + Cont vs JUND + GLT. In (D), P values were calculated by one-way ANOVA and significance of p < 0.05 denoted by # for comparison to ALBUMIN in control treatment group and $ for comparison to ALBUMIN in GLT treatment group.
To assess the expression of JUND targets during metabolic stress, we selected a panel of these genes with links to oxidative stress and inflammation for further study. Interestingly, almost all of these genes were significantly upregulated by metabolic stress in Min6 cells, and loss of JUND either blocked or blunted this induction by glucolipotoxicity (Figure 6C). To determine whether JUND binds to genomic sites near these genes, we examined JUND ChIP-seq data performed in mouse embryonic fibroblasts [45] and found at least one JUND binding peak that contained a JUND/AP1 binding motif (TGAGTCA) near all of these genes (Supplementary Table 3). Despite its cell-type-specific properties, we reasoned that at least some of these sites may also be occupied by JUND in β cells. Indeed, ChIP-qPCR in Min6 cells showed that JUND binds to a subset of these peaks, especially during metabolic stress (Figure 6D). To determine whether these JUND targets were also induced by metabolic stress in primary tissue, mouse islets were treated with high levels of glucose and palmitate, which caused a significant increase in expression for all of these genes (Figure 7A). Furthermore, β cell-specific depletion of JUND in mouse islets followed by culturing in glucolipotoxic conditions confirmed that most of these genes are regulated by JUND in primary β cells (Figure 7B). Together, these results indicate that JUND governs the induction of genes linked to deleterious effects in β cells, including pro-oxidant and pro-inflammatory genes, during metabolic stress.
Figure 7.
JUND targets are upregulated in several models of β cell dysfunction. (A) Increased transcript levels of JUND target genes in mouse islets cultured for 2 days in glucolipotoxic (GLT) conditions (n = 3). (B) RT-qPCR panel of select JUND target genes in mouse islets with shRNA-mediated depletion of JUND and cultured for 2 days in GLT conditions (n = 3). (C) Heatmap of the genes downregulated after JUND depletion with detectable expression in islets from db/db mice. The change in expression after JUND depletion (JUND KO) is compared to the change in expression in islets from db/db vs db/+ mice. Genes with an asterisk had a statistically significant change in both data sets. P values were calculated by unpaired two-tailed Student's t-test in (A) and (B). * = p < 0.05, unless otherwise noted.
We next compared our RNA-seq results to several publicly available data sets to determine if the identified JUND target genes are dysregulated in other models of β cell dysfunction. Strikingly, nearly half of the genes downregulated with depletion of JUND were upregulated in islets from diabetic db/db mice [46] (Figure 6F), constituting a statistically significant overlap (Figure 6G, p = 6.0 × 10−5). The JUND target genes also significantly overlapped with genes upregulated during PDX1 deficiency (p = 7.7 × 10−5) or after treatment of human islets with palmitate (p = 3.5 × 10−4) [47]. Together, these findings indicate that JUND regulates a set of genes that are commonly increased in models of β cell dysfunction, suggesting a broad role for JUND in the maladaptive response of β cells under stress.
4. Discussion
Oxidative stress has long been recognized as a major contributor to β cell demise in T2D. Thus, new insights into the molecular mechanisms controlling redox homeostasis in β cells are central to our understanding of diabetes pathophysiology. Here, we have used a translation-centric approach to uncover JUND as a stress-responsive gene and a novel regulator of β cell redox homeostasis. JUND was upregulated in isolated mouse and human islets exposed to metabolic stress, as well as islets from diabetic db/db mice, indicating that this response is conserved across species and relevant for in vivo stress conditions. In β cells, JUND depletion dampens oxidative stress caused by the presence of excess metabolic fuel, which is in contrast to reports in other cell types where loss of JUND enhances oxidative stress due to downregulation of antioxidant genes [14], [15]. Although this discrepancy was unexpected, the cell-type-specific function of JUND is consistent with its ability to dimerize with a variety of binding partners, which likely shapes its tissue-specific effects [48]. Further, the amelioration of ROS accumulation with JUND depletion agrees with our finding that JUND positively regulates several genes with the capacity to increase ROS production in β cells, including Ptgs2, Steap4, and Nos2, while having no impact on the expression of antioxidant genes. Additionally, the concordance between the effects on oxidative stress and apoptosis further strengthens our conclusion that JUND plays a maladaptive role in β cells during prolonged metabolic stress.
It is unclear whether one or several of the identified JUND targets contribute to its effect on oxidative stress and apoptosis in β cells. Interestingly, both Ptgs2 and Steap4 are upregulated in islets from db/db mice and have some genetic association with T2D in humans [49], [50]. Ptgs2, which encodes cyclooxygenase-2 (COX-2), imparts deleterious effects in β cells, including impaired insulin secretion, reduced cell proliferation, and increased free radical levels [41], [51], [52]. Steap4 encodes a metalloreductase involved in iron and copper reduction [53]. While Steap4−/− mice develop hyperglycemia likely due to adipose inflammation and insulin resistance [54], its function in β cells is unknown. In osteoclast differentiation and prostate carcinogenesis, however, increased STEAP4 levels cause an elevation in cellular ROS [43], [44]. Further, Nos2, or inducible nitric oxide synthase (iNOS), has strong connections to cytokine-mediated β cell dysfunction and death [55]. Several other genes downregulated by JUND depletion are implicated in islet inflammation, including Ccl2, Cxcl1, and Cxcl2, which have been linked to poor outcomes for islet transplantation [56], [57]. Thus, targeting JUND may provide a new avenue for dampening oxidative stress and inflammation in islets, which warrants further investigation for clinical applications such as diabetes therapy and islet transplantation.
Translational regulation plays critical roles in shaping gene expression programs, especially during stress conditions, yet changes in mRNA translation are often overlooked in gene expression analyses. Here, we have used TRAP-seq to identify changes in translation associated with deficiency of the transcription factor PDX1. Given the critical roles for PDX1 in shaping the β cell gene expression program [34], [58], we reasoned that this model may reveal translational regulation that is especially important in β cells rather than general stress response factors, such as components of the unfolded protein response. This approach was successful in that it indicated translational regulation of genes with important roles in β cells, such as Nkx2.2 and Srebf1 [36], [59] while also highlighting genes that have previously not been studied in β cells, such as Jund. While we used PDX1 deficiency to screen for translationally regulated genes, this approach could readily be applied to a variety of stress conditions to reveal additional factors or pathways with important roles in β cell adaptation to T2D-associated conditions.
Our work addresses a critical gap in knowledge of the translational regulation underlying the β cell response to metabolic stress. We implicate the translational induction of JUND in β cells as a maladaptive response to excess metabolic fuel, thus uncovering a new prospect for therapeutic intervention in diabetes.
Author contributions
A.L.G. conceived of, designed, and performed experiments, interpreted results, and drafted and reviewed the manuscript. C.E.C., M.W.H., and J.Y. designed and performed experiments. N.M.D. performed experiments. M.J.B conceived of experiments and reviewed the manuscript. D.A.S. conceived of and designed the studies, interpreted results, and edited and reviewed the manuscript.
Acknowledgments
We thank the University of Pennsylvania Diabetes Research Center (DRC) for the use of the Functional Genomics Core and the Mouse Phenotyping, Physiology and Metabolism Core (P30-DK19525). Specifically, we acknowledge Dr. J. Schug and S. Rao for aid in data analysis, Dr. W. Quinn for assistance with NEFA assays, and Dr. A. Rozo and A. Gonzalez for assistance with db/db mouse dissections. We also thank Dr. S. Liebhaber and Dr. K. Lynch for helpful discussions. This work was supported by National Institutes of Health Grants F30-DK105758 (to A.L.G.) and P01-DK49210 (to D.A.S.). We also would like to thank the Medical Scientist Training Program (MSTP) at the University of Pennsylvania for their support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.04.007.
Conflict of interest
The authors have no conflict of interest to declare.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kitamura T. The role of FOXO1 in β-cell failure and type 2 diabetes mellitus. Nature Reviews Endocrinology. 2013;9(10):615–623. doi: 10.1038/nrendo.2013.157. [DOI] [PubMed] [Google Scholar]
- 2.Cao S.S., Kaufman R.J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxidants and Redox Signaling. 2014;21(3):396–413. doi: 10.1089/ars.2014.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spriggs K.A., Bushell M., Willis A.E. Translational regulation of gene expression during conditions of cell stress. Molecular Cell. 2010;40(2):228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 4.Delépine M., Nicolino M., Barrett T., Golamaully M., Lathrop G.M., Julier C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nature Genetics. 2000;25(4):406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 5.Hatanaka M., Maier B., Sims E.K., Templin A.T., Kulkarni R.N., Evans-Molina C. Palmitate induces mRNA translation and increases ER protein load in islet β-cells via activation of the mammalian target of rapamycin pathway. Diabetes. 2014;63(10):3404–3415. doi: 10.2337/db14-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itoh N., Okamoto H. Translational control of proinsulin synthesis by glucose. Nature. 1980;283(5742):100–102. doi: 10.1038/283100a0. [DOI] [PubMed] [Google Scholar]
- 7.Kuersten S., Radek A., Vogel C., Penalva L.O.F. Translation regulation gets its “omics” moment. Wiley Interdisciplinary Reviews: RNA. 2013;4(6):617–630. doi: 10.1002/wrna.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sidrauski C., McGeachy A.M., Ingolia N.T., Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife. 2015 doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lerner A.G., Upton J.-P., Praveen P.V.K., Ghosh R., Nakagawa Y., Igbaria A. IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metabolism. 2012;16(2):250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou P., Zhang Y., Ma Q., Gu F., Day D.S., He A. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proceedings of the National Academy of Sciences. 2013;110(38):15395–15400. doi: 10.1073/pnas.1304124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatanaka M., Anderson-Baucum E., Lakhter A., Kono T., Maier B., Tersey S.A. Chronic high fat feeding restricts islet mRNA translation initiation independently of ER stress via DNA damage and p53 activation. Scientific Reports. 2017;7(1):102–111. doi: 10.1038/s41598-017-03869-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sendoel A., Dunn J.G., Rodriguez E.H., Naik S., Gomez N.C., Hurwitz B. Translation from unconventional 5' start sites drives tumour initiation. Nature. 2017;541(7638):494–499. doi: 10.1038/nature21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schafer S., Adami E., Heinig M., Rodrigues K.E.C., Kreuchwig F., Silhavy J. Translational regulation shapes the molecular landscape of complex disease phenotypes. Nature Communications. 2015;6:1–9. doi: 10.1038/ncomms8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerald D., Berra E., Frapart Y.M., Chan D.A., Giaccia A.J., Mansuy D. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell. 2004;118(6):781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 15.Paneni F., Osto E., Costantino S., Mateescu B., Briand S., Coppolino G. Deletion of the activated protein-1 transcription factor JunD induces oxidative stress and accelerates age-related endothelial dysfunction. Circulation. 2013;127(11):1229–1240. doi: 10.1161/CIRCULATIONAHA.112.000826. [DOI] [PubMed] [Google Scholar]
- 16.Mechta-Grigoriou F., Gerald D., Yaniv M. The mammalian Jun proteins: redundancy and specificity. Oncogene. 2001;20(19):2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- 17.Laurent G., Solari F., Mateescu B., Karaca M., Castel J., Bourachot B. Oxidative stress contributes to aging by enhancing pancreatic angiogenesis and insulin signaling. Cell Metabolism. 2008;7(2):113–124. doi: 10.1016/j.cmet.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Sastry L., Johnson T., Hobson M.J., Smucker B., Cornetta K. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Therapy. 2002;9(17):1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- 19.Claiborn K.C., Sachdeva M.M., Cannon C.E., Groff D.N., Singer J.D., Stoffers D.A. Pcif1 modulates Pdx1 protein stability and pancreatic β cell function and survival in mice. Journal of Clinical Investigation. 2010;120(10):3713–3721. doi: 10.1172/JCI40440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgenstern J.P., Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Research. 1990;18(12):3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jimenez-Moreno C.M., Herrera-Gomez I.G., Lopez-Noriega L., Lorenzo P.I., Cobo-Vuilleumier N., Fuente-Martin E. A simple high efficiency intra-islet transduction protocol using lentiviral vectors. Current Gene Therapy. 2015;15(4):436–446. doi: 10.2174/1566523215666150630121557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyle J.P., Dougherty J.D., Heiman M., Schmidt E.F., Stevens T.R., Ma G. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knott S.R.V., Maceli A.R., Erard N., Chang K., Marran K., Zhou X. A computational algorithm to predict shRNA potency. Molecular Cell. 2014;56(6):796–807. doi: 10.1016/j.molcel.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campeau E., Ruhl V.E., Rodier F., Smith C.L., Rahmberg B.L., Fuss J.O. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4(8):e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 28.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Juliana C.A., Yang J., Cannon C.E., Good A.L., Haemmerle M.W., Stoffers D.A. A PDX1-ATF transcriptional complex governs β cell survival during stress. Molecular Metabolism. 2018;17:39–48. doi: 10.1016/j.molmet.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan B.J., Krokowski D., Majumder M., Schmotzer C.L., Kimball S.R., Merrick W.C. Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2α. Journal of Biological Chemistry. 2014;289(18):12593–12611. doi: 10.1074/jbc.M113.543215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoffers D.A., Ferrer J., Clarke W.L., Habener J.F. Early-onset type-ll diabetes mellitus (MODY4) linked to IPF1. Nature Genetics. 1997;17(2):138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 33.Brissova M., Shiota M., Nicholson W.E., Gannon M., Knobel S.M., Piston D.W. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. Journal of Biological Chemistry. 2002;277(13):11225–11232. doi: 10.1074/jbc.M111272200. [DOI] [PubMed] [Google Scholar]
- 34.Sachdeva M.M., Sachdeva M.M., Claiborn K.C., Claiborn K.C., Khoo C., Khoo C. Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(45):19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle M.J., Sussel L. Nkx2.2 regulates beta-cell function in the mature islet. Diabetes. 2007;56(8):1999–2007. doi: 10.2337/db06-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutiérrez G.D., Bender A.S., Cirulli V., Mastracci T.L., Kelly S.M., Tsirigos A. Pancreatic β cell identity requires continual repression of non-β cell programs. Journal of Clinical Investigation. 2017;127(1):244–259. doi: 10.1172/JCI88017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piro S., Anello M., Di Pietro C., Lizzio M.N., Patan egrave G., Rabuazzo A.M. Chronic exposure to free fatty acids or high glucose induces apoptosis in rat pancreatic islets: possible role of oxidative stress. Metabolism. 2002;51(10):1340–1347. doi: 10.1053/meta.2002.35200. [DOI] [PubMed] [Google Scholar]
- 38.Poitout V., Robertson R.P. Glucolipotoxicity: fuel excess and β-cell dysfunction. Endocrine Reviews. 2008;29(3):351–366. doi: 10.1210/er.2007-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenzen S., Drinkgern J., Tiedge M. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radical Biology and Medicine. 1996;20(3):463–466. doi: 10.1016/0891-5849(96)02051-5. [DOI] [PubMed] [Google Scholar]
- 40.Xia Y., Zweier J.L. Superoxide and peroxynitrite generation from inducible nitric oxide synthase in macrophages. Proceedings of the National Academy of Sciences. 1997;94(13):6954–6958. doi: 10.1073/pnas.94.13.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabatabaie T., Vasquez-Weldon A., Moore D.R., Kotake Y. Free radicals and the pathogenesis of type 1 diabetes: beta-cell cytokine-mediated free radical generation via cyclooxygenase-2. Diabetes. 2003;52(8):1994–1999. doi: 10.2337/diabetes.52.8.1994. [DOI] [PubMed] [Google Scholar]
- 42.Maciag A. Mutant K-rasV12 increases COX-2, peroxides and DNA damage in lung cells. Carcinogenesis. 2004;25(11):2231–2237. doi: 10.1093/carcin/bgh245. [DOI] [PubMed] [Google Scholar]
- 43.Jin Y., Wang L., Qu S., Sheng X., Kristian A., Maelandsmo G.M. STAMP2 increases oxidative stress and is critical for prostate cancer. EMBO Molecular Medicine. 2015;7(3):315–331. doi: 10.15252/emmm.201404181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J., Ye S., Fujiwara T., Manolagas S.C., Zhao H. Steap4 plays a critical role in osteoclastogenesis in vitro by regulating cellular iron/reactive oxygen species (ROS) levels and cAMP response element-binding protein (CREB) activation. Journal of Biological Chemistry. 2013;288(42):30064–30074. doi: 10.1074/jbc.M113.478750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vierbuchen T., Ling E., Cowley C.J., Couch C.H., Wang X., Harmin D.A. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Molecular Cell. 2017;68(6):1067–1082. doi: 10.1016/j.molcel.2017.11.026. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neelankal John A., Ram R., Jiang F.X. RNA-seq analysis of islets to characterise the dedifferentiation in type 2 diabetes model mice db/db. Endocrine Pathology. 2018;15(4):207–221. doi: 10.1007/s12022-018-9523-x. [DOI] [PubMed] [Google Scholar]
- 47.Cnop M., Abdulkarim B., Bottu G., Cunha D.A., Igoillo-Esteve M., Masini M. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63(6):1978–1993. doi: 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- 48.Hai T., Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proceedings of the National Academy of Sciences. 1991;88(9):3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konheim Y.L., Wolford J.K. Association of a promoter variant in the inducible cyclooxygenase-2 gene (PTGS2) with type 2 diabetes mellitus in Pima Indians. Human Genetics. 2003;113(5):377–381. doi: 10.1007/s00439-003-1000-y. [DOI] [PubMed] [Google Scholar]
- 50.Sharma P.R., Mackey A.J., Dejene E.A., Ramadan J.W., Langefeld C.D., Palmer N.D. An islet-targeted genome-wide association scan identifies novel genes implicated in cytokine-mediated islet stress in type 2 diabetes. Endocrinology. 2015;156(9):3147–3156. doi: 10.1210/en.2015-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Persaud S.J., Muller D., Belin V.D., Kitsou-Mylona I., Asare-Anane H., Papadimitriou A. The role of arachidonic acid and its metabolites in insulin secretion from human islets of langerhans. Diabetes. 2007;56(1):197–203. doi: 10.2337/db06-0490. [DOI] [PubMed] [Google Scholar]
- 52.Oshima H., Taketo M.M., Oshima M. Destruction of pancreatic beta-cells by transgenic induction of prostaglandin E2 in the islets. Journal of Biological Chemistry. 2006;281(39):29330–29336. doi: 10.1074/jbc.M602424200. [DOI] [PubMed] [Google Scholar]
- 53.Scarl R.T., Lawrence C.M., Gordon H.M., Nunemaker C.S. STEAP4: its emerging role in metabolism and homeostasis of cellular iron and copper. Journal of Endocrinology. 2017;234(3):R123–R134. doi: 10.1530/JOE-16-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wellen K.E., Fucho R., Gregor M.F., Furuhashi M., Morgan C., Lindstad T. Coordinated regulation of nutrient and inflammatory responses by STAMP2 is essential for metabolic homeostasis. Cell. 2007;129(3):537–548. doi: 10.1016/j.cell.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zumsteg U., Frigerio S., Holländer G.A. Nitric oxide production and Fas surface expression mediate two independent pathways of cytokine-induced murine beta-cell damage. Diabetes. 2000;49(1):39–47. doi: 10.2337/diabetes.49.1.39. [DOI] [PubMed] [Google Scholar]
- 56.Piemonti L., Leone B.E., Nano R., Saccani A., Monti P., Maffi P. Human pancreatic islets produce and secrete MCP-1/CCL2: relevance in human islet transplantation. Diabetes. 2002;51(1):55–65. doi: 10.2337/diabetes.51.1.55. [DOI] [PubMed] [Google Scholar]
- 57.Citro A., Cantarelli E., Maffi P., Nano R., Melzi R., Mercalli A. CXCR1/2 inhibition enhances pancreatic islet survival after transplantation. Journal of Clinical Investigation. 2012;122(10):3647–3651. doi: 10.1172/JCI63089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao T., McKenna B., Li C., Reichert M., Nguyen J., Singh T. Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metabolism. 2014;19(2):259–271. doi: 10.1016/j.cmet.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takahashi A., Motomura K., Kato T., Yoshikawa T., Nakagawa Y., Yahagi N. Transgenic mice overexpressing nuclear SREBP-1c in pancreatic beta-cells. Diabetes. 2005;54(2):492–499. doi: 10.2337/diabetes.54.2.492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data that support the findings of this study have been deposited in NCBI's GEO under accession codes GSE115219 and GSE115239. Previously published sequencing data that were re-analyzed here are available under accession codes GSE107489 and GSE53949. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.