Understanding the immunopathology of acute HIV-1 infection will help to develop eradication strategies. We demonstrate here that a CD4+ T cell memory subset expands during acute HIV-1 infection, which is associated with disease progression.
KEYWORDS: CD4 T cell, Fas, SCM (stem-cell-like memory), acute infection, human immunodeficiency virus, latent reservoir, memory population
ABSTRACT
Acute HIV-1 infection is characterized by high viremia and massive depletion of CD4+ T cells throughout all tissue compartments. During this time the latent viral reservoir is established but the dynamics of memory CD4+ T cell subset development, their infectability and influence on disease progression during acute HIV-1 infection has not been carefully described. We therefore investigated the dynamics of CD4+ T cell memory populations in the RV217 (ECHO) cohort during the acute phase of infection. Interestingly, while we found only small changes in central or effector memory compartments, we observed a profound expansion of stem cell-like memory CD4+ T cells (SCM) (2.7-fold; P < 0.0001). Furthermore, we demonstrated that the HIV-1 integration and replication preferentially take place in highly differentiated CD4+ T cells such as transitional memory (TM) and effector memory (EM) CD4+ T cells, while naive and less mature memory cells prove to be more resistant. Despite the relatively low frequency of productively infected SCM, we suggest that their quiescent phenotype, increased susceptibility to HIV-1 integration compared to naive cells and extensive expansion make them one of the key players in establishment and persistence of the HIV-1 reservoir. Moreover, the expansion of SCM in acute HIV-1 infection was a result of Fas upregulation on the surface of naive CD4+ T cells. Interestingly, the upregulation of Fas receptor and expansion of SCM in acute HIV-1 infection was associated with the early viral set point and disease progression (rho = 0.47, P = 0.02, and rho = 0.42, P = 0.041, respectively). Taken together, our data demonstrate an expansion of SCM during early acute HIV-1 infection which is associated with disease outcome.
IMPORTANCE Understanding the immunopathology of acute HIV-1 infection will help to develop eradication strategies. We demonstrate here that a CD4+ T cell memory subset expands during acute HIV-1 infection, which is associated with disease progression.
INTRODUCTION
During the first weeks of HIV-1 infection, virus exponentially replicates to high levels and disseminates throughout all tissue compartments (1, 2). Moreover, during this period a preferential depletion of more than half of all memory CD4+ T cells in all tissue compartments occurs, suggesting major and irreparable damage to the immune system (3, 4). With limited control over viremia, CD4+ T cell count partially recovers but does not reach comparable levels prior to infection. Over the course of chronic HIV-1 infection, the CD4+ T count further slowly declines, with an average CD4+ T cell loss of 50 cells/μl/year, which ultimately leads to the clinical onset of AIDS (5, 6). The early start of antiretroviral treatment (ART) preserves CD4+ T cells and maintains important helper function (7, 8). Indeed, individuals infected with HIV-1 have almost comparable life expectancy to HIV-1-negative individuals when treated early and effectively (9). However, HIV-1 infection is still a chronic infection, and there is no promising approach for a cure. Even after decades of fully suppressed viremia HIV-1 remains integrated into the genome of a small reservoir of latently infected cells. Several studies have investigated the properties of the latent reservoir. It has been shown that seeding of a latent reservoir occurs surprisingly early, as soon as 3 days after the HIV-1 transmission (8, 10). The primary target of HIV-1 integration during this period are CD4+ T cells. It is important to consider that these cells do not represent a homogenous population but can be compartmentalized according to their function or maturation status. The maturational profile of CD4+ T cells has been conceptualized as a linear model in which long-lived immature cells gradually develop into short-lived memory CD4+ T cells with effector properties (10, 11). Currently, there are six generally accepted and well defined subsets (stated from least to most mature): naive cells, as well as stem cell-like memory (SCM), central memory (CM), transitional memory (TM), effector memory (EM), and terminal effector (TE) CD4+ T cells (12, 13). Indeed, not all CD4+ memory T cell subsets equally harbor HIV-1 in their genome, and some have a longer life span than the others. It has been shown that in chronic untreated HIV-1-positive individuals, CM harbor the largest amount of HIV-1 (14). This picture remains the same even after years of treatment (11, 15). Interestingly, however, while SCM make only minor contribution to the overall reservoir of HIV-1 in chronic infection, they appear to play a major role in long-term persistence of latent HIV-1 infection. The pronounced importance of SCM as a reservoir is based on their quiescent phenotype, high proliferative capacity, high per-cell HIV-1 DNA content, and the slow decay of HIV-1 DNA in these cells (10, 11, 16). The estimated half-life of HIV-1 DNA within SCM was shown to be almost two times longer than in CM, increasing the contribution of this subset to the reservoir with prolonged antiretroviral therapy (11). Overall, these findings suggest distinct roles of CD4+ T cell memory subsets in the establishment of latent infection, indicating the importance of investigating CD4+ T cell memory subsets during the early days of HIV-1 infection. Here, we analyzed CD4+ T cell memory subset dynamics in peripheral blood of acutely HIV-1-infected individuals and their susceptibility to HIV-1 infection. Furthermore, we demonstrate how these temporal changes in subset frequencies and differences in infectibility influence progression and persistence of the disease.
RESULTS
A rapid expansion of SCM during hyper-acute HIV-1 infection.
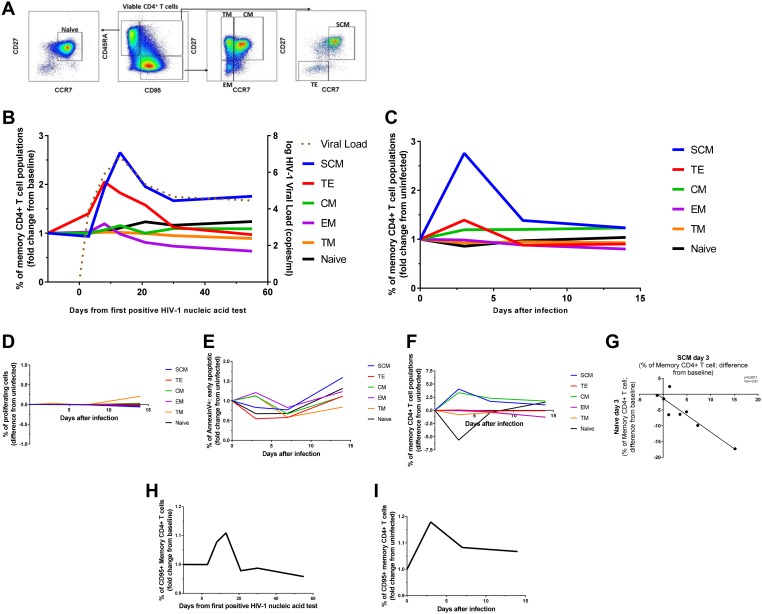
To understand the dynamics of long-term seeding of the HIV-1 reservoir in different T cell memory compartments, we first determined the kinetics and changes of memory subsets in a group of 24 individuals from the RV217 cohort (17). All individuals were HIV negative during enrollment, giving the opportunity to determine changes in memory subsets before and after infection. Moreover, having two time points prior to peak viremia, dynamic changes in T cell subsets could be assessed in greater detail. We therefore first assessed, longitudinally in all 24 subjects, changes in CD4+ T cell phenotype and memory subsets along the course of acute HIV-1 infection. Determination of probable time point of HIV-1 infection and viral load monitoring was assessed as previously described (18). We differentiated between the following CD4+ T cell subsets: naive cells (CCR7+, CD27+, CD95–, CD45RA+), SCM (CCR7+, CD27+, CD95+, CD45RA+), CM (CCR7+, CD27+, CD95+, CD45RA−), TM (CCR7–, CD27+, CD95+, CD45RA−), EM (CCR7–, CD27−, CD95+, CD45RA−), and TE (CCR7–, CD27−, CD95+, CD45RA+) (Fig. 1A). CD4+ T cell counts during acute HIV-1 infection dropped from a median of 855 to 472 cells/μl in the first 20 days of infection (P < 0.0001). Apart from decreased CD4+ T cell counts, flow-cytometric assessment of CD4+ T cell memory subsets before and during acute HIV-1 infection revealed dramatic changes in frequency of those subsets (Fig. 1B). In particular, we observed an average of 2.7-fold increase in SCM and a 1.2-fold increase in CM frequencies compared to baseline (P < 0.0001 and P = 0.0015, respectively) during peak viral load. Interestingly, in 17 of 24 individuals, we also observed a profound 2.4-fold (average of the 17 individuals) expansion of TE just before peak viremia, but this was overall not significantly different in all subjects. Moreover, our data demonstrated a significant increase in the frequency of naive cells at the early set point of viral load on day 55 (average of 1.2-fold; P = 0.026) and a gradual depletion of EM with progressing acute HIV-1 infection (average of 0.6-fold at the early set point; P < 0.0001). While TE decreased into the chronic phase of HIV-1 infection back to baseline levels, SCM and CM remained significantly expanded (average of 1.8-fold increase; P = 0.0007 and 1.1-fold increase; P = 0.012 at the early set point, respectively). In contrast, TM frequencies did not significantly change during acute HIV-1 infection. Taken together, in addition to CD4+ T cell count reduction, we observed significant changes in the CD4+ memory subsets with a notable expansion of SCM.
FIG 1.
Longitudinal changes in CD4+ T cell subset frequencies during the acute phase of HIV-1 infection. (A) Gating strategy used to delineate memory CD4+ T cell subsets. (B) Average fold change in CD4+ T cell subset frequency in peripheral blood of 24 acutely HIV-1-infected individuals. Superimposed is the course of viral load development. (C and F) Average fold change (C) and average absolute change (F) in CD4+ T cell subset frequency in HIV-1-infected cell cultures of healthy donor PBMC. (D) The frequencies of proliferating CD4+ T cells were not significantly higher in infected cell cultures compared to the uninfected controls. (E) Fold changes in the frequencies of early apoptotic CD4+ T cells showed no significant differences compared to the uninfected control. (G) The frequency of naive CD4+ T cells inversely correlates with the frequency of SCM on day 3 after the infection as confirmed by the Pearson’s correlation test. (H and I) Changes in prevalence of CD95+ CD4+ T cells along the course of acute HIV-1 infection closely resemble those of SCM in clinical settings (H), as well as in vitro (I). Statistical significance of changes in subset frequency at different time points was assessed by Kruskal-Wallis test.
Expansion of SCM during acute HIV-1 infection is characterized by CD95 upregulation.
To further study the phenomenon of SCM expansion in acute HIV-1 infection, we established an in vitro HIV-1-infected cell culture. For these experiments, we isolated peripheral blood mononuclear cells (PBMCs) from eight healthy donors and added inoculum containing infectious HIV-1 Ba-L virus in R10 media in the absence of any activation agent. We next monitored changes in CD4+ T cell memory subsets immediately before the infection and during the first 14 days after infection using the same phenotypic markers as described above. Strikingly, we observed almost identical subset dynamics in our in vitro experiments as observed ex vivo in the RV217 cohort (Fig. 1C). SCM peaked 3 days after infection with an average of 2.8-fold increase compared to baseline (P < 0.0094), while CM increased by 1.2-fold (P = 0.035) at the same time point. Both subsets remained elevated for the time of the experiment. TE, TM, and EM also followed similar patterns, as observed in the clinical data. Similarly, the frequency of naive cells contracted on day 3 but afterward slowly increased over time.
We wondered whether the expansion of SCM and CM could be explained by cell proliferation or enhanced resistance to apoptosis. We therefore analyzed annexin V binding by different CD4+ T cell subsets, which allows for the detection of early apoptotic cells (19). In addition, we labeled T cells prior to HIV-1 infection with cell-membrane-permeable amine-reactive dye to track the proliferation of the cells after infection. Interestingly, however, we did not observe proliferation of any cellular subset during the time of infection (Fig. 1D). In addition, we also did not observe a preferential loss or increase in apoptosis at any time point in any of these subsets (Fig. 1E). This indicates that the spike in SCM or CM is not due to a preferential loss of other subsets. Taken together, SC and CM expansion in culture mimics the expansion of CM and SCM observed in acute HIV-1 infection but is not due to the proliferative activity or depletion of CD4+ T cells.
SCM has been described as an important viral reservoir (10), and thus we further investigated the expansion of SCM ex vivo and in vitro. We noticed that the expansion of SCM was in parallel to a contraction in the naive T cell compartment (Fig. 1F) and that there is an inverse correlation between the frequencies of both subsets on day 3 after the infection of our cell culture (rho = –0.91, P = 0.0017) (Fig. 1G). Furthermore, we analyzed longitudinal changes in frequency of CD95+ CD4+ T cells during the acute HIV-1 infection and found that they closely resemble those of SCM in both clinical (average of 1.11-fold enrichment at its peak compared to baseline; P < 0.02) and in vitro settings (average of 1.18-fold enrichment at its peak compared to baseline; P < 0.017) (Fig. 1H and I, respectively). Indeed, the upregulation of the Fas receptor (CD95) on the surfaces of naive CD4+ T cells fully explained the expansion of the SCM compartment. Taken together, our data indicate that the early upregulation of Fas receptor in acute HIV-1 infection drives the development of SCM, an important part of the viral reservoir.
Highly differentiated memory CD4+ T cell subsets are the most susceptible to HIV-1 infection.
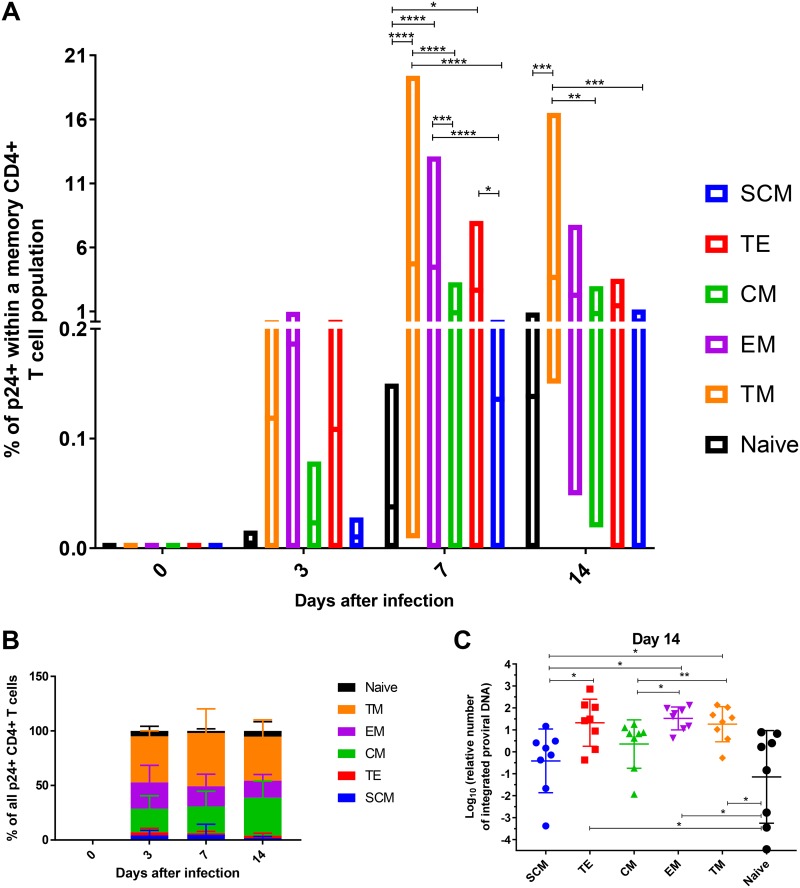
It has been previously described that CM, TM, and EM subsets make up most of the HIV-1 reservoir, while SCM seem to be less frequent but exclusively persistent (10). We therefore determined HIV-1 infection and replication in different CD4+ T cell subsets to assess whether this early expansion of SCM is critical for the immunopathogenesis of HIV. Identification of productively infected cells in our acute HIV-1 infection cell culture model was carried out by flow-cytometric detection of cytoplasmic p24 antigen and measured after 3, 7, and 14 days. As described above, CD4+ T cells were not activated or treated with any cytokines/chemokines. Three days after infection, we observed that the first CD4+ T cell subsets to become infected and produce HIV-1 virions were those exhibiting a higher degree of differentiation, namely, TM, EM, and TE, similar to those described in the literature (Fig. 2A). Differences in infectibility of CD4+ T memory cells between the subsets became significantly more apparent on day 7 after infection, with the p24+ cell frequencies reaching an average of 0.59% of the cell subsets CD4+ T cells. TM, EM, and TE subsets continued to harbor the highest percentage of p24+ cells, closely followed by CM, while naive cells and SCM proved to be much less prone to becoming productively infected. This pattern was maintained throughout HIV-1 infection until day 14 (Fig. 2A). The differences in HIV-1 infection of the different CD4+ T cell subsets became more apparent when we analyzed the overall contribution (Fig. 2B). Most of the productively infected cells arose from the TM subset, followed by EM and CM, while contributions of naive cells, SCM, and TE appeared to be modest, similar to previous ex vivo reports (14, 16, 20–23). Interestingly, the frequencies of p24+ CD4+ T cells were highly variable between donors (ranging from 0.06 to 0.002% on day 3, 2.14 to 0.001% on day 7, and 1.87 to 0.024% on day 14 after the infection), suggesting that the cells of some individuals are more susceptible to HIV-1 infection than others (Fig. 2A). Overall, however, although SCM did become infected, we mostly detected HIV-1 replication in CD4+ T cells that were differentiated into TM, TE, or CM.
FIG 2.
Susceptibility of CD4+ T cell subsets to HIV-1 infection increases with their maturation status. (A) Mean frequencies and ranges of p24+ cells within each of the analyzed subsets for every time point of monitoring. The data represent a summary from eight independent experiments. The statistical significance of differences between the subsets was determined by two-way analysis of variance (ANOVA) and is indicated by asterisks (*, P < 0.033; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001). (B) The pool of productively infected CD4+ T cells mainly consists of TM, EM, and CM, as demonstrated by flow-cytometric measurement of the p24 antigen. Shown are the proportions of p24+ cells stemming from different subsets at each time point of cell culture analysis. (C) Scatter plot depicting the frequency of HIV-1 genome integration into the DNA of cells belonging to different CD4+ T cell subsets, as measured by nested PCR. Statistical differences were determined by one-way ANOVA.
We next assessed whether there are differences in infectibility and susceptibility to productive infection among different CD4+ T cell subsets. We therefore measured the relative amount of integrated proviruses in the different CD4+ T cell subsets in our cell culture model. On day 14 after infection, we sorted CD4+ T cells into different subsets and isolated genomic DNA which was subsequently subjected to specific preamplification of integrated proviruses by Alu-gag PCR, as previously described (24). The relative amounts of resulting amplicons were then quantified by quantitative PCR (qPCR). The data revealed strikingly similar picture to the one derived from flow-cytometric p24 antigen detection (Fig. 2C). Subsets with the highest frequency of integrated provirus were TM, EM, and TE, followed by CM. Naive cells and SCM harbored the smallest amounts of the provirus (Fig. 2C). Collectively, our data suggest that HIV-1 provirus integration and viral replication preferentially take place in highly differentiated CD4+ T cells belonging to TM, EM, CM, and TE subsets. In contrast, naive cells and SCM retain much lower levels of detectable HIV-1 integration and replication.
SCM expansion is associated with the progression of HIV-1 infection.
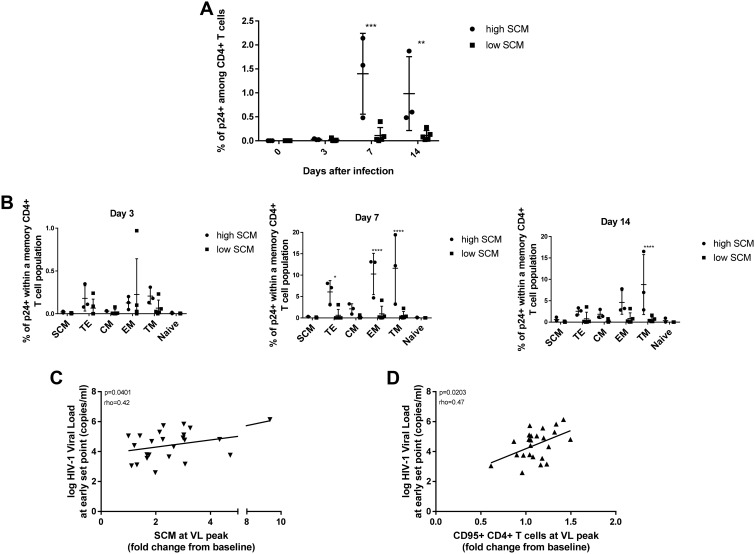
Given the infectibility of SCM in culture and previous reports of SCM being an important HIV-1 latent reservoir, we wanted to further assess the importance of SCM in the immunopathogenesis of HIV-1. This was particularly interesting, since we observed in the acute HIV-1 infection cell culture model two distinct groups of SCM expansion. One group had expansion of SCM of <1.5-fold (low SCM; n = 5), while a second group had a comparatively strong expansion of SCM of >3-fold (high SCM, n = 3) on day 3 after the in vitro infection. When we compared the frequency of productively infected CD4+ T cells in both groups, we found a striking difference on days 7 and 14 after infection, with significantly higher levels of p24+ CD4+ T cells in high SCM group (average of 1.4% versus 0.1% on day 7; average of 1.0% versus 0.1% on day 14), suggesting an association between the expansion of SCM and the degree of infectibility of CD4+ T cells (Fig. 3A). To obtain a deeper insight, we compared the frequencies of productively infected cells between the two groups for each CD4+ T cell subset at each time point of monitoring. While we did not observe any significant differences on day 3 after the infection, there was a clear trend on days 7 and 14 showing that the high SCM group tend to be more sensitive to HIV-1 infection, regardless of a subset, with the differences being most profound in highly susceptible subsets (Fig. 3B). Given the higher degree of productively infected CD4+ T cells in individuals with the more prominent expansion of SCM, we analyzed whether SCM expansion is also related to viremia during acute HIV-1 infection. Strikingly, we observed a significant positive correlation (rho = 0.42, P = 0.041) between the extent of SCM expansion and viral load at the early set point of acute infection (Fig. 3C), indicating that SCM expansion in acute HIV-1 infection is not only associated with a larger HIV-1 reservoir but also associated with a more rapid course of disease progression. Moreover, we found a significant positive correlation (rho = 0.47, P = 0.02) between the set point viral load and the frequency of bulk CD95+ CD4+ T cells at the time of SCM expansion (Fig. 3D). Taken together, we observed that SCM expansion occurring early in acute HIV-1 infection is significantly associated with productive HIV-1 infection and disease progression.
FIG 3.
HIV-1 replicates more effectively in the CD4+ T cells of individuals with profound SCM expansion. (A and B) Comparison of p24+ cell frequencies between the low and high SCM groups at each day of cell culture monitoring revealed striking differences within the entire CD4+ T cell population (A), as well as within the distinct CD4+ T cell subsets (B). The statistical significance of differences between the data sets depicted on bar graphs was analyzed using two-way ANOVA and is indicated by asterisks (*, P < 0.033; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001). (C and D) The degree of SCM (C) and CD95+ CD4+ T cell (D) expansion in the peripheral blood of infected individuals is positively associated with the viral load at the early set point of acute HIV-1 infection according to the Pearson’s correlation test.
DISCUSSION
Acute HIV-1 infection is characterized by a massive depletion of memory CD4+ T cells permanently damaging the immune system. It has been demonstrated that within few days after the transmission, HIV-1 latently inscribes into CD4+ T cells. The outcome is a small reservoir of CD4+ T cells harboring latent, replication-competent HIV-1 DNA. These cells persist during suppressive antiretroviral treatment and in case of interrupted treatment cause rapid rebound of viremia. However, CD4+ T cells do not represent a uniform population but can rather be divided into subsets with strikingly different roles in establishment and maintenance of HIV-1 latency. Especially interesting in that manner are SCM that due to their stem-like properties greatly contribute to the persistence HIV-1 reservoir. We show here that soon after the establishment of acute HIV-1 infection an expansion of SCM occurs that is associated with factors of disease progression and possibly serves as a substrate for seeding of the latent reservoir.
Studies conducted in nonhuman primates and humans demonstrated that HIV-1/SIV-1 preferentially infects highly mature subsets, causing a reduction in their frequencies during the acute phase of the disease. At the same time, less differentiated subsets, such as naive cells and CM, are less likely to be infected but can be stimulated to proliferate and expand (3, 25–28). In line with these findings, we found that the frequencies of naive cells, SCM, and CM remain elevated during the late acute HIV-1 infection, whereas those of EM and TE progressively decrease. Importantly, we were able to monitor CD4+ T cell frequencies already during the first few days after the establishment of infection, which allowed us to investigate the complexity of CD4+ T cell memory subset kinetics in peripheral blood of acutely HIV-1-infected individuals in more detail. Our data revealed a profound expansion of SCM. Expansion of SCM occurred simultaneously with the viral load peak, which may be a critical time period for the seeding of HIV-1’s reservoir in SCM (8).
Interestingly, our findings ex vivo were replicable in an infection assay in vitro, and we found that the expansion of SCM subset is due to an upregulation of Fas receptor on naive cells. Previous studies demonstrated increased CD95 density on the surfaces of CD4+ T cells during HIV-1 infection (29, 30), suggesting that CD95 upregulation on CD4+ T cells occurs as a result of increased tumor necrosis factor alpha and gamma interferon secretion by CD4+ T cells, which is in turn caused by HIV-1 envelope-mediated cross-linking of CD4+ molecules (31). However, it is unclear whether these CD95+ CD4+ T cells already encountered cognate antigen and can therefore be considered true antigen-experienced memory cells. This is also true for most of publications investigating SCM during HIV-1/SIV-1 infection using similar methods and will warrant further investigations (16, 23, 32, 33).
To further understand CD4+ T memory cell dynamics, we investigated the degree of HIV-1 replication within the individual subsets and observed substantial differences in HIV-1 infection of memory subsets. Similar to previous studies (3, 34–36), we found the highest levels of HIV-1 replication in EM and TM and the lowest level in naive cells. The main reason for the preferential infection of highly mature CD4+ T cells is, supposedly, different densities of CCR5 coreceptors on the surfaces of these cells (34). It has been demonstrated that the degree of CCR5 expression in each subset increases with the maturation status but slightly declines in TE (31, 34). Remarkably, we observed the exact same pattern of HIV-1 replication and integration in different memory subsets. When exposed to R5-tropic virus, naive cells proved to be the most resistant to infection. The frequency of infected cells gradually increased in SCM, CM, and TE and reached the highest level in TM and EM. Together with the effects of HIV-1 causing expansion of some subsets, a different susceptibility to infection offers a tempting explanation for observed CD4+ T cell subset dynamics, since we observed a slow but persistent decrease in the frequency of more susceptible subsets toward the end of the acute phase of infection. The EM depletion that took place during this phase may also be explained by Fas-mediated cell death since it has been previously demonstrated that EM show higher susceptibility to Fas-mediated apoptosis compared to CM or naive CD4+ T cells (37, 38).
Interestingly, we did not observe an equal expansion of SCM or Fas receptor upregulation in all donors. While the frequency of SCM greatly increased in some individuals, others showed only little SCM expansion. However, SCM expansion was highly associated with the level of productive HIV-1 infection. Furthermore, our clinical findings revealed a positive correlation between the degree of SCM expansion and viral load at the early set point of acute infection. It is surprising how well CD95 not only describes the level of productively infected CD4+ cells but also disease outcome. The mechanism of this association remains unclear but it is plausible that Fas-mediated killing of CD4+ T cells, which is a reason for massive depletion of CD4+ T cells during acute HIV-1 infection, induces apoptosis of preferentially uninfected cells and a mechanism of bystander cell death (34, 39). This will relatively increase the frequency of infected CD4+ T cells.
Indeed, previous studies have shown that the expression of Fas receptor indicates increased susceptibility to apoptosis (40, 41). It has been demonstrated that HIV-1 not only causes the upregulation of Fas receptor expression but also sensitizes CD4+ T cells to undergo Fas/FasL-mediated apoptosis through the action of Tat and Env proteins (42). Taken together, our results provide a detailed look at the dynamics of memory CD4+ T cell subsets during acute HIV-1 infection. Furthermore, we observed a significant expansion of SCM during acute HIV-1 infection which is associated with the level of productively infected CD4+ T cells and disease outcome.
MATERIALS AND METHODS
Study participants.
Twenty-four acute HIV-1-infected participants, identified from the RV217 early-capture HIV-1 cohort (ECHO), were selected for this study. The RV217 study is a multicenter, nonrandomized clinical observational study, designed to describe the biological characteristics of acute HIV-1 infection in high-risk volunteers from Africa and Southeast Asia. The main criteria for selection of the patients was the availability of samples collected before the infection and at minimum two time points after the infection and prior to peak viremia. Further details about the study are already described elsewhere (18).
For our in vitro experiments, PBMCs were obtained from the Blood Donation Center, Institute for Transfusion Medicine, University Hospital, University Duisburg-Essen, Essen, Germany.
Ethics statement.
All individuals participating in this study provided written informed consent. Ethical approval was obtained from institutional review boards in each country, as well as the Human Subjects Protection Branch at the Walter Reed Army Institute of Research, which approved the overall protocol. The local IRB of the University Duisburg-Essen approved the performed laboratory testing and in vitro studies.
Phenotypic analysis of clinical samples.
Cryopreserved PBMCs collected at seven different time points prior to and during the course of acute HIV-1 infection were thawed and used for this study. Cells were stained for viability (Live/Dead Fixable Aqua; Life Technologies) and phenotypic surface markers, including anti-CD4-Qdot605 (clone S3.5), anti-CD8a-PE-TR (clone 3B5), anti-CD14-Tri-Color (clone Tük4), and anti-CD19-Tri-Color (clone SJ25-C1) (all purchased from Life Technologies); anti-CD3-APC-eF780 (clone UCHT1) and anti-CD45R0-eFluor-650NC (clone UCHL1) (all purchased from e-Bioscience); anti-CD27-AF700 (clone O323), anti-CD28-biotin (clone CD28.2), anti-CD45RA-PerCP-Cy5.5 (clone HI100), anti-CD57-FITC (clone HCD57), anti-CD95-AF647 (clone DX2), and anti-CD127-BV421 (clone A019D5) (all purchased from BioLegend); and anti-CD56-PE-Cy5 (clone B159), anti-CCR7-PE-Cy7 (clone 3D12), and anti-CXCR3-PE (clone 1C6/CXCR3) (all purchased from BD Biosciences). Stained cells were then incubated with streptavidin-Qdot 800 conjugate (Life Technologies) and subsequently fixed with a 2% formaldehyde solution. Samples were acquired on BD LSRII (BD Biosciences) and analyzed using FlowJo (v9.7.5; TreeStar).
Virus propagation.
Infectious HIV-1 virus stocks for in vitro studies were generated as follows. Cryopreserved PBMCs from healthy donors were thawed and cultured for 3 days in R10 medium (RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, penicillin [100 U/ml], and streptomycin [100 μg/ml]) containing 1 μg/ml of phytohemagglutinin (PHA). A total of 40 million PHA-stimulated cells were resuspended in 4 ml of R10 medium, followed by the addition of 1 ml of HIV-1 Ba-L containing supernatant (NIH AIDS Reagent Program). Inoculated cell suspension was incubated at 37°C and 5% CO2 for 2 h and gently swirled every 20 min. After a wash with R+ medium (RPMI 1640 supplemented with 2 mM l-glutamine, penicillin [100 U/ml], and streptomycin [100 μg/ml]), the infection cycle was repeated with a fresh vial of viral inoculum. Infected PHA-blasts were then thoroughly washed and resuspended at a density of 2 million cells/ml. The levels of p24 protein in the culture supernatant were monitored every 3 days by enzyme-linked immunoassay (HIV-1 Gag p24 Quantikine ELISA kit; R&D). When the p24 concentration exceeded 50 ng/ml, cell-free supernatant was harvested and stored at –80°C.
HIV-1-infected cell cultures.
A total of 100 million of freshly isolated PBMCs from healthy donors were resuspended in RAB medium (RPMI 1640 supplemented with 10% heat-inactivated human male AB serum, 2 mM l-glutamine, penicillin [100 U/ml], streptomycin [100 μg/ml], and interleukin-2 [50 IU/ml]) and inoculated with the amount of HIV-1 Ba-L virus stock, corresponding to 100 ng of p24. The final volume of inoculated cell suspension was 5 ml. Uninfected controls were treated equally with exception of the infectious inoculum which was substituted by R10 media. After two infection cycles were performed as described in the virus propagation section, the cells were thoroughly washed with R+ medium, resuspended in 50 ml RAB medium, and incubated at 37°C and 5% CO2. Every 5 days, half of the supernatant was aspirated and replaced by fresh RAB medium.
Phenotypic analysis of HIV-1-infected cells.
A sample of 4 million cells was collected from our HIV-1-infected cell cultures, as well as uninfected controls, right before the infection was performed and on days 3, 7, and 14 after the infection. Collected cells were subjected to negative immunomagnetic CD4+ T cell isolation (CD4+ T cell isolation kit, human; Miltenyi Biotec) according to the manufacturer’s instructions. Cells were then washed in a serum-free medium and stained with Live/Dead dye (Zombie Aqua fixable viability kit; BioLegend) to exclude nonviable cells. Next, cells were stained with the following antibodies against surface antigens that allowed for the discrimination between CD4+ T cell memory subsets: anti-CD45RA-BV785 (clone HI100), anti-CCR7-PE-Cy7 (clone G043H7), anti-CD27-BV605 (clone O323), and anti-CD95-APC (clone DX2) (all purchased from BioLegend). Early apoptotic cells were labeled with annexin V-FITC (BioLegend) in a suitable buffer (annexin V binding buffer; BioLegend), washed with the same buffer to prevent dissociation of annexin V, and fixed and permeabilized (Fix/Perm kit; BD Biosciences). HIV-infected cells were detected using anti-p24-PE antibody (clone kc57; Beckman Coulter) and washed with phosphate-buffered saline (PBS). Samples were kept at 4°C throughout the procedure, incubation times for labeling were 15 min with exception of 30 min for p24 staining. All antibodies and other staining reagents were used in pretitrated concentrations. Labeled cells were acquired on BD FACS Celesta flow cytometer (BD Biosciences) and analyzed with FACSDiva software (BD Biosciences). Compensation was performed with single-stained capture beads (CompBeads; BD Biosciences). At each time point, possible fluctuations in laser intensity were checked using multi-fluorescence calibration beads (Rainbow calibration particles; BioLegend), and detection voltages were accordingly adjusted to maintain equivalent fluorescence readings throughout the experiment. Data were analyzed with FlowJo, v9.4.1 (TreeStar).
Proliferation assay.
To measure cellular proliferation, HIV-1-infected cell cultures were established as described above. After two infection cycles, the cells were washed with R+ medium, counted, and resuspended in warm PBS at a cell density of 4 million/ml. For each ml of cell suspension, 5 μl of proliferation dye (CellTrace Violet cell proliferation kit; Thermo Fisher) was added. Samples were incubated at room temperature for 20 min and subsequently washed with five times the staining volume of PBS containing 2% of heat-inactivated fetal calf serum. Labeled cells were finally resuspended in RAB medium and further cultured as already described. Detection of proliferating cells among memory CD4+ T cell subsets was achieved as defined in the previous section, with the exception of the additional fluorescence detection channel where proliferation dye was measured.
Measurement of integrated HIV-1 genomes by qPCR.
HIV-1-infected cell cultures were established as described above. On day 14 after the infection, cells were harvested and subsequently processed as described above, with some modifications. Labeled cells were subjected to fluorescence-activated cell sorting on BD FACSAria III cell sorter with FACSDiva software (BD Biosciences). Compensation was performed with single-stained capture beads (CompBeads; BD Biosciences). A maximum of 1 million cells was sorted for each CD4+ T cell subset.
Genomic DNA from the sorted cells was isolated using a DNA purification kit (QIAamp DNA Micro kit; Qiagen) according to the manufacturer’s protocol. A concentration of eluted DNA was determined spectrophotometrically. To measure the relative amount of integrated HIV-1 genomes, nested PCR was performed. Selective preamplification of HIV-1 genomes integrated into human DNA was achieved by Alu-gag PCR. In this step alu-oligonucleotide hybridizes with Alu repeats in the human genome, whereas the gag oligonucleotide hybridizes with the gag region of HIV-1’s genome, which results in exponential amplification of only integrated HIV-1 DNA. Reactions were carried out in a final volume of 20 μl under following conditions: sample DNA (2 ng/μl), alu oligonucleotide (600 nM), gag oligonucleotide (150 nM), MgCl2 (3 mM), deoxynucleoside triphosphates (dNTPs; 300 μM each), KB extender for long amplicons (5% by volume), 10× PCR buffer (10% by volume), and Platinum Taq DNA polymerase (0.1 U/μl). Thermal program was set to 8 min of initial denaturation at 95°C, followed by 25 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 1 min, elongation at 72°C for 10 min and completed with the final elongation step at 72°C for 15 min.
Real-time quantitative PCR was performed using 10 μl of the preamplification product in a 20-μl reaction with the following composition: LTR forward oligonucleotide (260 nM), LTR reverse oligonucleotide (260 nM), MgCl2 (3.5 mM), dNTPs (300 μM each), LTR TaqMan probe (200 nM), 10× PCR buffer (10% by volume), and Platinum Taq DNA polymerase (0.1 U/μl). Samples were analyzed on qTOWER2 real-time PCR thermal cycler (Analytik Jena), with the program initiated by 1 min of denaturation at 95°C, followed by 50 cycles of denaturation at 95°C for 20 s and annealing/extension at 60°C for 1 min. The fluorescence intensity was scanned at the end of each annealing/extension step.
As a normalization standard β-globin DNA levels were assessed. Reactions were carried out in a final volume of 20 μl, under following conditions: sample DNA (0.4 ng/μl), β-globin forward oligonucleotide (500 nM), β-globin reverse oligonucleotide (500 nM), and innuMIX Green PCR MasterMix (Analytik Jena) at 50% by volume. The thermal program was set to 2 min of initial denaturation at 95°C, followed by 50 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min.
Oligonucleotide and TaqMan probe sequences were as follows: alu (forward oligonucleotide), 5′-GCC TCC CAA AGT GCT GGG ATT ACA G-3′; gag (reverse oligonucleotide), 5′-GTT CCT GCT ATG TCA CTT CC-3′; LTR forward oligonucleotide, 5′-TTA AGC CTC AAT AAA GCT TGC C-3′; LTR reverse oligonucleotide, 5′-GTT CGG GCG CCA CTG CTA GA-3′; LTR TaqMan probe, 5′-(FAM)-CCA GAG TCA CAC AAC AGA CGG GCA CA-(BHQ)-3′; β-globin forward oligonucleotide, 5′-CCC TTG GAC CCA GAG GTT CT-3′; and β-globin reverse oligonucleotide, 5′-TCA TGG CAA GAA AGT GCT CG-3′.
ACKNOWLEDGMENTS
We thank volunteers that participated in the RV217 study. We also thank the RV217 study team: Jeffrey R. Currier, Peter Dawson, Fatim Jallow, Silvia Ratto-Kim, Eugene Kroon, Cornelia Lueer, Jennifer Malia, Mark Manak, Mary A. Marovich, Mark Milazzo, Robert O’Connell, Joseph Oundo, Donald Stablein, Erica Sanga, Somchai Sriplichien, and Rapee Trichavaroj.
The reagent HIV-1 Ba-L was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Suzanne Gartner, Mikulas Popovic, and Robert Gallo.
Financial support for this study was provided by the U.S. Army under cooperative agreement W81XWH-11-2-0174, the National Institute of Allergy and Infectious Diseases, U.S. National Institutes of Health and German Research Association DFG (Deutsche Forschungsgemeinschaft) through the research training group Graduiertenkolleg 1949/2, entitled Immunantwort in Infektionskrankheiten—Regulation Zwischen Angeborener und Erworbener Immunität.
The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army or the Department of Defense.
REFERENCES
- 1.Robb ML, Ananworanich J. 2016. Lessons from acute HIV infection. Curr Opin HIV AIDS 11:555–560. doi: 10.1097/COH.0000000000000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen MS, Shaw GM, McMichael AJ, Haynes BF. 2011. Acute HIV-1 infection. N Engl J Med 364:1943–1954. doi: 10.1056/NEJMra1011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grossman Z, Meier-Schellersheim M, Paul WE, Picker LJ. 2006. Pathogenesis of HIV infection: what the virus spares is as important as what it destroys. Nat Med 12:289–295. doi: 10.1038/nm1380. [DOI] [PubMed] [Google Scholar]
- 4.Mattapallil JJ, Douek DC, Buckler-White A, Montefiori D, Letvin NL, Nabel GJ, Roederer M. 2006. Vaccination preserves CD4 memory T cells during acute simian immunodeficiency virus challenge. J Exp Med 203:1533–1541. doi: 10.1084/jem.20060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams BG, Korenromp EL, Gouws E, Schmid GP, Auvert B, Dye C. 2006. HIV infection, antiretroviral therapy, and CD4+ cell count distributions in African populations. J Infect Dis 194:1450–1458. doi: 10.1086/508206. [DOI] [PubMed] [Google Scholar]
- 6.Holmes CB, Wood R, Badri M, Zilber S, Wang B, Maartens G, Zheng H, Lu Z, Freedberg KA, Losina E. 2006. CD4 decline and incidence of opportunistic infections in Cape Town, South Africa: implications for prophylaxis and treatment. J Acquir Immune Defic Syndr 42:464–469. doi: 10.1097/01.qai.0000225729.79610.b7. [DOI] [PubMed] [Google Scholar]
- 7.Herout S, Mandorfer M, Breitenecker F, Reiberger T, Grabmeier-Pfistershammer K, Rieger A, Aichelburg MC. 2016. Impact of early initiation of antiretroviral therapy in patients with acute HIV infection in Vienna, Austria. PLoS One 11:e0152910. doi: 10.1371/journal.pone.0152910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, Smith JY, Brinkman AL, Peter LE, Mathew SI, Smith KM, Borducchi EN, Rosenbloom DI, Lewis MG, Hattersley J, Li B, Hesselgesser J, Geleziunas R, Robb ML, Kim JH, Michael NL, Barouch DH. 2014. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katz IT, Maughan-Brown B. 2017. Improved life expectancy of people living with HIV: who is left behind? Lancet HIV 4:e324–e326. doi: 10.1016/S2352-3018(17)30086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulpa DA, Chomont N. 2015. HIV persistence in the setting of antiretroviral therapy: when, where and how does HIV hide? J Virus Erad 1:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee GQ, Lichterfeld M. 2016. Diversity of HIV-1 reservoirs in CD4+ T-cell subpopulations. Curr Opin HIV AIDS 11:383–387. doi: 10.1097/COH.0000000000000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E. 2013. The who’s who of T-cell differentiation: human memory T-cell subsets. Eur J Immunol 43:2797–2809. doi: 10.1002/eji.201343751. [DOI] [PubMed] [Google Scholar]
- 13.Geginat J, Paroni M, Maglie S, Alfen JS, Kastirr I, Gruarin P, De Simone M, Pagani M, Abrignani S. 2014. Plasticity of human CD4 T cell subsets. Front Immunol 5:630. doi: 10.3389/fimmu.2014.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol 78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM. 2014. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol 88:14070–14077. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. 2014. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med 20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robb ML, Eller LA, Kibuuka H, Rono K, Maganga L, Nitayaphan S, Kroon E, Sawe FK, Sinei S, Sriplienchan S, Jagodzinski LL, Malia J, Manak M, de Souza MS, Tovanabutra S, Sanders-Buell E, Rolland M, Dorsey-Spitz J, Eller MA, Milazzo M, Li Q, Lewandowski A, Wu H, Swann E, O’Connell RJ, Peel S, Dawson P, Kim JH, Michael NL, Team R. 2016. Prospective study of acute HIV-1 infection in adults in East Africa and Thailand. N Engl J Med 374:2120–2130. doi: 10.1056/NEJMoa1508952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eller MA, Goonetilleke N, Tassaneetrithep B, Eller LA, Costanzo MC, Johnson S, Betts MR, Krebs SJ, Slike BM, Nitayaphan S, Rono K, Tovanabutra S, Maganga L, Kibuuka H, Jagodzinski L, Peel S, Rolland M, Marovich MA, Kim JH, Michael NL, Robb ML, Streeck H. 2016. Expansion of inefficient HIV-specific CD8 T cells during acute infection. J Virol 90:4005–4016. doi: 10.1128/JVI.02785-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. 1995. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labeled annexin V. J Immunol Methods 184:39–51. doi: 10.1016/0022-1759(95)00072-I. [DOI] [PubMed] [Google Scholar]
- 20.Holl V, Schmidt S, Aubertin AM, Moog C. 2007. The major population of PHA-stimulated PBMC infected by R5 or X4 HIV variants after a single cycle of infection is predominantly composed of CD45RO+ CD4+ T lymphocytes. Arch Virol 152:507–518. doi: 10.1007/s00705-006-0873-1. [DOI] [PubMed] [Google Scholar]
- 21.Schnittman SM, Lane HC, Greenhouse J, Justement JS, Baseler M, Fauci AS. 1990. Preferential infection of CD4+ memory T cells by human immunodeficiency virus type I: evidence for a role in the selective T-cell functional defects observed in infected individuals. Proc Natl Acad Sci U S A 87:6058–6062. doi: 10.1073/pnas.87.16.6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groot F, van Capel TM, Schuitemaker J, Berkhout B, de Jong EC. 2006. Differential susceptibility of naive, central memory and effector memory T cells to dendritic cell-mediated HIV-1 transmission. Retrovirology 3:52. doi: 10.1186/1742-4690-3-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabler CO, Lucera MB, Haqqani AA, McDonald DJ, Migueles SA, Connors M, Tilton JC. 2014. CD4+ memory stem cells are infected by HIV-1 in a manner regulated in part by SAMHD1 expression. J Virol 88:4976–4986. doi: 10.1128/JVI.00324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liszewski MK, Yu JJ, O’Doherty U. 2009. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 47:254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okoye A, Meier-Schellersheim M, Brenchley JM, Hagen SI, Walker JM, Rohankhedkar M, Lum R, Edgar JB, Planer SL, Legasse A, Sylwester AW, Piatak M Jr, Lifson JD, Maino VC, Sodora DL, Douek DC, Axthelm MK, Grossman Z, Picker LJ. 2007. Progressive CD4+ central memory T cell decline results in CD4+ effector memory insufficiency and overt disease in chronic SIV infection. J Exp Med 204:2171–2185. doi: 10.1084/jem.20070567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Jiao YM, Wang R, Ji YX, Zhang HW, Zhang YH, Chen DX, Zhang T, Wu H. 2012. High CCR5 density on central memory CD4+ T cells in acute HIV-1 infection is mostly associated with rapid disease progression. PLoS One 7:e49526. doi: 10.1371/journal.pone.0049526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, Vinton C, Gordon SN, Bosinger SE, Francella N, Hallberg PL, Cramer E, Schlub T, Chan ML, Riddick NE, Collman RG, Apetrei C, Pandrea I, Else J, Munch J, Kirchhoff F, Davenport MP, Brenchley JM, Silvestri G. 2011. Low levels of SIV infection in sooty mangabey central memory CD(4)+ T cells are associated with limited CCR5 expression. Nat Med 17:830–836. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pastor L, Urrea V, Carrillo J, Parker E, Fuente-Soro L, Jairoce C, Mandomando I, Naniche D, Blanco J. 2017. Dynamics of CD4 and CD8 T-cell subsets and inflammatory biomarkers during early and chronic HIV infection in Mozambican adults. Front Immunol 8:1925. doi: 10.3389/fimmu.2017.01925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aries SP, Schaaf B, Muller C, Dennin RH, Dalhoff K. 1995. Fas (CD95) expression on CD4+ T cells from HIV-infected patients increases with disease progression. J Mol Med (Berl) 73:591–593. [DOI] [PubMed] [Google Scholar]
- 30.Bohler T, Wintergerst U, Linde R, Belohradsky BH, Debatin KM. 2001. CD95 (APO-1/Fas) expression on naive CD4+ T cells increases with disease progression in HIV-infected children and adolescents: effect of highly active antiretroviral therapy (HAART). Pediatr Res 49:101–110. doi: 10.1203/00006450-200101000-00021. [DOI] [PubMed] [Google Scholar]
- 31.Oyaizu N, McCloskey TW, Than S, Hu R, Kalyanaraman VS, Pahwa S. 1994. Cross-linking of CD4 molecules upregulates Fas antigen expression in lymphocytes by inducing interferon-gamma and tumor necrosis factor-alpha secretion. Blood 84:2622–2631. [PubMed] [Google Scholar]
- 32.Manganaro L, Hong P, Hernandez MM, Argyle D, Mulder LCF, Potla U, Diaz-Griffero F, Lee B, Fernandez-Sesma A, Simon V. 2018. IL-15 regulates susceptibility of CD4+ T cells to HIV infection. Proc Natl Acad Sci U S A 115:E9659–E9667. doi: 10.1073/pnas.1806695115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cartwright EK, McGary CS, Cervasi B, Micci L, Lawson B, Elliott ST, Collman RG, Bosinger SE, Paiardini M, Vanderford TH, Chahroudi A, Silvestri G. 2014. Divergent CD4+ T memory stem cell dynamics in pathogenic and nonpathogenic simian immunodeficiency virus infections. J Immunol 192:4666–4673. doi: 10.4049/jimmunol.1303193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okoye AA, Picker LJ. 2013. CD4+ T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev 254:54–64. doi: 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khoury G, Rajasuriar R, Cameron PU, Lewin SR. 2011. The role of naive T-cells in HIV-1 pathogenesis: an emerging key player. Clin Immunol 141:253–267. doi: 10.1016/j.clim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Veazey RS, Tham IC, Mansfield KG, DeMaria M, Forand AE, Shvetz DE, Chalifoux LV, Sehgal PK, Lackner AA. 2000. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4+ T cells are rapidly eliminated in early SIV infection in vivo. J Virol 74:57–64. doi: 10.1128/JVI.74.1.57-64.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramaswamy M, Cruz AC, Cleland SY, Deng M, Price S, Rao VK, Siegel RM. 2011. Specific elimination of effector memory CD4+ T cells due to enhanced Fas signaling complex formation and association with lipid raft microdomains. Cell Death Differ 18:712–720. doi: 10.1038/cdd.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riou C, Yassine-Diab B, Van Grevenynghe J, Somogyi R, Greller LD, Gagnon D, Gimmig S, Wilkinson P, Shi Y, Cameron MJ, Campos-Gonzalez R, Balderas RS, Kelvin D, Sekaly RP, Haddad EK. 2007. Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. J Exp Med 204:79–91. doi: 10.1084/jem.20061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poonia B, Pauza CD, Salvato MS. 2009. Role of the Fas/FasL pathway in HIV or SIV disease. Retrovirology 6:91. doi: 10.1186/1742-4690-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bohler T, Nedel S, Debatin KM. 1997. CD95-induced apoptosis contributes to loss of primed/memory but not resting/naive T cells in children infected with human immunodeficiency virus type 1. Pediatr Res 41:878–885. doi: 10.1203/00006450-199706000-00013. [DOI] [PubMed] [Google Scholar]
- 41.Sieg S, Huang Y, Kaplan D. 1997. Viral regulation of CD95 expression and apoptosis in T lymphocytes. J Immunol 159:1192–1199. [PubMed] [Google Scholar]
- 42.Westendorp MO, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debatin KM, Krammer PH. 1995. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature 375:497–500. doi: 10.1038/375497a0. [DOI] [PubMed] [Google Scholar]