Understanding how viruses of microbes interact with its hosts is not only important from a basic scientific point of view but also for a better comprehension of the evolution of life. Studies involving large and giant viruses have revealed original and outstanding mechanisms concerning virus-host relationships. Here, we report a mechanism developed by Vermamoeba vermiformis, a free-living amoeba, to reduce Faustovirus mariensis dissemination. Once infected, V. vermiformis cells release a factor that induces the encystment of neighbor cells, preventing infection of further cells and/or trapping the viruses and viral factories inside the cyst walls. This phenomenon reinforces the need for more studies regarding large/giant viruses and their hosts.
KEYWORDS: antiviral, cysts, faustovirus, vermoameba, virus control
ABSTRACT
Viruses depend on cells to replicate and can cause considerable damage to their hosts. However, hosts have developed a plethora of antiviral mechanisms to counterattack or prevent viral replication and to maintain homeostasis. Advantageous features are constantly being selected, affecting host-virus interactions and constituting a harsh race for supremacy in nature. Here, we describe a new antiviral mechanism unveiled by the interaction between a giant virus and its amoebal host. Faustovirus mariensis infects Vermamoeba vermiformis, a free-living amoeba, and induces cell lysis to disseminate into the environment. Once infected, the cells release a soluble factor that triggers the encystment of neighbor cells, preventing their infection. Remarkably, infected cells stimulated by the factor encyst and trap the viruses and viral factories inside cyst walls, which are no longer viable and cannot excyst. This unprecedented mechanism illustrates that a plethora of antiviral strategies remains to be discovered in nature.
IMPORTANCE Understanding how viruses of microbes interact with its hosts is not only important from a basic scientific point of view but also for a better comprehension of the evolution of life. Studies involving large and giant viruses have revealed original and outstanding mechanisms concerning virus-host relationships. Here, we report a mechanism developed by Vermamoeba vermiformis, a free-living amoeba, to reduce Faustovirus mariensis dissemination. Once infected, V. vermiformis cells release a factor that induces the encystment of neighbor cells, preventing infection of further cells and/or trapping the viruses and viral factories inside the cyst walls. This phenomenon reinforces the need for more studies regarding large/giant viruses and their hosts.
INTRODUCTION
Virion release and dissemination are sine qua non conditions for the maintenance of most of viral species in nature. Evolutionary constraints have shaped a variety of mechanisms promoting viral particle release and dissemination from infected cells; these range from mature virions budding through the cell membrane to virus-induced cell lysis. As a response, hosts have developed mechanisms to block or reduce viral particle propagation, replication, or both through host populations in a constant struggle against viruses. The Red Queen theory illustrates such a race for supremacy, in which advantageous features are selected, and this changes, at least temporarily and spatially, the balance of the interaction to one of the sides. Examples of hosts limiting viruses can be found in many groups of organisms, from bacterial antibacteriophage defenses (1), to plants silencing viral genes (2), and to the importance of pattern recognition receptors (PRRs) capable of triggering immune responses for multicellular host species (3). As a remarkable example, the interferon (IFN) system acts as a major player against viral propagation in vertebrates (4–6). Host cells recognize viral molecules through PRRs, resulting in signaling pathways that lead to the production of IFN molecules. These are secreted and act in paracrine and autocrine ways by activating a second round of signaling, this time responsible for establishing an antiviral state, which leads to cell death in the case of infection. Although some cells are infected and lysed, IFN signaling reduces virus propagation and total viral load (6).
In this context, comprehensive studies involving large and giant viruses have revealed original and outstanding mechanisms concerning virus-host relationships. A complex interaction involving three players has been described for the free-living protist Cafeteria roenbergensis, in which a provirophage (mavirus) integrated in the genome of the protist imparts a partial protection to C. roenbergensis populations in case of an eventual infection by a lytic giant virus called Cafeteria roenbergensis virus (CroV) (7). It has been also demonstrated that haploid cells (but not diploid cells) of Emiliania huxleyi, a free-living marine protist, are refractory to Emiliania huxleyi virus 86 infection in an evasion mechanism known as Cheshire Cat (8). Analogously, our team described the same phenomenon for free-living amoebas belonging to the genus Acanthamoeba, in which the cysts, but not the trophozoitic forms, are resistant to mimivirus infection (9, 10). We have demonstrated that, once infected by mimivirus, Acanthamoeba trophozoites are no longer able to encyst because mimivirus blocks the expression of a serine proteinase gene, a canonical element involved in the encystment process (10). Although it has been described that Acanthamoeba trophozoites are able to encyst in the presence of some intracellular bacteria (11), this phenomenon has never been described during amoebal virus infections.
Here, we report a mechanism developed by Vermamoeba vermiformis, a free-living amoeba, to reduce Faustovirus mariensis dissemination and consequently protect neighbor cells. Once infected, V. vermiformis cells release a nonproteic soluble factor that induces the encystment of neighbor cells, preventing infection of further cells, since Faustovirus is only able to infect trophozoites. Interestingly, if cells already infected are exposed to the soluble encystment factor, they encyst and trap the viruses and viral factories inside the cyst walls. Unlike what has been described for amoebal cysts containing intracellular bacteria, cysts enclosing Faustovirus particles, factories, or both are no longer viable and cannot excyst, thus trapping viral progeny irreversibly inside the thick cyst walls and promoting an effective reduction of viral load on, and dissemination to, the amoebal population.
RESULTS
Faustovirus mariensis: isolation and genomic analysis.
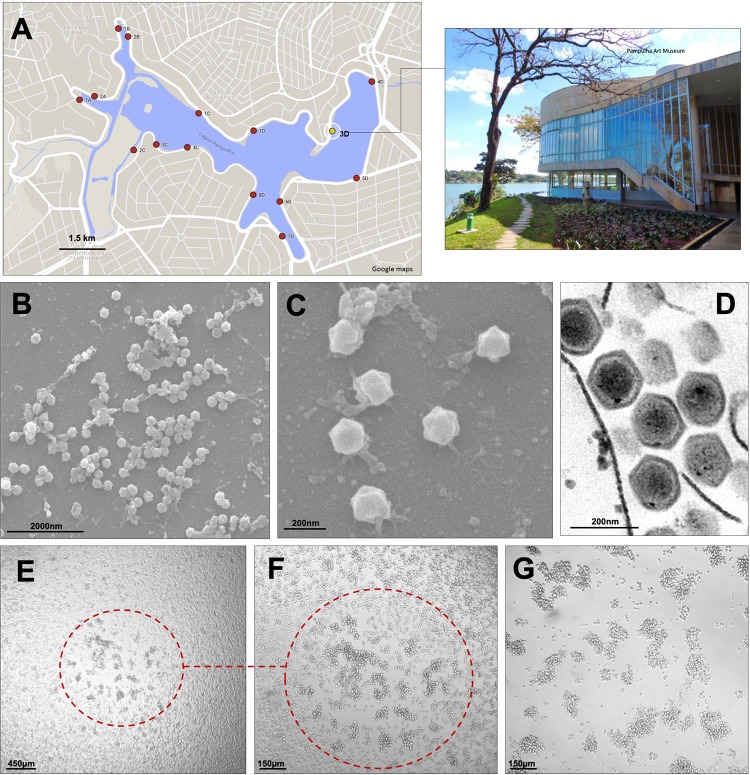
While attempting to isolate new amoebal viruses, we performed collections of surface water samples at Pampulha Lagoon in Belo Horizonte, Brazil. One of the samples, collected in front of the Pampulha Art Museum (Fig. 1A), induced a cytopathic effect (CPE) in V. vermiformis, a free-living amoeba that occurs worldwide and is already described as one of the hosts of some giant or large viruses, including Faustovirus, Tupanvirus, Kaumoebavirus, and Orpheovirus. Transmission and scanning electron microscopy (TEM and SEM, respectively) of cells presenting CPE revealed viral particles similar to faustoviruses, with approximately 190 nm, icosahedral symmetry, and a capsid containing an electrodense central region (genome) (Fig. 1B to D). We named this new isolate Faustovirus mariensis. During virus propagation and purification, we observed that F. mariensis induces the formation of plaque-forming units, a CPE not previously described for any other amoebal giant virus according to our knowledge (Fig. 1E and F). During the time course of infection, plaque units expand and coalesce in V. vermiformis monolayers (Fig. 1G).
FIG 1.
Faustovirus mariensis isolation sites, particle images, and cytopathic effects. (A) Pampulha Lagoon map with collection sites highlighted (dots). The yellow dot represents where F. mariensis was collected, in front of the Pampulha Art Museum (top right of photo). Map courtesy of Google Maps. (B to D) F. mariensis viral particles visualized by scanning (B and C) and transmission (D) electron microscopy. (E to G) Plaque-forming unit (PFU) induced by F. mariensis infection in a Vermamoeba vermiformis monolayer. (F) Closeup of a PFU shown in panel E, observed 24 h postinfection. (G) Forty-eight hours postinfection, the PFUs expand and coalesce.
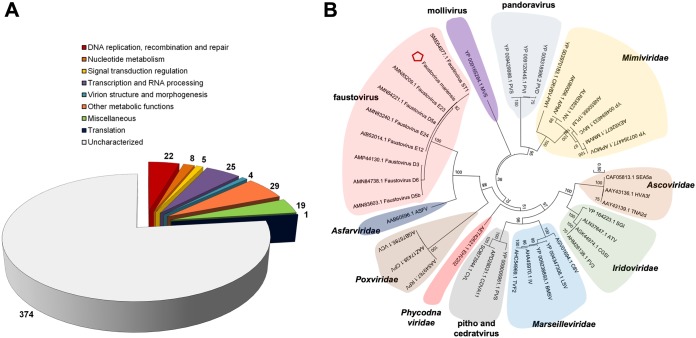
The genome of F. mariensis is a circular, double-stranded DNA molecule of 466,080 bp in length (see Fig. S1 at https://5c95043044c49.site123.me/my-blog/supp-material-trapping-the-enemy-vermamoeba-vermiformis-circumvents-faustovirus-mariensis-dissemination-by-enclosing-viral-progeny-inside-cysts). The GC content is 36%, and it was predicted to encode 483 genes (210 located at the negative strand; 273 at the positive strand), with a coding density of 90%. The predicted proteins had a mean length (plus or minus standard deviation) of 279 ± 258 amino acids (ranging from 53 to 2,980 amino acids). A total of 374 proteins (77.4%) had no known function (Fig. 2A). The major functional gene categories were represented by DNA replication, recombination, and repair, as well as by transcription and RNA processing, with 22 and 25 genes, respectively (Fig. 2A), with additional genes for DNA polymerase, D5 primase helicase, topoisomerase II, different subunits of DNA-directed RNA polymerase, mRNA capping enzymes, and transcription factors, among others. No tRNA was predicted and no open reading frames with no detectable homology to other open reading frames (ORFs) in a database (ORFans) were detected. Differently from other giant viruses, only one translation factor is encoded by F. mariensis, the translation initiation factor SUI1, which was the only translation-related gene also observed in Faustovirus E12, the first member of this new group of viruses to be described (12). Other genes previously described for faustoviruses were also encoded in F. mariensis. These include two adjacent polyproteins of 220 kDa and 60 kDa, a ribosomal protein acetyltransferase, and some genes belonging to large paralogous families, such as membrane occupation and recognition nexus (MORN) repeat-containing proteins and ankyrin repeat-containing proteins, although only two genes were found to have this repeat domain (Fig. 2A). Synteny analysis revealed a highly conserved genome organization among faustoviruses. However, the F. mariensis genome exhibited a small region (∼11.5 kb) that was rearranged in its genome in regard to those of the other described faustoviruses, i.e., the region was located after a conserved block of >30 kb in F. mariensis and before that block in other viruses (see Fig. S2 at https://5c95043044c49.site123.me/my-blog/supp-material-trapping-the-enemy-vermamoeba-vermiformis-circumvents-faustovirus-mariensis-dissemination-by-enclosing-viral-progeny-inside-cysts). This region comprised 18 genes, most of them with unknown functions. The analysis of DNA polymerase, a marker used for phylogenetic analysis of nucleocytoplasmic large-DNA viruses (NCLDVs) confirmed that F. mariensis was clustered with other Faustovirus isolates and was more related to the clade containing Faustovirus E12 (Fig. 2B).
FIG 2.
Gene categories and phylogeny. (A) F. mariensis gene set classified according to predicted gene categories. (B) DNA polymerase subunit B tree, constructed using the maximum likelihood evolution method and 1,000 replicates. Faustovirus mariensis (pentagon) clusters with other Faustovirus strains. Tree scale represents evolutionary distance.
Faustovirus mariensis replication cycle.
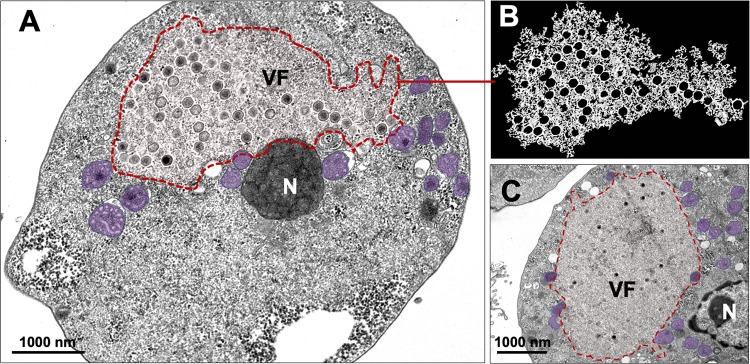
The analyses of the F. mariensis replication cycle in V. vermiformis were performed based on asynchronous infection (multiplicity of infection [MOI] of 0.1) starting from fresh trophozoites and purified viral particles. We analyzed whether V. vermiformis cysts would be permissive to F. mariensis infection, but neither cytopathic effects nor increases in viral loads were observed; therefore, Faustovirus, as has been described for Mimivirus, needs to infect amoebal trophozoites to start its replication cycle. In the cytoplasm of trophozoites, F. mariensis induced the formation of large electron-lucent viral factories (about 3.5 μm in length) frequently observed close to the cell nucleus (Fig. 3A to C). Viral infection induced the recruitment of mitochondria to the periphery of the viral factory, suggesting virus-induced energy optimization during viral morphogenesis (Fig. 3A and C). Curiously, intranuclear particles were also observed, but further studies are necessary to determine their role in the F. mariensis replication cycle (see Fig. S3 at https://5c95043044c49.site123.me/my-blog/supp-material-trapping-the-enemy-vermamoeba-vermiformis-circumvents-faustovirus-mariensis-dissemination-by-enclosing-viral-progeny-inside-cysts).
FIG 3.
Electron-lucent viral factory and cytoplasmic modifications induced by Faustovirus mariensis. (A to C) F. mariensis presents an electron-lucent viral factory (contoured in red and shown in detail in panel B), which was not easily distinguished from the rest of the cytoplasm and was observed at the perinuclear region. It is possible to visualize the abundant presence of mitochondria surrounding the viral factory (purple highlights in panels A and C). VF, viral factory; N, nucleus. Image B was obtained by TEM and graphically highlighted by using IOS image visualization software (Apple Technology Company).
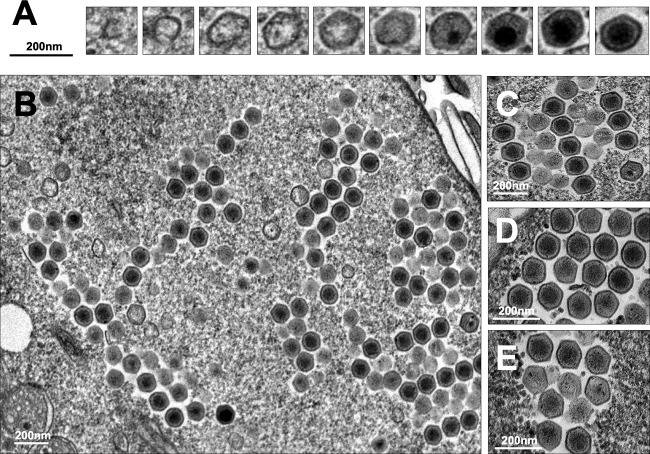
As observed for other viruses of free-living amoebae (13–16), F. mariensis morphogenesis began with crescents, open structures of approximately 50 nm, which grew as the electron-dense material of the viral factory filled them (Fig. 4A). Particles of almost 200 nm without genomic content were observed in the late phases of morphogenesis, when the genome was incorporated and centralized within several newly formed viral particles (Fig. 4A). Faustovirus mariensis progeny were organized in a honeycomb fashion inside viral factories, as previously described for other Faustovirus isolates (12) (Fig. 4B to E). Small honeycombs expanded as new mature viruses formed and coalesced with others in the cytoplasm (Fig. 4B). By the end of the replication cycle, the cytoplasm was fully taken by new F. mariensis particles (see Fig. S4 at https://5c95043044c49.site123.me/my-blog/supp-material-trapping-the-enemy-vermamoeba-vermiformis-circumvents-faustovirus-mariensis-dissemination-by-enclosing-viral-progeny-inside-cysts). Contrary to what has been described for Marseillevirus and Pandoravirus, exocytosis was not observed (16). Cellular lysis is likely the most important form of liberation of newly formed F. mariensis particles. Isolated particles or aggregates of F. mariensis associated with cellular structures were observed extracellularly, and aggregates became as large as a trophozoite of V. vermiformis (see Fig. S4 at https://5c95043044c49.site123.me/my-blog/supp-material-trapping-the-enemy-vermamoeba-vermiformis-circumvents-faustovirus-mariensis-dissemination-by-enclosing-viral-progeny-inside-cysts).
FIG 4.
Faustovirus mariensis morphogenesis and particle organization in honeycomb-like structures. (A) F. mariensis morphogenesis begins with crescents, open structures of approximately 50 nm, which grow as an electron-dense material of the viral factory fills them. Particles of almost 200 nm without genomic content are observed in late phases of morphogenesis, when the genome is incorporated and centralized within several newly formed viral particles. (B to E) F. mariensis progeny are organized in a honeycomb fashion inside viral factories. Small honeycombs expand as new mature viruses are formed and coalesce to others in the cytoplasm (B).
V. vermiformis cells may encyst and trap F. mariensis particles and factories inside cyst wall.
Interestingly, during routine production and titration of F. mariensis, we observed a substantial formation and accumulation of V. vermiformis cysts, especially when cells were infected at high MOIs (1 and 10). This curious effect drew our attention, and we decided to verify whether other viruses would be able to trigger encystment. However, V. vermiformis infection with Tupanvirus or Orpheovirus did not induce the formation and accumulation of cysts in the culture flasks, regardless of the MOI (data not shown). As such phenomena seemed to be a singular characteristic of F. mariensis, we decided to further characterize them.
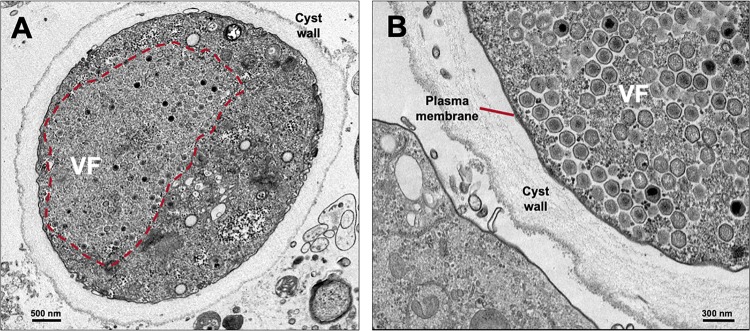
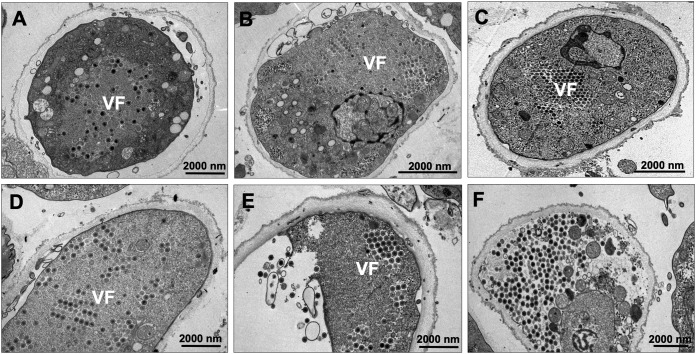
Hence, V. vermiformis cells were infected with F. mariensis at an MOI of 10 and prepared for transmission electron microscopy (TEM; 24 hours postinfection [hpi]). Remarkably, TEM images revealed F. mariensis particles and viral factories not only inside a few V. vermiformis trophozoites but also within the cytoplasm of many cysts and cells under encystment (Fig. 5). This phenomenon, previously described for some bacteria (Salmonella enterica, Listeria monocytogenes, Yersinia enterocolitica, and Escherichia coli) (11), had never been described, to our knowledge, for viruses. The observation of more than one hundred cysts revealed the presence of F. mariensis under distinct phases of the replication cycle, including the early viral factory formation, late morphogenesis, honeycomb formation/coalescence and, at last, cytoplasm filled with mature viral particles (Fig. 6A to F). In cases of cells filled with viral particles (late cycle), some cellular structures seemed to be degraded, including mitochondria and plasma membrane (Fig. 6F). Despite the observation of variations in wall thickness among infected cysts, the cyst walls appeared intact and with no damage (Fig. 6).
FIG 5.
Faustovirus mariensis particles and viral factories inside Vermamoeba vermiformis cysts. (A) Mature V. vermiformis cyst enclosing a large viral factory (highlighted with a red dashed line). (B) Plasma membrane is clearly visible below the thick cyst wall. For this experiment, V. vermiformis cells were infected with F. mariensis at an MOI of 10 and prepared for transmission electron microscopy (TEM; 24 hpi).
FIG 6.
Vermamoeba vermiformis cysts enclosing Faustovirus mariensis in different stages of viral replication cycle (A to F). The observation of more than 100 cysts revealed the presence of F. mariensis during distinct phases of the replication cycle, including early viral factory formation (A and B), late morphogenesis/honeycomb formation/coalescence (C to E), and cytoplasm filled with mature viral particles (F).
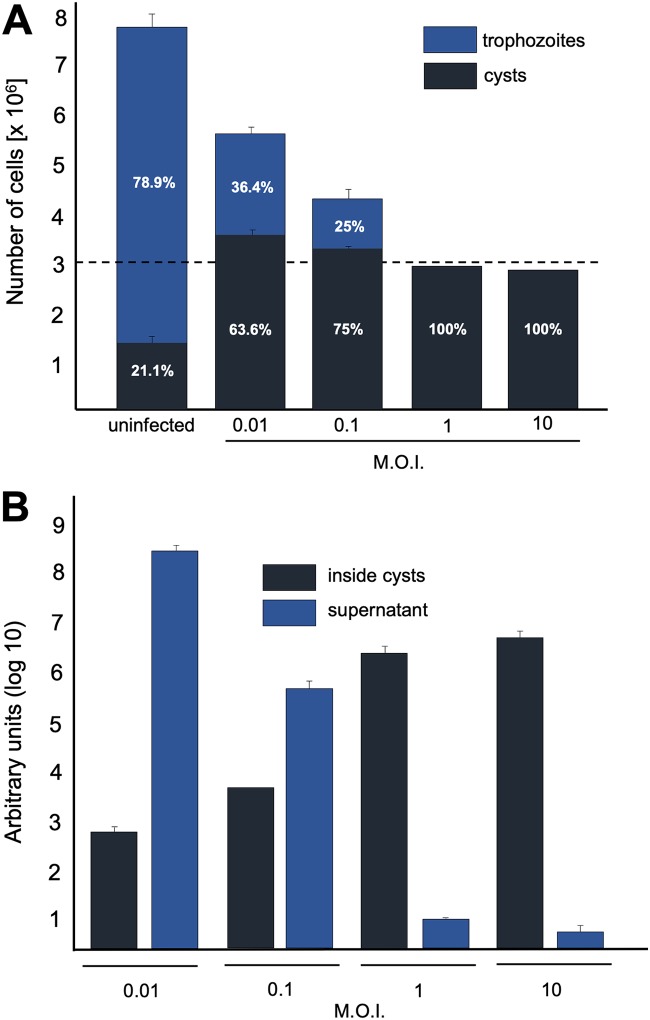
We then quantified the number of cysts of V. vermiformis formed after the inoculation of F. mariensis at different MOIs. A total of 3 × 106 trophozoites were added to T25 flasks with 5 ml of peptone yeast extract glucose (PYG) medium (this medium favors the propagation of amoebas) and inoculated at MOIs of 0.01, 0.1, 1, or 10. After adsorption, cells were washed and then incubated for 48 h with PYG medium. Uninfected cells were used as controls and, after 48 h of incubation, we measured the propagation of those cells, which reached approximately 7.8 × 106 cells; among those cells, we observed a natural encystment rate of approximately 21%, which is related to the reduction of nutrients in the PYG medium during the time of incubation (Fig. 7A). However, V. vermiformis trophozoites infected by F. mariensis presented a higher rate of cysts 48 hpi, and this number was positively related to the MOI. Flasks inoculated at MOIs of 0.01 or 0.1 resulted in 63.6% and 75% of cysts, respectively; while flasks inoculated at higher MOIs, 1 or 10, both presented 100% cysts at 48 hpi (Fig. 7A). Interestingly, infections with high MOIs (1 and 10) presented an encystment ratio (number of cysts obtained divided by the number of trophozoites imputed) of approximately 1:1, close to the initial input of trophozoites. These results indicated that trophozoites infected at high MOIs (1 or 10) with F. mariensis were almost totally converted to cysts (Fig. 7A). We also analyzed whether F. mariensis inactivated particles (same MOI as that described above) would be able to trigger V. vermiformis encystment, but we could not observe differences in the number of cysts between inoculated and noninoculated cells, indicating that the triggering of encystment is dependent on virus entry and replication.
FIG 7.
Cyst formation and viral replication at different multiplicities of infection (MOI). (A) Cyst and trophozoite quantification 48 h after the inoculation of F. mariensis at MOIs of 0.01, 0.1, 1, and 10. The dashed line represents the input of amoebae in the beginning of the experiment (3 × 106). Uninfected cells had a natural encystment rate of 21.1%. (B) F. mariensis genome quantification in the supernatants and cysts of V. vermiformis inoculated at different MOIs 1 week postinoculation. The supernatants and cysts were separated by centrifugation, and the viral genome load was quantified by qPCR (DNA polymerase subunit B). Error bars indicate standard deviations (SDs). These experiments were performed three times in triplicate.
The next step was to measure F. mariensis replication in V. vermiformis cells inoculated at different MOIs. Therefore, V. vermiformis cells were infected with F. mariensis at MOIs of 0.01, 0.1, 1, and 10, and incubated for 1 week. At this stage, trophozoites were no longer observed, regardless of the MOI, since they were either lysed (releasing viruses to supernatant) or converted to cysts. This longer time of incubation was designed to allow the lysis or encystment of all trophozoites in the system; therefore, at the end of the experiments, we would find viruses only in the supernatants (originated from lysis of infected trophozoites) or inside cysts. The supernatants and cysts were separated by centrifugation and viral genome load was quantified by quantitative PCR (qPCR). A substantial replication of the virus was correlated with low MOIs, with larger amounts of DNA detected in the supernatant at MOIs of 0.01 and 0.1 (Fig. 7B). Higher MOIs (1 and 10) however, presented relatively small amounts of viral DNA in the supernatant but a substantial amount inside cysts, indicating that the virus was able to initiate the infection, but not to release its progeny, after having its genome (and particles) imprisoned within the newly formed cysts (Fig. 7B).
Faustovirus infected cysts fail excystment.
Studies demonstrated that several intracellular bacteria are able to survive and take advantage of amoebal encystment (11, 17). It has been suggested this feature would guarantee protection of bacteria during adverse environmental conditions while inside the cyst. Once good environmental conditions are available, these bacteria could return to multiply and maintain their life cycle upon excystment. At a first glance, F. mariensis could possibly use this same stratagem to eventually return to replicate in new trophozoites.
To test this hypothesis, trophozoites of V. vermiformis were infected with different MOIs of F. mariensis diluted 10-fold from 0.01 to 10. Cysts produced after the infection were collected, washed to remove external viral particles, and then their potential of excystment was evaluated. Two different methods to trigger excystment were used, exposure of cysts to PYG media with 5% of fetal bovine serum (FBS) in T25 cell flasks and plating of cysts on petri dishes with Bacto agar covered with a monolayer of heat-inactivated Escherichia coli cells.
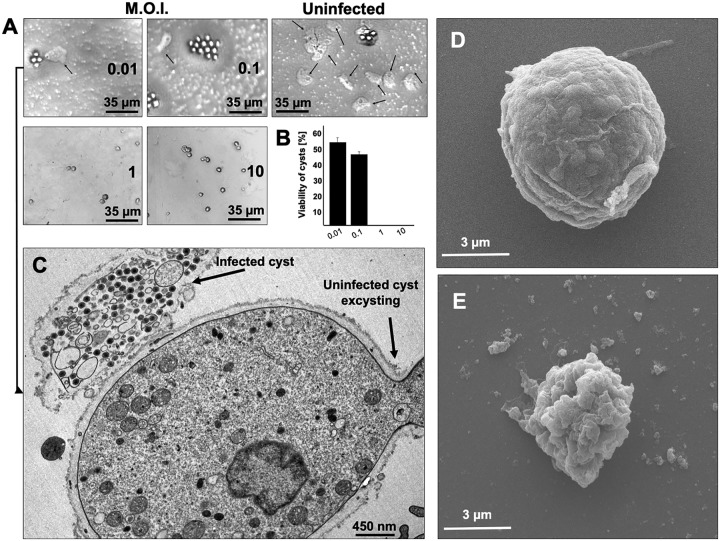
Surprisingly, cysts derived from the higher MOIs (1 and 10) did not excyst upon either method employed (Fig. 8A). Those cysts were then analyzed by TEM, revealing viral particles inside almost all of the cysts. Curiously, some of those cysts, in spite of appearing to have a typical thick wall, had a reduced diameter (about 3.5 μm) and had lost their rounded shape (Fig. 8C to E). For cysts derived from infections with MOIs of 0.01 and 0.1, excystment was observed for nearly 24% of cells. The TEM analysis revealed that only cysts not presenting viral particles, factories, or both in their cytoplasm were able to excyst (Fig. 8C). These data were confirmed by subculturing individual trophozoites obtained after excystment (from cultures inoculated at MOIs of 0.01 and 0.1), which did not show any cytopathic effect or detection of viral particles. As described for cells infected at MOIs of 1 and 10, we also observed infected cysts with reduced diameters and irregular shapes in flasks inoculated at MOIs of 0.01 and 0.1 (Fig. 8C to E).
FIG 8.
Excystment assays. Cysts were produced by the inoculation of Vermamoeba vermiformis trophozoites with Faustovirus mariensis at MOIs of 0.01, 0.1, 1, and 10, and then their excystment potential was evaluated. (A) Agar plate excystment assay demonstrating that few cysts obtained from infections at MOIs of 0.01 and 0.1 were able to become trophozoites (arrows). Excystment was not observed for cysts obtained from infections at MOIs of 1 and 10. (B) Viability of cysts produced from infections at different MOIs (trypan blue, 0.4%). Error bars indicate SDs. (C) Transmission electron microscopy representative image demonstrating that only uninfected cysts are able to excyst. This image corresponds to cysts obtained from infections at an MOI of 0.01, after excystment stimulus. (D and E) Cysts produced after infection can present either a regular shape (D) or reduced diameters and irregular shapes (E). These experiments were performed three times in triplicate.
When the viability of cysts was assayed with 0.4% trypan blue, it was revealed that 100% of cysts from cells inoculated at MOIs of 1 and 10 were no longer viable, while 46% and 57% of cysts from flasks inoculated at MOIs of 0.01 and 0.1 were not viable, respectively (Fig. 8B). Although not viable, infected cysts were as resistant as uninfected cysts against hydrochloric acid and heating treatments followed by 3 cycles of sonication. They kept their walls visually intact and preserved intracellular structures, including viruses and factories (see Fig. S5 at https://5c95043044c49.site123.me/my-blog/supp-material-trapping-the-enemy-vermamoeba-vermiformis-circumvents-faustovirus-mariensis-dissemination-by-enclosing-viral-progeny-inside-cysts). Taken together, these results indicate that V. vermiformis encystment was triggered by F. mariensis infection in an MOI-dependent way. Once encysted, infected cells were no longer able to excyst, thus irreversibly imprisoning the viral progeny within their thick walls.
Faustovirus mariensis is unable to circumvent V. vermiformis encystment.
Our team has previously shown that Mimivirus was capable of inhibiting the process of encystment in Acanthamoeba castellanii by downregulating the expression of a cellular serine protease gene, which is an essential enzyme responsible for triggering the encystment pathway in these cells (10). Once infected by Mimivirus, a given trophozoite of A. castellanii inevitably suffers from lysis due to viral replication, since the virus blocks encystment. In contrast, when A. castellanii trophozoites were incubated with an encystment stimulation factor (e.g., Neff solution) previous to Mimivirus inoculation, serine proteinase gene expression was stimulated, which triggered encystment and circumvented Mimivirus replication. Therefore, avoiding or blocking the triggering of encystment is critical for amoebal virus replication, since they are able to replicate only in nonencysting trophozoites. Herein, we investigated whether F. mariensis could be able to block V. vermiformis encystment.
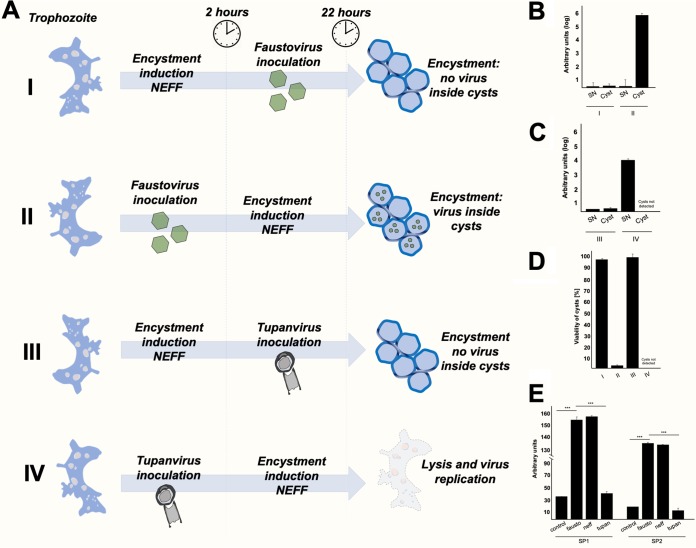
Briefly, V. vermiformis trophozoites were treated with Neff solution and this solution was removed 2 h later; the purified virus was inoculated at an MOI of 10, then flasks were added with PYG medium and incubated for 24 h. Virus genome replication was assayed by qPCR, and cells were analyzed by TEM and optical microscopy. Cell viability was analyzed using trypan blue. After 24 h, virus genome replication was not observed, and all trophozoites were converted into viable uninfected cysts (Fig. 9A to D; see also Fig. S6 at https://5c95043044c49.site123.me/my-blog/supp-material-trapping-the-enemy-vermamoeba-vermiformis-circumvents-faustovirus-mariensis-dissemination-by-enclosing-viral-progeny-inside-cysts). Alternatively, we inoculated V. vermiformis trophozoites with F. mariensis at an MOI of 10; then, after 2 h, we treated the cells with NEFF solution, and then cells were incubated for 24 h. In this case, we detected viral genome replication inside cysts (about 6 logs, arbitrary units); and most of the cysts (nonviable) had viral particles or factories in the cytoplasm (Fig. 9A to D and Fig. S6 at the above website). As an experimental control, we submitted Tupanvirus, a Mimivirus relative, to the same experiments (MOI of 10). It was not possible to detect Tupanvirus genome replication in V. vermiformis pretreated with NEFF; there were neither viral particles nor genomes inside cysts (Fig. 9A to D and Fig. S6 at the website given above). On the other hand, when V. vermiformis trophozoites were infected with Tupanvirus and then treated with NEFF, the virus genome was able to replicate (about four logs, arbitrary units), cells were lysed, and no cyst formation was observed (Fig. 9A to D and Fig. S6 at the above website).
FIG 9.
Faustovirus mariensis is not able to circumvent Vermamoeba vermiformis encystment. (A) Scheme highlighting the experimental strategy and results of an experiment testing the ability of F. mariensis and Tupanvirus to circumvent V. vermiformis encystment. In I and III, the encystment solution Neff was added prior to virus inoculation. In II and IV, the viruses were inoculated before Neff addition. (B) F. mariensis and (C) Tupanvirus genome quantification, in supernatant and cysts, for each experimental scenario analyzed (I to IV), by qPCR. The relative quantification was performed with the ΔCT method, and results were presented as arbitrary units (log10). (D) Quantification of the viability of cysts (0.4% trypan blue) produced from infections under different scenarios (I to IV). (E) Relative quantification of the expression of two serine proteinase mRNA isotypes present in V. vermiformis upon infection (MOI of 10) with F. mariensis or Tupanvirus or upon Neff treatment. The quantification was performed 5 h postinfection or after Neff inoculation. Error bars indicate SDs. The statistical significance was calculated using a two-tailed 2-way analysis of variance (ANOVA) test and Tukey’s range test, using GraphPad Prism. ***, P < 0.001. These experiments were performed three times in triplicate.
As previously mentioned, the encystment process is triggered after the expression of cellular serine proteinases. The catalytic site of such proteases is composed of a triad of three amino acids, His 57, Ser 195 and Asp 102. The importance of such enzymes for Acanthamoeba encystment has been demonstrated by measuring the levels of serine proteinase transcripts and protein during encystment process (9, 10). Inhibition of serine proteinase gene transcription negatively affects encystment. We measured (by qPCR) the expression levels of two serine proteinase mRNA isotypes present in V. vermiformis upon infection (MOI of 10) with F. mariensis, Tupanvirus, or NEFF treatment. Five hours postinfection, F. mariensis induced significant expression of both analyzed serine proteinase isotypes (P < 0.001), at levels comparable to those detected in cells treated with the encystment stimulation solution NEFF (Fig. 9E). In contrast, Tupanvirus circumvented the expression of V. vermiformis serine proteinases, keeping the levels similar to those of uninfected or untreated trophozoites (Fig. 9E). Altogether, these results suggest that F. mariensis was incapable of downregulating factors that trigger the encystment of V. vermiformis, as has been described for Mimivirus. Tupanvirus, however, demonstrated a behavior in V. vermiformis similar to that of Mimivirus in A. castellanii.
Infected trophozoites release soluble factors that trigger the encystment.
Previous studies have demonstrated that the secretion of soluble factors can modulate the process of encystment in A. castellanii (18). Although the nature of such factors remains to be better characterized, it has been suggested they could act as major players in a communication system among amoebas, stimulating cell encystment as a response to harsh conditions. To investigate whether V. vermiformis cells infected by F. mariensis secrete factors involved with encystment, the supernatant of a culture infected at an MOI of 10 was collected 4 days postinfection (dpi). The supernatant was filtered with a 0.1-μm pore size filter to remove all viral particles and subsequently diluted 2-fold to inoculate new cultures of V. vermiformis trophozoites.
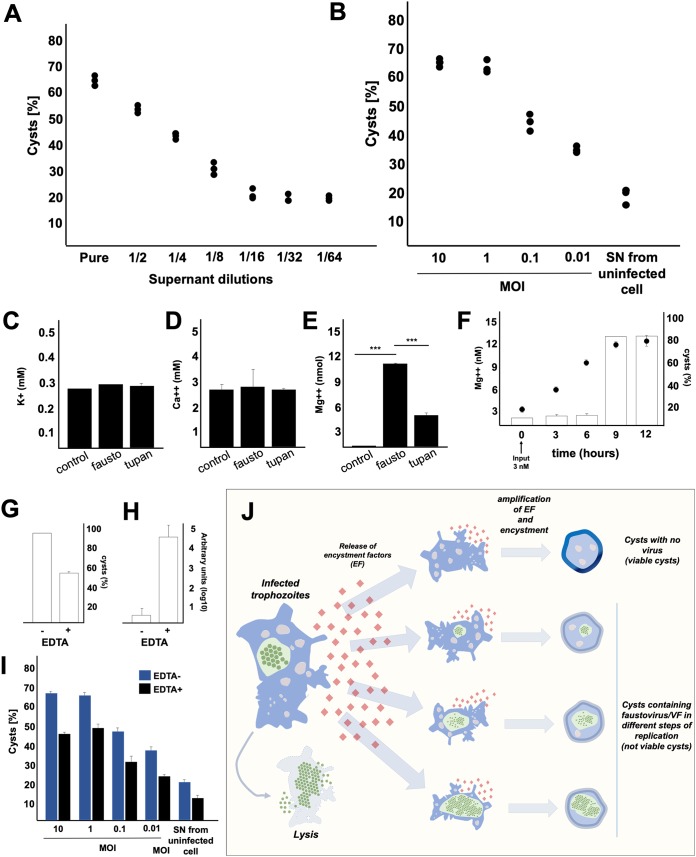
Remarkably, we observed a strong induction of encystment in V. vermiformis cells in a dose-dependent fashion by the virus-free supernatant from previous F. mariensis infections. Undiluted (pure) supernatant induced the encystment of approximately 67% of cells (Fig. 10A). Supernatants from V. vermiformis cells infected by F. mariensis at MOIs of 0.01, 0.1, and 1 were also collected, filtered, and inoculated (undiluted) into fresh V. vermiformis cells. Supernatants with MOIs of 0.01, 0.1, and 1 caused encystment of nearly 35%, 43%, and 65% of cells, respectively (Fig. 10B). To determine the nature of this encystment factor, the supernatant was treated with different concentrations of proteinase K or bromelain (to remove proteins) and inoculated into fresh cultures of V. vermiformis trophozoites. The enzymatic treatment did not affect the encystment of V. vermiformis, suggesting that such encystment factor(s) were not proteins.
FIG 10.
Investigation of encystment factors secreted by Vermamoeba vermiformis. (A) Induction of encystment (%) in V. vermiformis cells caused by the virus-free supernatant from a previous F. mariensis infection of a V. vermiformis culture (MOI of 10). Supernatant was diluted 2-fold from undiluted to 1/64. (B) Induction of encystment (%) of V. vermiformis cells caused by undiluted (pure) supernatant from a previous F. mariensis infection in V. vermiformis culture at different MOIs. (C to E) Quantification of the concentration of (C) K+, (D) Ca2+, and (E) Mg2+ in the supernatants of cells infected (MOI of 10, 10 hpi) with F. mariensis or Tupanvirus and in those of uninfected cells (control). The statistical significance was calculated using a two-tailed 2-way ANOVA test and a Tukey’s range test, using GraphPad Prism. ***, P < 0.001. (F) Evaluation of the potential of Mg2+ as an inductor of encystment in V. vermiformis. A total of 3 nmol of Mg2+ was added to a culture of 4 × 104 amoeba trophozoites, and the concentration of Mg2+ was measured, as well as the rate of cyst formation over time. (G and H) Evaluation of EDTA effect on (G) cyst formation and (H) viral genome replication during F. mariensis infection at an MOI of 10. (I) Evaluation of the inhibitory activity of 10 mM EDTA on the encystment process induced by supernatants of V. vermiformis infected by F. mariensis at different MOIs, 24 h postinoculation. For all graphs, error bars indicate SDs. These experiments were performed three times in triplicate. (J) Scheme summarizing the phenomenon described in this work. Once infected by F. mariensis, a V. vermiformis trophozoite can be lysed, as described for other amoebal giant viruses. However, during the first events of infection in a V. vermiformis population, encystment factors are released into the supernatant, which can induce the encystment of uninfected cells or even cause the encystment of infected trophozoites. As a result, we found cysts with no excystment ability during different stages of the replication cycle. The overall viral load in the supernatant is controlled by V. vermiformis trapping a substantial amount of viral progeny inside cysts.
We measured the concentration of different inorganic factors in the supernatants of infected cells (MOI of 10; 10 hpi) with F. mariensis or Tupanvirus and in those of uninfected cells (control). Supernatants of all studied groups had similar concentrations of potassium and calcium (Fig. 10C and D). However, we observed a significant increase in magnesium ion (Mg2+) concentrations in cells infected with F. mariensis (Fig. 10E). This result is consistent with previous studies that have shown Mg2+ as a factor able to trigger encystment in A. castellanii (9, 19). We then tested the potential of Mg2+ to induce encystment in V. vermiformis. First, 3 nmol of Mg2+ was put in a culture of 4 × 104 amoeba trophozoites, and the concentration of Mg2+ was measured, as well as the rate of cyst formation at different times. The Mg2+ input not only stimulated the encystment of 79% of the cells after 9 h, but it also promoted a gradual increase of Mg2+ concentration during the experimental period (Fig. 10F). These results suggest that once stimulated by Mg2+, V. vermiformis trophozoites trigger an encystment program and secrete more Mg2+ as an encystment stimulus for neighbor trophozoites.
Considering these results, we evaluated the effect of a bivalent cation inhibitor, ethylenediaminetetraacetic acid (EDTA), on cyst formation during F. mariensis infection, at an MOI of 10. EDTA is well known to act as an Mg2+ chelating agent. EDTA caused a significant reduction in cyst formation rates (Fig. 10G). Our results demonstrated that EDTA promoted a significant increase in the viral genome load in the supernatants of cultures, since the viral progeny succeeded in being released from infected trophozoites because they were not being trapped inside cysts (Fig. 10H). We also observed that EDTA affected the capacity of supernatant collected from cells infected with F. mariensis to induce encystment in fresh trophozoites in a dose-dependent way (Fig. 10I), reinforcing the idea that Mg2+ may be one of the soluble factors released by V. vermiformis to stimulate encystment in neighbor cells.
DISCUSSION
Viral sensing, as well as the molecular communication within multicellular organisms or among unicellular individuals, is an important factor for survival in the never-ending evolutionary struggle between viruses and hosts. The ability to sense that a virus is present and respond to an infection is crucial for the host population survival. Bacterial enzymes discriminate between endogenous and foreign nucleic acids by using epigenetic cues; Sulfolobus islandicus (Archaea) is able to enter dormancy in response to phage presence, and quorum sensing has been shown to be important for antiphage defense strategies in Vibrio anguillarum (20–22). PRRs are conserved molecules found in multicellular organisms, such as plants, insects, and vertebrates (23). The activation of these receptors by pathogens results in signaling pathways that lead to innate immune mechanisms aimed at controlling the infections. As the complexity of organisms increases, the immune responses become more robust. When a pathogen is sensed by PRRs in a vertebrate cell, soluble molecules (IFNs) are secreted and establish an antiviral state in any other cell that receives the IFN, besides acting on immune cells to mount an adaptive immune response (4). Although much is known about viral sensing and antiviral responses on multicellular organisms, the basis of pathogen recognition and response to infection in unicellular eukaryotes is still obscure. Understanding how viruses of microbes interact with their hosts is not only important from a basic scientific point of view but also for a better comprehension of the evolution of life, as well as for discovering novel mechanisms and molecules that could be used for applied science. In recent examples, communication between viruses has been shown and described to be mediated by small peptides, while the investigation of bacterial DNA-based acquired immunity led to the recent CRISPR revolution in science and biomedicine (24–26).
Here, we show that when V. vermiformis faces infection by Faustovirus, it is able to trap the viruses inside cysts. The virus-containing cysts are nonviable, so the trapping process protects the uninfected amoebal population and could be considered a novel type of antiviral strategy. Our data also show that the encystment response to F. mariensis was mediated by at least one unknown encystment factor released by infected cells in conjunction with Mg2+. An overview of the process is shown in Fig. 10J. The putative encystment factor was not a protein, since a proteinase K/bromelain treatment did not abolish the cyst-inducing ability of conditioned media. Secretion of the encystment factor was dependent on Faustovirus entry and replication, as conditioned media from V. vermiformis exposed to inactivated virions did not enhance cyst formation. Since it is unlikely that the encystment factor was derived from the virus due to its negative impact on viral success, it is tempting to assume that viruses are being recognized by the infected amoebal cell, which in turn produces the encystment factor as a response. When this factor is secreted, nearby cells are warned of viral presence and either reversibly encyst before the infection happens or trap the viruses inside damaged cysts, thus protecting the rest of the population. This could be considered analogous to the IFN response, in which virus sensing leads to molecular communication between cells and a subsequent sacrifice of those infected in order to minimize damage to the organism population. In the case of giant viruses, such as Tupanvirus and Mimivirus, escape mechanisms against the production of the encystment factor would have appeared, analogous to the myriad of evasion mechanisms possessed by viruses of vertebrates against the IFN system. Recently, Yoshikawa and coworkers demonstrated that a new, remarkable giant virus called Medusavirus is able to induce encystment of Acanthamoeba cells as well (27). However, the mechanisms involved in such phenomena remain to be investigated. New studies on amoebas (including V. vermiformis) cyst wall composition, synthesis, and dynamics would be welcome for a better understanding of such processes (28). Ecological approaches would be very interesting as well, concerning how encystment factors (as Mg+2) would act on lakes, rivers, and oceans, and their impacts on the communication of amoeba communities on narrow and broad scales.
The complexity of the amoebal host cell, along with the large genomes of giant viruses, makes a reductionist approach to study V. vermiformis and F. mariensis interactions challenging. The nonprotein nature of the encystment factor(s) makes isolating it from the complex metabolomics of an amoebal cell difficult, while the large genome and elaborate particle of the virus make pinpointing specific targets for recognition or other interactions challenging. However, our data can be considered a starting point for better comprehending ancient and unique virus-host interactions. The antiviral strategy described here is a new example of the constant race for supremacy between parasites and hosts. This unprecedented mechanism illustrates that many different antiviral strategies are yet to be unveiled in the biosphere.
MATERIALS AND METHODS
Virus isolation.
In August 2015, 15 water samples were collected from Pampulha Lagoon, Belo Horizonte, Brazil. The collection was performed with sterile tubes, and the samples were stored at 4°C until the inoculation process. The samples were then subjected to filtration through a paper filter and then through a 5-μm filter to remove large particles of sediment. For viral isolation, we used Acanthamoeba polyphaga (ATCC 30461), Acanthamoeba castellanii (ATCC 30234), and Vermamoeba vermiformis (ATCC CDC19). Amoebae were grown in 75-cm2 Nunc cell culture treated flasks (Thermo Fisher Scientific, USA) with 30 ml of peptone-yeast extract-glucose (PYG) medium (29) supplemented with 0.14 mg/ml penicillin (Sigma-Aldrich, USA), 50 mg/ml gentamicin (Thermo Fisher Scientific, USA), and 2.5 mg/ml amphotericin (Bristol-Myers Squibb, New York, USA) at 32°C. For virus isolation, amoebae were resuspended in 10 ml of PYG supplemented with an antibiotic mix containing 0.004 mg/ml ciprofloxacin (Cellofarm, Brazil), 0.004 mg/ml vancomycin (Sigma-Aldrich, USA), and 0.020 mg/ml doxycycline (Sigma-Aldrich, USA). Then, the suspension was diluted 1:10 in phosphate-buffered saline (PBS), and inoculated in 96-well plates containing 4 × 104 cells per well. The plates were incubated for 7 days at 32°C, and observations of the cytopathic effects were done daily using an inverted optical microscope (Quimis, Brazil). The well contents were then collected, frozen, and thawed three times to help release the viruses from intact amoeba cells. The samples were reinoculated for two new subcultures on fresh amoebae, as described above (blind passages). The contents of wells with cytopathic effects were collected and inoculated into new 25-cm2 Nunc cell culture treated flasks with Filter Caps (Thermo Fisher Scientific, USA) cultures containing 1 million cells. The cytopathic effects were confirmed, and these cultures were centrifuged at 1,200 × g for lysate clearance and further analyzed for giant viruses. Negative controls with no sample-inoculated amoeba were used in all microplates. From this collection of samples, we isolated two amoeba viruses, Niemeyer virus (Mimivirus), which was able to infect Acanthamoeba cells, and F. mariensis, which was able to replicate in V. vermiformis. These new isolates were registered at the Brazilian Biological Resources Bank under Sistema de Autorização e Inforação em Biodiversidade (SISBIO) number A237F1B.
Virus production, purification and cytopathic effect analysis.
For Faustovirus production and purification, twenty T175 flasks (Thermo Fisher Scientific, USA) containing 20 million V. vermiformis cells in PYG medium were inoculated with F. mariensis at an MOI of 0.01 and incubated for 5 days, at 28°C. The lysate was centrifuged at 1,200 × g. Then, the supernatant was collected, inoculated onto a 24% sucrose (Merck, Germany) cushion, and centrifuged at 8,000 × g for 2 h. The pellet was resuspended in PBS and stored in a −80°C freezer. Three aliquots of the virus stock were titrated to the 50% endpoint, which was calculated by the Reed-Muench method (30). Half of the viral production (600 μl) was used for biological assays, and the other half (600 μl) was used for genome sequencing. For plaque-forming unit (PFU) analysis, four million V. vermiformis cells were added to 6-well plates, forming cell monolayers. Then, the monolayers were inoculated with F. mariensis diluted 10-fold and observed for 48 h with an optical microscope in association with a digital camera (Moticam 2300; Quimis, Brazil). To test whether F. mariensis would be able to infect V. vermiformis cysts, T25 flasks containing 4 million cysts were inoculated with F. mariensis at MOIs of 0.01, 0.1, 1, and 10 in PBS and observed for 48 h. In addition, 1-ml aliquots of these cultures were collected at 0, 4, 8, 12, and 24 hpi and submitted to PCR targeting Faustovirus DNA polymerase to check virus replication. As previously mentioned, we had no evidence that F. mariensis was able to initiate its cycle by infecting V. vermiformis cysts.
Tupanvirus soda lake and Orpheovirus IHUMI LCC2 were propagated and purified as described for F. mariensis, except a 46% sucrose cushion was used during ultracentrifugation. The pellets were resuspended in PBS and stored in a −80°C freezer. Three aliquots of each virus stock were titrated to the endpoint, which was calculated by the Reed-Muench method (30).
Electron microscopy.
For transmission electron microscopy (TEM) assays, V. vermiformis cells were infected/encysted and then were prepared for microscopy. Briefly, the medium was discarded and the monolayer gently washed twice with 0.1 M phosphate buffer. Glutaraldehyde (2.5%, vol/vol) was added to the system, followed by incubation for 1 h at room temperature for fixation. The cells were then collected, centrifuged at 1,200 × g for 10 min, the medium discarded, and the cells stored at 4°C in phosphate buffer until electron microscopy analyses.
For the scanning electron microscopy (SEM) assays, the cells or virus particles were prepared on round glass blades covered by poly-l-lysine and fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 1 h at room temperature. Samples were then washed three times with 0.1 M cacodylate buffer and postfixed with 1.0% osmium tetroxide for 1 h at room temperature. After a second fixation, the samples were washed three times with 0.1 M cacodylate buffer and immersed in 0.1% tannic acid for 20 min. Samples were then washed in cacodylate buffer and dehydrated by serial passages in ethanol solutions with concentrations ranging from 35% to 100%. They were dried at the critical CO2 point, transferred onto stubs, and metalized with a 5-nm gold layer. The analyses were completed with scanning electronic microscopy (FEG Quanta 200 FEI) at the Center of Microscopy of UFMG, Brazil.
Genome sequencing and analyses.
The F. mariensis genome was sequenced using a MiSeq instrument (Illumina, Inc., San Diego, CA, USA) with the paired-end application. The sequence reads were assembled de novo using ABYSS and SPADES software, and the resulting contigs were ordered by the Python-based CONTIGuator.py software. The obtained draft genomes were mapped back to verify the read assembly and close gaps. Open reading frames were predicted by GeneMarkS. The tRNA genes were searched using tRNAscan-SE and ARAGORN software. Predicted proteins of fewer than 50 amino acids in length were discarded. Gene annotation was performed using Blast2GO software (31). A BLASTp search against the NCBI nonredundant (nr) database was performed, with hits being considered significant if expect (E) values were lower than 1 × 10−3. A BLASTp search was also performed with the same parameters against the clusters of orthologous groups (COGs) of proteins of the nucleocytoplasmic large DNA viruses (NCVOG) (32). In addition, we searched for conserved domains in different databases using InterProScan implemented in Blast2GO software. The genome annotation and functional classification were then manually revised and curated. Synteny analysis was performed using Mauve software (33). The DNA polymerase tree was constructed in MEGA 7.0 software using the maximum likelihood evolution method and 1,000 replicates (34).
Viral cycle and amoebal encystment characterization.
The analysis of the cycle of F. mariensis by TEM was performed in the context of asynchronous infection, at an MOI of 0.1. T175 flasks containing 40 million V. vermiformis cells were inoculated with F. mariensis, and then analyzed 24 hpi by TEM. A total of 250 images were evaluated in order to investigate the major steps of the viral cycle.
As mentioned, during F. mariensis replication we observed substantial formation and accumulation of V. vermiformis cysts after infection, especially when cells were infected at high MOIs (1 and 10). Therefore, we verified whether other V. vermiformis-infecting viruses would be able to trigger encystment as well. T125 flasks containing 40 million amoebas were inoculated with F. mariensis, Tupanvirus, or Orpheovirus at an MOI of 10 and compared to uninfected cells. After 24 hpi, trophozoites and cysts were quantified in a Neubauer chamber (Kasvi, Brazil), and remaining cells were submitted to TEM to check whether viruses were imprisoned inside cysts. This experiment was performed three times in triplicate.
Considering that only F. mariensis was able to remain inside V. vermiformis cysts, a new experiment was performed, in order to quantify cysts and trophozoites at different F. mariensis MOIs, namely 0.01, 0.1, 1, and 10. A total of 3 × 106 trophozoites were added to T25 flasks with 5 ml of PYG medium and inoculated at MOIs of 0.01, 0.1, 1, or 10. After adsorption, cells were washed and then incubated for 48 h with PYG medium. Uninfected cells were used as the control. Trophozoites and cysts were quantified in a Neubauer chamber (Kasvi, Brazil).
The next step was to measure F. mariensis replication in V. vermiformis inoculated at different MOIs. Therefore, V. vermiformis cells were infected with F. mariensis at MOIs of 0.01, 0.1, 1, and 10 and incubated for 1 week. At this stage, trophozoites were no longer observed, regardless of the MOI, since they were either lysed (releasing viruses to supernatant) or converted to cysts. This longer time of incubation was designed to allow the lysis or encystment of all trophozoites in the system; therefore, at the end of the experiments, we would find viruses only in the supernatant (originated from the lysis of infected trophozoites) or inside cysts. The supernatant and cysts were separated by centrifugation (1,200 × g, 10 min), and viral genome load was quantified by qPCR targeting the DNA polymerase gene (5′-AAAACGATTCCGTGCGCAAA-3′ and 5′-ACTAACATCGGGGCGGTTTT-3′). The primers were designed using a freely available primer design tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) at the National Center for Biotechnology Information (NCBI), USA.
PCR was preceded by DNA extraction using the proteinase K/phenol-chloroform method and DNA was used at the concentration of 50 μg/μg as a template for PCR assays. PCR assays were performed using 1 μl of extracted DNA (∼ 50 ng) in an amplification reaction mix containing 5 μl of SYBR green master mix and 0.4 μl (10 μM) of forward and reverse primers. The final volume of the reaction was adjusted with ultrapure water to 10 μl. The conditions of the StepOne thermal cycler reactions (Applied Biosystem, USA) were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min, which was followed by a final step of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. As negative controls, we used DNA extracted from uninoculated amoebas with purified virus or samples, and as a positive control, we used DNA from amoebae infected with purified virus. The relative quantification was performed with the cycle threshold (ΔCT) method, and the results were presented as arbitrary units (log10). This experiment was performed three times in triplicate.
Excystment assays.
A stock of uninfected V. vermiformis cysts was produced by treating ten T175 flasks containing amoebae with Neff solution (19) for 3 days. These cysts were quantified and stocked at room temperature. For assays of cysts containing virus, T175 flasks containing 40 million fresh trophozoites of V. vermiformis were infected with F. mariensis at MOIs of 0.01, 0.1, 1, and 10. One week postinfection, remaining cysts were collected and washed 10 times with PBS, which was followed by 1,200 × g centrifugation (10 min) to remove external viral particles. Cysts were then quantified in a Neubauer chamber, and then 105 cysts had their excystment competence evaluated after inoculation in T25 flasks containing PYG media with 5% fetal calf serum (FCS). As an excystment control, 105 noninfected cysts were added to a T25 flask containing PYG media with 5% FCS. In parallel, another excystment protocol was tested, namely, the nonnutrient agar plate. Briefly, 1 ml of fresh heat-inactivated (99°C, 2 h) Escherichia coli DH5-α (109 cells/ml) was added to the surface of 1% nonnutrient agar (Merck, Germany) and spread with a sterile glass Drigalski hook. After drying at room temperature, the bacterial monolayer was inoculated in 11 equidistant spots with V. vermiformis cysts produced at different MOIs, for a total of 105 cysts per agar plate. As an excystment control, 105 noninfected cysts were added to a control plate. The excystment rate was calculated after 3 days, by quantifying 1,000 cells three times in triplicate.
To investigate whether cells under excystment had viruses inside them, fresh excysted trophozoites at MOIs of 0.01 and 0.1 (excystment at MOIs of 1 and 10 was not observed) were individually collected from agar plates with sterile tips. The cells were transferred to 200-μl micro tubes, washed 3 times with 10 μl PBS, centrifuged at 1,200 × g for 10 min, and subcultivated onto agar nonnutrient plates containing fresh heat-inactivated (99°C, 2 h) Escherichia coli. Cells were observed for 5 days to determine the occurrence of any cytopathic effect or encystment. After reaching 80% confluence on the agar plate, trophozoites were transferred to T25 flasks containing PYG medium, and then the presence/replication of the virus was tested by qPCR (targeting the F. mariensis DNA polymerase gene) during five subcultivations.
To investigate whether cells under excystment had F. mariensis inside their cytoplasm, T175 flasks containing 40 million fresh trophozoites of V. vermiformis were infected with F. mariensis at MOIs of 0.01, 0.1, 1, and 10 (three flasks per MOI). One week postinfection, remaining cysts were collected and washed 10 times with PBS, followed by 1,200 × g centrifugation (10 min) to remove external viral particles. Cysts were then prepared for TEM and SEM. In parallel, those cysts were inoculated in T175 flasks containing fresh PYG medium supplemented with 5% FCS, and after 24 h, cells were prepared for TEM. The viability of cysts produced at different MOIs was assayed with 0.4% trypan blue (Sigma, USA), in a Neubauer chamber.
For cyst resistance assays, 106 cysts produced at MOIs of 1 and 10 (each) were submitted to 4% hydrochloric acid heated (99°C) treatments for 6, 12, 24, 48, 60, and 72 h, followed by 3 cycles of sonication at 50 kHz (30 s each), washed three times with PBS and then quantified in a Neubauer chamber. As a control, we used cysts produced under noninfectious conditions (Neff). Those samples were also prepared for TEM, with the aim of analyzing the presence and appearance of cyst walls.
Circumvention of encystment analyses.
To evaluate whether F. mariensis or Tupanvirus were able to circumvent V. vermiformis encystment, T25 flasks containing 4 million trophozoites were treated with a 5 ml Neff solution, and 2 h later, this solution was removed. The purified viruses (F. mariensis or Tupanvirus) were inoculated at an MOI of 10, 5 ml PYG medium was added to the flasks, which were then incubated for 24 h at 28°C. Virus genome replication was assayed by qPCR (targeting the DNA polymerase for both viruses) (Tupanvirus primers 5′-TCACTGGTTCGTCATGCACT-3′ and 5′-TCGCTTTGAGAGGTTTGGCT-3′) and cells were analyzed by TEM and optical microscopy. Cell viability was analyzed with 0.4% trypan blue in a Neubauer chamber. Alternatively, we inoculated T25 flasks containing 4 million V. vermiformis trophozoites with F. mariensis or Tupanvirus at an MOI of 10; after 2 h we treated the cells with 5 ml Neff solution, and then cells were incubated for 24 h at 28°C.
Using qPCR, we measured the expression of two serine proteinase mRNA isotypes present in V. vermiformis upon infection (MOI of 10) with F. mariensis or Tupanvirus or Neff treatment. T25 flasks containing 4 million trophozoites were inoculated, and 5 h postinfection, cells were collected and submitted to RNA extraction using the RNAeasy minikit (Qiagen). The samples were treated with DNase (Invitrogen) and reverse transcribed using M-MLV reverse transcriptase (200 U/liter; Thermo Fisher Scientific), according to the manufacturer’s instructions. The resulting cDNAs were used as a template in a StepOne thermocycler (Applied Biosystems) quantitative PCR (qPCR) assay to target two V. vermiformis serine proteinase mRNA isotypes (5′-GACTGGTGGACGAGCTATGG-3′ and 5′-TCTAGGCTCGTCAAGTCCCA-3′; 5′-GCTAAATCGTTCACCGTGGC-3′ and 5′-CAACGCGTATTCATCGGCTG-3′). PCR assays were performed using 1 μl of extracted DNA (50 ng) in an amplification reaction mix containing 5 μl of SYBR green master mix and 0.4 μl (10 μM) of forward and reverse primers. The final volume of the reaction was adjusted with ultrapure water to 10 μl. The conditions of the StepOne thermal cycler reactions (Applied Biosystem, USA) were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, which was followed by a final cycle of 95°C for 15 s, 60°C for 1 min and 95°C for 15 s. The relative quantification was performed with the ΔCT method, and results were presented as arbitrary units (log10). This experiment was performed three times in triplicate.
Analyses of secreted factors involved in encystment.
To investigate whether V. vermiformis cells infected by F. mariensis secretion factors were involved with the encystment, the supernatant of a culture infected at an MOI of 10 was collected 4 days postinfection (T175 flask; 40 million trophozoites). The supernatant was filtered with a 0.1-μm filter (Millipore, Brazil) to remove all viral particles and subsequently diluted 2-fold for inoculation in T25 flasks containing 4 million fresh V. vermiformis trophozoites. Similarly, supernatants from V. vermiformis cells infected by F. mariensis at MOIs of 0.01, 0.1, and 1 were also prepared, collected, filtered, and inoculated (undiluted) in T125 flasks containing 40 million fresh V. vermiformis trophozoites. To comprehend the nature of V. vermiformis factors related to encystment, the supernatant was treated with different concentrations of proteinase K (10, 20, 30, 40 mg/ml, 2 h of incubation, room temperature; Gibco, USA), followed by bromelain from pineapple stems (45 mg/ml, 24 h of incubation, room temperature; Sigma, USA) and enzyme inactivation. Bovine serum albumin (BSA) was used as the protease K/bromelain activity control. Treated supernatants were then inoculated into T25 flasks containing 4 million fresh V. vermiformis trophozoites, and their activity regarding cyst formation was evaluated.
The concentrations of potassium, calcium, and magnesium in the supernatants of cells infected with F. mariensis or Tupanvirus and in those of uninfected cells (control) were also assayed. For this, 96-well plates containing 4 × 104 V. vermiformis trophozoites were inoculated and, at 10 hpi, the concentrations of potassium, calcium, and magnesium were measured with a potassium assay kit (MyBioSource, USA.), a calcium detection assay kit (Abcam, USA), and a magnesium assay kit (Sigma, USA), respectively, following the manufacturers’ instructions. To test the potential of Mg2+ as an encystment inductor in V. vermiformis, a total of 3 nmol of Mg2+ (MgCl2, Merck, Germany) was put in a culture of 4 × 104 amoeba trophozoites, and the rate of cyst formation from 3 to 12 h was measured, as well as the concentration of Mg2+ (using a magnesium assay kit; Sigma, USA). The effect of EDTA (Merck, Germany) on cyst formation during F. mariensis infection was evaluated. For this, T25 flasks containing 4 million trophozoites were infected with F. mariensis at an MOI of 10, and 2 h postinfection, 10 mM EDTA was added. Flasks were incubated for 24 h at 28°C, followed by cyst counting and virus quantification by qPCR (targeting the DNA polymerase gene). Results were compared to those from an EDTA-free control. To evaluate the effect of EDTA on the inhibition of cyst formation induced by the supernatants of preinfected trophozoites, T25 flasks containing 4 million trophozoites were infected at MOIs of 0.01, 0.1, 1, and 10. After 24 h, the supernatants were collected, filtered (0.1 μm), and then inoculated into new T25 flasks containing 4 million fresh amoebae. One hour postinfection, 10 mM EDTA was added, and at 16 hpi, cysts and trophozoites were quantified in a Neubauer chamber. Results were compared to those from EDTA-free controls, corresponding to each MOI evaluated.
Data availability.
The genome sequence of Faustovirus mariensis has been submitted to GenBank under accession number MK506267.
ACKNOWLEDGMENTS
We are grateful to our colleagues from Laboratório de Vírus and Microscopy Center of Universidade Federal de Minas Gerais for their excelent technical support.
In addition, we acknowledge the financial support from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do estado de Minas Gerais (FAPEMIG), and the Ministério da Saúde (MS-DECIT).
E.G.K. and J.S.A. are CNPq researchers. E.G.K., B.L.S., and J.S.A. are members of a CAPES-COFECUB project.
REFERENCES
- 1.Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R. 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359:eaar4120. doi: 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ratcliff F, Harrison BD, Baulcombe DC. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 3.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. 2015. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Secombes CJ, Zou J. 2017. Evolution of interferons and interferon receptors. Front Immunol 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov 6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonjardim CA, Ferreira PCP, Kroon EG. 2009. Interferons: signaling, antiviral and viral evasion. Immunol Lett 122:1–11. doi: 10.1016/j.imlet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer MG, Hackl T. 2016. Genome integration and reactivation of the virophage mavirus in the marine protozoan Cafeteria roenbergensis. Nature 540:288–291. doi: 10.1038/nature20593. [DOI] [PubMed] [Google Scholar]
- 8.Frada M, Probert I, Allen MJ, Wilson WH, de Vargas C. 2008. The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc Natl Acad Sci U S A 105:15944–15949. doi: 10.1073/pnas.0807707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva LKDS, Boratto PVM, La Scola B, Bonjardim CA, Abrahão JS. 2016. Acanthamoeba and mimivirus interactions: the role of amoebal encystment and the expansion of the ‘Cheshire Cat’ theory. Curr Opin Microbiol 31:9–15. doi: 10.1016/j.mib.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Boratto P, Albarnaz JD, Almeida G, Botelho L, Fontes ACL, Costa AO, Santos DDA, Bonjardim CA, La Scola B, Kroon EG, Abrahão JS. 2015. Acanthamoeba polyphaga mimivirus prevents amoebal encystment-mediating serine proteinase expression and circumvents cell encystment. J Virol 89:2962–2965. doi: 10.1128/JVI.03177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambrecht E, Baré J, Sabbe K, Houf K. 2017. Impact of Acanthamoeba cysts on stress resistance of Salmonella enterica serovar Typhimurium, Yersinia enterocolitica 4/O:3, Listeria monocytogenes 1/2a, and Escherichia coli O:26. Appl Environ Microbiol 83:e00754-17. doi: 10.1128/AEM.00754-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, Klose T, Rossmann M, Colson P, Raoult D, La Scola B. 2015. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol 89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrade A, Rodrigues RAL, Oliveira GP, Andrade KR, Bonjardim CA, La Scola B, Kroon EG, Abrahão JS. 2017. Filling knowledge gaps for mimivirus entry, uncoating, and morphogenesis. J Virol 91:e01335-17. doi: 10.1128/JVI.01335-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arantes TS, Rodrigues RAL, dos Santos Silva LK, Oliveira GP, de Souza HL, Khalil JYB, de Oliveira DB, Torres AA, da Silva LL, Colson P, Kroon EG, da Fonseca FG, Bonjardim CA, La Scola B, Abrahão JS. 2016. The large marseillevirus explores different entry pathways by forming giant infectious vesicles. J Virol 90:5246–5255. doi: 10.1128/JVI.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva L, Andrade A, Dornas FP, Rodrigues RAL, Arantes T, Kroon EG, Bonjardim CA, Abrahaõ JS. 2018. Cedratvirus getuliensis replication cycle: an in-depth morphological analysis. Sci Rep 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira Andrade A, Boratto PVM, Rodrigues RAL, Bastos TM, Azevedo BL, Dornas FP, Oliveira DB, Drumond BP, Kroon EG, Abrahão JS. 2018. New isolates of pandoraviruses: contribution to the study of replication cycle steps. J Virol 93:e01942-18. doi: 10.1128/JVI.01942-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambrecht E, Baré J, Chavatte N, Bert W, Sabbe K, Houf K. 2015. Protozoan cysts act as a survival niche and protective shelter for foodborne pathogenic bacteria. Appl Environ Microbiol 81:5604–5612. doi: 10.1128/AEM.01031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouque E, Trouilhé M-C, Thomas V, Hartemann P, Rodier M-H, Héchard Y. 2012. Cellular, biochemical, and molecular changes during encystment of free-living amoebae. Eukaryot Cell 11:382–387. doi: 10.1128/EC.05301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neff RJ, Ray SA, Benton WF, Wilborn M. 1964. Induction of synchronous encystment in Acanthamoeba spp., p 56–83. In Methods in cell physiology, vol 1 Academic Press, New York, NY. [Google Scholar]
- 20.Seed KD. 2015. Battling phages: how bacteria defend against viral attack. PLoS Pathog 11:e1004847. doi: 10.1371/journal.ppat.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bautista MA, Zhang C, Whitaker RJ. 2015. Virus-induced dormancy in the archaeon Sulfolobus islandicus. mBio 6:e02565-14. doi: 10.1128/mBio.02565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan D, Svenningsen SL, Middelboe M. 2015. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. mBio 6:e00627. doi: 10.1128/mBio.00627-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mushegian A, Medzhitov R. 2001. Evolutionary perspective on innate immune recognition. J Cell Biol 155:705–710. doi: 10.1083/jcb.200107040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, Melamed S, Leavitt A, Savidor A, Albeck S, Amitai G, Sorek R. 2017. Communication between viruses guides lysis-lysogeny decisions. Nature 541:488–493. doi: 10.1038/nature21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 26.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna J, Charpentier E. 2012. A programmable dual-RNA–guided bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshikawa G, Blanc-Mathieu R, Song C, Kayama Y, Murata K, Ogata H, Takemura M, Ogata H. 2019. Medusavirus, a novel large DNA virus discovered from hot spring water. J Virol 93:e02130-18. doi: 10.1128/JVI.02130-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouque E, Yefimova M, Trouilhe M, Quellard N, Fernandez B, Rodier M, Thomas V, Humeau P, Hechard Y. 2015. Morphological study of the encystment and excystment of Vermamoeba vermiformis revealed original traits. J Eukaryot Microbiol 62:327–337. doi: 10.1111/jeu.12185. [DOI] [PubMed] [Google Scholar]
- 29.Schuster FL. 2002. Cultivation of pathogenic and opportunistic free-living amebas. Clin Microbiol Rev 15:342–354. doi: 10.1128/CMR.15.3.342-354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 31.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence of Faustovirus mariensis has been submitted to GenBank under accession number MK506267.