Zika virus (ZIKV) is a global health threat causing severe disease in humans, including microcephaly in newborns and Guillain-Barré syndrome in adults. Here, we analyzed the human monoclonal antibody response to acute ZIKV infection and found that neutralizing antibodies could not elicit Fc-mediated immune effector functions but could potentiate antibody-dependent enhancement of disease. We further identified critical epitopes involved with neutralization by generating and characterizing escape variants by whole-genome sequencing. We demonstrate that the lateral ridge region, particularly the S368 amino acid site, is critical for neutralization by domain III-specific antibodies.
KEYWORDS: Zika virus, antibody-dependent cellular cytotoxicity, monoclonal antibodies, neutralizing antibodies, prM-E, whole-genome sequencing
ABSTRACT
The mosquito-borne Zika virus (ZIKV) has been causing epidemic outbreaks on a global scale. Virus infection can result in severe disease in humans, including microcephaly in newborns and Guillain-Barré syndrome in adults. Here, we characterized monoclonal antibodies isolated from a patient with an active Zika virus infection that potently neutralized virus infection in Vero cells at the nanogram-per-milliliter range. In addition, these antibodies enhanced internalization of virions into human leukemia K562 cells in vitro, indicating their possible ability to cause antibody-dependent enhancement of disease. Escape variants of the ZIKV MR766 strain to a potently neutralizing antibody, AC10, exhibited an amino acid substitution at residue S368 in the lateral ridge region of the envelope protein. Analysis of publicly availably ZIKV sequences revealed the S368 site to be conserved among the vast majority (97.6%) of circulating strains. We validated the importance of this residue by engineering a recombinant virus with an S368R point mutation that was unable to be fully neutralized by AC10. Four out of the 12 monoclonal antibodies tested were also unable to neutralize the virus with the S368R mutation, suggesting this region to be an important immunogenic epitope during human infection. Last, a time-of-addition infection assay further validated the escape variant and showed that all monoclonal antibodies inhibited virus binding to the cell surface. Thus, the present study demonstrates that the lateral ridge region of the envelope protein is likely an immunodominant, neutralizing epitope.
IMPORTANCE Zika virus (ZIKV) is a global health threat causing severe disease in humans, including microcephaly in newborns and Guillain-Barré syndrome in adults. Here, we analyzed the human monoclonal antibody response to acute ZIKV infection and found that neutralizing antibodies could not elicit Fc-mediated immune effector functions but could potentiate antibody-dependent enhancement of disease. We further identified critical epitopes involved with neutralization by generating and characterizing escape variants by whole-genome sequencing. We demonstrate that the lateral ridge region, particularly the S368 amino acid site, is critical for neutralization by domain III-specific antibodies.
INTRODUCTION
Zika virus (ZIKV) is a mosquito-borne flavivirus causing epidemic outbreaks on a global scale. Infection with ZIKV is frequently asymptomatic, and clinical manifestations, including low-grade fever, rash, and arthralgia, occur in approximately 20% of cases (1). ZIKV infection is also associated with Guillain-Barré syndrome, a rare immune-mediated paralytic illness (2). Perhaps the most feared consequence of ZIKV infection is the effect it has on the developing fetus. Congenital infection is known to result in developmental abnormalities, including microcephaly, sensorineural hearing loss, and vision deficits, and, in some cases, fetal loss (3, 4).
There are currently no approved treatments or vaccines for ZIKV. A number of vaccines are in development and are designed to elicit antibodies to the ZIKV envelope (E) protein. These vaccine constructs include DNA, RNA, and purified-protein vaccines as well as live attenuated viruses (5–9). A major concern in the field of flavivirus vaccine development is the possibility of disease enhancement mediated by cross-reactive or poorly neutralizing antibodies (10). While there is currently little evidence of its occurrence in humans, antibody-dependent enhancement (ADE) of ZIKV infection was shown in vitro and in animal models using both polyclonal sera and monoclonal antibodies (MAbs) (11–18).
In order to characterize the immunological determinants of antibody-mediated protection, previous groups have isolated and characterized ZIKV-specific monoclonal antibodies from infected patients. Many groups have identified the envelope dimer epitope and the lateral ridge epitope of domain III as critical for a potent neutralizing response (11, 19, 20). Here, we add the characterization of 12 neutralizing monoclonal antibodies from an acutely ZIKV-infected patient. These antibodies represent the initial plasmablast response to acute infection and exhibit low levels of somatic hypermutation. Nevertheless, four of these antibodies (AC10, AC4, AC3, and GD12) were able to potently neutralize virus with half-maximal inhibitory concentrations at the nanogram-per-milliliter range. We next explored the Fc-mediated functions of these antibodies. We report that these monoclonal antibodies were able to elicit Fc-mediated ADE of infection in vitro. However, these antibodies did not elicit Fc-mediated activation of immune effector cells on ZIKV-infected cells as measured by an antibody-dependent cellular cytotoxicity (ADCC) reporter assay. By generating escape variants, we identified residue 368 of the E protein as critical for neutralization by a subset of the antibodies. A recombinant virus with an S368R mutation was resistant to neutralization by antibodies AC10, AC4, AC3, and GD12.
Additionally, we mapped escape mutations from five other less potently neutralizing antibodies and found a majority of the mutations to be on solvent-exposed residues located on domain II of the envelope protein. Finally, we performed a time-of-addition infection assay to determine the mechanism and kinetics by which these antibodies neutralize virus. We show that all of the neutralizing antibodies were able to inhibit viral infection when added prior to or at the infection step. However, these antibodies did not completely neutralize virus when added postinfection. We can therefore conclude that these antibodies inhibit binding of virion to the cell surface and/or fusion of the virus with the host cell membrane.
Our results underscore the importance of the lateral ridge region as a putatively immunologically dominant epitope on the Zika virion. In particular, the serine residue at site 368 is ZIKV specific and conserved among ZIKV strains, indicating this site to be a viable target for a ZIKV vaccine if the neutralization potency of antibodies against this site overcomes their potential ADE properties. Furthermore, we demonstrated that escape mutations in this region result in attenuated viral growth in vitro, suggesting that the 368 site is important for efficient virus replication.
RESULTS
Generation of human ZIKV antibodies from an infected patient.
To investigate the antibody response to acute ZIKV infection, we isolated peripheral blood mononuclear cells (PBMCs) from a patient who was infected by ZIKV while traveling in Central America. This patient was likely not preexposed to dengue virus, and these antibodies were likely associated with a primary immune response (21). We adapted a previously established protocol to generate fully human monoclonal antibodies specific to ZIKV (22). First, we single-cell-sorted plasmablasts isolated during acute infection and sequenced the variable regions of the immunoglobulin genes. Next, we cloned the genes into a human IgG1 expression vector and transfected HEK293F cells to generate fully human monoclonal antibodies.
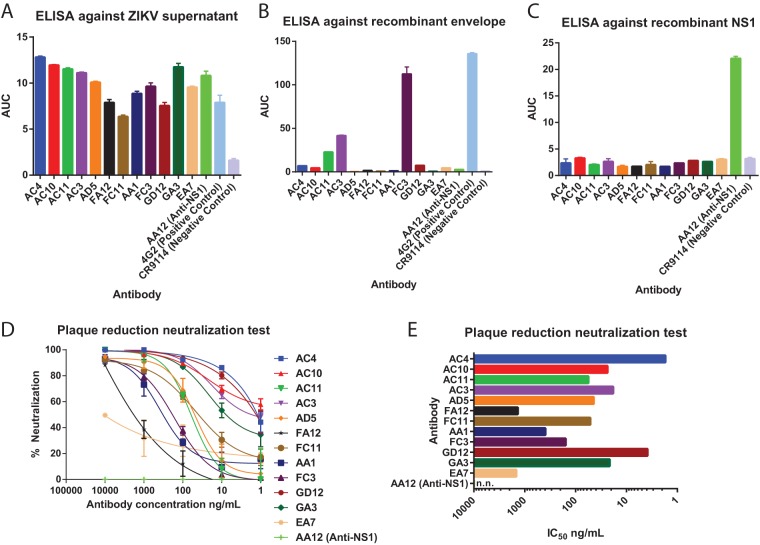
We previously reported four antibodies obtained from the same patient that bound the nonstructural protein NS1 and that are nonneutralizing in vitro but protected against ZIKV infection in the mouse model (21). Here, we report an additional 12 antibodies that bound and neutralized ZIKV. These antibodies were initially identified by their ability to bind to proteins present in supernatants of ZIKV-infected cells (Fig. 1A). Next, antibodies were tested for binding to recombinant envelope protein (Fig. 1B). Only one of the 12 antibodies, FC3, was able to potently bind recombinant envelope. Quaternary epitopes are known to play an important role in anti-flavivirus antibody responses, and we suspect that the other 11 antibodies are only able to bind the properly folded structural proteins on intact virions (11, 23–25). As we reported previously (21), antibodies to the nonstructural protein NS1 are also elicited by acute ZIKV infection. We confirmed that these 12 antibodies did not bind to recombinant NS1 protein by enzyme-linked immunosorbent assay (ELISA) (Fig. 1C). Finally, plaque reduction assays were performed in Vero cells to determine the potency of neutralization of these antibodies (Fig. 1D and E). We found antibodies neutralized to various degrees, with 50% inhibitory concentration (IC50) values ranging from 10 μg/ml to approximately 2 ng/ml. In contrast, the NS1-specific antibody AA12 was not able to neutralize infectious virus at the highest concentration tested, confirming our previous findings (21). All of these isolated antibodies were encoded by heavy/light-chain combinations with low degrees of somatic hypermutation (Table 1). Additionally, 9 of the 12 antibodies were of the IgG1 isotype, while antibody AC4 belongs to the IgG2 isotype, FA12 to IgA1, and FC3 to IgM.
FIG 1.
Neutralizing ZIKV-specific antibodies are induced by ZIKV-infection. (A to C) ELISAs were performed against MR766 ZIKV supernatant, recombinant MR766 envelope protein, or recombinant MR766 NS1 protein to assess binding activities. ELISAs were performed as duplicates, and results are reported as values of the area under the concentration-time curve (AUC). Error bars represent standard errors of the means (SEM). (D) Neutralization activity of 12 antibodies against MR766 ZIKV. (E) Neutralization activity shown as IC50 values. Nonneutralizing antibody AA12 is designated n.n.
TABLE 1.
Antibody characteristics
| Antibody | Heavy chain profilea |
Light chain profilea |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| V gene | J gene | CDR3 sequence | % germ line identity | Isotype | V gene | J gene | CDR3 sequence | % germ line identity | Isotype | |
| AC4 | IGHV1-2*06 | IGHJ6*02 | CARDRRSTWYFYYYGMDVW | 99.1 | IgG2 | IGLV2-8*01 | IGLJ2*01 | CSSYAGSNIYVVF | 99.7 | Lambda |
| AC10 | IGHV1-2*06 | IGHJ6*02 | CARDRRSSWYYYYYGMDVW | 99.1 | IgG1 | IGLV2-8*01 | IGLJ2*01 | CSSYAGSNNFRVF | 99.1 | Lambda |
| AC11 | IGHV5-10-1*01 | IGHJ4*02 | CARLGLTNYFDYW | 98.8 | IgG1 | IGLV3-21*02 | IGLJ1*01 | CQVWDSSSGGVF | 98.4 | Lambda |
| AC3 | IGHV1-2*06 | IGHJ4*02 | CARDHYGDSWHYVFDYW | 97.9 | IgG1 | IGKV3-15*01 | IGKJ2*01 | CQQYNNWPRYTF | 97.5 | Kappa |
| AD5 | IGHV1-18*01 | IGHJ4*02 | CAREYYDFWSGYYAYYFDYW | 99.7 | IgG1 | IGLV2-14*01 | IGLJ3*02 | CSSYTSSSTRVF | 99.1 | Lambda |
| FA12 | IGHV1-69*01 | IGHJ4*02 | CVRALYCSSTSCYAARGGYFDYW | 98.2 | IgA1 | IGLV2-14*01 | IGLJ3*02 | CSSYTSSSTWVF | 99.1 | Lambda |
| FC11 | IGHV1-18*01 | IGHJ4*02 | CAREYYDFWSGYSSYYFDYW | 99.4 | IgG1 | IGLV2-14*01 | IGLJ3*02 | CSSYTSSSTRVF | 99.7 | Lambda |
| AA1 | IGHV4-4*02 | IGHJ4*02 | CARDSQQLVPFDYW | 98.2 | IgG1 | IGLV2-8*01 | IGLJ3*02 | CSSYAGSNNLRVF | 99.4 | Lambda |
| FC3 | IGHV3-33*03 | IGHJ4*02 | CVREDYGIFTKFDYW | 90.5 | IgM | IGKV1-5*03 | IGKJ1*01 | CQQYNSDSWTF | 94.3 | Kappa |
| GD12 | IGHV1-2*06 | IGHJ6*02 | CARDRRSSWYYYYYGMDVW | 99.14 | IgG1 | IGLV2-8*01 | IGLJ2*01 | CSSYAGSNNFRVF | 99.4 | Lambda |
| GA3 | IGHV3-30-3*01 | IGHJ6*02 | CARDRSRDSSGYWLIYYYYYGMDVW | 98.2 | IgG1 | IGLV1-47*01 | IGLJ2*01 | CAAWDDSLSGVVF | 99.7 | Lambda |
| EA7 | IGHV3-21*01 | IGHJ6*02 | CARGGHRVDYYYNMDVW | 96.2 | NDb | IGKV1-9*01 | IGKJ1*01 | CQQLNSYPLTF | 95.3 | Kappa |
IMGT/V-QUEST software was used to assign the germ line reference for IGHV and IGLV genes and to determine percent identity to the germ line.
ND, not determined.
Neutralizing antibodies do not activate ADCC on infected cells but can enhance uptake of virions.
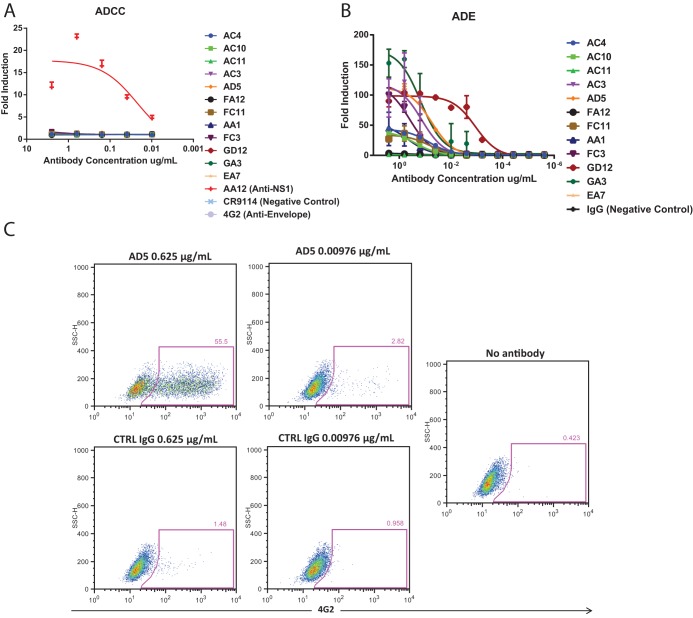
Besides neutralization, antibodies might be able to elicit effector functions via interactions between their Fc regions and Fcγ receptor (FcγR)-expressing myeloid cells. We previously reported that ZIKV NS1-specific antibodies were able to elicit Fc-mediated immune effector functions on infected cells (21). Additionally, these NS1-specific antibodies did not induce antibody-dependent enhancement of disease when tested on Zika viral particles. Here, we evaluated whether our panel of virion-binding antibodies could induce Fc-mediated immune effector functions on infected cells and if these antibodies could mediate virus internalization into susceptible cells.
To determine the activation of ADCC, we used a genetically modified Jurkat cell line that expresses human FcγRIIIa and a luciferase reporter gene under a nuclear factor of activated T cells (NFAT) promoter (21). This surrogate assay examines the ability of antibodies to engage and activate Fc-mediated effector functions. We infected Vero cells with the MR766 virus and found that none of the neutralizing ZIKV antibodies were able to activate effector functions of FcγR-expressing Jurkat cells. In contrast, the positive-control NS1-specific antibody AA12 was able to potently elicit these Fc-mediated effector functions (Fig. 2A). We posit that the inability of neutralizing antibodies to elicit these functions is due to the lack of their ability to bind virus-infected cells.
FIG 2.
Neutralizing antibodies cannot elicit ADCC but can elicit ADE in vitro. We examined the ability of neutralizing antibodies to elicit Fc-mediated effector functions either on the surface of infected cells or against the virion itself. (A) Vero cells were infected with MR766 ZIKV and used as targets for measuring Fc-FcγR effector functions with a genetically modified Jurkat cell line expressing human FcγRIIIa with an inducible luciferase reporter gene. Fold induction was measured in relative light units, and results were compared to infected cells with no antibody added. Error bars represent standard error of the means (SEM). (B) To examine whether enhancement of flavivirus infection in vitro is observed, monoclonal antibodies were incubated with ZIKV and added to FcγR-bearing K562 cells. All monoclonal antibodies were tested at a starting concentration of 3.3 μg per ml and serially diluted 4-fold. Both assays were run in duplicate, and fold induction was measured as the percentage of infected cells as determined by flow cytometry divided by the percentage of infected cells with no antibody added (virus alone). (C) Representative flow cytometry plots for antibody AD5 and control IgG are shown.
ADE of disease is commonly thought to be caused by antibodies binding to the surface of virions at nonneutralizing concentrations, which enhances viral uptake into Fc receptor-bearing myeloid cells (10). To determine whether our antibodies can enhance entry in myeloid cells, we utilized human leukemia K562 cells which are poorly permissive to ZIKV infection in the absence of virion-bound antibody (12). We measured the level of ADE of infection by using a flow cytometry-based assay in which serial dilutions of each of the 12 monoclonal antibodies was preincubated with MR766 ZIKV prior to addition to K562 cells, as done previously (12). The cells were fixed and intracellularly stained with the murine monoclonal antibody 4G2 at 48 h postinfection (hpi) to determine the proportion of infected cells. Fold induction was calculated as the percentage of infected cells in the presence of antibody divided by the percentage of infected cells in the absence of antibody. We found that most neutralizing antibodies were able to facilitate viral internalization to some degree (Fig. 2B). Antibodies AC11, FA12, and EA7 poorly mediated the internalization of Zika virus in K562 cells. Representative flow cytometry data of antibody AD5 and the control antibody are shown as well (Fig. 2C).
Generation of escape mutants.
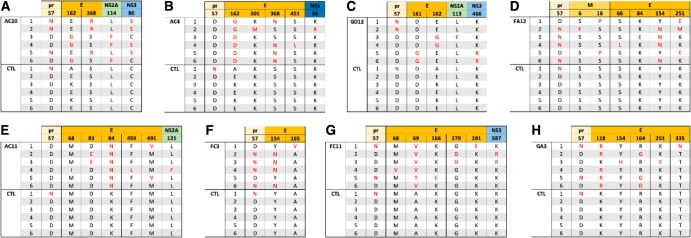
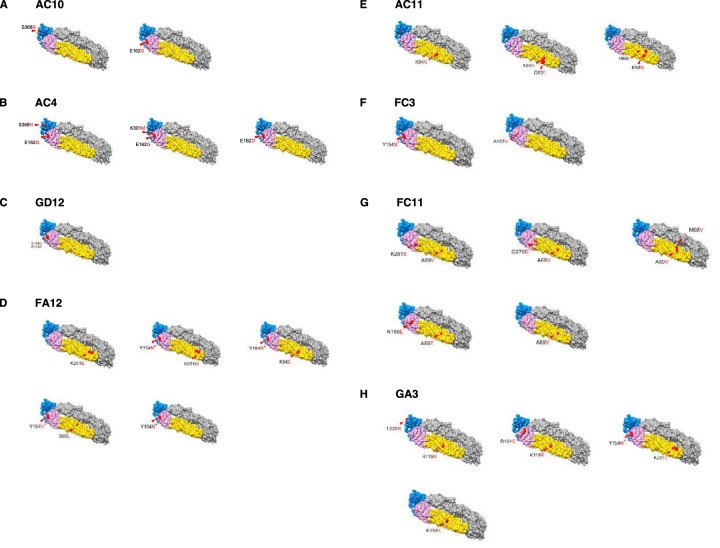
To identify residues critical for antibody neutralization, we generated ZIKV escape mutants. We serially passaged MR766 ZIKV with increasing concentrations beginning at 0.5 ng/ml to 500 ng/ml of antibodies AC10, GD12, AC4, AC11, GA3, FC11, FC3, and FA12 in duplicates. To account for adaptations to growth in cell culture, we serially passaged ZIKV with no antibody in parallel as a control. After 7 to 12 serial passages, we grew viruses from the last passage in 1 to 10 μg/ml of antibody and plaque purified six escape variant viruses from each monoclonal antibody (MAb) and another six from the control. We inoculated these plaques in Vero cells supplemented with the appropriate antibody at 1 to 10 μg/ml to obtain sufficient virus for RNA sequencing. Next, we isolated viral RNA and performed whole-genome sequencing from the six plaque-purified escape variants from the last passage. We then compared the sequences of the viruses passaged in the presence of antibody (escape variants) to the sequence of the control. Analysis of the E coding regions from the six plaque-purified escape variants to MAb AC10 or AC4 showed mutations in residues 162, 301, 368 and/or 451 (Fig. 3A and B). Residues 162, 301, and 368 are located on the ectodomain of the E protein, while site 451 resides within the transmembrane domain. Escape variants to AC10 had point mutations in site 162 located in domain I or in site 368 (Fig. 4A) in domain III. Escape variants to AC4 had point mutations in sites 162, 162, and 368 or in 162 and 301 (Fig. 4B). The majority of the escape variants to AC10 and AC4 had either a glutamic acid or a glycine at site 162 (domain I) and an arginine or asparagine at site 368 (domain III), which suggest that these positions had critical roles in neutralization. However, it is interesting that the wild-type virus (Fig. 3A) passaged in the absence of antibody also had amino acid variability in site 162. A survey of the escape variants to the other MAbs yielded a variety of E-specific sites that are putatively important for resistance. For example, GD12 variants had mutations at site 161 or 162 (domain I) (Fig. 3C and 4C), FA12 had a combination of mutations in domain II only (site 66, 84, or 251) or an additional mutation in domain I (site 154) (Fig. 3D and 4D), and AC11 had changes in sites in domain II (at residues 83/84, 84, or 68/84) (Fig. 3E and 4E). Escape variants to MAb FC3 had mutations only in domain I at site 165 or 154 (Fig. 3F and 4F), while FC11 escape variants yielded mutations in domains I and/or II in sites 68, 69, 166, 279, and/or 281 (Fig. 3G and 4G). Last, escape variants to GA3 had a number of mutations in domain I, domain II, and/or domain III (Fig. 3H and H).
FIG 3.
Whole-genome sequencing of escape variants. Panels A to H show sequence alignments of escape mutations generated by antibodies AC10, AC4, GD12, FA12, AC11, FC3, FC11, and GA3, respectively. The corresponding protein is labeled at the top with the relevant amino acid. Six plaque-purified viruses (numbered 1 to 6) were sequenced, and amino acids are shown in red (mutated) or black (control; CTL). Six plaque-purified wild-type viruses were sequenced in parallel after an identical number of passages in Vero cells. pr, premembrane.
FIG 4.
Escape mutants are mapped to a crystal structure of the ZIKV E protein. Panels A to H show graphical representations of the envelope protein with relevant mutations found on plaque-purified viruses 1 to 6 used for the experiment shown in Fig. 3 indicated in red. PDB accession number 5JHM was used to generate the three-dimensional model using UCSF Chimera.
We also detected other mutations outside the E protein, such as site 57 on the premembrane (prM region) (AC10, GD12, FA12, FC3, FC11, and GA3), residues 6 and 15 on membrane (M) (FA12), residues 113 (GD12), 114 (MAb AC10), and 121 (AC11) on NS2A, residues 80 (AC10), 466 (GD12), and 587 (FC11) on NS3, and residue 96 (AC4) on NS5. As these mutations arose outside the E protein, the importance of these mutations is still unknown.
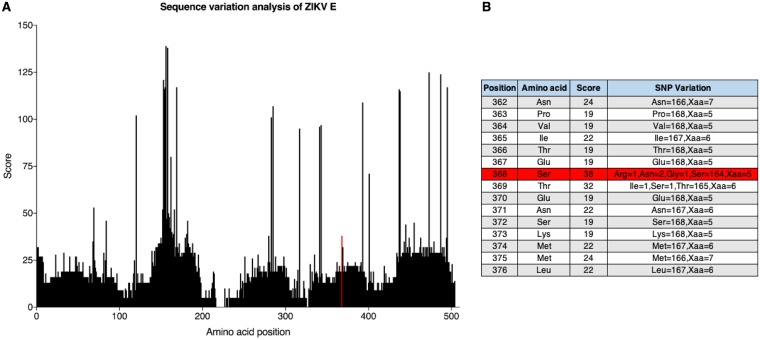
To further characterize how viruses escape antibody-mediated neutralization, we focused on site 368 (domain III) of the E protein, where mutations were found in the escape variants to AC10 and AC4, our most potent neutralizing MAbs. This amino acid residue is located in the lateral ridge epitope of domain III of the ZIKV E protein and is conserved among almost all circulating ZIKV strains (11, 26). We quantified the variation of amino acids in the ZIKV E protein by performing a sequence alignment of 173 publicly available sequences from the Virus Pathogen Resource (Fig. 5). We found that 164 of 168 sequences (97.6%) with identifiable amino acids had a serine at position 368 with a polymorphism score of 38, suggesting a very high level of sequence conservation. An asparagine occurred twice, arginine occurred once, and a glycine occurred once. Of note, a site with no polymorphism has a score of 0, while a maximum score of 439 equates to an equal distribution of 21 alleles (20 amino acids and an indel) at ∼4.7% each.
FIG 5.
Sequence alignment of ZIKV E protein. (A) A sequence alignment and a consensus sequence were generated from 173 publicly available sequences from the Virus Pathogen Resource. Each amino acid position was then assigned a polymorphism score based on Crooks et al. (46). The polymorphism score represents the normalized entropy of an observed allele distribution. Amino acid scores can have values of 0 (no observed polymorphism) to 439 (20 alleles and an indel, ∼4.7% frequency each). The red line indicates amino acid position 368. (B) The residue at site 368 (highlighted in red) has a polymorphism score of 38, where the majority of amino acids (n = 164) are serines. Of note, Xaa indicates a missing or ambiguous amino acid.
To determine whether the mutation at site 368 was sufficient for escape from antibody binding, we utilized an established plasmid-based rescue system for the MR766 ZIKV isolate that efficiently produces infectious virus upon transfection into human embryonic kidney (HEK) 293T cells (27). Using site-directed mutagenesis, we mutated the serine at envelope position 368 to an arginine. To generate recombinant virus, we transfected 293T cells with the wild-type or the S368R mutant plasmid, and supernatant was harvested 3 days posttransfection. The titer of the supernatant was then determined by plaque assay, and virus with the S368R escape mutant had a significantly lower rescue titer (5.2 × 106 PFU per milliliter) than the wild-type virus rescued in parallel (3.1 × 107 PFU per milliliter) (Fig. 6A). As the rescue titer of the mutant antibody was nearly 10-fold lower than that of the wild type, we considered the possibility of viral attenuation in the escape mutant.
FIG 6.
A recombinant MR766 ZIKV with an S368R point mutation escapes neutralization by AC10 in vitro. Serial passaging of MAb AC10 led to the identification of critical residue S368, which lies in the lateral ridge epitope of domain III of the ZIKV envelope protein. Site-directed mutagenesis was performed to generate recombinant MR766 ZIKV with the S368R point mutation. (A) Rescue titer after 72 h posttransfection demonstrates a reduction in viral titer. The dotted line represents the limit of detection. (B) Multicycle growth curves were performed to compare viral fitness between wild-type (WT) and S368R viruses. Supernatants were collected at the indicated time points, and titers were determined by plaque assays. Data points represent the means for two biological replicates, and error bars represent standard errors of the means (SEM). (C) To confirm that the S368R mutation is sufficient for escape from neutralization by MAb AC10, a plaque reduction neutralization test was performed with equivalent amounts of wild-type or S368R MR766 ZIKV. The assay was performed in duplicate, and error bars represent SEM. (D and E) Antibodies GA3 and AA1, which induce escape mutations in different sites, were tested by the same plaque reduction neutralization test. The assay was performed in duplicate, and error bars represent SEM. *, P < 0.05.
Next, we performed multicycle growth curves to compare viral fitness levels between the S368R mutant and wild-type MR766 (Fig. 6B). Vero cells were infected at a multiplicity of infection (MOI) of 0.001, and virus titers were determined at 24-h time points. Again, we found the S368R mutant to be attenuated in vitro, displaying significantly reduced viral titers at the 48-h and 72-h time points. Differences in the titers at the 96-h time point, however, were not statistically significant. Sequencing of the viral genome at the 96-h time point revealed that the S368R mutation remained intact, ruling out that the virus reverted to express the wild-type envelope protein. Nevertheless, there may be undetected compensatory mutations in other regions of the genome that could have arisen to increase viral fitness.
To determine the effect that the S368R mutation has on virus neutralization, we next performed plaque reduction assays on these recombinant viruses. We found that while antibody AC10 was able to neutralize wild-type virus, it was unable to neutralize the S368R variant (Fig. 6C). Antibodies GA3 and AA1, which are of a different germ line variant from AC10 and do not induce mutations at the S368 site, were also tested for their abilities to neutralize both wild-type and S368R variant viruses (Fig. 6D and E). We find that these two antibodies neutralize both viruses to the same degrees, suggesting that they bind an epitope independent of position 368. From this we can conclude that the antibody AC10 potently neutralizes ZIKV and that a critical residue at position 368 of the E protein is required for complete neutralization.
Time-of-addition infection assay.
Finally, we sought to determine the mechanism by which these antibodies neutralize the wild type and the S368R variant virus as described previously (28). To this end, we compared the neutralization activities of our antibodies at different stages of viral entry for either the wild-type MR766 or the S368R variant (Fig. 7). Briefly, we incubated virus with cells for 3 h at 4°C, a temperature at which the virion attaches to the surface but is not internalized. At time zero (0 h), unattached virus was removed from cell culture by two washes with phosphate-buffered saline (PBS), and the cells were moved to 37°C for internalization to occur. Antibodies were added at a concentration of 10 IC50 at either −3 h, 0 h, or 6 h in order to test inhibition of antibody at the stages of viral attachment, postattachment, or postentry, respectively. Once the antibody was added, it remained in culture for 24 h. Flow cytometry analysis was performed after 4G2 anti-envelope staining 48 h postinfection.
FIG 7.
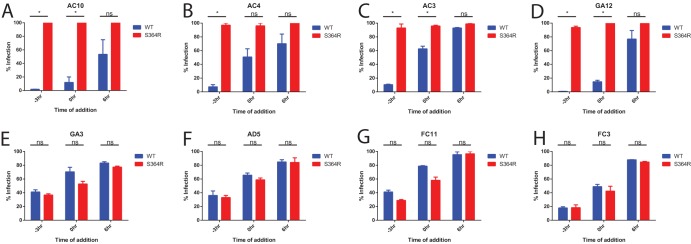
Neutralization occurs at a binding step. Analysis of the timing of antibody neutralization activity was performed. The results for synchronized infections of Vero cells with wild-type or S368R MR766 virus are shown. Vero cells were equilibrated to 4°C at −3 h, and virus plus antibody or virus alone was added to the cells. At 0 h Vero cells were washed twice with PBS, warm medium was added with the relevant antibody, and cells were moved to 37°C. For each assay, results are normalized to those of infections performed without any antibody added. Antibody was added at a concentration of 10 IC50s as determined by plaque reduction neutralization test. Infection was measured by 4G2 anti-envelope staining at 48 h postinfection. Assays were performed in duplicate, and error bars represent SEM. *, P < 0.05; ns, not significant.
All of our antibodies were able to neutralize wild-type virus when added at −3 h (Fig. 6), with about 1 to 30% infection. A lower degree of neutralization was observed when antibody was added at 0 h, with observed values of infection between 10% and 70%, which suggests that at least some of the neutralization activity was derived through blocking the binding of virus to host cells. Little to no neutralization was observed when the antibody was added at 6 h postinfection, with observed values between 70 and 100%, indicating that the inhibitory activity was derived mostly at the entry stage of the viral life cycle. Because of the stark difference between results for antibody added at −3 h and antibody added at 6 h postinfection, we concluded that all tested antibodies inhibited virus binding to the cell surface or fusion with the host endosomal membrane. We also were able to show that antibodies AC10, AC4, AC3, and GA12, all of which were derived from the same germ line, were unable to neutralize the S368R variant (Fig. 7A to D). On the other hand, antibodies GA3, AD5, FC11, and FC3 were able to neutralize the S368R variant at a level comparable to that of the wild-type virus (Fig. 7E to H).
DISCUSSION
With recent advancements in isolating human monoclonal antibodies (22), a number of groups have characterized the antibody response to ZIKV infection (11, 19, 26, 29). However, additional work is required to map the epitopes targeted by neutralizing antibodies and to understand whether a potent dominant neutralizing response correlates with a particular germ line rearrangement (30). In the current study, we characterized monoclonal antibodies generated from the PBMC compartment of a ZIKV-infected individual. In prior work, we identified four nonneutralizing but protective antibodies that bind to the ZIKV NS1 protein (21). Here, we examined the neutralizing antibody response from the same patient.
We isolated 12 antibodies with various neutralizing potencies and binding properties, as shown in Fig. 1 and Table 1. Numerous germ line rearrangements were found in the antibodies isolated from this patient, suggesting a polyclonal response targeting distinct epitopes. The rearrangement VH1-2/VL2-8 (where VH and VL are heavy and light variable regions, respectively) was most common and was found in 4 of the 12 antibodies: AC4, AC10, AC3, and GD12. These antibodies were all potently neutralizing, with IC50 values below 25 ng/ml. An additional potently neutralizing antibody, GA3, had a distinct rearrangement, using the VH3-21/VK1-9 genes (where VK is the kappa light-chain variable region). All of these potently neutralizing antibodies contained a motif of at least three tyrosine residues in the complementarity-determining region 3 (CDR3) fragment of the heavy chain. This unusual motif in the CDR3 sequence was reported in mouse anti-idiotypic monoclonal antibodies; however, the significance of this motif remains unknown (31, 32). AD5 and FC11 were moderately neutralizing, with IC50 values close to 50 ng/ml, and contained the same VH1-18/VL2-14 germ line rearrangements. Moderate to poorly neutralizing antibodies with IC50 values greater than 50 ng/ml included antibodies AC11, AA1, EA7, FC3, and FA12, all of which had distinct germ line rearrangements. This lack of high neutralizing activity may also be due to the absence of somatic hypermutation. Of note, antibody FC3 was the only antibody that bound soluble envelope protein by ELISA. With the finding that only 1 out of 12 antibodies isolated bound recombinant protein, we speculated that a large number of epitopes that elicit neutralizing antibodies are quaternary structures on the surface of the virion.
Similar work by Robbiani et al., was performed by isolating memory B cells from ZIKV-seropositive cohorts in Brazil and Mexico (20). In this study, the VH3-23/VKL1-5 rearrangement was found in five of the six donors and exhibited the largest degree of clonal expansion in three of the six donors. The authors were able to map the binding sites of these antibodies to the lateral ridge epitope of domain III of the envelope protein and found a similarly high potency of neutralization in the nanogram-per-milliliter range. We were not able to isolate any antibodies with the same rearrangement, despite mapping our antibodies with VH1-2/VL2-8 rearrangements to the lateral ridge of the envelope protein as well. This suggests that germ line rearrangements may vary from person to person and that distinct germ line rearrangements may target the same epitopes.
Next, we wanted to explore the role of Fc-mediated functions exhibited by these neutralizing antibodies. Fc-mediated functions can generally be divided into two categories. First, antibodies that target virally infected cells can direct the clearance of these infected cells by activating natural killer cells, macrophages, or neutrophils via Fc-FcγR interactions (33). Alternatively, antibodies can facilitate internalization of virions via Fc-mediated endocytosis into innate immune cells. In this context, antibody-mediated uptake of virus may increase viral replication and potentially enhance disease. The phenomenon of antibody-dependent enhancement (ADE) of disease was shown in vitro with Zika and dengue viruses and also for a number of other virus families, including the Filoviridae and Togaviridae (34, 35).
Antibodies targeting the ZIKV NS1 protein were shown previously to induce protective Fc-mediated effector functions by an in vitro reporter assay (21). This is likely due to the high abundance of NS1 expressed at the surface of virally infected cells (36). In contrast, the ZIKV envelope protein is less accessible at the cell surface as ZIKV particles bud internally from the Golgi apparatus rather than from the plasma membrane. Here, we demonstrated that neutralizing monoclonal antibodies were unable to elicit ADCC as measured by an in vitro reporter assay (Fig. 2). On the other hand, ADE of infection occurs when antibody binds virion and facilitates internalization into innate immune cells that allow for viral replication. By measuring ADE of infection in vitro, we were able to show that many neutralizing antibodies had the ability to induce ADE at low concentrations. Neutralizing antibodies are superior to NS1-binding antibodies in providing sterilizing immunity to ZIKV, but they also bear the potential risk of inducing ADE. Additionally, these neutralizing antibodies were unable to elicit protective Fc-mediated effector functions on infected cells, suggesting the limited utility of E-specific antibodies in clearing virally infected cells. Antibodies FA12 and EA7 were both poor neutralizers of ZIKV and exhibited very little activity in our ADE in vitro assay. Antibody AC11 was a rather potent neutralizer, with activity in the nanogram-to-milliliter range, but was a poor inducer of ADE. We speculate that the location of the epitope may impact the ability of an antibody to optimally engage and activate the Fcγ receptors, similar to our observations with antibodies that bind the influenza virus hemagglutinin (37). However, this would require further characterization.
We passaged wild-type virus with increasing concentrations of neutralizing antibodies to identify critical amino acid residues required for neutralization of virus. We passaged the antibodies with the MR766 strain of ZIKV as a reverse genetics system based on this strain was available to us (27). In Fig. 4, we show mutations of escape mutants from eight antibodies mapped onto crystal structures of the E protein. We found that, by and large, the moderately neutralizing antibodies tended to induce escape mutations in solvent-exposed residues in domain II, while potently neutralizing antibodies induced escape mutations in the lateral ridge region of domain III and domain I. We also found that for antibody AC10, the S368R mutation correlated with the appearance of a D57N mutation in the premembrane gene of the virus. This residue at site 57 may play a compensatory role in viral replication. Our hypothesis will require further investigation in future studies. We conclude that, consistent with previous reports, the lateral ridge region of the envelope protein tends to induce more potently neutralizing antibodies than domain II (30). To determine if the serine residue at site 368 was sufficient to mediate viral escape or whether compensatory mutations are required, we generated a recombinant virus displaying only the S368R point mutation. Upon analysis of publicly available sequences from the Virus Pathogen Resource, we found that 164 of 168 sequences (97.6%) with identifiable amino acids had a serine at site 368. However, one strain contained an S368R mutation, suggesting that this variant, although very uncommon, does occur in nature. We found that the MR766 variant with this mutation displayed reduced growth in vitro, suggesting that this mutation caused attenuation of the virus, which was confirmed by multicycle growth curves. By analyzing the whole genomes of plaque-purified escape mutants, we observed that the immunological pressure induced by MAbs did not always result in the same mutation although they were generally located in the same region. Furthermore, we identified escape variants with multiple mutations; we speculate that there may be mutations in the same epitope close to each other or that there are compensatory mutations that are required for proper viral replication. Notably, we also detected mutations outside the E coding region located in other structural domains, such as the prM, and in regions encoding nonstructural proteins NS2A, NS3, and NS5. More work is required to fully characterize the significance of these mutations in the overall fitness of these escape variants.
By using a time-of-addition infection assay, we were able to compare neutralization of this mutant MR766 with that of wild-type MR766 rescued in parallel. The goal of this experiment was 2-fold: we aimed to determine whether our antibodies could inhibit Zika virus replication after virion was bound to the cell surface, and we sought to further validate our escape variant ZIKV S368R. While the antibodies were able to neutralize virus before and at the time of infection, neutralization efficiency was substantially lower when antibody was added after viral entry. We could therefore conclude that our neutralizing antibodies inhibited viral binding and/or fusion and had significantly reduced potency after these events. Of note, there are slight variations in neutralization percentages in plaque reduction neutralization assays compared to those of a flow cytometry-based assay due to differences in viral quantification and assay sensitivity. However, we found a marked reduction in wild-type virus replication when virus was treated with AC10 compared to that of the S368R variant treated with AC10. Our results also showed that antibodies containing the same germ line rearrangements as AC10 were unable to neutralize the S368R variant and therefore likely recognized a similar epitope. However, our results do not fully explain why we did not recover any escape mutants to GD12 that possessed a mutation at site 368. We speculate that both sites 162 and 368 are involved in the formation of epitopes recognized by AC10, AC4, AC3, and GD12 or that mutations at site 368 are rarer in escape mutants to GD12. In contrast, the neutralizing antibodies GA3, AD5, FC11, and FC3, which had distinct germ line rearrangements, neutralized S368R and wild-type MR766 to the same degrees, suggesting that these antibodies recognized epitopes distinct from those of AC10.
In this study, we characterized monoclonal antibodies elicited by acute Zika virus infection. These antibodies neutralized virus by directly binding to the virion surface (38). We found that these antibodies enhanced infection of myeloid cells but did not appear to activate Fc-effector functions in vitro. Finally, we hypothesize that the majority of the antibodies we isolated recognize quaternary epitopes. One such epitope is putatively located on the lateral ridge region of the envelope protein, where mutation of the amino acid at position 368 inhibits neutralization by AC10 and antibodies with the same germ line rearrangements.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney (HEK) 293T cells (American Type Culture Collection [ATCC] CRL-1573) and African green monkey kidney cells (Vero; ATCC CCL-81) were grown in Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS) (HyClone) and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin [Pen-Strep]; Gibco). Human embryonic kidney Expi293F cells (Gibco) were grown in Expi293 expression medium. MR766 virus (Rhesus/1947/Uganda BEI NR-50065) was obtained from BEI Resources. FcγR-expressing K562 cells were obtained through the ATCC (catalog no. CCL-243). Pan-flavivirus antibody 4G2 was obtained from ATCC D1-4G2-4-15 cells (ATCC HB-112). ZIKV was propagated in Vero cells in DMEM supplemented with 2% FBS; after 96 h postinfection (hpi), cell culture supernatants were harvested, aliquoted, and stored at −80°C until use.
Human plasmablast isolation.
Plasmablasts were isolated from an infected patient at approximately 2 weeks after onset of symptoms. The patient was confirmed to have active ZIKV infection based on symptomatology and laboratory testing by reverse transcription-PCR (RT-PCR); however, we were not able to sequence the virus from the patient, and the exact ZIKV strain is unknown. Plasmablasts defined as CD19+ CD3− CD20− CD38high CD27high were isolated, and monoclonal antibodies were generated in accordance with the Icahn School of Medicine at Mount Sinai Institutional Review Board based on a previously published protocol (22). Briefly, Ficoll density (GE Healthcare) centrifugation was performed on whole-blood samples to isolate the buffy coat, and peripheral blood mononuclear cells (PBMCs) were single-cell sorted onto freshly prepared catch buffer (50 μl of 1 M Tris, pH 8, and 125 μl of RNasin in 5 ml of RNase-free water) on 96-well plates using a BD FACSAria III instrument. Reverse transcription reactions were performed to generate cDNA (22, 39). Two nested PCRs incorporating IgG-, IgA-, IgM-, kappa- and, lambda-specific primers were performed on the cDNA to amplify heavy and light chains. The International Immunogenetics Information System software (http://www.imgt.org/IMGT_vquest/vquest) was used to view productive immunoglobulin sequence rearrangements. Sixteen Zika virus antibodies were isolated from one patient, 4 of which are NS1 specific and were characterized previously (21), and the remaining 12 are reported here. Genetic characteristics of recombinant antibodies are reported in Table 1.
Recombinant human antibodies.
The human heavy (VH) and light (VL) variable regions of the isolated antibodies were amplified by PCR and cloned into human IgG1 and kappa or lambda mammalian expression vectors, respectively (pFUESss-CHIg-hIgG1, pFUESss-CLIg-hK, or pFUSEss-CLIg-hl2; Invivogen). To generate recombinant antibodies, 30 ml of Expi293 cells at 1 × 106 cells/ml was transfected with 30 μg of pFUESss-CHIg-hIgG1, pFUESss-CLIg-hK plasmids, and 81 μl of ExpiFectamine reagent (Gibco) as per the manufacturer’s instructions. After 120 h, supernatants were cleared by low-speed centrifugation and purified with protein G resin (Thermo Scientific).
Recombinant ZIKV NS1.
A mammalian expression plasmid expressing the NS1 of ZIKV MR766 (Rhesus/1947/Uganda BEI NR-50065) was generated by incorporating the last 24 amino acids of ZIKV envelope (NGSISLMCLALGGVLIFLSTAVSA) into the amino terminus of the NS1 coding region; the entire sequence was human codon optimized using the Integrated DNA Technologies Codon Optimization Tool (http://www.idtdna.com/CodonOpt). A PreScission Protease cleavage site (LEVLFNGPG) and a hexahistidine motif (HHHHHH) were added to the carboxy terminus of the NS1 coding region, resulting in pCAGGS NS1-His. To generate recombinant NS1 proteins, 30 ml of Expi293 cells at 1 × 106 cells/ml was transfected with 30 μg of pCAGGS-NS1-His plasmids and 81 μl of ExpiFectamine transfection reagent (Gibco) as per the manufacturer’s instructions. After 120 h, cells were pelleted by low-speed centrifugation and sonicated. Sonicated cells were pelleted again by centrifugation, and supernatant was removed and incubated with Ni-nitrilotriacetic acid (NTA) resin overnight at 4°C. The resin-supernatant mixture was then passed over 10-ml polypropylene columns (Qiagen). The retained resin was washed four times with 15 ml of washing buffer (50 mM Na2HCO3, 300 mM NaCl, 20 mM imidazole, pH 8), and protein was eluted with elution buffer (50 mM Na2HCO3, 300 mM NaCl, 300 mM imidazole, pH 8). The eluate was concentrated using Amicon Ultracell (Millipore) centrifugation units with a cutoff of 10 kDa, and buffer was exchanged to phosphate-buffered saline (PBS) of pH 7.4. Protein concentration was quantified using a Pierce bicinchoninic acid protein assay kit (Thermo Scientific) with a bovine serum albumin (BSA) standard curve.
ELISA.
Immulon 4 HBX enzyme-linked immunosorbent assay (ELISA) plates (Thermo Scientific) were coated with 1:25-diluted supernatant from Vero cells infected with MR766, 2 μg/ml recombinant envelope protein (catalog no. MBS319787; MyBioSource), or 2 μg/ml recombinant ZIKV MR766 NS1 protein (produced in-house) in pH 9.41 carbonate buffer overnight at 4°C. Plates were washed three times with PBS between each step. After plates were blocked with 5% nonfat milk powder for 1 h, antibodies were incubated at a starting concentration of 10 μg/ml and serially diluted 3-fold and incubated for 2 h at room temperature. Horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (AP504P; Millipore Sigma) was used to detect binding of IgG antibodies, followed by development with HRP substrate (Sigmafast OPD; Sigma-Aldrich). Reactions were stopped by addition of 3 M HCl, and absorbance was measured at 490 nm on a microplate spectrophotometer (Bio-Rad). Experiments were performed in duplicates. GraphPad Prism, version 6, was used to calculate values of the area under the concentration-time curve (AUC).
Plaque reduction neutralization tests.
A modified plaque reduction neutralization test was performed as described previously (40). Vero cells were plated in 24-well plates 1 day prior to infection to achieve 90% confluence the day of infection. On the day of infection, a dilution series of antibodies was preincubated with approximately 50 PFU of virus for 1 h at room temperature. The virus and antibody mixture was used to infect the Vero cells in duplicate, and cells were incubated for 1 h at 37°C, with rocking every 20 min. The virus-antibody mixture was aspirated, and a 1% methylcellulose overlay medium was added to each well with the appropriate antibody dilutions. At 96 h postinfection, or when plaques became visible by eye, the monolayer was fixed with 4% paraformaldehyde (PFA) in PBS for 1 h. The cells were washed with water and stained with 4G2 at 5 μg/ml overnight. After being washed, cells were incubated with HRP-conjugated goat anti-mouse IgG antibody (AP503P; Millipore Sigma) in 5% nonfat milk powder in PBS. Plaques were subsequently visualized using TrueBlue peroxidase substrate (KPL, Inc.). Experiments were performed twice as biological duplicates. A nonlinear curve was generated with GraphPad Prism, version 6, and the 50% inhibitory concentration (IC50) was calculated from the curve.
Antibody-dependent effector functions.
Vero cells were seeded into 96-well flat white-bottom plates (Corning) and infected after 24 h with Zika virus MR766 at an MOI of 0.01. At 40 h postinfection, the medium was removed, and 25 μl of assay buffer (RPMI 1640 medium with 4% low-IgG FBS) was added to each well. Then antibodies were added in a volume of 25 μl at a starting concentration of 10 μg/ml and serially diluted 3-fold in assay buffer in duplicate. The antibodies were then incubated with the infected cells for 30 min at 37°C. Genetically modified Jurkat cells expressing the human FcγRIIIa with a luciferase reporter gene under the transcriptional control of a nuclear factor of activated T cells (NFAT) promoter were added at 7.5 × 104 cells in 25 μl per well, which is approximately a 1:2 ratio of target cells to effector cells (Promega). Cells were then incubated for another 6 h at 37°C. Seventy-five microliters per well of Bio-Glo luciferase assay reagent was added, and luminescence was quantified using a microplate reader. Fold induction was measured in relative light units (RLU) and calculated by subtracting background signal from wells without effector cells and then dividing values for wells with antibody by values for wells with no antibody added. Specifically, fold induction was calculated as follows: (RLUinduced – RLUbackground)/(RLUuninduced – RLUbackground). The mean values and standard errors of the means (SEM) were reported, and a nonlinear regression curve was generated using GraphPad Prism, version 6.
Antibody-dependent enhancement of infection.
Enhancement of ZIKV infection was measured using a flow cytometry-based assay as previously described (12). Briefly, serial dilutions of purified monoclonal antibody were incubated with ZIKV (MR766) for 1 h at 37°C in RPMI 1640 medium supplemented with 10% FBS, 2 mM l-glutamine, and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin [Pen-Strep]; Gibco). As a negative control, purified human polyclonal antibody that was not reactive to ZIKV envelope protein was used. The virus-antibody complexes were then added to K562 cells in 96-well U-bottom plates at an MOI of 1. After 2 days at 37°C, cells were fixed with 4% PFA, permeabilized with PBS containing 0.2% BSA and 0.05% saponin, and stained with 4G2 pan-flavivirus anti-envelope antibody (1 μg per ml) for 1 h at room temperature. After a washing step, cells were then incubated with goat anti-mouse IgG conjugated to phycoerythrin (1 μg per ml; Invitrogen) for 1 h at room temperature. The number of infected cells was determined by flow cytometry using a FACSCalibur instrument (BD Biosciences) and analyzed using FlowJo2 software, version 10.1.r7.
Generation of escape mutants.
To generate escape variants, virus was serially passaged on a Vero cell monolayer in the presence of increasing amounts of antibody. In parallel, wild-type virus was passaged on the same days to account for potential adaptive mutations that may arise during passaging. Initially, neutralizing antibody at 0.1 IC50 was added to MR766 ZIKV (10 50% tissue culture infective doses [TCID50]) in 2 ml of minimum essential medium and incubated at room temperature for 15 min. The antibody and virus mixtures were added to six-well plates of Vero cells and moved to 37°C. After 1 h, the medium was removed and replaced with minimum essential medium with the antibody alone. The cells were observed daily for signs of cytopathic effect (CPE). When CPE was observed, 100 μl of supernatant was collected and added to a fresh monolayer of Vero cells in the presence of antibody. This process was repeated for 12 passages until the final antibody concentration was approximately 10,000-fold IC50. At this point, a serial dilution of escaped or wild-type virus was plated in the presence of 100× IC50 of antibody or no antibody. Escaped virus was then plaque purified and amplified on Vero cells, and RNA was extracted using an RNA isolation kit (Qiagen). RT-PCR was performed using overlapping envelope-specific primers, and cDNA was isolated by agarose gel electrophoresis and sequenced, at which point the mutation S368R was identified.
Whole-genome sequencing.
Viral RNA was extracted from plaque-purified cell culture supernatants as described above. cDNA was generated using Superscript III Supermix (ThermoFisher) using random hexamers, followed by PCR amplification using virus-specific primers (41). Each sample was double barcoded using a sequence-independent single primer amplification (42). Samples were sequenced using 300-bp paired-end reads on an Illumina MiSeq instrument. Consensus sequence of viruses were analyzed using CLC Genomics Workbench 11. Assemblies were annotated with the Viral Genome ORF reader (VIGOR), version 3, software (43) before submission to GenBank.
Sequence alignment of ZIKV E.
We performed a sequence variation analysis on available viral sequence data using Virus Pathogen Resource (44, 45). Briefly, we qualified our search by choosing only complete genomes and unique samples (by removing identical duplicate sequences), which brought the total number of sequences to 173. Of note, in certain cases, complete viral genomes may still contain indels in the middle of a gene, according to the National Center for Bioinformatics. A multiple-sequence alignment is first generated using MUSCLE, and a consensus sequence is determined (where the presence of an allele is greater than 50%). A polymorphism score is assigned for each amino acid position using a modified formula from Crooks et al. (46): score = −100 × sum(Pi × log Pi), where Pi is the frequency of the ith allele. The polymorphism score represents the normalized entropy of an observed allele distribution. Amino acid scores can have values of 0 (no observed polymorphism) to 439 (20 alleles and an indel, ∼4.7% frequency each).
Rescue of recombinant MR766.
Plasmid-based MR766 ZIKV was produced in 293T cells by transfection (27). An AGC-to-AGA mutation (S368R) was introduced into the MR766 viral genome and cloned by the use of overlapping primer sets encoding the S368R mutation (primer sequences available upon request). Of note, the glycan from MR766 was deleted from the plasmid by removal of the nucleotide sequence GTCAATGATACA, corresponding to amino acids VNDT, by site-directed mutagenesis in order to ensure a proper comparison to the MR766 virus obtained from BEI Resources (GenBank accession number KU963573.2). The PCR products were cloned into the pCDNA6.2 plasmid by use of an In-Fusion HD Cloning kit (TaKaRa Bio). 293T cells were seeded at a density of 1 × 106 cells per ml in six-well polylysine-coated plates. Cells were transfected with 5 μg of DNA per well with 100 μl of Opti-MEM (Gibco BRL Life Technologies, Gaithersburg, MD) and 15 μl of TransIT (Mirus Bio, Madison, WI) that had been incubated with the DNA for 30 min prior to transfection. At 72 h posttransfection virus was harvested and plaque purified. RT-PCR followed by sequencing was performed to confirm the S368R mutation. Quantification of virus in a multicycle growth curve on Vero cells was performed by plaque assay at 24-h time points performed in duplicate. For virus used in time-of-addition infection assays, ZIKV was harvested at 96 h postinfection to ensure similar viral titers.
Time-of-addition infection assays.
Time-of-addition infection assays were performed utilizing a flow cytometry-based readout (28). Vero cells were seeded on poly-l-lysine-coated 24-well tissue culture plates 24 h prior to infection. Cells were cooled to 4°C at −3.5 h. At −3 h, the cells were washed once with PBS, and medium was replaced with DMEM supplemented with 2% FBS and either wild-type or S368R MR766 virus at an MOI of 0.2 at 4°C. At 0 h, the cultures were washed twice with cold PBS, and fresh DMEM with 2% FBS was added. The cultures were then moved to 37°C. Antibodies were added at either −3 h, 0 h, or +6 h. Antibodies added at −3 h were replenished when the cells were washed with PBS and moved to 37°C. At 24 h postinfection the medium was replenished with fresh DMEM with 2% FBS. At 2 days postinfection cells were collected, fixed, and stained with 4G2 pan-flavivirus anti-envelope antibody in Perm/Wash buffer (BD). Cells were then incubated with goat anti-mouse IgG conjugated to Alexa Fluor 488 (1:2,000 dilution; Invitrogen). Cells were analyzed by flow cytometry to determine the percentage of infected cells compared to that in a no-antibody control. Percent infection values above 100% were set as 100%. All experiments were performed as duplicates.
Study approval.
An Institutional Review Board (IRB)-approved written informed consent document was obtained from the patient prior to participation in this study. No further demographic data are included in the manuscript in order to protect the participant’s privacy.
Statistical analysis.
Results from multiple experiments are presented as means ± SEM. Student’s t tests were used to test for statistical differences between mean values. Data were analyzed with GraphPad Prism, version 6, software, and P values below 0.05 were considered statistically significant (denoted by an asterisk).
Data availability.
Sequences are available in GenBank under accession numbers MH130094 to MH130109 and MH061852 to MH061915.
ACKNOWLEDGMENTS
We thank B. Fulton and P. Leon for valuable assistance in plasmablast isolation. We also acknowledge the Flow Cytometry Shared Resource Facility at the Icahn School of Medicine at Mount Sinai.
This work was supported by grants R01AI120998 (V.S), R21AI130299 (J.K.L.), and R21AI133649 (M.J.E.). M.J.E. holds an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. G.S.T. is supported by a J. Craig Venter Institute start-up fund and U19AI110819. This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under award number U19AI110819. M.J.B. was supported by the MSTP Training Grant NIH T32 GM007280. F.B. thanks the German Academy of Sciences Leopoldina for financial support through a postdoctoral stipend.
REFERENCES
- 1.Petersen LR, Jamieson DJ, Honein MA. 2016. Zika virus. N Engl J Med 375:294–295. doi: 10.1056/NEJMc1606769. [DOI] [PubMed] [Google Scholar]
- 2.Cao-Lormeau V-M, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial A-L, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra J-C, Despres P, Fournier E, Mallet H-P, Musso D, Fontanet A, Neil J, Ghawché F. 2016. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brasil P, Pereira JP, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai U-A, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baião AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. 2016. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. 2016. Zika virus and birth defects - Reviewing the evidence for causality. N Engl J Med 374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 5.Gaudinski MR, Houser KV, Morabito KM, Hu Z, Yamshchikov G, Rothwell RS, Berkowitz N, Mendoza F, Saunders JG, Novik L, Hendel CS, Holman LA, Gordon IJ, Cox JH, Edupuganti S, McArthur MA, Rouphael NG, Lyke KE, Cummings GE, Sitar S, Bailer RT, Foreman BM, Burgomaster K, Pelc RS, Gordon DN, DeMaso CR, Dowd KA, Laurencot C, Schwartz RM, Mascola JR, Graham BS, Pierson TC, Ledgerwood JE, Chen GL, Plummer S, Costner P, Zephir K, Casazza J, Ola A, Victorino M, Levinson C, Whalen W, Wang X, Cunningham J, Vasilenko O, Burgos Florez M, Hickman S, Pittman I, Le L, Larkin B, et al. 2018. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: randomised, open-label, phase 1 clinical trials. Lancet 391:552–562. doi: 10.1016/S0140-6736(17)33105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richner JM, Jagger BW, Shan C, Fontes CR, Dowd KA, Cao B, Himansu S, Caine EA, Nunes BTD, Medeiros DBA, Muruato AE, Foreman BM, Luo H, Wang T, Barrett AD, Weaver SC, Vasconcelos PFC, Rossi SL, Ciaramella G, Mysorekar IU, Pierson TC, Shi P-Y, Diamond MS. 2017. Vaccine-mediated protection against Zika virus-induced congenital disease. Cell 170:273–283.e12. doi: 10.1016/j.cell.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richner JM, Himansu S, Dowd KA, Butler SL, Salazar V, Fox JM, Julander JG, Tang WW, Shresta S, Pierson TC, Ciaramella G, Diamond MS. 2017. Modified mRNA vaccines protect against Zika virus infection. Cell 168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modjarrad K, Lin L, George SL, Stephenson KE, Eckels KH, De La Barrera RA, Jarman RG, Sondergaard E, Tennant J, Ansel JL, Mills K, Koren M, Robb ML, Barrett J, Thompson J, Kosel AE, Dawson P, Hale A, Tan CS, Walsh SR, Meyer KE, Brien J, Crowell TA, Blazevic A, Mosby K, Larocca RA, Abbink P, Boyd M, Bricault CA, Seaman MS, Basil A, Walsh M, Tonwe V, Hoft DF, Thomas SJ, Barouch DH, Michael NL. 2018. Preliminary aggregate safety and immunogenicity results from three trials of a purified inactivated Zika virus vaccine candidate: phase 1, randomised, double-blind, placebo-controlled clinical trials. Lancet 391:563–571. doi: 10.1016/S0140-6736(17)33106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giel-Moloney M, Goncalvez AP, Catalan J, Lecouturier V, Girerd-Chambaz Y, Diaz F, Maldonado-Arocho F, Gomila RC, Bernard M-C, Oomen R, Delagrave S, Burdin N, Kleanthous H, Jackson N, Heinrichs J, Pugachev KV. 2018. Chimeric yellow fever 17D-Zika virus (ChimeriVax-Zika) as a live-attenuated Zika virus vaccine. Sci Rep 8:13206. doi: 10.1038/s41598-018-31375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitehead SS, Blaney JE, Durbin AP, Murphy BR. 2007. Prospects for a dengue virus vaccine. Nat Rev Microbiol 5:518–528. doi: 10.1038/nrmicro1690. [DOI] [PubMed] [Google Scholar]
- 11.Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, Foglierini M, Pedotti M, Simonelli L, Dowall S, Atkinson B, Percivalle E, Simmons CP, Varani L, Blum J, Baldanti F, Cameroni E, Hewson R, Harris E, Lanzavecchia A, Sallusto F, Corti D. 2016. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- 12.Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, Stramer SL, García-Sastre A, Krammer F, Lim JK. 2017. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356:175–180. doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau V-M, Malasit P, Rey FA, Mongkolsapaya J, Screaton GR. 2016. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol 17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, Ahmed R, Suthar MS, Wrammert J. 2016. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A 113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willis E, Hensley SE. 2017. Characterization of Zika virus binding and enhancement potential of a large panel of flavivirus murine monoclonal antibodies. Virology 508:1–6. doi: 10.1016/j.virol.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown JA, Singh G, Acklin JA, Lee S, Duehr JE, Chokola AN, Frere JJ, Hoffman KW, Foster GA, Krysztof D, Cadagan R, Jacobs AR, Stramer SL, Krammer F, García-Sastre A, Lim JK. 2019. Dengue virus immunity increases Zika virus-induced damage during pregnancy. Immunity 50:751–762.e5. doi: 10.1016/j.immuni.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George J, Valiant WG, Mattapallil MJ, Walker M, Huang Y-J, Vanlandingham DL, Misamore J, Greenhouse J, Weiss DE, Verthelyi D, Higgs S, Andersen H, Lewis MG, Mattapallil JJ. 2017. Prior exposure to Zika virus significantly enhances peak dengue-2 viremia in rhesus macaques. Sci Rep 7:10498. doi: 10.1038/s41598-017-10901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerman MG, Quicke KM, O'Neal JT, Arora N, Machiah D, Priyamvada L, Kauffman RC, Register E, Adekunle O, Swieboda D, Johnson EL, Cordes S, Haddad L, Chakraborty R, Coyne CB, Wrammert J, Suthar MS. 2018. Cross-reactive dengue virus antibodies augment Zika virus infection of human placental macrophages. Cell Host Microbe 24:731–742.e6. doi: 10.1016/j.chom.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapparapu G, Fernandez E, Kose N, Bin Cao Fox JM, Bombardi RG, Zhao H, Nelson CA, Bryan AL, Barnes T, Davidson E, Mysorekar IU, Fremont DH, Doranz BJ, Diamond MS, Crowe JE. 2016. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature 540:443–447. doi: 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbiani DF, Bozzacco L, Keeffe JR, Khouri R, Olsen PC, Gazumyan A, Schaefer-Babajew D, Avila-Rios S, Nogueira L, Patel R, Azzopardi SA, Uhl LFK, Saeed M, Sevilla-Reyes EE, Agudelo M, Yao K-H, Golijanin J, Gristick HB, Lee YE, Hurley A, Caskey M, Pai J, Oliveira T, Wunder EA, Sacramento G, Nery N, Orge C, Costa F, Reis MG, Thomas NM, Eisenreich T, Weinberger DM, de Almeida ARP, West AP, Rice CM, Bjorkman PJ, Reyes-Teran G, Ko AI, MacDonald MR, Nussenzweig MC. 2017. Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell 169:597–609.e11. doi: 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey MJ, Duehr J, Dulin H, Broecker F, Brown JA, Arumemi FO, González MCB, Leyva-Grado VH, Evans MJ, Simon V, Lim JK, Krammer F, Hai R, Palese P, Tan GS. 2018. Human antibodies targeting Zika virus NS1 provide protection against disease in a mouse model. Nat Commun 9:4560. doi: 10.1038/s41467-018-07008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith K, Garman L, Wrammert J, Zheng N-Y, Capra JD, Ahmed R, Wilson PC. 2009. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala W, White LJ, Diamond MS, Baric RS, Crowe JE, de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci 109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG. 2010. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc Natl Acad Sci U S A 107:18950–18955. doi: 10.1073/pnas.1011036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney M-C, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, Girard-Blanc C, Petres S, Shepard WE, Desprès P, Arenzana-Seisdedos F, Dussart P, Mongkolsapaya J, Screaton GR, Rey FA. 2015. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 26.Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS, Fremont DH. 2016. Structural basis of Zika virus-specific antibody protection. Cell 166:1016–1027. doi: 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz MC, Sourisseau M, Espino MM, Gray ES, Chambers MT, Tortorella D, Evans MJ. 2016. Rescue of the 1947 Zika virus prototype strain with a cytomegalovirus promoter-driven cDNA clone. mSphere 1:e00246-16. doi: 10.1128/mSphere.00246-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers MT, Schwarz MC, Sourisseau M, Gray ES, Evans MJ. 2018. Probing Zika virus neutralization determinants with glycoprotein mutants bearing linear epitope insertions. J Virol 92:e00505-18. doi: 10.1128/JVI.00505-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Yang H, Liu X, Dai L, Ma T, Qi J, Wong G, Peng R, Liu S, Li J, Li S, Song J, Liu J, He J, Yuan H, Xiong Y, Liao Y, Li J, Yang J, Tong Z, Griffin BD, Bi Y, Liang M, Xu X, Qin C, Cheng G, Zhang X, Wang P, Qiu X, Kobinger G, Shi Y, Yan J, Gao GF. 2016. Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Sci Transl Med 8:369ra179. doi: 10.1126/scitranslmed.aai8336. [DOI] [PubMed] [Google Scholar]
- 30.Heinz FX, Stiasny K. 2017. The antigenic structure of Zika virus and its relation to other flaviviruses: implications for infection and immunoprophylaxis. Microbiol Mol Biol Rev 81:e00055-16. doi: 10.1128/MMBR.00055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armandola EA, Mariani SM, Ferrone S. 1993. Serological and molecular characterization of mouse anti-idiotypic monoclonal antibodies elicited with the syngeneic Anti-HLA-A2,28 monoclonal antibody CR11-351. Mol Immunol 30:287–300. doi: 10.1016/0161-5890(93)90057-I. [DOI] [PubMed] [Google Scholar]
- 32.Burlingham WJ, Jankowska-Gan E, DeVito-Haynes L, Fechner JH, Hogan KT, Claas FHJ, Mulder A, Wang X, Ferrone S. 1998. HLA (A*0201) mimicry by anti-idiotypic monoclonal antibodies. J Immunol 161:6705–6714. [PubMed] [Google Scholar]
- 33.Bournazos S, DiLillo DJ, Ravetch JV. 2015. The role of Fc-FcγR interactions in IgG-mediated microbial neutralization. J Exp Med 212:1361–1369. doi: 10.1084/jem.20151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor A, Foo S-S, Bruzzone R, Dinh LV, King NJC, Mahalingam S. 2015. Fc receptors in antibody-dependent enhancement of viral infections. Immunol Rev 268:340–364. doi: 10.1111/imr.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuzmina NA, Younan P, Gilchuk P, Santos RI, Flyak AI, Ilinykh PA, Huang K, Lubaki NM, Ramanathan P, Crowe JE, Bukreyev A. 2018. Antibody-dependent enhancement of Ebola virus infection by human antibodies isolated from survivors. Cell Rep 24:1802–1815.e5. doi: 10.1016/j.celrep.2018.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rastogi M, Sharma N, Singh SK. 2016. Flavivirus NS1: a multifaceted enigmatic viral protein. Virol J 13:131. doi: 10.1186/s12985-016-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leon PE, He W, Mullarkey CE, Bailey MJ, Miller MS, Krammer F, Palese P, Tan GS. 2016. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc Natl Acad Sci U S A 113:E5944–E5951. doi: 10.1073/pnas.1613225113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowd KA, Pierson TC. 2011. Antibody-mediated neutralization of flaviviruses: a reductionist view. Virology 411:306–315. doi: 10.1016/j.virol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho IY, Bunker JJ, Erickson SA, Neu KE, Huang M, Cortese M, Pulendran B, Wilson PC. 2016. Refined protocol for generating monoclonal antibodies from single human and murine B cells. J Immunol Methods 438:67–70. doi: 10.1016/j.jim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan GS, Krammer F, Eggink D, Kongchanagul A, Moran TM, Palese P. 2012. A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J Virol 86:6179–6188. doi: 10.1128/JVI.00469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shrivastava S, Puri V, Dilley KA, Ngouajio E, Shifflett J, Oldfield LM, Fedorova NB, Hu L, Williams T, Durbin A, Amedeo P, Rashid S, Shabman RS, Pickett BE. 2018. Whole genome sequencing, variant analysis, phylogenetics, and deep sequencing of Zika virus strains. Sci Rep 8:15843. doi: 10.1038/s41598-018-34147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, Afonso C, Zhang X, Anderson NG, Ghedin E, Spiro DJ. 2008. Viral genome sequencing by random priming methods. BMC Genomics 9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Sundaram JP, Stockwell TB. 2012. VIGOR extended to annotate genomes for additional 12 different viruses. Nucleic Acids Res 40:W186–W192. doi: 10.1093/nar/gks528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pickett BE, Greer DS, Zhang Y, Stewart L, Zhou L, Sun G, Gu Z, Kumar S, Zaremba S, Larsen CN, Jen W, Klem EB, Scheuermann RH. 2012. Virus pathogen database and analysis resource (ViPR): a comprehensive bioinformatics database and analysis resource for the coronavirus research community. Viruses 4:3209–3226. doi: 10.3390/v4113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z, Zhou L, Larson CN, Dietrich J, Klem EB, Scheuermann RH. 2012. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res 40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences are available in GenBank under accession numbers MH130094 to MH130109 and MH061852 to MH061915.