Retroviruses occasionally can infect host germ lines, forming endogenous retroviruses. Vertebrates, in turn, recruited retroviral genes for their own biological functions, a process formally known as co-option or exaptation. To date, co-opted retroviral gag genes have rarely been reported. In this study, we identified two co-opted retroviral gag genes, designated wucaishi1 (wcs1) and wucaishi2 (wcs2), in mammals. The co-option of wcs1 and wcs2 occurred before the origin of modern placentals and before the origin of modern marsupials, respectively. Our study indicates that retroviral gag gene co-option might have occurred more frequently than previously thought during the evolutionary course of vertebrates.
KEYWORDS: co-option, endogenous retrovirus, paleovirology, phylogenetics
ABSTRACT
Endogenous retroviruses, records of past retroviral infections, are ubiquitous in vertebrate genomes. On occasion, vertebrate hosts have co-opted retroviral genes for their own biological functions. Here, we perform a phylogenomic survey of retroviral gag gene homologs within vertebrate genomes and identify two ancient co-opted retroviral gag genes, designated wucaishi1 (wcs1) and wucaishi2 (wcs2), in mammals. Conserved synteny and evolutionary analyses suggest that the wcs1 and wcs2 co-options occurred before the origin of modern placental mammals (∼100 million years ago) and before the origin of modern marsupials (∼80 million years ago), respectively. We found that the wcs genes were lost or pseudogenized multiple times during the evolutionary course of mammals. While the wcs1 gene is mainly subject to negative selection in placental mammals (except in Perissodactyla), the wcs2 gene underwent positive selection in marsupials. Moreover, analyses of transcriptome-sequencing (RNA-seq) data suggest that the wcs1 and the wcs2 genes are expressed in a wide range of tissues. The convergent wcs co-option in mammals implies the retroviral gag gene might have been repurposed more frequently than previously thought.
IMPORTANCE Retroviruses occasionally can infect host germ lines, forming endogenous retroviruses. Vertebrates, in turn, recruited retroviral genes for their own biological functions, a process formally known as co-option or exaptation. To date, co-opted retroviral gag genes have rarely been reported. In this study, we identified two co-opted retroviral gag genes, designated wucaishi1 (wcs1) and wucaishi2 (wcs2), in mammals. The co-option of wcs1 and wcs2 occurred before the origin of modern placentals and before the origin of modern marsupials, respectively. Our study indicates that retroviral gag gene co-option might have occurred more frequently than previously thought during the evolutionary course of vertebrates.
INTRODUCTION
Retroviruses infect a variety of vertebrates and cause many diseases, such as AIDS and cancers. Uniquely among RNA viruses, retroviruses replicate through reverse transcription of viral RNA into DNA and integration of newly synthesized DNA into the host chromosome. While retrovirus infection usually takes place in host somatic cells, retrovirus occasionally infects host germ line cells. The integrated retroviral copies in the germ line (known as endogenous retroviruses [ERVs]) begin to be vertically inherited as host genetic elements (1, 2). ERVs are ubiquitously present and highly abundant in vertebrate genomes (3–5); for example, ERVs make up around 8% of the human genome (6). Recent comparative genomic studies have uncovered many nonretroviral sequences endogenized in diverse eukaryotes (7–10). Endogenous viral elements (EVEs), including ERVs, record past viral infections and thus provide molecular fossils to study the origin and deep history of viruses, laying the foundation of an emerging field, paleovirology (11, 12).
Most ERVs accumulate deleterious mutations and become degraded over time (2, 5). On occasion, vertebrate hosts can recruit ERVs for their own biological functions, a process formally termed co-option or exaptation (13, 14). Co-opted retroviral genes mediate a variety of host biological processes, such as protecting hosts from exogenous retrovirus infection (e.g., the Fv1 gene, a co-opted gag gene in rodents), regulating placenta formation (e.g., the syncytin genes, co-opted env genes in placentals), and regulating the expression of host genes by co-opting retroviral regulatory sequences (12–21). While the retroviral env gene has been frequently co-opted in placentals, few cases of retroviral gag co-option have been reported, with the Fv1 gene the best-known example (13, 14). As a restriction factor, the Fv1 gene blocks the replication of various retroviruses, such as murine leukemia virus, lentiviruses, and foamy viruses (22, 23). Sequence analysis has revealed similarity between Fv1 and the Gag protein of murine endogenous retrovirus L (MuERV-L) (22). Conserved synteny analysis dates the integration of the Fv1 progenitor into the genomes of Muroidea, a superfamily of rodents, back to ∼45 million years ago (MYA) (24, 25). In addition to retroviral gag genes, retrotransposon gag genes have also been found to be repurposed in mammals (20); for example, the Arc gene, a co-opted gag gene derived from Ty3/gypsy retrotransposons, mediates mRNA transfer between different nerve cells (20, 21). With the recent development of next-generation sequencing, an increasing number of vertebrate genomes have been sequenced, which provide important resources for identifying more cases of retroviral gene co-option.
As illustrated by the “Red Queen” hypothesis, the virus-host conflicts result in recurrent cycles of an arms race, where hosts evolve resistance to viral infection and viruses, in turn, develop countermeasures to evade or block host defenses (26–30). Both host and virus genes involved in the genetic conflicts have been found to be subject to positive selection (26–30). Nearly all known restriction factors, host proteins that inhibit the replication of viruses (29), exhibit signatures of positive selection, including the Fv1 gene (23–25, 29, 31, 32).
In this study, we performed a phylogenomic analysis of retroviral gag gene co-option events and identified two co-opted retroviral gag genes in mammals. The co-option events date back to the origin of modern placentals and the origin of modern marsupials, respectively. We also analyzed the evolutionary fingerprints and expression patterns of the co-opted gag genes.
RESULTS
Co-option of retroviral gag genes in mammals.
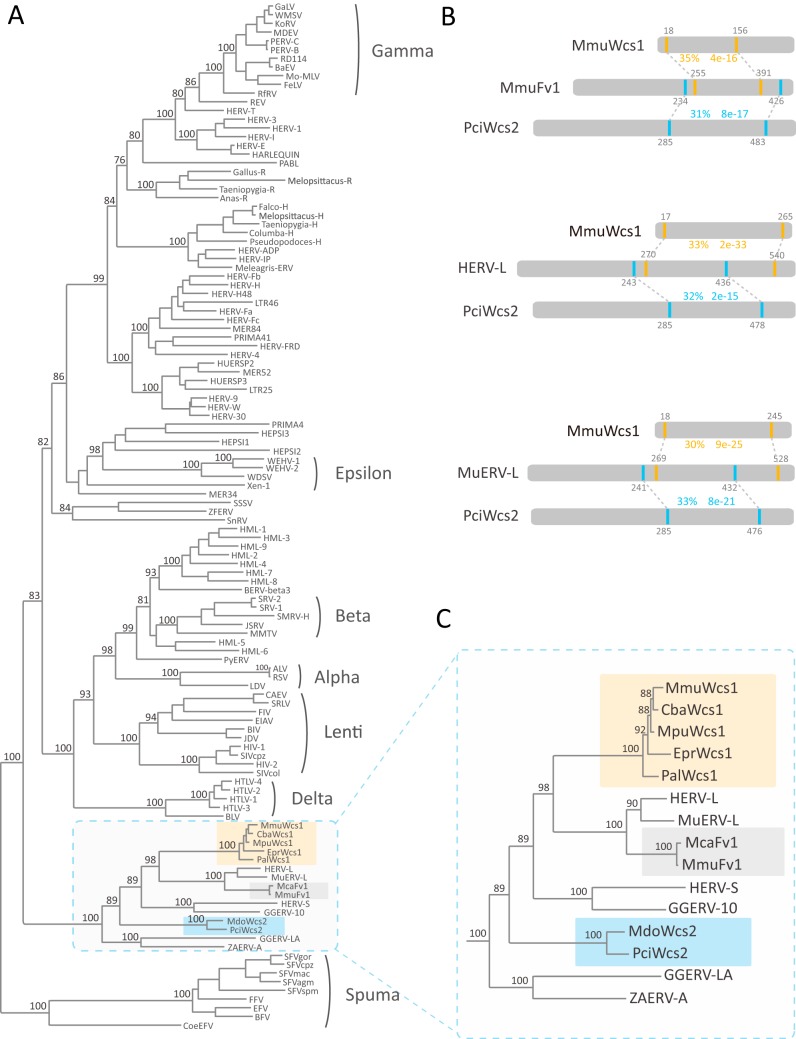
Initially, to explore the evolutionary history of the Fv1 gene, we employed a combined similarity search and phylogenetic analysis approach to identify homologs of MuERV-L Gag protein, the closest relative of Fv1, within the vertebrate proteomes. To our surprise, we identified several proteins that share significant similarity with the Gag proteins of MuERV-L in mammals but do not share high identity with Fv1 (Fig. 1; see Table S1 in the supplemental material). We designated those Gag-like proteins Wucaishi (Wcs) proteins. In Chinese mythology, wucaishi (five-colored stone) was repurposed and used by the goddess Nüwa to patch up the sky after the pillars of heaven were broken. To further explore the relationship among Wcs, Fv1, and retroviral Gag proteins (see Table S2 in the supplemental material), we performed phylogenetic analysis and found that the newly identified Wcs proteins do not group with the Fv1 proteins. The Wcs proteins cluster into two monophyletic groups, termed Wcs1 and Wcs2, with strong support (bootstrap values of 100% for both groups) (Fig. 1A and C). The Wcs1 protein is more closely related to Fv1, MuERV-L, and human endogenous retrovirus L (HERV-L) than the Wcs2 protein. These results suggest that the wcs1 and wcs2 genes arose through two co-option events independently of Fv1 co-option.
FIG 1.
Relationships among the Wcs proteins, the Fv1 proteins, and retrovirus Gag proteins. (A) Phylogenetic relationships among the Wcs proteins, the Fv1 proteins, and representative retrovirus Gag proteins. The numbers near selected nodes represent bootstrap values. The Wcs1 and Wcs2 proteins are highlighted in orange and blue, respectively. (B) Similarities among the Wcs proteins, the Fv1 protein, and HERV-L and MuERV-L Gag proteins. The dashed lines indicate regions that share significant similarity between two proteins. The gray numbers indicate residue positions. The numbers in blue or orange indicate sequence identities and BLASTp E values. (C) Phylogenetic relationships among the Wcs proteins, the Fv1 protein, and HERV-L and MuERV-L Gag proteins (enlargement of the boxed area in panel A). Mca, Mus caroli; Mmu, Mus musculus; Mpu, Mustela putorius; Cba, Camelus bactrianus; Pal, Pteropus alecto; Epr, Equus przewalskii; Mdo, Monodelphis domestica; Pci, Phascolarctos cinereus.
Evolutionary history of the wcs1 locus.
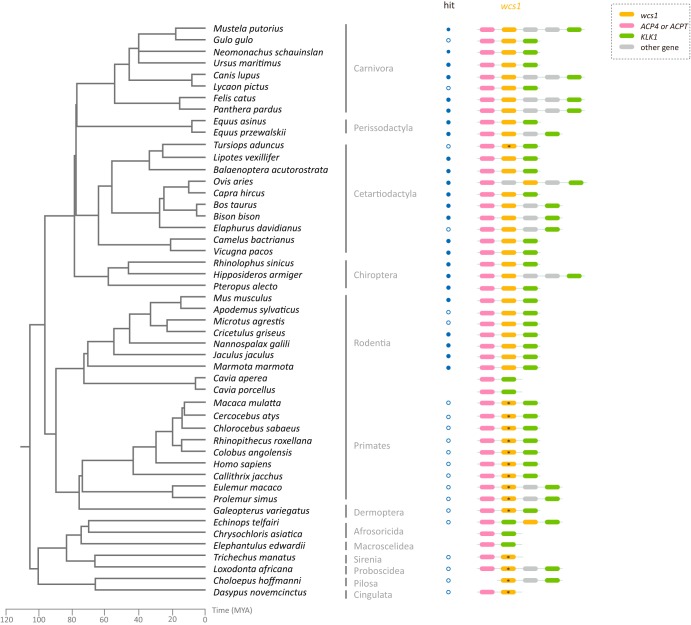
A similarity search based on vertebrate proteomes and phylogenetic analysis suggested that the Wcs1 protein is present within at least five mammalian orders, Carnivora, Perissodactyla, Cetartiodactyla, Chiroptera, and Rodentia (Fig. 1 and 2; see Table S5 in the supplemental material). The wcs1 gene is located between Acid phosphatase 4 (ACP4) or Acid phosphatase T (ACPT) (ACP4 and ACPT form a monophyletic group and are essentially orthologs) (see Fig. S1 and Table S6 in the supplemental material) and Kallikrein 1 (KLK1) (Fig. 2). The ACP4- or ACPT-KLK1 synteny is conserved across placentals (Fig. 2). To further explore the evolutionary history of the wcs1 gene, we used a combined gene synteny and similarity search approach and found sequences that share significant similarity with the wcs1 gene and are located between ACP4 and KLK1 in other orders of placentals, including Primates, Dermoptera, Afrosoricida, Sirenia, and Proboscidea. Most of the wcs1 sequences in these orders appear to be pseudogenized (Fig. 2). The wcs1 gene appears to be lost in some species of Rodentia, Afrosoricida, and Macroscelidea. The wcs1 sequence is located only together with KLK1 in Choloepus hoffmanni (order: Pilosa) and is located only together with ACP4 in Dasypus novemcinctus (order: Cingulata) and Trichechus manatus (order: Sirenia), which might be due to the shortness of the contigs. Nevertheless, the conserved synteny of ACP4-wcs1-KLK1 among major orders of placentals suggests that the insertion of the wcs1 gene occurred at least in the most recent common ancestor of modern placentals, around 100 MYA (33, 53).
FIG 2.
Conserved synteny of the wcs1 genes. The phylogenetic relationships among placentals and the evolutionary time scale are based on TimeTree (33, 53). The solid and open circles indicate that the wcs1 gene homologs were identified by BLASTp and BLASTn/tBLASTn, respectively. The gene synteny of wcs1 is shown near the corresponding species. Genes with premature stop codons or frameshift mutations are labeled with asterisks.
Evolutionary history of the wcs2 locus.
Our initial similarity search and phylogenetic analysis suggested the Wcs2 protein is present in the genomes of Phascolarctos cinereus and Monodelphis domestica. For both species, the wcs2 gene is located between Transgelin-3 (TAGLN3) and Abhydrolase domain containing 10 (ABHD10) (Fig. 3). The TAGLN3-ABHD10 synteny is conserved across vertebrates ranging from zebrafish (Danio rerio) to human. The different syntenies between the wcs1 and wcs2 loci also support the notion that the wcs1 and the wcs2 genes arose through independent co-option events. To further explore the evolutionary history of the wcs2 gene, we used a combined gene synteny and similarity search approach and found sequences that share significant similarity with the wcs2 gene and are located between TAGLN3 and Abdh10 in three marsupial orders, that is, Diprotodontia, Dasyuromorphia, and Didelphimorphia. In Notamacropus eugenii, the wcs2 gene is located only together with Abdh10, which might be due to the shortness of the contig. The wcs2 gene has been pseudogenized in some marsupial species (Fig. 3; see Table S5). Nevertheless, the conserved synteny of TAGLN2-wcs2-ABHD10 across marsupials suggests that the co-option of wcs2 occurred in the common ancestor of modern marsupials, around 80 MYA (33, 53).
FIG 3.
Conserved synteny of the wcs2 genes. The phylogenetic relationships among marsupials and the evolutionary time scale are based on TimeTree (33, 53). The solid and open circles indicate that the wcs2 gene homologs were identified by BLASTp and BLASTn/tBLASTn, respectively. The gene synteny of wcs2 is shown near the corresponding species. A gene with a premature stop codon or frameshift mutation is labeled with an asterisk.
Selection analyses of the wcs genes.
The selection pressure that has acted on a gene can be inferred by comparing the number of nonsynonymous substitutions per nonsynonymous site (dN) and the number of synonymous substitutions per synonymous site (dS) (27–30). A dN/dS ratio of greater than one indicates positive selection. We characterized the evolutionary fingerprints in the wcs1 gene for five placental orders and the wcs2 gene for marsupials. First, we used the single-likelihood ancestor-counting (SLAC) method (34) to estimate the dN/dS ratios for the wcs genes of five placental orders and marsupials and observed generally small dN/dS values (most around 0.1), except for Perissodactyla (dN/dS = 1.95) (Table 1). The dN/dS ratios suggest the wcs genes might mainly undergo negative selection in mammals, except Perissodactyla. Because the dN/dS ratio appears to be a conservative statistic (35, 36), we used site models to detect positively selected sites in the wcs genes. The neutral model (M8a) was not rejected in favor of the model with positive selection (M8) in four placental orders. The M8a model was rejected in the wcs genes of Perissodactyla (P = 0.03) and marsupials (P = 0.01) (Table 1). We detected 17 sites and 9 sites under positive selection in Perissodactyla and marsupials, respectively. Moreover, we also used a fixed effects likelihood (FEL) method to detect positively selected sites (34). For all five mammalian orders, no site of the wcs1 gene was found to be under positive selection at a significance level of 0.05, while many sites (up to 45.07%) were found to be under negative selection (Table 1). For marsupials, there was only one site (0.16%) under positive selection but 34 sites (5.27%) under negative selection. Finally, we employed the adaptive branch site random-effects likelihood (aBSREL) method to test whether positive selection had occurred on a proportion of branches (36, 37). We found only three branches subject to positive selection for three mammalian lineages, that is, Perissodactyla (1 out of 3 branches), Cetartiodactyla (only 1 out of 41 branches), and Rodentia (only 1 out of 47 branches). Taken together, our analyses suggest that the wcs1 gene is mainly subjected to negative selection in placentals, except Perissodactyla, and that the wcs2 gene might undergo positive selection.
TABLE 1.
Selection analysis of wcs genes in mammals
| Order/class | No. of species | No. of sites | dN/dSa | No. of branches under positive selection/totalb | No. (%) of sites under positive selectionc | No. (%) of sites under negative selectionc | 2Δld | P valued | No. (%) of sites under positive selectiond | Codons with dN/dS value of >1 |
|---|---|---|---|---|---|---|---|---|---|---|
| Carnivora | 22 | 291 | 0.07 | 0/41 | 83 (28.52) | 2.41 | 0.12 | |||
| Perissodactyla | 3 | 259 | 1.95 | 1/3 | 4.79 | 0.03 | 17 (6.56) | T9e, H20e, V26e, S27e, Q31e, L38e, T88e, W94e, R97e, L137e, V146e, D170e, Q174e, V182e, Q186e, T188e, R255e | ||
| Cetartiodactyla | 22 | 288 | 0.08 | 1/41 | 72 (25.00) | 0.00 | 1.00 | |||
| Chiroptera | 6 | 289 | 0.18 | 0/9 | 56 (19.38) | 0.00 | 1.00 | |||
| Rodentia | 25 | 284 | 0.07 | 1/47 | 128 (45.07) | 0.00 | 0.95 | |||
| Marsupialia | 4 | 645 | 0.65 | 0/5 | 1 (0.16) | 34 (5.27) | 6.73 | 0.01 | 9 (1.40) | L233e, V246e, S305e, G336e, K343e, E353e, T369e, S389e, Q591e |
The dN/dS values of the wcs genes were calculated using the SLAC method in HyPhy.
Branches under positive selection with P values of <0.05 were detected using the aBSREL method in HyPhy.
Sites under positive or negative selection with P values of <0.05 were detected using the FEL method in HyPhy.
2Δl represents twice the difference in the natural log values of the likelihoods of the two models (M8a versus M8) being compared. The P value indicates the confidence with which the neutral model (M8a) can be rejected in favor of the positive-selection model (M8).
Codon under positive selection with a posterior probability of >95% by Bayes empirical Bayes (BEB) analysis (54).
Expression patterns of the wcs genes.
To explore the expression patterns of the wcs genes, we retrieved transcriptome-sequencing (RNA-seq) data for seven mammal species, that is, Mus musculus (order: Rodentia), Mustela putorius (order: Carnivora), Bos taurus (order: Cetartiodactyla), Hipposideros armiger (order: Chiroptera), Equus caballus (order: Perissodactyla), Monodelphis domestica (order: Didelphimorphia), and Phascolarctos cinereus (order: Diprotodontia) (Fig. 4; see Table S3 in the supplemental material). Similar to the Fv1 gene, the wcs1 gene was expressed in nearly all the tissues studied (Fig. 4). However, the wcs2 gene was expressed in only some of the tissues (Fig. 4). Nevertheless, our results show that both the wcs1 and wcs2 genes are expressed in mammals.
FIG 4.
Expression patterns of the wcs genes. The solid and open circles indicate that the wcs1 gene is expressed or is not expressed, respectively. NA indicates that there are no RNA-seq data for the organ (tissue) of the species. Mmu, Mus musculus; Mpu, Mustela putorius; Bta, Bos Taurus; Har, Hipposideros armiger; Eca, Equus caballus; Mdo, Monodelphis domestica; Pci, Phascolarctos cinereus.
DISCUSSION
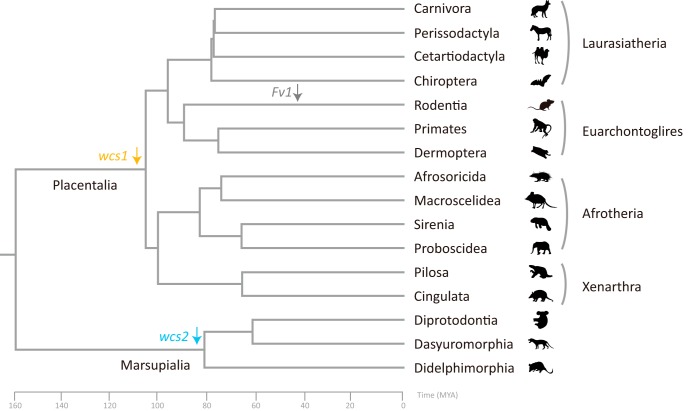
Retroviral env genes have been frequently captured and repurposed for placentation in placental mammals and the viviparous placental Mabuya lizard and are known as the syncytin genes (17, 38, 39). Syncytins arose independently more than 10 times in placental mammals (17, 38, 39). Moreover, a captured env gene was found to be conserved in spiny-rayed fishes (Acanthomorpha) for more than 110 million years (40). However, unlike retroviral env genes, few co-opted retroviral gag genes have been identified to date (22, 24, 25). In this study, we identified two new co-opted retroviral gag genes in mammals, that is, the wcs1 and wcs2 genes, which arose convergently in two major linages of mammals. The wcs1 and wcs2 co-options occurred before the origin of modern placentals (∼100 MYA) and before the origin of modern marsupials (∼80 MYA), respectively (Fig. 5). Both wcs genes are much older than the Fv1 gene, which integrated into the genomes of Muroidea ∼45 MYA (24, 25). The convergent co-option during the early evolution of mammals suggests that co-option of retroviral gag genes might occur more frequently than previously thought.
FIG 5.
Evolutionary history of the wcs genes. The phylogenetic relationships among mammals and the evolutionary time scale are based on TimeTree (33). The wcs1 (orange) and wcs2 (blue) genes originated before the emergence of modern placentals (∼100 MYA) and marsupials (∼80 MYA), respectively. The Fv1 gene (labeled in gray) originated in rodents ∼45 MYA.
Some of the Wcs proteins, for example, the Wcs1 protein of M. putorius (accession no. XP_004767240) and the Wcs1 protein of Camelus bactrianus (accession no. XP_010945334), were annotated as Fv1 proteins, probably because Fv1 and Wcs proteins have significant similarity to each other (Fig. 1B). However, the Wcs proteins arose independently from Fv1, and it might not be appropriate to annotate the Wcs proteins as Fv1. Therefore, caution should be exercised when annotating retroviral gene homologs in vertebrate genome-sequencing projects. We would do better to name a retroviral Gag protein homolog an “uncharacterized protein derived from retroviral Gag protein” rather than Fv1.
Synteny analysis suggests that the wcs1 and wcs2 genes arose once through co-opting retroviral gag genes in placentals and marsupials, respectively. However, the wcs1 gene has been lost or pseudogenized multiple times during the course of placental evolution, and the wcs2 gene was pseudogenized in Dasyuromorphia. Frequent loss or pseudogenization of the wcs genes in some linages, such as primates and Dasyuromorphia, suggests that the wcs genes might not work as essential genes in these linages. On the other hand, strong purifying selection acted on the wcs genes in some mammalian orders, implying that the wcs genes were recruited for important host functions. Moreover, the wcs genes are expressed in a wide range of tissues. All these lines of evidence suggest that the wcs genes might be functional co-opted retroviral gag genes in mammals.
The Fv1 protein blocks the replication of various retroviruses. Historically, the term “restriction factor” was coined following the characterization of the Fv1 gene (29). Intuitively, it can be hypothesized that the gag-derived wcs genes might act as restriction factors, like the Fv1 gene. The evolutionary arms race is expected to drive positive selection in host genes involved in host-virus conflicts (27–30). Nearly all known restriction factors exhibit strong signatures of positive selection (29). On one hand, we found some significant evidence that positive selection might have acted on the wcs1 gene in Perissodactyla and on the wcs2 gene in marsupials, suggesting the corresponding Wcs proteins might be involved in host-virus interactions. However, this result is based on a limited number of sequences and should be confirmed with a larger data set. On the other hand, we found no strong evidence for positive selection in the wcs1 gene in other placental mammal lineages. The wcs gene seems to undergo mainly negative selection in placental mammals, suggesting that some Wcs1 proteins might mediate some biological functions other than antiviral host defense. Indeed, not all Gag-derived proteins are involved in host-virus interactions. For example, the Arc protein, which is derived from the Gag protein of a Ty3/gypsy retrotransposon (different from a retrovirus in the strict sense), mediates RNA transportation across synaptic boutons (20, 21). It is possible that the Wcs proteins mediate some biological processes in mammals other than antiviral host defense, and further work is needed to characterize the functions of the wcs genes.
MATERIALS AND METHODS
Identification of retroviral Gag homologs.
We employed a combined similarity search and phylogenetic analysis approach to screen 261 vertebrate proteomes for homologs of the retroviral Gag proteins (see Table S4 in the supplemental material). First, we performed a similarity search against 261 vertebrate proteomes using the MuERV-L Gag protein (accession no. CAA73250.1) as the query and an E value cutoff of 10−5. Next, the significant hits, Fv1, and the Gag proteins of representative retrotransposons and retroviruses were aligned using MAFFT 7 and then manually refined (41, 42). Initial phylogenetic analyses were performed using an approximate-maximum-likelihood method implemented in FastTree 2 (43). The significant hits that clustered together with retroviral Gag proteins were retrieved for synteny analysis. Among these hits, we found only two clusters of proteins, that is, Wcs1 and Wcs2, which shared conserved synteny.
Phylogenetic analyses.
To further explore the phylogenetic relationships among Wcs, Fv1, and retrovirus Gag proteins, we used 7 Wcs protein sequences, 2 Fv1 protein sequences, and 109 representative retrovirus Gag protein sequences to perform a phylogenetic analysis (see Tables S1 and S2). All the protein sequences were aligned using MAFFT 7 with the L-INS-i strategy (42). The alignment was manually refined and trimmed using TrimAl 1.2b with a gt value of 0.19 to exclude ambiguous regions (44). A phylogenetic tree was inferred using a maximum-likelihood method implemented in IQ-TREE 1.6.0 with an LG-plus-F-plus-G4 amino acid substitution model (45). The best model was chosen using ProtTest 3.4 (46). The branch supports were assessed using ultrafast bootstrap with 1,000 replications (47).
Distribution and synteny of wcs in mammals.
To further explore the distribution and synteny of the wcs genes in mammals, we used the BLASTn or tBLASTn algorithm with an E value cutoff of 10−5 to identify the wcs gene homologs in representative species covering a broad diversity of mammals (Fig. 2 and 3). The synteny of the wcs genes was identified based on gene annotation and a similarity search.
Selection analyses.
To characterize the selection pressure on the wcs genes, we choose five mammalian orders and marsupials, including 78 wcs1 genes and 4 wcs2 genes without premature stop codons or frameshift mutations. We performed selection pressure analysis for each mammal order and marsupials. All the wcs sequences were aligned using MAFFT 7 and then manually refined (42). The gene trees of each order were reconstructed using a maximum-likelihood method implemented in PhyML 3.1 (48). The best-fit substitution models were chosen using jModelTest 2.1.10 (48, 49). The wcs gene trees are generally similar to the species trees. First, we used the SLAC method in the HyPhy package (34, 50) to calculate the dN/dS value for the wcs genes. Next, we used codeml in PAML 4.9 (51) and FEL in the HyPhy package (34, 50) to detect the codons under positive selection. For the PAML analyses, likelihood ratio tests were performed to compare two pairs of site-specific models (a neutral model versus a positive-selection model): M8a versus M8. χ2 analyses were performed using R. For the FEL analyses, we used a P value of 0.05 as the cutoff value to summarize the number of sites under positive selection or negative selection. Finally, we employed the aBSREL method in the HyPhy package to detect branches under positive selection (36, 37, 50).
Expression pattern of the wcs genes.
RNA-seq raw read sequences from 13 tissues of seven species were retrieved for analysis of the expression patterns of the wcs genes (see Table S3). The 13 tissues were brain, liver, kidney, lung, heart, spleen, muscle, testis, blood, lymph node, skin, placenta, and pancreas. The short reads were mapped on the wcs genes with an identity cutoff value of 99%. The wcs genes were defined as being expressed if more than one read was mapped (52).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31701091), the Natural Science Foundation of Jiangsu Province (BK20161016), the Program for Jiangsu Excellent Scientific and Technological Innovation Team (17CXTD00014), and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00542-19.
REFERENCES
- 1.Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 2.Johnson WE. 2015. Endogenous retroviruses in the genomics era. Annu Rev Virol 2:135–159. doi: 10.1146/annurev-virology-100114-054945. [DOI] [PubMed] [Google Scholar]
- 3.Hayward A, Grabherr M, Jern P. 2013. Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc Natl Acad Sci U S A 110:20146–20151. doi: 10.1073/pnas.1315419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayward A, Cornwallis CK, Jern P. 2015. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc Natl Acad Sci U S A 112:464–469. doi: 10.1073/pnas.1414980112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X, Zhao H, Gong Z, Han GZ. 2018. Endogenous retroviruses of non-avian/mammalian vertebrates illuminate diversity and deep history of retroviruses. PLoS Pathog 14:e1007072. doi: 10.1371/journal.ppat.1007072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han GZ, Worobey M. 2012. An endogenous foamy-like viral element in the coelacanth genome. PLoS Pathog 8:e1002790. doi: 10.1371/journal.ppat.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet 6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes EC. 2011. The evolution of endogenous viral elements. Cell Host Microbe 10:368–377. doi: 10.1016/j.chom.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 10.Aiewsakun P, Katzourakis A. 2015. Endogenous viruses: connecting recent and ancient viral evolution. Virology 479-480:26–37. doi: 10.1016/j.virol.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Emerman M, Malik HS. 2010. Paleovirology—modern consequences of ancient viruses. PLoS Biol 8:e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katzourakis A. 2013. Paleovirology: inferring viral evolution from host genome sequence data. Philos Trans R Soc Lond B Biol Sci 368:20120493. doi: 10.1098/rstb.2012.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin EV, Krupovic M. 2018. The depths of virus exaptation. Curr Opin Virol 31:1–8. doi: 10.1016/j.coviro.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Frank JA, Feschotte C. 2017. Co-option of endogenous viral sequences for host cell function. Curr Opin Virol 25:81–89. doi: 10.1016/j.coviro.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewannieux M, Heidmann T. 2013. Endogenous retroviruses: acquisition, amplification and taming of genome invaders. Curr Opin Virol 3:646–656. doi: 10.1016/j.coviro.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Aswad A, Katzourakis A. 2012. Paleovirology and virally derived immunity. Trends Ecol Evol 27:627–636. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Malfavon-Borja R, Feschotte C. 2015. Fighting fire with fire: endogenous retrovirus envelopes as restriction factors. J Virol 89:4047–4050. doi: 10.1128/JVI.03653-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV, McCormick J, Yoder N, Belnap DM, Erlendsson S, Morado DR, Briggs JAG, Feschotte C, Shepherd JD. 2018. The neuronal gene Arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell 172:275–288. doi: 10.1016/j.cell.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson T. 2018. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 172:262–274.e11. doi: 10.1016/j.cell.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best S, Le Tissier P, Towers G, Stoye JP. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 23.Yap MW, Colbeck E, Ellis SA, Stoye JP. 2014. Evolution of the retroviral restriction gene Fv1: inhibition of non-MLV retroviruses. PLoS Pathog 10:e1003968. doi: 10.1371/journal.ppat.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boso G, Buckler-White A, Kozak CA. 2018. Ancient evolutionary origin and positive selection of the retroviral restriction factor Fv1 in muroid rodents. J Virol 92:e00850-18. doi: 10.1128/JVI.00850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young GR, Yap MW, Michaux JR, Steppan SJ, Stoye JP. 2018. Evolutionary journey of the retroviral restriction gene Fv1. Proc Natl Acad Sci U S A 115:10130–10135. doi: 10.1073/pnas.1808516115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worobey M, Bjork A, Wertheim JO. 2007. Point, counterpoint: the evolution of pathogenic viruses and their human hosts. Annu Rev Ecol Evol Syst 38:515–540. doi: 10.1146/annurev.ecolsys.38.091206.095722. [DOI] [Google Scholar]
- 27.Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 28.Sironi M, Cagliani R, Forni D, Clerici M. 2015. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat Rev Genet 16:224–236. doi: 10.1038/nrg3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duggal NK, Emerman M. 2012. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han GZ. 2019. Origin and evolution of the plant immune system. New Phytol 222:70–83. doi: 10.1111/nph.15596. [DOI] [PubMed] [Google Scholar]
- 31.Qi CF, Bonhomme F, Buckler-White A, Buckler C, Orth A, Lander MR, Chattopadhyay SK, Morse HC III.. 1998. Molecular phylogeny of Fv1. Mamm Genome 9:1049–1055. doi: 10.1007/s003359900923. [DOI] [PubMed] [Google Scholar]
- 32.Yan Y, Buckler-White A, Wollenberg K, Kozak CA. 2009. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc Natl Acad Sci U S A 106:3259–3263. doi: 10.1073/pnas.0900181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol 32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosakovsky Pond SL, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol 32:1342–1353. doi: 10.1093/molbev/msv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosakovsky Pond SL, Murrell B, Fourment M, Frost SD, Delport W, Scheffler K. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol 28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cornelis G, Funk M, Vernochet C, Leal F, Tarazona OA, Meurice G, Heidmann O, Dupressoir A, Miralles A, Ramirez-Pinilla MP, Heidmann T. 2017. An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc Natl Acad Sci U S A 114:E10991–E11000. doi: 10.1073/pnas.1714590114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. 2013. Paleovirology of 'syncytins', retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci 368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henzy JE, Gifford RJ, Kenaley CP, Johnson WE. 2017. An intact retroviral gene conserved in spiny-rayed fishes for over 100 My. Mol Biol Evol 34:634–639. doi: 10.1093/molbev/msw262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Llorens C, Fares MA, Moya A. 2008. Relationships of gag-pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis. BMC Evol Biol 8:276. doi: 10.1186/1471-2148-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 7:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 49.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 51.Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 52.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR. 2007. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z, Wong WS, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.