Flaviviruses such as yellow fever virus (YFV), Zika virus (ZIKV), and dengue virus (DENV) are important human pathogens. A common flavivirus trait is the antagonism of interferon (IFN) signaling to enhance viral replication and spread. We report that like ZIKV NS5 and DENV NS5, YFV NS5 binds human STAT2 (hSTAT2) but not mouse STAT2 (mSTAT2), a type I IFN (IFN-α/β) pathway component. Additionally, we show that contrary to what has been demonstrated with ZIKV NS5 and DENV NS5, YFV NS5 is unable to interact with hSTAT2 in murine cells. We demonstrate that mSTAT2 restricts YFV replication in mice and that this correlates with a lack of IFN-α/β-induced YFV NS5 ubiquitination in murine cells. The lack of suitable animal models limits flavivirus pathogenesis, vaccine, and drug research. These data serve as further impetus for the development of an immunocompetent mouse model that can serve as a disease model for multiple flaviviruses.
KEYWORDS: NS5, STAT2, flavivirus, host tropism, interferon, interferon antagonism, yellow fever virus
ABSTRACT
The recent yellow fever virus (YFV) epidemic in Brazil in 2017 and Zika virus (ZIKV) epidemic in 2015 serve to remind us of the importance of flaviviruses as emerging human pathogens. With the current global flavivirus threat, there is an urgent need for antivirals and vaccines to curb the spread of these viruses. However, the lack of suitable animal models limits the research questions that can be answered. A common trait of all flaviviruses studied thus far is their ability to antagonize interferon (IFN) signaling so as to enhance viral replication and dissemination. Previously, we reported that YFV NS5 requires the presence of type I IFN (IFN-α/β) for its engagement with human signal transducer and activator of transcription 2 (hSTAT2). In this manuscript, we report that like the NS5 proteins of ZIKV and dengue virus (DENV), YFV NS5 protein is able to bind hSTAT2 but not murine STAT2 (mSTAT2). Contrary to what has been demonstrated with ZIKV NS5 and DENV NS5, replacing mSTAT2 with hSTAT2 cannot rescue the YFV NS5-STAT2 interaction, as YFV NS5 is also unable to interact with hSTAT2 in murine cells. We show that the IFN-α/β-dependent ubiquitination of YFV NS5 that is required for STAT2 binding in human cells is absent in murine cells. In addition, we demonstrate that mSTAT2 restricts YFV replication in vivo. These data serve as further impetus for the development of an immunocompetent mouse model that can serve as a disease model for multiple flaviviruses.
IMPORTANCE Flaviviruses such as yellow fever virus (YFV), Zika virus (ZIKV), and dengue virus (DENV) are important human pathogens. A common flavivirus trait is the antagonism of interferon (IFN) signaling to enhance viral replication and spread. We report that like ZIKV NS5 and DENV NS5, YFV NS5 binds human STAT2 (hSTAT2) but not mouse STAT2 (mSTAT2), a type I IFN (IFN-α/β) pathway component. Additionally, we show that contrary to what has been demonstrated with ZIKV NS5 and DENV NS5, YFV NS5 is unable to interact with hSTAT2 in murine cells. We demonstrate that mSTAT2 restricts YFV replication in mice and that this correlates with a lack of IFN-α/β-induced YFV NS5 ubiquitination in murine cells. The lack of suitable animal models limits flavivirus pathogenesis, vaccine, and drug research. These data serve as further impetus for the development of an immunocompetent mouse model that can serve as a disease model for multiple flaviviruses.
INTRODUCTION
Yellow fever virus (YFV) is the causative agent of yellow fever (YF), a hemorrhagic disease that is transmitted by several Aedes mosquito species (1). YFV has caused major epidemics since the 17th century, but the disease incidence declined drastically with the introduction of YFV-17D, a highly efficacious live-attenuated vaccine strain. Despite the success of YFV-17D, adverse effects such as vaccine-associated viscerotropic and neurotropic disease have been reported (2, 3). In addition, decreased vaccination in countries where YFV is endemic results in YFV epidemics with high rates of mortality. An estimated 51,000 to 380,000 cases of yellow fever occurred in Africa in 2013 (4). Between December 2015 and October 2016, Angola and the Democratic Republic of Congo had their largest yellow fever outbreak in thirty years, with 962 confirmed infections including 393 reported deaths (5, 6). This outbreak almost obliterated international vaccine stockpiles. This was followed by the resurgence of YFV in Brazil between December 2016 and June 2017 with 777 confirmed cases and a 33.6% fatality ratio for severe YF cases (7). The epidemic continued in Brazil with 464 confirmed human cases with 154 deaths in Brazil between July 2017 and February 2018 (8). Development of YFV-specific antivirals and vaccine alternatives has been hampered by the lack of a tractable small animal model of YF. Both the Asibi wild-type (WT) and 17D vaccine strains of YFV replicate in mice with defects in type I IFN (IFN-α/β) signaling but are unable to efficiently replicate in WT mice (9–11).
IFN-α/β receptor (IFNAR)-deficient mice and signal transducer and activator of transcription 1 (STAT1)-deficient mice, unlike their WT counterparts, exhibit viremia, viscerotropic disease, and mortality when infected with YFV-Asibi (9). This suggests that the IFN-α/β response serves as the major barrier to mouse infection. We have shown that YFV antagonizes IFN-α/β signaling in human and nonhuman primate cells and that the NS5 proteins of both YFV-Asibi and YFV-17D bind and sequester human STAT2 (hSTAT2) after binding of IFN-α/β to its receptor (12, 13). In this paper, we show that YFV NS5 is unable to bind murine STAT2 (mSTAT2) and to inhibit murine IFN-α/β signaling. We also show that mouse STAT2 restricts YFV replication in vivo by showing that YFV-Asibi can replicate in STAT2−/− mice. In contrast to dengue virus (DENV) and Zika virus (ZIKV) (14, 15), YFV NS5 is unable to bind hSTAT2 in murine cells. In addition, we show that YFV NS5 is ubiquitinated in an IFN-α/β-dependent manner in human cells but not murine cells and that this ubiquitination correlates with binding to hSTAT2. This indicates that in addition to STAT2, there are other human-specific factors, most likely involved in YFV NS5 ubiquitination, that are required for the inhibition of IFN-α/β signaling by YFV.
RESULTS
YFV does not inhibit IFN-α/β signaling in murine cells.
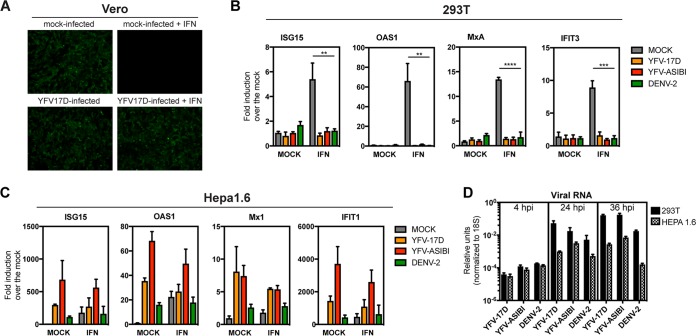
YFV can replicate and inhibit type I IFN signaling in primate cells (12). For example, YFV-17D rescues the replication of the IFN-sensitive green fluorescent protein (GFP)-tagged Newcastle disease virus (NDV) in Vero (African green monkey kidney) cells (Fig. 1A). However, productive infection of YFV-17D and YFV-Asibi in mice requires a defective IFN-α/β signaling pathway. A likely explanation for this is that YFV, though capable of antagonizing human IFN-α/β signaling, is unable to antagonize murine IFN-α/β signaling (9–11). To test if YFV was capable of antagonizing IFN-α/β signaling in mouse cells, 293T (a human cell line) and Hepa1.6 (a mouse cell line) cells were infected with YFV-Asibi or YFV-17D at a multiplicity of infection (MOI) of 10 for 20 h, followed by treatment with 100 units of universal type I IFN for 12 h. We then assessed interferon-stimulated gene (ISG) mRNA expression by reverse transcription-quantitative PCR (qRT-PCR). The experiment was carried out in tandem with DENV2 (16681 strain), which cannot inhibit murine IFN-α/β signaling (14). Both YFV-Asibi and YFV-17D strains as well as DENV2 were able to inhibit ISG expression in human cells (Fig. 1B). However, expression of ISG15, OAS1, Mx1, and IFIT1 was not impaired in type I IFN-treated murine Hepa1.6 cells infected with YFV-Asibi, YFV-17D, or DENV (Fig. 1C). Importantly, 293T and Hepa1.6 cells were able to support viral entry and replication as shown by the increasing levels of intracellular viral RNA at 4, 24, and 36 h after infection (Fig. 1D). This indicates that YFV, like DENV2, is unable to inhibit murine IFN-α/β signaling.
FIG 1.
YFV does not inhibit IFN-α/β signaling in murine cells. (A) Vero cells were either mock infected or infected with YFV-17D at an MOI of 1. At 14 h postinfection, cells were mock treated or treated with 1,000 U/ml universal type I IFN for 12 h and then infected with NDV-GFP. NDV-GFP replication was monitored by fluorescence microscopy 14 h later. 293T (B) or Hepa1.6 (C) cells were infected with YFV-Asibi and YFV-17D at an MOI of 10 for 20 h and then treated with 100 U of universal type I IFN for 12 h. DENV2 (16681 strain) was used as a positive control. ISGs were measured by qRT-PCR as described in the Materials and Methods section. Fold change of the gene expression over the mock is plotted on the graphs. Statistical analysis was performed using unpaired, two-tailed t test versus mock-infected cells (**, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). (D) Levels of Intracellular viral RNA over time by qRT-PCR following infection of 293T or Hepa1.6 cells with the indicated viruses.
YFV NS5 binds hSTAT2 but not mSTAT2 in an IFN-α/β-dependent manner in human cells.
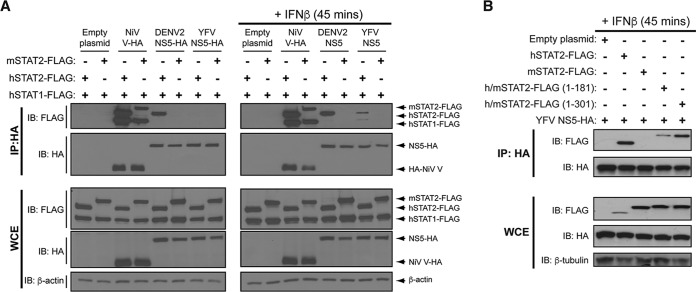
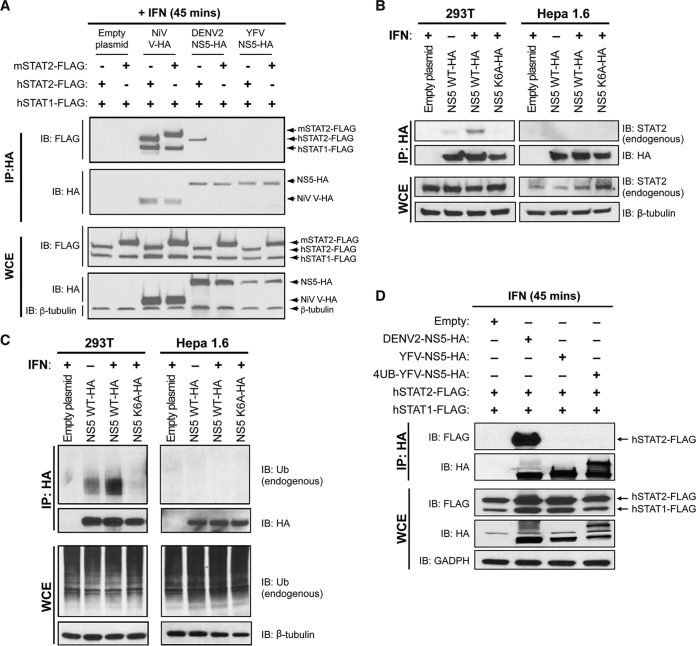
YFV NS5 inhibits the IFN-α/β signaling pathway in human cells by binding STAT2 in an IFN-α/β-dependent manner (12). This allows YFV to replicate optimally in IFN-α/β-treated human cells. The NS5 proteins of DENV and ZIKV bind and degrade hSTAT2 but not mSTAT2, resulting in enhanced DENV and ZIKV replication in human cells but not mouse cells (14–16). As DENV and YFV have a similar host tropism and both inhibit hSTAT2, albeit by different mechanisms, a likely explanation for the failure of YFV to antagonize murine IFN-α/β signaling was an inability of YFV NS5 to bind murine STAT2. YFV NS5, DENV2 NS5, and Nipah virus (NiV) V were overexpressed in U6A cells (a STAT2-deficient human cell line [17]) coexpressing either mSTAT2 or hSTAT2 along with human STAT1 (hSTAT1). Nipah virus binds both mouse and human STAT2 and STAT1 (18), and we observed this in our assay (Fig. 2A). DENV2 NS5 bound hSTAT2 but not mSTAT2 in both untreated and IFN-α/β-treated U6A cells, while YFV NS5 bound hSTAT2 but not mSTAT2 in IFN-α/β-treated U6A cells (Fig. 2A). We also expressed chimeric STAT2 proteins to identify which region of hSTAT2 was necessary for YFV NS5 binding. As has been shown before for DENV NS5 (14), when the first 300 amino acids of mSTAT2 were replaced by those of hSTAT2, mSTAT2 was able to bind YFV NS5 in IFN-α/β-treated cells (Fig. 2B); a chimeric protein with the first 180 amino acids of hSTAT2 also bound YFV NS5 but to a lesser extent (Fig. 2B).
FIG 2.
YFV NS5 associates with hSTAT2 but not mSTAT2. (A) U6A cells were cotransfected with the indicated HA-tagged plasmids and STAT2-FLAG constructs. At 24 h posttransfection, cells were mock stimulated or stimulated with 1,000 U/ml IFN-β for 45 min. Cells were lysed, and immunoprecipitation was performed against the HA epitope followed by Western blot analysis with the indicated antibodies. (B) U6A cells were cotransfected with plasmids expressing YFV NS5-HA and h/mSTAT2-FLAG chimeras as indicated. At 24 h posttransfection, cells were treated as in part A and immunoprecipitation was performed against the HA epitope followed by Western blot analysis with the indicated antibodies.
Murine STAT2 restricts YFV-17D replication in vitro.
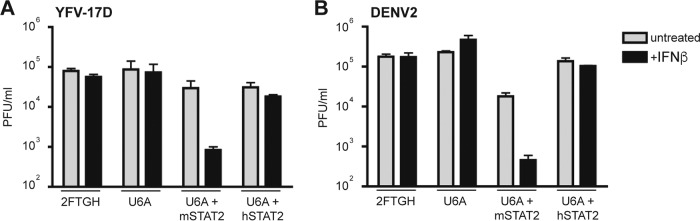
Like DENV, YFV does not bind mSTAT2. In the case of DENV, there is a clear biological consequence; DENV replication is inhibited by IFN-α/β in human cells that express mSTAT2 (14). We therefore tested if mSTAT2 would also inhibit YFV replication in IFN-α/β-treated human cells. Reconstitution of U6A cells with either hSTAT2 or mSTAT2 restores IFN-α/β signaling (14, 19). U6A cells that express exogenous hSTAT2 or mSTAT2, mock-transfected 2fTGH cells, and mock-transfected U6A cells were infected with YFV-17D or DENV2 (MOI 1) and then treated with 100 units of IFN-β 6 h later. In cells that lacked STAT2 (U6A cells) or that contained hSTAT2 (U6A-hSTAT2 and 2fTGH cells), neither DENV2 nor YFV-17D was inhibited by IFN-β treatment. However, both viruses were inhibited by IFN-β in cells that contained mSTAT2 (U6A-mSTAT2) (Fig. 3).
FIG 3.
YFV-17D replication is inhibited by mSTAT2 but not hSTAT2 in an IFN-α/β-dependent manner. 2fTGH, U6A, or U6A cells stably expressing mSTAT2-FLAG or hSTAT2-FLAG (U6A + mSTAT2-FLAG and U6A + hSTAT2-FLAG, respectively) were infected at an MOI of 1 with either YFV-17D (A) or DENV2 (B). The cells were treated with 100 U/ml IFN-β at 6 h postinfection. The cells were harvested 24 h postinfection and viral titers were measured by plaque assay on BHK-21 cells.
Murine STAT2 restricts YFV-Asibi replication in vivo.
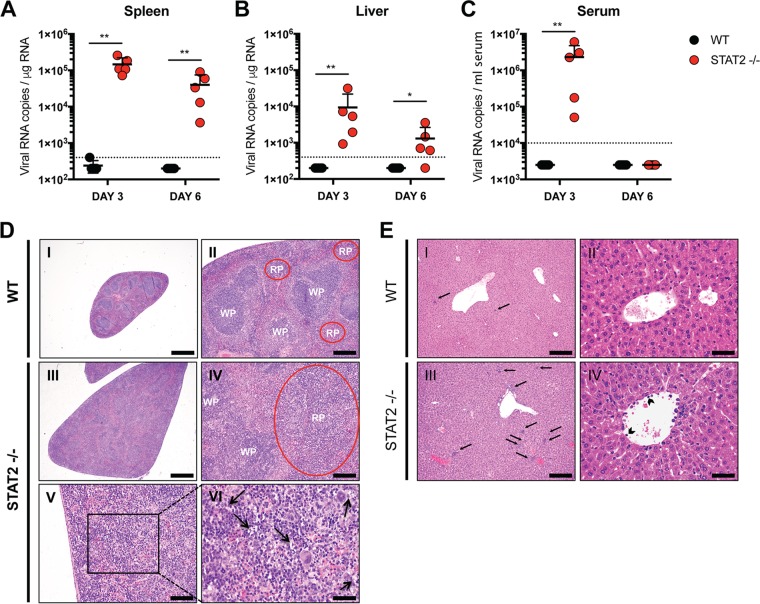
To test what effect STAT2 deficiency would have in vivo, we infected WT and STAT2-deficient mice with 104 PFU of YFV-Asibi. Viral RNA levels in the spleen, liver, and serum were measured at days 3 and 6 postinfection by qRT-PCR. Virus was detected in the spleens (Fig. 4A) and livers (Fig. 4B) of STAT2 knockout mice on days 3 and 6 but was undetectable in WT mice at any time point tested. Virus was also observed in the serum of STAT2-deficient mice at day 3, but the viral RNA declined to undetectable levels by day 6 (Fig. 4C). No virus was detected in the serum of WT mice. No mice died by day 6 postinfection with the dose of virus injected, though enlarged spleens were clearly observed in STAT2-deficient mice (Fig. 4D). Tissues harvested from STAT2−/− mice were histologically distinguishable from WT mouse tissues (Fig. 4D and E). In contrast to WT mouse spleens, STAT2−/− mouse spleens lacked distinct marginal zones and germinal centers. In addition, there was marked extramedullary hematopoiesis (EMH) and tingible body macrophages containing scant basophilic cell debris in the expanded red pulp (Fig. 4D). The STAT2−/− livers revealed moderate EMH composed of erythroid and myeloid lineages as well as megakaryocytes. Leukocytes were often observed lining and adherent to the endothelium of central veins and portal vasculature of the STAT2−/− mice (Fig. 4E). These results indicate that mSTAT2 restricts YFV-Asibi replication in vivo.
FIG 4.
Murine STAT2 restricts YFV-Asibi replication in vivo. C57BL/6 WT and STAT2−/− mice were infected via hock injection with 104 PFU of YFV-Asibi. Spleen (A), liver (B), and serum (C) were collected at the indicated times, and viral RNA was quantified by qRT-PCR as described in the Materials and Methods section (n = 5, Mann-Whitney test; *, P < 0.05; **, P < 0.01). Dotted lines represent the limit of detection of the assay. (D and E) Histological findings in YFV-Asibi-infected mice. WT and STAT2−/− mice were infected as described above. H&E-stained sections of spleen (D) and liver (E) at day 6 postinfection are shown. (D) No significant microscopic lesions are noted in the WT spleens (n = 5) (I and II). Splenomegaly was observed in all STAT2−/− spleens (n = 5) (III). No distinct marginal zone and germinal centers are observed in the spleens of STAT2−/− mice (IV). All STAT2−/− spleens exhibit marked expansion of red pulp (RP) by marked EMH, which is likely to be a reactive change to the infection. There are frequent tingible-body macrophages (black arrows) containing scant basophilic cell debris in the expanded RP (V and VI). (E) No significant microscopic lesions are noted in the WT livers (n = 5) (I and II). All STAT2−/− livers (n = 5) exhibit moderate EMH (black arrows) composed of erythroid and myeloid lineages as well as megakaryocytes. Scant necrotic debris is rarely present within these foci (III). Leukocytes are often noted lining and adherent to the endothelium of central veins and portal vasculature of the STAT2−/− mice (arrowheads) (IV).
YFV NS5 does not bind hSTAT2 in murine cells.
Though mSTAT2 functions similarly to hSTAT2 in response to type I IFN treatment in both human and murine cells, these orthologs interact with different proteins (19). It was therefore possible that expression of mSTAT2 in human cells could yield a different result from expression of mSTAT2 in mouse cells. When we expressed hSTAT2 and hSTAT1 in mouse Hepa1.6 cells, DENV2 NS5 bound hSTAT2, and NiV V bound both mSTAT2 and hSTAT2, as well as hSTAT1, which is consistent with published studies (18, 20). However, no interaction between YFV NS5 and hSTAT2 was observed even when the cells were treated with universal type I IFN (Fig. 5A). Though hSTAT1 was overexpressed in conjunction with hSTAT2 in this assay, YFV NS5 was unable to bind hSTAT2 in IFN-α/β-treated murine cells. This indicates that other human-specific factors are required for YFV NS5 to inhibit IFN-α/β signaling.
FIG 5.
YFV NS5 does not associate with hSTAT2 in murine cells. Hepa1.6 cells (A), 293T and Hepa1.6 cells (B, C), and Hepa1.6 cells (D) were cotransfected with the indicated HA-tagged plasmids and STAT-FLAG constructs. Twenty-four hours posttransfection, cells were stimulated with 1,000 U/ml universal type I IFN for 45 min. Cells were lysed, and immunoprecipitation was performed against the HA epitope followed by Western blot analysis with the indicated antibodies.
IFN-α/β-dependent K6-polyubiquitination of YFV NS5 occurs in human cells but not murine cells and is necessary for the YFV NS5-STAT2 interaction.
We previously showed that IFN-α/β-induced K63-linked polyubiquitination at a lysine position 6 in the N-terminal region of YFV NS5 is necessary for YFV NS5-hSTAT2 interaction (12). A YFV NS5 protein lacking the lysine residue at position 6 is unable to interact with hSTAT2 (12). We evaluated whether any differences in IFN-induced YFV NS5 ubiquitination in murine cells compared to that in human cells could account for the lack of YFV NS5 binding to hSTAT2 in murine cells. 293T cells and murine Hepa1.6 cells were transfected with empty vectors or plasmids expressing YFV NS5 or the YFV NS5-K6A mutant, which is unable to bind hSTAT2 (12). Cells were then mock treated or treated with universal type I IFN. As expected, coimmunoprecipitation assays demonstrated that ectopically expressed YFV NS5 but not NS5-K6A bound endogenous STAT2 in human cells in an IFN-dependent fashion (Fig. 5B). In addition, we observed an increase in ubiquitination of YFV NS5 but not NS5-K6A in human cells upon the addition of IFN-α/β, which is consistent with published studies (12). However, this type I IFN-dependent increase in YFV NS5 ubiquitination as well as endogenous STAT2 binding was not observed in murine cells (Fig. 5B and C).
We previously showed that covalently attaching an ubiquitin chain to the N terminus of the YFV NS5 K6R protein resulted in the ability of the YFV NS5 K6R mutant protein to rescue binding to human STAT2 in a type I IFN-dependent manner in human cells (12). This covalently attached ubiquitin chain may compensate for the lack of NS5 K6 ubiquitination, thus allowing mSTAT2 binding, or it may provide additional lysine residues that would act as a scaffold for NS5 K63 polyubiquitination. To examine whether this YFV NS5 covalently attached to ubiquitin would rescue hSTAT2 binding in murine cells, Hepa1.6 cells were transfected with empty vectors or plasmids expressing YFV NS5 and 4UB YFV NS5 along with hSTAT2 and hSTAT1. DENV2 NS5 that is able to bind hSTAT2 in murine cells was used as a positive control. As expected, DENV2 NS5 was able to coimmunoprecipitate hSTAT2 in murine cells; however, expression of the 4UB YFV NS5 construct did not rescue hSTAT2 binding in cells treated with universal type I IFN (Fig. 5D). This further suggests that human-specific factors are required for efficient YFV NS5 K6 ubiquitination and STAT2 binding.
DISCUSSION
Flaviviruses like DENV, YFV, and ZIKV are restricted to primates, while others such as West Nile virus (WNV) and tick-borne encephalitis (TBEV) have the ability to replicate in murine species. The ability of DENV and ZIKV NS5 to bind and degrade hSTAT2 and not the mSTAT2 protein provides the basis for their host tropism (14, 15). Specifically, the coiled-coil domain is required for this interaction of DENV NS5 and hSTAT2. All STAT proteins contain an N-terminal region that includes a coiled-coil domain, which is important for protein-protein interaction; a central DNA binding domain; and a C-terminal region containing the SH2 and transcriptional activation domains. Overall, human and murine STAT proteins are highly identical at the amino acid level, with about 90% similarity for the STAT1 protein. However, hSTAT2 and mSTAT2 are less than 70% identical at the amino acid level, with most of these differences due to the highly divergent C terminus.
In this manuscript, we have shown that YFV NS5, like DENV NS5 and ZIKV NS5, inhibits IFN signaling in a species-specific manner. YFV NS5 binds hSTAT2 after treatment with type I IFN but is unable to bind mSTAT2. In addition, we show that YFV NS5 is unable to bind hSTAT2 in mouse cells and that YFV NS5 is not ubiquitinated in a type I IFN-dependent manner in murine cells. This suggests the need for additional host factors for efficient YFV NS5 ubiquitination and STAT2 binding in mouse cells. In addition, we used chimeric human-mouse STAT2 proteins to map the region of STAT2 required for interaction with YFV NS5. Our experiments revealed that YFV NS5, like DENV NS5, requires the coiled-coil domain of hSTAT2 located at its N terminus for association. The sequences of the first 301 residues from hSTAT2 and mSTAT2 are 65% identical, and their predicted 3D structures are quite similar. mSTAT2 is predicted to contain an interruption in one of the alpha helices that is absent in hSTAT2. However, when we replaced mouse STAT2 residues 69 to 80 with the human STAT2 alpha helix, we could not rescue YFV NS5 binding, and a human STAT2 chimera with the interrupted mouse alpha helix was still able to bind YFV NS5 (data not shown). Taken together these data suggest that the residues that contribute to STAT2 binding lie between amino acid 80 to 181 in the coiled-coil domain of hSTAT2.
We show that the YFV-17D strain is sensitive to the effects of type I IFN in human cells that express mSTAT2 but not hSTAT2. Hence, mSTAT2 inhibits the YFV-17D strain in an IFN-dependent manner. Additional in vivo experiments using STAT2-deficient mice confirmed that mouse STAT2 limits YFV-Asibi replication in mice. Similar observations were made with DENV2 (14). These data suggest that the interaction of YFV, ZIKV, and DENV NS5 proteins with hSTAT2 but not murine STAT2 restrict the host tropism of these viruses to primates.
The study of host tropism determinants can inform the design of animal models of viral disease. In some cases, the expression of a missing human host factor can lead to productive viral infection and disease as is the case for the poliovirus mouse model (21). In other cases, expression of host factors can allow limited viral replication that is hampered by the lack of other components. This is the case for hepatitis C virus that can infect mice that express various human receptors but cannot achieve a full replication cycle unless parts of the innate immune system are removed (22, 23). In the case of DENV, it is predicted that a mouse that expresses hSTAT2 in place of mSTAT2 would allow some amount of viral replication, as the IFN-α/β response of the mouse would be targetable by these viruses, though other aspects of the murine immune system would prevent robust replication (14, 24). Recent work involving our group has shown that mouse-adapted Zika virus replicates in hSTAT2 gene knock-in mice (25). A hSTAT2 knock-in mouse does not appear to be a viable option for an immunocompetent YFV mouse model, as YFV NS5 is incapable of binding hSTAT2 in murine cells, indicating that other host-specific factors are necessary for this interaction to occur.
MATERIALS AND METHODS
Cell lines and viruses.
The 293T (a human embryonic kidney line), Hepa1.6 (a mouse liver line), previously described 2fTGH (a human fibrosarcoma line), and the 2fTGH derivative, U6A (17), as well as the previously described U6A-hSTAT2-FLAG and U6A-mSTAT2-FLAG cell lines (14) were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. High titer stocks of DENV2 (DENV2 16681) were obtained by passage in C6/36 cells. High titer stocks of the wild-type YFV Asibi strain (YFV-Asibi) and live-attenuated YFV-17D strain were obtained by passage in BHK-21 cells.
Plasmids.
All flavivirus genes were cloned into the mammalian expression vector pCAGGS (chicken-β-actin promoter). Primer sequences used in the generation of the constructs are available upon request. The HA-NiV V and mSTAT2 plasmids were gifts from Megan Shaw and David Levy, respectively. The STAT chimeras were described previously (14).
Antibodies and cytokines.
The following antibodies were utilized in this study: anti-hemagglutinin (HA) (H9658; Sigma-Aldrich, USA), anti-FLAG (F7425; Sigma-Aldrich), anti-STAT2 (C-20) (sc-476; Santa Cruz Biotechnology, USA), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (G-9) (sc-365062; Santa Cruz Biotechnology), anti-ubiquitin (P4D1) (BML-PW0930; Enzo, USA), anti-β-tubulin (T8328; Sigma-Aldrich), and anti-β-actin (A5441; Sigma-Aldrich, USA). Universal type I IFN and human IFN-β were obtained from PBL InterferonSource, USA.
Transfections.
All transfections were performed using Lipofectamine 2000 (Invitrogen, USA) in Opti-MEM (Invitrogen). HEK293T cells were transfected using a 1:1 plasmid DNA (μg) to Lipofectamine 2000 (μl) ratio. Hepa1.6 cells, as well as the 2fTGH and 2fTGH derivatives, were transfected using a 1:3 DNA to Lipofectamine 2000 ratio.
Cell culture infections.
For virus infection, monolayers of cells were initially adsorbed with virus at the indicated multiplicity of infection (MOI) for 1 h at 37°C. After adsorption, unbound virus was removed from cells, and the cells were subsequently maintained in DMEM with 10% FBS at 37°C. Viral RNA levels were measured by quantitative PCR (qPCR). The YFV NS5 primers were described previously (12). Quantification of YFV-Asibi RNA copies per microgram of total RNA was performed against a standard curve of in vitro transcribed YFV NS5 RNA.
Mouse infections.
Twenty microliters of virus inoculum containing 1 × 104 PFU YFV-Asibi was administered via hock injection into 20 WT C57BL/6 and 20 STAT2−/− C57BL/6 mice. Mock-infected mice received 20 μl phosphate-buffered saline (PBS). Organs were harvested at day 3 and day 6 after infection. Animal research was conducted under the guidance of the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee.
Histopathology.
Mice were euthanized on day 6 postinfection, and liver and spleen tissues were fixed in 10% neutral buffered formalin for 48 h. Paraffin-embedded hematoxylin and eosin (H&E)-stained sections were evaluated by an experienced comparative pathologist from the Center of Comparative Medicine and Surgery in the Icahn School of Medicine at Mount Sinai. The pathologist was blinded to the groups in order to prevent bias.
Quantitative RT-PCR for YFV replication.
Total RNA was isolated from tissues lysed and homogenized in TRIzol reagent (Invitrogen) using the Direct-zol RNA MiniPrep Plus kit (Zymo Research). Reverse transcription was performed with the high-capacity cDNA reverse transcription kit (Applied Biosystems). qPCR was done in triplicate using SYBR green I master mix (Roche) in a Roche LightCycler 480. The YFV NS5 primers were described previously (12). Quantification of YFV-Asibi RNA copies per microgram of total RNA was performed against a standard curve of in vitro transcribed YFV NS5 RNA.
Quantitative RT-PCR for interferon-stimulated gene transcription.
Total RNA was isolated from cells lysed in TRIzol reagent (Invitrogen) using the Direct-zol RNA MiniPrep Plus kit (Zymo Research). Reverse transcription was performed with the high-capacity cDNA reverse transcription kit (Applied Biosystems). qPCR was carried out in triplicate using SYBR green I master mix (Roche) in a Roche LightCycler 480. Relative mRNA values were calculated using the ΔΔCT method with 18S as an internal control and shown as fold change by normalizing to mock control. The following primers were used: hISG15 Fwd, 5′-TCCTGGTGAGGAATAACAAGGG-3′; hISG15 Rev, 5′-GTCAGCCAGAACAGGTCGTC-3′; mISG15 Fwd, 5′-GGTGTCCGTGACTAACTCCAT-3′; mISG15 Rev, 5ʹ-TGGAAAGGGTAAGACCGTCCT-3′; hOAS1 Fwd, 5′-GATCTCAGAAATACCCCAGCCA-3′; hOAS1 Rev, 5′-AGCTACCTCGGAAGCACCTT-3′; mOAS1 Fwd, 5′-ATGGAGCACGGACTCAGGA-3′; mOAS1 Rev, 5′-TCACACACGACATTGACGGC-3′; hMXA Fwd, 5′-GTGGCTGAGAACAACCTGTG-3′; hMXA Rev, 5′-GGCATCTGGTCACGATCCC-3′; mMX1 Fwd, 5′-GACCATAGGGGTCTTGACCAA-3′; mMX1 Rev, 5′-AGACTTGCTCTTTCTGAAAAGCC-3′; hIFIT3 Fwd, 5′-AGAAAAGGTGACCTAGACAAAGC-3′; hIFIT3 Rev, 5′-CCTTGTAGCAGCACCCAATCT-3′; mIFIT1 Fwd, 5′-CTGAGATGTCACTTCACATGGAA-3′; mIFIT1 Rev, 5ʹ-GTGCATCCCCAATGGGTTCT-3′; 18S-rRNA Fwd, 5′-GTAACCCGTTGAACCCCATT-3′; 18S-rRNA Rev, 5′-CCATCCAATCGGTAGTAGCG-3′.
Immunoprecipitation and immunoblot analysis.
Twenty-four hours after transfection, cells were mock stimulated or stimulated with 1,000 U/ml IFN for 45 min. Lysis and washes were performed using buffer containing 50 mM Tris (pH 8.0), 280 mM NaCl, 0.2 mM EDTA, 0.5% NP-40, 10% glycerol, 1 mM sodium orthovanadate, and a protease inhibitor mixture (Complete; Roche Diagnostics, Germany) at 4°C. Whole-cell lysates were used for immunoprecipitation with the indicated antibodies. In general, 1 to 2 μg of antibody was added to 1 ml of cell lysate and incubated overnight at 4°C followed by incubation with protein A/G agarose beads for 2 h. Immunoprecipitates were washed extensively and eluted from the beads by boiling with Laemmli sample buffer, separated on a polyacrylamide gel, and transferred onto a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked for 1 h at room temperature in Tris-buffered saline (TBS) containing 0.5% Tween 20 and either 5% nonfat milk or 5% bovine serum albumin (BSA) and then incubated overnight at 4°C in this buffer containing the appropriate primary antibody. PVDF membranes were washed, incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature, washed 3 times, and finally developed with ECL (Amersham Biosciences, USA).
ACKNOWLEDGMENTS
We thank Brandy Russell for providing the YFV-Asibi virus and Christian Schindler for providing the original STAT2−/− mice. We thank Virginia Gillespie and the Center for Comparative Medicine and Surgery, Icahn School of Medicine at Mount Sinai, for their excellent assistance. The HA-NiV V and mSTAT2 plasmids were gifts from Megan Shaw and David Levy, respectively.
These studies were partly funded by NIAID grants U19AI118610 and R21AI129486 to A.G.-S.
REFERENCES
- 1.Butler D. 2016. Fears rise over yellow fever's next move. Nature 532:155–156. doi: 10.1038/532155a. [DOI] [PubMed] [Google Scholar]
- 2.Seligman SJ, Casanova JL. 2016. Yellow fever vaccine: worthy friend or stealthy foe? Expert Rev Vaccines 15:681–691. doi: 10.1080/14760584.2016.1180250. [DOI] [PubMed] [Google Scholar]
- 3.de Menezes Martins R, da Luz Fernandes Leal M, Homma A. 2015. Serious adverse events associated with yellow fever vaccine. Hum Vaccin Immunother 11:2183–2187. doi: 10.1080/21645515.2015.1022700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garske T, Van Kerkhove MD, Yactayo S, Ronveaux O, Lewis RF, Staples JE, Perea W, Ferguson NM, Yellow Fever Expert Committee . 2014. Yellow fever in Africa: estimating the burden of disease and impact of mass vaccination from outbreak and serological data. PLoS Med 11:e1001638. doi: 10.1371/journal.pmed.1001638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. 2016. Situation report: yellow fever. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.World Health Organization. 2018. Yellow fever fact sheet. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs100/en/. [Google Scholar]
- 7.Faria NR, Kraemer MUG, Hill SC, Goes de Jesus J, Aguiar RS, Iani FCM, Xavier J, Quick J, Du Plessis L, Dellicour S, Thézé J, Carvalho RDO, Baele G, Wu C-H, Silveira PP, Arruda MB, Pereira MA, Pereira GC, Lourenço J, Obolski U, Abade L, Vasylyeva TI, Giovanetti M, Yi D, Weiss DJ, Wint GRW, Shearer FM, Funk S, Nikolay B, Fonseca V, Adelino TER, Oliveira MAA, Silva MVF, Sacchetto L, Figueiredo PO, Rezende IM, Mello EM, Said RFC, Santos DA, Ferraz ML, Brito MG, Santana LF, Menezes MT, Brindeiro RM, Tanuri A, Dos Santos FCP, Cunha MS, Nogueira JS, et al. 2018. Genomic and epidemiological monitoring of yellow fever virus transmission potential. Science 361:894–899. doi: 10.1126/science.aat7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. 2018. Yellow fever – Brazil. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/don/27-february-2018-yellow-fever-brazil/en/. Accessed 14 May 2018. [Google Scholar]
- 9.Meier KC, Gardner CL, Khoretonenko MV, Klimstra WB, Ryman KD. 2009. A mouse model for studying viscerotropic disease caused by yellow fever virus infection. PLoS Pathog 5:e1000614. doi: 10.1371/journal.ppat.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thibodeaux BA, Garbino NC, Liss NM, Piper J, Blair CD, Roehrig JT. 2012. A small animal peripheral challenge model of yellow fever using interferon-receptor deficient mice and the 17D-204 vaccine strain. Vaccine 30:3180–3187. doi: 10.1016/j.vaccine.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson AK, Pfeiffer JK. 2013. Dynamic viral dissemination in mice infected with yellow fever virus strain 17D. J Virol 87:12392–12397. doi: 10.1128/JVI.02149-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurent-Rolle M, Morrison J, Rajsbaum R, Macleod JML, Pisanelli G, Pham A, Ayllon J, Miorin L, Martínez-Romero C, tenOever BR, García-Sastre A. 2014. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe 16:314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miorin L, Maestre AM, Fernandez-Sesma A, García-Sastre A. 2017. Antagonism of type I interferon by flaviviruses. Biochem Biophys Res Commun 492:587–596. doi: 10.1016/j.bbrc.2017.05.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, García-Sastre A. 2010. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe 8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, García-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LCF, Fernandez-Sesma A, García-Sastre A. 2013. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog 9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung S, Qureshi SA, Kerr IM, Darnell JE Jr, Stark GR. 1995. Role of STAT2 in the alpha interferon signaling pathway. Mol Cell Biol 15:1312–1317. doi: 10.1128/MCB.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez JJ, Parisien JP, Horvath CM. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J Virol 76:11476–11483. doi: 10.1128/JVI.76.22.11476-11483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park C, Lecomte MJ, Schindler C. 1999. Murine Stat2 is uncharacteristically divergent. Nucleic Acids Res 27:4191–4199. doi: 10.1093/nar/27.21.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashour J, Laurent-Rolle M, Shi P-Y, García-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren RB, Costantini F, Gorgacz EJ, Lee JJ, Racaniello VR. 1990. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell 63:353–362. doi: 10.1016/0092-8674(90)90168-E. [DOI] [PubMed] [Google Scholar]
- 22.Dorner M, Horwitz JA, Donovan BM, Labitt RN, Budell WC, Friling T, Vogt A, Catanese MT, Satoh T, Kawai T, Akira S, Law M, Rice CM, Ploss A. 2013. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature 501:237–241. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, Law M, Rice CM, Ploss A. 2011. A genetically humanized mouse model for hepatitis C virus infection. Nature 474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. 2011. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog 7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorman MJ, Caine EA, Zaitsev K, Begley MC, Weger-Lucarelli J, Uccellini MB, Tripathi S, Morrison J, Yount BL, Dinnon KH III, Ruckert C, Young MC, Zhu Z, Robertson SJ, McNally KL, Ye J, Cao B, Mysorekar IU, Ebel GD, Baric RS, Best SM, Artyomov MN, Garcia-Sastre A, Diamond MS. 2018. An immunocompetent mouse model of Zika virus infection. Cell Host Microbe 23:672–685. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]