Abstract
The Streptococcus mutans Cid/Lrg system represents an ideal model for studying this organism’s ability to withstand various stressors encountered in the oral cavity. The lrg and cid operons display distinct and opposite patterns of expression in response to growth phase and glucose levels, suggesting that the activity and regulation of these proteins must be tightly coordinated in the cell and closely associated with metabolic pathways of the organism. Here, we demonstrate that expression of the cid and lrg operons is directly mediated by a global transcriptional regulator CcpA in response to glucose levels. Comparison of the cid and lrg promoter regions with the conserved CcpA binding motif revealed the presence of two potential cre sites (for CcpA binding) in the cid promoter (designated cid-cre1 and cid-cre2), which were arranged in a similar manner to those previously identified in the lrg promoter region (designated lrg-cre1 and lrg-cre2). We demonstrated that CcpA binds to both the cid and lrg promoters with a high affinity, but has an opposing glucose-dependent effect on the regulation of cid (positive) and lrg (negative) expression. DNase I footprinting analyses revealed potential binding sequences for CcpA in both cid and lrg promoter regions. Collectively, these data suggest that CcpA is a direct regulator of cid and lrg expression, and are suggestive of a potential mechanism by which Cid/Lrg-mediated virulence and cellular homeostasis is integrated with signals associated with both the environment and cellular metabolic status.
Keywords: Streptococcus mutans, CcpA, glucose metabolism, Cid and Lrg
Introduction
Development of a mature biofilm on the tooth surface is the central event in the pathogenesis of dental caries [1]. This process primarily requires that cariogenic organisms, including Streptococcus mutans, withstand the limited resources and extreme environmental fluctuations experienced in the oral cavity [2–4]. Recent studies have revealed that the survival and persistence of micro-organisms during the development of biofilms may be mediated by regulated cell death and lysis processes, consequently eliminating bacterial cells damaged by adverse environments and benefiting the rest of the population within the biofilm [5–7]. The lysis of a small sub-population also releases DNA that glues the extracellular matrix together, augmenting biofilms. Although the underlying mechanisms for regulated cell death and lysis remain unclear, it is likely that understanding such altruistic behaviours provides valuable insight into inhibiting the initiation and/or progression of biofilm diseases. In this regard, the two paralogously related dicistronic operons, lrgAB (SMU.575c/574c) and cidAB (SMU.1701c/1700c), represent an ideal model system to study and appreciate the complexity of biofilm development and pathogenicity in the context of dental plaque, which is heterogeneous and embodies many different environmental stresses [8–10]. The cid and lrg operons encode predicted membrane-associated proteins CidA, CidB, LrgA and LrgB, and notably their single or combinatory mutations affect comprehensive virulence traits, such as autolysis, biofilm development, oxidative and heat stress responses, antibiotic resistance and genetic competence in S. mutans [8–12], all of which are required for successful colonization and persistence of this organism in the oral cavity. The CidA and LrgA proteins also share structural features with the bacteriophage-encoded holin family of proteins [9, 13–15]. Holins control the timing of host cell lysis during bacteriophage lytic infection and anti-holins function to inhibit holin action [13–16]. Therefore, it has been hypothesized that CidA and LrgA may control cell death and lysis in a manner analogous to effector and inhibitor holins, respectively [6, 17]. Moreover, they may impact bacteria at the community level by coordinating differentiation of a biofilm community into distinct functional subpopulations, diversification which may render the biofilm more resistant to environmental stress [5–7]. This idea is in agreement with the previous observation that lrgA/B were among the most highly upregulated genes associated with thicker S. mutans biofilms [18]. A holin-like role for CidA and antiholin-like role for LrgA in mediating cell death and autolysis was originally proposed in Staphylococcus aureus [19–21], but the molecular details of how Cid and Lrg function to control cell death and lysis have not yet been completely elucidated. In addition, our previous analysis of several cid and lrg mutant phenotypes in S. mutans strongly suggests a more complex regulation than previously anticipated, including interplay between the gene products of these operons [9, 11, 12].
Another notable feature of the S. mutans Cid/Lrg system is that expression levels of lrg and cid are counterbalanced throughout the growth cycle and in response to the availability of oxygen and glucose [9]. The lrg genes are highly induced in cultures containing lower levels of glucose (≤15 mM) but almost completely repressed in cultures containing glucose at concentrations of 20 mM and higher. In contrast, expression of cid genes is negligible when cells are cultured in the presence of lower glucose concentrations (≤20 mM), but increases at higher glucose concentrations (>20 mM). In this previous study, we also demonstrated that CcpA (catabolite response protein A), a global regulator of carbon metabolism involved in carbon catabolite repression (CCR) [22–24], is involved in the expression of cid and lrg [9, 12]. CcpA can repress or activate gene expression by direct interaction with catabolite-responsive element (cre) sequences (WTGNAANCGNWNNCW) in the promoter regions of its target genes [25]. In the presence of a preferred carbon source, such as glucose, CcpA typically represses expression of genes involved in utilization of secondary carbon sources while activating carbon overflow pathways [26, 27]. In most of the low-G+C Gram-positive bacteria, CcpA forms a complex with a phosphorylated (HPr-Ser46-P) form of HPr, stimulating CcpA binding to cre sites. HPr is phosphorylated by HPr kinase, which is stimulated by elevated levels of particular glycolytic intermediates, usually fructose-1,6-bisphosphate (FBP) or glucose-6-phosphate (G6P), when cells are under conditions that trigger CCR [28–30]. In our previous study, we identified two cre-like consensus elements in the DNA sequence immediately upstream of lrgAB only, but not upstream of cidAB [9]. However, detailed genetic investigations of CcpA-mediated cid and lrg expression have not been performed. In this study, we show that CcpA is directly involved in the regulation of cid and lrg operons and is, at least partly, responsible for the inversely correlated regulation of cid and lrg expression in response to glucose levels. We demonstrate herein that the cid and lrg promoter regions are direct targets of S. mutans CcpA binding, using electrophoretic mobility shift assays (EMSAs). The potential binding sequences in each promoter region were also identified by DNase I footprinting analyses. Mutagenesis analysis of the identified binding sequences provides further insights into how CcpA modulates cid and lrg expression. The data presented suggest that the opposing expression patterns of cid and lrg in response to carbohydrate availability and nutritional status of the organism are coordinated by CcpA.
Methods
Bacterial strains, plasmids and growth conditions
All Escherichia coli strains were cultured in Luria–Bertani (LB) medium at 37 °C. Streptococcus mutans UA159 and its derivatives were cultured in brain–heart infusion (BHI) medium (Difco Laboratories, Detroit, MI) or chemically defined medium FMC [31] containing 11 mM (or 45 mM) glucose at 37 °C in a 5 % CO2 incubator. Antibiotics were used to supplement growth media in the following concentrations: ampicillin (100 µg ml−1 for E. coli), spectinomycin (50 µg ml−1 for E. coli and 1 mg ml−1 for S. mutans); kanamycin (50 µg ml−1 for E. coli and 1 mg ml−1 for S. mutans); and erythromycin (300 µg ml−1 for E. coli and 10 µg ml−1 for S. mutans).
Construction of reporter gene fusions and DNA manipulation
All the strains and plasmids used in this study are listed in Table 1. The Plrg-gfp and Pcid-gfp reporter strains were constructed by replacing the promoter region of our previous gfp reporter strains [32, 33] with Pcid or Plrg. Briefly, a region of about 200 bp comprising Pcid or Plrg was PCR-amplified with primers, incorporated HindIII and SpeI sites, respectively, and was cloned in front of the superfolder green fluorescent protein (sGFP) gene in the shuttle vector pDL278. The resulting construct was transformed into S. mutans strains: wild-type UA159 and ccpA-deficint mutant [34]. All constructs were Sanger sequenced to confirm cid and lrg promoter sequence fidelity.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotypes and/or descriptions | Source or reference |
|---|---|---|
| S. mutans strains | ||
| UA159 | Wild-type | ATCC 700610 |
| ΔccpA | ΔccpA :: NPEmr | [34] |
| UA159/Plrg-gfp | UA159 carrying pDL278 :: Plrg-gfp | This study |
| UA159/Pcid-gfp | UA159 carrying pDL278 :: Pcid-gfp | This study |
| ΔccpA/Plrg-gfp | ΔccpA carrying pDL278 :: Plrg-gfp | This study |
| ΔccpA/Pcid-gfp | ΔccpA carrying pDL278 :: Pcid-gfp | This study |
| UA159/Pcidcre1-gfp | UA159 carrying pDL278 :: cre1 site mutated Pcid-gfp | This study |
| UA159/Pcidcre2-gfp | UA159 carrying pDL278 :: cre2 site mutated Pcid-gfp | This study |
| UA159/Pcidcre12-gfp | UA159 carrying pDL278 :: cre1 and cre2 sites mutated Pcid-gfp | This study |
| UA159/Plrgcre1-gfp | UA159 carrying pDL278 :: cre1 site mutated Plrg-gfp | This study |
| UA159/Plrgcre2-gfp | UA159 carrying pDL278 :: cre2 site mutated Plrg-gfp | This study |
| UA159/Plrgcre12-gfp | UA159 carrying pDL278 :: cre1 and cre2 sites mutated Plrg-gfp | This study |
| E. coli strains | ||
| His6-CcpA | ccpA coding region cloned into pQE30, Kmr, Ampr | [26] |
| Plasmid | ||
| pDL278 | E. coli/Streptococcus shuttle vector | [44] |
Microplate reporter assay
GFP fluorescence of the S. mutans strains harbouring the Plrg-gfp or Pcid-gfp promoter fusion constructs was measured using a Synergy microplate reader (BioTek) controlled by Gen5 software. Overnight cultures were diluted 1 : 50 into 1.5 ml of FMC media and grown to an OD600=0.5. At this point, these cultures were diluted 1 : 50 into 175 μl FMC in individual wells of a 96-well plate (black walls, clear bottoms; Corning). The optical density at 600 nm (OD600) and green fluorescence were monitored (sensitivity=45; excitation=485 nm; emission=520 nm) at 30 min intervals. To calculate relative expression, the fluorescence of wild-type harbouring plasmid without the reporter gene fusion was subtracted from fluorescence readings of the S. mutans strains harbouring the Plrg-gfp or Pcid-gfp gene fusion.
5′ RACE
The cid and lrg transcription start sites (TSSs) were mapped using a classical 5′ rapid amplification of cDNA ends (5′ RACE) on total mRNA extracted from S. mutans UA159 culture, grown in BHI medium to an OD600 of 0.5. We conducted 5′ RACE according to a protocol provided by Life Technologies (Carlsbad, CA, USA). Briefly, cDNA was generated with 5′ RACE-GSP (for cid, 5′-GGACAACTAAAGCAATCACTGCAA-3′; for lrg, 5′-GCGACCAACTGCAATCCTTC-3′) using the iScript Select cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). Product was purified via the Zymo (Irvine, CA, USA) Clean and Concentrator-5 kit and used in a homopolymeric tailing reaction with 2 mM dCTP and 15 units of TdT (Invitrogen). The tailing reaction product was then further amplified using several nested gene-specific primers and anchor primers as specified in the supplier’s protocol. PCR products were analysed on 1 % agarose gels by electrophoresis. When a suitable amount of product could be visualized after electrophoresis, PCR products were purified and Sanger sequenced by Genewiz (South Plainsfield, NJ, USA). The TSSs were then determined from sequencing results by locating the beginning of the homopolymeric tailing sequence.
Electrophoretic mobility shift assays (EMSAs)
EMSAs were performed as described elsewhere [35, 36]. Briefly, DNA probes containing the promoter regions of lrg or cid were PCR-amplified by primers labelled with biotinylated nucleotides on their 5′ end. To introduce mutations into one or both predicted cre site(s) in each of the cid and lrg promoter regions, biotinylated DNA probes were amplified with primers with the desired base changes in the promoter sequence. All mutated PCR products were confirmed by DNA sequencing. His6-tagged CcpA [26] was overexpressed in E. coli by induction with isopropyl-β-D-thiogalactopyranoside (IPTG) and purified as a soluble protein using Ni-NTA agarose (Qiagen) as recommended by the supplier. For EMSA reactions, each biotin-labelled DNA probe was used with different concentrations of purified CcpA in a 10 µl reaction mixture containing 10 mM HEPES (pH 7.8), 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM EDTA, 1 µg poly(dI-dC), 1 µg bovine serum albumin (BSA) and 10 % glycerol. The binding reaction was performed at room temperature for 30 min, followed by separation of the DNA-protein samples in a 4.5 % non-denaturing, low-ionic strength polyacrylamide gel. The samples were transferred to a Nylon membrane (Thermo Scientific) using a semi-dry transfer apparatus (Bio-Rad). The membrane was air-dried and then exposed to UV light to cross-link biological materials on the membrane. DNA signals were detected using a chemiluminescent nucleic acid detection module kit (Thermo Scientific) as recommended by the supplier.
DNase I footprinting assay
DNase I footprinting assays were carried out using a non-radiochemical capillary electrophoresis method [37]. To detect CcpA binding site(s) in the promoter regions of lrg and cid, DNA probes containing the promoter regions of lrg or cid were amplified by primers labelled with 6-FAM fluorescence on their 5′ end and biotinylated nucleotides on their 3’ end. The amplified labelled DNA fragments were incubated with His6-tagged CcpA at room temperature for 30 min in a 50 µl reaction mixture identical to that used for EMSAs. Digestion of DNase I (0.1 unit) was carried out at 37 °C for 2 min, the enzyme reaction was inhibited by adding EDTA to a final concentration of 60 mM and the samples were heated at 80 °C for 10 min. The samples were purified using a MinElute Reaction Cleanup Kit (Qiagen), vacuum-dried, re-suspended in 10 µl Hi-Di formamide (Applied Biosystems, USA) and subjected to capillary electrophoresis by loading into a 3730xl DNA Analyzer with GeneScan 600 LIZ dye (Applied Biosystems, USA) for size standard. Peak Scanner Software v1.0 was used to analyse electropherograms.
Measurement of extracellular glucose levels
S. mutans UA159 was grown in FMC medium, supplemented by 11 mM glucose. For time course measurements of extracellular glucose during growth, samples (200 µl) were taken at 1–2 h intervals. Half of the harvested volume was used to measure its optical density at 600 nm in a spectrophotometer for monitoring of culture growth. The other half (100 µl) was centrifuged for 2 min at 13 400 r.p.m. to remove the cells, and the supernatant was used to determine the concentration of glucose with a glucose (HK) assay kit (Sigma-Aldrich), according to the manufacturer’s instructions.
Results
Promoter activities of lrg and cid display distinct expression patterns in a glucose-dependent manner
Previous studies in our laboratory revealed that lrg and cid RNA levels were regulated in an opposing manner in response to growth phase and glucose levels [9]. To better understand this expression pattern in a continuous fashion during growth, expression of cid and lrg was monitored via gfp trancriptional fusions in the wild-type UA159 strain. Real-time measurement of gfp expression was performed in a plate reader. We first confirmed the opposing response of these lrg and cid promoter fusions to initial culture glucose levels. The UA159/Plrg- gfp strain produced gfp fluorescence only when grown in media containing a low level (11 mM) of glucose (Fig. S1a, available in the online version of this article), whereas the UA159/Pcid - gfp strain produced gfp only in a high-glucose (45 mM) culture (Fig. S1b). Remarkably, lrg promoter activity was sharply induced in the low-glucose culture at the onset of the stationary phase (Fig. S1a). A similar pattern was also observed in cultures containing 5 mM glucose (Fig. S2a). Sharp stationary phase induction of lrg promoter activity was reduced in cultures containing 20 mM glucose (Fig. S2b), and no induction was observed at ≥30 mM glucose (Fig. S2c–d). In contrast, cid promoter activity steadily increased as cells grew in the high-glucose (45 mM) medium (Fig. S1b). This pattern was also obvious in cultures grown with ≥30 mM glucose (Fig. S2c, d). Intriguingly, in cultures grown with glucose at a concentration of 20 mM, both lrg and cid were simultaneously induced (Fig. S2b) but at lower levels than in the presence of low and high glucose, respectively (Fig. S1). These observations confirm a strict dependence of cid and lrg promoter activity on both initial glucose levels in the media, as well as growth phase.
Stationary phase lrgAB induction in low-glucose cultures coincides with depletion of glucose
The fact that the promoter of lrgAB is dramatically activated in low-glucose cultures as cells enter stationary phase (Fig. S1a) implies that LrgAB may be important for survival of cells when exogenous carboydrate has been depleted. When we measured the concentration of extracellular glucose during growth of the wild-type strain in the low-glucose (11 mM) medium, most glucose was rapidly depleted during exponential growth (Fig. S3). Strong induction of lrgAB expression in BHI (low-glucose) cultures is also demonstrated by differential expression analysis of previously published RNA microarray data (GEO Accession #GSE39470), comparing wild-type expression profiles between early- and late-exponential growth phases [11]. When the expression data were reconstituted for comparison of early- vs late-exponential growth phases in the wild type, it was shown that lrgAB (SMU.575c-574c) was dramatically upregulated (about >900-fold) at late-exponential phase, compared to that of early-exponential growth phase (Table S1). A four-gene operon (SMU.1421–SMU.1424), encoding the components of the pyruvate dehydrogenase complex (PDH), was also remarkably upregulated (by >284-fold) at late-exponential phase, compared to that of early-exponential phase (Table S1), suggesting that LrgAB may be related to pyruvate metabolism.
Both lrg and cid are regulated by direct binding with CcpA
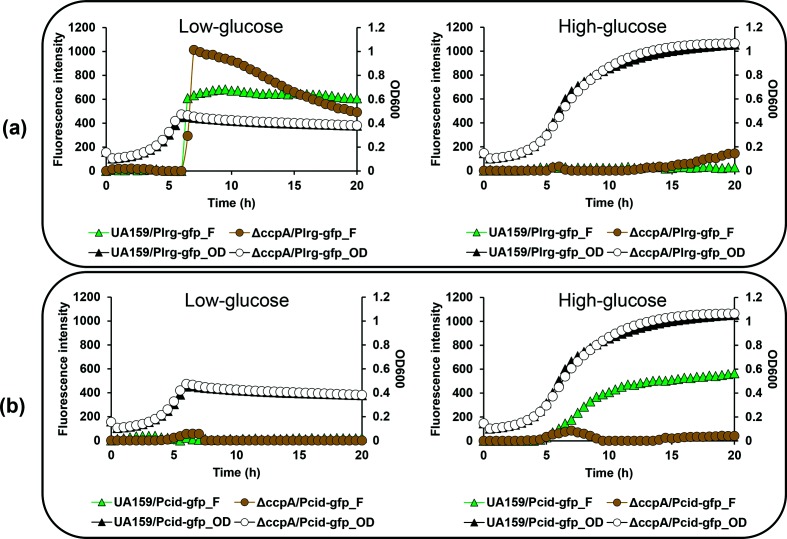
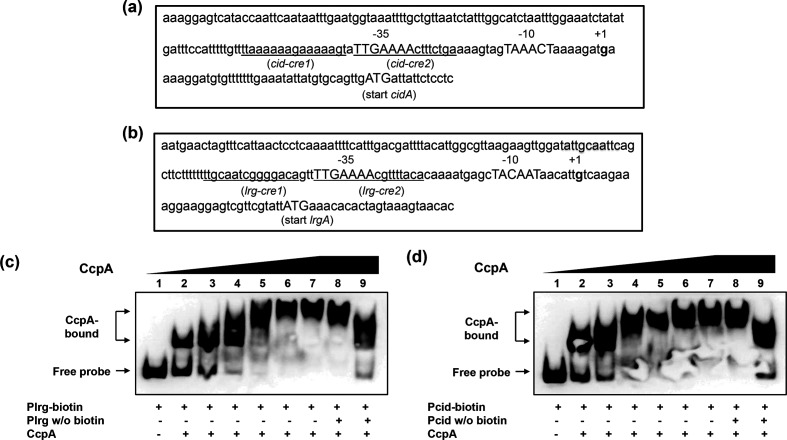
Our previous northern blot analysis [9] demonstrated that lrg and cid expression is regulated by CcpA, the master regulator of carbon metabolism [11, 26]. Two catabolite-responsive element (cre)-like consensus sites, predicted binding sequences for CcpA [25], were previously predicted in the lrgAB promoter region but no well-conserved cre sequences were found in the promoter region of cidAB [9]. Consistent with this observation, the bioinformatics program RegPrecise (http://regprecise.lbl.gov/RegPrecise/regulon.jsp?regulon_id=35145) does not predict cidAB (SMU1701c/1700c) as belonging to the CcpA regulon of S. mutans UA159. Nevertheless, the fact that expression of cid was completely repressed by the presence of ≤10 mM glucose still leaves open the potential for involvement of glucose-mediated, CcpA-dependent catabolite repression. This idea was further supported by our recent real-time qPCR data, showing that cid expression is regulated by CcpA [12]. In that study, however, lack of CcpA had no significant influence on the regulation of lrg over growth [12]. To further investigate the relationship between cid and lrg promoter activity and CcpA, the pDL278 Plrg - gfp and Pcid- gfp constructs were transformed into a ccpA mutant strain (Table 1), and GFP fluorescence from each promoter was monitored in low- and high-glucose cultures (Fig. 1). Expression of lrg promoter activity rose sharply at early-stationary phase in both the wild-type and ccpA mutant backgrounds. However, this induction was increased by about 60 % in the ccpA mutant low-glucose culture compared to expression in the wild-type strain under the same growth condition (Fig. 1a, left). No obvious difference in lrg promoter activity between wild-type and ccpA mutant was observed in the high-glucose culture (Fig. 1a, right). In contrast, cid promoter activity in the high-glucose culture was dramatically repressed in the ccpA mutant strain (Fig. 1b, right) and no significant induction was observed in the low-glucose culture (Fig. 1b, left). These data confirm that lack of CcpA has an effect on cid promoter activity, although the detailed mechanism remains to be elucidated. It is possible that CcpA may indirectly play a role in regulation of cid, or that a less-conserved CcpA binding site may be present in the cid promoter region. When we re-searched for the potential target(s) for CcpA in the cid promoter region, by using the cre consensus sequence, WTGNAANCGNWNNCW [25] and the CLUSTALW program (http://www.genome.jp/tools-bin/clustalw), two potential CcpA binding sites were identified, one at −75 bp (designated cid-cre1) and the other at −59 bp (designated cid-cre2) upstream of the ATG start codon of cidA (Fig. 2a). Although these two potential binding sites are relatively less conserved with respect to the consensus sequence (Fig. 2a), they are arranged in a similar manner to those previously identified in the lrg promoter region (Fig. 2b) [9]. For example, each pair of two predicted cre sites is separated by 1 nt and 3 nt in the cid and lrg promoter regions, respectively. To further characterize the predicted cre sites, we mapped the 5′ termini of the mRNAs of cid and lrg genes using the 5′-RACE technique and determined the potential transcription start sites (TSSs, denoted as +1) and −10/–35 elements in the cid (Fig. 2a) and lrg (Fig. 2b) promoter regions. Intriguingly, the sites closer to the ATG start codon (cid-cre2 and lrg-cre2) overlap the predicted −35 regions of their respective promoters (Fig. 2a, b). We next examined whether CcpA can directly bind to the cid and lrg promotor regions by performing electrophoretic mobility shift assays (EMSAs) with increasing amounts of purified CcpA-His and biotin-labelled cid or lrg promoter regions each containing their respective predicted cre sites. As shown in Fig. 2c, d (lanes 2–7, respectively), a relatively low concentration of CcpA protein (>1 pmol) was able to impede migration of both labelled promoter fragments. The addition of unlabelled (cold) probes to each reaction mixture was able to reduce the presence of the CcpA-shifted cid and lrg promoters, (Fig. 2c, d, lanes 8–9, respectively), confirming that CcpA binds specifically to the promoter regions of lrg and cid.
Fig. 1.
Effect of CcpA on S. mutans lrg and cid promoter activity over the growth in the low- and high-glucose cultures. The Plrg - gfp (a) and Pcid- gfp (b) constructs in pDL278 were created in the S. mutans UA159 (wild-type) and ∆ccpA (ccpA-deficient) strains. The strains were grown in a chemically defined medium (FMC) supplemented by 11 mM (left) or 45 mM (right) glucose. Relative gfp fluorescence intensity (coloured lines; F) and OD600 (black lines; OD) were monitored on a plate reader (see Methods for details). The results are representative of three independent experiments.
Fig. 2.
The role of CcpA in promoters cid and lrg. a–b; The promoter regions of cid (a) and lrg (b). The 5′ termini of the cid and lrg coding region transcripts were mapped using 5′ RACE. The potential transcriptional start sites (TSSs) are numerically denoted (+1) in bold, and −10/–35 sequences are indicated by upper case letters. The hypothetical cre sites underlined. c–d; Electrophoretic mobility shift assays (EMSAs) of CcpA binding with the promoter region of the lrg (c) or cid (d) gene. Biotin-labelled promoter regions (4 fmol) of lrg (a) and cid (b) were incubated with increasing amounts of purified His-CcpA protein (0, 1.5625, 3.125, 6.25, 12.5, 25 and 50 pmol). Unlabelled promoter regions (0.1 and 1 pmol) were added to the binding reaction mixtures (lanes 8 and 9). The reactions were run on a non-denaturing polyacrylamide gel and the signal observed via chemiluminescence.
Identification of binding sites for CcpA in the cid promoter region
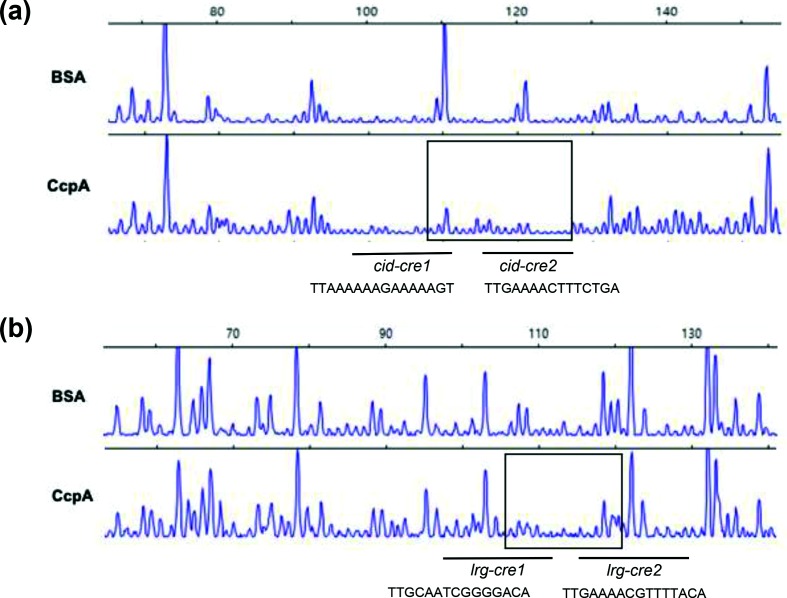
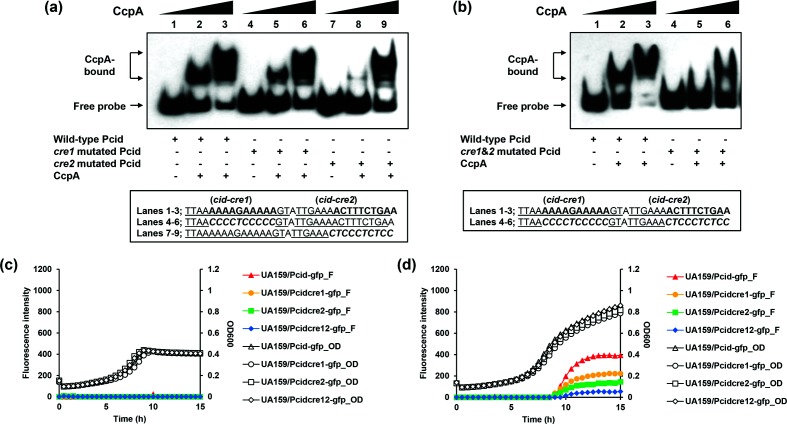
EMSAs showed that CcpA could directly bind to the promoter region of cid and lrg (Fig. 2c, d). To identify the precise location of binding by CcpA on the cid promoter region, DNase I footprinting experiments using capillary electrophoresis (fragment analysis) were conducted using a PCR-amplified 6-carboxyfluorescein (6-FAM)-labelled DNA probe of the cid promoter region (201 bp) bound to CcpA under the same conditions as used for EMSAs (Fig. 3a). Sites protected from DNase I digestion by CcpA were visualized as regions lacking discernable peaks compared to a control reaction mixture containing BSA (bovine serum albumin). Reaction mixtures containing CcpA yielded only a single region of protection spanning from −77 to −59, with respect to the ATG start site of the cidA gene. This protected region largely corresponds to the predicted cid-cre2 site (nt −73/–59) (Figs 2a and 3a), implicating its involvement in binding with CcpA. To investigate the requirement of this protected region in DNA binding by CcpA, mutations were introduced into each cid-cre site by substituting 10 nucleotides in the 3′ end of cid-cre1 and cid-cre2, followed by DNA mobility shift assays with CcpA. The DNA fragment containing the mutated cid-cre2 sequence affected the proportion of DNA shifted by CcpA more than the DNA fragment containing the mutated cid-cre1 sequence, relative to the wild-type cid promoter sequence, suggesting that cid-cre2 may be a higher-affinity cre site for CcpA (Fig. 4a). When both cid-cre1 and cid-cre2 were mutated, the shift of the DNA-CcpA complex was much less than that observed in each single cre mutation, suggesting that both cid-cre1 and cid-cre2 are required for full CcpA binding (Fig. 4b). To further evaluate whether less efficient binding of CcpA to the mutated cid promoter impacts its promoter activity in vivo, cid promoter regions were constructed containing mutated cid-cre1 or cid-cre2 alone, or both mutated sequences together. Each of these promoter constructs was used to replace the wild-type cid promoter (Pcid) in the UA159/Pcid-gfp reporter strain, generating UA159/Pcidcre1-gfp, UA159/Pcidcre2-gfp and UA159/Pcidcre12-gfp. In low-glucose cultures, all cid cre mutant promoters (as measured by GFP fluorescence) showed a level of activity comparable to the unmodified cid promoter, indicating that the cre elements do not influence cid expression during low-glucose growth (Fig. 4c). However, in high-glucose cultures, cid promoter activity was greatly reduced in the cid-cre2 mutant promoter, and to a lesser extent in the cid-cre1 mutant promoter (Fig. 4d). When both cid-cre1 and cid-cre2 were mutated, the reduction in cid promoter activity was greater than that observed in each single cre mutation (Fig. 4d). Therefore, these results suggest that CcpA binding to both cid-cre1 and cid-cre2 sites may be required for full induction of cid expression in high-glucose cultures. Taken together, the data show that the cid-cre2 site seems more important in regard to CcpA binding relative to the cid-cre1 site. Nevertheless, the observation that cre mutations alone did not completely inhibit CcpA binding suggests that additional allosteric effector(s) may be needed for full binding of CcpA to the cid promoter region.
Fig. 3.
DNase I footprinting assays of CcpA binding to the cid and lrg promoter regions. Solid boxes depict the regions protected from DNase I digestion upstream of cid (a) and lrg (b) by CcpA. The electropherograms represent control DNA with BSA (bovine serum albumin) in the upper panels and footprints with of CcpA in the lower panels.
Fig. 4.
Effects of putative cre site mutations in the cid promoter region. (a–b); EMSA. Underlined letters indicate two putative CcpA-binding (cid-cre1 and cid-cre2) sequences. Italic letters indicate the mutated sequence regions. EMSA was performed with 1 fmol biotinylated cid promoter (Pcid) regions with mutation in the putative cre sequences and various amounts of purified His-CcpA (0, 0.78125 or 1.5625 pmol). The reactions were run on a non-denaturing polyacrylamide gel and the signal observed by chemiluminescence. (a) Lanes 1–3 contain biotinylated Pcid wild-type DNA probes; lanes 4–6 contain biotinylated Pcid-cre1 mutated DNA probes; and lanes 7–9 contain biotinylated Pcid-cre2 mutated DNA probes. (b) Lanes 1–3 contain biotinylated Pcid wild-type DNA probes; lanes 4–6 contain biotinylated Pcid-cre1&2 mutated DNA probes. (c–d); Effects of cre mutations on cid expression over growth. The S. mutans UA159 (wild-type) strains contained pDL278 carrying the gfp gene driven by wild-type and mutated Pcid. The strains were grown in a chemically defined medium (FMC) supplemented by either a low [11 mM, (c)] or high level [45 mM, (d)] of glucose. Relative gfp fluorescence intensity (coloured lines; F) and OD600 (black lines; OD) were monitored on a plate reader (see Methods for details). The results are the average values for at least three biological replicates performed in triplicate.
Identification of binding sites for CcpA in the lrg promoter region
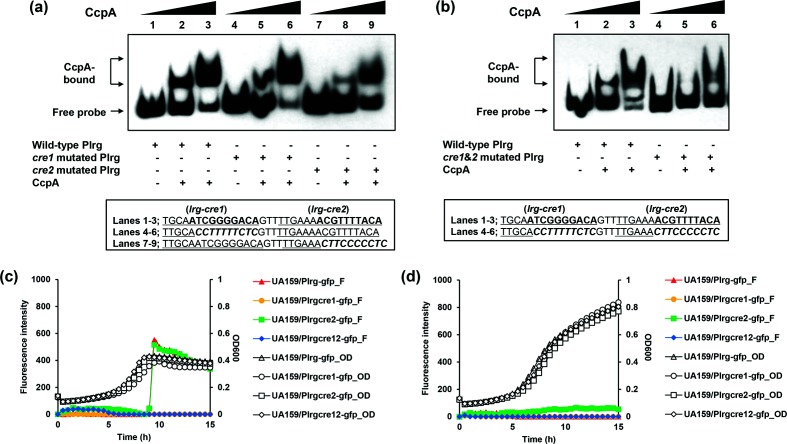
Unlike the cid-cre sites (Fig. 2a), two cre-like consensus elements, previously identified in the lrg promoter region [9] and designated lrg-cre1 (TTGCAATCGGGGACA) and lrg-cre2 (TTGAAAACGTTTTACA), appeared to possess more sequence conservation (Fig. 2b) relative to the cre consensus sequence (WTGNAANCGNWNNCW) [25]. However, the DNase I footprinting experiment on the lrg promoter region (201 bp) revealed that the region protected by CcpA was less distinctive than that of the cid promoter region, spanning the 3′ region of the predicted lrg-cre1 site and 5′ region of the lrg-cre2 site (from −70 to −55 with respect to the ATG start site of the lrgA gene; Fig. 3b). Similar to the cid-cre2 element, the 5′ region of lrg-cre2 also overlapped the predicted −35 element (Fig. 2b). When the 3′ regions of lrg-cre1 and lrg-cre2 were each mutated and subjected to DNA mobility shift assays with CcpA, the lrg-cre2 mutation strongly decreased the affinity of CcpA for the lrg promoter while the effect of lrg-cre1 mutation was minimal (Fig. 5a), suggesting that lrg-cre2 functions as a primary binding site for CcpA. Importantly, however, when both lrg-cre1 and lrg-cre2 were mutated, the shift of the DNA-CcpA complex was more profoundly affected than that observed in each single cre mutation (Fig. 5b), similar to the observation in the cid operon (Fig. 4b). To evaluate whether less binding of CcpA to the mutated lrg promoter impacted its activity in vivo, mutant lrg promoter regions that have mutations in either cre1, cre2 or both were cloned in the same gfp transcriptional reporter as used above (Fig. S1). Real-time measurement of GFP expression in UA159 was performed in the presence of low and high glucose levels. Interestingly, although the cre2 mutated lrg promoter showed less binding of CcpA than the wild-type promoter by EMSA (Fig. 5a), there was no difference in the promoter activity of lrg compared with the wild-type promoter in the presence of low glucose (Fig. 5c). In contrast, when the mutated promoter on lrg-cre1 (which did not appear to be necessary for CcpA binding by EMSA) was used, the promoter activity dramatically decreased in the presence of low glucose (Fig. 5c). In the presence of high glucose, the lrg-cre2 mutant promoter showed slightly increased activity compared with wild-type and lrg-cre1 mutated promoters (Fig. 5d). Therefore, CcpA may repress the expression of lrg by binding to the lrg-cre2 site in the presence of high glucose, and the lrg-cre1 site may affect binding of another transcriptional regulator that acts as an activator of the lrg promoter in the presence of low glucose.
Fig. 5.
Effects of putative cre site mutations in the lrg promoter region. (a–b); EMSA. Underlined letters indicate two putative CcpA-binding (lrg-cre1 and lrg-cre2) sequences. Italic letters indicate the mutated sequence regions. The EMSA was performed with 1 fmol biotinylated lrg promoter (Plrg) region with mutation in the putative cre sequences and various amounts of purified His-CcpA (0, 0.78125 or 1.5625 pmol). The reactions were run on a non-denaturing polyacrylamide gel and the signal observed by chemiluminescence. (a) Lanes 1–3 contain biotinylated Plrg wild-type DNA probes; lanes 4–6 contain biotinylated Plrg-cre1 mutated DNA probes; and lanes 7–9 contain biotinylated Plrg-cre2 mutated DNA probes. (b) Lanes 1–3 contain biotinylated Plrg wild-type DNA probes; lanes 4–6 contain biotinylated Plrg-cre1&2 mutated DNA probes. (c–d); Effects of cre mutations in the lrg expression over the growth. The S. mutans UA159 (wild-type) strains contain pDL278 carrying the gfp gene driven by wild-type and mutated Plrg. The strains were grown in a chemically defined medium (FMC) supplemented by low [11 mM, (c)] or high levels (45 mM, (d)] of glucose. Relative gfp fluorescence intensity (coloured lines; F) and OD600 (black lines; OD) were monitored on a plate reader (see Methods for details). The results are the average values from at least three biological replicates performed in triplicate.
Discussion
In this study, the regulation of S. mutans cid and lrg operons by CcpA and culture glucose levels was investigated. Due to the high sensitivity and opposite expression patterns that these operons exhibit in response to growth phase and glucose concentration, as well as oxygenation [9], it has been generally assumed that lrg and cid expression is tightly regulated and closely associated with metabolic pathways of the organism. We have previously demonstrated the potential involvement of CcpA in the regulation of cid and lrg [9, 12], though the mechanisms were not elucidated. In this present study, we showed that CcpA is involved in the regulation of both cid and lrg expression in an opposite manner, and this regulation is exerted by direct binding of CcpA to the promoter regions of cid and lrg. More interestingly, the predicted binding sequences for CcpA are arranged in a similar manner in both cid and lrg promoter regions and overlap the predicted −35 regions of both cid and lrg promoters, suggesting that CcpA regulates expression of the cid and lrg operons, possibly by direct interaction with RNA polymerase, and further supporting a potential functional interplay between cid and lrg.
It is notable that CcpA efficiently bound both the cid and lrg promoters but has the opposite effect on the regulation of cid (positive) and lrg (negative). Particularly in the cid promoter region, binding of CcpA to both cid-cre1 and cid-cre2 sites appears be required for full induction of cid expression during growth in high-glucose culture, because deficiency of either CcpA itself or deletion of both its binding sites almost completely abolishes cid promoter activity in this culture condition. Given that mutation of the cid-cre2 site led to a greater reduction of CcpA binding in EMSA assays relative to mutation of cid-cre1, CcpA may regulate cid expression primarily through the binding to cid-cre2 and then a less-conserved cid-cre1 site. However, mutation of either cre site decreases cid promoter activity in S. mutans high-glucose cultures. Furthermore, given that the cid-cre2 site overlaps the −35 element of the cid promoter, it still remains to be elucidated how CcpA could positively regulate cid expression. In contrast, elevated lrg promoter activity observed in low-glucose ccpA mutant cultures (and to a lesser extent in the high-glucose culture) suggests that CcpA functions as a repressor when bound to the lrg promoter region. In fact, increased lrg expression in the high-glucose culture of the ccpA-deficient strain was more evident in our previous northern blot analysis [9], which could indicate an effect of CcpA on in vivo lrg promoter activity and/or mRNA stability, especially as cells transition to stationary growth phase. It is also noteworthy that the lrg-cre2 mutation, showing a greater reduction for CcpA binding by EMSA, had little impact on lrg promoter activity, while the lrg-cre1 mutation, showing a negligible influence on CcpA binding, could completely repress lrg expression in low-glucose culture conditions. One possible explanation for this discrepancy could be difficulty in mimicking in vivo conditions that modulate cre-dependent CcpA promoter binding using in vitro assays (i.e. EMSA), such as the possible requirement for HPr-Ser46-P, or certain allosteric effector molecule(s). Alternatively, the observation that the lrg-cre1 site was required for lrg expression, but not for CcpA binding, suggests that the lrg-cre1 site may be actually involved in the regulation by LytST, previously shown to positively regulate lrg expression [9]. This idea is further supported by a previous generic phylogenic footprinting/shadowing analysis, predicting a specific operator motif associated with the LytTR-family of response regulators [38, 39]. In this study, LytT was predicted to bind a DNA sequence composed of direct repeats separated by 10–11 nt and, notably, a predicted second LytT binding motif (TGCAATCGGG) overlaps the lrg-cre1site. We are currently investigating whether the lrg-cre1 site mediates binding with LytT and how LytST contributes to regulation of cid and lrg expression in S. mutans. Nevertheless, the promoter experiments presented here suggest that binding of CcpA to its target sequence (probably lrg-cre2) may be optimal when pools of intermediates of glycolysis, such as fructose-1,6-bisphosphate (F-1,6-BP) and glucose-6-phosphate (G6P), are increased. These glycolytic intermediates could activate an HPr kinase/phosphorylase that uses ATP to phosphorylate HPr [23, 30], a histidine protein that functions as a binding partner of CcpA for regulation [40, 41]. Taken together, these observations suggest that CcpA binding is an important mediator of cid and lrg transcriptional responses to glucose levels. Nevertheless, we recognize that additional experiments are required to fine-tune our understanding of the involvement of CcpA in regulating cid and lrg expression, because the effects of CcpA on cid and lrg promoter activities appear to be sensitive to the metabolic status of the cell and growth parameters. For example, CcpA was observed to function as a repressor for cid expresson in the low-glucose condition in our previous northern blot analysis [9], which may be due to different culture media (complex Todd–Hewitt broth vs. chemically defined FMC), or to an effect of CcpA on cid RNA stability vs. cid promoter activity. However, this study offers a potential insight toward understanding metabolic linkages with cid and lrg operons.
In conclusion, our data suggest that the Cid/Lrg system integrates signals associated with the metabolic status of the cell and surrounding environments through direct interaction with CcpA. Consequently, it contributes to the ability of S. mutans to preserve normal cellular homeostasis during periods of stress, e.g. during periods of ‘feast of famine’ in the oral cavity. In fact, involvement of CcpA adds another layer of complexity in the regulation of cid and lrg, because we previously showed that at least two TCSs (LytST and VicRK) can also regulate cid and lrg expression [9, 12]. Thus, an interesting future study will be the investigation of how these regulators coordinate cid and lrg expression, and which signals, probably metabolic intermediates or end products, drive the regulatory interactions between these TCS. These signals may be linked to the bacterial cell death and lysis process, determining the fate of the cell. In this regard, it is noteworthy that in recent studies, the B. subtilis ysbA/pftA (a homologue of lrgA) and lytS genes were involved in pyruvate utilization [42, 43]. In this study, pyruvate was hypothesized to be excreted as an overflow metabolite, and possibly consumed after glucose is depleted, when ysbA (lrgA) expression is maximal. Consistent with this observation is that lrg promoter activity is dramatically induced as cells transition to stationary phase in the low-glucose media (Fig. S1a and Table S1). In our preliminary experiments, we have found that S. mutans does not utilize pyruvate as its sole carbon source (data not shown). However, the fact that pdh genes were simultaneously drastically induced with lrg at stationary phase implies that Lrg may be related to pyruvate metabolism at a certain point of this metabolic pathway. We further envision that cid and lrg may respond to pyruvate as a metabolic determinant that influences the fate of cells within a population to lyse under unfavourable conditions, which is currently under investigation. Taken together, this study provides new insights into how the regulation of cid and lrg expression mediates the response of S. mutans to the unpredictable and constantly changing oral cavity environment, as well as improves our understanding of potential functional links between cell death/lysis and metabolism.
Supplementary Data
Funding information
This work was supported by the National Institutes of Health (NIH) National Institute of Dental and Craniofacial Research (NIDCR) grant R01 DE025237 (S. -J. A.).
Acknowledgements
We thank Professor Robert A. Burne (Department of Oral Biology, University of Florida) for providing all the resources needed. We also thank Dr Jung-Nam Kim (Department of Microbiology, Pusan National University, Pusan, South Korea) for technical assistance with DNase I footprinting analysis.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No human/animal experiments are involved in this study.
Footnotes
Abbreviations: BHI, brain heart infusion; BSA, bovine serum albumin; CcpA, catabolite response protein A; Cre, catabolite-responsive element; EMSA, electrophoretic mobility shift assay; 6-FAM, 6-carboxyfluorescein; FBP, fructose-1,6-bisphosphate; FMC, chemically defined medium; G6P, glucose-6-phosphate; LB, Luria-Bertani; TCS, two-component signal transduction system.
One supplementary table and three supplementary figures are available with the online version of this article.
Edited by: M. Vickerman and J. Stülke
References
- 1.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burne RA, Ahn SJ, Wen ZT, Zeng L, Lemos JA, et al. Opportunities for disrupting cariogenic biofilms. Adv Dent Res. 2009;21:17–20. doi: 10.1177/0895937409335593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burne RA, Zeng L, Ahn SJ, Palmer SR, Liu Y, et al. Progress dissecting the oral microbiome in caries and health. Adv Dent Res. 2012;24:77–80. doi: 10.1177/0022034512449462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26 doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayles KW. Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol. 2003;11:306–311. doi: 10.1016/S0966-842X(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 6.Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 7.Bayles KW. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol. 2014;12:63–69. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn SJ, Gu T, Koh J, Rice KC. Remodeling of the Streptococcus mutans proteome in response to LrgAB and external stresses. Sci Rep. 2017;7:14063. doi: 10.1038/s41598-017-14324-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahn SJ, Rice KC, Oleas J, Bayles KW, Burne RA. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology. 2010;156:3136–3147. doi: 10.1099/mic.0.039586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice KC, Turner ME, Carney OV, Gu T, Ahn SJ. Modification of the Streptococcus mutans transcriptome by LrgAB and environmental stressors. Microb Genom. 2017;3:e000104. doi: 10.1099/mgen.0.000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn SJ, Qu MD, Roberts E, Burne RA, Rice KC. Identification of the Streptococcus mutans LytST two-component regulon reveals its contribution to oxidative stress tolerance. BMC Microbiol. 2012;12:187. doi: 10.1186/1471-2180-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn SJ, Rice KC. Understanding the Streptococcus mutans Cid/Lrg System through CidB Function. Appl Environ Microbiol. 2016;82:6189–6203. doi: 10.1128/AEM.01499-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young R. Bacteriophage holins: deadly diversity. J Mol Microbiol Biotechnol. 2002;4:21–36. [PubMed] [Google Scholar]
- 15.Young R, Bläsi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 16.Bayles KW. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 2000;8:274–278. doi: 10.1016/S0966-842X(00)01762-5. [DOI] [PubMed] [Google Scholar]
- 17.Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. 2008;72:85–109. doi: 10.1128/MMBR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shemesh M, Tam A, Kott-Gutkowski M, Feldman M, Steinberg D. DNA-microarrays identification of Streptococcus mutans genes associated with biofilm thickness. BMC Microbiol. 2008;8:236. doi: 10.1186/1471-2180-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol. 2000;182:1794–1801. doi: 10.1128/JB.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice KC, Bayles KW. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol Microbiol. 2003;50:729–738. doi: 10.1046/j.1365-2958.2003.t01-1-03720.x. [DOI] [PubMed] [Google Scholar]
- 21.Rice KC, Firek BA, Nelson JB, Yang SJ, Patton TG, et al. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol. 2003;185:2635–2643. doi: 10.1128/JB.185.8.2635-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henkin TM. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 23.Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 25.Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 2000;28:1206–1210. doi: 10.1093/nar/28.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, et al. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol. 2008;190:2340–2349. doi: 10.1128/JB.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 28.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 29.Deutscher J, Saier MH. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci USA. 1983;80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saier MH. Cyclic AMP-independent catabolite repression in bacteria. FEMS Microbiol Lett. 1996;138:97–103. doi: 10.1111/j.1574-6968.1996.tb08141.x. [DOI] [PubMed] [Google Scholar]
- 31.Terleckyj B, Shockman GD. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect Immun. 1975;11:656–664. doi: 10.1128/iai.11.4.656-664.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol. 2012;86:258–272. doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Son M, Ghoreishi D, Ahn SJ, Burne RA, Hagen SJ. Sharply tuned pH Response of genetic competence regulation in Streptococcus mutans: a microfluidic study of the environmental sensitivity of comX. Appl Environ Microbiol. 2015;81:5622–5631. doi: 10.1128/AEM.01421-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen ZT, Burne RA. Analysis of cis- and trans-acting factors involved in regulation of the Streptococcus mutans fructanase gene (fruA) J Bacteriol. 2002;184:126–133. doi: 10.1128/JB.184.1.126-133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JN, Burne RA. CcpA and CodY coordinate acetate metabolism in Streptococcus mutans. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.03274-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng L, Dong Y, Burne RA. Characterization of cis-acting sites controlling arginine deiminase gene expression in Streptococcus gordonii. J Bacteriol. 2006;188:941–949. doi: 10.1128/JB.188.3.941-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yindeeyoungyeon W, Schell MA. Footprinting with an automated capillary DNA sequencer. Biotechniques. 2000;29:1034–1036, 1038, 1040–1031. doi: 10.2144/00295st05. [DOI] [PubMed] [Google Scholar]
- 38.de Been M, Bart MJ, Abee T, Siezen RJ, Francke C. The identification of response regulator-specific binding sites reveals new roles of two-component systems in Bacillus cereus and closely related low-GC Gram-positives. Environ Microbiol. 2008;10:2796–2809. doi: 10.1111/j.1462-2920.2008.01700.x. [DOI] [PubMed] [Google Scholar]
- 39.Francke C, Kerkhoven R, Wels M, Siezen RJ. A generic approach to identify Transcription Factor-specific operator motifs; inferences for LacI-family mediated regulation in Lactobacillus plantarum WCFS1. BMC Genomics. 2008;9:145. doi: 10.1186/1471-2164-9-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weickert MJ, Chambliss GH. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HM, Park YH, Yoon CK, Seok YJ. Histidine phosphocarrier protein regulates pyruvate kinase A activity in response to glucose in Vibrio vulnificus. Mol Microbiol. 2015;96:293–305. doi: 10.1111/mmi.12936. [DOI] [PubMed] [Google Scholar]
- 42.Charbonnier T, Le Coq D, McGovern S, Calabre M, Delumeau O, et al. Molecular and physiological logics of the pyruvate-induced response of a novel transporter in Bacillus subtilis. MBio. 2017;8 doi: 10.1128/mBio.00976-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Esker MH, Kovács ÁT, Kuipers OP. YsbA and LytST are essential for pyruvate utilization in Bacillus subtilis. Environ Microbiol. 2017;19:83–94. doi: 10.1111/1462-2920.13454. [DOI] [PubMed] [Google Scholar]
- 44.LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid. 1992;28:130–145. doi: 10.1016/0147-619X(92)90044-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.