Abstract
Ubiquitination regulates nearly every aspect of cellular events in eukaryotes. It modifies intracellular proteins with 76-amino acid polypeptide ubiquitin (Ub) and destines them for proteolysis or activity alteration. Ubiquitination is generally achieved by a tri-enzyme machinery involving ubiquitin activating enzymes (E1), ubiquitin conjugating enzymes (E2) and ubiquitin ligases (E3). E1 activates Ub and transfers it to the active cysteine site of E2 via a transesterification reaction. E3 coordinates with E2 to mediate isopeptide bond formation between Ub and substrate protein. The E1-E2-E3 cascade can create diverse types of Ub modifications, hence effecting distinct outcomes on the substrate proteins. Dysregulation of ubiquitination results in severe consequences and human diseases. There include cancers, developmental defects and immune disorders. In this review, we provide an overview of the ubiquitination machinery and discuss the recent progresses in the ubiquitination-mediated regulation of embryonic stem cell maintenance and cancer biology.
Keywords: ubiquitination, embryonic stem cells, colorectal cancer, osteosarcoma
1. Introduction
Ubiquitination is an energy-dependent enzymatic process to modify proteins with a small-sized polypeptide ubiquitin (Ub) at the post-translational level. Its name was derived from the Latin “ubique”, which means "everywhere", describing its ubiquitous expression pattern in diverse types of eukaryotic cells [1]. Ub is highly conserved in sequence and structure across animals, plants and fungi [2,3]. It modifies thousands of intracellular proteins either as a "death label" for degradation or non-proteolytic signal governing protein activity [4,5,6,7]. This modification is of great biological significance since it encompasses nearly all aspects of cellular events. Dysregulation in ubiquitination leads to severe consequences and human diseases, such as cancers, degenerative diseases and immune disorders [8]. This review provides overview of the enzymatic machinery mediating ubiquitination and surveys the roles of ubiquitination in regulating embryonic stem (ES) cell maintenance and cancer development.
2. The Ubiquitination Machinery
Canonical ubiquitination is an ATP-dependent enzymatic process during which an isopeptide bond is formed between the C-terminal carboxy group of the Ub residue glycine-76 (Ub-G76) and the ε-amino group of lysine (K) residues in proteins. In a small number of instances, Ub can be conjugated to the N-terminal methionine (M1) and other nonlysine residues, such as cysteine (C), serine (S), threonine (T) or tyrosine (Y) [9,10,11,12,13,14]. For instance, members of the SidE family, which serve as effectors of the pathogen Legionella pneumophila, can directly ubiquitinate the S residues of substrates via the phosphor-ribosyl linkage [15].
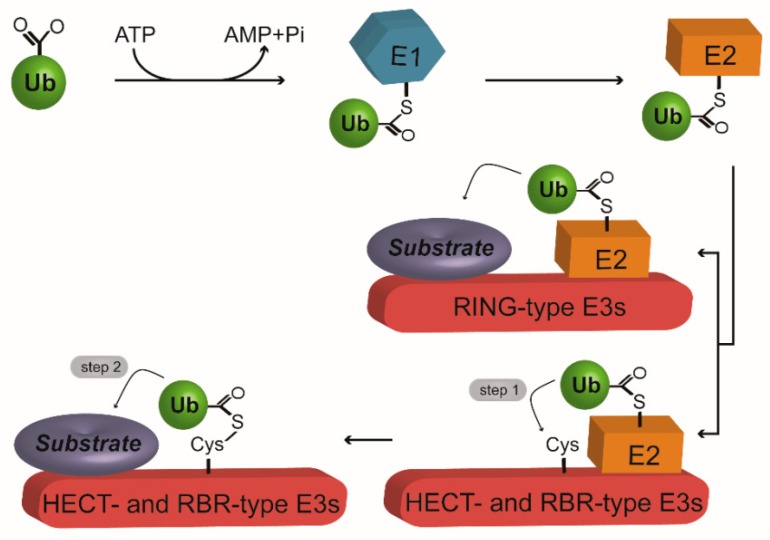
Ubiquitination is generally achieved by the machinery comprising three types of enzymes (Figure 1). They include Ub-activating enzyme (Uba, E1), Ub-conjugating enzyme (UBC, E2) and Ub ligase (E3) [4]. The catalytic activities of these enzymes are sequentially organized in the reaction cascade. Firstly, E1 activates the Ub-G76 residue in ATP hydrolysis-dependent manner to produce a Ub-adenylate intermediate. Subsequently, AMP is released and the Ub is transferred to the active C site of E1 via a thioester bond [16]. Next, the E1-Ub conjugate interacts with E2 for a transthiolation reaction during which the E1- activated Ub is transferred to the active C site of E2 to form an E2-Ub conjugate. At the final step, E3 concurrently associates with the E2-Ub conjugate and a substrate protein to mediate isopeptide bond formation between the Ub moiety and substrate [17]. In general, E2s dominantly determine the exact K residue of Ub for ubiquitination, the number of Ub moieties added and the linkage type of poly-Ub chains, whereas E3s regulate the specificity of substrate recognition [18,19,20,21]. Although the E1-E2-E3 cascade is adopted by most intracellular processes, some instances require an additional E4 ligase for poly-Ub chain extension or multi-Ub chain assembly [22,23,24,25,26,27,28,29]. For example, E3 mouse double minute 2 homolog (MDM2) modifies p53 with a single Ub moiety (monoubiquitination). Following that, p300 serves as an E4 ligase, adding more Ub moieties to the monoubiquitinated residue of p53 to form a poly-Ub chain [29]. Of note, ubiquitination can also be accomplished by a single E3, in place of a multi-enzyme machinery. Such fact has been reported for the mono-ADP-ribosyltransferase SdeA. SdeA is secreted by Legionella pneumophila in the host cells. To directly ubiquitinate host's substrate proteins, it utilizes nicotinamide adenine dinucleotide (NADH) to form ADP-ribosylated Ub [30].
Figure 1.
The ubiquitination machinery. Ubiquitination is initiated by E1-mediated ubiquitin (Ub) activation. Next, Ub is transferred to E2 to form an E2-Ub conjugate. At the final step, E3 mediates isopeptide bond formation between the Ub and the substrate. Really interesting new gene (RING)-type E3s serve as a scaffold to directly transfer the Ub from E2 to the substrate. On the other hand, homologous to E6-AP COOH terminus (HECT)- and RING between RING (RBR)-type E3s require a two-step reaction to achieve Ub ligation with the substrate. In the first step, Ub is transferred from E2 to E3, producing an E3-Ub thioester intermediate. At the second step, Ub is finally handed over to the substrate. Arrows represent the next steps during the process of ubiquitination.
Compared with E1s, there is a wider variety of E2 and E3 enzymes in eukaryotes. The human genome encodes only two E1s, but 40 E2s and over 600 E3s [20,21,31]. All E2s contain a conserved catalytic UBC domain with the active site C. The UBC domain has about 150 amino acids and constitutes the full-length sequence of class I E2s. In addition, other E2s possess extended sequences at either the C- (class II) or the N-terminus (class III). Meanwhile, E2s with extension regions at both the N- and C-terminus are grouped as class IV. The extension regions are involved in the determination of cellular localization and protein-protein interaction [31,32].
E3s are the most abundant enzymes involved in ubiquitination. According to their catalytic domains and Ub transfer mechanisms, E3s are classified into three groups. These comprise of the Really Interesting New Gene (RING)-type, homologous to E6-AP COOH terminus (HECT)-type and RING between RING (RBR)-type E3s [33]. The RING-type E3 family members are characterized by its RING or U-box domain. These two domains exhibit similar RING finger fold in structure. However, the activity of RING domain requires chelation of two zinc ions (Zn2+), whereas the U-box domain is Zn2+-independent. During ubiquitination, RING-type E3s serve as a scaffold for the binding of the E2s and their substrates. This allosterically stimulates a direct transfer of Ub moiety from the E2-Ub conjugate to the substrates [33]. Compared with the other types of E3s, RING-type E3s represent the most abundant ligases with over 500 family members [33]. Notably, some RING-type E3s, also known as the Cullin-RING ligases (CRLs), form a large complex with multiple subunits to mediate ubiquitination [34]. In spite of its diversity in subunit assembly, all CRLs possess at least four common subunits, including an E2-binding catalytic RING finger, a scaffold comprising seven Cullins (CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5, and CUL7), a receptor for substrate recognition and an adaptor arm responsible for the linkage between the receptor and the Cullin scaffold [34]. Two typical CRLs are the anaphase-promoting complex/cyclosome (APC/C) and the Skp1/Cul1/F-box (SCF). 1.2 MDa-sized APC/C is a large ligase complex which consists of 19 subunits, such as the Apc11 (RING subunit), Apc2 (Cullin scaffold) and coactivator subunit Cdc20/Cdh1 [35,36]. Apc11 and Apc2 form the catalytic center, while Cdc20/Cdh1 is involved in substrate recognition and enhancement of the catalytic activity of Apc11 [35,37]. The HECT-type E3s possess a conserved catalytic HECT domain with the active site C at the C-terminus and a variable N-terminal extension that largely determines the specificity of its substrate recognition [34]. There are about 28 HECT-type E3s encoded by the human genome [38]. According to the variable N-terminal extensions, these HECT-type E3s can be further classified into three subfamilies, including the WW domain-containing Nedd4/Nedd4-like E3s, HECT and RCC1-like (HERC)- and RCC1-like domains (RLD)-containing E3s, and the HECT-type E3s without WW and RLD domains [39]. Distinct from the RING-type ligases, HECT E3s require a two-step reaction to ligate Ub with sustrates. In the first step, the Ub moiety from the Ub-E2 conjugate is transferred to the catalytic C site of HECT-type E3 to form a HECT-Ub thioester intermediate. Subsequently, the Ub is relocated from the intermediate to the substrates [33]. There are about 14 RBR-type E3s encoded in the human genome [40]. These ligases possess Zn2+-binding RING domains (RING1 and RING2). The RING2 domain contains an active site C which alike the HECT-type E3s, is absent in the RING-type E3s. Thus, RBR-type E3s appear to be RING-HECT hybrid in its sequence and domain structure. Catalytically, it adopts similar two-step mechanism as the HECT-type E3s to ligate Ub to substrate proteins [41]. Specifically, the RING1 provides a binding site for the E2-Ub conjugate, and the Ub moiety is firstly transferred to the active C site of RING2 to form a covalent E3-Ub intermediate. During the second step, RING2 mediates ligation of the Ub moiety to the substrate [41].
3. The Types of Ubiquitination
The canonical ubiquitination linkage types include monoubiquitination (addition of one Ub monomer at a single K residue), multi-monoubiquitination (simultaneous monoubiquitination at multiple K residues) and polyubiquitination (addition of a Ub chain in which Ub moieties are sequentially linked to a K residue of the existing Ub) [42]. Ub altogether possesses seven K residues (K6, K11, K27, K29, K33, K48 and K63), and any of these Ks can be a site of chain linkage [5,43]. In some cases, the M1 residue of Ub could also be used to form a linear poly-Ub chain. Generally, a poly-Ub chain is homotypic since all Ub moieties in the chain provide the same residue for linkage. Notwithstanding, multiple linkage types can simultaneously arise in a single poly-Ub chain. On the other hand, atypical branched linkages can be formed in which more than one K residues of a single Ub moiety are involved in linkage formation at the same time [44,45]. To date, all the linkage types have been described in eukaryotic cells [7,46]. They are implicated in regulating the fate and activities of the substrate proteins [7,46]. The best-known role of ubiquitination is to serve as a "death label" for substrate protein degradation. Mono-Ub, K6-, K11-, K29- and K48-linked poly-Ub chains are signals which drive proteins for proteasome or lysosome-mediated proteolysis [47,48]. In addition, ubiquitination could also serve as a nonproteolytic signal to regulate the activity and sub-cellular localization of substrate proteins. For instance, receptor interacting protein 1 (RIP1) can be modified by K63-linked poly-Ub chain. This modification is required for the interaction of RIP1 with the Transforming growth factor β-activated kinase 1 (TAK1)/TGF-beta-activated kinase 1 and MAP3K7-binding protein (TAB) complex and inhibitors of I-κB kinase (IKK) to activate signal transduction in the NFκB pathway [49,50,51]. Of note, modification by the same linkage type could produce differing outcomes. For example, it is generally believed that K63 linkage acts as a non-degradative signal to modify protein activity. However, the same modification labels Octamer-binding transcription factor-4 (Oct4) for 26S proteasome-mediated degradation in pluripotent stem cells [52,53,54]. K11-linked poly-Ub chain exhibits similar dual roles. It marks substrates for their degradation during cell-cycle process and stem cell differentiation [55,56,57]. In contrast, K11 linkage increases the stability of β-Catenin, hence contributing to the accumulation of the oncogenic β-Catenin in human colon cancer cells [57,58]. On the other hand, a single protein can be modified by different types of ubiquitination, resulting in diverse outcomes. For instance, DNA polymerase processivity factor PCNA (proliferating cell nuclear antigen) can be either monoubiquitinated by the E2-E3 complex RAD6-RAD18 at the K164 residue or further modified with K63-linked poly-Ub chain by another E2-E3 complex MMS2-UBC13-RAD5 [59,60]. These modifications on PCNA determine which DNA damage tolerance (DDT) pathways will be utilized by cells to bypass DNA lesions during replication. Monoubiquitinated PCNA promotes DNA polymerase ζ-dependent (mutagenic) or DNA polymerase η-dependent (error-free) translesion DNA synthesis (TLS), while polyubiquitinated PCNA can initiate template switching (TS) for an error-free lesion bypass [59,61].
4. Ubiquitination and Embryonic Stem Cells
Stem cells exhibit unique "stemness" state that is defined by the ability to self-renew and differentiate to germ lineages. These specialized cells can be found in both adult and embryonic tissues, performing vital functions in cell regenerations, growth and embryo development. Based on their capacity to differentiate, stem cells can be distinguished into four types, namely the totipotent, pluripotent, multipotent or unipotent stem cells. Totipotent stem cells alone can give rise to an entire organism. Such developmental potential resembles that of the fertilized zygote and the blastomeres up till the eight-cell stage [62]. Pluripotent stem cells are not able to produce an organism by themselves. Nevertheless, they can differentiate into all the cell types in an organism. They are best represented by the embryonic stem (ES) cells, embryonic germ (EG) cells or the embryonic carcinoma (EC) cells. Multipotent stem cells can give rise to certain specialized lineage cells. Most of the adult stem cells are multipotent. They include hematopoietic stem cells, mesenchymal stem cells (MSCs), and other adult progenitor cells. The differentiation capacity of unipotent stem cells is restricted since they can only give rise to one type of cells, such as the myoblast.
Pluripotent ES cells can be derived from the inner cell mass (ICM) of early blastocysts [63]. The embryonic origin and pluripotent potential provides ES cells as a great model for the study of gene function, early embryogenesis and directed differentiation for future cell replacement therapy in clinic. A panel of proteins, including signaling pathway mediators, transcription factors (TFs) and epigenetic regulators, cooperate tightly to form precise regulatory networks orchestrating the stemness of ES cells. Any dosage or activity alteration in these proteins via ubiquitination could impact on ES cell self-renewal and differentiation capacity. As expected, increasing numbers of ubiquitination-related factors have been identified for their important roles in ES cell regulation.
4.1. Regulation of Stemness-Related TFs by Ubiquitination in ES Cells
TFs are the most abundant group of proteins encoded by the mammalian genome [64]. They are characterized by the capability of directly interacting with DNA elements to regulate transcription. In ES cells, a group of TFs comprise a delicate regulatory circuitry that pivotally monitors the stemness-specific gene expression profile. Among them are the core stemness regulator trinity, Oct4, Sex determining region Y (SRY)-related high-mobility group (HMG) Box 2 (Sox2) and Nanog homeobox (Nanog) [65,66,67].
Oct4 belongs to the Pit-Oct-Unc (POU) TF family. It contains two main DNA-binding domains, the POU-specific domain (POUs) and POU homeo-domain (POUh), which are separated by a flexible α-helix linker that enables both POUs and POUh to independently interact with DNA targets. These two domains exhibit high evolutionary conservation in sequence and can also mediate Oct4 interaction with other transcriptional regulators [68]. Two other regions, including the N- and C-terminus, are required for transactivation [69]. ES cells are highly sensitive to the dosage of Oct4. Either two-fold induction or reduction of Oct4 results in ES cell differentiation [70]. Hence, stemness maintenance requires a precise regulation of Oct4 level. HECT-type E3 Wwp2 is the first E3 identified capable of ubiquitinating both mouse and human Oct4, promoting the 26S proteasome-mediated degradation [71,72]. Wwp2 belongs to the WW domain-containing Nedd4 subtype of HECT E3s. Moreover, the C-terminal HECT domain, it contains one N-terminal C2 domain and four tandem WW domains in the middle portion [54,72]. Unexpectedly, Wwp2 and Oct4 exhibit a similar, rather than opposite, expression profile during ES cell differentiation [71,73]. This could be because the repression of Oct4 transcription surmounts Wwp2 downregulation-resulted Oct4 induction in the differentiation process. Importantly, Wwp2-mediated Oct4 repression could impede stemness re-establishment since both Wwp2 knockout and mutation in the Oct4 ubiquitination site increase the efficiency of somatic cell reprogramming into induced pluripotent stem cells (iPSCs) [74]. Another interesting observation is that the Wwp2-mediated regulation of Oct4 varies among ES cells originated from different sources. In human ES cells (hESCs), WWP2 downregulates OCT4 in a dosage-dependent manner, whereas in mouse ES cells (mESCs), Wwp2 fails in reducing Oct4 unless differentiation occurs [54,71]. Itch is a second HECT-type E3 mediating Oct4 ubiquitination and degradation by 26S proteasome [75]. Although there is evidence showing that Itch depletion impairs self-renewal and reduces the efficiency of iPSC formation, whether Oct4 serves as the dominant downstream effector of Itch is unclear [75]. Of note, both Itch and Wwp2 ubiquitinate Oct4 with K63-linked polymer, which serves as classical examples for K63 linkage-driven substrate degradation [54,75]. RING-type E3 TRIM32 is a third ligase capable of mediating Oct4 ubiquitination [76]. However, it is rather unexpected that it regulates Oct4 independent of its enzymatic RING domain [76].
Sox2 belongs to the HMG-domain containing Sox TF family [77]. It is important for embryogenesis and ES cell maintenance. Similar with Oct4, Sox2 has dosage-dependent role in ES cells. Either the reduction or induction of its expression results in the loss of ESC stemness [78,79] [80]. Moreover, Sox2 and Oct4 form a binary complex to activate stemness-related genes, while repressing differentiation-promoted transcripts [65,81]. Their close association extends to ubiquitination-mediated regulation. The E3 Wwp2 for Oct4 could also target Sox2 for ubiquitination; hence, resulting in its degradation in mESCs [82]. However, this modification requires a Set7-catalyzed monomethyl signal at Sox2-K119 [82]. Since the activity of Wwp2 toward Oct4 exhibits inconsistency among different-originated ES cells, how WWP2 regulates SOX2 in hESCs remains unclear. A study by Wang et al. shows that APC/C coordinates with a priming E2 UbcH5/UbcH10 and an elongating E2 ubiquitin-conjugating Enzyme E2S (Ube2s) to modify Sox2 with K11-linked poly-Ub chain at Sox2-K123 residue for 26S proteasome-mediated degradation [57]. Furthermore, Ube2s has been shown to reinforce the stemness state through the fine-tuning of the precise level of Sox2 [57]. Under specific induction condition, Ube2s-APC/C-mediated Sox2 modification can increase the efficiency of mesoendoderm formation, while blocking neuroectodermal lineage commitment [82]. It will be of great interest to decipher how Wwp2 and Ube2s-APC/C exert concerted action to fine-tune Sox2 and Oct4 in the process of ES cell maintenance and cell fate commitment.
Nanog was first identified as “ENK” (early embryo specific NK) based on the homolog of its homeodomain to NK protein family [83]. Besides the DNA-binding homeodomain, Nanog contains two additional transactivation domains at the N- and C-terminus, respectively [10]. The C-terminus can be further divided into three sub-regions, namely CD1, tryptophan repeat (WR) and CD2 [10]. In ES cells, Nanog plays a pivotal role in maintaining stemness-specific genetic and epigenetic landscape [77,84,85,86,87]. Nanog knockout leads to loss in pluripotency and self-renewal, while Nanog-elevated mESCs impedes chemical-induced differentiation. In hESCs, NANOG promotes pluripotency and inhibits neuroectoderm differentiation [80,84,87,88]. NANOG exhibits high sensitivity to 26S proteasome inhibitor and can be modified by K48- and K63-linked poly-Ub chain in hESCs. Furthermore, F-box and WD40 domain-containing protein 8 (FBXW8) is identified as an E3 ligase for Nanog ubiquitination and degradation. However, this activity is restricted to phosphorylated Nanog at S52/71/78 [89]. FBXW8 depletion results in impaired self-renewal [89]. On the other hand, ES cells employ compensatory mechanisms to prevent the excessive degradation of Nanog. For example, H2A.Z directly interacts with Nanog to prevent Nanog ubiquitination in mESCs [90]. The deubiquitinase USP21 is able to remove the degradative Ub signal on human and mouse Nanog, hence preventing its degradation [91,92,93].
4.2. Regulation of Signal Transduction Pathways by Ubiquitination in ES Cells
Precise regulation of self-renewal and pluripotency requires concerted action between extra- and intracellular signals. In general, the external molecules bind with cell-surface receptors to evoke signal transduction in the cytoplasm and modulate gene activity in the nucleus. Human and mouse ES cells dissimilarly respond to external signals and adopt distinct pathways to maintain their properties [94,95,96]. For instance, Leukemia inhibitory factor (LIF) is specifically required by in vitro culture of mESCs, while the basic fibroblast growth factor (FGF2) signal is employed for human ES cell growth [97,98,99,100,101,102]. Yet, some signaling pathways, such as the bone morphogenetic protein (BMP) and Wnt pathways are required by both types of ES cells.
BMPs belong to the transformation growth factor beta (TGFβ) family, which is widely involved in cell proliferation, differentiation and apoptosis [103]. There are two main types of BMP receptors for over 20 BMPs: type I (including Alk2, Alk3, and Alk6) and type II (BmprII) [85]. Interaction between different receptors determines the specificity and consequences of BMP functions [85]. In general, BMPs bind to receptors, resulting in phosphorylation of downstream effectors, Smad1, Smad5 or Smad8 (receptor-regulated Smad, R-Smad). Two of these phosphorylated R-Smads form a heterotrimer with a common Smad protein, Smad4 (co-Smad), to translocate into the nucleus for transcription regulation [104,105,106,107]. The BMP signals can be inhibited by two inhibitory Smads (I-Smads), Smad6 and Smad7 [108,109]. In mESCs, the BMP/Smad signaling coordinates with the LIF stimulus to sustain stemness. Loss of BMP signal leads to impaired pluripotency and neuroectodermal differentiation [110]. In hESCs, the BMP signal cooperates with OCT4 and the FGF2 signal, respectively, to govern cell fate commitment [80]. Several studies have investigated how ubiquitination regulates the BMP signaling pathway in embryos and multiple types of adult stem cells [111]. Two HECT-type E3 ligases, Smad ubiquitination regulatory factor 11 (Smurf1) and Smurf2 are involved in ubiquitinating Smads. Smurf1 can modify Smad1 and Smad5 for degradation via poly-Ub chain, while Smurf2 preferentially ubiquitinates Smad1 [112,113,114]. The C-terminus of Hsc70-interacting protein (CHIP) is a third E3 ligase regulating Smad degradation. CHIP belongs to U box-type E3s. It can mediate poly-Ub chain formation specifically on Smad1 and Smad5 for 26S proteasome-mediated degradation [50,115]. Moreover, another two HECT-type E3s, WWP1 and NEDD4–2, are also identified for their capability of ubiquitinating and degrading Smad proteins [116]. However, how these E3s regulate ES cell maintenance is unknown. A study by Zhang et al., revealed the mechanism underlying Smad7 ubiquitination in mESCs. It reported that RING-type E3 RNF12 promotes Smad7 degradation via polyubiquitination. Inhibition of RNF12 results in Smad7 accumulation, which rescues BMP-triggered neuroectodermal differentiation [117].
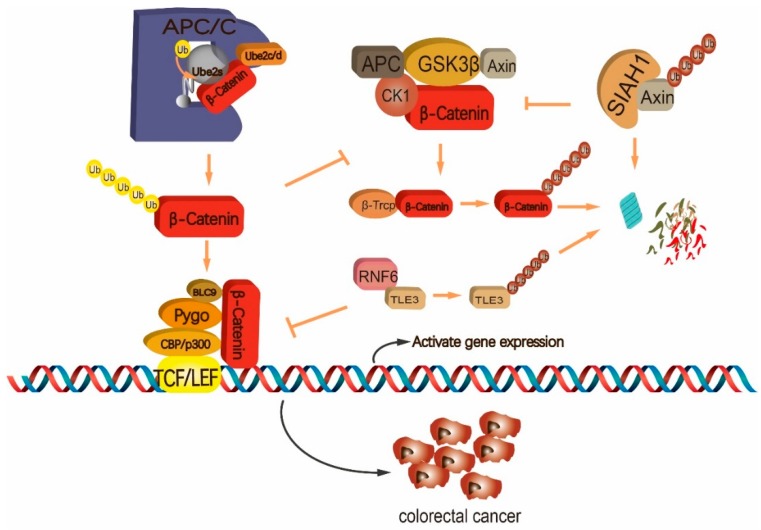
The Wnt signaling cascade is activated by the binding of Wnt ligands to Frizzled receptor and its co-receptor, low-density-lipoprotein-related protein 5/6 (LRP5/6). Upon ligand binding, β-Catenin is dissociated from the Axin destructive complex and enters the nucleus to interact with T-cell factor/lymphoid enhancer factors (Tcf/Lef) for transcription regulation [118,119,120]. Under the normal serum culture condition, activation of the Wnt pathway by CHIR99021, an inhibitor against glycogen synthase kinase 3 (GSK3), confers mESCs resistance to LIF withdrawal-induced differentiation [121]. Consistently, increased β-catenin accumulation leads to impaired ES cell differentiation [122]. In addition, in a serum-free culture medium, CHIR99021 combines with the MEK1/2 inhibitor PD0325901 and/or LIF to endow mESCs with a naive pluripotency [123]. In hESCs, either excessive induction or repression of the Wnt/β-catenin signaling results in loss of stemness [124]. As the key effector of the Wnt signal, β-catenin is precisely regulated to guarantee appropriate activation status of the pathway. Without Wnt stimulus, β-Catenin is associated with the Axin destructive complex which is composed of multiple factors, mainly including Axin, adenomatous polyposis coli (APC), GSK3 and casein kinase 1 (CK1). β-Catenin is modified by CK1- and GSK3-mediated sequential phosphorylation at its S45, S33, S37 and T41 residues [125,126,127]. Consequently, the phosphorylated β-Catenin is recognized by the E3 complex Skp1/Cul1/F-boxβ-TrCP for K48-linked polyubiqutination and proteasomal degradation [118,128,129,130,131]. Upon Wnt ligand binding, LRP6 is phosphorylated, which leads to GSK3 inhibition, collapse of the Axin complex and β-Catenin dissociation. Axin is further captured by RING-type E3 SIAH1/2 for ubiquitination and degradation (Figure 2) [132]. Dissociated β-Catenin is modified by Ube2s-APC/C-mediated polyubiquitination with a K11 linkage (Figure 2) [58]. This modification allows β-Catenin to avoid the β-TrCP-mediated degradative ubiquitination, hence enhancing its stability [58]. Importantly, the activity of Ube2s-APC/C toward β-Catenin promotes mESC commitment to the mesoendoderm lineage [58].
Figure 2.
Ubiquitination-monitored regulation of the Wnt/β-Catenin pathway in colorectal cancer (CRC) cells. UBE2S coordinates with the E3 complex anaphase-promoting complex/cyclosome (APC/C) to mediate K11-linked ubiquitin polymer on β-Catenin, which prevents beta-transducin repeat containing E3 ubiquitin protein ligase (β-TrCP)-orchestrated degradation of β-Catenin. RING-type E3 ring finger protein 6 (RNF6) ubiquitinates the inhibitor of β-Catenin, transducin-like enhancer of split 3 (TLE3), for degradation, which enhances the transcriptional activity of β-Catenin. SIAH1 ubiquitinates Axin for degradation, and thus blocks β-TrCP-mediated recognition and degradation of β-Catenin. Arrows represent enhancement and T-bars represent inhibition.
4.3. Regulation of ES Cell-Related Epigenetic Regulators by Ubiquitination
ES cells exhibit unique chromatin architecture, which contributes to the stemness maintenance and at the same time, allows ES cells to rapidly respond to differentiation signals [133,134]. The ES cell-specific chromatin landscape is largely monitored by histone modifications, such as methylation, acetylation and ubiquitination [135]. For example, the nucleosomes of ES cell chromatin contain a characteristic bivalent domain simultaneously possessing transcriptional active H3K4 and the repressive H3K27 methylation signals [136]. The K120 residue of Histone H2B can be monoubiquitylated (H2B-K120Ub1) by the RING-type E3 RNF20/RNF40 in mammalian cells, which is generally related with gene activation [137,138,139]. Depletion of RNF20 can confer mESC resistance to induced differentiation [140]. H2B-K119 is another monoubiquitination site which is closely related to the ES cell identity. H2B-K119Ub1 is dominantly achieved by the Ring1A and Ring1B subunits of Polycomb repressive complex 1 (PRC1). Knockdown of Ring1A/1B leads to reduction in H2B-K119Ub1 and ES cell differentiation [141,142,143,144,145]. RING-type E3 Dzip3 can also mediate H2B-K119Ub1 formation. However, its activity appears to be restricted to the promoter regions of several differentiation-related genes [146]. Interestingly, a cross-talk exists between the H2B-K119Ub1 signal and the bivalent domain to co-operatively monitor gene activity. They co-occupy differentiation-related genes for its enhanced repression, resulting in the reinforcement of the stemness state [146]. Reduction of the H2B-K119Ub1 signal upon Ring1A/1B depletion results in inefficient inhibition of bivalent genes and the lost of pluripotency [147].
5. Ubiquitination-Mediated Regulation of Cancer Development
Malignant cancers display uncontrolled growth, capability of invasion and mobility, intratumoral heterogeneity, and high recurrence rate [148]. Mounting evidences show the critical roles of ubiquitination-related factors in regulating tumorigenesis and malignancy. This part of the review focuses on two human cancers, the colorectal cancer (CRC) and osteosarcoma (OS), highlighting recent the recent progresses in the field.
Malignant CRC displays a high rate of incidence and mortality [147]. The lesion sites can be found in colon, rectum and appendix. The molecular signatures governing CRC occurrence and progression can mainly be grouped into three categories: (1) Genomic instability induced by inactivation of tumor suppressors (APC, TP53, SMAD4) or activation of proto-oncogene KRAS; (2) Microsatellite instability (MSI) caused by abnormal DNA mismatch repair (MMR); (3) Abnormal transcription due to hypermethylation in CpG islands [149,150,151,152]. Moreover, other mutations can also be observed in CRC. For example, about 10% of CRC patients bear mutation in tumor suppressor von Hippel-Lindau (VHL) [153]. VHL forms a CRL2VHL E3 complex with elongin B/C, RBX1, ROC1 and CUL2 to ubiquitinate Hypoxia Inducible Factor 1α (HIF-1α) for proteolysis in an oxygen-dependent manner [154,155,156]. HIF-1α promotes angiogenesis, cell metabolism and survival under hypoxia condition and VHL mutation exhibits a close correlation with abnormal accumulation of HIF-1α in CRC cells [153,157,158]. Of note, among all these mutations, APC mutation affected about 90% of CRC patients [151,159]. Since APC is required by β-TrCP-mediated degradative ubiquitination of β-Catenin in the Wnt pathway, abnormal β-Catenin accumulation is always detected in the CRC patients. Moreover, about 10% of CRC patients contain genetic mutation in β-Catenin whose product maintains the ability in mediating Wnt signal transduction but fails to be modified and degraded by the Axin-β-TrCP cascade [160]. Therefore, excessive β-Catenin accumulation is considered as a fundamental event in CRC. However, genetic mutation-caused inefficiency in β-Catenin degradation could be conquered by RING-type E3 TNF receptor-associated factor 6 (TRAF6). It ubiquitinates autophagy sensor LC3B via K63 linkage, which allows LC3B to recognize β-Catenin and drive it for autophagic degradation, independent on the activity of APC and β-TrCP ligase (Figure 2) [161]. Besides genetic mutations, some ubiquitination factors contribute to the excessive activation of the Wnt/β-Catenin pathway in CRC cells (Figure 2). E2 UBE2S coordinates with the E3 complex APC/C to mediate K11-linked ubiquitin polymer on intact β-Catenin, which prevents β-TrCP-orchestrated degradation and enhances β-Catenin accumulation in CRC cells (Figure 2) [58,162]. Moreover, RING-type E3 ring finger protein 6 (RNF6) indirectly enhances the activity of β-Catenin through suppressing its inhibitor, transducin-like enhancer of split 3 (TLE3) [162]. TLE3 inhibits the transcriptional activity of β-Catenin [163]. RNF6 modifies TLE3 via ubiquitination for proteolysis, which promotes CRC malignancy and recurrence (Figure 2) [162]. On the other hand, ubiquitination-mediated regulation of CRC progression is not restricted to the Wnt signaling, but widely involved in various important cellular processes, such as stem cell differentiation, autophagy, epithelial–mesenchymal transition (EMT) and epigenetic regulation of transcription [164,165,166]. The E3 ubiquitin ligase FBXW7 is involved in regulating stem cell proliferation and commitment in normal intestine and colon tissues [167,168]. FBXW7 deletion in mouse intestine impairs stem cell differentiation and promotes tumorigenesis [167]. In human CRC cells, FBXW7 represses EMT-mediated metastasis through ubiquitinating transcription factor ZEB2 for degradation [169]. However, disabled FBXW7 mutation is detected in CRC patients and FBXW7 exhibits a decreased expression in lesion sites, which deprives of FBXW7-mediated CRC repression [170,171,172]. In addition, FBXW7 mutation results in accumulation of myeloid cell leukemia 1 (MCL1) that is another substrate of FBXW7-mediated ubiquitination [173,174]. MCL1 belongs to pro-survival BCL2 family and is involved in mitochondrial apoptosis [175]. FBXW7 mutation-induced MCL1 accumulation results in chemotherapy insensitivity of CRC cells in clinic [176]. HECT-type E3 HECTH9 exhibits a very low expression in normal gut epithelium and induced expression in CRC cells [177]. It modifies stem cells-related regulator C-MYC with K63-linked poly-Ub chain to monitor C-MYC transcriptional activity and thus promotes CRC cell proliferation [177]. Contradictorily, Hecth9 does not promote tumor growth but acts as a repressor of CRC development in mice [178,179]. On the other hand, several studies provide clues for uiquitination-monitored epigenetic regulation of CRC progression. First of all, most CRC patients possess decreased RNF20/RNF40 expression and global loss in H2BK120ub1 signal accompanying poor therapeutic outcome [180,181]. Defect in H2BK120ub1 results in series of disorders, such as proto-oncogene activation, replication stress and impaired DNA damage repair genome instability and subsequent tumorigenesis [182,183,184,185]. Therefore, abnormity in H2BK120ub1 signal could be employed to develop novel strategy for CRC treatment in future. Chromatin-remodeling factor special AT-rich sequence-binding protein-1 (SATB1) promotes colon tumorigenesis [186,187]. SMURF2 ligase mediates SATB1 ubiquitination and promotes its degradation [188]. SMURF2-mediated SATB1 modification effectively inhibits CRC progression and confers sensitivity of CRC cells to conventional chemotherapy agents [188]. Recently, SMURF2 is suggested as a putative prognostic marker for MSI-free CRC patients [189]. Of note, besides the factors which were discussed above, additional ubiquitination-related factors have been identified to play critical roles in the regulation of CRC. They are summarized in Table 1.
Table 1.
Ubiquitination factors involved in regulating CRC.
| E2 | E3 | Function | Reference |
|---|---|---|---|
| UBC3 /UBC4 | β-TrCP | 1. β-TrCP ubiquitinates phosphorylated IκB for degradation, which enhances the NF-κB signaling. Increased β-TrCP is associated with an enhanced NF-κB signaling in CRC. 2. β-TrCP ubiquitinates β-Catenin via K48-linked poly-Ub chain for proteasomal degradation, which suppresses CRC progression. |
[190,191,192] |
| UBCH5 /UBCH10/UBE2S | APC/C | 1. APC/Ccdc20 ubiquitinates Conductin for degradation during mitotic exit, which regulates the Wnt/β-catenin signaling and CRC cell growth. 2. UBE2S collaborates with the APC/C complex to stablize β-Catenin via K11-linked polyubiqutination. This activity enhances CRC proliferation and metastasis. |
[58,193] |
| UBCH5 | HECTH9 | HECTH9 modifies C-MYC with K63-linked poly-Ub chain to promote CRC cell proliferation. The expression of HECTH9 is increased in the cancer tissues of CRC patients. | [177] |
| UBCH5 | S-phase kinase protein 2 (SKP2) | SKP2 ubiquitinates p27Kip1 for degradation in CRC cells. Elevated expression of SKP2 and reduced expression of p27Kip1 is associated with poor prognosis and decreased survival of CRC patients. | [194,195,196,197] |
| UBCH5B/ UBE2S |
von Hippel-Lindau protein (pVHL) | VCB-Cul2 ubiquitinates HIF-1α for degradation under hypoxic conditions, which suppresses CRC malignancy. | [156,198,199,200] |
| UBCH5B | X-chromosome-linked IAP (XIAP), | 1. XIAP ubiquitinates active caspase-3 for degradation to suppress apoptosis. Inhibition of XIAP increases the sensitivity of PIK3CA-mutated CRC cells for induced cell death. 2. XIAP monoubiquitinates TLE, which promotes β-catenin-TCF association and enhances activation of the Wnt pathway in CRC cells. |
[201,202,203] |
| UBCH6 | RNF20/RNF40 | RNF20/RNF40 monoubiquitinates H2B-K120, which is required by transcription regulation. Loss of H2BK120ub1 is associated with poor therapeutic outcome in CRC. | [181,182,204] |
| UBC9 | E6-AP | E6-AP coordinates with UBC9 to ubiquitinate SOX9 for degradation, which may repress Sox9-enhanced CRC malignancy. | [205] |
| UBC13/ UEV1A |
Tumor necrosis factor receptor-associated factor 6 (TARF6) | 1. TARF6 ubiquitinates IKK via K63-linked poly-Ub chain to promote the NF-κB signaling pathway. High expression of TRAF6 is associated with a decreased survival of CRC patients. 2. TARF6 stabilizes hypoxia-inducible factor (HIF)–1a through K63-linked polyubiquitination, which promotes angiogenesis and growth of CRC. 3. TRAF6 ubiquitinates LC3B via K63 linkage, which allows LC3B to recognize β-Catenin and drive it for autophagic degradation. This activity is involved in inhibiting the metastasis of CRC cells. |
[161,206,207,208,209] |
| c-IAP | c-IAP is upregulated in CRC patients with a reduced survival. | [210] | |
| FBXW7 | FBXW7 ubiquitinates ZEB2 and MCL1 for degradation, which is involved in regulating the malignancy and therapy resistance of CRC cells. Somatic mutations in FBXW7 is detected in CRC patients. | [169,171,172,176,211,212] | |
| Human upstream regulatory element binding protein 1 (hUREB1) | hUREB1 down-regulates p53 through ubiquitination in CRC cells. Increased expression of hUREB1 is correlated with p53 destabilization in CRC patients. | [213] | |
| MDM2 | MDM2 ubiquitinates p53 for degradation. Increased expression of MDM2 is correlated with negative expression of p53 in CRC patients. | [214,215] | |
| RNF4 | RNF4 ubiquitinates and stabilizes multiple oncoproteins, such as c-Myc and β-Catenin. Elevated expression of RNF4 is correlated with CRC tumorigenesis. | [216,217,218] | |
| RNF6 | RNF6 enhances the interaction between β-Catenin and TCF4/LEF through ubiquitinating transducin-like enhancer of split 3 (TLE3) for degradation, which promotes CRC cell growth and metastasis. | [162] | |
| RNF14 | RNF14 activates the Wnt pathway through interacting with TCFs to promote β-Catenin recruitment, which promotes CRC cell growth. | [219,220] | |
| Ubiquitin E3 ligase ring finger 43 (RNF43) | RNF43 ubiquitinates Frizzled (FZD) and LRP6 for degradation. About 18% of CRC patients bear RNF43 truncating mutation. | [221,222,223] | |
| Tripartite motif (TRIM3) | TRIM3 enhances the stability of p53 and suppresses CRC development. | [224] | |
| TRIM15 | TRIM15 serves as a putative CRC suppressor, which inhibits CRC cell growth and metastasis. | [225] | |
| TRIM29 | TRIM29 is upregulated in aberrant crypt foci in human colon and serves as a putative biomarker for CRC diagnosis. | [226] |
OS is a malignant bone tumor with high incidence in children and adolescents [227]. In spite of relatively low prevalence, OS results in high death rate due to poor therapeutic effect in clinic [228]. OS displays intratumoral heterogeneity and stem cell properties possibly due to defect in osteoblast differentiation from MSCs [229,230,231]. In the normal commitment process from MSC to terminal osteocytes, different types of mid-term cells are transiently produced at corresponding differentiation stages, including committed osteoprogenitor, proosteoblast, early osteoblast, mature osteoblast and osteocyte. These cells can be characterized by marker gene expression. For example, in undifferentiated MSCs, the BMP/SMAD signal is highly activated that dominantly induces the expression of inhibitors of differentiation (IDs) to support cell proliferation [232,233,234]. Upon differentiation, the BMP/SMAD pathway is inactivated while the markers of pro-osteoblasts, RUNX2 and OSTERIX, are induced [232,233,234,235]. Mature osteoblasts and osteocytes can be featured by the expression of Osteocalcin (OC) and Osteopontin (OPN, SPP1) [232]. Although there are evidences showing OSs can be originated from terminally differentiated osteoblast, immature cells can always be detected in OS expressing semidifferentiated cell markers and even ES cell markers, such as Oct4 and Sox2 [236,237,238]. To suppress the poor differentiation properties through eliminating the excessive expression of stem cell markers could serve as an avenue for a better treatment of OS. This is well supported by the study of Zhang et al. [238]. They exploit SMURF1 to inhibit the BMP/SMAD signal, which successfully drives OS cells to re-enter the process of differentiation. More importantly, the differentiated OS cells are conferred sensitivity to chemotherapeutic agents [238]. In details, SMURF1 cooperates with E2 complex UBCH5B-UEV1A to modify SMAD1 with poly-Ub chain, which destines SMAD1 for 26S proteasome-mediated degradation [238]. Interestingly, SMURF1 can also serve as the E3 of RUNX2. It mediates RUNX2 ubiquitination for degradation, which is important for osteoblast differentiation [113]. Therefore, SMURF1 possibly possess double effect on OS repression especially for the subtypes highly expressing both SMAD1 and RUNX2. Interestingly, HECT-type E3 WWP1 is also involved in regulating RUNX2. It is recruited by zinc finger-containing adaptor Schnurri-3 (SHN3) to interact with RUNX2 and mediate its polyubiquitination for degradation [239]. This activity is involved in regulating extracellular matrix mineralization in the process of adult bone formation [239]. However, their impact on OS growth and metastasis is unclear. E3 MDM2 is involved in maintaining the stem cell properties of OS cells. It ubiquitinates retinoic acid receptor alpha (RARα) for proteasomal degradation, which impedes retinoic acid (RA)-induced OS differentiation and promotes malignancy [240] Therefore, inhibition of MDM2 could serve as a possible avenue to effectively suppress OS progression. On the other hand, increasing numbers of ubiquitination factors have been identified for their roles in regulating OSs independent of cell differentiation. For instance, CUL4B displays an elevated expression in OS cells to promote proliferation and inhibit apoptosis [241]. It cooperates with three additional proteins, RING-box protein 1 (RBX1), DNA damage binding protein 1 (DDB1), and DDB1- and CUL4-associated factor 13 (DCAF13), to form the CRL4BDCAF13 E3 complex. Via the DCAF13 subunit, this complex can specifically recognize the tumor repressor, phosphatase and tensin homolog deleted on chromosome 10 (PTEN), and exert CUL4/RBX1-mediated ubiquitination of PTEN for degradation [242]. Another study shows that CUL4B/RBX1/ DDB1 can coordinate with DCAF11 to form the CRL4BDCAF11 E3 complex that ubiquitinates the cyclin-dependent kinase (CDK) inhibitor p21Cip1 and promotes its degradation. This activity is required by the proliferation of OS cells [243]. The RING-type E3 c-Cbl serves as an OS repressor [244]. It is downregulated in OS cells, and targets receptor tyrosine kinase (RTK) for degradation via ubiquitination. Overexpression of c-Cbl results in excessive degradation of RTK, which inhibits the proliferation and metastasis of OS cells [244]. Tryptophane-aspartic acid (WD) repeat and SOCS box containing 1 (WSB1) ligase, elongin B, C-Cullin5 and Rbx1 form a multiple-unite E3 complex, involved in regulating multiple types of cancers [245,246,247]. In OS cells, this E3 complex promotes hypoxia-enhanced metastasis through modifying Rho guanosine diphosphate dissociation inhibitor 2 (RhoGDI2) with poly-Ub chain for proteasome-mediated degradation [248]. Moreover, other ubiquitination factors have also been reported for their involvement in regulating OS biology, such as UBE2T, Adaptor Speckle-type pox virus and zinc finger protein (POZ) protein (SPOP) and HECT domain and ankyrin-repeat-containing E3 ubiquitin-protein ligase 1 (HACE1) [249,250,251]. These factors could form a network to globally regulate the onset and progression of OS cells.
6. Concluding Remarks and Perspectives
Ubiquitination monitors the longevity and activity of proteins. A myriad of protein-dependent cellular processes, including cell cycle progression, gene transcription, chromatin remodeling, signaling transduction and endocytosis, are precisely dominated by the ubiquitination machinery. Mounting evidences show the close correlation between ubiquitination dysregulation and human diseases. Recent studies start a promising paradigm to utilize the "death label" produced by the ubiquitination system to remove disease-causing proteins. Ubiquitination mediators, especially E3s, act as putative targets to develop novel therapeutic approaches with higher effectiveness and fewer side effects [252]. Excitingly, the inhibitors against several E3s, such as APC/C, MDM2 and SKP2, are being evaluated for cancer treatment in the preclinical or clinical stages [253,254,255,256,257,258]. Moreover, proteasome inhibitor drugs, Bortezomib and Carfilzomib, have been successfully applied in the clinic for the treatment of human cancers [259]. However, to extend this paradigm for more types of cancers requires a comprehensive understanding of the ubiquitination-mediated regulatory mechanisms. We need to identify dominant ubiquitination factors related to tumorigenesis and malignancy. Moreover, it is also challenging to develop effective therapeutic molecules specifically targeting these factors. In pluripotent ES cells, previous studies largely focused on the transcriptional regulatory network, while the information about how ubiquitination regulates the self-renewal and pluripotency is much more limited. Specific questions of interest include how ubiquitination coordinates with other types of post-translational modifications, such as methylation, acetylation and phosphorylation, to globally monitor the properties of ES cells and cell fate specification. To dissect these puzzles could open a new arena for ES cell application and disease therapy in future.
Acknowledgments
We apologize to the authors whose work could not be cited in this review due to space constraints. We appreciate the great work about ubiquitination from all of the past and present lab members.
Abbreviations
| APC | Adenomatous polyposis coli |
| APC/C | Anaphase-promoting complex/cyclosome |
| BMP | Bone morphogenetic protein |
| CDK | Cyclin-dependent kinase |
| CHIP | Hsc70-interacting protein |
| CK1 | Casein kinase 1 |
| CRC | Colorectal cancer |
| CRLs | Cullin-RING ligases |
| DCAF13 | DDB1- and CUL4-associated factor 13 |
| DDB1 | DNA damage binding protein 1 |
| DTT | DNA damage tolerance |
| E3 | Ub ligase |
| EC cells | embryonic carcinoma cells |
| EG cells | Embryonic germ cells |
| EMT | Epithelial–mesenchymal transition |
| ENK | Arly embryo specific NK |
| ES cells | Embryonic stem cells |
| FBXW8 | F-box and WD40 domain-containing protein 8 |
| FGF2 | Fibroblast growth factor |
| FZD | ubiquitinates Frizzled |
| GSK3 | Glycogen synthase kinase 3 |
| H2B-K120Ub1 | K120 residue of Histone H2B can be monoubiquitylated |
| HACE1 | HECT domain and ankyrin-repeat-containing E3 ubiquitin-protein ligase 1 |
| HECT | homologous to E6-AP COOH terminus |
| hESCs | Human ES cells |
| HIF-1α | Hypoxia inducible factor 1α |
| hUREB | Human upstream regulatory element binding protein |
| ICM | Inner cell mass |
| IDs | Inhibitors of differentiation |
| IKK | Inhibitors of I-κB kinase |
| iPSCs | Induced pluripotent stem cells |
| LIF | Leukemia inhibitory factor |
| LRP5/6 | low-density-lipoprotein-related protein 5/6 |
| MCL1 | Myeloid cell leukemia 1 |
| MDM2 | Mouse double minute 2 |
| mESCs | Mouse ES cells |
| MMR | Mismatch repair |
| MSCs | Mesenchymal stem cells |
| MSI | Microsatellite instability |
| NADH | Nicotinamide adenine dinucleotide |
| OC | Osteocalcin |
| Oct4 | Octamer-binding transcription factor-4 |
| OPN/SPP1 | Osteopontin |
| OS | Osteosarcoma |
| PCNA | Proliferating cell nuclear antigen |
| POUh | POU homeo-domain |
| POUs | POU-specific domain |
| PRC1 | Polycomb repressive complex 1 |
| PTEN | Phosphatase and tensin homolog deleted on chromosome 10 |
| pVHL | von Hippel-Lindau protein |
| RA | Retinoic acid |
| RARα | Retinoic acid receptor alpha |
| RBR | RING between RING |
| RBX1 | RING-box protein 1 |
| RhoGDI2 | Rho guanosine diphosphate dissociation inhibitor 2 |
| RING | Really Interesting New Gene |
| RIP1 | Receptor interacting protein 1 |
| RTK | Receptor tyrosine kinase |
| RNF6 | RING-type E3 ring finger protein 6 |
| RNF43 | Ubiquitin E3 ligase ring finger 43 |
| SATB1 | Special AT-rich sequence-binding protein-1 |
| SCF | Skp1/Cul1/F-box |
| SHN3 | Zinc finger-containing adaptor Schnurri-3 |
| Smurf1 | Smad ubiquitination regulatory factor 11 |
| Sox2 | SRY-related HMG Box 2 |
| SPOP | Speckle-type POZ protein |
| Tcf/Lef | T-cell factor/lymphoid enhancer factors |
| TLS | Translesion DNA synthesis |
| TRAF6 | NF receptor-associated factor 6 |
| Ub | Ubiquitin |
| Uba/E1 | Ub-activating enzyme |
| UBC/E2 | Ub-conjugating enzyme |
| Ube2s | E2 ubiquitin-conjugating Enzyme E2S |
| WSB1 | WD repeat and SOCS box containing 1 |
Author Contributions
D.W.: Figure 2, Table, manuscript writing and final approval of manuscript; F.B. Figure 1 and final approval of manuscript; W.Z.: conception and design, financial support, manuscript writing and final approval of manuscript.
Funding
Our work is supported by the NSFC fund (01018210011048) and Scientific Research Program of Beijing Education Commission (SQKM201810028010).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hicke L., Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 2.Vijay-Kumar S., Bugg C.E., Wilkinson K., Vierstra R.D., Hatfield P., Cook W.J. Comparison of the three-dimensional structures of human, yeast, and oat ubiquitin. J. Biol. Chem. 1987;262:6396–6399. [PubMed] [Google Scholar]
- 3.Callis J., Vierstra R.D. Ubiquitin and ubiquitin genes in higher plants. Oxford Surv. Plant Mol. Cell. Biol. 1989;6:1–30. [Google Scholar]
- 4.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 6.Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner S.A., Beli P., Weinert B.T., Nielsen M.L., Cox J., Mann M., Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteomics. 2011;10:M111.013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukasawa H., Fujigaki Y., Yamamoto T., Hishida A., Kitagawa M. Protein Degradation by the Ubiquitin-Proteasome Pathway and Organ Fibrosis. Curr. Med. Chem. 2012;19:893–900. doi: 10.2174/092986712799034941. [DOI] [PubMed] [Google Scholar]
- 9.Aviel S., Winberg G., Massucci M., Ciechanover A. Degradation of the epstein-barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 2000;275:23491–23499. doi: 10.1074/jbc.M002052200. [DOI] [PubMed] [Google Scholar]
- 10.Pan G.J., Pei D.Q. Identification of two distinct transactivation domains in the pluripotency sustaining factor nanog. Cell Res. 2003;13:499–502. doi: 10.1038/sj.cr.7290193. [DOI] [PubMed] [Google Scholar]
- 11.Ciechanover A. N-terminal ubiquitination: More protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Breitschopf K., Bengal E., Ziv T., Admon A., Ciechanover A. A novel site for ubiquitination: The N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadwell K., Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 14.Bhogaraju S., Kalayil S., Liu Y., Bonn F., Colby T., Matic I., Dikic I. Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell. 2016;167:1636–1649. doi: 10.1016/j.cell.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Bhogaraju S., Dikic I. Ubiquitination without E1 and E2 enzymes. Nature. 2016;533:43. doi: 10.1038/nature17888. [DOI] [PubMed] [Google Scholar]
- 16.Ciechanover A., Heller H., Katz-Etzion R., Hershko A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc. Natl. Acad. Sci. USA. 1981;78:761–765. doi: 10.1073/pnas.78.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershko A., Heller H., Elias S., Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 18.Hershko A. Ubiquitin-mediated protein degradation. J. Biol. Chem. 1988;263:15237–15240. [PubMed] [Google Scholar]
- 19.Ye Y., Rape M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varshavsky A. The ubiquitin system, an immense realm. Annu. Rev. Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 21.Metzger M.B., Hristova V.A., Weissman A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman S.R., Deato M.E., Brignone C., Chan H.M., Kung A.L., Tagami H., Nakatani Y., Livingston D.M. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 23.Hoppe T. Multiubiquitylation by E4 enzymes: ‘one size’ doesn't fit all. Trends Biochem. Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Baranes-Bachar K., Levy-Barda A., Oehler J., Reid D.A., Soria-Bretones I., Voss T.C., Chung D., Park Y., Liu C., Yoon J.-B., et al. The Ubiquitin E3/E4 Ligase UBE4A Adjusts Protein Ubiquitylation and Accumulation at Sites of DNA Damage, Facilitating Double-Strand Break Repair. Mol. Cell. 2018;69:866–878. doi: 10.1016/j.molcel.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C., Liu W., Ye Y., Li W. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat. Commun. 2017;8:14274. doi: 10.1038/ncomms14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H., Pomeroy S.L., Ferreira M., Teider N., Mariani J., Nakayama K.I., Hatakeyama S., Tron V.A., Saltibus L.F., Spyracopoulos L., et al. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat. Med. 2011;17:347–355. doi: 10.1038/nm.2283. [DOI] [PubMed] [Google Scholar]
- 27.Wu H., Leng R.P. UBE4B, a ubiquitin chain assembly factor, is required for MDM2-mediated p53 polyubiquitination and degradation. Cell Cycle. 2011;10:1912–1915. doi: 10.4161/cc.10.12.15882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ackermann L., Schell M., Pokrzywa W., Kevei É., Gartner A., Schumacher B., Hoppe T. E4 ligase-specific ubiquitination hubs coordinate DNA double-strand-break repair and apoptosis. Nat. Struct. Mol. Biol. 2016;23:995–1002. doi: 10.1038/nsmb.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koegl M., Hoppe T., Schlenker S., Ulrich H.D., Mayer T.U., Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/S0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 30.Qiu J., Sheedlo M.J., Yu K., Tan Y., Nakayasu E.S., Das C., Liu X., Luo Z.Q. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature. 2016;533:120–124. doi: 10.1038/nature17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart M.D., Ritterhoff T., Klevit R.E., Brzovic P.S. E2 enzymes: More than just middle men. Cell Res. 2016;26:423–440. doi: 10.1038/cr.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plafker S.M., Plafker K.S., Weissman A.M., Macara I.G. Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J. Cell Biol. 2004;167:649–659. doi: 10.1083/jcb.200406001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morreale F.E., Walden H. Types of Ubiquitin Ligases. Cell. 2016;165:248. doi: 10.1016/j.cell.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Deshaies R.J., Joazeiro C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 35.Chang L.F., Zhang Z., Yang J., McLaughlin S.H., Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min M., Mevissen T.E.T., De Luca M., Komander D., Lindon C. Efficient APC/C substrate degradation in cells undergoing mitotic exit depends on K11 ubiquitin linkages. Mol. Biol. Cell. 2015;26:4325–4332. doi: 10.1091/mbc.E15-02-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly A., Wickliffe K.E., Song L., Fedrigo I., Rape M. Ubiquitin Chain Elongation Requires E3-Dependent Tracking of the Emerging Conjugate. Mol. Cell. 2014;56:232–245. doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Scheffner M., Staub O. HECT E3s and human disease. BMC. Biochem. 2007;8(Suppl. 1):S6. doi: 10.1186/1471-2091-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rotin D., Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 40.Marin I., Lucas J.I., Gradilla A.C., Ferrus A. Parkin and relatives: The RBR family of ubiquitin ligases. Physiol. Genomics. 2004;17:253–263. doi: 10.1152/physiolgenomics.00226.2003. [DOI] [PubMed] [Google Scholar]
- 41.Dove K.K., Klevit R.E. RING-Between-RING E3 Ligases: Emerging Themes amid the Variations. J. Mol. Biol. 2017;429:3363–3375. doi: 10.1016/j.jmb.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hochstrasser M. Ubiquitin-dependent protein degradation. Ann. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 43.Xu P., Peng J. Characterization of Polyubiquitin Chain Structure by Middle-down Mass Spectrometry. Anal. Chem. 2008;80:3438–3444. doi: 10.1021/ac800016w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yau R., Rape M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 46.Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pickart C.M., Fushman D. Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Livneh I., Kravtsova-Ivantsiv Y., Braten O., Kwon Y.T., Ciechanover A. Monoubiquitination joins polyubiquitination as an esteemed proteasomal targeting signal. Bioessays. 2017;39 doi: 10.1002/bies.201700027. [DOI] [PubMed] [Google Scholar]
- 49.Ea C.K., Deng L., Xia Z.P., Pineda G., Chen Z.J. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 50.Li L., Xin H., Xu X., Huang M., Zhang X., Chen Y., Zhang S., Fu X.Y., Chang Z. CHIP mediates degradation of Smad proteins and potentially regulates Smad-induced transcription. Mol. Cell Biol. 2004;24:856–864. doi: 10.1128/MCB.24.2.856-864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanayama A., Seth R.B., Sun L., Ea C.-K., Hong M., Shaito A., Chiu Y.-H., Deng L., Chen Z.J. TAB2 and TAB3 Activate the NF-κB Pathway through Binding to Polyubiquitin Chains. Mol. Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Skaug B., Jiang X., Chen Z.J. The role of ubiquitin in NF-kappaB regulatory pathways. Annu. Rev. Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 53.Zhao S., Ulrich H.D. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc. Natl. Acad. Sci. USA. 2010;107:7704–7709. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao B., Jin Y. Wwp2 mediates Oct4 ubiquitination and its own auto-ubiquitination in a dosage-dependent manner. Cell Res. 2010;20:332–344. doi: 10.1038/cr.2009.136. [DOI] [PubMed] [Google Scholar]
- 55.Garnett M.J., Mansfeld J., Godwin C., Matsusaka T., Wu J., Russell P., Pines J., Venkitaraman A.R. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu T., Merbl Y., Huo Y., Gallop J.L., Tzur A., Kirschner M.W. UBE2S drives elongation of K11-linked ubiquitin chains by the Anaphase-Promoting Complex. Proc. Natl. Acad. Sci. USA. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J., Zhang Y., Hou J., Qian X., Zhang H., Zhang Z., Li M., Wang R., Liao K., Wang Y., et al. Ube2s regulates Sox2 stability and mouse ES cell maintenance. Cell Death Differ. 2016;23:393–404. doi: 10.1038/cdd.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z., Wang Y., Li Y., Yin W., Mo L., Qian X., Zhang Y., Wang G., Bu F., Zhang Z., et al. Ube2s stabilizes β-Catenin through K11-linked polyubiquitination to promote mesendoderm specification and colorectal cancer development. Cell Death Dis. 2018;9:456. doi: 10.1038/s41419-018-0451-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang W., Qin Z., Zhang X., Xiao W. Roles of sequential ubiquitination of PCNA in DNA-damage tolerance. FEBS Lett. 2011;585:2786–2794. doi: 10.1016/j.febslet.2011.04.044. [DOI] [PubMed] [Google Scholar]
- 60.Hedglin M., Benkovic S.J. Regulation of Rad6/Rad18 Activity During DNA Damage Tolerance. Annu. Rev. Biophys. 2015;44:207–228. doi: 10.1146/annurev-biophys-060414-033841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leung W., Baxley R., Moldovan G.-L., Bielinsky A. Mechanisms of DNA Damage Tolerance: Post-Translational Regulation of PCNA. Genes. 2018;10:10. doi: 10.3390/genes10010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weissman I.L. Stem cells: Units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/S0092-8674(00)81692-X. [DOI] [PubMed] [Google Scholar]
- 63.Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 64.Levine M., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 65.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silva J., Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brehm A., Ohbo K., Scholer H. The carboxy-terminal transactivation domain of Oct-4 acquires cell specificity through the POU domain. Mol. Cell Biol. 1997;17:154–162. doi: 10.1128/MCB.17.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeineddine D., Hammoud A.A., Mortada M., Boeuf H. The Oct4 protein: More than a magic stemness marker. Am. J. Stem Cells. 2014;3:74–82. [PMC free article] [PubMed] [Google Scholar]
- 70.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 71.Xu H., Wang W., Li C., Yu H., Yang A., Wang B., Jin Y. WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 2009;19:796. doi: 10.1038/cr.2009.63. [DOI] [PubMed] [Google Scholar]
- 72.Pirozzi G., McConnell S.J., Uveges A.J., Carter J.M., Sparks A.B., Kay B.K., Fowlkes D.M. Identification of Novel Human WW Domain-containing Proteins by Cloning of Ligand Targets. J. Biol. Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 73.Xu H.M., Liao B., Zhang Q.J., Wang B.B., Li H., Zhong X.M., Sheng H.Z., Zhao Y.X., Zhao Y.M., Jin Y. Wwp2, an E3 Ubiquitin Ligase That Targets Transcription Factor Oct-4 for Ubiquitination. J. Biol. Chem. 2004;279:23495–23503. doi: 10.1074/jbc.M400516200. [DOI] [PubMed] [Google Scholar]
- 74.Li S., Xiao F., Zhang J., Sun X., Wang H., Zeng Y., Hu J., Tang F., Gu J., Zhao Y., et al. Disruption of OCT4 Ubiquitination Increases OCT4 Protein Stability and ASH2L-B-Mediated H3K4 Methylation Promoting Pluripotency Acquisition. Stem Cell Rep. 2018;11:973–987. doi: 10.1016/j.stemcr.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao B., Zhong X., Xu H., Xiao F., Fang Z., Gu J., Chen Y., Zhao Y., Jin Y. Itch, an E3 ligase of Oct4, is required for embryonic stem cell self-renewal and pluripotency induction. J. Cell Physiol. 2013;228:1443–1451. doi: 10.1002/jcp.24297. [DOI] [PubMed] [Google Scholar]
- 76.Bahnassawy L., Perumal T.M., Gonzalez-Cano L., Hillje A.L., Taher L., Makalowski W., Suzuki Y., Fuellen G., del Sol A., Schwamborn J.C. TRIM32 modulates pluripotency entry and exit by directly regulating Oct4 stability. Sci. Rep. 2015;5:13456. doi: 10.1038/srep13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Novo C.L., Tang C., Ahmed K., Djuric U., Fussner E., Mullin N.P., Morgan N.P., Hayre J., Sienerth A.R., Elderkin S., et al. The pluripotency factor Nanog regulates pericentromeric heterochromatin organization in mouse embryonic stem cells. Genes Dev. 2016;30:1101–1115. doi: 10.1101/gad.275685.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chew J.L., Loh Y.H., Zhang W., Chen X., Tam W.L., Yeap L.S., Li P., Ang Y.S., Lim B., Robson P., et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kopp J.L., Ormsbee B.D., Desler M., Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26:903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 80.Wang Z., Oron E., Nelson B., Razis S., Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 81.Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J., et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 82.Fang L., Zhang L., Wei W., Jin X., Wang P., Tong Y., Li J., Du J.X., Wong J. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell. 2014;55:537–551. doi: 10.1016/j.molcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 83.Wang S.H., Tsai M.S., Chiang M.F., Li H. A novel NK-type homeobox gene, ENK (early embryo specific NK), preferentially expressed in embryonic stem cells. Gene Expr. Patterns. 2003;3:99–103. doi: 10.1016/S1567-133X(03)00005-X. [DOI] [PubMed] [Google Scholar]
- 84.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 85.Mishina Y. Function of bone morphogenetic protein signaling during mouse development. Front Biosci. 2003;8:855–869. doi: 10.2741/1097. [DOI] [PubMed] [Google Scholar]
- 86.Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 87.Heurtier V., Owens N., Gonzalez I., Mueller F., Proux C., Mornico D., Clerc P., Dubois A., Navarro P. The molecular logic of Nanog-induced self-renewal in mouse embryonic stem cells. Nat.Commun. 2019;10:1109. doi: 10.1038/s41467-019-09041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 89.Pei D. Deubiquitylating Nanog: Novel role of USP21 in embryonic stem cell maintenance. Signal Transduct. Target Ther. 2017;2:17014. doi: 10.1038/sigtrans.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang J., Qiao M., He Q., Shi R., Loh S.J., Stanton L.W., Wu M. Pluripotency Activity of Nanog Requires Biochemical Stabilization by Variant Histone Protein H2A.Z. Stem Cells. 2015;33:2126–2134. doi: 10.1002/stem.2011. [DOI] [PubMed] [Google Scholar]
- 91.Kwon S.K., Lee D.H., Kim S.Y., Park J.H., Choi J., Baek K.H. Ubiquitin-specific protease 21 regulating the K48-linked polyubiquitination of NANOG. Biochem. Biophys. Res. Commun. 2017;482:1443–1448. doi: 10.1016/j.bbrc.2016.12.055. [DOI] [PubMed] [Google Scholar]
- 92.Jin J., Liu J., Chen C., Liu Z., Jiang C., Chu H., Pan W., Wang X., Zhang L., Li B., et al. The deubiquitinase USP21 maintains the stemness of mouse embryonic stem cells via stabilization of Nanog. Nat. Commun. 2016;7:13594. doi: 10.1038/ncomms13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu X., Yao Y., Ding H., Han C., Chen Y., Zhang Y., Wang C., Zhang X., Zhang Y., Zhai Y., et al. USP21 deubiquitylates Nanog to regulate protein stability and stem cell pluripotency. Signal Transduct. Target Ther. 2017;2:16046. doi: 10.1038/sigtrans.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pera M.F., Tam P.P.L. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 95.Nichols J., Smith A. The origin and identity of embryonic stem cells. Development. 2011;138:3–8. doi: 10.1242/dev.050831. [DOI] [PubMed] [Google Scholar]
- 96.Yu J., Thomson J.A. Pluripotent stem cell lines. Genes Dev. 2008;22:1987–1997. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Niwa H., Burdon T., Chambers I., Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hirai H., Karian P., Kikyo N. Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. Biochem. J. 2011;438:11–23. doi: 10.1042/BJ20102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matsuda T., Nakamura T., Nakao K., Arai T., Katsuki M., Heike T., Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoshida K., Chambers I., Nichols J., Smith A., Saito M., Yasukawa K., Shoyab M., Taga T., Kishimoto T. Maintenance of the pluripotential phenotype of embryonic stem cells through direct activation of gp130 signalling pathways. Mech. Dev. 1994;45:163–171. doi: 10.1016/0925-4773(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 101.Vallier L., Alexander M., Pedersen R.A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 102.Xu C., Rosler E., Jiang J., Lebkowski J.S., Gold J.D., O'Sullivan C., Delavan-Boorsma K., Mok M., Bronstein A., Carpenter M.K. Basic fibroblast growth factor supports undifferentiated human embryonic stem cell growth without conditioned medium. Stem Cells. 2005;23:315–323. doi: 10.1634/stemcells.2004-0211. [DOI] [PubMed] [Google Scholar]
- 103.Massague J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 104.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 105.Weissman A.M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 106.Miyazono K., ten Dijke P., Heldin C.H. TGF-beta signaling by Smad proteins. Adv. Immunol. 2000;75:115–157. doi: 10.1016/s0065-2776(00)75003-6. [DOI] [PubMed] [Google Scholar]
- 107.Moustakas A., Souchelnytskyi S., Heldin C.H. Smad regulation in TGF-beta signal transduction. J. Cell Sci. 2001;114:4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- 108.Hata A., Lagna G., Massagué J., Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hanyu A., Ishidou Y., Ebisawa T., Shimanuki T., Imamura T., Miyazono K. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J. Cell Biol. 2001;155:1017–1027. doi: 10.1083/jcb.200106023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ying Q.L., Nichols J., Chambers I., Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/S0092-8674(03)00847-X. [DOI] [PubMed] [Google Scholar]
- 111.Lee T.H., Shank J., Cusson N., Kelliher M.A. The kinase activity of Rip1 is not required for tumor necrosis factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J. Biol. Chem. 2004;279:33185–33191. doi: 10.1074/jbc.M404206200. [DOI] [PubMed] [Google Scholar]
- 112.Zhu H., Kavsak P., Abdollah S., Wrana J.L., Thomsen G.H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 113.Zhao M., Qiao M., Harris S.E., Oyajobi B.O., Mundy G.R., Chen D. Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. J. Biol. Chem. 2004;279:12854–12859. doi: 10.1074/jbc.M313294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Y., Chang C., Gehling D.J., Hemmati-Brivanlou A., Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang L., Liu Y.T., Hao R., Chen L., Chang Z., Wang H.R., Wang Z.X., Wu J.W. Molecular mechanism of the negative regulation of Smad1/5 protein by carboxyl terminus of Hsc70-interacting protein (CHIP) J. Biol. Chem. 2011;286:15883–15894. doi: 10.1074/jbc.M110.201814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moren A., Imamura T., Miyazono K., Heldin C.H., Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J. Biol. Chem. 2005;280:22115–22123. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- 117.Zhang L., Huang H., Zhou F., Schimmel J., Pardo C.G., Zhang T., Barakat T.S., Sheppard K.A., Mickanin C., Porter J.A., et al. RNF12 Controls Embryonic Stem Cell Fate and Morphogenesis in Zebrafish Embryos by Targeting Smad7 for Degradation. Mol. Cell. 2012;46:650–661. doi: 10.1016/j.molcel.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 118.Kim S.-E., Huang H., Zhao M., Zhang X., Zhang A., Semonov M.V., MacDonald B.T., Zhang X., Abreu J.G., Peng L., et al. Wnt Stabilization of β-Catenin Reveals Principles for Morphogen Receptor-Scaffold Assemblies. Science. 2013;340:867. doi: 10.1126/science.1232389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moon R.T., Bowerman B., Boutros M., Perrimon N. The Promise and Perils of Wnt Signaling Through β-Catenin. Science. 2002;296:1644. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- 120.van Es J.H., Barker N., Clevers H. You Wnt some, you lose some: Oncogenes in the Wnt signaling pathway. Curr. Opin. Genet. Dev. 2003;13:28–33. doi: 10.1016/S0959-437X(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 121.Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 122.Kielman M.F., Rindapaa M., Gaspar C., van Poppel N., Breukel C., van Leeuwen S., Taketo M.M., Roberts S., Smits R., Fodde R. Apc modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nat. Genet. 2002;32:594–605. doi: 10.1038/ng1045. [DOI] [PubMed] [Google Scholar]
- 123.Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Blauwkamp T.A., Nigam S., Ardehali R., Weissman I.L., Nusse R. Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat. Commun. 2012;3:1070. doi: 10.1038/ncomms2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu C., Li Y., Semenov M., Han C., Baeg G.H., Tan Y., Zhang Z., Lin X., He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/S0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 126.He X., Semenov M., Tamai K., Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: Arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 127.Pez F., Lopez A., Kim M., Wands J.R., Caron de Fromentel C., Merle P. Wnt signaling and hepatocarcinogenesis: Molecular targets for the development of innovative anticancer drugs. J. Hepatol. 2013;59:1107–1117. doi: 10.1016/j.jhep.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 128.Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. β-catenin is a target for the ubiquitin–proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ha N.C., Tonozuka T., Stamos J.L., Choi H.J., Weis W.I. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol. Cell. 2004;15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 130.Su Y., Fu C., Ishikawa S., Stella A., Kojima M., Shitoh K., Schreiber E.M., Day B.W., Liu B. APC is essential for targeting phosphorylated beta-catenin to the SCFbeta-TrCP ubiquitin ligase. Mol. Cell. 2008;32:652–661. doi: 10.1016/j.molcel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 131.Wu G., Xu G., Schulman B.A., Jeffrey P.D., Harper J.W., Pavletich N.P. Structure of a β-TrCP1-Skp1-β-Catenin Complex: Destruction Motif Binding and Lysine Specificity of the SCFβ-TrCP1 Ubiquitin Ligase. Mol. Cell. 2003;11:1445–1456. doi: 10.1016/S1097-2765(03)00234-X. [DOI] [PubMed] [Google Scholar]
- 132.Ji L., Jiang B., Jiang X., Charlat O., Chen A., Mickanin C., Bauer A., Xu W., Yan X., Cong F. The SIAH E3 ubiquitin ligases promote Wnt/β-catenin signaling through mediating Wnt-induced Axin degradation. Genes Dev. 2017;31:904–915. doi: 10.1101/gad.300053.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meshorer E., Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 134.Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 135.Campos E.I., Reinberg D. Histones: Annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 136.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 137.Thorne A.W., Sautiere P., Briand G., Crane-Robinson C. The structure of ubiquitinated histone H2B. EMBO J. 1987;6:1005–1010. doi: 10.1002/j.1460-2075.1987.tb04852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Minsky N., Shema E., Field Y., Schuster M., Segal E., Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 139.Kim J., Hake S.B., Roeder R.G. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 140.Fuchs G., Shema E., Vesterman R., Kotler E., Wolchinsky Z., Wilder S., Golomb L., Pribluda A., Zhang F., Haj-Yahya M., et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol. Cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.de Napoles M., Mermoud J.E., Wakao R., Tang Y.A., Endoh M., Appanah R., Nesterova T.B., Silva J., Otte A.P., Vidal M., et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 142.Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 143.Stock J.K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A.G., Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 144.van der Stoop P., Boutsma E.A., Hulsman D., Noback S., Heimerikx M., Kerkhoven R.M., Voncken J.W., Wessels L.F., van Lohuizen M. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PloS ONE. 2008;3:2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Endoh M., Endo T.A., Endoh T., Fujimura Y., Ohara O., Toyoda T., Otte A.P., Okano M., Brockdorff N., Vidal M., et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 146.Inoue D., Aihara H., Sato T., Mizusaki H., Doiguchi M., Higashi M., Imamura Y., Yoneda M., Miyanishi T., Fujii S., et al. Dzip3 regulates developmental genes in mouse embryonic stem cells by reorganizing 3D chromatin conformation. Sci. Rep. 2015;5:16567. doi: 10.1038/srep16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Singh M.P., Rai S., Suyal S., Singh S.K., Singh N.K., Agarwal A., Srivastava S. Genetic and epigenetic markers in colorectal cancer screening: Recent advances. Expert Rev. Mol. Diagn. 2017;17:665–685. doi: 10.1080/14737159.2017.1337511. [DOI] [PubMed] [Google Scholar]
- 148.Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 149.Kinzler K.W., Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 150.Markowitz S.D., Bertagnolli M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Toyota M., Ahuja N., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.P. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lynch H.T., Lynch J.F. Genetics of colonic cancer. Digestion. 1998;59:481–492. doi: 10.1159/000007525. [DOI] [PubMed] [Google Scholar]
- 153.Kuwai T., Kitadai Y., Tanaka S., Hiyama T., Tanimoto K., Chayama K. Mutation of the von Hippel-Lindau (VHL) gene in human colorectal carcinoma: Association with cytoplasmic accumulation of hypoxia-inducible factor (HIF)-1alpha. Cancer Sci. 2004;95:149–153. doi: 10.1111/j.1349-7006.2004.tb03196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lisztwan J., Imbert G., Wirbelauer C., Gstaiger M., Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Iwai K., Yamanaka K., Kamura T., Minato N., Conaway R.C., Conaway J.W., Klausner R.D., Pause A. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ohh M., Park C.W., Ivan M., Hoffman M.A., Kim T.Y., Huang L.E., Pavletich N., Chau V., Kaelin W.G. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 157.Maxwell P.H., Wiesener M.S., Chang G.W., Clifford S.C., Vaux E.C., Cockman M.E., Wykoff C.C., Pugh C.W., Maher E.R., Ratcliffe P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–2753. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 158.Semenza G.L. Regulation of Mammalian O2 Homeostasis by Hypoxia-Inducible Factor 1. Annu. Rev. Cell Dev. Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 159.Miyoshi Y., Nagase H., Ando H., Horii A., Ichii S., Nakatsuru S., Aoki T., Miki Y., Mori T., Nakamura Y. Somatic mutations of the APC gene in colorectal tumors: Mutation cluster region in the APC gene. Hum. Mol. Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 160.Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K.W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 161.Wu H., Lu X.X., Wang J.R., Yang T.Y., Li X.M., He X.S., Li Y., Ye W.L., Wu Y., Gan W.J., et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/beta-catenin degradation and is targeted for GSK3B/GSK3beta-mediated phosphorylation and degradation. Autophagy. 2019;26:1554–8627. doi: 10.1080/15548627.2019.1586250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu L., Zhang Y., Wong C.C., Zhang J., Dong Y., Li X., Kang W., Chan F.K.L., Sung J.J.Y., Yu J. RNF6 Promotes Colorectal Cancer by Activating the Wnt/beta-Catenin Pathway via Ubiquitination of TLE3. Cancer Res. 2018;78:1958–1971. doi: 10.1158/0008-5472.CAN-17-2683. [DOI] [PubMed] [Google Scholar]
- 163.Daniels D.L., Weis W.I. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 164.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 165.Barker N., van de Wetering M., Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Datta K., Suman S., Kumar S., Fornace A.J., Jr. Colorectal Carcinogenesis, Radiation Quality, and the Ubiquitin-Proteasome Pathway. J. Cancer. 2016;7:174–183. doi: 10.7150/jca.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sancho R., Jandke A., Davis H., Diefenbacher M.E., Tomlinson I., Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 168.Babaei-Jadidi R., Li N., Saadeddin A., Spencer-Dene B., Jandke A., Muhammad B., Ibrahim E.E., Muraleedharan R., Abuzinadah M., Davis H., et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J. Exp. Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li N., Babaei-Jadidi R., Lorenzi F., Spencer-Dene B., Clarke P., Domingo E., Tulchinsky E., Vries R.G.J., Kerr D., Pan Y., et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis. 2019;8:13. doi: 10.1038/s41389-019-0125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kemp Z., Rowan A., Chambers W., Wortham N., Halford S., Sieber O., Mortensen N., von Herbay A., Gunther T., Ilyas M., et al. CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res. 2005;65:11361–11366. doi: 10.1158/0008-5472.CAN-05-2565. [DOI] [PubMed] [Google Scholar]
- 171.Miyaki M., Yamaguchi T., Iijima T., Takahashi K., Matsumoto H., Mori T. Somatic mutations of the CDC4 (FBXW7) gene in hereditary colorectal tumors. Oncology. 2009;76:430–434. doi: 10.1159/000217811. [DOI] [PubMed] [Google Scholar]
- 172.Iwatsuki M., Mimori K., Ishii H., Yokobori T., Takatsuno Y., Sato T., Toh H., Onoyama I., Nakayama K.I., Baba H., et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: Clinical significance. Int. J. Cancer. 2010;126:1828–1837. doi: 10.1002/ijc.24879. [DOI] [PubMed] [Google Scholar]
- 173.Inuzuka H., Shaik S., Onoyama I., Gao D., Tseng A., Maser R.S., Zhai B., Wan L., Gutierrez A., Lau A.W., et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wertz I.E., Kusam S., Lam C., Okamoto T., Sandoval W., Anderson D.J., Helgason E., Ernst J.A., Eby M., Liu J., et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 175.Bhola P.D., Letai A. Mitochondria-Judges and Executioners of Cell Death Sentences. Mol. Cell. 2016;61:695–704. doi: 10.1016/j.molcel.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tong J., Tan S., Zou F., Yu J., Zhang L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene. 2017;36:787–796. doi: 10.1038/onc.2016.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 178.Myant K.B., Cammareri P., Hodder M.C., Wills J., Von Kriegsheim A., Gyorffy B., Rashid M., Polo S., Maspero E., Vaughan L., et al. HUWE1 is a critical colonic tumour suppressor gene that prevents MYC signalling, DNA damage accumulation and tumour initiation. EMBO Mol. Med. 2017;9:181–197. doi: 10.15252/emmm.201606684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dominguez-Brauer C., Khatun R., Elia A.J., Thu K.L., Ramachandran P., Baniasadi S.P., Hao Z., Jones L.D., Haight J., Sheng Y., et al. E3 ubiquitin ligase Mule targets beta-catenin under conditions of hyperactive Wnt signaling. Proc. Natl. Acad. Sci. USA. 2017;114:1148–1157. doi: 10.1073/pnas.1621355114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tarcic O., Pateras I.S., Cooks T., Shema E., Kanterman J., Ashkenazi H., Boocholez H., Hubert A., Rotkopf R., Baniyash M., et al. RNF20 Links Histone H2B Ubiquitylation with Inflammation and Inflammation-Associated Cancer. Cell Rep. 2016;14:1462–1476. doi: 10.1016/j.celrep.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang Z., Zhu L., Guo T., Wang Y., Yang J. Decreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinoma. Hum. Pathol. 2015;46:1006–1014. doi: 10.1016/j.humpath.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 182.Espinosa J.M. Histone H2B ubiquitination: The cancer connection. Genes Dev. 2008;22:2743–2749. doi: 10.1101/gad.1732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chernikova S.B., Razorenova O.V., Higgins J.P., Sishc B.J., Nicolau M., Dorth J.A., Chernikova D.A., Kwok S., Brooks J.D., Bailey S.M., et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 2012;72:2111–2119. doi: 10.1158/0008-5472.CAN-11-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Carter S.L., Eklund A.C., Kohane I.S., Harris L.N., Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 185.Thompson L.L., Jeusset L.M., Lepage C.C., McManus K.J. Evolving Therapeutic Strategies to Exploit Chromosome Instability in Cancer. Cancers. 2017;9:151. doi: 10.3390/cancers9110151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Brocato J., Costa M. SATB1 and 2 in colorectal cancer. Carcinogenesis. 2014;36:186–191. doi: 10.1093/carcin/bgu322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mir R., Pradhan S.J., Patil P., Mulherkar R., Galande S. Wnt/beta-catenin signaling regulated SATB1 promotes colorectal cancer tumorigenesis and progression. Oncogene. 2016;35:1679–1691. doi: 10.1038/onc.2015.232. [DOI] [PubMed] [Google Scholar]
- 188.Yu L., Dong L., Wang Y., Liu L., Long H., Li H., Li J., Yang X., Liu Z., Duan G., et al. Reversible regulation of SATB1 ubiquitination by USP47 and SMURF2 mediates colon cancer cell proliferation and tumor progression. Cancer Lett. 2019;448:40–51. doi: 10.1016/j.canlet.2019.01.039. [DOI] [PubMed] [Google Scholar]
- 189.Klupp F., Giese C., Halama N., Franz C., Lasitschka F., Warth A., Schmidt T., Kloor M., Ulrich A., Schneider M. E3 ubiquitin ligase Smurf2: A prognostic factor in microsatellite stable colorectal cancer. Cancer Manag. Res. 2019;11:1795–1803. doi: 10.2147/CMAR.S178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ougolkov A., Zhang B., Yamashita K., Bilim V., Mai M., Fuchs S.Y., Minamoto T. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. J. Natl. Cancer Inst. 2004;96:1161–1170. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- 191.Kitagawa M., Hatakeyama S., Shirane M., Matsumoto M., Ishida N., Hattori K., Nakamichi I., Kikuchi A., Nakayama K., Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu C., Kato Y., Zhang Z., Do V.M., Yankner B.A., He X. beta-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc. Natl. Acad. Sci. USA. 1999;96:6273–6278. doi: 10.1073/pnas.96.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hadjihannas M.V., Bernkopf D.B., Bruckner M., Behrens J. Cell cycle control of Wnt/beta-catenin signalling by conductin/axin2 through CDC20. EMBO Rep. 2012;13:347–354. doi: 10.1038/embor.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Uddin S., Ahmed M., Bavi P., El-Sayed R., Al-Sanea N., AbdulJabbar A., Ashari L.H., Alhomoud S., Al-Dayel F., Hussain A.R., et al. Bortezomib (Velcade) induces p27Kip1 expression through S-phase kinase protein 2 degradation in colorectal cancer. Cancer Res. 2008;68:3379–3388. doi: 10.1158/0008-5472.CAN-07-6109. [DOI] [PubMed] [Google Scholar]
- 195.Shapira M., Ben-Izhak O., Linn S., Futerman B., Minkov I., Hershko D.D. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer. 2005;103:1336–1346. doi: 10.1002/cncr.20917. [DOI] [PubMed] [Google Scholar]
- 196.Bochis O.V., Irimie A., Pichler M., Berindan-Neagoe I. The role of Skp2 and its substrate CDKN1B (p27) in colorectal cancer. J. Gastrointestin Liver Dis. 2015;24:225–234. doi: 10.15403/jgld.2014.1121.242.skp2. [DOI] [PubMed] [Google Scholar]
- 197.Hao B., Zheng N., Schulman B.A., Wu G., Miller J.J., Pagano M., Pavletich N.P. Structural Basis of the Cks1-Dependent Recognition of p27Kip1 by the SCFSkp2 Ubiquitin Ligase. Mol. Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 198.Okuda H., Saitoh K., Hirai S., Iwai K., Takaki Y., Baba M., Minato N., Ohno S., Shuin T. The von Hippel-Lindau tumor suppressor protein mediates ubiquitination of activated atypical protein kinase C. J. Biol. Chem. 2001;276:43611–43617. doi: 10.1074/jbc.M107880200. [DOI] [PubMed] [Google Scholar]
- 199.Cockman M.E., Masson N., Mole D.R., Jaakkola P., Chang G.-W., Clifford S.C., Maher E.R., Pugh C.W., Ratcliffe P.J., Maxwell P.H. Hypoxia Inducible Factor-α Binding and Ubiquitylation by the von Hippel-Lindau Tumor Suppressor Protein. J. Biol. Chem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 200.Jung C.R., Hwang K.S., Yoo J., Cho W.K., Kim J.M., Kim W.H., Im D.S. E2-EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat. Med. 2006;12:809–816. doi: 10.1038/nm1440. [DOI] [PubMed] [Google Scholar]
- 201.Suzuki Y., Nakabayashi Y., Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc. Natl. Acad. Sci. USA. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ehrenschwender M., Bittner S., Seibold K., Wajant H. XIAP-targeting drugs re-sensitize PIK3CA-mutated colorectal cancer cells for death receptor-induced apoptosis. Cell Death Dis. 2014;5:e1570. doi: 10.1038/cddis.2014.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hanson A.J., Wallace H.A., Freeman T.J., Beauchamp R.D., Lee L.A., Lee E. XIAP monoubiquitylates Groucho/TLE to promote canonical Wnt signaling. Mol. Cell. 2012;45:619–628. doi: 10.1016/j.molcel.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhu B., Zheng Y., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 205.Hattori T., Kishino T., Stephen S., Eberspaecher H., Maki S., Takigawa M., de Crombrugghe B., Yasuda H. E6-AP/UBE3A protein acts as a ubiquitin ligase toward SOX9 protein. J. Biol. Chem. 2013;288:35138–35148. doi: 10.1074/jbc.M113.486795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Heng S., Xue-Bing L., Ya M., Li F., Min L., Jing F.J.C.R. TRAF6 upregulates expression of HIF-1α and promotes tumor angiogenesis. Cancer Res. 2013;73:4950–4959. doi: 10.1158/0008-5472.CAN-13-0370. [DOI] [PubMed] [Google Scholar]
- 207.Zhang T., Wang H., Han L. Expression and Clinical Significance of Tumor Necrosis Factor Receptor-Associated Factor 6 in Patients with Colon Cancer. Iran. Red. Crescent Med. J. 2016;18:e23931. doi: 10.5812/ircmj.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu L., Wong C.C., Gong B., Yu J. Functional significance and therapeutic implication of ring-type E3 ligases in colorectal cancer. Oncogene. 2017;37:148. doi: 10.1038/onc.2017.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z.J. Activation of the IkappaB Kinase Complex by TRAF6 Requires a Dimeric Ubiquitin-Conjugating Enzyme Complex and a Unique Polyubiquitin Chain. Cell. 2000;103:351–361. doi: 10.1016/S0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 210.Krajewska M., Kim H., Kim C., Kang H., Welsh K., Matsuzawa S., Tsukamoto M., Thomas R.G., Assa-Munt N., Piao Z., et al. Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin. Cancer Res. 2005;11:5451–5461. doi: 10.1158/1078-0432.CCR-05-0094. [DOI] [PubMed] [Google Scholar]
- 211.Welcker M., Clurman B.E. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 212.Nateri A.S., Riera-Sans L., Da Costa C., Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 213.Yoon S.Y., Lee Y., Kim J.H., Chung A.S., Joo J.H., Kim C.N., Kim N.S., Choe I.S., Kim J.W. Over-expression of human UREB1 in colorectal cancer: HECT domain of human UREB1 inhibits the activity of tumor suppressor p53 protein. Biochem. Biophys. Res. Commun. 2005;326:7–17. doi: 10.1016/j.bbrc.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 214.Kubbutat M.H., Jones S.N., Vousden K.H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 215.Abdel-Fattah G., Yoffe B., Krishnan B., Khaoustov V., Ltani K. MDM2/p53 protein expression in the development of colorectal adenocarcinoma. J. Gastrointest Surg. 2000;4:109–114. doi: 10.1016/S1091-255X(00)80041-4. [DOI] [PubMed] [Google Scholar]
- 216.Thomas J.J., Abed M., Heuberger J., Novak R., Zohar Y., Beltran Lopez A.P., Trausch-Azar J.S., Ilagan M.X.G., Benhamou D., Dittmar G., et al. RNF4-Dependent Oncogene Activation by Protein Stabilization. Cell Rep. 2016;16:3388–3400. doi: 10.1016/j.celrep.2016.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Gonzalez-Prieto R., Cuijpers S.A., Kumar R., Hendriks I.A., Vertegaal A.C. c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle. 2015;14:1859–1872. doi: 10.1080/15384101.2015.1040965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hakli M., Lorick K.L., Weissman A.M., Janne O.A., Palvimo J.J. Transcriptional coregulator SNURF (RNF4) possesses ubiquitin E3 ligase activity. FEBS Lett. 2004;560:56–62. doi: 10.1016/S0014-5793(04)00070-5. [DOI] [PubMed] [Google Scholar]
- 219.Wu B., Piloto S., Zeng W., Hoverter N.P., Schilling T.F., Waterman M.L. Ring Finger Protein 14 is a new regulator of TCF/β-catenin-mediated transcription and colon cancer cell survival. EMBO Rep. 2013;14:347–355. doi: 10.1038/embor.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cantù C., Valenta T., Basler K. A RING finger to wed TCF and β-catenin. EMBO Rep. 2013;14:295–296. doi: 10.1038/embor.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Jiang X., Charlat O., Zamponi R., Yang Y., Cong F. Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol. Cell. 2015;58:522–533. doi: 10.1016/j.molcel.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 222.Koo B.K., Spit M., Jordens I., Low T.Y., Stange D.E., van de Wetering M., van Es J.H., Mohammed S., Heck A.J., Maurice M.M., et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 223.Giannakis M., Hodis E., Jasmine Mu X., Yamauchi M., Rosenbluh J., Cibulskis K., Saksena G., Lawrence M.S., Qian Z.R., Nishihara R., et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 2014;46:1264–1266. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Piao M.Y., Cao H.L., He N.N., Xu M.Q., Dong W.X., Wang W.Q., Wang B.M., Zhou B. Potential role of TRIM3 as a novel tumour suppressor in colorectal cancer (CRC) development. Scand. J. Gastroenterol. 2016;51:572–582. doi: 10.3109/00365521.2015.1124285. [DOI] [PubMed] [Google Scholar]
- 225.Lee O.H., Lee J., Lee K.H., Woo Y.M., Kang J.H., Yoon H.G., Bae S.K., Songyang Z., Oh S.H., Choi Y. Role of the focal adhesion protein TRIM15 in colon cancer development. Biochim. Biophys. Acta. 2015;1853:409–421. doi: 10.1016/j.bbamcr.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 226.Glebov O.K., Rodriguez L.M., Soballe P., DeNobile J., Cliatt J., Nakahara K., Kirsch I.R. Gene expression patterns distinguish colonoscopically isolated human aberrant crypt foci from normal colonic mucosa. Cancer Epidemiol. Biomarkers Prev. 2006;15:2253–2262. doi: 10.1158/1055-9965.EPI-05-0694. [DOI] [PubMed] [Google Scholar]
- 227.Coulie P.G., Van den Eynde B.J., van der Bruggen P., Boon T. Tumour antigens recognized by T lymphocytes: At the core of cancer immunotherapy. Nat. Rev. Cancer. 2014;14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 228.Botter S.M., Neri D., Fuchs B. Recent advances in osteosarcoma. Curr. Opin. Pharmacol. 2014;16:15–23. doi: 10.1016/j.coph.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 229.Kaste S.C., Pratt C.B., Cain A.M., Jones-Wallace D.J., Rao B.N. Metastases detected at the time of diagnosis of primary pediatric extremity osteosarcoma at diagnosis: Imaging features. Cancer. 1999;86:1602–1608. doi: 10.1002/(SICI)1097-0142(19991015)86:8<1602::AID-CNCR31>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 230.Haydon R.C., Luu H.H., He T.C. Osteosarcoma and osteoblastic differentiation: A new perspective on oncogenesis. Clin. Orthop. Relat. Res. 2007;454:237–246. doi: 10.1097/BLO.0b013e31802b683c. [DOI] [PubMed] [Google Scholar]
- 231.Thomas D.M., Johnson S.A., Sims N.A., Trivett M.K., Slavin J.L., Rubin B.P., Waring P., McArthur G.A., Walkley C.R., Holloway A.J., et al. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J. Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Luo X., Chen J., Song W.X., Tang N., Luo J., Deng Z.L., Sharff K.A., He G., Bi Y., He B.C., et al. Osteogenic BMPs promote tumor growth of human osteosarcomas that harbor differentiation defects. Lab. Invest. 2008;88:1264–1277. doi: 10.1038/labinvest.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Peng Y., Kang Q., Cheng H., Li X., Sun M.H., Jiang W., Luu H.H., Park J.Y., Haydon R.C., He T.-C. Transcriptional Characterization of Bone Morphogenetic Proteins (BMPs)-Mediated Osteogenic Signaling. J. Cell Biochem. 2003;90:1149–1165. doi: 10.1002/jcb.10744. [DOI] [PubMed] [Google Scholar]
- 234.Peng Y., Kang Q., Luo Q., Jiang W., Si W., Liu B.A., Luu H.H., Park J.K., Li X., Luo J., et al. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J. Biol. Chem. 2004;279:32941–32949. doi: 10.1074/jbc.M403344200. [DOI] [PubMed] [Google Scholar]
- 235.Ducy P., Schinke T., Karsenty G. The osteoblast: A sophisticated fibroblast under central surveillance. Science. 2000;289:1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 236.Wang L., Park P., Lin C.Y. Characterization of stem cell attributes in human osteosarcoma cell lines. Cancer Biol. Ther. 2009;8:543–552. doi: 10.4161/cbt.8.6.7695. [DOI] [PubMed] [Google Scholar]
- 237.Levings P.P., McGarry S.V., Currie T.P., Nickerson D.M., McClellan S., Ghivizzani S.C., Steindler D.A., Gibbs C.P. Expression of an exogenous human Oct-4 promoter identifies tumor-initiating cells in osteosarcoma. Cancer Res. 2009;69:5648–5655. doi: 10.1158/0008-5472.CAN-08-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhang W., Zhuang Y., Zhang Y., Yang X., Zhang H., Wang G., Yin W., Wang R., Zhang Z., Xiao W. Uev1A facilitates osteosarcoma differentiation by promoting Smurf1-mediated Smad1 ubiquitination and degradation. Cell Death Dis. 2017;8:2974. doi: 10.1038/cddis.2017.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jones D.C., Wein M.N., Oukka M., Hofstaetter J.G., Glimcher M.J., Glimcher L.H. Regulation of Adult Bone Mass by the Zinc Finger Adapter Protein Schnurri-3. Science. 2006;312:1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- 240.Ying M., Zhang L., Zhou Q., Shao X., Cao J., Zhang N., Li W., Zhu H., Yang B., He Q. The E3 ubiquitin protein ligase MDM2 dictates all-trans retinoic acid-induced osteoblastic differentiation of osteosarcoma cells by modulating the degradation of RARα. Oncogene. 2016;35:4358. doi: 10.1038/onc.2015.503. [DOI] [PubMed] [Google Scholar]
- 241.Chen Z., Shen B.L., Fu Q.G., Wang F., Tang Y.X., Hou C.L., Chen L. CUL4B promotes proliferation and inhibits apoptosis of human osteosarcoma cells. Oncol. Rep. 2014;32:2047–2053. doi: 10.3892/or.2014.3465. [DOI] [PubMed] [Google Scholar]
- 242.Chen Z., Zhang W., Jiang K., Chen B., Wang K., Lao L., Hou C., Wang F., Zhang C., Shen H. MicroRNA-300 Regulates the Ubiquitination of PTEN through the CRL4B(DCAF13) E3 Ligase in Osteosarcoma Cells. Mol. Ther. Nucleic Acids. 2018;10:254–268. doi: 10.1016/j.omtn.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang C., Chen B., Jiang K., Lao L., Shen H., Chen Z. Activation of TNF-α/NF-κB axis enhances CRL4B(DCAF)(11) E3 ligase activity and regulates cell cycle progression in human osteosarcoma cells. Mol. Oncol. 2018;12:476–494. doi: 10.1002/1878-0261.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Severe N., Dieudonne F.X., Marty C., Modrowski D., Patino-Garcia A., Lecanda F., Fromigue O., Marie P.J. Targeting the E3 ubiquitin casitas B-lineage lymphoma decreases osteosarcoma cell growth and survival and reduces tumorigenesis. J. Bone. Miner. Res. 2012;27:2108–2117. doi: 10.1002/jbmr.1667. [DOI] [PubMed] [Google Scholar]
- 245.Archange C., Nowak J., Garcia S., Moutardier V., Calvo E.L., Dagorn J.C., Iovanna J.L. The WSB1 gene is involved in pancreatic cancer progression. PLoS ONE. 2008;3:2475. doi: 10.1371/journal.pone.0002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Dentice M., Bandyopadhyay A., Gereben B., Callebaut I., Christoffolete M.A., Kim B.W., Nissim S., Mornon J.P., Zavacki A.M., Zeold A., et al. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nat. Cell Biol. 2005;7:698–705. doi: 10.1038/ncb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Choi D.W., Seo Y.M., Kim E.A., Sung K.S., Ahn J.W., Park S.J., Lee S.R., Choi C.Y. Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J. Biol. Chem. 2008;283:4682–4689. doi: 10.1074/jbc.M708873200. [DOI] [PubMed] [Google Scholar]
- 248.Cao J., Wang Y., Dong R., Lin G., Zhang N., Wang J., Lin N., Gu Y., Ding L., Ying M., et al. Hypoxia-Induced WSB1 Promotes the Metastatic Potential of Osteosarcoma Cells. Cancer Res. 2015;75:4839–4851. doi: 10.1158/0008-5472.CAN-15-0711. [DOI] [PubMed] [Google Scholar]
- 249.El-Naggar A.M., Clarkson P.W., Negri G.L., Turgu B., Zhang F., Anglesio M.S., Sorensen P.H. HACE1 is a potential tumor suppressor in osteosarcoma. Cell Death Dis. 2019;10:21. doi: 10.1038/s41419-018-1276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Chen L., Pei H., Lu S.J., Liu Z.J., Yan L., Zhao X.M., Hu B., Lu H.G. SPOP suppresses osteosarcoma invasion via PI3K/AKT/NF-kappaB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:609–615. doi: 10.26355/eurrev_201802_14275. [DOI] [PubMed] [Google Scholar]
- 251.Zhao M., Su Z., Zhang S., Zhuang L., Xie Y., Li X. Suppressive Role of MicroRNA-148a in Cell Proliferation and Invasion in Ovarian Cancer Through Targeting Transforming Growth Factor-β-Induced 2. Oncol. Res. 2016;24:353–360. doi: 10.3727/096504016X14685034103275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Moon S., Lee B.H. Chemically Induced Cellular Proteolysis: An Emerging Therapeutic Strategy for Undruggable Targets. Mol. Cells. 2018;41:933–942. doi: 10.14348/molcells.2018.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zeng X., Sigoillot F., Gaur S., Choi S., Pfaff K.L., Oh D.C., Hathaway N., Dimova N., Cuny G.D., King R.W. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sackton K.L., Dimova N., Zeng X., Tian W., Zhang M., Sackton T.B., Meaders J., Pfaff K.L., Sigoillot F., Yu H., et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature. 2014;514:646–649. doi: 10.1038/nature13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zhang B., Golding B.T., Hardcastle I.R. Small-molecule MDM2-p53 inhibitors: Recent advances. Future Med. Chem. 2015;7:631–645. doi: 10.4155/fmc.15.13. [DOI] [PubMed] [Google Scholar]
- 256.Zhao Y., Aguilar A., Bernard D., Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 Inhibitors) in clinical trials for cancer treatment. J. Med. Chem. 2015;58:1038–1052. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Chan C.-H., Morrow J.K., Li C.-F., Gao Y., Jin G., Moten A., Stagg L.J., Ladbury J.E., Cai Z., Xu D., et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Wu L., Grigoryan A.V., Li Y., Hao B., Pagano M., Cardozo T.J. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem. Biol. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Veggiani G., Gerpe M.C.R., Sidhu S.S., Zhang W. Emerging drug development technologies targeting ubiquitination for cancer therapeutics. Pharmacol. Ther. 2019;19 doi: 10.1016/j.pharmthera.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]