BK polyomavirus (BKPyV) is an emerging pathogen that reactivates in immunosuppressed organ transplant patients. We wanted to understand why BKPyV-induced activation of the DNA damage response (DDR) enhances viral titers and prevents host DNA damage. Here, we show that the virus activates the DNA damage response in order to keep the infected cells in S phase to replicate the viral DNA. The source of DNA damage was due to actively replicating cells with uncondensed chromosomes entering directly into mitosis when the DDR was inhibited in BKPyV-infected cells. This study clarifies the previously enigmatic role of the DDR during BKPyV infection by demonstrating that the virus activates the DDR to maintain the cells in S phase in order to promote viral replication and that disruption of this cell cycle arrest can lead to catastrophic DNA damage for the host.
KEYWORDS: ATM, ATR, BK polyomavirus, cell cycle, DNA damage, DNA damage response, premature mitosis, Wee1
ABSTRACT
BK polyomavirus (PyV) is a major source of kidney failure in transplant recipients. The standard treatment for patients with lytic BKPyV infection is to reduce immunosuppressive therapy, which increases the risk of graft rejection. PyVs are DNA viruses that rely upon host replication proteins for viral genome replication. A hallmark of PyV infection is activation of the DNA damage response (DDR) to prevent severe host and viral DNA damage that impairs viral production by an unknown mechanism. Therefore, we sought to better understand why BKPyV activates the DDR through the ATR and ATM pathways and how this prevents DNA damage and leads to increased viral production. When ATR was inhibited in BKPyV-infected primary kidney cells, severe DNA damage occurred due to premature Cdk1 activation, which resulted in mitosis of cells that were actively replicating host DNA in S phase. Conversely, ATM was required for efficient entry into S phase and to prevent normal mitotic entry after G2 phase. The synergistic activation of these DDR kinases promoted and maintained BKPyV-mediated S phase to enhance viral production. In contrast to BKPyV infection, DDR inhibition did not disrupt cell cycle control in uninfected cells. This suggests that DDR inhibitors may be used to specifically target BKPyV-infected cells.
IMPORTANCE BK polyomavirus (BKPyV) is an emerging pathogen that reactivates in immunosuppressed organ transplant patients. We wanted to understand why BKPyV-induced activation of the DNA damage response (DDR) enhances viral titers and prevents host DNA damage. Here, we show that the virus activates the DNA damage response in order to keep the infected cells in S phase to replicate the viral DNA. The source of DNA damage was due to actively replicating cells with uncondensed chromosomes entering directly into mitosis when the DDR was inhibited in BKPyV-infected cells. This study clarifies the previously enigmatic role of the DDR during BKPyV infection by demonstrating that the virus activates the DDR to maintain the cells in S phase in order to promote viral replication and that disruption of this cell cycle arrest can lead to catastrophic DNA damage for the host.
INTRODUCTION
BK polyomavirus (BKPyV) persistently infects the genitourinary tract of >90% of all humans (1). Following immunosuppression, viral reactivation can cause diseases, such as nephropathy, which is ultimately responsible for approximately 10% of kidney transplant rejections (1). Despite the high carriage rate of BKPyV in the human population, BKPyV-associated cancer is rare though high-grade, intrarenal, and transitional cell carcinomas have been linked to clonal integration of replication-incompetent BKPyV (2). There are no FDA-approved therapies to combat BKPyV reactivation, its associated diseases, or PyV-linked malignancies.
PyVs are the group of DNA tumor viruses with the smallest genome, encoding only five to eight proteins (1). Because of this, PyVs rely on host DNA replication machinery to amplify the viral genome. To hijack the host S-phase proteins, PyVs drive the host into S phase by expressing large tumor antigen (TAg) and a splice variant, small TAg (tAg). TAg inactivates the S-phase suppressor, retinoblastoma protein (pRb) (3, 4), while tAg inactivates protein phosphatase 2A (PP2A), a master phosphatase that is essential for cell cycle progression (5, 6). Together, these oncogenes commandeer the host cell, promote S-phase entry, and induce multiple rounds of viral and host chromatin replication (7). In addition to promoting S phase, all studied PyVs activate the DNA damage response (DDR) during infection, which is required for efficient amplification of the viral genome, production of virions, and protection of the host from severe DNA damage linked to infection (7–9).
The DDR is a cellular response to genotoxic stress that governs three processes: DNA repair, cell cycle arrest, and cell death (10). The DDRs that impact PyV infection are regulated by ataxia-telangiectasia mutated (ATM) and ATM-Rad 3-related (ATR). ATM coordinates homologous recombination to repair double-stranded breaks. Replication factor A (RPA) association with single-stranded DNA (ssDNA) activates the ATR pathway, causing replication to slow, and mediates recovery from replication fork collapse (11). Both ATR and ATM mediate cell cycle arrest by activating the downstream checkpoint kinases (Chk), Chk1 and Chk2, respectively. Together, ATM and ATR phosphorylate potentially hundreds of proteins in response to DNA damage (12). Recent proteomic profiling of the nuclear compartment revealed that most of the cellular pathways that are upregulated during BKPyV infection are involved in DNA repair and cell cycle arrest (13). The current paradigm is that ATM and ATR are important for promoting viral chromatin replication by preventing the accumulation of viral replication intermediates (9). However, when the DDR is inactivated, only a small fraction of the total viral genomes are affected, which cannot account for the dramatic decrease in viral titers. Furthermore, TAg expression from a mutant virus that was not competent for viral DNA replication still induced DNA damage and failed to activate the DDR despite triggering DNA damage. This suggests that neither ATM nor ATR is activated by DNA damage during a BKPyV infection. Instead, BKPyV viral chromatin replication triggers DDR activation early in infection (14), suggesting that activation of the DDR by viral DNA replication may be to prevent the DNA damage rather than to mediate its repair. These published findings suggest that there is an alternative mechanism by which DDR activation enhances viral infection that can be characterized by identifying the origin of host DNA damage that occurs when the DDR is inhibited during infection.
Here, we identified the source of host DNA damage that occurs during BKPyV infection when the DDR is not activated. We found that the DDR was required to prevent host DNA damage rather than to repair existing damage caused intrinsically by BKPyV-induced replication stress. Specifically, we found that ATR prevented premature mitosis during BKPyV infection by blocking activation of cyclin-dependent kinase 1 (Cdk1). Consistent with this finding, blocking premature mitosis by Cdk1-depletion prevented DNA damage and rescued viral titers that were attenuated by both ATM and ATR inhibition. These findings suggest that the DDR was required late during infection when the majority of viral replication and assembly occurs to prolong S phase and prevent mitotic entry, thus extending the window for viral production. These studies also revealed differences between the roles of ATM and ATR during a BKPyV infection. ATM was required for efficient S-phase entry as well as for prolonging the S phase since inhibiting ATM drove cells into regulated mitosis following S-phase termination. In contrast, inhibition of ATR resulted in a dramatic shift of the population of cells that entered mitosis while actively synthesizing DNA, resulting in severe DNA damage. Taken together, these findings demonstrate that ATR and ATM function synergistically to maintain cells in S phase for BKPyV replication.
RESULTS
BKPyV activation of the DDR induced cell cycle arrest.
Primary renal proximal tubule epithelial (RPTE) cells were used to study lytic BKPyV host-pathogen interactions in cells with intact cell cycle control pathways (15, 16). This avoids complications with using immortalized cells that commonly harbor mutations and epigenetic modifications in the DDR and cell cycle control pathways.
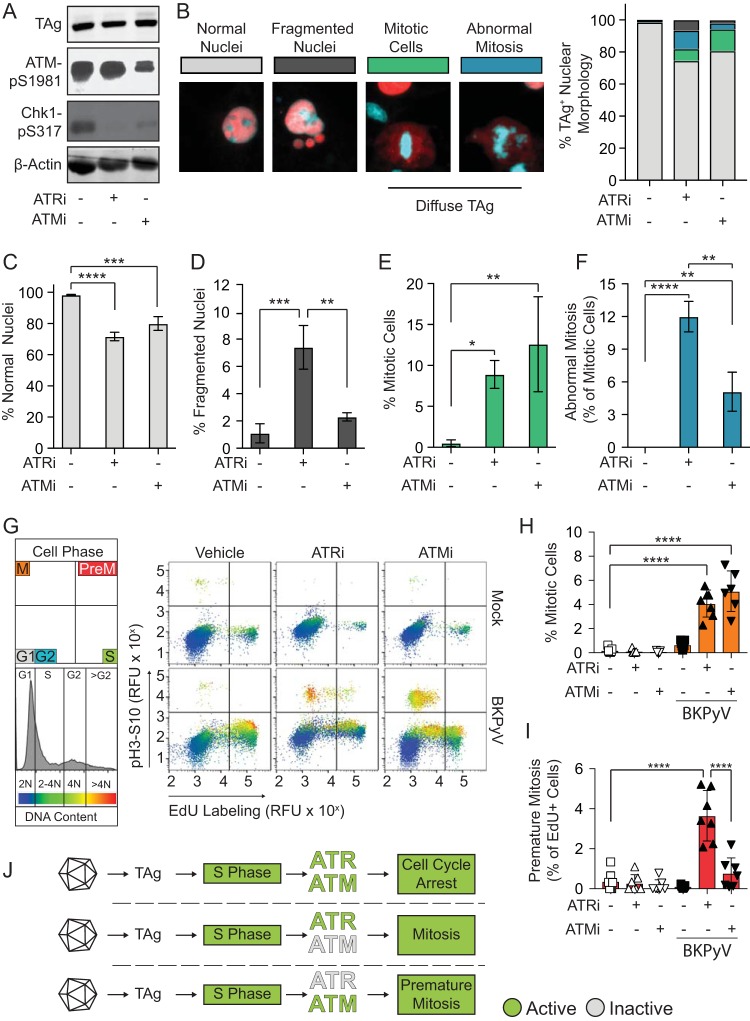
To understand the origin of the BKPyV-dependent DNA damage that occurs on host chromatin when the DDR is inactivated, we used potent and specific chemical inhibitors (17, 18). As expected, ATM and ATR inhibition (ATMi and ATRi) reduced the BKPyV-induced DDR activation as measured by reduced Chk1 phosphorylation at Ser317 (pChk1S317) and ATM autophosphorylation at Ser1981 (pATMS1981) (Fig. 1A). The decreased Chk1 phosphorylation observed in the ATMi treatment group is likely due to cross talk between the ATM and ATR pathways (19).
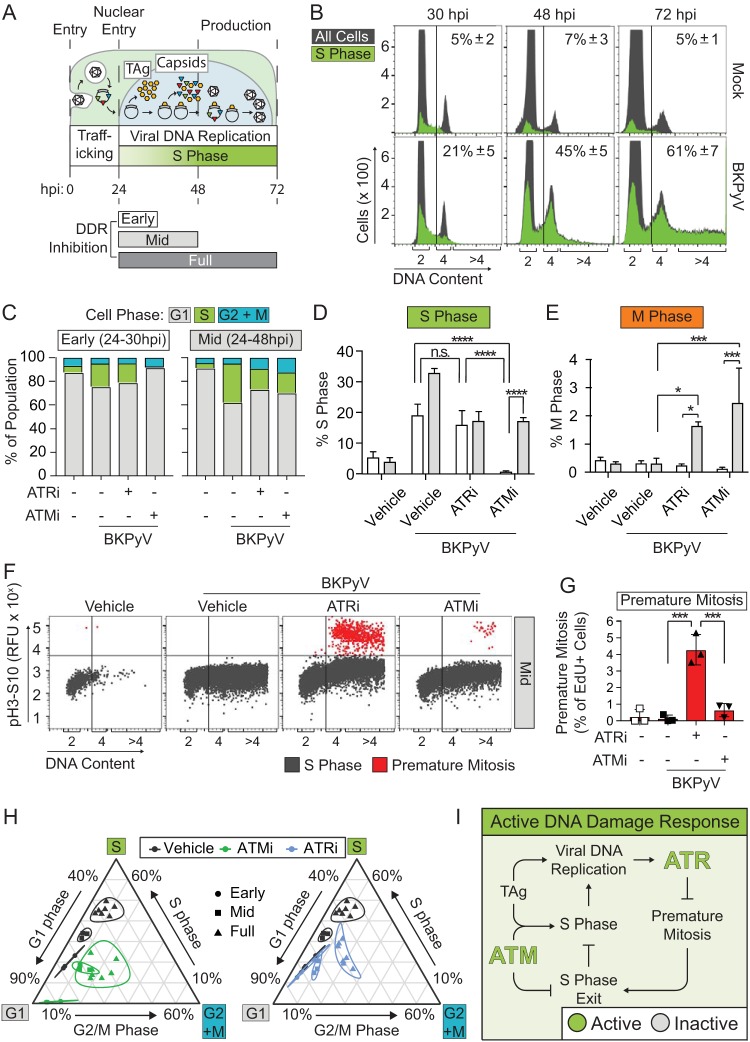
FIG 1.
BKPyV activation of the DDR induced cell cycle arrest. DNA damage and effects on cell cycle arrest were measured during BKPyV (multiplicity of infection of 0.5) infection in RPTE cells treated with ATRi (5 μM VE-821), ATMi (10 μM KU-55933), or vehicle control (DMSO) at 24 hpi and analyzed at 72 hpi. (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (A) Western blotting for markers of DDR activation (pChk1 and pATM), viral infection (TAg), and β-actin (loading control). A representative of n = 3 biological replicates is shown. (B) DNA damage was assessed by nuclear morphology. At left are representative IFA images of nuclear morphologies (blue, DAPI) in BKPyV-infected cells (red, TAg). Percentages of TAg-positive cells (>100 per n = 3) with normal (gray), fragmented (dark gray), or mitotic (green) nuclei or abnormal mitosis (blue) phenotypes are shown as a percentage of the total population. (C to F) Normal, fragmented, mitotic, and abnormal mitosis phenotypes from the experiment described in panel B were graphed separately to show differences between groups. Values are the means ± standard deviations for n = 3 biological replicates. (G) To determine if DDRi affected cell cycle arrest, mitosis and premature mitosis events were measured by cell cycle analysis (FACS). S phase (EdU pulse-labeling) versus M phase (pH3S10) and DNA content (FxCycle Violet) are indicated by blue to red pseudocoloring. Results are representative for n = 7 biological repeats. RFU, relative fluorescence units. (H) Quantification of mitotic cells (pH3S10) from the experiment shown in panel G. The means ± standard deviations for n = 7 biological replicates are shown. Symbols are as follows: white, mock infection; black BKPyV infection; square, vehicle; upward triangle, ATRi; downward triangle, ATMi. (I) The percentages of premature mitotic events (EdU and pH3S10 positive) from the experiment shown in panel G were quantified as the percentage of cells positive for mitosis from the total cells in S phase. Error bars represent standard deviations for n = 7 biological replicates. Symbols are as described for panel H. (J) Working model. Activation of the DDR during a BKPyV infection arrests the cell cycle and prevents entry into mitosis. ATRi, but not ATMi, causes premature mitosis.
Examination of the nuclear morphologies of BKPyV-infected cells revealed that ATR and ATM inhibition reduced the percentage of normal nuclei and increased the fraction of cells with diffuse TAg staining in the cytoplasm (Fig. 1B to F, mitotic cells and abnormal mitosis), as previously reported (7). Closer examination of the cells with diffuse TAg staining revealed alignment of chromatin on the metaphase plate, characteristic of mitosis (Fig. 1B). Nuclear envelope breakdown during mitosis likely explains the presence of cytoplasmic TAg, which is a nuclear protein. Additionally, ATRi caused an ∼7-fold increase in the number of cells with DNA damage in the form of nuclear fragmentation compared to the level in the control and an ∼3-fold increase compared to the level in the ATMi-treated group (Fig. 1D). Under normal conditions PyVs arrest the cell cycle (8), but mitosis was increased by DDR inhibition. This suggested that DDR activation might be required for cell cycle arrest during infection, such as at the G2/M checkpoint (20). Interestingly, a subset of these mitotic cells possessed chromatin morphologies that are characteristic of severely impaired mitotic progression, such as lagging and misaligned chromatin as well as anaphase bridges, which we collectively refer to as abnormal mitosis (Fig. 1F). Activation of the ATR pathway prevents metaphase shattering, which is similar to what is observed when an S-phase cell goes into mitosis, here referred to as premature mitosis (7, 21). Therefore, we hypothesized that ATR may be important during infection to prevent premature mitotic entry.
To determine if BKPyV requires ATR or ATM activation to arrest the cell cycle and prevent premature mitosis, we examined the cell cycle distribution of BKPyV-infected cells when ATM or ATR was inhibited (Fig. 1G). Both ATR and ATM inhibition significantly increased mitotic entry of BKPyV-infected cells but had no effect on uninfected cells (Fig. 1H). ATRi increased premature mitosis 30-fold in BKPyV-infected cells (Fig. 1I), while premature mitosis was rarely observed for uninfected cells under any condition (<0.5% of S-phase cells). Interestingly, although both ATRi and ATMi induced mitosis during BKPyV infection, only ATRi significantly induced premature mitosis (Fig. 1H and I). Thus, premature mitosis caused by ATRi was not simply a consequence of increased mitotic entry because ATMi did not induce premature mitosis but did increase overall mitotic entry. Rather, inhibition of ATR resulted in abnormal mitotic entry. These data suggest that the DNA damage observed when the DDR is inhibited during a BKPyV infection may be caused by abnormal mitotic entry during S phase (Fig. 1J).
Blocking mitosis prevented BKPyV-dependent host DNA damage when ATR was inactive.
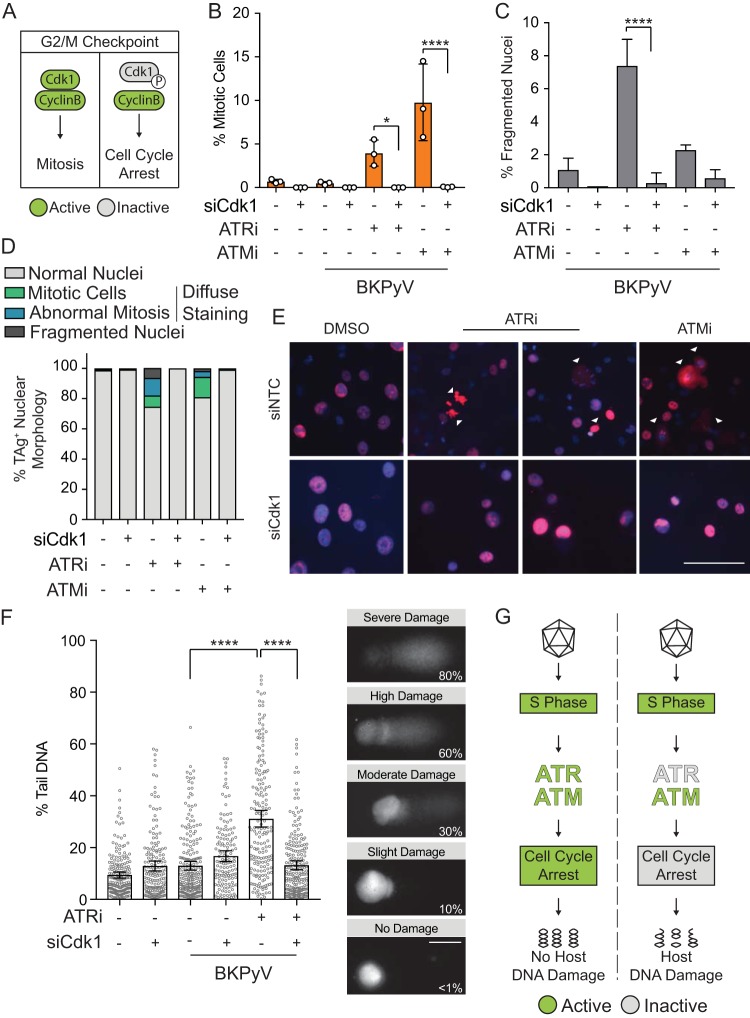
Normally, ATR activation phosphorylates Chk1, which leads to cell cycle arrest at the G2/M checkpoint through inactivation of Cdk1 by phosphorylation (Fig. 2A) (22). To directly test whether the DDR is required to prevent DNA damage by blocking cell cycle progression, we blocked mitosis by knocking down Cdk1 of the mitosis-promoting factor ([MPF] Cdk1/cyclin B1) to see if DNA damage could be prevented during a BKPyV infection when the DDR was inhibited. Knockdown of Cdk1 prevented mitotic entry when the DDR was inhibited (Fig. 2B). Abnormal mitosis and fragmented nuclei that resulted from ATRi or ATMi during BKPyV infection were dramatically reduced by Cdk1 knockdown, indicating that DNA damage caused by DDR inhibition (DDRi) was dependent upon mitosis (Fig. 2C to E). Additionally, cytoplasmic TAg was absent when Cdk1 was depleted, indicating that the cells did not enter mitosis.
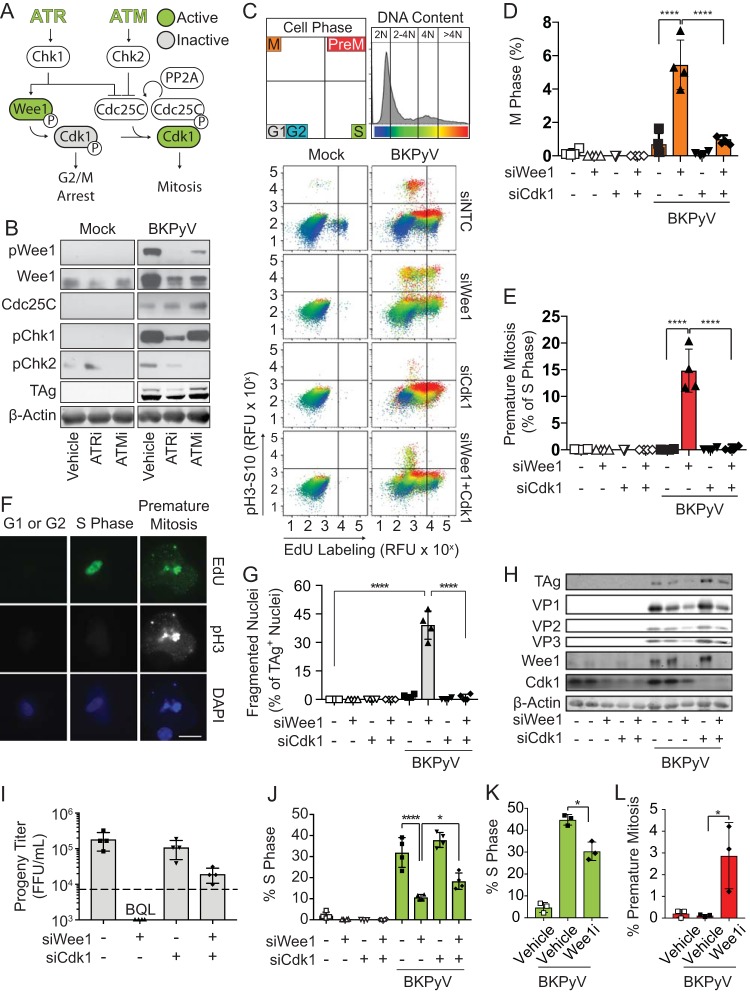
FIG 2.
Blocking mitosis prevented BKPyV-dependent host DNA damage when ATR was inactive. To determine if mitosis is required for DNA damage to occur during BKPyV infection when the DDR is inhibited, RPTE cells were transfected with siCdk1 or a control siRNA (siNTC) and then infected with BKPyV (multiplicity of infection of 0.5). At 24 hpi cells were treated with ATRi (5 μM VE-821), ATMi (10 μM KU-55933), or vehicle control (DMSO) and harvested at 72 hpi. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (A) Diagram of the G2/M checkpoint. DDR activation phosphorylates Cdk1 (inactive; gray) and arrests the cell cycle. In the absence of DDR activation, Cdk1 is active (green), and it complexes with cyclin B to promote mitosis. (B) To test if Cdk1 silencing blocked mitosis, mitotic cells were enumerated by FACS when the DDR was inhibited (ATMi or ATRi) with and without Cdk1 knockdown. Values are means ± standard deviations for n = 3 biological replicates. (C to E) Nuclear morphology was used to assess DNA damage to determine if Cdk1 silencing reduced DNA damage caused by DDRi. Quantification of fragmented nuclei (C) and quantification of nuclear morphology for >100 nuclei per biological replicate (n = 3) (D) are shown. Data from the experiment shown in Fig. 1B are shown again here for a direct comparison to siCdk1 nuclear morphology, which was determined as described for the experiments shown in Fig. 1B. Representative IFA with TAg (red) and DAPI (blue) is also shown (E). Scale bar, 100 μm. Values are the means ± standard deviations for n = 3 biological replicates. (F) DNA damage in BKPyV-infected (multiplicity of infection of 1.0) RPTE cells was quantified by alkaline comet assay. Comets from all biological replicates (n = 3) were combined. Error bars represent the 95% confidence intervals. Images are representative comets with various levels of DNA damage determined by the percentage of comet DNA in the tail. Scale bar, 50 μm. (G) Working model. Mitotic entry leads to DNA damage during BKPyV infection when ATR is inhibited.
Since fragmented nuclei are a product of the nuclear envelope forming around severely damaged DNA following mitosis, we sought to determine whether DNA damage occurred independent of mitosis (23). We utilized an alkaline comet assay, in which DNA damage is visualized with single-cell resolution by a comet-like appearance of broken DNA migrating away from the genomic pellet in an electric field (24). As previously reported, BKPyV infection itself did not cause observable DNA damage since the comet tails were similar to those with mock infection (Fig. 2F) (14). However, inhibition of ATR in a BKPyV-infected cell increased the amount of DNA within the comet tail, signifying a significant increase in DNA damage (Fig. 2F). This DNA damage was rescued by Cdk1 knockdown. Therefore, BKPyV activation of the ATR pathway is required to prevent Cdk1 activation and subsequent mitotic entry, which is associated with DNA damage (Fig. 2G).
DDR activation enhanced BKPyV titers by arresting the cell cycle.
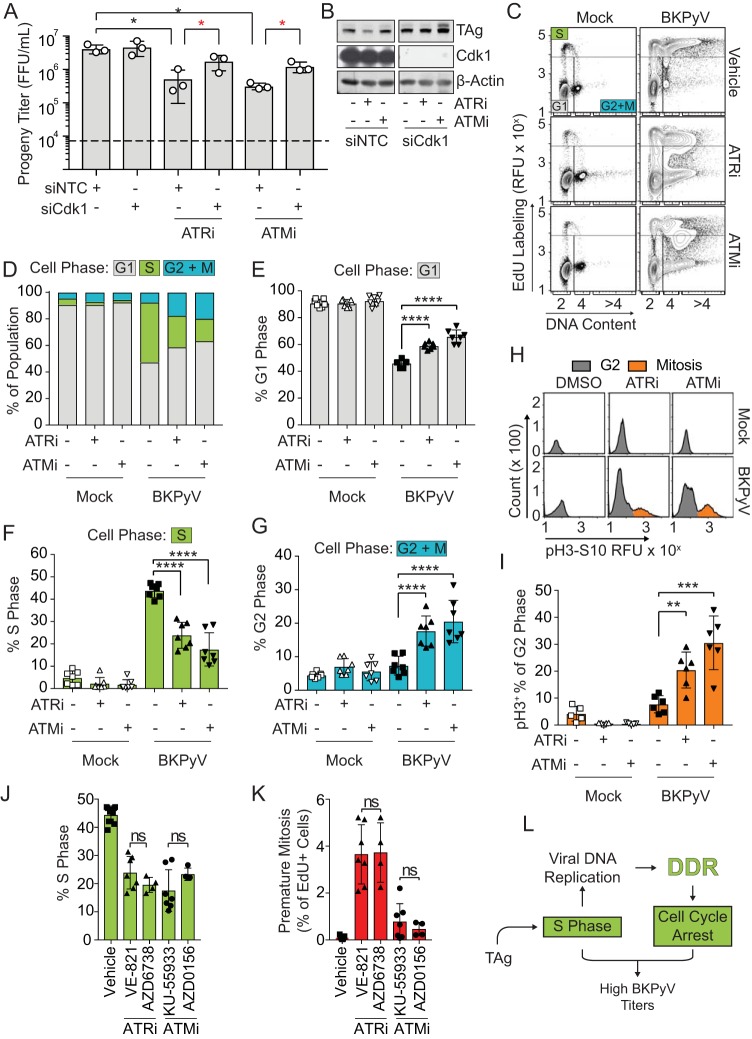
Because preventing mitotic entry by knocking down Cdk1 in DDRi-treated cells during a BK infection prevented DNA damage (Fig. 2D to F), we next sought to determine if blocking premature mitosis rescued viral titers that were decreased by ATRi (Fig. 3A and B). Blocking mitosis by silencing Cdk1 rescued viral titers that were decreased by both ATRi and ATMi (Fig. 3A and B). However, since only ATRi was significantly linked to premature mitosis, this suggests that premature mitosis was not responsible for diminished viral titers but, rather, that overall mitotic entry reduced viral production (Fig. 1H and I).
FIG 3.
DDR activation enhanced BKPyV titers by arresting the cell cycle. To determine if DDR inhibition reduced BKPyV productive infection due to defective cell cycle arrest, RPTE cells were infected with BKPyV (multiplicity of infection of 0.5). At 24 hpi cells were treated with either ATRi (5 μM VE-821), ATMi (10 μM KU-55933), or vehicle control (DMSO). Cells were harvested at 72 hpi for analysis. Symbols are as described in the legend of Fig. 1. The numbers of biological replicates were n = 3 for Western blots and n = 7 for viral titers and cell cycle analysis. (A) Viral titers were determined by FFU assay. Values are the means ± standard deviations. Significant differences from the value for the NTC (black asterisk, P < 0.05) were determined by one-way ANOVA with Dunnett’s posttest. Significant differences from the result for the NTC under the treatment conditions (red asterisk, P < 0.05) were determined by Student's t test. (B) Representative Western blot of TAg (viral infection) and Cdk1 knockdown. (C) To determine how DDR activation influences the cell cycle profile of a BKPyV infection, cell cycle analysis was performed by FACS of mock- or BKPyV-infected RPTE cells treated with ATRi or ATMi, and results are shown as contour plots (5%). (D) The percentages of cells in G1 (gray), S (green), and G2+M (blue) phases from the experiment shown in panel C were quantified and reported as the percentage of the total population. (E to G) The average percentages of cells in G1 phase, S phase, and G2+M phase, as indicated, were regraphed from panel D to show the differences in the populations. Values are the means ± standard deviations. (H and I) G2-and M-phase population of cells from the experiment shown in panel C were further separated into nonmitotic (gray) and mitotic (orange) cells by pH3S10 expression (H), and the average percentages of mitotic cells were then quantified as percentages of total G2- and M-phase cells (I). Values are the means ± standard deviations. (J and K) Comparison of the average proportion of cells in S phase and premature mitosis caused by chemical inhibition with structurally different inhibitors of ATM (5 μM AZD0156) and ATR (5 μM AZD6738) compared to results with KU-55933 and VE-821, respectively. VE-821 and KU-55933 data are regraphed from panel C to visually compare the data. Values are the means ± standard deviations for n = 4 to 7 biological repeats. (L) Working model. If ATR or ATM is inhibited during infection, this prevents cell cycle arrest and allows progression into mitosis, which decreases BKPyV titers and reduces S-phase levels. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Cell cycle profiling was performed to determine if ATR and ATM activation reprogramed the cell cycle during BKPyV infection (Fig. 3C and D). As expected, BKPyV infection significantly increased the fraction of cells in S phase and correspondingly decreased the number of cells that were in G1 phase compared to the levels in the uninfected control (Fig. 3E and F). A portion of BKPyV-infected S-phase cells were marked by >4N DNA content, reflecting the reduplication of host chromatin (Fig. 3C). Also, since BKPyV infection induces cell cycle arrest, infection alone did not significantly increase the portion of cells in G2 or M phase compared to the level in mock-infected cells (Fig. 3G). In mock-infected cells, neither ATRi nor ATMi significantly altered the distribution of the cells in G1, S, or G2-M phase (Fig. 3D to G). In BKPyV-infected cells, however, both ATRi and ATMi significantly decreased the percentage of cells in S phase while increasing the percentages of cells in G1 and G2-M phases (Fig. 3E to G). Further characterization of the G2-M population revealed that a significant number of the cells had entered mitosis (Fig. 3H and I). Similar results were obtained using structurally different inhibitors of ATR and ATM to rule out off-target or nonspecific effects from using a single set of DDR inhibitors (Fig. 3J and K) (25, 26). These findings suggest that activation of both ATR and ATM is required synergistically to prevent progression from S phase during BKPyV infection (Fig. 3L).
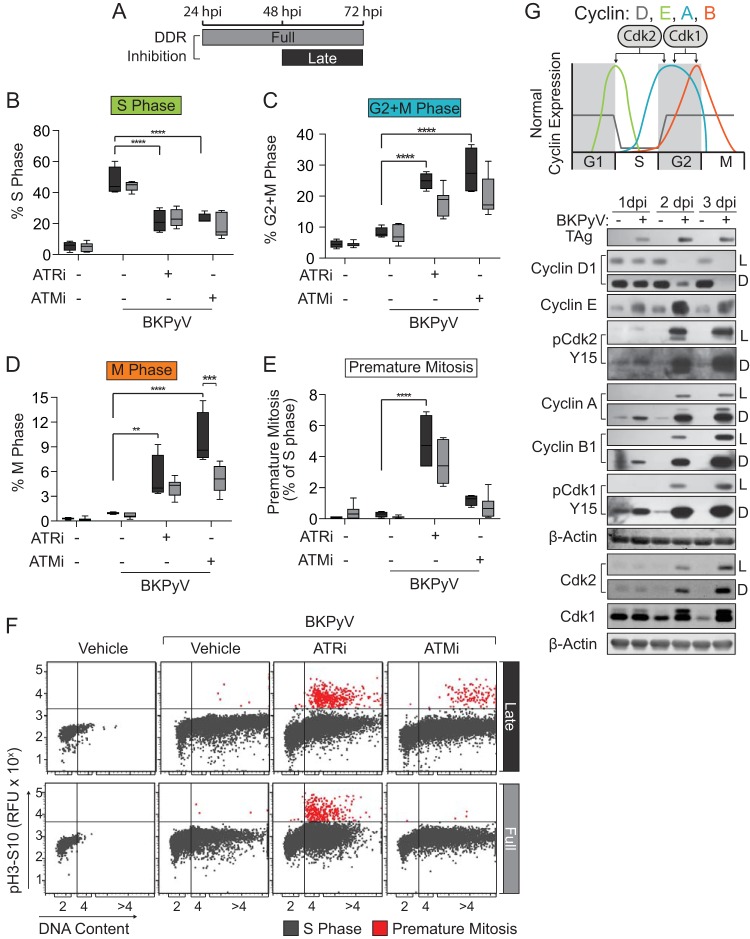
An elevated G1 population may reflect cells that failed to enter S phase or proceeded through mitosis due to loss of BKPyV-mediated cell cycle arrest. Since the DDRi treatments were applied after the virus reached the nucleus and since TAg was expressed driving the cells into S phase, it is more likely that this cell cycle distribution was due to increased mitosis rather than to failure to enter S phase. In fact, inhibiting the DDR late during infection well after S-phase induction by the virus (48 to 72 h postinfection [hpi]) also decreased S-phase levels and increased both mitosis and premature mitosis (for ATRi) (Fig. 4A to F). Thus, constitutive DDR activation is required throughout infection to block cell cycle progression.
FIG 4.
Constitutive ATM and ATR activation was required to prolong BKPyV-induced S phase. To determine if DDR activation is required throughout infection by BKPyV to alter the cell cycle, RPTE cells were infected with BKPyV (multiplicity of infection of 0.5) and treated at 48 hpi (as indicated) with ATRi (5 μM VE-821), ATMi (10 μM KU-55933), or DMSO (control) and harvested at 72 hpi. Full (24 to 72 hpi) treatment data were regraphed from the experiment shown in Fig. 3C for direct comparison. Significant differences were determined by two-way ANOVA with Tukey’s post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (A) DDRi treatment diagram to indicate the late (48 to 72 hpi; black) and full (24 to 72 hpi; dark gray) treatment windows. (B to D) The percentages of cells in S (B), G2+M (C), or M (D) phase were determined by flow cytometry for late or full DDR inhibition windows. Box-whisker plots indicate late (black) or full (dark gray) inhibition windows from the experiment shown in panel A. Values shown are the means ± standard deviations for n = 4 to 7 biological repeats. (E and F) The average percentage of cells undergoing premature mitosis (EdU+ and pH3S10+) from late (black) and full (dark gray) treatment windows were quantified from the experiment shown in panel F and graphed in panel E as box-whisker plots ± standard deviations for n = 4 to 7 biological replicates. EdU+ cells were plotted to separate the occurrence of premature mitosis (red) from S-phase cells (gray) by DNA content (x axis) for full and late DDRi treatment windows. Representatives of n = 4 to 7 biological replicates are shown. (G) Cell cycle protein expression profile for uninfected and infected RPTE cells. The diagram shows the expression levels of cyclins D, E, A, and B, according to the color legend, during the phases of a normal cell cycle (x axis) (top). Western blotting of cyclin protein levels during BKPyV (multiplicity of infection of 1.0) or mock infection was performed at 1, 2, and 3 days postinfection (dpi). Shown are light (L) and dark (D) exposure times, when appropriate, to accurately reflect the relative protein abundance. A representative of n = 3 biological replicates is shown.
Characterization of the cell cycle using cyclin expression levels during a BKPyV infection revealed that BKPyV infection globally upregulated the expression of the S- and G2-phase cyclins E, A, and B1 along with their partners, Cdk2 and Cdk1 (Fig. 4G). DDR inhibition decreased the expression of the cyclin E and Cdk2, supporting the cell cycle analyses that showed that cells were exiting S phase (data not shown). Taken together, these data suggest that ATR and ATM activation was required during BKPyV infection to prevent cell cycle progression from S phase into mitosis.
ATM enhanced S-phase entry while ATR induced cell cycle arrest during BKPyV infection.
S phase is a tightly regulated process during which the 2N DNA content of the cell is replicated only once to 4N before entering the G2 phase. Following nuclear entry of the viral genome at 24 hpi, TAg expression drives the host cell into S phase and, when coupled to cell cycle arrest, leads to host polyploidization (>4N DNA content) (7). This results in viral production that peaks at 72 hpi (Fig. 5A) (15, 27). Early during infection (30 hpi) most cells in S phase had <4N DNA content, suggesting that host DNA rereplication had not begun for the majority of the cells (Fig. 5B). By 48 hpi there was a prominent accumulation of cells that were positive for 5-ethynyl-2-deoxyuridine (EdU+) at 4N, with some cells having >4N DNA content. This suggested that BKPyV induced a prolonged S phase by 48 hpi and that rereplication of the host genome had begun. By 72 hpi, the majority of the BKPyV-infected cells had >4N DNA content, which is clearly distinguishable from the 4N peak.
FIG 5.
ATM enhanced S-phase entry while ATR induced cell cycle arrest during BKPyV infection. To determine if ATM and ATR are required early during BKPyV infection to alter the cell cycle, RPTE cells were infected with BKPyV (multiplicity of infection of 0.5) and treated at 24 hpi with either ATRi (5 μM, VE-821), ATMi (10 μM, KU-55933), or vehicle control (DMSO). Samples were harvested at either 30 or 48 hpi for cell cycle analysis. Also, data from the experiment shown in Fig. 3C (24- to 72-h inhibition) are presented again here for a direct comparison. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (A) A model of the viral life cycle with a DDR inhibition scheme. BKPyV traffics to the nucleus following viral entry. By 24 hpi TAg (yellow) is expressed in the nucleus, which induces host cell entry into S phase and promotes viral replication and expression of capsid proteins (triangles). Viral progeny can be detected by 48 hpi and peaks by 72 hpi. Bars represent the drug treatment windows for DDR inhibitors during viral infection, defined as early (white, 24 to 30 hpi), mid (light gray, 24 to 48 hpi), and full (dark gray, 24 to 72 hpi). (B) A time course to reveal the proportion of cells in S phase and their DNA content during a normal BKPyV infection. Cell cycle analyses were performed at 30, 48, and 72 hpi. S-phase (EdU+) (green) cells were superimposed on the total population (gray). The average percentages of cells with ≥4N DNA content were quantified, and mean values ± standard deviations for n = 3 to 7 biological replicates are shown. (C) To determine how ATRi or ATMi affected the cell cycle in early or mid-infection, cell cycle analysis was performed by FACS on BKPyV-infected cells. The average percentages of cells in G1 (gray), S (green), and G2 and/or M (blue) phases were quantified and reported as the percentages of total cells for early or mid-DDRi treatments for n = 3 biological replicates. (D and E) The average fractions of S phase or mitotic (M) cells (pH3S10) after 30 hpi (white) or 48 hpi (light gray) were quantified by FACS. The mean values ± standard deviations for n = 3 biological replicates are shown. Significant differences were determined by two-way ANOVA with Tukey’s post hoc test. (F and G) To determine the effect of ATR or ATM inhibition on the incidence of premature mitosis (red), all S-phase cells (gray) were plotted based on DNA content and mitosis (pH3S10). The average percentage of premature mitosis was quantified from the data shown in panel F. The mean values ± standard deviations for n = 3 biological replicates are shown. (H) To determine how ATRi or ATMi affected the distribution of cells in the cell cycle over time during a BKPyV infection, a ternary analysis was utilized to represent the proportional relationship of G1, S, and G2+M phases for each of 3 to 7 replicates from the early (circle), mid (square), and full (triangle) DDRi treatment windows quantified from the experiments shown in panel C and in Fig. 3C. The 95% confidence interval is represented for each treatment population (line). Treatments are indicated as follows: black, vehicle; green, ATMi; blue, ATRi. (I) Working model. ATM is required for efficient S-phase entry early during infection while ATR activation prevents mitotic entry of actively replicating cells later during infection. Together, this results in a prolonged S phase during infection.
Inhibition of either ATM or ATR throughout the infection cycle (24 to 72 hpi) reduced the proportion of cells in S phase, which could be due to a delay in either S-phase entry or entry into mitosis (Fig. 3). To better understand how ATR and ATM differentially contributed to S-phase maintenance, we inhibited them at different time points during infection and performed cell cycle analysis. Interestingly, ATMi reduced the number of S-phase cells at early time points (30 hpi), but ATRi did not affect S-phase levels early (Fig. 5C and D, compare ATMi to ATRi). Because both forms of DDR induce mitosis when applied throughout infection (Fig. 1H), we examined mitotic entry early during infection to determine if S-phase levels in ATMi-treated cells were decreased due to increased S-phase exit and subsequent mitotic entry. Neither ATRi nor ATMi increased mitotic entry during the early treatment window (Fig. 5E, white bars); thus, the reduction in S-phase cells when ATM was inhibited was not due to enhanced mitotic entry. Taken together, these data indicate that ATM activity, but not ATR activity, was required for cells to efficiently enter S phase during a BKPyV infection. Furthermore, since entry into S phase was delayed when ATM was inhibited, this suggests that ATM must be active prior to the onset of DNA replication in order for BKPyV to efficiently induce the cells to enter S phase. By 48 hpi, ATMi-treated cells eventually entered S phase, indicating that although ATM was required for efficient S-phase entry early in infection, it was not strictly required for S-phase entry overall (Fig. 5C and D). Interestingly, S-phase levels remained unchanged and were significantly lower from 48 hpi to 72 hpi than those in infected cells with an uninhibited DDR, which continued to increase the proportion of cells in S phase (data not shown). Failure to accumulate high S-phase levels at these later time points (from 24 to 48 hpi) was likely due to defective cell cycle arrest and exit from S phase since there was a corresponding increase in the number of cells in M phase (Fig. 5E). These data suggest that ATM activity is required at two different time points during infection: early during infection to induce efficient S-phase entry and then later to prevent entry into mitosis. However, it is possible that S-phase reentry must occur during infection when cells reach 4N DNA content, and thus increased mitotic entry due to ATMi may result from a failure to reenter S phase.
ATR inhibition did not affect S-phase levels at the early time point (30 hpi) (Fig. 5C and D) but failed to keep pace with the percentage of S-phase cells at later time points. This suggests that ATR may be required later during infection to prevent mitotic entry but is not required for S-phase entry early during infection. In fact, mitosis levels at 30 hpi were not affected, but by 48 hpi both ATRi and ATMi increased overall mitotic entry (Fig. 5E). Interestingly, at 48 hpi cells that were undergoing premature mitosis had ≥4N DNA content when ATR was inhibited, but ATMi did not induce premature mitosis over the control level at any time point (Fig. 4F and 5F and G). Because the ≥4N DNA content population of cells accumulated during late BKPyV infection when the virus induced rereplication of the cellular DNA (Fig. 5B) and because premature mitosis was only enriched in the ≥4N DNA content cells (Fig. 5F), these data suggest that ATR was required late during BKPyV infection to prevent premature mitotic entry.
The mitosis and S-phase analyses suggested that ATM and ATR were required synergistically but at different time points to maintain S-phase levels and block mitotic entry during infection (Fig. 5C to E). However, it is difficult to interpret the effect of DDRi on the overall cell cycle profile over time compared to that of the vehicle control with these analyses. To statistically characterize the cell cycle profile of a BKPyV infection that had DDR inhibited, a ternary analysis was used to show significant shifts in the cell cycle populations (Fig. 5H) (28). The ternary analysis characterized each cell cycle replicate as a single point with three coordinates (percent G1 phase, percent S phase, and percent G2 plus M [G2+M] phase). From this data transformation, a confidence interval was calculated to determine if populations of points were statistically dissimilar to each other. The ternary analysis demonstrated that inhibiting ATM early during infection significantly reduced the population of cells in S phase compared to that in untreated cells (Fig. 5H, left), whereas early ATRi and vehicle control levels were completely overlapping, with no significant difference (Fig. 5H, right). This suggests that ATM, but not ATR, is required early during infection. Late during BKPyV infection both ATM and ATR inhibition significantly reduced S phase and increased mitosis compared to the level with the vehicle control. Taken together with our analysis of premature mitosis and mitotic entry, these data suggest that ATR and ATM are required synergistically but have somewhat different roles in reprogramming the cell cycle during BKPyV infection. Specifically, ATM is required for efficient S-phase entry and blocking mitosis after DNA replication is completed while ATR is required later in infection for cell cycle arrest to prevent entry into mitosis in actively replicating cells (Fig. 5I).
Premature mitosis was the source of DNA damage due to ATR inhibition during BKPyV infection.
The DDR activates the G2/M checkpoint and temporarily blocks cell cycle progression, which allows DNA repair to occur in the G2 phase (Fig. 6A) (29). Both ATM and ATR activities were required to block overall mitotic entry, but only ATR inhibition significantly increased premature mitosis (Fig. 1H and I). Thus, we determined if there was a difference in abundances or activation levels of the G2/M checkpoint proteins during BKPyV infection following ATR versus ATM inhibition. ATR inhibition reduced Chk1 (pChk1) and Wee1 kinase (pWee1) activation, but ATM inhibition did not (Fig. 6B) (30, 31). Total levels of Wee1 were decreased by both ATR and ATM inhibition, which was likely due to rapid degradation of Wee1 following mitotic entry (32). Cdc25C, which is required for reactivation of Cdk1 (Fig. 6A), was unaffected by either ATR or ATM inhibition (Fig. 6B). Unlike ATM, ATR activity disproportionally contributed to Wee1 activation, suggesting that the ATR-Wee1 axis is required to block premature mitotic entry by inactivating Cdk1 during ongoing DNA replication (33, 34). Supporting this, Wee1 knockdown in BKPyV-infected cells with an uninhibited DDR dramatically increased the number of cells that entered mitosis or premature mitosis in a manner that was reversible by Cdk1 silencing (Fig. 6C to E).
FIG 6.
Premature mitosis was the source of DNA damage due to ATR inhibition during BKPyV infection. To determine if ATR activates the Wee1 pathway to block premature mitosis during BKPyV infection (multiplicity of infection of 0.5), Wee1 and/or Cdk1 was silenced with siRNAs to induce and/or block premature mitosis, respectively. At 72 hpi cells were harvested to assess the amount of premature mitosis and DNA damage during BKPyV infection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (A) A diagram showing how ATM and ATR regulate the G2/M checkpoint. ATR and ATM activation stimulates Chk1 and Chk2, respectively. Activation of the Wee1 kinase inhibits Cdk1 through phosphorylation leading to G2/M arrest. Simultaneously, DDR activation inhibits Cdc25C, the phosphatase that reactivates Cdk1 to promote mitosis. Cdc25C turnover requires protein PP2A. (B) Western blot of G2/M checkpoint control proteins in BKPyV-infected (multiplicity of infection of 1.0) or mock-infected cells treated with ATRi, ATMi, or a vehicle control (DMSO) from 48 to 72 hpi. Data shown are representative of n = 3 biological replicates. (C) Levels of mitosis and premature mitosis measured by FACS as described in the legend of Fig. 1G. Data representative of n = 4 replicates are shown. (D and E) Quantification of cells from the experiment shown in panel C that are mitotic or undergoing premature mitosis. Values are the means ± standard deviations of n = 4 biological replicates. Symbols are as follows: white, mock-infected cells; black, BKPyV-infected cells; square, siNTC treatment; upward triangle, siWee1 treatment; downward triangle, siCdk1 treatment; diamond double-knockdown treatment. Significant differences were determined by two-way ANOVA with Tukey’s post hoc test. (F) To determine if cells undergoing premature mitosis acquire DNA damage, siWee1 samples stained for FACS (C) were analyzed by IFA for evidence of BKPyV-induced DNA damage. Results shown are representative of >20 cells from G1, S, or premature mitosis from the experiment shown in panel C for n = 3 biological replicates. EdU incorporation (green), pH3S10 (white), and chromatin (blue) are represented. Scale bar, 20 μm. (G) DNA damage was assessed by nuclear morphology using immunofluorescent microscopy of TAg and DAPI staining. The percentage of DNA-damaged nuclei (fragmented) was quantified from at least 100 nuclei per condition per replicate. Data are means ± standard deviations for n = 4 biological replicates. Significant differences were determined by two-way ANOVA with Tukey’s post hoc test. (H) Western analysis of markers of viral infection and knockdown efficiency for Wee1 and Cdk1. Values representative of n = 4 biological replicates are shown. (I) The impact of Wee1 silencing on BKPyV titers was determined by focus-forming assays. The means ± standard deviations for n = 4 biological replicates are shown. The dotted line represents the quantifiable limit of detection for the assay. BQL, below the quantifiable limit. (J) The percentages of cells in S phase from the experiment shown in panel C were quantified and are presented as means ± standard deviations for n = 4 biological replicates. Significant differences were determined by two-way ANOVA with Tukey’s post hoc test. (K and L) RPTE cells were mock or BKPyV infected (multiplicity of infection of 0.5) and then at 24 hpi treated with Wee1i (300 nM MK1775). Cell cycle analysis to identify S phase (EdU) and premature mitosis based on pH3S10 expression was performed by FACS at 72 hpi. The mean percentage of cells in each phase ± standard deviation is shown for n = 3 biological replicates. Symbols are as follows: white, mock infection; black, BKPyV black infection; square, vehicle control; diamond, Wee1i.
One study suggested that the DDR is required to repair DNA damage that accumulates throughout BKPyV infection (7). However, our data suggest that host DNA damage is caused by premature entry into mitosis. To differentiate between these two possibilities, cell cycle arrest was blocked by Wee1 knockdown in cells with active DDR to repair DNA damage. Single cells from either the G1/G2, S, or premature mitosis fractions from the experiment shown in Fig. 6C were evaluated for evidence of DNA damage. Cells undergoing premature mitosis had evidence of DNA breaks, with chromatin dispersed throughout the cytoplasm, indicating complete misalignment of the chromatin (Fig. 6F). Furthermore, these orphaned fragments occurred adjacent to sites of DNA synthesis (EdU+) and costained for mitosis (pH 3). Quantification of DNA damage by nuclear fragmentation showed that Wee1 silencing induced nuclear fragmentation in 40% of Tag-positive nuclei compared to <1% in the small interfering RNA (siRNA) nontargeting control (siNTC) (Fig. 6G). Silencing Cdk1 along with Wee1 reversed DNA damage and premature mitosis but not mitotic entry all together (Fig. 6C to F). DNA damage caused by Wee1 silencing was specific to BKPyV infection, consistent with what has been shown for ATR depletion (7). This suggests that BKPyV-induced DNA damage caused by DDR inhibition is due to premature Cdk1 activation rather than to inefficient DNA repair.
Since Wee1 silencing induced both premature mitosis and DNA damage to a greater extent than had been previously observed by ATR inhibition, we investigated the impact of Wee1 silencing on BKPyV productive infection. The abundances of TAg and capsid proteins were reduced by Wee1 silencing and rescued by cosilencing Cdk1, indicating that viral protein levels were dependent upon cell cycle arrest by Wee1 (Fig. 6H). Similarly, viral titers were reduced to unquantifiable levels in the absence of Wee1 but were partially restored by Cdk1 silencing, suggesting that cell cycle arrest by Wee1 is essential for productive infection (Fig. 6I). To address this possibility, cell cycle profiling of BKPyV-infected cells with Wee1 silenced or inhibited revealed that Wee1 was required to prolong S phase and prevent premature mitosis during BKPyV infection (Fig. 6C and E to J). Because blocking mitosis with Cdk1 cosilencing with Wee1 knockdown partially restored S-phase levels, this demonstrates that Wee1 inactivated Cdk1 during infection to prevent mitotic entry (Fig. 6C, E, and H to L).
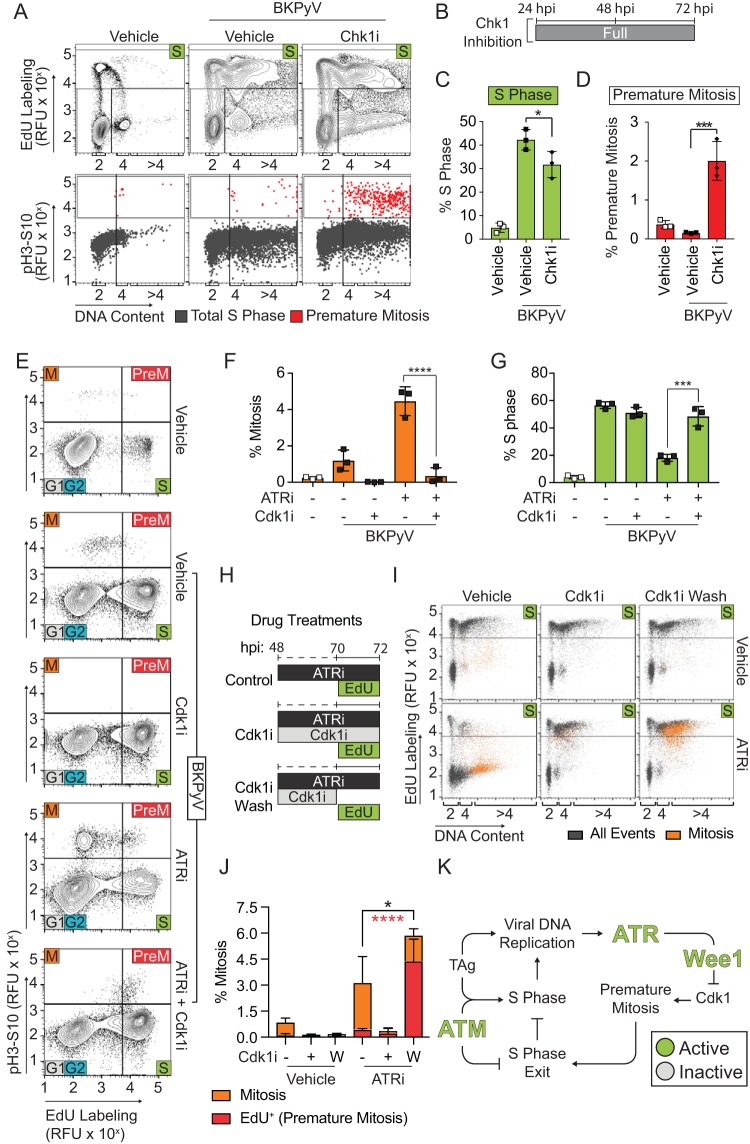
To fully delineate the molecular pathway leading to ATR-mediated cell cycle arrest, we performed cell cycle analysis of a BKPyV infection with Chk1 inhibition (Chk1i), a possible intermediate in the ATR-Wee1 pathway. We found that Chk1i reduced S phase and induced premature mitosis similar to the effects observed with ATRi and Wee1i (Fig. 7A to D). Thus, our findings indicated that productive BKPyV infection is dependent upon cell cycle arrest mediated by the ATR-Chk1-Wee1-Cdk1 axis, which blocks premature mitotic entry. To test this, we performed cell cycle analysis of BKPyV-infected cells treated with ATRi in the presence Cdk1 inhibition (Cdk1i) and observed that Cdk1i blocked ATRi-associated mitotic entry (Fig. 7E and F). Reciprocally, S-phase levels that were decreased with ATRi were restored to levels approximating the those of the vehicle control (Fig. 7G). Thus, ATR activation during a BKPyV infection prevents mitosis through activation of the Wee1 pathway to block Cdk1 activation. Furthermore, when the Cdk1 inhibitor was washed from ATR-inhibited cells, mitosis occurred primarily in cells actively replicating DNA (75% versus 14% in contrast to ATRi alone) (Fig. 7H to J). Taken together, these findings strongly suggest that the ATR pathway is activated during BKPyV infection to prevent the activation of Cdk1 during active DNA replication, thereby blocking premature mitotic entry and expanding the window for viral replication by prolonging S phase (Fig. 7K).
FIG 7.
Activated Cdk1 is required for S-phase exit during BKPyV infection. To determine if the ATR pathway is activated during a BKPyV infection to prevent S-phase exit and mitotic entry of actively replicating cells, the contribution of Chk1i (2 μM; MK8776), ATRi (5 μM, VE-821), and Cdk1i (10 μM, RO-3306) to premature mitosis and cell cycle arrest were measured by FACS analysis of BKPyV-infected cells (multiplicity of infection of 0.5) at 72 hpi against a vehicle control (DMSO). (A) Cell cycle analysis was performed on Chk1i-treated cells (24 to 72 hpi) by FACS to identify S (EdU), G1, and G2 (based on DNA content staining) phases and M (pH3S10) phase. Representative contour plots (5%) are shown (top). The fractions of S-phase cells from the experiment shown in the top panel were graphed based on their DNA content and mitosis (pH3S10) to differentiate premature mitosis (red) from S phase (gray) (bottom). Data are representative of n = 3 biological replicates. (B) Diagram of Chk1i treatment during BKPyV infection. (C and D) The average percentages ± standard deviations of cells in S phase or premature mitosis were quantified for n = 3 biological replicates. Symbols are as follows: white, mock infection; black, BKPyV infection; square, vehicle; diamond, Chk1i. (E to G) Cdk1i was added to ATRi-treated, BKPyV-infected cells to determine if Cdk1 activity was required for ATRi to reduce S-phase levels. Briefly, mock-infected or infected RPTE cells were treated at 48 hpi with ATRi (5 μM, VE-821), Cdk1i (10 μM, RO-3306), or vehicle control (DMSO). Cells were pulse-labeled with EdU at 70 to 72 hpi to assess DNA synthesis and analyzed by FACS at 72 hpi for cell cycle distribution. Contour plots (5%) are representative of n = 3 biological replicates (E). The percentages of mitotic (pH3S10) (F) or S-phase (G) cells are presented as the means ± standard deviations for n = 3 biological repeats. Symbols are as follows: white, mock infection; black, BKPyV infection. (H and I) Cdk1i was used to synchronize BKPyV RPTE cells treated with ATRi, and then Cdk1 was washed out to determine if cells entered mitosis immediately (premature mitosis) or completed S phase prior to entering mitosis. Small-molecule inhibitor treatment and EdU labeling schema are shown for each ATRi treatment condition (control, Cdk1i, and Cdk1i wash) (H). The control, no-ATRi treatment, scheme is not shown. Cell cycle analysis (FACS) with (bottom) and without (top) ATRi was performed (I). Data shown are representative of n = 3 biological replicates. All events (mitosis, pH3S10-positive cells) were plotted as S phase (EdU+) versus DNA content. (J) Quantification for panel I, where Cdk1i was added (+) or not (−) or washed out (W) at 70 hpi, as diagrammed in panel H. The percentages of cells in mitosis (orange, positive for pH3S10) or premature mitosis (red, EdU+ and pH3S10+) were quantified as means ± standard deviations of n = 3 biological replicates. The fraction of premature mitosis is shown as a portion of overall mitosis in each group. Asterisks denote significant differences determined by one-way ANOVA between the overall mitosis population (black) and the premature mitosis population (red). (K) Working model. ATR prolongs S phase by activating Wee1 to inhibit mitotic entry by inhibiting Cdk1. Thus, ATR activation prevents premature mitosis, which in turn maintains S phase, prevents DNA damage, and enhances reduced viral titers. When ATR is inhibited, Wee1 is no longer activated to inhibit Cdk1. Activated Cdk1 in S phase induces premature mitosis, leading to DNA damage and reducing viral titers. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
BKPyV replication activates the DDR (14) and protects the host from DNA damage. Our studies here showed that the DNA damage resulted not from a lack of DNA repair but, rather, from a failure to arrest the cell cycle to allow cells in S phase to enter mitosis prematurely (Fig. 6). Interestingly, our findings suggest separate and synergistic roles for ATR and ATM during a BKPyV infection (Fig. 5). Inhibiting ATR resulted in cells entering mitosis when S-phase DNA synthesis was ongoing (Fig. 1G). This premature mitosis correlated with severe host DNA damage as measured by a comet assay (Fig. 2F). In addition, if the block to mitosis was removed in ATR-inhibited BKPyV-infected cells, there was a dramatic and immediate induction of premature mitosis (Fig. 7H to J). When ATM was inhibited, the cells entered S phase at a reduced rate early in infection but still entered mitosis later during infection at a higher rate than cells with an active DDR (Fig. 5C to E). In contrast to ATRi, ATMi resulted in cessation of DNA synthesis prior to entry into mitosis (Fig. 1G), which also correlated with a reduction in the DNA damage (Fig. 2D and E). Finally, we induced mitosis in cells with an active DDR without altering the DNA repair capacity of these cells by silencing Wee1. When Wee1 was silenced during infection, cells acquired DNA damage due to mitotic entry despite an otherwise fully functional DDR (Fig. 6C to F). Together, these data suggest that the source of the DNA damage is premature entry into mitosis. In fact, when mitosis was inhibited in cells that did not have an active ATR, premature mitosis and DNA damage were prevented. This demonstrated that cell cycle arrest, not DNA repair, is critical to prevent host DNA damage during a BKPyV infection.
We demonstrated that activation of both ATR and ATM was required nonredundantly to prevent BKPyV-infected cells from entering mitosis (Fig. 1). This suggests at least two distinct mechanisms by which the DDR promotes cell cycle arrest during infection. Inhibiting the function of ATR or ATM during defined windows of the BKPyV replication cycle revealed that Wee1 was required to arrest the cell cycle to prevent entry into mitosis and to prolong S phase (Fig. 6C to E). Recently, ATR has been found to mediate the S/G2 phase transition by monitoring DNA replication in an unperturbed cell cycle by blocking Cdk1 activation (34). Thus, DNA replication itself is an essential signal to restrict premature Cdk1 activation (35). This model of ATR activity agrees with our observations that ATR inhibition resulted in premature mitosis of S-phase cells with a ≥4N DNA content. However, our findings revealed that for uninfected cells or infected cells with <4N DNA content during infection, ATR was not necessary to block premature mitosis through Cdk1 inactivation (Fig. 5F). This may suggest that in certain primary cells there are additional layers of regulation in S phase that prevent progression into G2 phase in the absence of ATR. Although our data suggest that ATR inhibition did not induce premature mitosis in uninfected cells, ATR mutations are lethal in utero, and the Chk1 response is required to prevent mitotic entry in trophoblast giant cells; therefore, it is possible that the ATR pathway regulation is cell type specific and is important for genomic reduplication events such as those involved in embryogenesis, cellular differentiation, and tissue repair (36–38). ATM activation was required to block mitotic entry, but premature mitosis was not linked to ATM inhibition during BKPyV infection, which is likely due to intact ATR activity in the ATM-inhibited cells.
While ATR was not required for efficient S-phase entry, ATM inhibition during the early window of BKPyV infection prevented expedient entry into S phase (Fig. 5D and E). An earlier report revealed a physical interaction between PCNA and ATM that enhances DNA replication, which supports our finding that ATM was required for S-phase entry during infection (39). The mechanism by which ATM induces S-phase entry during infection is unknown. One possibility is that ATM is required for replication fork licensing because ATM targets numerous proteins at the replication fork, including the GINS and minichromosome maintenance (MCM) complexes (19, 40). Another possibility is that ATM is required to form the viral replication compartment, as has been observed with other PyVs; thus, without ATM activation, replication is delayed (41). Additional studies will be required to fully understand how ATM facilitates S-phase entry during a PyV infection.
The current paradigm for how ATM and ATR contribute to PyV replication is that they are directly required to replicate the circular PyV genome and resolve viral DNA damage. ATM has been suggested to be required to prevent rolling circle replication of the viral DNA, which must undergo theta replication to produce genomes that are packaged (9, 41). In contrast, ATR prevents replication fork collision of the two forks that fire from the PyV origin of replication on the circular genome (9, 41). However, in those studies ATM or ATR inhibition sharply decreased overall viral DNA replication 60% to 80%, yet only 10 to 20% of the viral genomes had replication defects (9, 41). The relatively small fraction of damaged viral genomes does not account for the substantial decrease in viral DNA levels. Consistent with our findings, the same study showed that DDR inhibition reduces the fraction of cells in S phase during simian virus 40 (SV40) infection (41) but did not provide any insight into whether S-phase entry or exit was affected. Our findings showed that the DDR was primarily required to prolong S phase by demonstrating that efficient viral production was restored by blocking S-phase exit in DDR-inhibited BKPyV-infected cells (Fig. 3A). Other studies have suggested that cell cycle arrest is important for viral titers (42), but it was unknown how and whether the DDR contributed to cell cycle arrest. Overall, our studies do not rule out a role for the DDR in DNA repair but suggest that DDR activation is primarily required during BKPyV infection for cell cycle arrest.
A key difference between activation of ATR and that of ATM during BKPyV infection was activation of the Wee1 pathway by ATR but not by ATM (Fig. 6B). Wee1 is one of two inhibitory kinases that are important for governing the G2/M transition by inactivating Cdk1 (43). As such, Wee1 is being explored as a target for chemotherapy to induce mitotic catastrophe (33, 43, 44). Although Cdk1 knockdown rescued DNA damage during BKPyV infection in Wee1-silenced cells, our investigation revealed that viral production and S-phase levels were not fully rescued by blocking mitosis, suggesting multiple roles for Wee1 during infection (Fig. 6). Since Wee1 also inhibits Cdk2 (45) and since Cdk2 remained inhibited throughout infection, it is possible that Wee1 is required for inhibition of Cdk2 during a BKPyV infection as well.
PyV-associated cancers are not concomitant with a productive infection but are driven by clonal integration of TAg with mutations in the helicase domain of TAg or in the viral origin of replication (2, 46). In these tumors, episomal viral DNA is depleted, and viral replication is limited, suggesting that oncogenic transformation does not occur during a normal productive PyV infection (47). Given that DNA amplification is the trigger for PyV-mediated DDR activation and that nonpermissive tissue culture models similarly limit DDR activation (14, 48), our results suggest that this would lead to a failure to arrest the cell cycle during infection. In fact, both SV40 and BKPyV cause cancer in a nonpermissive host when viral replication is attenuated (49, 50). Our observations, in combination with those in the field, provide a framework to understand the cause of oncogenic transformation by PyVs and how DDR activation may be a countermeasure to oncogenesis. Our findings support a model by which viral replication activates the DDR to inhibit cellular proliferation and prevent DNA damage by blocking entry into mitosis. While additional evidence is required to fully support this model, this interpretation underscores the role of the DDR during PyV infection not only to increase viral production but also as a mechanism to counteract the oncogenic potential of TAg. Future studies focusing on the mechanism of DDR activation during PyV infection will be necessary to test this model and connect tumorigenesis by PyVs to the broader fields of cell cycle control, replication licensing, and oncogenesis.
MATERIALS AND METHODS
Tissue culture.
RPTE cells were purchased from Lonza (Basel, Switzerland) and maintained according to the supplier’s recommendation in renal epithelial growth medium (REGM) (CC-3190). Cells were expanded in T75 flasks twice from the original vial of cells, and approximately 2.25 × 106 cells were frozen in REGM plus 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen (51). All studies utilized these expanded RPTE cell stocks, which can be passaged one additional time.
Viral stocks.
BKPyV (Dunlop strain) was maintained on the pBR322 vector (catalog no. 25466; Addgene) (a gift from Peter Howley) ligated into the BamHI site (52). Original stocks of infectious BKPyV were prepared from a pBR322:Dunlop genomic clone which was digested and purified prior to transfection into a T75 flask of RPTE cells at 60% confluence with Lipofectamine 2000 (ThermoFisher) (7). The cells were scraped in medium after 21 days and subjected to three freeze-thaw cycles, and the viral titer was determined by focus-forming assay.
Viral infection.
For infection, RPTE cells were chilled at 4°C for 15 min and then incubated with BKPyV in REGM at one-fourth the volume of the dish at 4°C for 1 h with intermittent shaking before the inoculum was replaced with REGM at 37°C (7). Viral infections were performed at 0.5 focus-forming units (FFU)/cell unless otherwise specified.
siRNA knockdown.
Silencer Select siRNAs for Wee1 (s21) and Cdk1 (s464) and a nontargeting siRNA (Silencer Select negative control no. 1) were purchased from Ambion (ThermoFisher). Reverse transfection into RPTE cells was performed using 10.8 μl/ml Lipofectamine RNAiMax (ThermoFisher) and 10 nM siRNA using a protocol adapted from Jiang et al. (7). Briefly, siRNA and Lipofectamine were preincubated for 20 min in one half of the final volume of the tissue culture plate with REGM in the dish prior to the addition of cells. Subsequently, RPTE cells were suspended at 1.5 × 105 cells/ml and added to the lipid-siRNA mixture in one half of the final volume of the well. In the case of double knockdowns, single-knockdown and control conditions were supplemented with an additional nontargeting control siRNA such that all conditions were transfected with equal concentrations of siRNA.
Inhibitor treatment.
All inhibitors were added to RPTE cells after nuclear entry of the virus. ATR and ATM were treated with the inhibitors VE-821 (5 μM in DMSO) and KU-55933 (10 μM in DMSO), respectively, or with AZD6738 (5 μM in DMSO) and AZD0156 (5 μM in DMSO), respectively, as in the experiment shown in Fig. 3. Cdk1 was inhibited by the potent and selective inhibitor RO-3306 (10 μM in DMSO) (53). Chk1 and Wee1 were treated with the inhibitors MK-8766 (2 μM in DMSO) and MK-1775 (0.3 μM in DMSO), respectively. All inhibitors were purchased from Selleck Chemicals (Houston, TX), and final concentrations of the vehicle (DMSO) were kept constant between conditions.
IFA.
For immunofluorescence assays (IFA), RPTE cells grown on coverslips were fixed in 250 μl of 4% paraformaldehyde (PFA) for 20 min as described in Jiang et al. (52). Subsequently, the samples were washed in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4-7H2O, 1.4 mM KH2PO4) prior to permeabilization (0.1% Triton X-100 in PBS). Blocking was performed in PBS with 5% goat serum for 1 h. TAg expression (pAb416 at 1:250 in block) was used to mark BKPyV-infected cells. Coverslips were mounted onto glass microscopy slides with ProLong Gold antifade reagent (Thermo) with 4′,6-diamidino-2-phenylindole (DAPI) and stored overnight prior to quantification. Analysis was performed on a Nikon Eclipse Ti-S inverted microscope using a Nikon 40×/0.60 S Plan Fluor ELWD AMD objective.
Viral titers were determined using a focus-forming unit assay (52). Briefly, RPTE cells were infected as described above in duplicate with undiluted viral stocks and with 10−1 and 10−2 dilutions of viral stocks in REGM. Cells were fixed at 48 hpi and stained for TAg as described above. The average numbers of Tag-positive nuclei in seven fields of view (FOV) at ×10 magnification per technical replicate were counted for dilutions that showed TAg staining for between 20 and 200 cells per FOV. This number was multiplied by the dilution factor and number of FOV per surface area of a 12-well plate (determined by dividing the area of the well by the area of one FOV); then technical duplicates were averaged to determine viral titers. All titers were determined in biological triplicates of technical duplicates.
DNA damage was visualized by IFA for the fragmented nucleus assay to assess nuclear fragmentation of BKPyV-infected RPTE cells at 72 hpi (7). At least 100 TAg-positive (TAg+) nuclei per biological replicate were scored on the basis of nuclear morphology. Nuclear classifications were either the following: normal (rounded), fragmented (multiple smaller nuclei in which DAPI and TAg staining overlapped), or diffuse (TAg staining is dispersed throughout the cytoplasm). The diffuse TAg staining pattern was then subdivided into two categories, normal mitosis and abnormal mitosis; abnormal mitosis was marked by aberrant DAPI staining such as that showing misaligned chromatin in reference to the metaphase plate, lagging chromatin in anaphase, and anaphase bridges connecting separating chromatids.
Western analysis.
Protein lysates were collected in E1A lysis buffer (50 mM HEPES, pH 7, 250 mM NaCl, 0.1% NP-40) with protease inhibitors (52). An equal amount of protein lysates (minimum, 20 μg) was loaded on either an 8% or 10% polyacrylamide gel for SDS-PAGE. Gel electrophoresis was performed at either 45 V overnight or at 120 V for 6 h and then transferred using a wet-transfer method at 60 V overnight to a polyvinylidene difluoride (PVDF) membrane. Membranes were cut on the basis of molecular weight based on the protein marker. Membranes were blocked in either 2% fat free dry milk or 2% fetal bovine serum (FBS) in 1× PBS with 0.1% Triton X-100 and probed with primary antibodies in blocking buffer (Table 1). Secondary antibody was added depending upon the application. Quantitative Western analysis was performed using a Li-Cor Odyssey platform; however, when the Li-Cor system was not capable of detection of the primary antibody, then secondary antibody conjugated to horseradish peroxidase (HRP) and Immobilon Forte Western HRP substrate were utilized with radiography film. Multiple exposures of each target were collected and presented to best represent the relative abundance of the target protein.
TABLE 1.
List of antibodies used for Western analysis, immunofluorescent microscopy analysis, and fluorescence-activated cell sorting
| Reagent or resource | Source | Identifier | Analysis method(s) (dilution)a |
|---|---|---|---|
| Mouse anti-SV40 T antigen (pAb416) | Abcam | pAb16879 | IFA (1:250), WB (1:2,000) |
| Mouse anti-SV40 VP1 (P5G6) | Abcam | pAb53977 | WB (1:2,000) |
| Rabbit anti-SV40 VP2/VP3 | Abcam | Ab53983 | WB (1:2,000) |
| Rabbit anti-β-actin | Cell Signaling | 4968 | WB (1:2,000) |
| Rabbit anti-pChk1Ser10 | Cell Signaling | 2344 | WB (1:500) |
| Rabbit anti-pChk2Thr68 | Cell Signaling | 2661 | WB (1:500) |
| Rabbit anti-pATMThr1981 | Abcam | GR125270 | WB (1:2,000) |
| Mouse anti-pCdk1Tyr15 | Santa Cruz | sc-136014 | WB (1:1,000) |
| Rabbit anti-pCdk2Tyr15 | Cell Signaling | 9111 | WB (1:1,000) |
| Rabbit anti-pWee1S642 | Cell Signaling | 4910 | WB (1:1,000) |
| Mouse anti-Cdk1 | Abcam | Ab18 | WB (1:1,000) |
| Mouse anti-Cdk2 | Santa Cruz | sc-6248 | WB (1:1,000) |
| Moue anti-Cdc25C | Santa Cruz | sc-7389 | WB (1:1,000) |
| Mouse anti-Wee1 | Santa Cruz | sc-5285 | WB (1:1,000) |
| Mouse anti-cyclin D1 | Santa Cruz | sc-8396 | WB (1:1,000) |
| Mouse anti-cyclin E | Santa Cruz | sc-247 | WB (1:1,000) |
| Mouse anti-cyclin A | Santa Cruz | sc-27682 | WB (1:1,000) |
| Mouse anti-cyclin B1 | Santa Cruz | sc-245 | WB (1:1,000) |
| Alexa Fluor 647-rabbit anti-pH3Ser10 | Cell Signaling | 3458 | FACS 1:100 |
| HRP anti-mouse IgG | General Electric | NA931 | WB (1:3,000 to 1:5,000) |
| HRP anti-rabbit IgG | General Electric | NA934 | WB (1:3,000 to 1:5,000) |
| IRDye 800CW anti-mouse IgG | Li-Cor | 926-332210 | WB (1:10,000 to 1:20,000) |
| IRDye 680RD anti-rabbit IgG | Li-Cor | 926-68071 | WB (1:10,000 to 1:20,000) |
| Alexa Fluor 594 goat anti-mouse IgG(H + L) secondary antibody | Invitrogen | A11005 | IFA (1:250) |
IFA, immunofluorescence assay; WB, Western blotting; FACS, fluorescence-activated cell sorting.
FACS analysis.
A 10-cm2 dish of RPTE cells was labeled by adding a thymidine analogue, 10 mM 5-ethynyl-2′-deoxyuridine (EdU) (Click Chemistry Tools; Scottdale, AZ) to the medium for 3 h to detect newly synthesized DNA prior to fixation in 4% PFA for 20 min at room temperature (22°C) (54). Samples were washed in wash buffer (1× PBS with 2% FBS) two times with centrifugation at 1,200 × g for 8 min and then permeabilized in 0.3% Triton X-100 in wash buffer for 15 min on ice. Then cells were incubated with Click-IT staining solution (20 μM Alexa Fluor 488 azide, 2 mM CuSO4, 10 mM Na-ascorbate) to conjugate EdU to the fluorophore (Alexa Fluor 488; Click Chemistry Tools). To determine which cells were in mitosis, cells were stained with anti-pH3S10 (histone H3 phosphorylated at S10) antibody (55). To stain for DNA content (separating G1 and G2 phases), cells were incubated for >30 min with 1 μg/ml FxCycle Violet (ThermoFisher) in wash buffer. Fluorescence-activated cell sorting (FACS) analysis was performed on an LSR II flow cytometer using the 405, 488, and 647 laser lines and the FACSDIVA software and then analyzed using FlowJo, version 10. Gating was applied identically to each sample within the experiment. It should be noted that our DNA stain cannot differentiate between cellular and viral DNA; thus, viral DNA may contribute to the DNA signal in cells. However, the viral chromatin accounts for only <5% of total DNA in the cell at the 72-hpi time point and thus only minimally affects the analysis of the data (56, 57).
In certain cases, samples that were prepared for flow cytometry as described above were fixed onto microscopy slides by addition of Prolong Gold antifade reagent with DAPI at 1:1 (vol/vol) with the sample and covered with a glass coverslip. After an overnight incubation, samples were analyzed using a Nikon Eclipse Ti-S inverted microscope using a Nikon 100×/1.45 oil S Plan APO λ objective. At least 10 images of cells in each of the G1/G2 phase, S phase, and premature mitosis phase were collected per biological replicate. Images were analyzed on the basis of nuclear morphology.
Comet assay.
The comet assay detects DNA damage with single-cell resolution by detecting the comet-like appearance of broken DNA migrating away from the unbroken genomic pellet of cells embedded in agarose and pulsed in an electric field. A Trevigen comet assay kit was utilized to prepare samples for analysis as per the manufacturer’s instructions for 30 min. Following staining with 1× SYBR Gold nucleic acid stain (ThermoFisher), samples were analyzed on a Nikon Eclipse Ti-S inverted microscope using a Nikon 40×/0.60 S Plan Fluor ELWD AMD objective. At least 50 comets in random FOV were captured for each biological repeat (n = 3), totaling no fewer than 150 comets for the overall analysis. Comets were scored using the Open Comet plug-in for ImageJ on the basis of the percentage of DNA in the tail, which is a reflection of the amount of damaged DNA (58). Significant differences were determined using one-way analysis of variance (ANOVA), which is the standard in the field (59).
Statistical analysis.
Unless otherwise noted, tests of significance herein are one-way ANOVA with Dunnett’s posttest for multiple comparisons. In the event of a significant Bartlett’s test, a Kruskal-Wallace test was utilized instead. Significant difference was determined against the result for the BKPyV-infected DMSO control unless otherwise noted by brackets on the graph. The GraphPad Prism analysis suite was utilized for all statistical analysis.
Ternary plots allow visualization of proportional data from three conditions or states and the visual and mathematical depiction of distinct populations from the data (28). Ternary analyses were performed using the ggtern package within the R software environment (version 3.4.3) using RStudio (version 1.1.447). Briefly, the percentages of cells in G1, S, and G2 (or M) phases under various conditions were determined by flow cytometry and plotted. The 95% confidence interval was determined by the Mahalanobis distance and log-ratio transformation. Ternary plot visualization was performed using the ggplot2 package, version 3.1.0.
ACKNOWLEDGMENTS
We thank the members of the Mengxi Jiang and Thompson laboratories for helpful discussions. We thank Peter Prevelige for careful readings of the manuscript.
This work was supported by the NIH (grant R01AI123162 to S.R.T.) and the training program in cell, molecular, and developmental biology T32 (grant T32 GM008111 to J.L.J.) led by Bradley Yoder at the University of Alabama at Birmingham.
REFERENCES
- 1.Bennett SM, Broekema NM, Imperiale MJ. 2012. BK polyomavirus: emerging pathogen. Microbes Infect 14:672–683. doi: 10.1016/j.micinf.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nickeleit V, Singh HK, Kenan DJ, Mieczkowski PA. 2018. The two-faced nature of BK-polyomavirus: Lytic infection or non-lytic large-T positive carcinoma. J Pathol 246:7–11. doi: 10.1002/path.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An P, Saenz Robles MT, Pipas JM. 2012. Large T antigens of polyomaviruses: amazing molecular machines. Annu Rev Microbiol 66:213–236. doi: 10.1146/annurev-micro-092611-150154. [DOI] [PubMed] [Google Scholar]
- 4.Bartek J, Vojtesek B, Grand RJ, Gallimore PH, Lane DP. 1992. Cellular localization and T antigen binding of the retinoblastoma protein. Oncogene 7:101–108. [PubMed] [Google Scholar]
- 5.Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. 1990. Polyoma small and middle T antigens and SV40 small T antigen form stable complexes with protein phosphatase 2A. Cell 60:167–176. doi: 10.1016/0092-8674(90)90726-U. [DOI] [PubMed] [Google Scholar]
- 6.Guergnon J, Godet AN, Galioot A, Falanga PB, Colle JH, Cayla X, Garcia A. 2011. PP2A targeting by viral proteins: a widespread biological strategy from DNA/RNA tumor viruses to HIV-1. Biochim Biophys Acta 1812:1498–1507. doi: 10.1016/j.bbadis.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Jiang M, Zhao L, Gamez M, Imperiale MJ. 2012. Roles of ATM and ATR-mediated DNA damage responses during lytic BK polyomavirus infection. PLoS Pathog 8:e1002898. doi: 10.1371/journal.ppat.1002898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Justice JL, Verhalen B, Jiang M. 2015. Polyomavirus interaction with the DNA damage response. Virol Sin 30:122–129. doi: 10.1007/s12250-015-3583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sowd GA, Li NY, Fanning E. 2013. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog 9:e1003283. doi: 10.1371/journal.ppat.1003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giglia-Mari G, Zotter A, Vermeulen W. 2011. DNA damage response. Cold Spring Harb Perspect Biol 3:a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marechal A, Zou L. 2013. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ. 2007. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A 104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Justice JL, Verhalen B, Kumar R, Lefkowitz EJ, Imperiale MJ, Jiang M. 2015. Quantitative proteomic analysis of enriched nuclear fractions from BK polyomavirus-infected primary renal proximal tubule epithelial cells. J Proteome Res 14:4413–4424. doi: 10.1021/acs.jproteome.5b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verhalen B, Justice JL, Imperiale MJ, Jiang M. 2015. Viral DNA replication-dependent DNA damage response activation during BK polyomavirus infection. J Virol 89:5032–5039. doi: 10.1128/JVI.03650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low J, Humes HD, Szczypka M, Imperiale M. 2004. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology 323:182–188. doi: 10.1016/j.virol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 16.Gordon K, Clouaire T, Bao XX, Kemp SE, Xenophontos M, de Las Heras JI, Stancheva I. 2014. Immortality, but not oncogenic transformation, of primary human cells leads to epigenetic reprogramming of DNA methylation and gene expression. Nucleic Acids Res 42:3529–3541. doi: 10.1093/nar/gkt1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charrier JD, Durrant SJ, Golec JM, Kay DP, Knegtel RM, MacCormick S, Mortimore M, O'Donnell ME, Pinder JL, Reaper PM, Rutherford AP, Wang PS, Young SC, Pollard JR. 2011. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem 54:2320–2330. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 18.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 19.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 20.Barnum KJ, O'Connell MJ. 2014. Cell cycle regulation by checkpoints. Methods Mol Biol 1170:29–40. doi: 10.1007/978-1-4939-0888-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao PN, Johnson RT. 1970. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature 225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- 22.Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J. 2004. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol 6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 23.Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P. 2011. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26:125–132. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- 24.Langie SA, Azqueta A, Collins AR. 2015. The comet assay: past, present, and future. Front Genet 6:266. doi: 10.3389/fgene.2015.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vendetti FP, Lau A, Schamus S, Conrads TP, O'Connor MJ, Bakkenist CJ. 2015. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget 6:44289–44305. doi: 10.18632/oncotarget.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pike KG, Barlaam B, Cadogan E, Campbell A, Chen Y, Colclough N, Davies NL, de-Almeida C, Degorce SL, Didelot M, Dishington A, Ducray R, Durant ST, Hassall LA, Holmes J, Hughes GD, MacFaul PA, Mulholland KR, McGuire TM, Ouvry G, Pass M, Robb G, Stratton N, Wang Z, Wilson J, Zhai B, Zhao K, Al-Huniti N. 2018. The identification of potent, selective, and orally available inhibitors of ataxia telangiectasia mutated (ATM) kinase: the discovery of AZD0156 (8-{6-[3-(dimethylamino)propoxy]pyridin-3-yl}-3-methyl-1-(tetrahydro-2 H-pyran-4-yl)-1,3-dihydro-2 H-imidazo[4,5- c]quinolin-2-one). J Med Chem 61:3823–3841. doi: 10.1021/acs.jmedchem.7b01896. [DOI] [PubMed] [Google Scholar]
- 27.Bennett SM, Zhao L, Bosard C, Imperiale MJ. 2015. Role of a nuclear localization signal on the minor capsid proteins VP2 and VP3 in BKPyV nuclear entry. Virology 474:110–116. doi: 10.1016/j.virol.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu B, Feng X, Burdine RD. 2010. Categorical data analysis in experimental biology. Dev Biol 348:3–11. doi: 10.1016/j.ydbio.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaltiel IA, Krenning L, Bruinsma W, Medema RH. 2015. The same, only different—DNA damage checkpoints and their reversal throughout the cell cycle. J Cell Sci 128:607–620. doi: 10.1242/jcs.163766. [DOI] [PubMed] [Google Scholar]
- 30.Perry JA, Kornbluth S. 2007. Cdc25 and Wee1: analogous opposites?. Cell Div 2:12. doi: 10.1186/1747-1028-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Kumagai A, Dunphy WG. 2001. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol Biol Cell 12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith A, Simanski S, Fallahi M, Ayad NG. 2007. Redundant ubiquitin ligase activities regulate wee1 degradation and mitotic entry. Cell Cycle 6:2795–2799. doi: 10.4161/cc.6.22.4919. [DOI] [PubMed] [Google Scholar]
- 33.Aarts M, Sharpe R, Garcia-Murillas I, Gevensleben H, Hurd MS, Shumway SD, Toniatti C, Ashworth A, Turner NC. 2012. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov 2:524–539. doi: 10.1158/2159-8290.CD-11-0320. [DOI] [PubMed] [Google Scholar]
- 34.Saldivar JC, Hamperl S, Bocek MJ, Chung M, Bass TE, Cisneros-Soberanis F, Samejima K, Xie L, Paulson JR, Earnshaw WC, Cortez D, Meyer T, Cimprich KA. 2018. An intrinsic S/G2 checkpoint enforced by ATR. Science 361:806–810. doi: 10.1126/science.aap9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemmens B, Hegarat N, Akopyan K, Sala-Gaston J, Bartek J, Hochegger H, Lindqvist A. 2018. DNA replication determines timing of mitosis by restricting CDK1 and PLK1 activation. Mol Cell 71:117–128.e113. doi: 10.1016/j.molcel.2018.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullah Z, de Renty C, DePamphilis ML. 2011. Checkpoint kinase 1 prevents cell cycle exit linked to terminal cell differentiation. Mol Cell Biol 31:4129–4143. doi: 10.1128/MCB.05723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gjelsvik KJ, Besen-McNally R, Losick VP. 2018. Solving the polyploid mystery in health and disease. Trends Genet 35:6–14. doi: 10.1016/j.tig.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown EJ, Baltimore D. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 39.Gamper AM, Choi S, Matsumoto Y, Banerjee D, Tomkinson AE, Bakkenist CJ. 2012. ATM protein physically and functionally interacts with proliferating cell nuclear antigen to regulate DNA synthesis. J Biol Chem 287:12445–12454. doi: 10.1074/jbc.M112.352310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cortez D, Glick G, Elledge SJ. 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci U S A 101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sowd GA, Mody D, Eggold J, Cortez D, Friedman KL, Fanning E. 2014. SV40 utilizes ATM kinase activity to prevent non-homologous end joining of broken viral DNA replication products. PLoS Pathog 10:e1004536. doi: 10.1371/journal.ppat.1004536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orba Y, Suzuki T, Makino Y, Kubota K, Tanaka S, Kimura T, Sawa H. 2010. Large T antigen promotes JC virus replication in G2-arrested cells by inducing ATM- and ATR-mediated G2 checkpoint signaling. J Biol Chem 285:1544–1554. doi: 10.1074/jbc.M109.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szmyd R, Niska-Blakie J, Diril MK, Renck Nunes P, Tzelepis K, Lacroix A, van Hul N, Deng LW, Matos J, Dreesen O, Bisteau X, Kaldis P. 2019. Premature activation of Cdk1 leads to mitotic events in S phase and embryonic lethality. Oncogene 38:998–1018. doi: 10.1038/s41388-018-0464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Witt Hamer PC, Mir SE, Noske D, Van Noorden CJF, Würdinger T. 2011. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin Cancer Res 17:4200–4207. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
- 45.Li C, Andrake M, Dunbrack R, Enders GH. 2010. A bifunctional regulatory element in human somatic Wee1 mediates cyclin A/Cdk2 binding and Crm1-dependent nuclear export. Mol Cell Biol 30:116–130. doi: 10.1128/MCB.01876-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer N, Brandner J, Fuchs F, Moll I, Grundhoff A. 2010. Detection of Merkel cell polyomavirus (MCPyV) in Merkel cell carcinoma cell lines: cell morphology and growth phenotype do not reflect presence of the virus. Int J Cancer 126:2133–2142. doi: 10.1002/ijc.24877. [DOI] [PubMed] [Google Scholar]
- 48.Tikhanovich I, Nasheuer HP. 2010. Host-specific replication of BK virus DNA in mouse cell extracts is independently controlled by DNA polymerase alpha-primase and inhibitory activities. J Virol 84:6636–6644. doi: 10.1128/JVI.00527-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shah KV, Daniel RW, Strandberg JD. 1975. Sarcoma in a hamster inoculated with BK virus, a human papovavirus. J Natl Cancer Inst 54:945–950. [PubMed] [Google Scholar]
- 50.Eddy BE, Borman GS, Grubbs GE, Young RD. 1962. Identification of the oncogenic substance in rhesus monkey kidney cell culture as simian virus 40. Virology 17:65–75. doi: 10.1016/0042-6822(62)90082-X. [DOI] [PubMed] [Google Scholar]
- 51.Abend JR, Low JA, Imperiale MJ. 2007. Inhibitory effect of gamma interferon on BK virus gene expression and replication. J Virol 81:272–279. doi: 10.1128/JVI.01571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang M, Abend JR, Tsai B, Imperiale MJ. 2009. Early events during BK virus entry and disassembly. J Virol 83:1350–1358. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vassilev LT. 2006. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell. Cell Cycle 5:2555–2556. doi: 10.4161/cc.5.22.3463. [DOI] [PubMed] [Google Scholar]
- 54.Kotogany E, Dudits D, Horvath GV, Ayaydin F. 2010. A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6:5. doi: 10.1186/1746-4811-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 56.Bernhoff E, Gutteberg TJ, Sandvik K, Hirsch HH, Rinaldo CH. 2008. Cidofovir inhibits polyomavirus BK replication in human renal tubular cells downstream of viral early gene expression. Am J Transplant 8:1413–1422. doi: 10.1111/j.1600-6143.2008.02269.x. [DOI] [PubMed] [Google Scholar]
- 57.International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 58.Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement MV. 2014. OpenComet: an automated tool for comet assay image analysis. Redox Biol 2:457–465. doi: 10.1016/j.redox.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moller P, Loft S. 2014. Statistical analysis of comet assay results. Front Genet 5:292. doi: 10.3389/fgene.2014.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]