Abstract
The alarmone (p)ppGpp mediates the stringent response and has a recognized role in bacterial virulence. We previously reported a stringent response-like state in Enterococcus faecalis isolated from a rabbit foreign body abscess model and showed that E. faecalis mutants with varying levels of cellular (p)ppGpp [Δrel, ΔrelQ and the (p)ppGpp0 ΔrelΔrelQ] had differential abilities to persist within abscesses. In this study, we investigated whether (p)ppGpp contributes to the pathogenesis of E. faecalis infective endocarditis (IE), a biofilm infection of the heart valves. While the stringent response was not activated in heart valve-associated E. faecalis, deletion of the gene encoding the bifunctional (p)ppGpp synthetase/hydrolase Rel significantly impaired valve colonization. These results indicate that the presence of (p)ppGpp is dispensable for E. faecalis to cause IE, whereas the ability to regulate (p)ppGpp levels is critical for valve colonization. Next, we characterized how basal (p)ppGpp levels affect processes associated with IE pathogenesis. Despite being defective in binding to BSA-coated polystyrene surfaces, the Δrel strain bound to collagen- and fibronectin-coated surfaces and ex vivo porcine heart valves as well as the parent and ΔrelΔrelQ strains, ruling out the possibility that the impaired IE phenotype was due to an attachment defect. Moreover, differences in cellular (p)ppGpp levels did not affect extracellular gelatinase activity but significantly impaired enterococcal invasion of human coronary artery endothelial cells. Taken together, this study uncovers for the first time the fact that differences in basal (p)ppGpp levels, rather than the stringent response, differentially affect processes that contribute to the pathogenesis of IE.
Keywords: vegetation, stringent response, heart valve, biofilm, Gram positive bacterial infection
Introduction
Bacterial infective endocarditis (IE) is a serious infection of the cardiac endothelium that is associated with an annual incidence of 7.7 cases per 100 000 persons and a mortality rate of approximately 25% [1, 2]. IE is most often characterized by the formation of a vegetation on the surface of a damaged heart valve. Valvular injury leads to the exposure of basement membrane matrix components, such as collagen, which serve as signals for subsequent platelet deposition at the site of damage [3]. Vegetation formation proceeds with the activation of fibrin, the production of fibronectin and the eventual binding of blood-borne bacteria to the accumulated host molecules [3]. Vegetations enlarge as the attached microorganisms replicate, and additional platelets, circulating immune cells and erythrocytes become entrapped in the fibrin meshwork. The complex amalgamation of bacteria and host factors that comprise a vegetation has been described as a biofilm [4] and protects the bacteria from clearance by phagocytosis [5]. IE is frequently associated with life-threatening sequelae, such as congestive heart failure and embolization [3]. The treatment regimens for IE are usually lengthy, often involve multiple antibiotics, and may require surgery [6].
Enterococcus species account for approximately 30% of bacterial IE cases worldwide [7, 8], with Enterococcus faecalis causing up to 90% of these infections [8]. Experimental models of IE and in vitro studies have identified several cell surface and secreted factors that contribute to E. faecalis IE pathogenesis, including the processes of binding to eukaryotic matrix proteins [9–12], interacting with platelets [13, 14] and facilitating embolization [15, 16]. Specifically, the endocarditis- and biofilm-associated pili (Ebp), the collagen-binding protein Ace and the fibronectin-binding protein EfbA are cell wall-anchored proteins that have been shown to contribute to virulence in rat models of IE [12, 17, 18]. In addition, gelatinase (GelE), a secreted metalloprotease controlled by the Fsr quorum sensing system [19], was shown to enable bacterial dissemination from the heart valve to other tissues in a rabbit IE model [15].
In addition to causing IE, E. faecalis is a leading cause of healthcare-associated infections, including central line-associated bloodstream infection, catheter-associated urinary tract infection and surgical site infection [20]. The expression of many E. faecalis stress response phenotypes, including tolerance to antibiotics [21, 22], biofilm formation [23] and metal homeostasis [24], has been associated with the molecular alarmones guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), collectively known as (p)ppGpp. Rapid accumulation of (p)ppGpp activates the stringent response, a conserved stress response that results from nutrient deprivation and other conditions that are unfavourable for growth [25]. During a stringent response, (p)ppGpp broadly orchestrates alterations in transcription, translation and replication to promote cell survival. Over the past decade, it has been demonstrated that basal (p)ppGpp levels, which are well below those required to activate the stringent response, can influence cellular homeostasis and virulence in E. faecalis and several other bacterial species [22, 26, 27].
(p)ppGpp is synthesized in E. faecalis by the bifunctional synthetase/hydrolase Rel and the small alarmone synthetase RelQ [21, 28]. The Rel enzyme is essential for stringent response activation in E. faecalis [21]. Basal levels of (p)ppGpp are increased in a Δrel mutant, and this is associated with a slow growth phenotype in rich medium and growth delays when challenged with osmotic shock, low pH and hydrogen peroxide [21, 22]. RelQ is primarily important for homeostatic growth [21, 22, 29]. A ΔrelQ mutant has normal basal (p)ppGpp levels and delayed onset of the stringent response [21, 22].
We reported previously that a transcriptional programme resembling a classic stringent response (i.e. down-regulation of rRNA genes and activation of stress survival genes) is activated in E. faecalis OG1RF during the early hours of infection in a rabbit subcutaneous foreign body abscess model [30]. In the same model, the Δrel mutant was significantly more resistant to initial clearance, while the ΔrelQ mutant and an E. faecalis strain that completely lacks (p)ppGpp (ΔrelΔrelQ mutant) displayed persistence defects [30]. These observations provided the first evidence that (p)ppGpp is associated with E. faecalis infection in a mammalian host [30]. We also showed that the ΔrelΔrelQ mutant has a reduced ability to survive in mouse macrophages and is impaired for growth and survival in human serum and whole blood ex vivo [29, 30]. In contrast, the survival of the Δrel and ΔrelQ strains in human serum and whole blood was similar to that of the wild-type strain [30]. These observations, combined with the prominence of enterococci as IE pathogens, led us to investigate the role of (p)ppGpp in the pathogenesis of E. faecalis IE in this work.
Methods
Bacterial strains, growth conditions and oligonucleotides
E. faecalis OG1RF and the (p)ppGpp mutant strains OG1RFΔrel (called OG1RFΔrelA or OG1RFΔrsh in previous publications), OG1RFΔrelQ and OG1RFΔrelΔrelQ were used in this study [21, 31]. E. faecalis strains were routinely cultivated in brain heart infusion broth (BHI; BD Bacto, Becton, Dickinson and Company, Sparks, MD, USA) under static conditions or on BHI agar at 37°C in ambient air. Cultures for rabbit IE and porcine heart valve attachment experiments were grown in trypsinized beef heart dialysate (BH) medium [32] under static conditions at 37°C with 5–7% CO2. Oligonucleotides were purchased from Life Technologies (Carlsbad, CA, USA), Integrated DNA Technologies (Coralville, IA, USA) and Eurofins Genomics (Louisville, KY, USA). The oligonucleotide sequences are listed in Table 1.
Table 1. List of oligonucleotides used in this study (written 5′ to 3′).
| Oligonucleotide | Sequence | Source |
|---|---|---|
| 5-EF0886RT | CAAGGAAGCGCCAATAGTGT | [30] |
| 3-EF0886RT | CCCTGTATCAGCCAAACCAT | [30] |
| sigA/rpoDF-RT | AAGACCAAGATGCTACCAGTCC | This study |
| sigA/rpoDR-RT | GTACGTGTCCGACCATCATCTA | This study |
| tsfF-RT | CGCTACTTCTTCGTCAGTTGTG | This study |
| tsfR-RT | CAGCTTCCGTCGTTTTGAAG | This study |
| relA-F | CAAGATTTACGGGTCATTATGG | [30] |
| relA-R | GACTAATCCCTAAGCGATGTG | [30] |
| relQ-F | GACGGCTATTCGGCATATTCC | [30] |
| relQ-R | AAGTGCGACTACCTGGTAAATG | [30] |
| ebpA qRT-PCR forward | CAACAACACCAGGGCTTTTTG | [57] |
| ebpA qRT-PCR reverse | ACCGGACCAGTCAACGACTAAG | [57] |
| AceQF1 | GGAGAGTCAAATCAAGTACGTTGGTT | [11] |
| AceQR1 | TGTTGACCACTTCCTTGTCGAT | [11] |
| efbA RT-F | GCTCCATTGGGTTTATGCAC | This study |
| efbA RT-R | ACTGAAGCCGCACTTCTAGC | This study |
| gelE forward | CTTTTTGGGATGGAAAAGCA | [35] |
| gelE reverse | CCGGCAGTATGTTCCGTTAC | [35] |
| ace ORF F | ATGACAAAAAGTGTAAAATTTTTA | This study |
| ace ORF R | TTAATTCTTTCTGATTTGTAGAT | This study |
| efbA ORF F | ATGTCATTTGATGGCGTATTT | This study |
| efbA ORF R | TTAGGAAGTAGAAGCCTTATT | This study |
Rabbit model of experimental infective endocarditis
The animal experiments were approved by The University of Minnesota Institutional Animal Care and Use Committee (protocol number 0910A73332). The IE infections were carried out as described previously [33, 34]. The inocula were prepared as described [35] and contained 3×108–1.3×109 c.f.u. ml−1 for OG1RF and 8×108–3×109 c.f.u. ml−1 for the (p)ppGpp mutant strains. Two millilitres of each prepared inoculum was injected into the marginal ear vein of each rabbit.
Six rabbits per strain were infected with the OG1RFΔrel, OG1RFΔrelQ and OG1RFΔrelΔrelQ strains. Rabbits were euthanized on the fourth day after infection with the following exceptions: (1) one animal infected with the ΔrelQ strain was euthanized due to moribundity on the third day post-infection according to the provisions of the approved protocol, and (2) one animal infected with the OG1RFΔrelΔrelQ strain was found dead on the morning of the fourth day post-infection. Following death or euthanasia, the hearts were removed and dissected to expose the aortic valve. Vegetations, valve leaflets and attached blood clots were harvested, weighed and homogenized in 1 ml of Todd–Hewitt broth (THB) or potassium phosphate-buffered saline (KPBS). The homogenates were serially diluted and plated on BHI agar to quantify bacteria. The valve bacterial load was defined as the log10-transformed number of c.f.u. recovered from each aortic valve homogenate.
Sixteen rabbits were infected with the OG1RF strain. Five rabbits were euthanized on the second day after infection and a sixth rabbit succumbed to infection within 3 hours prior to the scheduled endpoint at 2 days. The remaining rabbits were euthanized on the fourth day after infection with the following exceptions: (1) one animal was found dead on the morning of the third day after infection, and (2) one animal was found dead on the morning of the fourth day post-infection. Aortic valve tissues were collected and homogenized as described above for the (p)ppGpp mutants. An aliquot (0.05–0.075 ml) of each homogenate was used to quantify bacteria for determination of the valve bacterial load as described above. The remainder of each homogenate was added to two volumes of RNAprotect Bacteria Reagent (Qiagen, Inc., Valencia, CA, USA) and processed according to the manufacturer’s instructions. Pellets were flash frozen in a dry ice-chilled ethanol bath and stored at −80°C until RNA extraction. Aliquots of OG1RF grown as described above for endocarditis infection inoculum were similarly prepared for RNA extraction.
Measurement of gene expression in aortic valve homogenates
RNA was extracted from valve homogenates exactly as described previously for rabbit subdermal aspirates [30]. The same procedure was used for inoculum-condition OG1RF pellets, with the exception that each sample was not split into two duplicate extractions. Contaminating DNA was removed using a TURBO DNA-free kit (Life Technologies) following the manufacturer’s protocol for rigorous DNA removal. DNase-treated total RNA was reverse-transcribed with random primers using the SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies). qPCR was carried out on an iCycler equipped with an iQ5 real-time detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with iQ SYBR Green Supermix (Bio-Rad Laboratories, Inc.) or a CFX96 Touch Real-Time PCR detection system (Bio-Rad Laboratories, Inc.) with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Inc.). DNase-treated RNA was used as a template in control reactions to confirm the removal of DNA. EF0886 was used as a reference gene for normalization as previously described [30]. Each reaction was performed in triplicate and the average C t value of the technical replicates was used in calculations. The fold change for each sample was calculated using the Pfaffl method [36]. Insufficient starting amounts of RNA from three valve homogenates precluded their use in the final analysis.
Expression of ace and efbA in planktonic and biofilm cells
Three colonies each of OG1RF, Δrel and ΔrelΔrelQ were inoculated into 2 ml BHI and incubated overnight. The cultures were diluted 1 : 100 into tryptic soy broth without added dextrose (Becton, Dickinson and Company) containing 40% (v/v) horse serum (Remel, Thermo Fisher Scientific, Waltham, MA, USA) to induce the expression of ace and efbA as shown by others [11, 12]. Three millilitre aliquots were placed into six-well dishes (Costar 3516; Corning, Inc., Corning, NY, USA) that were shaken at 120 r.p.m. for 6.7 h at 37°C. After incubation, planktonic cells were pooled and aliquots were collected for quantitative culture or treated with RNAprotect for subsequent RNA extraction. Biofilms were washed gently twice with 1 ml KPBS and then manually removed with a cell scraper into 1 ml KPBS, which was transferred to each successive well in order to pool all of the biofilms that were grown in a single dish. Six wells were pooled for each strain per biological replicate. Due to loss of volume during transfer, the final volume of pooled cells was adjusted to 1 ml with KPBS. A 100 µl aliquot was used for quantitative cultures and the remainder was added to RNAprotect. Three biological replicates were collected on separate days.
RNA extraction, DNase-treatment and cDNA generation were performed as previously described [35]. qPCR reactions were carried out with undiluted cDNA on a CFX96 Touch Real-Time PCR detection system with SsoAdvanced Universal SYBR Green Supermix. The primers used in qPCR and for generating copy number standards are listed in Table 1. Absolute quantification of transcript copy number and preparation of the standard curves were carried out as previously described [35]. DNase-treated RNA was used as a template in control reactions to confirm the removal of DNA. Each reaction was performed in triplicate and the average copy number per µl of the technical replicates was used in calculations. The copy number per µl was divided by the corresponding number of c.f.u. from which RNA was extracted.
E. faecalis attachment assays on coated polystyrene surfaces
Ninety-six-well microtitre plates (Costar, Corning, Inc.) containing bovine serum albumin (Thermo Fisher Scientific), 1% rat tail collagen type-1 (Sigma-Aldrich, St Louis, MO, USA), or 20 µg ml−1 bovine fibronectin (Sigma) were incubated overnight at 4°C. Alternatively, wells were coated with 100 µl PBS (pH 7) as a control. Prior to use, the wells were washed three times with PBS and blocked with 5% bovine serum albumin for 90 min at 37°C. Unbound protein was removed by washing with PBS plus 0.01% Tween.
E. faecalis OG1RF, Δrel and ΔrelΔrelQ were grown overnight at 37°C in BHI supplemented with 40% horse serum (Lonza, Walkersville, MD, USA). Overnight cultures were washed once and resuspended in PBS to one-quarter of the original volume. Then, 100 µl aliquots of each concentrated cell suspension were added to at least three wells of the prepared microtitre plate and incubated at 37°C for 2 h. Quantitative cultures were also performed to enumerate the number of viable cells used. Following incubation, wells were washed three times with PBST and fixed by incubation at 55°C for at least 30 min. Bound cells were stained with 200 µl of 0.05 % crystal violet solution for 1 min at room temperature and then washed five times with PBS to remove excess stain. Bound cells in stained wells were reconstituted by vigorous pipetting in 200 µl of 7% acetic acid. Final optical density measurements at 575 nm (OD575) were obtained using a plate reader. The OD575 values were normalized to viable cell counts for each strain. Seven to eight biological replicates were collected in a total of three experiments.
Ex vivo porcine heart valve attachment assays
Porcine valve tissue sections that remained following the completion of a previous study [37] were kindly provided by Dr Anne-Marie Leuck (University of Minnesota). Valve sections that had been recovered, prepared and frozen for that study as previously described [37] were used in these experiments. Three colonies each of E. faecalis OG1RF, Δrel and ΔrelΔrelQ were inoculated into BH as described above for rabbit IE experiment inocula and incubated for approximately 19 h. Bacterial cells and valve sections were prepared exactly as described previously [37]. Bacteria were incubated with valve sections for 2 h at 37°C with 5–7% CO2 and gentle shaking. Following incubation, planktonic cells were collected for enumeration by plating of serial dilutions on BHI. The valve sections were washed in KPBS as described [37] and then placed in new tubes containing KPBS for processing to remove adherent bacteria. The tubes were vortexed for 10 s, sonicated in a bath sonicator for 3 min and treated with a motorized pestle for 1 min. Serial dilutions of the detached cells were plated on BHI for enumeration. The percentage of valve binding was calculated as follows: (c.f.u./ml recovered from heart valves)/(c.f.u./ml recovered from the planktonic phase culture)×100. Three biological replicates were performed on separate days.
E. faecalis invasion of human coronary artery endothelial cells (HCAECs)
The ability of E. faecalis OG1RF, Δrel and ΔrelΔrelQ to invade primary HCAECs was assessed as previously described [38, 39]. Briefly, 24-well tissue culture plates containing monolayers of HCAECs were incubated with ~5×107 c.f.u. in 1 ml of endothelial basal medium supplemented with 5% foetal bovine serum (EBM-2) to achieve a multiplicity of infection (m.o.i.) of 100 : 1. After 2 h, the wells were washed gently three times with prewarmed PBS and incubated for 3 h in EBM-2 containing 350 µg ml−1 gentamicin, 50 µg ml−1 penicillin G and 100 µg ml−1 vancomycin to kill extracellular bacteria. The HCAECs were lysed with 1 ml of sterile water. The released bacteria were serially diluted and plated onto tryptic soy agar plates to determine the number of intracellular bacteria. The percentage of invasion was calculated based on the total c.f.u. of the initial inoculum and the number of intracellular bacteria recovered from HCAEC lysates. Six biological replicates were collected in two independent experiments.
Assessment of relative gelatinase production following growth in serum
Gelatin protease assay plates were prepared as described [40]. Wells were created in the solidified agar by using the wide end of a sterile P200 pipette tip to punch out agar plugs. Gelatinase was assessed in the supernatants of E. faecalis OG1RF, Δrel and ΔrelΔrelQ cultures that were grown overnight in BHI supplemented with 40% horse serum at 37°C. The cells were pelleted, and 35 µl of each cleared supernatant was pipetted into an agar well. The plates were incubated upright at 37°C overnight. Following incubation, the plates were photographed, and the area of each zone of clearance was determined with ImageJ [41]. The zone of clearance areas for Δrel and ΔrelΔrelQ were calculated relative to that of OG1RF, which was set to 100% in each biological replicate. Four biological replicates were performed on separate days.
Statistical analyses
Statistical calculations were performed in GraphPad Prism version 7.03 (GraphPad Software, Inc., La Jolla, CA, USA). The BSA-coated polystyrene, collagen and fibronectin attachment assays were analysed for outliers using ROUT analysis [42] with Q=0.1% in GraphPad Prism; one biological replicate in the Δrel BSA-coated polystyrene attachment data set was deemed to be an outlier and was removed from the final analysis.
Ordinary one-way analysis of variance (ANOVA) with post hoc testing, as specified below, was performed to test for statistically significant differences between means in datasets as specified. A P-value <0.05 was considered statistically significant in all analyses. To determine the statistical significance in aortic valve homogenate gene expression experiments, the geometric mean and 95% confidence interval were calculated for each dataset. Confidence intervals that do not cross y=100 (representing no change in transcription between the inoculum control and the aortic valve homogenate sample) are significantly different (P<0.05) from the inoculum control.
Results
An E. faecalis rel deletion strain shows impaired valve colonization in an experimental model of IE
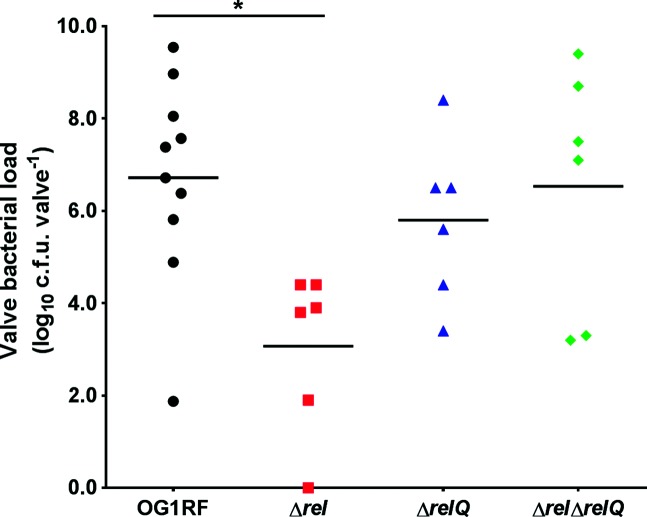
The E. faecalis OG1RF Δrel mutant is unable to activate the stringent response but has constitutively high levels of (p)ppGpp due to the presence of the (p)ppGpp synthetase RelQ [22]. Our previous work showed that the Δrel strain was more resistant to clearance at early time points in a rabbit subcutaneous foreign body abscess model, yet persisted similarly to the wild-type strain at later time points [30]. In contrast, an OG1RF relQ in-frame deletion mutant (ΔrelQ), which is able to activate a delayed stringent response [22], and an OG1RF (p)ppGpp0 strain (ΔrelΔrelQ), which is unable to synthesize (p)ppGpp and mount a stringent response [21, 22], persisted at lower levels than the wild-type strain in the later part of infection [30]. In this study, we evaluated the same strains for virulence in a rabbit model of IE. Virulence was measured as the ability of a strain to colonize damaged aortic valves up to 4 days after injection into the bloodstream. One rabbit in each of the groups infected with OG1RF, ΔrelQ and ΔrelΔrelQ succumbed to infection prior to the scheduled endpoint. The mean valve bacterial loads for OG1RF, ΔrelQ and ΔrelΔrelQ were 6.7, 5.8 and 6.5 log10 c.f.u./valve, respectively, which were not significantly different (Fig. 1). By comparison, all rabbits infected with the Δrel strain lived until the scheduled endpoint, and the mean valve bacterial load, 3.1 log10 c.f.u./valve, was significantly reduced relative to OG1RF (Fig. 1; P=0.0151, Tukey’s multiple comparison post hoc test). Since neither the Δrel nor the ΔrelΔrelQ strain can activate the stringent response [21], but only the Δrel strain is attenuated in this model, these results suggest that activation of the stringent response is not required for E. faecalis IE virulence. Unexpectedly, the complete absence of basal (p)ppGpp levels in the ΔrelΔrelQ strain does not impair colonization of heart tissues. Instead, increased basal levels of (p)ppGpp in the Δrel mutant are associated with a significant defect in aortic valve colonization following valvular injury.
Fig. 1.
The ∆rel strain is impaired in valve colonization in an experimental model of E. faecalis infective endocarditis. E. faecalis OG1RF and the isogenic (p)ppGpp mutant strains ∆rel, ∆relQ and ∆rel∆relQ were tested in a rabbit model of experimental infective endocarditis. Each symbol represents the aortic valve bacterial load in each animal at the experiment endpoint. The horizontal bars show the arithmetic mean of the log10-transformed values. Ordinary one-way ANOVA was used to determine whether the difference among the means was statistically significant (P=0.0180). *, P=0.0151 by Tukey’s multiple comparison post hoc test.
Valve-associated E. faecalis OG1RF cells are not in a stringent response-like state in the rabbit IE model
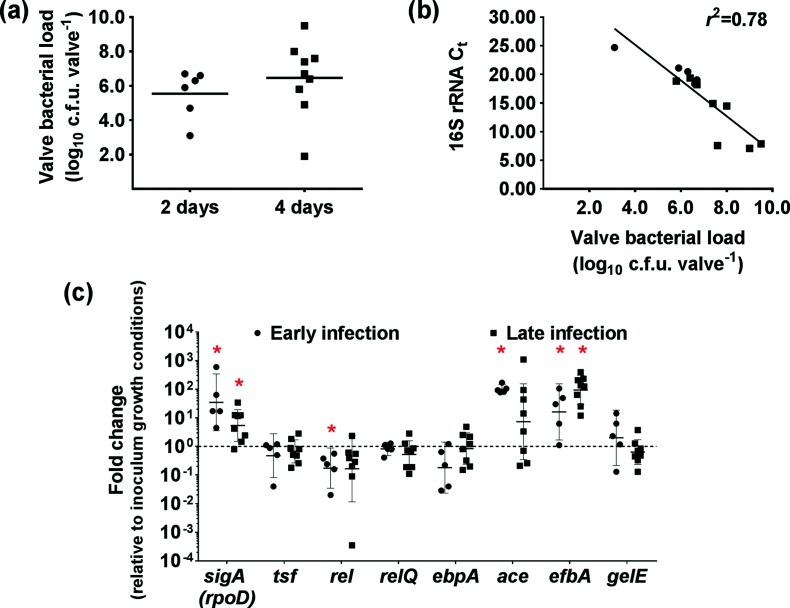
We previously found that OG1RF enters a stringent response-like state within the first 8 hours of infection in a rabbit subcutaneous foreign body abscess model [30]. The disparate IE virulence phenotypes observed between the Δrel and ΔrelΔrelQ strains (Fig. 1), neither of which can activate a stringent response [21], suggest that activation of the stringent response may not occur during aortic valve colonization in the rabbit IE model. Since the stringent response typically halts growth, we enumerated the bacteria recovered from heart valve homogenates at either 2 or 4 days post-infection. The mean valve bacterial load increased by 1.1 log10 c.f.u./valve between the two time points (Fig. 2a). While this increase was not statistically significant, it is suggestive of bacterial replication taking place at the aortic valve between the two points in IE progression. In E. coli, high levels of ppGpp are associated with reduced rRNA synthesis [43, 44]. Therefore, we reasoned that 16S rRNA transcript abundance in RNA samples collected from aortic valve homogenates of OG1RF-infected rabbits would correlate with valve bacterial loads if the cells were in an actively growing state. There was indeed a linear correlation between valve bacterial loads and 16S rRNA transcript abundance, as represented by the C t values generated from the qPCR analysis (r 2=0.78; Fig. 2b).
Fig. 2.
The stringent response is not activated in vivo in heart valve-associated E. faecalis cells. (a) Each symbol represents the aortic valve bacterial load in an animal from which a heart was collected at 2 or 4 days post-infection. The mean valve bacterial load (horizontal bar) increased from 5.6 log10 c.f.u./valve at 2 days to 6.7 log10 c.f.u./valve at 4 days, suggesting active bacterial replication at the aortic valve. (b) RNA was extracted from aortic valve homogenates collected from rabbits during early infection (2 days) or late infection (3 or 4 days; see the Methods section). RT-qPCR was used to measure the abundance of 16S rRNA transcripts in DNase-treated RNA from each valve homogenate. 16S rRNA transcript abundance, as represented by the C t value, correlates with valve bacterial load (r 2=0.78), further suggesting that E. faecalis cells at the aortic valve are in an active growth state. Circles, early infection (n=5). Squares, late infection (n=8). (c) RT-qPCR was used to measure the relative expression of the genes encoding the E. faecalis vegetative sigma factor RpoD (sigA), the translation elongation factor Ts (tsf), the (p)pGpp synthesis proteins (rel and relQ) and several known IE virulence factors (ebpA, ace, efbA and gelE) in early and late infection aortic valve homogenates relative to IE inoculum growth conditions. EF0886 was used as the reference gene. The horizontal bars and error bars represent the geometric means and 95% confidence intervals, respectively. Confidence intervals that do not cross y=100 (which represents no fold change in expression) are significantly different from the inoculum control (*, P<0.05). Transcription of sigA and tsf would be expected to be down-regulated if E. faecalis were in a stringent response state.
The genes encoding the E. faecalis vegetative sigma factor RpoD (rpoD; also called sigA) and the translation elongation factor Ts (tsf) are down-regulated in E. coli during the stringent response [45], and both were strongly down-regulated in E. faecalis cells in the abscess environment when the stringent response-like state was activated [30]. We compared the expression of sigA (rpoD), tsf and the (p)ppGpp synthetase-encoding genes, rel and relQ, in E. faecalis OG1RF cells from the IE inoculum and aortic valve homogenates (Fig. 2c). The transcription of sigA (rpoD) was significantly up-regulated in the early (2 days) and late (3–4 days) valve homogenates, which is consistent with cells in an actively growing state. There was no change in the average expression of tsf at either time point. The transcription of rel was only significantly down-regulated in the early valve homogenates, whereas relQ transcription was unchanged at both time points sampled.
Finally, we used the OG1RF early and late infection valve homogenate RNA to measure the change in in vivo transcriptional activity, relative to IE inoculum conditions, of three adhesin-encoding genes (ebpA, ace and efbA) and the gelatinase-encoding gene, gelE, which all have documented roles in E. faecalis IE virulence (Fig. 2c). Transcription of the gene encoding the Ebp pili tip protein, EbpA, which was chosen as a representative gene for ebp pili locus expression, was unchanged from the inoculum conditions in both early (2 days) and late (3–4 days) infection time points. On the other hand, the transcription of ace was highly up-regulated (P<0.05) in early infection homogenates, and efbA was strongly up-regulated (P<0.05) in the valve homogenates evaluated from both early and late infections. The transcription of gelE was unchanged between the inoculum and the two infection time points.
Taken together, the data in Fig. 2 do not indicate that the stringent response is triggered in valve-associated E. faecalis cells. Therefore, we conclude from these experiments that E. faecalis cells remain in an active state of growth at the heart valve surface during the course of the 4-day rabbit IE infection and that the attenuation of the Δrel strain in this model is not due to the inability to trigger the stringent response. These results suggest that sustained in vivo up-regulation of ace and efbA expression may be important for virulence in this model.
The absolute transcript levels of ace and efbA are higher in Δrel and ΔrelΔrelQ compared to OG1RF during in vitro biofilm growth
Rel is the only (p)ppGpp hydrolase in E. faecalis [21], making it the central regulator of cellular (p)ppGpp levels in E. faecalis. The results in Figs 1 and 2 suggest that the regulation of basal (p)ppGpp levels may be important for the pathogenesis of E. faecalis IE. Therefore, in an attempt to explain the unexpected phenotypes of the Δrel and ΔrelΔrelQ strains in the IE model (Fig. 1), the remainder of this study characterized how the aberrant basal (p)ppGpp levels in these two strains affect processes associated with IE pathogenesis. The ΔrelQ strain was not included in these investigations since it both lacks a phenotype in the IE animal model (Fig. 1) and has basal (p)ppGpp levels that are similar to those of the wild-type strain [22].
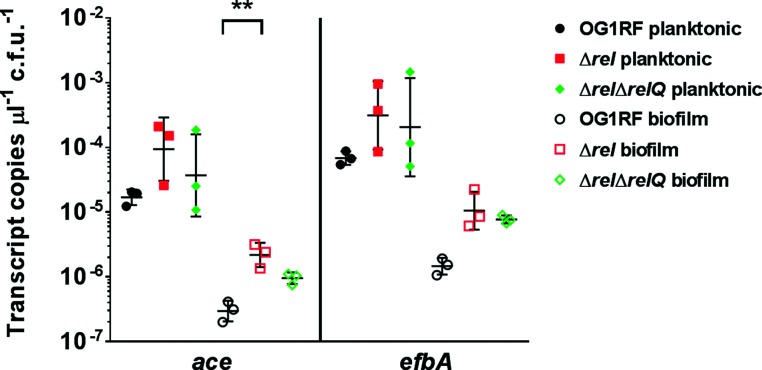
The Δrel and ΔrelΔrelQ strains show unique transcriptional profiles compared to OG1RF in stressed and unstressed conditions [22, 29]. Thus, we hypothesized that the expression of genes that contribute to virulence in the rabbit IE model may be dysregulated in the Δrel strain compared to the OG1RF or ΔrelΔrelQ strains, and that this may provide a possible explanation for the impaired colonization phenotype shown in Fig. 1. Based on the strong in vivo up-regulation of ace and efbA (Fig. 2), we measured the expression of these adhesin-encoding genes when OG1RF, Δrel and ΔrelΔrelQ were grown under planktonic or biofilm conditions in vitro. Due to the lack of a documented housekeeping gene that is constitutively expressed at equal levels in E. faecalis planktonic and biofilm cells, we measured the transcript copy number by absolute quantitation and normalized the values by the number of c.f.u. from which RNA was extracted [35]. The absolute transcript levels of ace and efbA were higher in planktonic cells (Fig. 3, solid symbols) compared to biofilm cells (Fig. 3, open symbols). In biofilms, the absolute transcript levels of ace and efbA were consistently higher in Δrel and ΔrelΔrelQ compared to OG1RF; however, only the difference between ace transcripts in OG1RF and Δrel was statistically significant (Fig. 3; P=0.0094, Tukey’s multiple comparison post hoc test). Overall, the similar expression patterns of ace and efbA in both of the (p)ppGpp mutant strains do not support our hypothesis that the dysregulation of adhesin-encoding gene expression may contribute to the impaired colonization phenotype of the Δrel strain in the rabbit IE model.
Fig. 3.
The transcript levels of the infective endocarditis virulence genes ace and efbA are dysregulated in strains with altered (p)ppGpp levels during in vitro biofilm growth. RNA was harvested from planktonic and biofilm cells of the strains OG1RF, ∆rel and ∆rel∆relQ that were grown under conditions known to induce in vitro expression of ace and efbA. The transcript copy number detected per microlitre of cDNA was divided by the corresponding number of c.f.u. from which RNA was extracted to obtain the absolute transcript level. Each symbol represents the absolute transcript level of the indicated gene in one biological replicate collected on separate days. The horizontal bars and error bars represent the geometric mean±geometric standard deviation. **, P=0.0094 by Tukey’s multiple comparison post hoc test after ordinary one-way ANOVA testing for differences in ace expression between OG1RF, ∆rel and ∆rel∆relQ biofilm cells. The differences in the means for the other conditions were not significant by one-way ANOVA.
The Δrel strain is impaired in attachment to BSA-coated polystyrene, but not collagen- or fibronectin-coated surfaces or ex vivo porcine heart valves
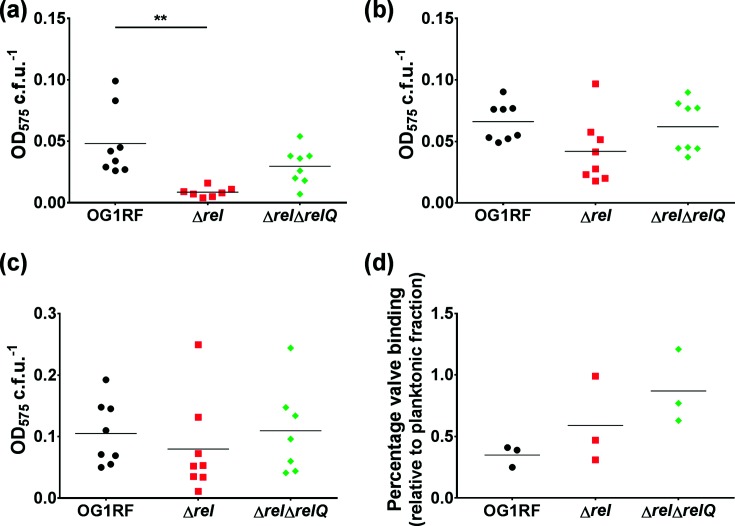
We next compared the ability of OG1RF, Δrel and ΔrelΔrelQ to attach to different types of surfaces. We first evaluated how well the strains attached to abiotic surfaces following growth in conditions to induce in vitro expression of ace and efbA. The average attachment of Δrel to BSA-coated polystyrene wells following 2 hours of incubation was 80% lower than the average attachment of OG1RF (Fig. 4a; P=0.0011 by Dunnett’s multiple comparison post hoc test). The attachment of ΔrelΔrelQ was reduced by approximately 40% relative to OG1RF (P=0.1091 by Dunnett’s multiple comparison post hoc test for the absolute values shown in Fig. 4a). Next, we evaluated the attachment of the strains to polystyrene surfaces coated with known substrates of Ace and EfbA, namely collagen and fibronectin, respectively. Although the average attachment of Δrel to either collagen (Fig. 4b) or fibronectin (Fig. 4c) was lower than the average attachment of the other two strains, the differences observed were not statistically significant. Taken together, these results indicate that the deletion of rel may have a negative impact on cell surface attachment, but that the defect becomes negligible when host matrix molecules that are relevant to endocarditis pathogenesis are available as binding substrates.
Fig. 4.
Deletion of rel impairs cellular attachment to BSA-coated polystyrene but not collagen, fibronectin, or ex vivo porcine heart valve tissue sections. For the experiments shown in panels (a–c), E. faecalis OG1RF, ∆rel and ∆rel∆relQ were grown in conditions known to induce in vitro expression of ace and efbA. Bacterial attachment to polystyrene wells that were (a) coated with BSA, (b) coated with collagen, or (c) coated with fibronectin was quantified by crystal violet staining and normalized by c.f.u. Each symbol represents one biological replicate. The horizontal bars show the mean. (a) Ordinary one-way ANOVA was used to determine whether the difference between the means of ∆rel or ∆rel∆relQ and OG1RF was statistically significant (P=0.0023). **, P=0.0011 by Dunnett’s multiple comparison post hoc test. (b, c) The differences in the means were not significant by one-way ANOVA. (d) The percentages of E. faecalis OG1RF, ∆rel and ∆rel∆relQ cells that attached to ex vivo porcine heart valve sections relative to the number of planktonic bacteria in the same well after 2 hours of incubation. Each symbol represents one biological replicate collected on a separate day. The horizontal bars show the mean. The means were not significantly different by statistical analysis (one-way ANOVA).
To further discern how cellular (p)ppGpp levels affect attachment to a more relevant biotic surface, we compared OG1RF, Δrel and ΔrelΔrelQ in an ex vivo porcine heart valve tissue section attachment assay [37, 46]. Following growth in conditions identical to those used for the rabbit IE model inoculum, the bacterial strains were incubated with heart valve sections for 2 hours. The mean percentage of valve binding for both the Δrel and ΔrelΔrelQ strains was higher than that for OG1RF (Fig. 4d), albeit the differences among the means were not statistically significant (P=0.1456, one-way ANOVA). The results of this experiment confirm that strains with disrupted levels of (p)ppGpp can functionally bind to heart valve tissues as well as the parent strain. Collectively, the results in Fig. 4 suggest that the impaired IE phenotype of the Δrel strain is not linked to a defect in attachment.
Mutants with disrupted (p)ppGpp levels retain high levels of gelatinase activity but are associated with reduced HCAEC invasion
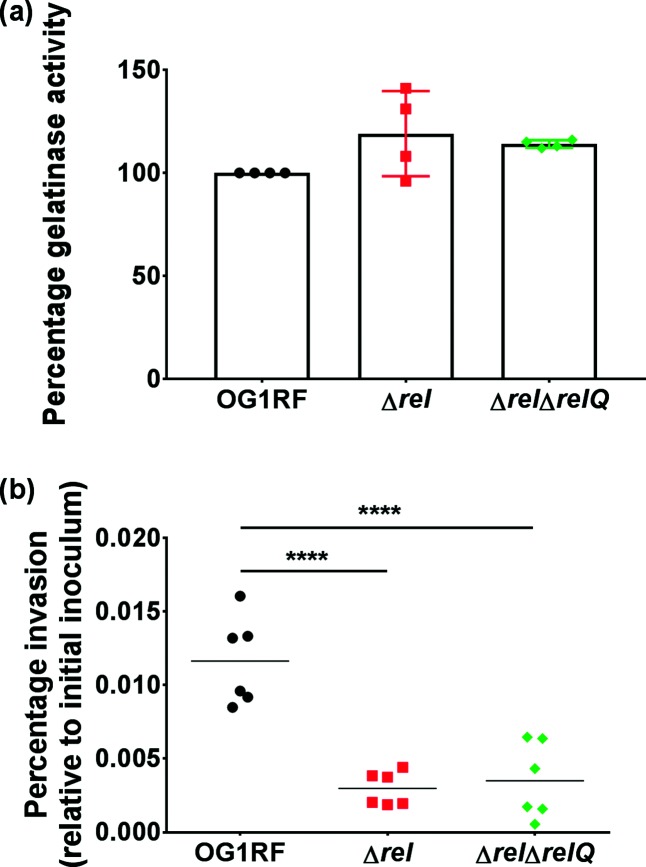
Finally, we investigated how the perturbation of (p)ppGpp levels affects the properties that enable E. faecalis to spread beyond the surface of the heart valve during IE. Notably, the reduced valve bacterial load of the Δrel strain in the rabbit IE model could derive from an increase in dissemination from the valve surface or invasion into the cardiac endothelium. We tested strains for production of the extracellular protease gelatinase (GelE), which is a central virulence factor in E. faecalis that was previously shown to be associated with the colonization of tissues away from the heart valve in a model of enterococcal IE [15]. The overall average relative gelatinase production was higher in both of the (p)ppGpp mutant strains than in OG1RF (Fig. 5a), but the differences in the means were not statistically significant (P=0.1198, one-way ANOVA). The invasion of cardiac endothelial cells was assessed by incubating OG1RF, Δrel and ΔrelΔrelQ with HCAECs for 2 hours. Both (p)ppGpp mutant strains had statistically significant defects in HCAEC invasion (Fig. 5b; P=0.0001 by Dunnett’s multiple comparison post hoc test for each strain compared to OG1RF). The results in Fig. 5 demonstrate that (p)ppGpp, or the ability to regulate its levels, is important for invasion but is dispensable for gelatinase production.
Fig. 5.
Disruption of cellular (p)ppGpp levels does not impact on gelatinase production but is associated with diminished invasion of human coronary artery endothelial cells (HCAECs). (a) Supernatants of OG1RF, ∆rel, or ∆rel∆relQ cultures grown overnight in medium supplemented with 40% horse serum were tested for gelatinase activity in a zone-of-clearance assay. The area of the zone of clearance for OG1RF was set to 100% in each biological replicate, and this was used to calculate the relative gelatinase activity based on the zone areas for the other strains tested. The symbols represent biological replicates measured on separate days; the bars show the means. The means were not significantly different by one-way ANOVA. (b) HCAECs were infected with OG1RF, ∆rel or ∆rel∆relQ at an m.o.i. of 100 : 1 for 2 hours. The ability of each E. faecalis strain to invade is reported as the percentage of intracellular bacteria recovered after 2 hours relative to the total c.f.u. of the initial inoculum. Each symbol represents one biological replicate. The horizontal bars show the mean. Ordinary one-way ANOVA was used to determine whether the difference between the means of ∆rel or ∆rel∆relQ and OG1RF was statistically significant (P<0.0001). ****, P=0.0001 by Dunnett’s multiple comparison post hoc test.
Discussion
Cashel and Gallant first detected what would later be identified as ppGpp [47] by thin layer chromatographic analysis of acid-soluble metabolites from amino acid-starved Escherichia coli cells in 1969 [48]. It is now established that ppGpp and pppGpp are powerful molecular mediators of adaptation to stress [25], which extends well beyond nutrient limitation to include virulence in many bacterial species [27]. An association between (p)ppGpp and E. faecalis virulence was first reported in invertebrate infection models [21, 28, 29], whereby the ΔrelΔrelQ [(p)ppGpp0] strain was significantly attenuated. More recently, we showed that alteration of (p)ppGpp levels in E. faecalis differentially affected clearance and persistence in a mammalian subcutaneous foreign body abscess model [30]. However, the localized nature of the foreign body abscess infection is not representative of the serious systemic infections, such as bacteraemia and IE, that E. faecalis is capable of causing. Therefore, the goal of this study was to define the relationship between (p)ppGpp and E. faecalis IE virulence. This is the first report to investigate the role of (p)ppGpp in the pathogenesis of IE in any bacterial species.
Our data reveal that deletion of the gene encoding the bifunctional (p)ppGpp synthetase/hydrolase Rel is detrimental to valve colonization in the rabbit IE model, whereas relQ and any cellular presence of (p)ppGpp are dispensable for in vivo colonization of damaged heart valves (Fig. 1). The significant IE defect of Δrel and the absence of a virulence defect for the (p)ppGpp0 strain (Fig. 1) indicate that, rather than the stringent response, tight regulation of basal levels of (p)ppGpp is important for E. faecalis IE virulence. Indeed, the data in Figs 1 and 2 collectively demonstrate that the activation of a stringent response is not required, and likely does not occur, as part of the pathogenesis of valve colonization in the E. faecalis IE rabbit model. In fact, the data shown in Fig. 2 support that E. faecalis cells undergo active growth in the valve environment. This makes sense when we consider that blood flow through the heart provides a constant stream of nutrients for bacteria that have attached to and formed a biofilm on damaged endocardial surfaces.
Considering that the virulence of the (p)ppGpp0 strain, but not the Δrel strain, was attenuated in two invertebrate infection models (Caenorhabditis elegans and Galleria mellonella) [21, 29], and that only the (p)ppGpp0 strain showed increased sensitivity to a mouse-derived macrophage cell line [29], the virulence attenuation of the Δrel mutant and the full virulence phenotype of the (p)ppGpp0 mutant in the IE model were unexpected. However, rel single mutants of Staphylococcus aureus, Streptococcus pneumoniae and Listeria monocytogenes have been shown to have impaired virulence in murine models of kidney abscess infection, pneumonia and listeriosis, respectively [49–51]. Therefore, attenuation of the E. faecalis Δrel strain in a systemic model of infection is not an unprecedented result. Collectively, these results support the notion that (p)ppGpp levels need to be tightly controlled to avoid deleterious effects during the infection process. More specifically, while the absence of (p)ppGpp reduced the virulence of E. faecalis in invertebrate hosts, high intracellular accumulation of basal (p)ppGpp is detrimental to IE in a mammalian host.
Of note, the Δrel strain grows more slowly than the parent and (p)ppGpp0 strains in BHI [21]. We also showed previously that the growth of the Δrel strain was slightly lower than that of the parent strain in human serum after 9 hours of incubation, but higher than that of the parent strain in human whole blood at the same time point [30]. However, both the parent and Δrel strains reached the same final optical density after 24 h and remained equally viable for up to 72 h in either serum or whole blood. The growth and survival of the (p)ppGpp0 strain, on the other hand, were dramatically impaired in serum, a phenotype later shown to be linked to the loss of metal homeostasis in the (p)ppGpp0 strain [24]. Because of the discrepant initial growth phenotype of the Δrel strain in serum or blood, we revisited this experiment using horse serum. Again, the Δrel strain grew slightly more slowly, but its final growth yields and overall survival were identical to those obtained with the parent strain (data not shown). Therefore, we conclude that the valve colonization defect of the Δrel strain cannot be linked to a defect in growth or survival after cells enter the bloodstream.
The adhesion of bacterial cells to damaged cardiac tissue is a crucial early step in the pathogenesis of endocarditis. In attempt to understand the stringent response-independent IE attenuation of the Δrel strain, we evaluated adhesin gene expression and then assessed the ability of the (p)ppGpp mutants to adhere to surfaces relevant to endocarditis. The genes encoding two important host ECM-binding adhesins, Ace and EfbA, were highly expressed in valve-associated E. faecalis OG1RF cells (Fig. 2c), underscoring the importance of bacterial attachment to host matrix proteins in the pathogenesis of IE. The quantitation of ace and efbA transcript levels in the (p)ppGpp mutant strains compared to the parent strain during biofilm growth suggested that cellular (p)ppGpp affects the expression of these genes during biofilm formation (Fig. 3). Despite this, the ability of the Δrel strain to bind to host ECM-coated abiotic surfaces and ex vivo porcine valves (Fig. 4) suggests that high basal pools of (p)ppGpp do not strongly influence E. faecalis cell attachment to exposed ECM components on damaged valve surfaces.
The detachment of bacteria from a biofilm is an essential step for the perpetuation of infection [52]. Embolization and tissue invasion are two mechanisms by which enterococci could disseminate beyond a biofilm localized on a heart valve. E. faecalis gelatinase mediates evasion of the host innate immune response [15, 53], tissue dissemination [15] and destruction of epithelial barriers [54, 55]. The gelatinase activity of the two mutant strains was comparable to that of the parent strain (Fig. 5a). Thus, our data do not support a relationship between (p)ppGpp and the propensity of a strain to embolize, as measured through extracellular gelatinase activity. In contrast to the strikingly different valve colonization phenotypes that the Δrel and (p)ppGpp0 strains displayed in the IE model (Fig. 1), both strains were equally deficient in their ability to invade coronary artery endothelial cells (Fig. 5b). These disparate phenotypes demonstrate that, while (p)ppGpp strongly influences the ability of E. faecalis to invade HCAECs, endocardial cell invasion is independent of a strain’s ability to robustly colonize a valve.
In summary, this study demonstrates a role for basal (p)ppGpp pools in the pathogenesis of E. faecalis endocarditis. We have shown that, while (p)ppGpp is not absolutely required for E. faecalis to cause IE in a rabbit model, the ability to regulate basal (p)ppGpp levels is critical for valve colonization in the model. Our data indicate that the processes for colonizing and forming vegetations on damaged valves in the rabbit IE model are not dependent on stringent response activation. Furthermore, the defective colonization phenotype of the Δrel strain cannot be explained by the altered expression of genes encoding fibronectin- and collagen-binding proteins or defects in binding to biologically relevant surfaces. Interestingly, the magnitude of the Δrel valve colonization defect in the rabbit IE model is similar to those previously determined for the biofilm mutant strains ΩahrC and ∆eep, which had the greatest degrees of attenuation that we have measured in the rabbit IE model following the disruption of any one single gene in the E. faecalis OG1RF strain background [34, 35]. While an explanation for the ΩahrC IE phenotype was recently proposed [56], the mechanism underlying the ∆eep IE phenotype, as with the Δrel strain, also remains to be elucidated. Additional studies will be necessary to determine why high basal levels of (p)ppGpp, as observed in the Δrel strain, impact negatively on valve colonization. Collectively, the data presented here demonstrate that cellular (p)ppGpp levels differentially affect distinct processes (e.g. the transcription of IE-associated adhesins, valve colonization and endocardial cell invasion) in the pathogenesis of E. faecalis IE.
Funding information
This work was supported by American Heart Association awards 10POST3290026 and 17SDG33350092 and Uniformed Services University start-up award R0733973 to K. L. F., American Heart Association award 16PRE29860000 to C. C.-W. and NIH award AI135158 to J. A. L. NIH grants AI058134 to Gary Dunny and AI74283 to Patrick Schlievert at the University of Minnesota also supported the early stages of this work. A. O. G. was supported by NIH/NIDCR training grant 1T90DE021985 at the University of Rochester. The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense, the Uniformed Services University of the Health Sciences, the National Institute of Allergy and Infectious Diseases, the National Institute of Dental and Craniofacial Research, the National Institutes of Health, or any other agency of the US Government, or the American Heart Association. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
The authors gratefully acknowledge Gary Dunny and Patrick Schlievert for scientific and financial support of this project during its development, Anne-Marie Leuck for providing reagents and protocols, and Adam Spaulding, Joseph Merriman, Yuqing Chen, Biko McMillan, Candace Rouchon and Arielle Weinstein for technical assistance. We thank Cara Olsen at the USUHS Biostatistics Consulting Center for valuable input on the statistical analysis of in vivo qPCR data.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: BH, beef heart dialysate; BHI, brain heart infusion; Ebp, endocarditis- and biofilm-associated pili; IE, infective endocarditis; KPBS, potassium phosphate-buffered saline; ppGpp, guanosine tetraphosphate; pppGpp, guanosine pentaphosphate; (p)ppGpp, guanosine tetra- and pentaphosphate; THB, Todd–Hewitt broth.
Edited by: M. Vickerman and J. Stülke
References
- 1.Toyoda N, Chikwe J, Itagaki S, Gelijns AC, Adams DH, et al. Trends in infective endocarditis in California and New York State, 1998–2013. JAMA. 2017;317:1652–1660. doi: 10.1001/jama.2017.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrosioni J, Hernandez-Meneses M, Téllez A, Pericàs J, Falces C, et al. The changing epidemiology of infective endocarditis in the twenty-first century. Curr Infect Dis Rep. 2017;19:21. doi: 10.1007/s11908-017-0574-9. [DOI] [PubMed] [Google Scholar]
- 3.Bashore TM, Cabell C, Fowler V. Update on infective endocarditis. Curr Probl Cardiol. 2006;31:274–352. doi: 10.1016/j.cpcardiol.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durack DT. Experimental bacterial endocarditis. IV. Structure and evolution of very early lesions. J Pathol. 1975;115:81–89. doi: 10.1002/path.1711150204. [DOI] [PubMed] [Google Scholar]
- 6.Baddour LM, Wilson WR, Bayer AS, Fowler VG, Tleyjeh IM, et al. Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications: a scientific statement for healthcare professionals from the American Heart Association. Circulation. 2015;132:1435–1486. doi: 10.1161/CIR.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 7.Murdoch DR, Corey GR, Hoen B, Miró JM, Fowler VG, et al. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the International Collaboration on Endocarditis-prospective cohort study. Arch Intern Med. 2009;169:463–473. doi: 10.1001/archinternmed.2008.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chirouze C, Athan E, Alla F, Chu VH, Ralph Corey G, et al. Enterococcal endocarditis in the beginning of the 21st century: analysis from the International Collaboration on Endocarditis-prospective cohort study. Clin Microbiol Infect. 2013;19:1140–1147. doi: 10.1111/1469-0691.12166. [DOI] [PubMed] [Google Scholar]
- 9.Rozdzinski E, Marre R, Susa M, Wirth R, Muscholl-Silberhorn A. Aggregation substance-mediated adherence of Enterococcus faecalis to immobilized extracellular matrix proteins. Microb Pathog. 2001;30:211–220. doi: 10.1006/mpat.2000.0429. [DOI] [PubMed] [Google Scholar]
- 10.Nallapareddy SR, Singh KV, Sillanpää J, Zhao M, Murray BE. Relative contributions of Ebp pili and the collagen adhesin Ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect Immun. 2011;79:2901–2910. doi: 10.1128/IAI.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nallapareddy SR, Murray BE. Ligand-signaled upregulation of Enterococcus faecalis ace transcription, a mechanism for modulating host-E. faecalis interaction. Infect Immun. 2006;74:4982–4989. doi: 10.1128/IAI.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh KV, La Rosa SL, Somarajan SR, Roh JH, Murray BE. The fibronectin-binding protein EfbA contributes to pathogenesis and protects against infective endocarditis caused by Enterococcus faecalis . Infect Immun. 2015;83:4487–4494. doi: 10.1128/IAI.00884-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasmussen M, Johansson D, Söbirk SK, Mörgelin M, Shannon O. Clinical isolates of Enterococcus faecalis aggregate human platelets. Microbes Infect. 2010;12:295–301. doi: 10.1016/j.micinf.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Nallapareddy SR, Sillanpää J, Mitchell J, Singh KV, Chowdhury SA, et al. Conservation of Ebp-type pilus genes among enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets. Infect Immun. 2011;79:2911–2920. doi: 10.1128/IAI.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, et al. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis . Infect Immun. 2010;78:4936–4943. doi: 10.1128/IAI.01118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters CM, Antiporta MH, Murray BE, Dunny GM. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J Bacteriol. 2003;185:3613–3623. doi: 10.1128/JB.185.12.3613-3623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nallapareddy SR, Singh KV, Sillanpää J, Garsin DA, Höök M, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis . J Clin Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh KV, Nallapareddy SR, Sillanpää J, Murray BE. Importance of the collagen adhesin Ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathog. 2010;6:e1000716. doi: 10.1371/journal.ppat.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin X, Singh KV, Weinstock GM, Murray BE. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J Bacteriol. 2001;183:3372–3382. doi: 10.1128/JB.183.11.3372-3382.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 21.Abranches J, Martinez AR, Kajfasz JK, Chávez V, Garsin DA, et al. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis . J Bacteriol. 2009;191:2248–2256. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaca AO, Kajfasz JK, Miller JH, Liu K, Wang JD, et al. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. MBio. 2013;4:e00646-13. doi: 10.1128/mBio.00646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chávez de Paz LE, Lemos JA, Wickström C, Sedgley CM. Role of (p)ppGpp in biofilm formation by Enterococcus faecalis . Appl Environ Microbiol. 2012;78:1627–1630. doi: 10.1128/AEM.07036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colomer-Winter C, Gaca AO, Lemos JA. Association of metal homeostasis and (p)ppGpp regulation in the pathophysiology of Enterococcus faecalis . Infect Immun. 2017;85:e00260-17. doi: 10.1128/IAI.00260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 26.Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol. 2015;197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan X, Zhao C, Budin-Verneuil A, Hartke A, Rincé A, et al. The (p)ppGpp synthetase RelA contributes to stress adaptation and virulence in Enterococcus faecalis V583. Microbiology. 2009;155:3226–3237. doi: 10.1099/mic.0.026146-0. [DOI] [PubMed] [Google Scholar]
- 29.Gaca AO, Abranches J, Kajfasz JK, Lemos JA. Global transcriptional analysis of the stringent response in Enterococcus faecalis . Microbiology. 2012;158:1994–2004. doi: 10.1099/mic.0.060236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank KL, Colomer-Winter C, Grindle SM, Lemos JA, Schlievert PM, et al. Transcriptome analysis of Enterococcus faecalis during mammalian infection shows cells undergo adaptation and exist in a stringent response state. PLoS One. 2014;9:e115839. doi: 10.1371/journal.pone.0115839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roggiani M, Schlievert PM. Purification of streptococcal pyrogenic exotoxin A. In: Evans TJ, editor. Septic Shock Methods and Protocols (Methods in Molecular Medicine Series vol. 36) Totowa, NJ: Humana Press; 2000. pp. 59–66. (editor) [DOI] [PubMed] [Google Scholar]
- 33.Chuang ON, Schlievert PM, Wells CL, Manias DA, Tripp TJ, et al. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect Immun. 2009;77:539–548. doi: 10.1128/IAI.01034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank KL, Barnes AM, Grindle SM, Manias DA, Schlievert PM, et al. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect Immun. 2012;80:539–549. doi: 10.1128/IAI.05964-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank KL, Guiton PS, Barnes AM, Manias DA, Chuang-Smith ON, et al. AhrC and Eep are biofilm infection-associated virulence factors in Enterococcus faecalis . Infect Immun. 2013;81:1696–1708. doi: 10.1128/IAI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leuck AM, Johnson JR, Dunny GM. A widely used in vitro biofilm assay has questionable clinical significance for enterococcal endocarditis. PLoS One. 2014;9:e107282. doi: 10.1371/journal.pone.0107282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abranches J, Zeng L, Bélanger M, Rodrigues PH, Simpson-Haidaris PJ, et al. Invasion of human coronary artery endothelial cells by Streptococcus mutans OMZ175. Oral Microbiol Immunol. 2009;24:141–145. doi: 10.1111/j.1399-302X.2008.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avilés-Reyes A, Miller JH, Simpson-Haidaris PJ, Lemos JA, Abranches J. Cnm is a major virulence factor of invasive Streptococcus mutans and part of a conserved three-gene locus. Mol Oral Microbiol. 2014;29:11–23. doi: 10.1111/omi.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montville TJ. Dual-substrate plate diffusion assay for proteases. Appl Environ Microbiol. 1983;45:200–204. doi: 10.1128/aem.45.1.200-204.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression – a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potrykus K, Murphy H, Philippe N, Cashel M. ppGpp is the major source of growth rate control in E. coli . Environ Microbiol. 2011;13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernandez VJ, Bremer H. Characterization of RNA and DNA synthesis in Escherichia coli strains devoid of ppGpp. J Biol Chem. 1993;268:10851–10862. [PubMed] [Google Scholar]
- 45.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli . Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuang-Smith ON, Wells CL, Henry-Stanley MJ, Dunny GM. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLoS One. 2010;5:e15798. doi: 10.1371/journal.pone.0015798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cashel M, Kalbacher B. The control of ribonucleic acid synthesis in Escherichia coli. V. Characterization of a nucleotide associated with the stringent response. J Biol Chem. 1970;245:2309–2327. [PubMed] [Google Scholar]
- 48.Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli . Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 49.Kazmierczak KM, Wayne KJ, Rechtsteiner A, Winkler ME. Roles of rel(Spn) in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol Microbiol. 2009;72:590–611. doi: 10.1111/j.1365-2958.2009.06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor CM, Beresford M, Epton HA, Sigee DC, Shama G, et al. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J Bacteriol. 2002;184:621–628. doi: 10.1128/JB.184.3.621-628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geiger T, Goerke C, Fritz M, Schäfer T, Ohlsen K, et al. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus . Infect Immun. 2010;78:1873–1883. doi: 10.1128/IAI.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park SY, Kim KM, Lee JH, Seo SJ, Lee IH. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect Immun. 2007;75:1861–1869. doi: 10.1128/IAI.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steck N, Hoffmann M, Sava IG, Kim SC, Hahne H, et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology. 2011;141:959–971. doi: 10.1053/j.gastro.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 55.Maharshak N, Huh EY, Paiboonrungruang C, Shanahan M, Thurlow L, et al. Enterococcus faecalis gelatinase mediates intestinal permeability via protease-activated receptor 2. Infect Immun. 2015;83:2762–2770. doi: 10.1128/IAI.00425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manias DA, Dunny GM. Expression of adhesive pili and the collagen-binding adhesin Ace is activated by ArgR family transcription factors in Enterococcus faecalis . J Bacteriol. 2018:JB.00269-18. doi: 10.1128/JB.00269-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao P, Pinkston KL, Nallapareddy SR, van Hoof A, Murray BE, et al. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. J Bacteriol. 2010;192:5489–5498. doi: 10.1128/JB.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]