Hepatitis C virus (HCV) is a member of the Flaviviridae family, and its infection causes chronic hepatitis, liver cirrhosis, and even hepatocellular carcinoma. No vaccine is available. Many host factors may be implicated in the pathogenesis of HCV-related diseases. This study discloses a new host factor that binds to the HCV 5′ UTR and promotes HCV replication. Sam68 may play an important role in HCV-related diseases, and further investigation is highly encouraged to explore its specific actions in HCV pathogenesis.
KEYWORDS: 5′ untranslated region, HCV, Sam68, replication, viral genome
ABSTRACT
The Src-associated in mitosis 68-kDa (Sam68) protein is a highly conserved nuclear protein and is involved in a series of cellular processes, including transcription and signal transduction. Sam68 is comprised of 443 amino acids and contains an RGG box domain, a KH domain, and a tyrosine-rich domain. Its role in hepatitis C virus (HCV) replication is unknown. Here, we find that Sam68 promotes HCV replication without affecting viral translation. The RNA immunoprecipitation experiments show that the positive strand of HCV RNA interacts with Sam68. HCV infection triggers the translocation of the Sam68 protein from the nucleus to the cytoplasm, where it interacts with the HCV genome. Further study shows that the region of Sam68 spanning amino acids 1 to 157 is the pivotal domain to interact with the stem-loop 2 of the HCV 5′ untranslated region (5′ UTR) and is responsible for the enhancement of HCV replication. These data suggested that Sam68 may serve as a proviral factor of HCV to facilitate viral replication through interaction with the viral genome.
IMPORTANCE Hepatitis C virus (HCV) is a member of the Flaviviridae family, and its infection causes chronic hepatitis, liver cirrhosis, and even hepatocellular carcinoma. No vaccine is available. Many host factors may be implicated in the pathogenesis of HCV-related diseases. This study discloses a new host factor that binds to the HCV 5′ UTR and promotes HCV replication. Sam68 may play an important role in HCV-related diseases, and further investigation is highly encouraged to explore its specific actions in HCV pathogenesis.
INTRODUCTION
Hepatitis C virus (HCV) is an important human pathogen that causes worldwide health problems, including chronic hepatitis, liver cirrhosis, and even hepatocellular carcinoma (1, 2). There is no vaccine for HCV. HCV is a member of the genus Hepacivirus within the Flaviviridae family and is a positive-sense, single-stranded RNA genome of about 9.6 kb that contains one open reading frame (ORF), which is flanked by a 5′ untranslated region (5′ UTR) as well as a 3′ untranslated region (3′ UTR). The ORF encodes a large polyprotein of approximately 3,010 amino acids (aa), which is processed into three structural (Core, E1, and E2) and seven nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins by host and viral proteases (3). The 5′ UTR and 3′ UTR are involved in the control of virus translation and replication. The 5′ UTR contains the internal ribosome entry site (IRES) that mediates viral translation of the viral RNA (4, 5). Computer analysis and structure probing suggest that the 5′ UTR of HCV consists of four highly conserved structural domains (stem-loop 1 [SL1], stem-loop 2 [SL2], stem-loop 3 [SL3], and stem-loop 4 [SL4]) (6). Ribosome binding to an IRES spans most of the 5′ UTR, which covers SL2 to SL4, together with the first 24 to 40 nucleotides of the core coding region. The SL1 formed by ribonucleotides 5 to 20 is not required for IRES activity yet (7). Several reports have revealed that SL1 and SL2 of the 5′ UTR are necessary for HCV replication, resembling the 3′-terminal region of the negative strand (8–10).
The 5′ UTR of HCV functions as a platform to recruit viral and cellular proteins, which directs IRES-dependent protein synthesis and regulates viral RNA replication. Some of the host factors regulate HCV RNA replication either by participating in the formation of RNA replication complex or by binding to viral RNA (11). PCBP2 binds to the 5′ and 3′ UTR of HCV RNA, thereby enhancing HCV translation and/or replication (12). La antigen interacts with the 5′ UTR of the HCV RNA genome and enhances HCV IRES-dependent translation (13). Besides, eIF3, PTB, hnRNP L, and HMGB1 have been reported to bind to the 5′ and/or 3′ UTR of HCV RNA and regulate translation and/or replication (14–20).
The Src-associated in mitosis 68-kDa protein (Sam68) is a RNA-binding protein, which belongs to the STAR protein family. The Sam68 protein is composed of 443 amino acids and contains an RGG box domain, a KH domain, and a tyrosine-rich domain. Sam68 has been reported to participate in many cellular processes, including transcription, signal transduction, cell cycle, etc. Upon poliovirus (PV) infection, Sam68 interacts with viral RNA-dependent RNA polymerase and colocalizes with the PV 2C protein (21). Infection with rhinovirus induces the redistribution of Sam68 (22). Sam68 interacts with the IRES within the 5′ UTR of the foot-and-mouth disease virus (FMDV) genome and enhances virus translation (23). Sam68 plays a role in the translational regulation of HIV-1 RNA (24).
In this study, we aimed to explore the Sam68 functions on HCV replication. We found that Sam68 enhances HCV replication. Upon HCV infection, the Sam68 protein is translocated from nucleus to cytoplasm to interact with viral RNA. Further studies showed that the region of Sam68 spanning amino acids 1 to 157 binds to the SL2 of the HCV 5′ UTR, which facilitates viral RNA replication. In summary, we demonstrated that Sam68 serves as a proviral factor of HCV to facilitate viral replication through interaction with viral genome.
RESULTS
Sam68 is associated with PCBP2 to be an interactome in the context of HCV infection.
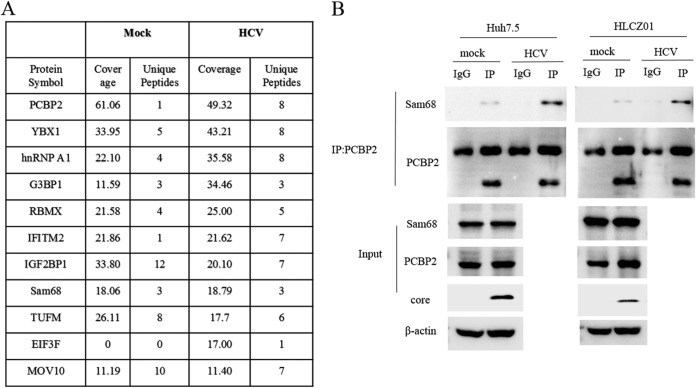
We used a mass spectrometry approach to identify PCBP2-associated host proteins. Data from mass spectrometry revealed Sam68 with an efficient high score as a PCBP2-binding partner. Moreover, the human proteins, including hnRNPA1 and IGF2BP1, were demonstrated to have high scores as the PCBP2 interactome (Fig. 1A), both of which have been reported to participate in regulation of HCV replication and translation (25, 26). We verified the interaction between PCBP2 and Sam68 using anti-PCBP2 immunoprecipitation of lysate from Huh7.5 or HLCZ01 cells with or without HCV infection. The interaction between PCBP2 and Sam68 was observed in all cases and enhanced upon HCV infection (Fig. 1B). Given that PCBP2 plays a crucial role in regulating HCV replication and translation in previous study (12), we turn to explore the role of Sam68 in the HCV life cycle.
FIG 1.
Sam68 is associated with PCBP2 to be an interactome upon HCV infection. (A) Flag-tagged vector transfected HLCZ01 cells and Flag-tagged PCBP2 transfected HLCZ01 cells with or without HCV infection were sequentially immunoprecipitated with anti-Flag antibody. The immunoprecipitates were fractionated by SDS-PAGE and visualized by Coomassie blue staining followed by mass spectrometry analysis. Numbers of candidate proteins and percentage of protein coverage of PCBP2 partners identified are shown in the table. (B) Huh7.5 or HLCZ01 cells were infected with HCV at an MOI of 0.1 for 3 days. Co-IP and immunoblotting were performed with the indicated antibodies.
Sam68 promotes HCV replication without affecting viral translation.
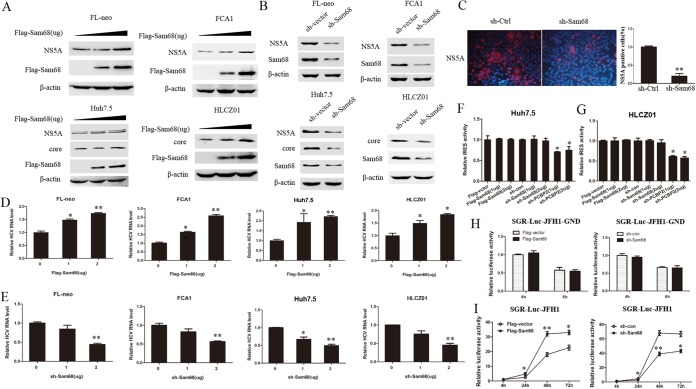
We utilized various cell lines, including FCA1, FL-neo, Huh7.5, and HLCZ01, to test whether Sam68 regulates the HCV life cycle. We overexpressed or knocked down Sam68 in FL-neo (HCV full-length replicon cell line) and FCA1 (HCV subgenomic replicon cell line) cells (Fig. 2A and B). Overexpression and silencing of Sam68 were confirmed by Western blotting (Fig. 2A and B). Overexpression of Sam68 significantly increased the level of viral core and NS5A protein (Fig. 2A) and intracellular viral RNA level (Fig. 2D). Silencing Sam68 by short hairpin RNA (shRNA) markedly reduced viral protein (Fig. 2B) and intracellular viral RNA levels compared to those of control shRNA-transfected cells (Fig. 2E). We observed similar results in HCV-infected Huh7.5 and HLCZ01 cells. Immunofluorescence staining with HCV NS5A antibody showed a markedly decreased percentage of virus-infected cells in the Sam68-silenced cell line compared to that of the control (Fig. 2C). Overexpression of Sam68 in virus-infected cells augmented the level of viral protein and intracellular viral RNA (Fig. 2A [lower] and Fig. 2D [right]). In contrast, viral protein and HCV RNA level in Sam68-silenced cells were notably decreased (Fig. 2B [lower] and Fig. 2E [right]). Collectively, these data suggested that Sam68 indeed promotes HCV replication.
FIG 2.
Sam68 promotes HCV replication without affecting viral translation. (A) FL-neo or FCA1 cells were transfected with increasing amounts of plasmid encoding Flag-tagged Sam68 for 48 h. Huh7.5 or HLCZ01 cells were infected with HCV at an MOI of 0.1 for 12 h and then transfected with an increasing amount of pFlag-tagged Sam68 for 60 h. All cell lysates were detected by immunoblotting with the indicated antibodies. β-Actin was used as an internal control. Triangles indicate increasing amounts of plasmid. (B) FL-neo or FCA1 cells were transfected with either the control shRNA or the Sam68 shRNA plasmid for 48 h. Huh7.5 or HLCZ01 cells were preinfected with HCV at an MOI of 0.1 for 12 h and then transfected with either the control shRNA plasmid or the Sam68 shRNA plasmid for 60 h. All cell lysates were detected by the indicated antibodies. (C) Huh7.5 cells expressing either scrambled shRNA (sh-Ctrl) or Sam68-targeting shRNA (sh-Sam68) were infected with HCV for 72 h followed by immunostaining using an anti-HCV NS5A antibody (red). DAPI was used to counterstain nuclei (blue). (D) The intracellular HCV RNA was measured by real-time PCR assay (normalized to GAPDH); meanwhile, the cells were under the same treatment as Fig. 2A. *, P < 0.05; **, P < 0.01. (E) FL-neo or FCA1 cells were transfected with a gradient of the Sam68 shRNA plasmid (1 μg, 2 μg) for 48 h. Huh7.5 or HLCZ01 cells were preinfected with HCV at an MOI of 0.1 for 12 h and then transfected with a gradient of the Sam68 shRNA plasmid (1 μg, 2 μg) for 60 h. The intracellular HCV RNA was analyzed by real-time PCR and normalized to GAPDH. (F, G) Huh7.5 (F) or HLCZ01 (G) cells were transfected with the indicated plasmids for 24 h together with the pRL-HL for an additional 24 h. Cell extracts were harvested to conduct a dual-luciferase reporter assay. (H, I) Huh7.5 cells were transfected with pFlag-vector or pFlag-Sam68 or control shRNA or Sam68 shRNA plasmid and then electroporated with the in vitro transcribed RNA (SGR-Luc-JFH1-GND [H] or SGR-Luc-JFH1 [I]). The luciferase assay was performed at the indicated time point postelectroporation.
The impact of Sam68 on viral protein level is possibly caused by the regulation of viral translation by Sam68. To verify our assumption, we used a pRL-HL plasmid containing HCV bicistronic RNAs. In the pRL-HL system, renilla luciferase gene expression is driven by Cap element, and firefly luciferase gene expression is driven by HCV IRES. This design allows the analysis of Sam68 requirements for HCV RNA translation. When we cotransfected the pRL-HL plasmid with different doses of the p3×FLAG-CMV-Sam68 or pSilencer-Sam68 plasmid into Huh7.5 cells, we found that the overexpression and silencing of Sam68 did not affect the translation activity of HCV IRES, although silencing of PCBP2, known as a positive factor for HCV translation, significantly reduced the activity of viral IRES translation (Fig. 2F). To further verify the effect of Sam68 on viral translation, we performed the above experiments in HLCZ01 cells and observed similar results (Fig. 2G). In addition, we used the replication-defective SGR-Luc-JFH1-GND, which has a mutated GDD to GND within the active-site motif of the NS5B polymerase, to analyze the influence of Sam68 on HCV IRES activity. No significant difference in luciferase activity was observed between vector- and Sam68-transfected Huh7.5 cells or between shcon- and shSam68-transfected Huh7.5 cells when we transfected SGR-Luc-JFH1-GND into these cells (Fig. 2H). The replication of HCV RNA was monitored by assessing luciferase activity at the indicated time points postelectroporation with HCV subgenomic RNA (SGR-Luc-JFH1). Overexpressing or silencing Sam68 increased or decreased luciferase activity, respectively, compared with that of control at 24 h, 48 h, and 72 h postelectroporation (Fig. 2I). These results indicated that Sam68 has no effect on HCV RNA translation, and the regulation of viral protein level by Sam68 may be caused by the modulation of viral RNA replication by Sam68.
Relocalization of Sam68 upon HCV infection.
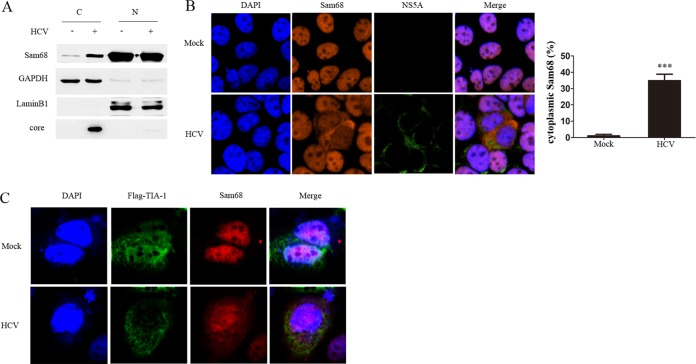
Sam68 is reported to be mainly located in the nucleus, whereas HCV replication occurs in the cytoplasm. We suspected that the intracellular localization of Sam68 may be altered during HCV infection. To verify this hypothesis, we separated the nuclear and cytoplasmic extracts and examined the localization of Sam68. Our data showed that the nuclear Sam68 protein is much more than cytoplasmic abundance in the cells without HCV infection, supporting that endogenous Sam68 is majority present in the nucleus. Upon HCV infection, the abundance of Sam68 protein in the nucleus decreased while its abundance in the cytoplasm increased, suggesting that Sam68 may shuttle from the nucleus to the cytoplasm during HCV infection (Fig. 3A). Furthermore, we monitored the intracellular distribution of Sam68 using immunofluorescence staining. Sam68 was mainly observed in the nucleus in uninfected cells, while it was relocalized to the cytoplasm in HCV-infected cells (Fig. 3B). To explore whether the translocation is due to the induction of stress, we detected the formation of stress granules upon HCV infection using a stress granule marker (TIA-1) (27). We did not see stress granules upon HCV infection (Fig. 3C). These findings indicate that HCV infection may cause the relocalization of Sam68 from nucleus to cytoplasm.
FIG 3.
Relocalization of Sam68 in HCV-infected cells. (A) Huh7.5 cells were mock or infected with HCV at an MOI of 0.1 for 72 h and cytoplasmic extracts (C) or nuclear extracts (N) were analyzed by immunoblotting with anti-Sam68, anti-HCV core, anti-GAPDH (cytoplasmic marker), and anti-LaminB (nuclear marker) antibodies. (B) Huh7.5 cells were infected with or without HCV for 72 h, followed by immunostaining using an anti-Sam68 antibody (red) and anti-HCV NS5A antibody (green). DAPI was used to counterstain nuclei (blue). (C) Huh7.5 cells were infected with or without HCV for 24 h, then transfected with pFlag-tagged TIA-1 for 48 h followed by immunostaining using an anti-Sam68 antibody (red) and anti-Flag antibody (green). DAPI was used to counterstain nuclei (blue).
Sam68 directly binds to HCV RNA.
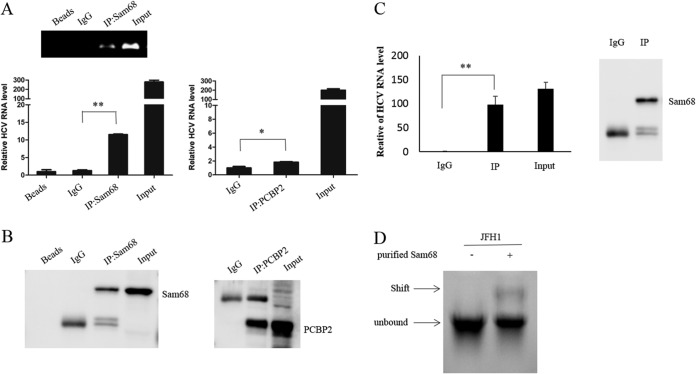
To analyze the mechanism of enhancement of HCV replication by endogenous Sam68, we detected the interaction between Sam68 and HCV RNA. We performed RNA immunoprecipitation (RIP) of the lysate of HCV-infected Huh7.5 cells using either an antibody specific to Sam68 or isotype anti-IgG. The precipitant complex was analyzed by reverse transcriptase PCR (RT-PCR) and immunoblotting. The common cell lysate was subjected to RIP with PCBP2 antibodies as a positive control. As shown in RT-PCR detection using primers specific to HCV, the amount of HCV RNA in precipitant with an anti-Sam68 antibody was much more than that in precipitant with IgG (Fig. 4A). Sam68 or PCBP2 protein enriched in RNA-protein complexes was confirmed by Western blotting (Fig. 4B). These results indicated that Sam68 specifically binds to HCV RNA in virus-infected cells.
FIG 4.
Sam68 directly binds to HCV RNA. (A and B) Huh7.5 cells were infected with HCV at an MOI of 0.1 for 72 h. The cells were harvested and immunoprecipitated with control IgG or anti-Sam68 antibody or anti-PCBP2 antibody. (A) RNA was extracted from the precipitate complex and subjected to RT-PCR or qRT-PCR analysis. (B) Protein from the precipitate complex was detected by immunoblotting with the anti-Sam68 and PCBP2 antibodies. (C) Purified Sam68 protein (0.5 μg) and in vitro transcribed JFH1 RNA (1 μg) were incubated in binding buffer for 20 min at room temperature followed by RIP analysis. (D) Gel shift analysis of complex formation between 30 pmol of purified Sam68 protein and 10 pmol of in vitro transcribed JFH1 RNA. Arrows denote the positions of unbound RNA and RNA-Sam68 complexes.
To further verify whether Sam68 directly binds to HCV RNA, we performed a binding assay with purified human Sam68 protein and HCV RNA in vitro. Purified Sam68 protein (0.5 μg) and in vitro transcribed HCV RNA (1 μg) were incubated in binding buffer for 20 min at room temperature, followed by RIP analysis and gel shift analysis. HCV RNA was precipitated with Sam68 protein by anti-Sam68-conjugated protein G beads (Fig. 4C). HCV RNA showed its slower migration in the presence of Sam68 protein (Fig. 4D). Taken together, these data demonstrated that Sam68 protein directly interacts with the HCV genome.
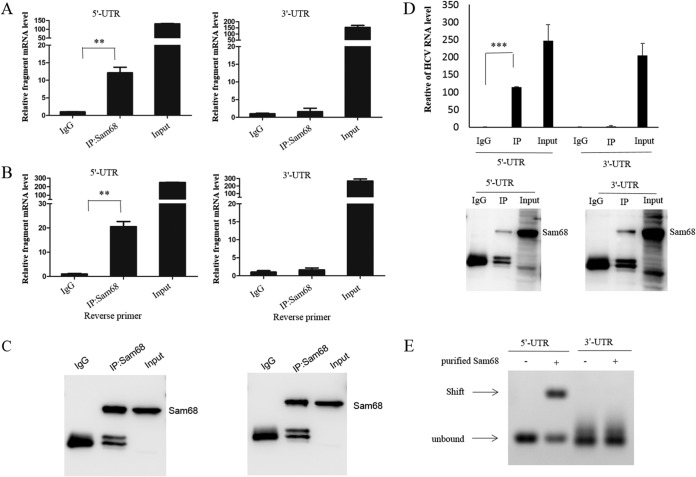
Sam68 is identified to directly interact with the HCV 5′ UTR.
Because the 5′ and 3′ UTR are considered to be involved in HCV replication, we investigated whether Sam68 directly interacts with the HCV 5′ and/or HCV 3′ UTR. For this purpose, we transfected the p3×FLAG-CMV-5′-UTR or p3×FLAG-CMV-3′-UTR plasmid into Huh7.5 cells, followed by RIP assay with either an antibody specific to Sam68 or isotype anti-IgG. The fragments of the HCV genome were amplified with specific primers and quantified by real-time PCR (Fig. 5A). HCV possesses a positive-sense, single-stranded RNA genome. We used a reverse primer to amplify the indicated target genes, such as positive-strand RNA of HCV or fragments in a reverse transcription experiment in immunoprecipitation (IP)-RT-PCR analyses. Meanwhile, the reverse primers of either HCV 5′ UTR or HCV 3′ UTR were used to perform RIP-RT-PCR analysis (Fig. 5B). Sam68 protein concentrated and separated by RIP was confirmed by Western blotting (Fig. 5C). Then, we analyzed the fractions of RNAs in the co-IP complexes. The results demonstrated that the level of 5′-UTR RNA was significantly higher in the anti-Sam68 antibody co-IP group than that in the control group (Fig. 5D). However, tests with the 3′ UTR of HCV RNA yielded negative results (Fig. 5D). The data suggested that Sam68 may bind to the 5′ UTR of HCV rather than the 3′ UTR. To further verify the association between the HCV 5′ UTR and Sam68, the purified human Sam68 protein and in vitro transcribed HCV 5′ UTR or 3′ UTR were used for the binding assay. Moreover, gel shift assay revealed that Sam68 protein directly bound to the HCV 5′-UTR fragment rather than the 3′ UTR (Fig. 5E). Taken together, these data supported that Sam68 protein directly interacts with the HCV 5′ UTR but not the 3′ UTR.
FIG 5.
Sam68 is identified to directly interact with the HCV 5′ UTR. (A to C) Huh7.5 cells were transfected with p3×FLAG-CMV-5′-UTR or p3×FLAG-CMV-3′-UTR for 48 h. The cells were harvested and co-IP experiments using IgG or anti-Sam68 antibody were performed. RNA was extracted from the precipitate complexes. (A) Different fragments of HCV RNA were measured by real-time PCR. (B) IP-RT-PCR with the reverse primers specific to either JFH1 5′ UTR or 3′ UTR were performed. (C) Protein from the precipitate complex was detected by immunoblotting with anti-Sam68 antibody. (D) Purified Sam68 protein (0.5 μg) and in vitro transcribed RNA (1 μg) were incubated in binding buffer for 20 min at room temperature, followed by RIP analysis and qRT-PCR. (E) Gel shift analysis of complex formation between 30 pmol of purified Sam68 protein and 10 pmol of in vitro transcribed RNA. Arrows denote the positions of unbound RNA and RNA-Sam68 complexes.
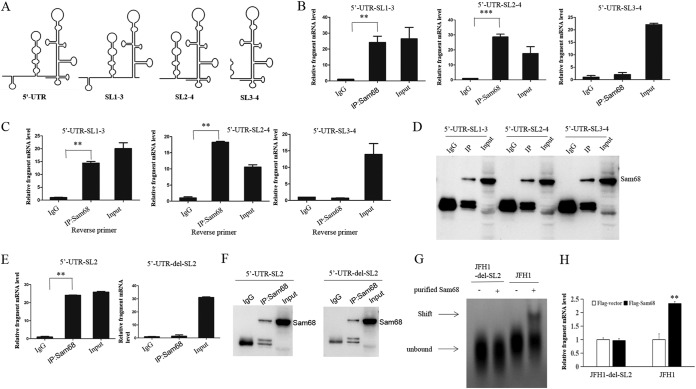
Sam68 binds to SL2 of HCV 5′ UTR.
HCV 5′ UTR contains four highly conserved structural domains including SL1, SL2, SL3, and SL4 (6). Our previous study revealed that SL4 of HCV 5′ UTR plays an important role in HCV RNA replication (15). To narrow down the sequence elements responsible for Sam68 binding, we constructed a series of truncated forms of the HCV 5′ UTR (Fig. 6A). These recombinant plasmids were transfected into Huh7.5 cells for 48 h, and the assays of co-IP and IP-RT-PCR using anti-Sam68 antibody or IgG were performed subsequently. The cells were harvested and subjected to RIP assay. The fragments of the HCV 5′ UTR were amplified with specific primers and quantified by real-time PCR (Fig. 6B). Meanwhile, the reverse primers specific to different fragments were used to perform reverse transcription in RIP-RT-PCR detection (Fig. 6C). Sam68 protein that was concentrated and separated by RIP was confirmed by Western blotting (Fig. 6D). The results showed the significant differences between IgG and anti-Sam68 antibody treatment in the groups of 5′-UTR-SL1-3 and 5′-UTR-SL2-4 assays rather than in the 5′-UTR-SL3-4 assay (Fig. 6C). Consistently, we utilized the fragments of the HCV 5′ UTR, including SL2 and the SL2 deletion mutant, to further verify the binding region of HCV 5′ UTR with the Sam68 protein. The result showed that SL2 is indeed necessary and sufficient for Sam68 binding (Fig. 6E and F). Moreover, a gel shift assay revealed that the Sam68 protein directly binds to the HCV JFH1 RNA but not to HCV JFH1-del-SL2 RNA (Fig. 6G). Overexpression of Sam68 enhanced HCV JFH1 RNA but had no effect on HCV JFH1-del-SL2 RNA (Fig. 6H). Taken together, all the data suggested that SL2 of the HCV 5′ UTR is the major region for the interaction between the HCV 5′ UTR and Sam68.
FIG 6.
Sam68 binds to SL2 of HCV 5′ UTR. (A) Scheme for HCV 5′-UTR truncations. (B to D) Huh7.5 cells were transfected with p3×FLAG-CMV-5′-UTR-SL1-3, p3×FLAG-CMV-5′-UTR-SL2-4, or p3×FLAG-CMV-5′-UTR-SL3-4 for 48 h. The cells were harvested and subjected to co-IP experiments using IgG or anti-Sam68 antibody. RNA was extracted from the precipitate complexes. (B) Different fragments of HCV 5′ UTR were measured by real-time PCR. (C) Different fragments of the HCV 5′ UTR were quantified by IP-RT-PCR with the indicated reverse primers. (D) Protein from the precipitate complex was detected by immunoblotting with anti-Sam68 antibody. (E) We amplified SL2 and del-SL2(SL1-SL3-SL4) fragments from p3×FLAG-CMV-5′-UTR and inserted them into p3×FLAG-CMV-vector. Huh7.5 cells were transfected with p3×FLAG-CMV-5′-UTR-SL2 or p3×FLAG-CMV-5′-UTR-del-SL2 for 48 h. SL2 or deletion of SL2 of the HCV 5′ UTR was quantified by IP-RT-PCR with the indicated reverse primers. (F) Protein from the precipitate complex (E) was detected by immunoblotting with anti-Sam68 antibody. (G) Gel shift analysis of complex formation between 30 pmol of purified Sam68 protein and 20 pmol of in vitro transcribed JFH1 RNA or JFH1-del-SL2 RNA. Arrows denote the positions of unbound RNA and RNA-Sam68 complexes. (H) Huh7.5 cells were cotransfected with Flag-tagged Sam68 and pJFH1 or Flag-tagged Sam68 and pJFH1-del-SL2, and the intracellular HCV RNA was measured by real-time PCR assay (normalized to GAPDH).
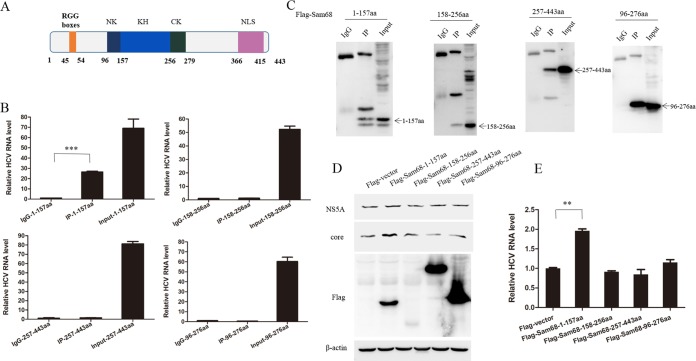
The region of Sam68 spanning amino acids 1 to 157 is crucial for HCV genome binding and responsible for the enhancement of viral replication.
Sam68 is a RNA-binding protein consisting of an RGG box domain, a tyrosine-rich domain (NLS), and a KH domain, which is flanked by NK (N-terminal region of KH) and CK (C-terminal region of KH) regions. The RGG boxes and KH domain spanning amino acids 45 to 54 and amino acids 157 to 256, respectively, are function domains of RNA binding (28). The tyrosine-rich domain, spanning amino acids 366 to 415, can modulate protein-protein interactions and is mapped to the C-terminal of Sam68 (29) (Fig. 7A). To identify the domain crucial for the interaction between Sam68 and HCV genome, we constructed a series of Sam68 truncations into the FLAG-tagged vector and transfected these plasmids into Huh7.5 cell, followed by HCV infection. The cells were harvested and subjected to RIP assay. The results showed that amino acids 1 to 157 of Sam68 could pull down the HCV genome (Fig. 7B and C), whereas other truncations, including amino acids 158 to 256, amino acids 257 to 443, as well as amino acids 96 to 279 of Sam68, did not associate with the HCV genome. Moreover, the overexpression of the Sam68 1 to 157 aa rather than other constructs of Sam68 enhanced viral protein level (Fig. 7D) and increased the amount of viral RNA in HCV-infected cells (Fig. 7E). These data suggested that amino acids 1 to 157 are necessary for the interaction between Sam68 and the HCV genome and are responsible for the enhancement of viral replication.
FIG 7.
The region of Sam68 spanning amino acids 1 to 157 is crucial for HCV genome binding and responsible for the enhancement of viral replication. (A) The scheme of Sam68 and its truncated forms. (B, C) The plasmid encoding Flag-tagged Sam68 1 to 157 aa, 158 to 256 aa, 257 to 443 aa, or 96 to 276 aa was transfected into Huh7.5 cells for 48 h. The cells were harvested and used for co-IP with the IgG or anti-FLAG antibody. (B) RNA was extracted from the precipitate complexes and subjected to qRT-PCR. (C) Protein from the precipitate complex was detected by immunoblotting with anti-Flag antibody. (D, E) Huh7.5 cells were infected with HCV for 24 h and then transfected with Flag-tagged Sam68 truncations for 48 h followed by immunoblotting with the indicated antibodies (D). HCV RNA was determined by real-time PCR and normalized to GAPDH (E).
DISCUSSION
The HCV genome contains a single long open reading frame flanked by 5′ and 3′ UTR, which are important for viral RNA replication and translation (4, 5). Additional RNA elements have also been identified in the negative-strand RNA replicative intermediate (30). Similar to other positive-strand RNA viruses, the noncoding regions of HCV RNA, 5′ UTR, and 3′ UTR required for viral RNA replication and translation often overlap, and the regulatory mechanisms can be separated or shared (31, 32). HCV often uses a series of self-nonstructural proteins or recruits host cellular proteins to facilitate its replication and make new viruses. Therefore, identification of viral and host proteins associated with viral genomic RNA is seen to play an important role in clarifying viral genomic RNA requiring efficient machinery for replication and translation. In this study, we found that Sam68 functions to interact with SL2 of the HCV 5′ UTR. Knockdown of Sam68 downregulated viral RNA and protein while overexpression of Sam68 increased viral RNA and protein, suggesting that Sam68 enhances HCV replication. Our data showed that Sam68 is not involved in HCV RNA translation. Upon HCV infection, Sam68 relocated from the nucleus to the cytoplasm to implement its interaction of the HCV genome, which conjuncts with the discovery that enterovirus 71 (EV71) and FMDV induce the redistribution of Sam68 (23, 33). Interestingly, we observed a weak interaction between Sam68 and PCBP2 in physiology, and the association appeared as upregulation upon HCV infection. We speculate that Sam68 binding to HCV RNA may be able to bend and change the conformation of viral RNA, which forms a new platform to recruit a series of essential proteins or long noncoding RNAs for supporting the high efficiency of HCV replication. A detailed mechanism for how Sam68 enhances HCV RNA replication requires further investigation.
The 5′ and 3′ UTR are involved in the initiation of both translation and RNA replication. The HCV RNA replication complexes consist of viral and host protein complexes (4, 34). IRES in the 5′ UTR of HCV RNA bypasses the host’s Cap-dependent translation and directly recruits the translation equipment to the viral RNA to start viral translation (20, 35, 36). Several host proteins are recruited to the HCV 5′ UTR and/or 3′ UTR and may regulate translation and/or replication. La autoantigen binds to the HCV IRES structure and significantly augments the IRES-directed translation (13). PTB interacts with the HCV 5′ UTR to enhance both HCV translation and replication (20, 37, 38). PCBP2 interacts with the HCV 5′ UTR and 3′ UTR and modulates both translation and RNA replication (12). Our recent study demonstrated that HMGB1 directly binds to the HCV 5′ UTR, thereby enhancing HCV RNA replication. Sam68 can be regarded as an important cellular factor participating in the regulation of HCV replication. The binding of Sam68 to the 5′ UTR may modulate the secondary structure of HCV RNA to facilitate its recognition by the replication complex.
Previous studies revealed that viral infections, including poliovirus, FMDV, rhinovirus, and EV71, induce the redistribution of Sam68 from nucleus to cytoplasm (21–23, 33, 39). Since Sam68 is mainly localized in the nucleus while HCV infection occurs in cytoplasm, we wonder how Sam68 is connected to the HCV genome and enhances HCV replication. We speculate that Sam68 may alter its subcellular location to the cytoplasm. Actually, our data show that Sam68 is primarily located in the nucleus in uninfected cells, while it is relocalized to the cytoplasm of HCV-infected cells. This finding is consistent with the fact that Sam68 plays an important role in HCV replication. Notably, we find that these viruses, including poliovirus, FMDV, rhinovirus, EV71, and HCV, are considered single-stranded positive-sense RNA viruses. We wonder whether Sam68 recognizes one kind of RNA by its specific structure, and the mechanism involved in the subcellular location of Sam68 during viral infection requires further investigation. Whether Sam68 interacts with other RNA viruses and regulates their replication needs to be explored. The mechanism of Sam68 translocation has been explored recently. FMDV stimulates the redistribution of Sam68 to the cytoplasm through cleaving the C terminus of Sam68, which functions as a protein nuclear localization signal, by viral 3C protease (23). Enterovirus 71 induces stress granule formation to recruit Sam68, as an SGs component, removal to the cytoplasm in a microtubule-dependent manner (27). The detailed mechanism of Sam68 translocation upon HCV infection is unknown. To explore the mechanism of HCV-induced Sam68 translocation, we detected the formation of stress granules using a stress granule marker (TIA-1). We did not observe stress granules upon HCV infection (Fig. 3C). We hypothesize whether the redistribution of Sam68 to the cytoplasm needs NS3/4A polymerase cleavage or posttranslational modifications containing acetylation, sumoylation, and phosphorylation of Sam68.
The 5′ UTR of HCV interacting either directly or indirectly with cellular proteins is most likely involved in the regulation of translation or RNA replication. The HCV 5′ UTR contains four major SL structures. SL1 is important for viral replication, while SL2 to SL4 comprise the HCV IRES to regulate translation. Recent studies show that the SL2z′, a structure residing in the 3′ end of viral negative strand RNA, is required for HCV RNA replication. An alternative stem-loop, which partially resembled SL2z′, includes parts of SL2 (8, 9). Our current study demonstrated that SL2 of the HCV 5′ UTR is the crucial region for Sam68 binding, which is consistent with our finding that the interaction between Sam68 and the HCV 5′ UTR facilitates viral replication. Since the function of SL2 of the HCV 5′ UTR in regulating HCV life cycle focuses on either regulation or translation, based on its different binding region, we ought to explore the narrow region of SL2 that connects to Sam68 in a future study. Previous reports show that the KH domain of Sam68 is required for the interaction of RNA virus (33). Our data support that the RGG box domain is necessary for the interaction between Sam68 and the HCV genome. It may result from the different secondary or tertiary structure of the virus genome itself.
In the present study, we found a new function of Sam68 in viral replication and elucidated the mechanism of Sam68 interaction with the HCV genome. Our data show that overexpression of Sam68 increases the level of viral RNA and protein, and knockdown of Sam68 decreases viral RNA and protein level, supporting that Sam68 enhances viral replication. HCV infection leads to the relocalization of Sam68 from the nucleus to the cytoplasm, where virus replication occurs inside the cell. Sam68 specifically binds to the 5′ UTR of the HCV genome. Further study demonstrated that SL2 of the HCV 5′ UTR is the major region for the interaction between HCV 5′ UTR and Sam68. The RGG box domain of Sam68 is necessary for their interaction. The region of Sam68 spanning amino acids 1 to 157 is crucial for HCV genome binding and responsible for the enhancement of HCV replication. In summary, our study revealed that Sam68 acts as a pivotal host factor for HCV highly efficient replication. Sam68 appears to play an important role in HCV-related diseases. Further investigation is greatly encouraged to elucidate the specific actions of Sam68 in HCV pathogenesis.
MATERIALS AND METHODS
Cell culture and reagents.
FCA1, a HCV subgenomic replicon cell line, was a gift from Christoph Seeger (Fox Chase Cancer Center, Philadelphia, PA). FL-neo (HCV 1b full-length replicon cell line) and Huh7.5 cell lines were kindly provided by Charles Rice (Rockefeller University, New York, NY). HLCZ01 cells were established in our laboratory. HLCZ01 is a novel human hepatoma cell line supporting the entire life cycle of both hepatitis B virus (HBV) and HCV (40). HLCZ01 cells were cultured in collagen-coated tissue culture plates and with Dulbecco’s modified Eagle medium (DMEM)–F-12 medium supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Gibco), 40 ng/ml of dexamethasone (Sigma), insulin-transferrin-selenium (ITS) (Lonza), penicillin, and streptomycin. FL-neo, FCA1, and Huh7.5 cells were propagated in DMEM supplemented with 10% FBS, l-glutamine, nonessential amino acids, penicillin, and streptomycin.
Plasmids and antibodies.
The pRL-HL plasmid was a gift from Kui Li (University of Tennessee Health Science Center, Tennessee). pJFH1 and pJFH1/GND plasmids were generously provided by Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan). Full-length and different truncated forms of Sam68 were synthesized from RNA from HLCZ01 cells by standard RT-PCR and subsequently cloned into the p3×FLAG-CMV vector. The shRNAs targeting Sam68 were constructed in the pSilencer-neo plasmid (Ambion). The target sequence of the Sam68 shRNA was 5′-GGCTACGAAGGCTATTACA-3′. Sam68 was subcloned into pGEX-4T-1 for glutathione S-transferase (GST)-fusion protein expression in Escherichia coli. 5′-UTR and 3′-UTR fragments were amplified from the pJFH1 plasmid and cloned into the p3×FLAG-CMV vector. Multiple domains of 5′ UTR were amplified and cloned into the p3×FLAG-CMV vector. The primers for the above-described genes or the amplification of domains are shown in Tables 1 and 2. Rabbit polyclonal antibody against Sam68 was obtained from Abcam. Monoclonal antibody against PCBP2 was purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-NS5A and anticore antibodies were gifts from Chen Liu (Rutgers University, Newark, NJ). Mouse monoclonal anti-FLAG and anti-β-actin antibodies were obtained from Sigma-Aldrich. Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody and anti-LaminB1 antibody were purchased from Merck Millipore and Cell Signaling Technology, respectively.
TABLE 1.
Primers for amplification of genes
| Construct | Primer sequence (direction) |
|---|---|
| p3×FLAG-CMV-Sam68 | CGGAATTCATGCAGCGCCGGGACGACCC (forward) |
| GGGGTACCGAATAACGTCCATATGGGTG (reverse) | |
| pGEX-4T-Sam68 | CGGAATTCATGCAGCGCCGGGACGACCC (forward) |
| ACGCGTCGACATAACGTCCATATGGGTGCT (reverse) | |
| p3×FLAG-CMV-PCBP2 | GGGGTACCATGGACACCGGTGTGATTGA (forward) |
| CGGGATCCGCTGCTCCCCATGCCACCCG (reverse) |
TABLE 2.
Primers for amplification of Sam68 domains
| Domain (position [aa]) | Primer sequence (direction) |
|---|---|
| p3×FLAG-CMV-Sam68 (1 to 157) | CGGAATTCCAGCGCCGGGACGACCC (forward) |
| CCCAAGCTTTTATTTCAGTTTCATGTTCTTATGA (reverse) | |
| p3×FLAG-CMV-Sam68 (158 to 256) | CGGAATTCGAGCGAGTGCTGATACCTGT (forward) |
| CCCAAGCTTTTATACTAGAAATTTCTTGACTT (reverse) | |
| p3×FLAG-CMV-Sam68 (257 to 443) | CGGGATCCCCGGATATGATGGATGATATCT (forward) |
| CGGAATTCTTAATAACGTCCATATGGGTGCT (reverse) | |
| p3×FLAG-CMV-Sam68 (96 to 276) | CGGAATTCAAGATGGAGCCAGAGAACAA (forward) |
| CCCAAGCTTTTATTCAGGTACTCCATTCAAGT (reverse) |
Production of HCV stocks.
The linearized DNA from the pJFH1 and pJFH1-del-SL2 plasmids was purified and used as the template for in vitro transcription using a MEGAscript kit (Ambion, Austin, TX). In vitro transcribed genomic JFH1 or JFH1/GND RNA was delivered into Huh7.5 cells by electroporation. The transfected cells were transferred to complete DMEM and cultured for the indicated periods. Cells were passaged every 3 to 5 days, and the corresponding supernatants were collected and filtered with a 0.45-μm filter device. The viral titers are presented as focus-forming units per milliliter, determined by the average number of NS5A-positive foci detected in Huh7.5 cells.
Real-time PCR assay.
RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol and reverse-transcribed by PrimeScript RT reagent kit gDNAEraser (Perfect Real Time) (TaKaRa). SYBR RT-PCR kit (Roche) was used for real-time reverse transcription-quantitative PCR (qRT-PCR) analysis. The specific primers for amplification of HCV RNA or fragments are listed in Table 3.
TABLE 3.
Primers used for amplification of HCV genome fragments
| Fragment (size [bp]) | Primer sequence (5′–3′) |
|---|---|
| 3′ UTR (236) | CGGAATTCAGCGGCACACACTAGGTACA (forward, 9443–9462) |
| GCTCTAGAACATGATCTGCAGAGAGACC (reverse, 9678–9659) | |
| 5′ UTR (340) | CGGAATTCACCTGCCCCTAATAGGGGCG (forward, 1–20) |
| GCTCTAGAGGTGCACGGTCTACGAGACC (reverse, 340–321) | |
| SL1-3 (296) | CGGAATTCACCTGCCCCTAATAGGGGCG (forward, 1–20) |
| GCTCTAGAATCAGGCAGTACCACAAGGC (reverse, 296–277) | |
| SL2-4 (301) | CGGAATTCCCCCTGTGAGGAACTACTGT (forward, 40–59) |
| GCTCTAGAGGTGCACGGTCTACGAGACC (reverse, 340–321) | |
| SL3-4 (241) | CGGAATTCGTCGTACAGCCTCCAGGCCC (forward, 100–119) |
| GCTCTAGAGGTGCACGGTCTACGAGACC (reverse, 340–321) |
Luciferase reporter assay.
Luciferase assays were performed by using a luciferase assay kit (Promega) according to the instructions. The luciferase activity was normalized to protein concentration as determined by Bradford assays.
Immunofluorescence staining.
Cells were seeded on glass coverslips and fixed with 4% paraformaldehyde fix solution for 15 min at room temperature. The cells were washed with phosphate-buffered saline (PBS), blocked with 1:50 goat serum for 30 min at room temperature, and then incubated for 1 h with mouse monoclonal anti-NS5A antibody (diluted in PBS to 1:100). The cells were stained with fluorescence-labeled secondary antibodies (Invitrogen) (diluted in PBS to 1:200) for 1 h at room temperature. The coverslips were extensively washed, and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories Inc., Burlingame, CA). Fluorescent images were obtained under fluorescence microscope (Olympus, Japan).
Coimmunoprecipitation.
Cells were washed thrice with 1 ml of ice-cold PBS and lysed in lysis buffer (25 mM Tris-HCl [pH 7.4] containing 150 mM NaCl, 1% NP-40, 1 mM EDTA, and 5% glycerol) supplemented with a protease inhibitors cocktail at 4°C. Cell lysates were incubated at 4°C for 30 min and centrifuged at 12,000 × g for 15 min. The cell lysate containing 300 μg total protein was incubated with protein G beads precoated with the indicated antibodies at 4°C overnight. The protein bound to the beads was boiled in 2× Laemmli sample buffer and then subjected to SDS-PAGE.
RNA immunoprecipitation.
Cells in a 60-mm culture dish were washed thrice with 1 ml of ice-cold PBS and lysed in 100 μl of lysis buffer (25 mM Tris-HCl [pH 7.4] containing 150 mM NaCl, 1% NP-40, 1 mM EDTA, and 5% glycerol) supplemented with a protease inhibitors cocktail at 4°C. Cell lysates were incubated at 4°C for 30 min, and centrifuged at 12,000 × g for 15 min. The cell lysate containing 300 μg total protein was incubated with protein G beads precoated with the indicated antibodies at 4°C overnight. The cells lysates were diluted to a concentration of 2 μg/μl, and the protein-RNA complexes binding to beads were eluted in lysis buffer at 70°C for 45 min. RNA was extracted from the precipitate complex using TRIzol reagent and reverse-transcribed by PrimeScript RT reagent kit gDNAEraser (Perfect Real Time) (TaKaRa). SYBR RT-PCR kit (Roche) was used for real-time qRT-PCR analysis. The protein from the precipitate complex was boiled in 2× Laemmli sample buffer and then subjected to SDS-PAGE.
Expression and purification of Sam68 protein.
The pGEX-4T-1-Sam68 plasmid was transformed into Escherichia coli BL21(DE3) cells, and the GST-tagged Sam68 fusion protein was expressed. Cells were grown in LB medium and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Sam68 protein was purified using glutathione Sepharose beads. The protein was eluted with PBS and identified using an anti-Sam68 antibody via Western blotting.
Nuclear and cytoplasmic extraction.
Nuclear and cytoplasmic extracts were obtained with the NE-PER nuclear and cytoplasmic extraction reagents kit (Thermo Fisher) according to the manufacturer’s instructions.
Gel shift assay.
To detect the direct interaction between Sam68 and HCV RNA in vitro, 30 pmol of purified Sam68 protein and 10 pmol of in vitro transcribed RNA were incubated in binding buffer (20 mM Tris-HCl [pH 8.0], 1.5 mM MgCl2, 1.5 mM dithiothreitol) for 15 min at 37°C. The samples were then mixed with native PAGE sample buffer (25 mM Tris-HCl [pH 6.8] and 0.02% bromophenol blue in 60% glycerol) and subjected to electrophoresis on a 1% agarose gel. The gel was stained with a SYBR green II RNA gel stain kit (Invitrogen).
Immunoblotting.
The procedure for immunoblotting was reported previously (41).
ACKNOWLEDGMENTS
We thank Charles M. Rice for the FL-neo and Huh7.5 cell lines, Takaji Wakita for pJFH1, Kui Li for pRL-HL, and Chen Liu for sharing research materials.
This work was supported by the National Natural Science Foundation of China (81730064, 81571985) and National Science and Technology Major Project (2017ZX10202201).
REFERENCES
- 1.Seeff LB. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35–S46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH. 2002. Course and outcome of hepatitis C. Hepatology 36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 3.Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. 2004. Structural biology of hepatitis C virus. Hepatology 39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 4.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 5.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci U S A 101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friebe P, Bartenschlager R. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol 76:5326–5338. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci U S A 104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schult P, Roth H, Adams RL, Mas C, Imbert L, Orlik C, Ruggieri A, Pyle AM, Lohmann V. 2018. microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat Commun 9:2613. doi: 10.1038/s41467-018-05053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friebe P, Bartenschlager R. 2009. Role of RNA structures in genome terminal sequences of the hepatitis C virus for replication and assembly. J Virol 83:11989–11995. doi: 10.1128/JVI.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reigadas S, Ventura M, Sarih-Cottin L, Castroviejo M, Litvak S, Astier-Gin T. 2001. HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus-strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. Eur J Biochem 268:5857–5867. doi: 10.1046/j.0014-2956.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 11.Gao L, Aizaki H, He JW, Lai MM. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol 78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Jeng KS, Lai MM. 2011. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5′ untranslated region and directs HCV RNA replication through circularizing the viral genome. J Virol 85:7954–7964. doi: 10.1128/JVI.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali N, Siddiqui A. 1997. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci U S A 94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao H, Zhao K, Yao Y, Guo J, Gao X, Yang Q, Guo M, Zhu W, Wang Y, Wu C, Chen J, Zhou Y, Hu X, Lu M, Chen X, Pei R. 2018. RNA binding protein 24 regulates the translation and replication of hepatitis C virus. Protein Cell 9:930–944. doi: 10.1007/s13238-018-0507-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu R, Yang D, Lei S, Wang X, Meng X, Xue B, Zhu H. 2015. HMGB1 promotes hepatitis C virus replication by interaction with stem-loop 4 in the viral 5′ untranslated region. J Virol 90:2332–2344. doi: 10.1128/JVI.02795-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang B, Lim JH, Hahm B, Jang SK, Lee SW. 2009. hnRNP L is required for the translation mediated by HCV IRES. Biochem Biophys Res Commun 378:584–588. doi: 10.1016/j.bbrc.2008.11.091. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Lai MM. 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 18.Ali N, Siddiqui A. 1995. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol 69:6367–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol 9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 20.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol 72:4775–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McBride AE, Schlegel A, Kirkegaard K. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc Natl Acad Sci U S A 93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gustin KE, Sarnow P. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J Virol 76:8787–8796. doi: 10.1128/JVI.76.17.8787-8796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence P, Schafer EA, Rieder E. 2012. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology 425:40–52. doi: 10.1016/j.virol.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 24.He JJ, Henao-Mejia J, Liu Y. 2009. Sam68 functions in nuclear export and translation of HIV-1 RNA. RNA Biol 6:384–386. doi: 10.4161/rna.6.4.8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim CS, Seol SK, Song OK, Park JH, Jang SK. 2007. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J Virol 81:3852–3865. doi: 10.1128/JVI.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinlich S, Huttelmaier S, Schierhorn A, Behrens SE, Ostareck-Lederer A, Ostareck DH. 2009. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3′UTR. RNA 15:1528–1542. doi: 10.1261/rna.1578409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Chen N, Li P, Pan Z, Ding Y, Zou D, Li L, Xiao L, Shen B, Liu S, Cao H, Cui Y. 2016. The nuclear protein Sam68 is recruited to the cytoplasmic stress granules during enterovirus 71 infection. Microb Pathog 96:58–66. doi: 10.1016/j.micpath.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Yang JP, Reddy TR, Truong KT, Suhasini M, Wong-Staal F. 2002. Functional interaction of Sam68 and heterogeneous nuclear ribonucleoprotein K. Oncogene 21:7187–7194. doi: 10.1038/sj.onc.1205759. [DOI] [PubMed] [Google Scholar]
- 29.Ishidate T, Yoshihara S, Kawasaki Y, Roy BC, Toyoshima K, Akiyama T. 1997. Identification of a novel nuclear localization signal in Sam68. FEBS Lett 409:237–241. doi: 10.1016/S0014-5793(97)00455-9. [DOI] [PubMed] [Google Scholar]
- 30.Lindenbach BD, Rice CM. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 31.Song Y, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M. 2006. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol 80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradrick SS, Walters RW, Gromeier M. 2006. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res 34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Song L, Cong H, Tien P. 2015. Nuclear protein Sam68 interacts with the enterovirus 71 internal ribosome entry site and positively regulates viral protein translation. J Virol 89:10031–10043. doi: 10.1128/JVI.01677-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buratti E, Tisminetzky S, Zotti M, Baralle FE. 1998. Functional analysis of the interaction between HCV 5′UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res 26:3179–3187. doi: 10.1093/nar/26.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito T, Lai MM. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol 71:8698–8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domitrovich AM, Diebel KW, Ali N, Sarker S, Siddiqui A. 2005. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology 335:72–86. doi: 10.1016/j.virol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Rai DK, Lawrence P, Kloc A, Schafer E, Rieder E. 2015. Analysis of the interaction between host factor Sam68 and viral elements during foot-and-mouth disease virus infections. Virol J 12:224. doi: 10.1186/s12985-015-0452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang D, Zuo C, Wang X, Meng X, Xue B, Liu N, Yu R, Qin Y, Gao Y, Wang Q, Hu J, Wang L, Zhou Z, Liu B, Tan D, Guan Y, Zhu H. 2014. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proc Natl Acad Sci U S A 111:E1264–E1273. doi: 10.1073/pnas.1320071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin Y, Xue B, Liu C, Wang X, Tian R, Xie Q, Guo M, Li G, Yang D, Zhu H. 2017. NLRX1 mediates MAVS degradation to attenuate hepatitis C virus-induced innate immune response through PCBP2. J Virol 91:e01264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]