Human herpesvirus 6 is highly prevalent and maintains chronic infection in immunocompetent individuals, with the potential to replicate widely in settings of immunosuppression, leading to clinical disease. Antiviral compounds may be ineffective and/or pose dose-limiting toxicity, and therefore, immune-based therapies have garnered increased interest in recent years. Attempts at addressing this unmet medical need begin with understanding the cellular response to HHV-6 at the individual and population levels. The present study provides a comprehensive assessment of HHV-6-specific T-cell responses that may inform the development of cell-based therapies directed at this virus.
KEYWORDS: CD4 T cell, antigen, epitope, human herpesvirus 6
ABSTRACT
Human herpesvirus 6 (HHV-6) and cytomegalovirus (CMV) are population-prevalent betaherpesviruses with intermittent lytic replication that can be pathogenic in immunocompromised hosts. Elucidation of the adaptive immune response is valuable for understanding pathogenesis and designing novel treatments. Knowledge of T-cell antigens has reached the genome-wide level for CMV and other human herpesviruses, but study of HHV-6 is at an earlier stage. Using rare-cell enrichment combined with an HLA-agnostic, proteome-wide approach, we queried HHV-6B-specific CD4 T cells from 18 healthy donors with each known HHV-6B protein. We detected a low abundance of HHV-6-specific CD4 T cells in blood; however, the within-person CD4 T-cell response is quite broad: the median number of open reading frame (ORF) products recognized was nine per person. Overall, the data expand the number of documented HHV-6B CD4 T-cell antigens from approximately 11 to 60. Epitopes in the proteins encoded by U14, U90, and U95 were mapped with synthetic peptides, and HLA restriction was defined for some responses. Intriguingly, CD4 T-cell antigens newly described in this report are among the most population prevalent, including U73, U72, U95, and U30. Our results indicate that selection of HHV-6B ORFs for immunotherapy should consider this expanded panel of HHV-6B antigens.
IMPORTANCE Human herpesvirus 6 is highly prevalent and maintains chronic infection in immunocompetent individuals, with the potential to replicate widely in settings of immunosuppression, leading to clinical disease. Antiviral compounds may be ineffective and/or pose dose-limiting toxicity, and therefore, immune-based therapies have garnered increased interest in recent years. Attempts at addressing this unmet medical need begin with understanding the cellular response to HHV-6 at the individual and population levels. The present study provides a comprehensive assessment of HHV-6-specific T-cell responses that may inform the development of cell-based therapies directed at this virus.
INTRODUCTION
Human herpesvirus 6 (HHV-6) is a ubiquitous virus that causes roseola upon primary infection and persists throughout life in immunocompetent carriers (1, 2). It exists as two closely related species, HHV-6A and HHV-6B (3), that have 90% genomic identity (4) but differ by tropism and epidemiology (5, 6). About 1% of humans harbor inherited chromosomally integrated HHV-6 (iciHHV-6), which is inherited in a Mendelian fashion (7). Lytic infection in immunocompetent adults in the chronic phase of infection appears to be relatively limited to the oropharynx, with high levels of HHV-6 DNA detectable in the saliva (8). Viral replication can be more widespread in immunocompromised settings such advanced HIV infection (9). Reactivation, defined as detection of HHV-6B DNA in blood, occurs in nearly half of post-hematopoietic stem cell transplant (HCT) patients and has been definitively linked to encephalitis. Adverse outcomes such as fever and graft-versus-host disease (1, 10, 11) may also be related to HHV-6 replication. Investigators have suggested associations with Hashimoto’s thyroiditis (12), multiple sclerosis (13–19), and a variety of other medical conditions (1).
Although no antiviral compounds are approved specifically for HHV-6 treatment, some anticytomegalovirus (anti-CMV) compounds, including ganciclovir, foscarnet, and cidofovir, inhibit HHV-6 (5, 20–22). These can have dose-limiting toxicities or lack efficacy (23–25). T-cell immunity is likely important for the control of HHV-6 based on the increase in HHV-6 replication detected in persons with advanced immunodeficiency (9, 26, 27). Based on this hypothesis, cell-based therapies are considered a promising alternative. In particular, detailed knowledge of which HHV-6 antigens rank highest in immunodominance and immunoprevalence will allow selectively expanded virus-specific T-cell (VST) or transgenic T-cell receptor (tgTCR) T-cell products to be optimized for effectiveness and HLA compatibility. While most emphasis has been placed on cytotoxic CD8 T cells for adoptive T-cell therapy, VST products are usually a mixture of CD4 and CD8 T cells (28), and adoptive therapy with antigen-specific CD4 T cells can be active against cancer (29). Therefore, continuing definition of HHV-6 CD4 T-cell targets may contribute to antiviral therapy.
The discovery and ranking of HHV-6 T-cell antigens are complicated by the low frequency of HHV-6-specific T cells in blood (30) and by the complex viral proteome. Because HHV-6 and CMV are genetically related betaherpesviruses, some studies have focused on the HHV-6 homologs of known CMV antigenic proteins. These studies have identified HHV-6 U11, U14, U54, and U90, the homologs of CMV UL32, UL25, UL82/83, and IE1, respectively, as targets for CD4 or CD8 T cells (31–34). Other studies have taken a cross-sectional approach by testing libraries of peptides that were predicted to bind to a particular HLA allele: DRB1*0101 for CD4 T cells (30, 35) or HLA-B*0801 for CD8 T cells (36). These studies expanded the number of known T-cell antigens considerably but were limited to single HLA allelic variants.
The present study uses a high-throughput approach to antigen discovery that was originally developed for large-genome pathogens, including vaccinia virus, herpes simplex virus 1 (HSV-1), HSV-2, varicella-zoster virus (VZV), and Mycobacterium tuberculosis (37–41). Enriched whole-virus-reactive CD4 T cells are initially tested against a library of all full-length viral proteins, with downstream deconvolution to the epitope level for selected antigens. As this workflow relies on sequential antigen processing of whole virus and long polypeptides by antigen-presenting cells (APCs), epitopes identified by this workflow are likely to be physiologically relevant. This genome-wide, HLA-agnostic approach allows antigen immunoprevalence ranking in the population to provide a comprehensive view of HHV-6B-specific CD4 T-cell responses.
RESULTS
Peripheral blood of healthy donors contains HHV-6B-specific CD4 T cells that can be enriched in vitro.
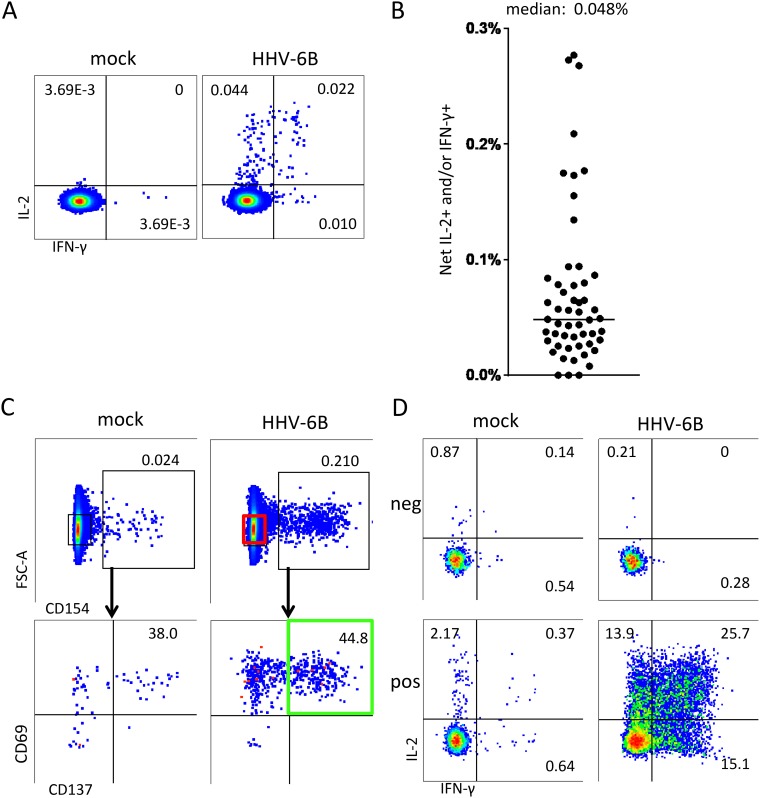
The initial cohort of 53 healthy donors was 34% male, and the median age at the time of blood draw was 40 years (range, 21 to 68 years). To ascertain the frequency of HHV-6B-specific CD4 T cells in peripheral blood, peripheral blood mononuclear cells (PBMCs) were assayed by an interleukin-2 (IL-2)/interferon gamma (IFN-γ) intracellular cytokine cytometry (ICC) assay (Fig. 1A). Donors had virus-specific cell populations that were of low abundance but clearly discernible in most subjects. Responses were typically less than 0.1% of total CD4 T cells, with an overall median of 0.048% (Fig. 1B).
FIG 1.
Isolation and enrichment of HHV-6B-specific CD4 T cells. (A) PBMCs from a representative donor were stimulated for 18 h with mock or HHV-6B lysate and tested by intracellular cytokine staining (ICS) for IL-2 and IFN-γ expression. Gated CD3+ and CD4+ cells are shown. (B) Results from similarly tested ex vivo PBMCs of 53 donors. For each, HHV-6B-treated cells expressing either cytokine, or both, were totaled, and mock values were subtracted for the net IL-2 and/or IFN-γ HHV-6B-specific T-cell frequency. The horizontal bar is the median (0.048%). (C) Sorting scheme for HHV-6B-specific T cells. Live cells from the same donor as in panel A were gated for CD3 and CD4 expression; from these, cells expressing CD154, CD69, and CD137 (green) were sorted and expanded in culture for downstream assays. Cells negative for CD154 (red gate) were sorted as a negative control. FSC, forward scatter. (D) Expanded polyclonal T cells from the same donor as in panel A originating from CD154-negative cells (“neg”) or CD154/CD137/CD69 triple-positive cells (“pos”) were evaluated to document enrichment for HHV-6B specificity among the AIM-positive cells. Numbers are percentages of gated cells expressing the indicated pattern of cytokines or activation markers.
Given the low abundance of HHV-6-reactive CD4 T cells in PBMCs, we enriched these cells from 42 donors prior to interrogation of the HHV-6B protein library. These donors were chosen based on PBMC availability and a detectable response to HHV-6B ex vivo. To sort virus-specific T cells with optimal specificity, we added CD69 and CD154 as activation-induced markers (AIMs) to CD137, the principal AIM in our previous work (37, 39, 40). Live CD4 T cells expressing triple AIMs were bulk sorted and expanded (Fig. 1C). As a negative control, CD3+ CD4+ T cells lacking CD154 expression were tested in parallel. The polyclonal AIM-positive (AIM+) cell lines showed strong HHV-6 reactivity and were generally 5% to 50% specific for HHV-6B antigen (representative data are shown in Fig. 1D), while AIM-negative (AIM−) cells showed no specific reactivity.
HLA-agnostic genome-wide screening reveals novel HHV-6B antigens for CD4 T cells.
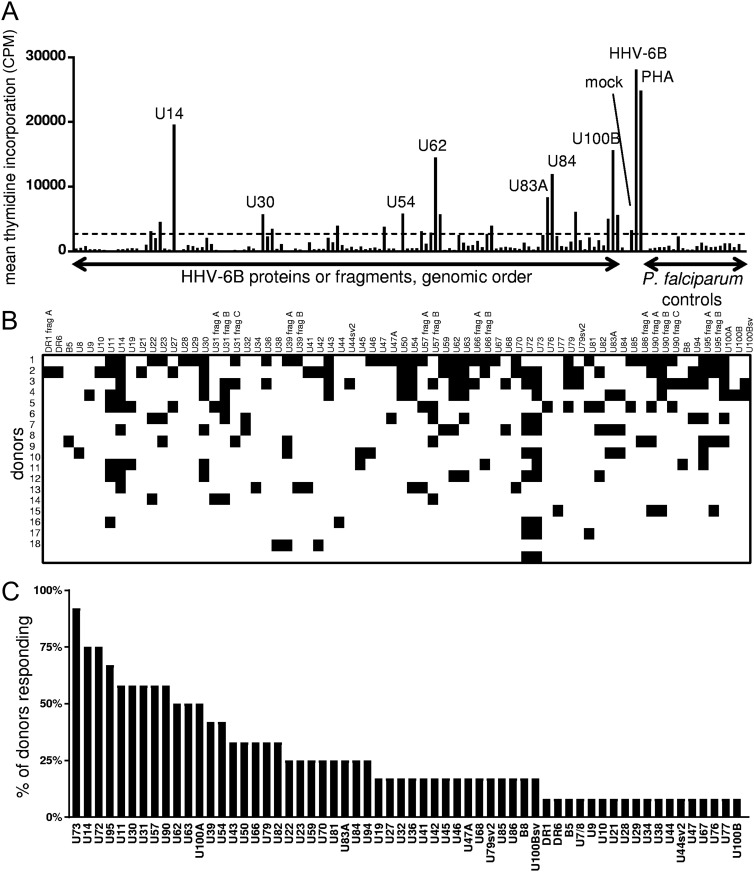
A set of 101 in vitro-expressed proteins covering curated and candidate novel HHV-6B open reading frames (ORFs) were tested with [3H]thymidine uptake assays. Five mRNA splice variants with amino acid sequences differing from the ORF set under NCBI accession number NC_000898.1 were also tested. Polyclonal HHV-6B-reactive CD4 T-cell lines showed discrete positive responses to specific HHV-6B proteins (Fig. 2A). We focused on 18 donors with sufficient PBMC availability, a proliferative response to whole HHV-6B at least twice that of mock, and a positive response to at least one HHV-6B protein. This group was 33% male, and the median age at the time of blood draw was 38 years (range, 25 to 54 years).
FIG 2.
Genome-wide HHV-6B CD4 T-cell antigen screens. (A) Representative data from the same donor as in Fig. 1A, C, and D. Shown are responses of polyclonal HHV-6B-reactive CD4 T cells to individual HHV-6B polypeptides, or controls, including mock or HHV-6B-infected lysate, PHA, or irrelevant Plasmodium falciparum proteins. CPM, counts per minute. The dotted horizontal line is the statistical cutoff for positive responses. (B) Summary of CD4 T-cell responses to the HHV-6B proteome. Positive responses are denoted by black squares. frag, fragment. (C) Population prevalence hierarchy of CD4 T-cell responses to HHV-6B proteins. Responses to >1 fragment for proteins studied in segments are collapsed to one positive response. sv, splice variant, as detailed in Table S1 in the supplemental material.
The median number of HHV-6B genes or gene fragments recognized per donor was 9, and 57% of all proteins were recognized by at least one donor (Fig. 2B). Of these, 5 are annotated as having immediate early, 14 are annotated as having early, and 18 are annotated as having late expression kinetics. Eleven of these HHV-6B ORFs have not, to our knowledge, previously been described as HHV-6B CD4 T-cell antigens (30, 31, 35). Unlike the originally annotated U83 (NCBI accession number NC_000898.1), the reannotated ORF “U83A” was transcribed in the reverse direction and contained one intron, resulting in an entirely new amino acid sequence; interestingly, the full-length U83A protein elicited responses from three donors, supporting its status as a novel HHV-6B ORF.
The U73 protein was the most frequently recognized, stimulating CD4 T cells from over half of the final cohort. The U14- and U72-encoded proteins were recognized by half of the final cohort. Among the dozen proteins recognized by at least one-third of donors, half, to our knowledge, have not previously been described as CD4 T-cell targets: U73, U72, U95, U30, U62, and U63 (Fig. 2C).
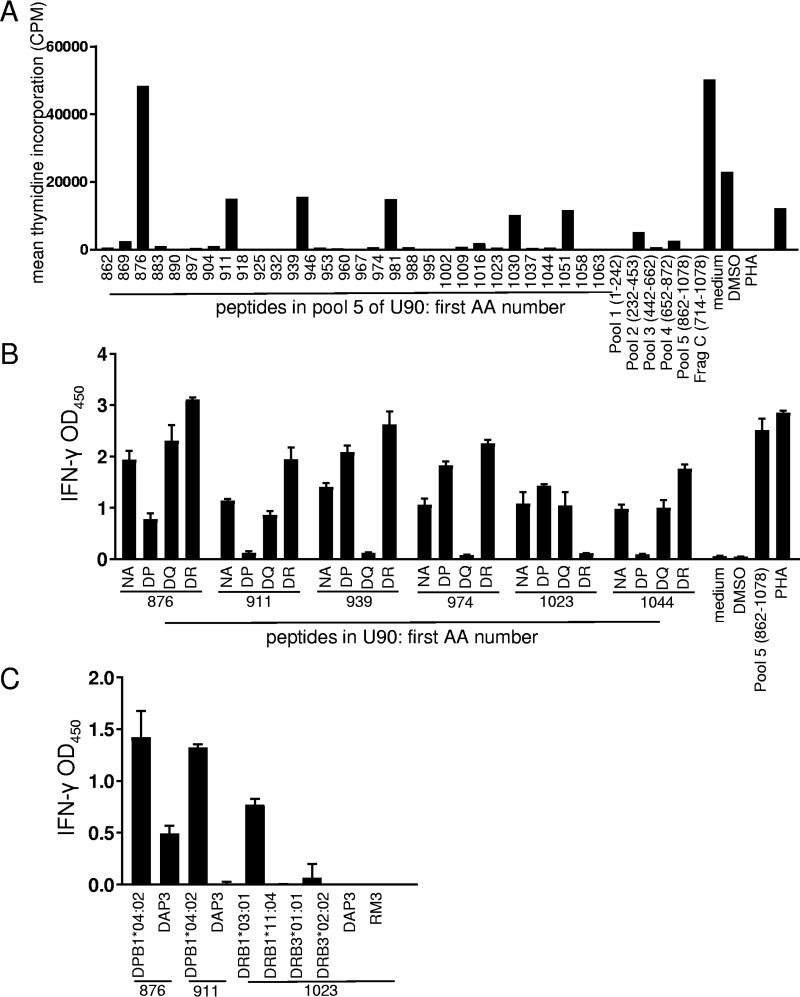
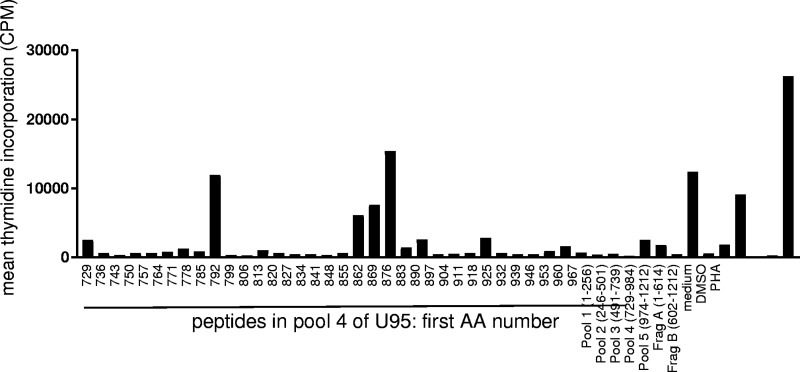
Validation of selected CD4 T-cell antigens at the peptide epitope level.
We confirmed selected positive responses to HHV-6 polypeptides with synthetic peptides. Bulk polyclonal HHV-6-reactive CD4 T-cell lines were again used as responder cells, and autologous PBMCs were used as APCs. In initial assays (data not shown), we tested pools of 30 to 35 overlapping peptides within the reactive U14, U90, and U95 ORFs for preliminary epitope mapping. Further testing with individual peptides from U90 (representative data are shown in Fig. 3A) and U95 (Fig. 4) peptide pools eliciting robust T-cell responses showed that the response to these HHV-6B proteins can include several epitopes per person. Data from several donors are summarized in Table 1. Protein BLAST revealed that all of these epitopes had homologs in HHV-6A, and nine had homologs in HHV-7 (see Table S2 in the supplemental material). No sequence homologs were detected in human CMV (HCMV) or any other human herpesviruses. Interestingly, 21 of the epitopes had perfect identity to a recently reported iciHHV-6B genomic sequence (GenBank accession number KY315520.1).
FIG 3.
Detection of CD4 T-cell epitopes in HHV-6B U90. (A) Polyclonal HHV-6B-reactive CD4 T cells from a representative donor were tested against U90 peptides. Controls at the right include responses to U90 peptide pools and the U90 fragment C IVTT polypeptide, which corresponds to part of pool 4, and pool 5. Numbers on the x axis indicate starting amino acid (AA) positions of 18-mer peptides or amino acid positions covered by peptide pools. Values are means of data from duplicates. DMSO, dimethyl sulfoxide. (B) Reactive U90 peptides were retested with HLA locus-specific blocking monoclonal antibody specific for HLA-DP, HLA-DQ, or HLA-DR or no antibody (NA). After overnight stimulation, supernatants were tested for IFN-γ by an ELISA. Values are means and standard deviations of data from triplicates. OD450, optical density at 450 nm. (C) Selected peptides from panel B were retested using single-HLA class II-expressing cell lines as APCs with ELISA readouts in triplicate. U90 peptides are indicated at the bottom. HLA class II-negative parental cell lines DAP3 and RM3 were used as controls.
FIG 4.
Detection of CD4 T-cell epitopes in HHV-6B U95. Polyclonal HHV-6B-reactive CD4 T cells from the same donor as in Fig. 3A were tested against U95 peptides as described in the legend of Fig. 3A. Controls at the right include responses to U95 peptide pools and U95 fragment IVTT polypeptides A and B.
TABLE 1.
HHV-6B peptides with CD4 T-cell responses detected in this study
| ORF | Positions | Amino acid sequencea | HLA restriction | Previously reportedb |
|---|---|---|---|---|
| U14 | 372–389 | EYDEDKPPIQVDPGRVDN | Yes | |
| U14 | 379–396 | PIQVDPGRVDNVLTDSDF | Yes | |
| U90 | 281–298 | KQELLESRNEIIENHVKN | Yes | |
| U90 | 309–326 | QMNQIFMDNsDKTFLKIH | No | |
| U90 | 316–333 | DNsDKTFLKIHINsKNLI | Yes | |
| U90 | 330–347 | KNLITAAKNIGIAVLQSI | Yes | |
| U90 | 344–361 | LQSIVLsSNEFSWQYLKP | Yes | |
| U90 | 365–382 | QFKITMMNMITHAsEsIE | Yes | |
| U90 | 876–893 | KTHKVDNsVIHSSAKMNV | DPB1*04:02 | Yes |
| U90 | 904–921 | HsFINHFVPIKTDDEEYE | Yes | |
| U90 | 911–928 | VPIKTDDEEYEKENVSYT | DPB1*04:02 | Yes |
| U90 | 939–956 | LEDITPTKKLITEMVMEN | Yes | |
| U90 | 946–963 | KKLITEMVMENFMDLTDI | DQ | Yes |
| U90 | 967–984 | GIAKHsQDLSSKYTVITH | Yes | |
| U90 | 974–991 | DLSSKYTVITHTAsEKNL | DQ | No |
| U90 | 981–998 | VITHTAsEKNLNVANSQN | No | |
| U90 | 1002–1019 | AETQIFDPQGTGNNSPIL | Yes | |
| U90 | 1009–1026 | PQGTGNNSPILNIINDTT | Yes | |
| U90 | 1016–1033 | SPILNIINDTTsQNDENR | Yes | |
| U90 | 1023–1040 | NDTTsQNDENRsTEGTSN | DRB1*03:01 | No |
| U90 | 1037–1054 | GTSNDNEKsTIRSDsNSD | Yes | |
| U90 | 1044–1061 | KsTIRSDsNSDKMEVFKL | DP | Yes |
| U95 | 1–18 | MSSNLEDLLWQQILSMDP | No | |
| U95 | 8–25 | LLWQQILSMDPAELLSDN | No | |
| U95 | 15–32 | SMDPAELLSDNAISSTSD | No | |
| U95 | 85–102 | TGLSLESINNQINVQPTQ | No | |
| U95 | 92–109 | INNQINVQPTQMTFQPIS | No | |
| U95 | 99–116 | QPTQMTFQPISPPMQGQN | No | |
| U95 | 729–746 | KRMHSEIGISEDGRVREE | No | |
| U95 | 785–802 | QDASGGSSSGTKKGEKLQ | No | |
| U95 | 848–865 | NPDYRQAKRLLADIPYRR | No | |
| U95 | 855–872 | KRLLADIPYRRWIPDTFN | No | |
| U95 | 862–879 | PYRRWIPDTFNMEEHEGP | No | |
| U95 | 876–893 | HEGPFLPIVTRPPTVFMG | No | |
| U95 | 904–921 | SVTSIGPLSKLTYFKELL | No | |
| U95 | 939–956 | AKHRVYIMSEEKLGYNHI | No |
To validate detection of canonical responses, HLA restriction was interrogated. Responses of HHV-6B-specific T-cell lines to six individual peptides of U90 were attenuated by antibody blockage of HLA molecules at one of the three DR, DP, or DQ loci, indicating locus-level restriction (Fig. 3B). Finally, cells expressing a single HLA allele were able to present selected peptides and elicit an IFN-γ response from HHV-6B-specific T cells, revealing the exact allelic restriction of these peptides (Fig. 3C). We documented examples of HLA restriction at the DR, DP, and DQ genetic loci and of restriction by the HLA DPB1*04:02 and HLA DRB1*03:01 allelic variants. Together, these results corroborated that T-cell responses detected with full-length proteins show the typical properties of HLA class II-restricted CD4 T cells.
DISCUSSION
Severe HHV-6 infections can occur in settings of compromised cellular immunity, yet even in immunocompetent individuals, HHV-6-specific T cells in blood are scarce. Compared to CMV, the frequency of CD4 T cells in blood is about 100 times higher for CMV than for HHV-6B; we found a median abundance for HHV-6B of approximately 0.05%, while others have found CMV-specific frequencies of up to 10% or higher (42). The acquired immune response coexists with anatomically limited, lytic viral replication in the oropharynx, as reflected by the detection of large amounts of HHV-6 DNA in most saliva samples (43). It is currently unknown how the low overall magnitude of the CD4 T-cell response to HHV-6, as detected previously (30) and confirmed in this report, is conditioned by constant antigen exposure. HHV-6B is CD4 T-cell tropic and may shape the immune response by killing HHV-6B-specific T cells during primary infection, similar to HIV and measles virus (44). In the present work, as in previous reports (30, 34, 35), the very low ex vivo frequency of HHV-6B-specific T cells required in vitro expansion for detailed study. The use of methods with high sensitivity helped to detect these rare cells and reveal the novel HHV-6B antigens described here.
The present study greatly expands the number of known CD4 T-cell antigens in HHV-6B via a high-throughput genome-wide method. Polyclonal HHV-6B-specific T-cell lines tested against full-length viral proteins revealed adaptive responses to both previously known and novel antigens of all viral kinetic classes. Responses could be confirmed with peptides. The accrual of new specificity data is not surprising, given the difficulties that attend study of HHV-6-specific T cells, as reviewed previously (45), and the application of sensitive methods. Some investigators have managed the complexity inherent in the large HHV-6 proteome by limiting query to epitopes restricted by HLA allelic variants with well-understood peptide binding motifs (30, 35). Our methods can access any epitope regardless of HLA type while still allowing definition of HLA restriction using a two-step approach (as outlined in Fig. 3). This approach has expanded the diversity of known HHV-6B CD4 T-cell antigens from approximately 11 to 60 ORF products.
Interestingly, some of the novel antigens found here are among the most prevalent, underscoring the importance of these findings. The most prevalent response was to novel antigen U73 (61% of donors), which has no HCMV homolog, suggesting that the responses that we detected were not due to cross-reactivity with CMV. Both U14 (previously described [30, 31]) and U72 (previously described [36]) had 50% response rates, and both have HCMV homologs (UL25 and UL100, respectively); however, U72 bears only 40% amino acid sequence identity to UL100, scattered irregularly throughout, so HCMV cross-reactivity seems unlikely to contribute substantially to U72 responses. The novel prevalent antigen U95, recognized by 44% of participants and confirmed by peptides, has no known HCMV homolog but is a highly expressed immediate early gene (46). Therefore, it may also be a rational target for cell-based immunotherapies alongside the immediate early U90 protein, a previously described antigen (31, 32, 34). Peptide epitopes confirmed here for U14, U90, and U95 had sequence homology to the related betaherpesviruses HHV-6A (all epitopes) and HHV-7 (nine epitopes) (see Table S2 in the supplemental material), suggesting possible cross-reactivity. Twenty-one epitopes had identity with the iciHHV-6B genomic sequence (GenBank accession number KY315520.1), and albeit beyond the purview of this study, it would be interesting to test if these epitopes are recognized by HHV-6B-specific T cells in a cohort with iciHHV-6B (47–49).
There are several advantages of the present approach. First, whereas previous peptide-based CD4 T-cell studies were limited to epitopes restricted by DRB1*0101 (30, 35), the present study allows genome-wide coverage that captures epitopes of any HLA restriction. Second, the whole HHV-6B antigen and full-length viral protein used prior to peptide confirmation require processing of complex forms of antigen, similar to in vivo conditions. This is in contrast to some epitope discovery methods reliant on peptides alone. Finally, [3H]thymidine-based proliferation assays are quite sensitive. Together, these methods provide the most comprehensive picture yet of HHV-6-specific T-cell responses among the human population.
The study also has certain limitations. The whole HHV-6B antigen is made with SupT1 cells, which may express viral proteins at different levels, with different splice variants, or even with different start and stop sites than cells infected in vivo. This could influence the likelihood of virus-specific T cells being activated and captured with our triple-AIM sorting technique. Moreover, differential T-cell division rates could skew the expansion of HHV-6B-specific clonotypes in vitro prior to readout assays. Therefore, we consider our assay results dichotomous and suitable for calling responses to individual proteins positive or negative and not suitable for establishing within-person immunodominance. False-negative responses to HHV-6B proteins are also possible. T-cell recognition of some epitopes could require posttranslational modifications, which would be not be recapitulated by in vitro transcription and translation (IVTT) proteins or by peptides.
Also, the bulk-expanded HHV-6B-specific T-cell lines from many donors showed high background signals in proliferation assays, possibly obscuring positive antigen calls and even completely invalidating data from several donors. One explanation could be that the frequency of HHV-6B-specific T cells in PBMCs is so low as to be only slightly higher than that of autoreactive T cells. In this scenario, autoreactive T cells, which exist at a low frequency in all donors, as shown in Fig. 1C (response to mock antigen), would be coenriched with HHV-6B-reactive cells and react to autologous PBMCs in proliferation assays, creating high background signals. The triple-AIM selection pathway used does not result in a pure population of HHV-6B-reactive CD4 T cells. In addition to the enrichment of autoreactive cells, this could be related to bystander activation of CD4 T cells in PBMCs in response to cytokines present after HHV-6B recall antigen addition. Triple-AIM sorting also excludes double-AIM-positive T cells; including this compartment at the sorting step would probably capture more HHV-6B-specific clonotypes and reveal additional antigens, provided that adequate enrichment of specific T cells could still be achieved.
Knowledge of HHV-6 T-cell specificity may have clinical implications. VST therapies are emerging as a promising alternative to small-molecule antiviral drugs in the post-HCT setting, during which nearly half of patients experience HHV-6 reactivation (1, 50). For HHV-6B, two proof-of-concept studies used U11, U14, and U90 (51, 52), and one study used U54 and U90 (53). However, it is still not clear if these antigens are optimal for broad applicability. The hierarchy of HHV-6B antigen prevalence provided here suggested that CD4 T-cell antigens U73, U14, U72, and U95 are among the most prevalently recognized and may therefore be rational targets in future VST trials, although further study with a larger sample size would be informative on this point. U95 may be particularly effective since it is a highly expressed immediate early protein, similar to U90. The most frequently used VST approach restimulates PBMCs with ORF-covering peptides and seeks to treat with both CD8 and CD4 T cells. Additional work is required to determine if the population-prevalent HHV-6 CD4 T-cell antigens uncovered in the present report are also prominent CD8 T-cell targets.
In summary, the CD4 T-cell response to HHV-6 has been challenging to study in detail due to the low abundance of the integrated PBMC response. In the present report, a combination AIM approach has allowed significant enrichment of HHV-6B polyclonal CD4 T cells from PBMCs, at the possible expense of coenriching autoreactive CD4 T cells. These cell lines have allowed us to establish a population prevalence hierarchy of responses in a small population of healthy donors across the entire viral proteome, including candidate novel ORFs and variants. In addition to confirming the identity of previously characterized CD4 T-cell antigens, new specificities have been detected that may prove useful in future efforts to study HHV-6 pathogenesis or provide immune-based therapies.
MATERIALS AND METHODS
Participants and specimens.
Healthy adults were recruited at the University of Washington Virology Research Clinic. Peripheral venous blood was collected with heparin, and peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation (54). PBMCs not immediately used after processing were cryopreserved until use. All donor PBMCs were tested for iciHHV-6B (55) and found to be negative. Donors gave informed consent, and all research was conducted according the principles of the Declaration of Helsinki.
Virus and cell culture.
HHV-6B strain Z29 was cultured in SupT1 cells (NIH AIDS Reagent Program) with RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Infected cells were periodically cocultured with uninfected SupT1 cells to maintain infection. Whole HHV-6 antigen was prepared by collection of Z29-SupT1 at 4+ cytopathic effect (CPE), sonication, and UV irradiation for 30 min. Control preparations were made from uninfected SupT1 cells. T-cell stimulation, expansion, and readout assays were performed by using T-cell medium (TCM) (RPMI 1640 with 5% FBS, 5% human serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine).
Enrichment and expansion of virus-reactive T cells from PBMCs.
In a 24-well plate, 7 × 107 PBMCs were incubated overnight at 5 × 106 cells per well with the HHV-6B-infected SupT1 lysate or mock-infected lysate (1:100 dilution). To prevent reuptake of CD154 due to ligation of CD40, anti-CD40 antibody (clone HB14; Miltenyi) was also added at 5 μl/ml. Cells were pooled and stained with 7-amino actinomycin D for viability and fluorochrome-labeled monoclonal antibody (mAb) specific for human CD3, CD4, CD69, CD137, and CD154 in 100 μl TCM. After two washes, cells were resuspended in 1 ml TCM and sorted using a FACSAria II instrument (BD Biosciences). After initial gating for live CD3+ CD4+ lymphocytes, cells gating positive for CD154, CD69, and CD137 were isolated, as were 5 × 104 live CD3+ CD4+ CD154-negative cells (as a negative control). Bulk populations were separately expanded using irradiated allogeneic PBMCs (3,300 rads) and 1.6 μg/ml of phytohemagglutinin (PHA-P; Remel) in 200 μl of TCM in a 96-well U-bottom plate. Natural human IL-2 (hIL-2) (32 U/ml; Hemagen) was added on day 2 and maintained for 14 days, typically yielding 7 × 106 to 1 × 107 cells. A portion of these initially expanded cells was then reexpanded with anti-CD3 mAb OKT3 as the mitogen, a combination of irradiated allogeneic PBMCs and Epstein-Barr virus (EBV)-transformed B lymphocytes as feeder cells, and recombinant hIL-2 (56) for an additional 14 to 18 days. This typically yielded a 1,000-fold increase in the number of T cells, which were cryopreserved in aliquots.
HHV-6B transcriptome analysis and cloning of open reading frames.
RNA sequencing and mass-spectrometry-based confirmation of viral protein expression were carried out on HHV-6B-infected SupT1 cells as described previously (57, 58). Based on these data and the HHV-6B strain Z29 GenBank sequence (accession number NC_000898.1), PCR primers were designed to amplify each annotated HHV-6B ORF and ORFs identified by RNA sequencing as having amino acid sequence disparities with respect to GenBank annotations (i.e., truncation, extension, and/or altered splicing) or as being entirely novel potential ORFs (see Table S1 in the supplemental material), here labeled U7A, U17A, U19A, U47A, U83A, U100A, U100B, and B9A. Total DNA was prepared from HHV-6B-infected SupT1 cells using the QIAamp DNA blood minikit (Qiagen) cultured-cell protocol. Whenever possible, full-length ORFs were amplified. Longer ORFs were amplified in fragments with several overlapping amino acids (Table S1). Where relevant, fragments are labeled fragment A, B, or C in the N- to C-terminal direction. For genes DR6, U12, U15, U79, and U91, which have predicted introns, total RNA was isolated from HHV-6B-infected SupT1 cells using the RNeasy minikit (Qiagen) and used to create a cDNA library by priming with random hexamers, which was used as the PCR template. For intron-containing ORFs, including DR1, U7, U17, and U66, each exon was amplified from genomic DNA primers designed to encode amino acids overlapping splice junctions, and fragments were labeled as described above.
For certain HHV-6B ORFs, RNA sequence data revealed discrepancies in splicing patterns or even translation start and/or stop sites in SupT1 cells vis-à-vis the HHV-6B Z29 NCBI reference genome (accession number NC_000898.1), and ORFs were amplified to reflect these modifications (57). The C-terminal end of U8 and the N-terminal end of U7 are conjoined by a splicing event to form one continuous ORF in the original reading frame, labeled here U7A; primers for these fragments therefore contained overlapping amino acid codons at the splice junction. RNA sequence data showed deep reads in the region between U17 and U18, with two clearly spliced-out introns; the region between predicted start and stop codons is labeled U17A. The C-terminal amino acid codon of U19 was spliced to a downstream region, thereby deleting the annotated stop codon and adding 15 new amino acid codons followed by a new stop codon; this modified ORF is labeled U19A. U47 appeared to have a C-terminal truncation in the RNA sequence data and is labeled U47A. Nearly all transcripts aligning to the U83 ORF region run in the reverse direction to the currently annotated U83, include a start and stop codon, and are predicted to encode a protein with 83 amino acid (aa) residues, as described previously (57). In U100, exons 7 and 8 were not spliced, leaving the intervening intron intact, thereby adding 28 amino acid residues to the end of exon 7 followed by a stop codon. The resulting altered protein is labeled U100A here. Exons 8 through 10 were labeled U100B, with a portion of the upstream intron extending to the first start codon 5′ to exon 8.
Primers included 5′ adaptor sequences for recombinase-mediated integration of PCR amplicons into pDONR221 (Invitrogen). HHV-6 sequences were then moved to pDEST203 (57). As an alternative to this two-step Gateway cloning, some HHV-6 sequences were PCR amplified with flanking restriction endonuclease sites and cloned directly into pDEST203. This included some sequences obtained as synthetic genes (GeneArt; Invitrogen). Once completed, the entire set of pDEST203 plasmids was pooled, diluted to 1 ng/μl, shotgun sequenced using Nextera XT tagmentation (59), and aligned to the NCBI reference sequence for HHV-6B (accession number NC_000898.1).
Intracellular cytokine cytometry.
HHV-6B specificity was tested by intracellular cytokine cytometry (ICC) as previously described (60). For direct ex vivo PBMC tests, 2 × 106 thawed PBMCs were incubated with mock or HHV-6B antigen (1:100 dilution) and medium or PHA controls. To test bulk-expanded T cells, 2 × 105 expanded T cells were cocultured overnight with an equal number of autologous PBMCs prelabeled with CellTrace violet (CTV; Thermo Fisher), along with the mock or HHV-6B antigen preparation or PHA as a positive control. Anti-CD28 and anti-CD49d mAbs were added at assay setup, and brefeldin A was used to reduce cytokine secretion. Cells were stained with Live/Dead Near-IR dye (Thermo Fisher), permeabilized, and stained with fluorochrome-labeled mAbs specific for CD3 (Miltenyi), CD4 (Life Technologies), IFN-γ (BD Biosciences), and IL-2 (BD Pharmingen). The frequency of HHV-6B-specific T cells was measured as the percentage of live CD3+ CD4+ lymphocytes that were positive for IFN-γ and/or IL-2, less mock values. For bulk-expanded responder cells, the CTV-labeled PBMCs used as APCs were dump gated prior to analysis.
T-cell activation assays.
HHV-6B and control microbial genes cloned into pDEST203 were expressed by E. coli in vitro transcription and translation (IVTT) as described previously (60). Proliferative responses of bulk-expanded T-cell lines were tested for each IVTT-expressed protein (60, 61). Briefly, in 96-well U-bottom plates, equal numbers of bulk responder T cells and γ-irradiated autologous PBMCs, used as APCs, were plated in the range of 5 × 104 to 1 × 105 cells per well in duplicate along with each HHV-6B protein diluted 1:2,000 in a total of 200 μl TCM. Controls included 35 IVTT-expressed P. falciparum proteins, empty pDEST203, whole HHV-6B antigen, mock antigen, PHA, and TCM only. On the fourth day after setup, cells were pulsed with 0.5 μCi of [3H]thymidine per well, and proliferation was measured 6 h later. The statistical cutoff for positive responses was described previously (62, 63). Peptides from selected HHV-6B ORFs (18 aa long, overlapping by 11 aa) were pooled at 30 or fewer peptides per pool and tested at a final concentration of 1 μg/ml of each peptide. Individual peptides from reactive pools were retested at 1 μg/ml. In selected cases, inhibitory mAbs that recognize all known HLA DR, HLA DP, or HLA DQ allelic variants were included (54). For these blocking assays, responder cells and APCs were incubated with single peptides and mAb overnight, and T-cell activation was determined by measuring IFN-γ in collected supernatants by an enzyme-linked immunosorbent assay (ELISA) (56). Determination of HLA restriction was also done by using HLA single-antigen lines (SALs) expressing known HLA class II heterodimers or their parental cell nonrecombinant cell lines (64). The SALs were cultured in selective medium and documented as Mycoplasma negative and as positive for surface HLA class II using the same mAbs used in blocking experiments.
Sequential comparison of confirmed HHV-6B epitopes between herpesviruses.
A protein BLAST search was performed with default settings on each confirmed epitope sequence in Table 1 (after reversing cysteine substitutions) against all human herpesvirus species, including HSV-1 (taxonomy identification number [taxid] 10298), HSV-2 (taxid 10310), VZV (taxid 10335), EBV (taxid 10376), HCMV (taxid 10359), HHV-6A (taxid 32603), HHV-6B (taxid 32604), HHV-7 (taxid 10372), and HHV-8 (taxid 37296). Comparison to iciHHV-6B was also performed by manually searching a representative genome sequence (GenBank accession number KY316046.1). All sequence matches with >50% identity to the HHV-6B epitope are compiled in Table S2.
Data availability.
Sequencing reads for the HHV-6B ORFeome are available under BioProject accession number PRJNA525305.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the research participants and the University of Washington Virology Research Clinic. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: SupT1 cells from Dharam Ablashi. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HHV-6B Z29-infected SupT1 cells from Dharam Ablashi and the DAIDS, NIAID. We thank Mauricio Calvo-Calle and Laurence Stern for kindly sharing HHV-6B peptides. We also thank Anuiska Becerra for advice and provision of supplemental virus stock. Meei-li Huang performed iciHHV-6 testing for the study.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number T32AI118690. Funding support also comes from NIH grant 5P50GM115305 and the HHV-6 Foundation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00321-19.
REFERENCES
- 1.Zerr DM, Corey L, Kim HW, Huang ML, Nguy L, Boeckh M. 2005. Clinical outcomes of human herpesvirus 6 reactivation after hematopoietic stem cell transplantation. Clin Infect Dis 40:932–940. doi: 10.1086/428060. [DOI] [PubMed] [Google Scholar]
- 2.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065–1067. [DOI] [PubMed] [Google Scholar]
- 3.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Hollsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T. 2014. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol 159:863–870. doi: 10.1007/s00705-013-1902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol 73:8040–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agut H, Bonnafous P, Gautheret-Dejean A. 2016. Human herpesviruses 6A, 6B, and 7. Microbiol Spectr 4(3):DMIH2-0007-2015. doi: 10.1128/microbiolspec.DMIH2-0007-2015. [DOI] [PubMed] [Google Scholar]
- 6.Tang H, Mori Y. 2015. Determinants of human CD134 essential for entry of human herpesvirus 6B. J Virol 89:10125–10129. doi: 10.1128/JVI.01606-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood ML, Royle NJ. 2017. Chromosomally integrated human herpesvirus 6: models of viral genome release from the telomere and impacts on human health. Viruses 9:E184. doi: 10.3390/v9070184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cone RW, Huang ML, Ashley R, Corey L. 1993. Human herpesvirus 6 DNA in peripheral blood cells and saliva from immunocompetent individuals. J Clin Microbiol 31:1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ablashi DV, Marsh S, Kaplan M, Whitman JE Jr, Pearson GR. 1998. HHV-6 infection in HIV-infected asymptomatic and AIDS patients. Intervirology 41:1–9. doi: 10.1159/000024909. [DOI] [PubMed] [Google Scholar]
- 10.Phan TL, Pritchett JC, Leifer C, Zerr DM, Koelle DM, Di Luca D, Lusso P. 2018. HHV-6B infection, T-cell reconstitution, and graft-vs-host disease after hematopoietic stem cell transplantation. Bone Marrow Transplant 53:1508–1517. doi: 10.1038/s41409-018-0225-2. [DOI] [PubMed] [Google Scholar]
- 11.Phan TL, Carlin K, Ljungman P, Politikos I, Boussiotis V, Boeckh M, Shaffer ML, Zerr DM. 2018. Human herpesvirus-6B reactivation is a risk factor for grades II to IV acute graft-versus-host disease after hematopoietic stem cell transplantation: a systematic review and meta-analysis. Biol Blood Marrow Transplant 24:2324–2336. doi: 10.1016/j.bbmt.2018.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caselli E, Zatelli MC, Rizzo R, Benedetti S, Martorelli D, Trasforini G, Cassai E, Degli Uberti EC, Di Luca D, Dolcetti R. 2012. Virologic and immunologic evidence supporting an association between HHV-6 and Hashimoto’s thyroiditis. PLoS Pathog 8:e1002951. doi: 10.1371/journal.ppat.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fillet AM, Lozeron P, Agut H, Lyon-Caen O, Liblau R. 1998. HHV-6 and multiple sclerosis. Nat Med 4:537 (Reply, 4:538.) doi: 10.1038/nm0598-537a. [DOI] [PubMed] [Google Scholar]
- 14.Coates AR, Bell J. 1998. HHV-6 and multiple sclerosis. Nat Med 4:537–538. doi: 10.1038/nm0598-537b. [DOI] [PubMed] [Google Scholar]
- 15.Ablashi DV, Lapps W, Kaplan M, Whitman JE, Richert JR, Pearson GR. 1998. Human herpesvirus-6 (HHV-6) infection in multiple sclerosis: a preliminary report. Mult Scler 4:490–496. doi: 10.1177/135245859800400606. [DOI] [PubMed] [Google Scholar]
- 16.Challoner PB, Smith KT, Parker JD, MacLeod DL, Coulter SN, Rose TM, Schultz ER, Bennett JL, Garber RL, Chang M, Schad PA, Stewart PM, Nowinski RC, Brown JP, Burmer GC. 1995. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc Natl Acad Sci U S A 92:7440–7444. doi: 10.1073/pnas.92.16.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soldan SS, Berti R, Salem N, Secchiero P, Flamand L, Calabresi PA, Brennan MB, Maloni HW, McFarland HF, Lin HC, Patnaik M, Jacobson S. 1997. Association of human herpes virus 6 (HHV-6) with multiple sclerosis: increased IgM response to HHV-6 early antigen and detection of serum HHV-6 DNA. Nat Med 3:1394–1397. doi: 10.1038/nm1297-1394. [DOI] [PubMed] [Google Scholar]
- 18.Friedman JE, Lyons MJ, Cu G, Ablashl DV, Whitman JE, Edgar M, Koskiniemi M, Vaheri A, Zabriskie JB. 1999. The association of the human herpesvirus-6 and MS. Mult Scler 5:355–362. doi: 10.1177/135245859900500509. [DOI] [PubMed] [Google Scholar]
- 19.Voumvourakis KI, Kitsos DK, Tsiodras S, Petrikkos G, Stamboulis E. 2010. Human herpesvirus 6 infection as a trigger of multiple sclerosis. Mayo Clin Proc 85:1023–1030. doi: 10.4065/mcp.2010.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agut H, Aubin JT, Huraux JM. 1991. Homogeneous susceptibility of distinct human herpesvirus 6 strains to antivirals in vitro. J Infect Dis 163:1382–1383. doi: 10.1093/infdis/163.6.1382. [DOI] [PubMed] [Google Scholar]
- 21.De Bolle L, Naesens L, De Clercq E. 2005. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev 18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prichard MN, Whitley RJ. 2014. The development of new therapies for human herpesvirus 6. Curr Opin Virol 9:148–153. doi: 10.1016/j.coviro.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isegawa Y, Hara J, Amo K, Osugi Y, Takemoto M, Yamanishi K, Fukunaga R, Shibata M, Ohshima A, Horiguchi Y, Sugimoto N. 2009. Human herpesvirus 6 ganciclovir-resistant strain with amino acid substitutions associated with the death of an allogeneic stem cell transplant recipient. J Clin Virol 44:15–19. doi: 10.1016/j.jcv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Isegawa Y, Matsumoto C, Nishinaka K, Nakano K, Tanaka T, Sugimoto N, Ohshima A. 2010. PCR with quenching probes enables the rapid detection and identification of ganciclovir-resistance-causing U69 gene mutations in human herpesvirus 6. Mol Cell Probes 24:167–177. doi: 10.1016/j.mcp.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Bounaadja L, Piret J, Goyette N, Boivin G. 2013. Analysis of HHV-6 mutations in solid organ transplant recipients at the onset of cytomegalovirus disease and following treatment with intravenous ganciclovir or oral valganciclovir. J Clin Virol 58:279–282. doi: 10.1016/j.jcv.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 26.Clark DA, Ait-Khaled M, Wheeler AC, Kidd IM, McLaughlin JE, Johnson MA, Griffiths PD, Emery VC. 1996. Quantification of human herpesvirus 6 in immunocompetent persons and post-mortem tissues from AIDS patients by PCR. J Gen Virol 77(Part 9):2271–2275. doi: 10.1099/0022-1317-77-9-2271. [DOI] [PubMed] [Google Scholar]
- 27.Greco R, Crucitti L, Noviello M, Racca S, Mannina D, Forcina A, Lorentino F, Valtolina V, Rolla S, Dvir R, Morelli M, Giglio F, Barbanti MC, Lupo Stanghellini MT, Oltolini C, Vago L, Scarpellini P, Assanelli A, Carrabba MG, Marktel S, Bernardi M, Corti C, Clementi M, Peccatori J, Bonini C, Ciceri F. 2016. Human herpesvirus 6 infection following haploidentical transplantation: immune recovery and outcome. Biol Blood Marrow Transplant 22:2250–2255. doi: 10.1016/j.bbmt.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 28.Muftuoglu M, Olson A, Marin D, Ahmed S, Mulanovich V, Tummala S, Chi TL, Ferrajoli A, Kaur I, Li L, Champlin R, Shpall EJ, Rezvani K. 2018. Allogeneic BK virus-specific T cells for progressive multifocal leukoencephalopathy. N Engl J Med 379:1443–1451. doi: 10.1056/NEJMoa1801540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, Parkhurst MR, Yang JC, Rosenberg SA. 2014. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nastke MD, Becerra A, Yin L, Dominguez-Amorocho O, Gibson L, Stern LJ, Calvo-Calle JM. 2012. Human CD4+ T cell response to human herpesvirus 6. J Virol 86:4776–4792. doi: 10.1128/JVI.06573-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerdemann U, Keukens L, Keirnan JM, Katari UL, Nguyen CT, de Pagter AP, Ramos CA, Kennedy-Nasser A, Gottschalk SM, Heslop HE, Brenner MK, Rooney CM, Leen AM. 2013. Immunotherapeutic strategies to prevent and treat human herpesvirus 6 reactivation after allogeneic stem cell transplantation. Blood 121:207–218. doi: 10.1182/blood-2012-05-430413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halawi M, Khan N, Blake N. 2015. Identification of novel CD8+ T cell epitopes in human herpesvirus 6B U11 and U90. Immun Inflamm Dis 3:118–131. doi: 10.1002/iid3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iampietro M, Morissette G, Gravel A, Dubuc I, Rousseau M, Hasan A, O’Reilly RJ, Flamand L. 2014. Human herpesvirus 6B immediate-early I protein contains functional HLA-A*02, HLA-A*03, and HLA-B*07 class I restricted CD8(+) T-cell epitopes. Eur J Immunol 44:3573–3584. doi: 10.1002/eji.201444931. [DOI] [PubMed] [Google Scholar]
- 34.Martin LK, Schub A, Dillinger S, Moosmann A. 2012. Specific CD8(+) T cells recognize human herpesvirus 6B. Eur J Immunol 42:2901–2912. doi: 10.1002/eji.201242439. [DOI] [PubMed] [Google Scholar]
- 35.Becerra-Artiles A, Dominguez-Amorocho O, Stern LJ, Calvo-Calle JM. 2015. A simple proteomics-based approach to identification of immunodominant antigens from a complex pathogen: application to the CD4 T cell response against human herpesvirus 6B. PLoS One 10:e0142871. doi: 10.1371/journal.pone.0142871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin LK, Hollaus A, Stahuber A, Hubener C, Fraccaroli A, Tischer J, Schub A, Moosmann A. 2018. Cross-sectional analysis of CD8 T cell immunity to human herpesvirus 6B. PLoS Pathog 14:e1006991. doi: 10.1371/journal.ppat.1006991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jing L, Chong TM, Byrd B, McClurkan CL, Huang J, Story BT, Dunkley KM, Aldaz-Carroll L, Eisenberg RJ, Cohen GH, Kwok WW, Sette A, Koelle DM. 2007. Dominance and diversity in the primary human CD4 T cell response to replication-competent vaccinia virus. J Immunol 178:6374–6386. doi: 10.4049/jimmunol.178.10.6374. [DOI] [PubMed] [Google Scholar]
- 38.Jing L, Haas J, Chong TM, Bruckner JJ, Dann GC, Dong L, Marshak JO, McClurkan CL, Yamamoto TN, Bailer SM, Laing KJ, Wald A, Verjans GM, Koelle DM. 2012. Cross-presentation and genome-wide screening reveal candidate T cells antigens for a herpes simplex virus type 1 vaccine. J Clin Invest 122:654–673. doi: 10.1172/JCI60556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jing L, Laing KJ, Dong L, Russell RM, Barlow RS, Haas JG, Ramchandani MS, Johnston C, Buus S, Redwood AJ, White KD, Mallal SA, Phillips EJ, Posavad CM, Wald A, Koelle DM. 2016. Extensive CD4 and CD8 T cell cross-reactivity between alphaherpesviruses. J Immunol 196:2205–2218. doi: 10.4049/jimmunol.1502366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jing L, Schiffer JT, Chong TM, Bruckner JJ, Davies DH, Felgner PL, Haas J, Wald A, Verjans GM, Koelle DM. 2013. CD4 T-cell memory responses to viral infections of humans show pronounced immunodominance independent of duration or viral persistence. J Virol 87:2617–2627. doi: 10.1128/JVI.03047-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nayak K, Jing L, Russell RM, Davies DH, Hermanson G, Molina DM, Liang X, Sherman DR, Kwok WW, Yang J, Kenneth J, Ahamed SF, Chandele A, Murali-Krishna K, Koelle DM. 2015. Identification of novel Mycobacterium tuberculosis CD4 T-cell antigens via high throughput proteome screening. Tuberculosis (Edinb) 95:275–287. doi: 10.1016/j.tube.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harnett GB, Farr TJ, Pietroboni GR, Bucens MR. 1990. Frequent shedding of human herpesvirus 6 in saliva. J Med Virol 30:128–130. doi: 10.1002/jmv.1890300209. [DOI] [PubMed] [Google Scholar]
- 44.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 45.Hanson DJ, Hill JA, Koelle DM. 2018. Advances in the characterization of the T-cell response to human herpesvirus-6. Front Immunol 9:1454. doi: 10.3389/fimmu.2018.01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takemoto M, Shimamoto T, Isegawa Y, Yamanishi K. 2001. The R3 region, one of three major repetitive regions of human herpesvirus 6, is a strong enhancer of immediate-early gene U95. J Virol 75:10149–10160. doi: 10.1128/JVI.75.21.10149-10160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strenger V, Kayser S, Witte KE, Lassner D, Schwinger W, Jahn G, Urban C, Feuchtinger T. 2016. Individuals with inherited chromosomally integrated human herpes virus 6 (ciHHV-6) have functionally active HHV-6 specific T-cell immunity. Clin Microbiol Infect 22:209.e5–209.e8. doi: 10.1016/j.cmi.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Barozzi P, Riva G, Vallerini D, Quadrelli C, Lagreca I, Eccheli R, Forghieri F, Coluccio V, Maccaferri M, Paolini A, Colaci E, Morselli M, Marasca R, Narni F, Comoli P, Campioli D, Trenti T, Potenza L, Luppi M. 2016. Circulating functional T cells specific to human herpes virus 6 (HHV6) antigens in individuals with chromosomally integrated HHV6. Clin Microbiol Infect 22:893–895. doi: 10.1016/j.cmi.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Strenger V, Caselli E, Lautenschlager I, Schwinger W, Aberle SW, Loginov R, Gentili V, Nacheva E, DiLuca D, Urban C. 2014. Detection of HHV-6-specific mRNA and antigens in PBMCs of individuals with chromosomally integrated HHV-6 (ciHHV-6). Clin Microbiol Infect 20:1027–1032. doi: 10.1111/1469-0691.12639. [DOI] [PubMed] [Google Scholar]
- 50.Hill JA, Zerr DM. 2014. Roseoloviruses in transplant recipients: clinical consequences and prospects for treatment and prevention trials. Curr Opin Virol 9:53–60. doi: 10.1016/j.coviro.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papadopoulou A, Gerdemann U, Katari UL, Tzannou I, Liu H, Martinez C, Leung K, Carrum G, Gee AP, Vera JF, Krance RA, Brenner MK, Rooney CM, Heslop HE, Leen AM. 2014. Activity of broad-spectrum T cells as treatment for AdV, EBV, CMV, BKV, and HHV6 infections after HSCT. Sci Transl Med 6:242ra83. doi: 10.1126/scitranslmed.3008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzannou I, Papadopoulou A, Naik S, Leung K, Martinez CA, Ramos CA, Carrum G, Sasa G, Lulla P, Watanabe A, Kuvalekar M, Gee AP, Wu MF, Liu H, Grilley BJ, Krance RA, Gottschalk S, Brenner MK, Rooney CM, Heslop HE, Leen AM, Omer B. 2017. Off-the-shelf virus-specific T cells to treat BK virus, human herpesvirus 6, cytomegalovirus, Epstein-Barr virus, and adenovirus infections after allogeneic hematopoietic stem-cell transplantation. J Clin Oncol 35:3547–3557. doi: 10.1200/JCO.2017.73.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishiyama-Fujita Y, Kawana-Tachikawa AI, Ono T, Tanaka Y, Kato T, Heslop HE, Morio T, Takahashi S. 2018. Generation of multivirus-specific T cells by a single stimulation of peripheral blood mononuclear cells with a peptide mixture using serum-free medium. Cytotherapy 20:1182–1190. doi: 10.1016/j.jcyt.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Koelle DM, Corey L, Burke RL, Eisenberg RJ, Cohen GH, Pichyangkura R, Triezenberg SJ. 1994. Antigenic specificities of human CD4+ T-cell clones recovered from recurrent genital herpes simplex virus type 2 lesions. J Virol 68:2803–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill JA, HallSedlak R, Magaret A, Huang ML, Zerr DM, Jerome KR, Boeckh M. 2016. Efficient identification of inherited chromosomally integrated human herpesvirus 6 using specimen pooling. J Clin Virol 77:71–76. doi: 10.1016/j.jcv.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koelle DM, Chen HB, Gavin MA, Wald A, Kwok WW, Corey L. 2001. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. J Immunol 166:4049–4058. doi: 10.4049/jimmunol.166.6.4049. [DOI] [PubMed] [Google Scholar]
- 57.Greninger AL, Knudsen GM, Roychoudhury P, Hanson DJ, Sedlak RH, Xie H, Guan J, Nguyen T, Peddu V, Boeckh M, Huang ML, Cook L, Depledge DP, Zerr DM, Koelle DM, Gantt S, Yoshikawa T, Caserta M, Hill JA, Jerome KR. 2018. Comparative genomic, transcriptomic, and proteomic reannotation of human herpesvirus 6. BMC Genomics 19:204. doi: 10.1186/s12864-018-4604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greninger AL, Roychoudhury P, Makhsous N, Hanson D, Chase J, Krueger G, Xie H, Huang ML, Saunders L, Ablashi D, Koelle DM, Cook L, Jerome KR. 2018. Copy number heterogeneity, large origin tandem repeats, and interspecies recombination in human herpesvirus 6A (HHV-6A) and HHV-6B reference strains. J Virol 92:e00135-18. doi: 10.1128/JVI.00135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greninger AL, Langelier C, Cunningham G, Keh C, Melgar M, Chiu CY, Miller S. 2015. Two rapidly growing mycobacterial species isolated from a brain abscess: first whole-genome sequences of Mycobacterium immunogenum and Mycobacterium llatzerense. J Clin Microbiol 53:2374–2377. doi: 10.1128/JCM.00402-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jing L, Davies DH, Chong TM, Chun S, McClurkan CL, Huang J, Story BT, Molina DM, Hirst S, Felgner PL, Koelle DM. 2008. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J Virol 82:7120–7134. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jing L, McCaughey SM, Davies DH, Chong TM, Felgner PL, De Rosa SC, Wilson CB, Koelle DM. 2009. ORFeome approach to the clonal, HLA allele-specific CD4 T-cell response to a complex pathogen in humans. J Immunol Methods 347:36–45. doi: 10.1016/j.jim.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung N, Zhang XD, Kreamer A, Locco L, Kuan PF, Bartz S, Linsley PS, Ferrer M, Strulovici B. 2008. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J Biomol Screen 13:149–158. doi: 10.1177/1087057107312035. [DOI] [PubMed] [Google Scholar]
- 63.Laing KJ, Russell RM, Dong L, Schmid DS, Stern M, Magaret A, Haas JG, Johnston C, Wald A, Koelle DM. 2015. Zoster vaccination increases the breadth of CD4+ T cells responsive to varicella zoster virus. J Infect Dis 212:1022–1031. doi: 10.1093/infdis/jiv164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKinney DM, Southwood S, Hinz D, Oseroff C, Arlehamn CS, Schulten V, Taplitz R, Broide D, Hanekom WA, Scriba TJ, Wood R, Alam R, Peters B, Sidney J, Sette A. 2013. A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population. Immunogenetics 65:357–370. doi: 10.1007/s00251-013-0684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, Peters B. 2015. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads for the HHV-6B ORFeome are available under BioProject accession number PRJNA525305.