Abstract
The needle structures of type III secretion (T3S) systems are formed by the secretion and polymerization of a needle subunit protein, YscF in Yersinia pestis. A subset of T3S systems employ unique heterodimeric chaperones, YscE and YscG in Y. pestis, to prevent the polymerization of needle subunits within the bacterial cell. We demonstrate that the YscE/YscG chaperone is also required for stable YscF expression and for secretion of YscF. Overexpression of a functional maltose-binding protein (MBP)–YscG hybrid protein stabilized cytoplasmic YscF but YscF was not secreted in the absence of YscE. Furthermore, a YscE mutant protein was identified that functioned with YscG to stabilize cytosolic YscF; however, YscF was not secreted. These findings confirm a role for the YscE/YscG chaperone in YscF secretion and suggest that YscE may have a specific role in this process. Recent studies have shown that YscF deleted of its N-terminal 15 residues is still secreted and functional, suggesting that YscF may not require an N-terminal secretion signal. However, we demonstrate that YscF contains an N-terminal secretion signal and that a functional N-terminal signal is required for YscF secretion.
Keywords: Yersinia pestis, plague, type III secretion, chaperone, protein secretion
Introduction
Plague is a widespread zoonotic disease caused by the Gram-negative pathogen Yersinia pestis [1]. Transmission of Y. pestis between its natural, primarily rodent, hosts occurs almost exclusively through infected fleas; however, infection of domestic cats and other carnivores is often due to the ingestion of infected rodents [2]. Human plague can be acquired via the bite of infected fleas (bubonic or septicaemic plague) or via aerosolized bacteria (pneumonic plague) [3]. Infection of animal or human hosts with Y. pestis normally leads to an invasive and systemic infection that is often fatal in the absence of prompt antimicrobial treatment. In contrast, the other human pathogenic yersiniae (Y. enterocolitica and Y. pseudotuberculosis) are transmitted via the faecal–oral route and normally cause more localized infections (gastroenteritis) that are associated with low mortality rates compared to plague [4].
To survive and multiply in the blood and tissues of its host, Y. pestis employs a type III secretion (T3S) system to mediate the injection of at least seven Yersinia outer-protein (Yop) effector proteins into host phagocytic cells [5]. The injected Yops interfere with host signalling pathways that are required for bacterial phagocytosis and the production of pro-inflammatory cytokines. The T3S apparatus is a complex multicomponent structure that spans both the bacterial inner and outer membranes and is topped by a needle structure that extends 40 to 60 nm from the bacterial surface [6, 7]. The proteins that assemble the T3S apparatus are termed Yersinia secretion (Ysc) components. The external needle is assembled from a single secreted 87 amino acid residue protein termed YscF and is capped by a LcrV needle tip complex [8, 9]. Prior to export, YscF interacts with the YscE/YscG chaperone, which prevents YscF polymerization within the bacterial cell [10, 11]. After the assembly of the minimal functional T3S apparatus, the apparatus initially secretes the needle protein YscF and the rod protein YscI, as well as YscP [12]. The length of the needle is controlled by YscP, a molecular ruler, which works in conjunction with the inner-membrane T3S component YscU and likely the inner-rod component YscI [7, 13, 14]. YscP and YscU function together to direct a switch in the substrate specificity of the apparatus from needle-type proteins to translocator and/or Yop effector proteins upon the completion of needle assembly. After the switch in substrate specificity, premature release of the YopB/YopD translocators and Yop effectors is prevented by a YopN/SycN/YscB/TyeA complex [15, 16]. Upon contact with a eukaryotic cell, the secretion of YopN, YopB, YopD and the effector Yops is triggered [17, 18]. Secretion of YopB and YopD facilitates the assembly of the YopB/YopD translocon at the needle tip, which leads to the formation of a pore in the eukaryotic membrane for entry of the effector proteins [19]. The assembled YscF needle has also been demonstrated to play a specific role in Yop translocation [20] and in the regulation of the T3S process in response to calcium and/or cell-contact [20, 21].
A key event in the assembly of the T3S apparatus is the selective recognition, secretion and assembly of the external YscF needle structure; however, little is known about how needle-type substrates are recognized by the T3S apparatus. In the bacterial cytosol YscF is found in complex with the heterodimeric YscE/YscG chaperone [10]. The crystal structure of the Y. pestis heterotrimer YscEFG complex (Fig. 1a) [10] as well as the homologous Pseudomonas aeruginosa PscEFG complex [22] and the Aeromonas hydrophila AscEG complex [23] have been determined. YscG, PscG and AscG are tetratricopeptide repeat family proteins that directly bind the C-terminal half of YscF, PscF and AscF, respectively. YscE, PscE and AscE are small, primarily helical, peptides that interact directly with the N-terminal region of YscG, PscG or AscG, but make little contact with the needle subunit protein (YscF, PscF or AscF). PscE and AscE are thought to function primarily as co-chaperones that stabilize the structure of PscG and AscG [23, 24]. The N-terminal 49 residues of YscF are not directly complexed with the YscE/YscG chaperone, appear to lack significant secondary structure and appear to be disordered in the YscEFG complex [10]. In this study, we examine the role of the YscE/YscG chaperone and YscF N-terminal amino acid residues in the secretion of the Y. pestis YscF needle protein. The results indicate that both the YscE/YscG chaperone and a YscF N-terminal secretion signal are required for the efficient recognition and secretion of YscF.
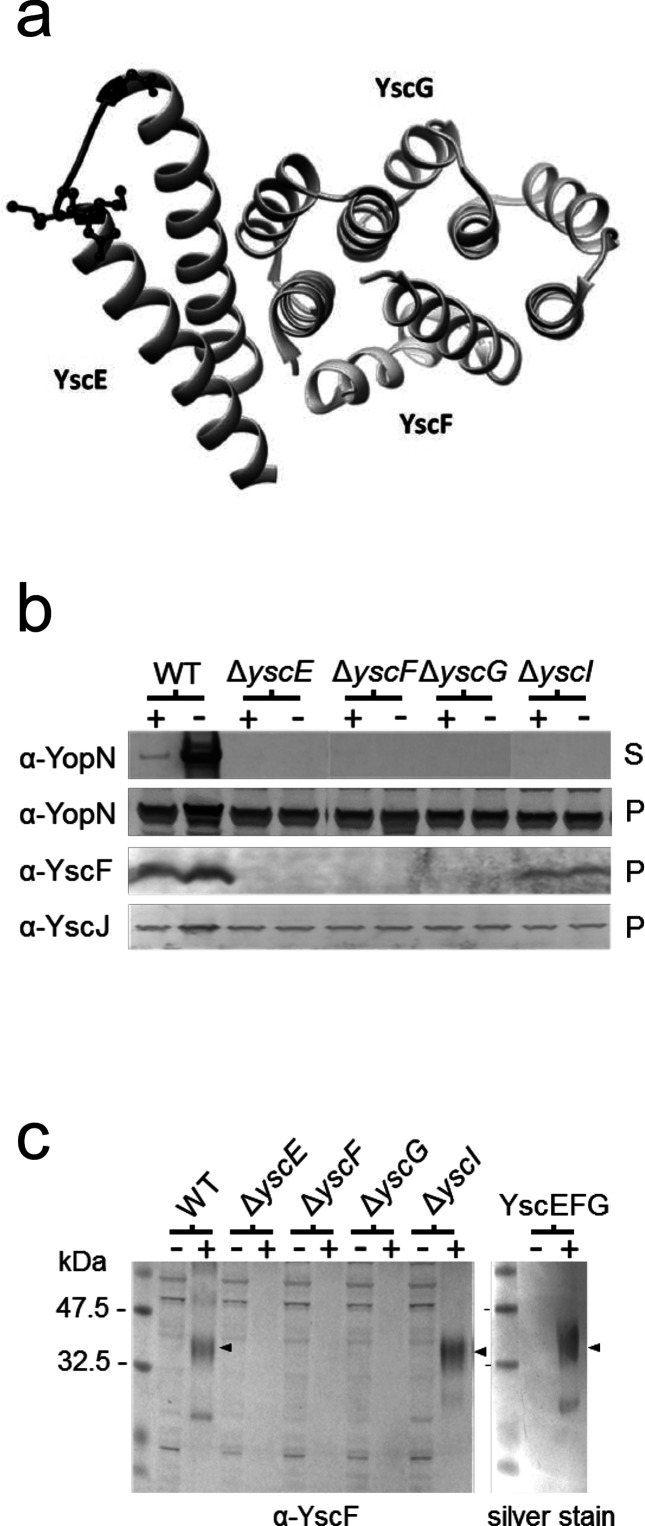
Fig. 1.
The YscE/YscG chaperone is required for stable expression of YscF. (a) Diagram of the YscEFG complex [10]. Residues mutated in YscE-6xHis mutant #11 (G35K G36H D37K) are highlighted in black and side chains are shown. The figure was generated using UCSF Chimera (Pettersen et al. [46]). (b) Production of YopN, YscF and YscJ and secretion of YopN by Y. pestis ΔyscE, ΔyscF, ΔyscG and ΔyscI deletion mutants determined by SDS-PAGE and immunoblotting. Y. pestis ΔyscE, ΔyscF, ΔyscG and ΔyscI deletion mutants were grown in the presence (+) and absence (−) of 2.5 mM CaCl2, and cell pellets (P) and culture supernatants (S) were analysed by SDS-PAGE and immunoblotting. (c) Identification of the YscEFG complex by BS3 crosslinking, SDS-PAGE and immunoblotting with anti-YscF (α-YscF) antibodies. The location of the crosslinked YscEFG complex (arrowheads) was determined by crosslinking the purified YscEFG complex, SDS-PAGE and silver staining (right panel; silver stain).
Methods
Bacterial strains and growth conditions
All of the Y. pestis strains used in this study are derivatives of Y. pestis KIM5-3001 [25] and carry a deletion of the pgm locus (pgm-), and are thus avirulent via peripheral routes of infection [26]. The E. coli and Y. pestis strains were routinely grown in heart infusion broth (HIB) or on tryptose blood agar (TBA) plates (Difco Laboratories) at 37 °C or 27 °C, respectively. For the growth and secretion assays, the Y. pestis strains were grown in thoroughly modified Higuchi’s (TMH) medium [27] overnight at 27 °C, and diluted the next day to an optical density at 620 nm (OD620) of 0.20 in 2 ml of fresh media with or without 2.5 mM CaCl2. The cultures were grown for 1 h at 27 °C and then shifted to 37 °C for 5 h of growth. Mix and Go Zymo 5α chemically competent E. coli (Zymo Research) or E. coli DH5α were used for routine cloning experiments. Bacteria with resistance markers were cultured in the presence of the appropriate antibiotic(s) at a final concentration of 25 µg ml–1 (chloramphenicol and kanamycin) or 50 µg ml–1 (ampicillin and streptomycin).
Construction of a ΔyscE ΔyscG deletion strain of Y. pestis
A deletion in the yscE gene (codons 13 to 53) of ΔyscG Y. pestis KIM5 was constructed using in vivo lambda Red recombination [28] and oligonucleotides YscE-P1 and YscE-P2 (Table S1, available in the online version of this article). The deletion in yscE of plasmid pCD1 was confirmed by polymerase chain reaction (PCR) with oligonucleotide primers YscE-CTL-F and YscE-CTL-R.
Construction of a Y. pestis KIM5 ΔyscI deletion strain
An in-frame deletion in yscI that removed DNA sequences encoding residues 9 to 64 of YscI was constructed using the PCR ligation PCR technique [29] with the oligonucleotide primer pairs YscI-SmaI-F and YscI-R, as well as YscI-F and YscI-SmaI-R (Table S1). The resulting DNA fragments were purified, phosphorylated and ligated together. The ligated products were used as a template for a second PCR reaction using the outside primers YscI-SmaI-F and YscI-SmaI-R. The final amplified DNA fragment carrying a deletion in yscI and flanked by approximately 700 bp of upstream and downstream DNA was inserted into SmaI-digested plasmid pRE112 [30], generating suicide vector pRE112-ΔYscI. Suicide vector pRE112-ΔYscI was used to move the in-frame deletion in yscI into plasmid pCD1 of Y. pestis KIM5-3001, generating strain KIM5-3001.P91 (ΔyscI). The deletion in yscI was confirmed by PCR with oligonucleotide primers YscI-CTL-F and YscI-CTL-R.
Construction of plasmids encoding C-terminal truncated YscF
Plasmid pBAD-YscF, encoding full-length YscF, was generated by the amplification of yscF from plasmid pCD1 using oligonucleotide primers YscF-XbaI-F and YscF-HindIII-R. The resultant DNA fragment was digested with XbaI and HindIII and inserted into XbaI- and HindIII-digested pBAD18 [31], generating plasmid pBAD-YscF. Plasmids pBAD-YscF1-86, pBAD-YscF1-85 and pBAD-YscF1-80 were generated by the amplification of yscF sequences from pBAD-YscF using oligonucleotide primer YscF-XbaI-F in conjunction with primers YscF-86-HindIII-R, YscF-85-HindIII-R and YscF-80-HindIII-R, respectively. The resultant PCR products were digested with XbaI and HindIII and inserted into pBAD18, generating plasmids pBAD-YscF1-86, pBAD-YscF1-85 and pBAD-YscF1-80.
Construction of plasmids encoding N-terminal truncated YscF
Plasmids pBAD-YscFΔ2-5, pBAD-YscFΔ2-10, pBAD-YscFΔ2-15 and pBAD-YscFΔ2-20 were generated from plasmid pBAD-YscF by whole-plasmid PCR [32] using oligonucleotide primer pairs YscF-1-R and YscF-6-F (pBAD-YscFΔ2-5), YscF-1-R and YscF-11-F (pBAD-YscFΔ2-10), YscF-1-R and YscF-16-F (pBAD-YscFΔ2-15) and YscF-1-R and YscF-21-F (pBAD-YscFΔ2-20). Amplification products were treated with DpnI to digest template DNA, ligated, and electroporated into E. coli DH5α. In-frame deletions were confirmed by DNA sequencing.
Construction of pFLAG-MAC-YscG-YscE-6xHis and pBAD33-YscE-6xHis
pFLAG-YscG-YscE-6xHis was constructed via the PCR ligation PCR technique [29]. A DNA fragment encoding yscG was amplified from plasmid pCD1 using oligonucleotide primers YscG-HindIII-F and YscG-R (Table S1). Similarly, DNA sequences encoding YscE were amplified using oligonucleotide primers YscE-F and YscE-6xHis-BglII-R. The resulting DNA fragments were purified, phosphorylated and ligated together. The ligated products were used as a template for a second PCR reaction using the outside primers YscG-HindIII-F and YscE-6xHis-BglII-R carrying HindIII and BglII restriction endonuclease sites, respectively. These fragments were digested and inserted into HindIII- and BglII-digested pFLAG-MAC (Sigma), generating plasmid pFLAG-MAC-YscG-YscE-6xHis. A YscE-6xHis-encoding DNA fragment of pFLAG-MAC-YscG-YscE-6xHis was amplified using oligonucleotide primers YscE-6xHis-KpnI-F and YscE-6xHis-XbaI-R, digested with KpnI and XbaI and inserted into KpnI- and XbaI-digested pBAD33, generating plasmid pBAD33-YscE-6xHis.
Generation of plasmids encoding YscE-His point mutants
Mutations in yscE of plasmid pYscE-6xHis (see Table S2) were generated by whole-plasmid PCR [32] using oligonucleotide primer pairs (see Table S1). The amplification products were treated with DpnI to digest template DNA, ligated and electroporated into E. coli DH5α. The point mutations in YscE of pYscE-6xHis were confirmed by DNA sequencing.
Construction of plasmids encoding YscF with N-terminal synthetic serine/isoleucine sequences
Deletion of plasmid pBAD-YscF DNA sequences encoding YscF residues 3 to 8 and insertion of DNA sequences encoding synthetic N-terminal functional amphipathic (ISSISI; SIISSI) as well as non-functional hydrophilic (SSSSSS) or hydrophobic (IIIIII) sequences was accomplished using whole-plasmid PCR [32] with oligonucleotide primer pairs (see Table S1). The amplification products were treated with DpnI to digest template DNA, ligated and electroporated into E. coli DH5α. The deletions and inserted DNA sequences in pBAD-YscF were confirmed by DNA sequencing.
Construction of plasmids encoding YscF-YopM30-STOP hybrid proteins
Plasmids encoding 77 bp upstream of the yscF start codon and including DNA sequences encoding YscF residues 1, 1–3, 1–5, 1–7, 1–9, 1–12, 1–15, 1–20, 1–30, 1–40 or 1–50 joined in-frame to DNA sequences encoding YopM residues 31 to 409 were constructed via the PCR ligation PCR technique [29]. DNA fragments encoding yscF fragments were amplified from plasmid pCD1 using oligonucleotide primer YscF-KpnI-F and a series of reverse primers that corresponded to the various amplified yscF coding sequences (see Table S1). DNA sequences encoding YopM residues 31 to 409 were PCR-amplified from pCD1 DNA using oligonucleotide primers YopM30-F and YopM-STOP-HindIII-R. The resulting PCR fragments were purified and then phosphorylated using T4 polynucleotide kinase, and the individual yscF DNA fragments were each ligated to the yopM fragment with T4 DNA ligase. The ligated products were used as a template for a second PCR reaction using the outside primers YscF-KpnI-F and YopM-STOP-HindIII-R carrying KpnI and HindIII restriction endonuclease sites, respectively. These fragments were digested and inserted into KpnI- and HindIII-digested pBAD18 [31], generating plasmids pYscF1-YopM30-STOP, pYscF3-YopM30-STOP, pYscF5-YopM30-STOP, pYscF7-YopM30-STOP, pYscF9-YopM30-STOP, pYscF12-YopM30-STOP, pYscF15-YopM30-STOP, pYscF20-YopM30-STOP, pYscF30-YopM30-STOP, pYscF40-YopM30-STOP and pYscF50-YopM30-STOP. The sequences of the constructs were confirmed by DNA sequencing.
BS3 crosslinking of the YscEFG complex
Y. pestis cultures were grown in the presence or absence of calcium in TMH medium for 5 h at 37 °C, centrifuged to pellet the bacteria (10 000 g for 5 min), resuspended in 50 mM HEPES, pH 7.4, and lysed by passage through a French pressure cell at 20 000 p.s.i. The resulting lysates were centrifuged at 100 000 g for 40 min to remove the remaining whole bacterial cells, cell membranes and cell debris. The cleared lysates were crosslinked with BS3 (1 mM final concentration) for 30 min at RT. The crosslinking reaction was quenched with Tris-HCl (50 mM), pH 8.0, for 15 min and the crosslinked lysates were analysed by SDS-PAGE and immunoblot analysis with an antibody specific for YscF. In addition, purified YscEFG complex (stoichiometry 1 : 1 : 1; 10 µg; gift from David Waugh) was crosslinked to serve as a control for the migration of the YscEFG complex by SDS-PAGE [10].
SDS-PAGE and immunoblotting
Volumes of cellular fractions corresponding to equal numbers of bacteria were mixed 1 : 1 (v/v) with 2× electrophoresis sample buffer and analysed by SDS-PAGE and immunoblot analysis as described previously [16] with rabbit polyclonal antisera specific for YopM, YopN, YscF, YscJ and MBP [15, 33] or mouse monoclonal antibodies specific for the FLAG epitope tag or polyhistidine tag (Sigma). Proteins were visualized using an alkaline phosphatase-conjugated secondary antibody (Sigma) and 1-Step NBT/BCIP Development Solution (Thermo Scientific) according to the manufacturer’s directions.
Results
The YscE/YscG chaperone is required for stable expression of YscF
The stability of the P. aeruginosa PscF needle subunit protein in the bacterial cytosol is dependent on the PscE/PscG chaperone [34]. To determine whether the stability of cytosolic YscF is dependent on the YscE/YscG chaperone, we evaluated the expression of YscF by immunoblot analysis of Y. pestis strains that either express or do not express a functional YscE/YscG chaperone (Fig. 1b). The parent Y. pestis KIM5-3001 strain expressed readily detectable levels of YscF in both secretion-non-permissive (+calcium) and secretion-permissive (−calcium) conditions (Fig. 1b). In contrast, KIM5-3001 strains carrying a deletion in yscF, yscE or yscG had no detectable level of YopN secretion (Fig. 1b), YscF expression (Fig. 1b) or YscEFG complex assembly (Fig. 1c). The assembled YscEFG complex was detected by BS3 crosslinking of bacterial extracts and analysis of crosslinked samples by SDS-PAGE and immunoblot analysis with antisera specific for YscF. A ΔyscI strain that, like the ΔyscE, ΔyscF and ΔyscG strains, does not assemble a functional T3S system (Fig. 1b) expressed detectable levels of YscF (Fig. 1b) and the YscEFG complex (Fig. 1c), showing that the lack of detectable YscF in the ΔyscE and ΔyscG strains was not due to the downregulation of T3S system gene expression that is associated with a non-functional T3S system. Furthermore, all of the strains, including the ΔyscE and ΔyscG mutants, expressed normal levels of another Ysc component, YscJ, a protein whose gene is encoded within the same operon as yscE, yscF, yscG and yscI (yscBCDEFGHIJKL operon) (Fig. 1b). Overall, these results indicate that the YscE/YscG chaperone is required for the stable expression of cytosolic YscF.
The C-terminal region of YscF is required for stable expression of YscF
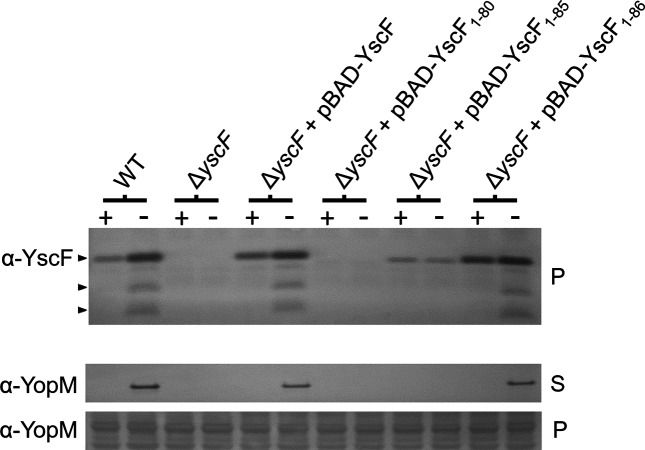
To further investigate the role of the YscE/YscG chaperone in the stable expression of YscF, a series of small deletions were generated in yscF of pBAD-YscF that removed the coding sequence for the C-terminal 1 (YscF1-86), 2 (YscF1-85) or 7 (YscF1-80) residues of the 87-residue YscF protein. Hydrophobic residues in the C-terminal region of YscF (Ile82, Leu83 and Phe86) mediate contact between YscF and YscG [10]. Derivatives of plasmid pBAD-YscF expressing YscF, YscF1-80, YscF1-85 or YscF1-86 were moved into ΔyscF Y. pestis KIM5-3001 and evaluated for YscF production, as well as T3S system function (YopM secretion; Fig. 2). Removal of the C-terminal proline residue (Pro87; YscF1-86) had no effect on YscF production or T3S system function, as measured by secretion of YopM; in contrast, the removal of 2 C-terminal residues (Pro87 and Phe86; YscF1-85) resulted in reduced YscF production and no T3S system activity. Furthermore, YscF1-80 lacking 7 C-terminal residues could not be detected and ΔyscF Y. pestis expressing the truncated protein failed to secrete YopM. Overall, these results suggest that the C-terminal region of YscF may be required for the YscF-YscE/YscG chaperone interaction and thus for stable expression of YscF.
Fig. 2.
The C-terminal 7 residues of YscF are required for stable YscF expression. Y. pestis KIM5-3001, an isogenic ΔyscF strain, and the ΔyscF mutant complemented with plasmids pBAD-YscF, pBAD-YscF1-86, pBAD-YscF1-85 and pBAD-YscF1-80 were grown in the presence (+) and absence (−) of 2.5 mM CaCl2 and analysed by SDS-PAGE and immunoblotting with α-YscF antibodies. The expression (P) and secretion (S) of YopM was determined by SDS-PAGE and immunoblotting with anti-YopM (α-YopM) antisera. The location of YscF and the plasminogen activator protease-processed products of YscF present on the bacterial surface is shown by arrowheads.
Expression of a MBP-YscG hybrid protein stabilizes YscF in the absence of YscE
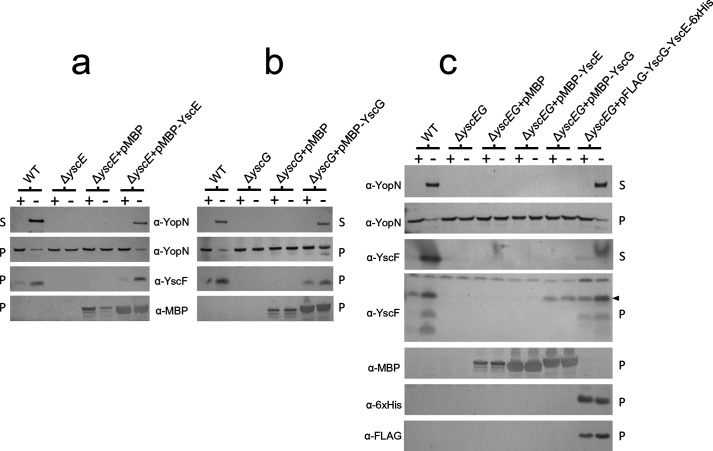
To further evaluate the role of YscE and YscG in the stable expression and secretion of YscF, we examined the effect of overexpressing YscE and/or YscG as functional MBP-YscE or MBP-YscG hybrid proteins in Y. pestis ΔyscE, ΔyscG or ΔyscE ΔyscG deletion mutants (Fig. 3) [11]. The expression of MBP-YscE but not MBP alone in ΔyscE Y. pestis restored YscF expression as well as YopN expression and secretion (Fig. 3a). Similarly, providing MBP-YscG to ΔyscG Y. pestis restored YscF expression and T3S system function (Fig. 3b). Surprisingly, providing pMBP-YscG, but not pMBP-YscE, to the ΔyscE ΔyscG deletion mutant restored stable YscF expression but not YscF or YopN secretion (Fig. 3c), suggesting that high-level expression of functional cytosolic MBP-YscG is able to bind and stabilize YscF in the absence of YscE, but is unable to target YscF for secretion and subsequent needle assembly in the absence of YscE. These results suggest that YscE likely plays a role in targeting the YscEFG complex to the T3S system.
Fig. 3.
A functional MBP–YscG hybrid protein functions to stabilize YscF, but YscF is not secreted in the absence of YscE. Y. pestis KIM5-3001, isogenic ΔyscE, ΔyscG, ΔyscEG mutants, and mutants complemented with pMAL-c2 (pMBP), pMBP-YscE, pMBP-YscG and/or pFLAG-MAC-YscG-YscE-6xHis were grown in the presence (+) or absence (−) of 2.5 mM CaCl2 and the cell pellets and culture supernatants were analysed by SDS-PAGE and immunoblot analysis with antibodies specific for YopN, YscF, MBP, penta-His and the FLAG epitope. (a) Expression of MBP-YscE in ΔyscE Y. pestis restores YscF expression and YopN secretion. (b) Expression of MBP-YscG in ΔyscG Y. pestis restores YscF expression and YopN secretion. (c) Expression of MBP-YscG but not MBP or MBP-YscE in ΔyscE ΔyscG Y. pestis restores stable YscF expression but not YscF secretion. The location of full-length YscF is shown by an arrowhead.
Identification of a YscE mutant that functions with YscG to stabilize YscF but not secrete YscF
To further investigate the role of YscE in the expression and secretion of YscF, we generated a series of YscE site-directed mutants. Mutations in yscE of plasmid pYscE-6xHis were designed to target residues throughout the length of the 66-residue protein, including YscE residues that were conserved among the various YscE family members [10, 35]. The residues that were mutated were on the exposed face of the YscE protein structure, potentially mediating contact of the YscEFG complex with a target on the T3S apparatus. Amino acid residues that mediate the interaction of YscE with YscG were not targeted, with the exception of YscE-6xHis mutant #14, which targeted three YscE residues (Ala50, Leu51 and Ala54) that mediate contact with YscG. The YscE-6xHis mutants are listed in Table S2. Plasmids encoding the YscE-6xHis mutants were moved into Y. pestis KIM5-3001 ΔyscE and evaluated for YscE, YscF and YopM expression, as well as for YopM secretion (Fig. S1). Y. pestis KIM5-3001 ΔyscE expressing His-tagged wild-type YscE, as well as the majority of the YscE-6xHis mutants (#2–9, #12, #13 and #15), expressed functional YscE-6xHis proteins that restored YscF expression and YopM secretion (Figs 4a and S1). YscE-6xHis mutants #1, #10 and #14 did not express a functional or stable YscE-6xHis or YscF protein, and did not restore YopM secretion. Interestingly, YscE-6xHis mutant #11 (YscE G35K G36H D37K) expressed a stable YscE-6xHis protein that migrated aberrantly by SDS-PAGE and restored stable YscF expression but not YopM secretion. To determine whether YscF was being exported to the bacterial surface, we examined the plasminogen activator (Pla) protease-dependent cleavage of YscF that results in several prominent degradation products and occurs post-secretion at the bacterial surface (Fig. S1). No YscF degradation products were observed in Y. pestis KIM5-3001 ΔyscE expressing YscE-6xHis mutant #11, suggesting that the mutant YscE/YscG chaperone can bind and stabilize YscF; however, the bound YscF is not secreted to the bacterial surface (Fig. S1). To confirm these findings, the secretion studies were repeated with Y. pestis KIM5-3001 ΔyscE carrying pYscE-6xHis or pYscE-6xHis #11 and probed directly for YscF in the culture supernatant (Fig. 4a). YscE-6xHis mutant #11 was expressed in the presence and absence of calcium at the same level as YscE-6xHis under conditions that were non-permissive for secretion (+calcium); however, no YscF was secreted from bacteria expressing the mutant YscE-6xHis protein. To verify that YscE-6xHis mutant #11 actually forms a complex with YscF and YscG, we repeated the BS3-based crosslinking experiments shown in Fig. 1(c) using a yscE deletion strain producing YscE-6xHis #11. An appropriate-sized complex representing the ~32 kDa YscEFG complex was identified in the parent strain, regardless of growth in the presence or absence of calcium and in the yscE deletion mutants producing YscE-6xHis or YscE-6xHis #11 (Fig. 4b). This complex was not observed in the yscE or yscF deletion mutants. These studies provide additional evidence that YscE plays a role in targeting YscF present in the cytosolic YscEFG complex for secretion.
Fig. 4.
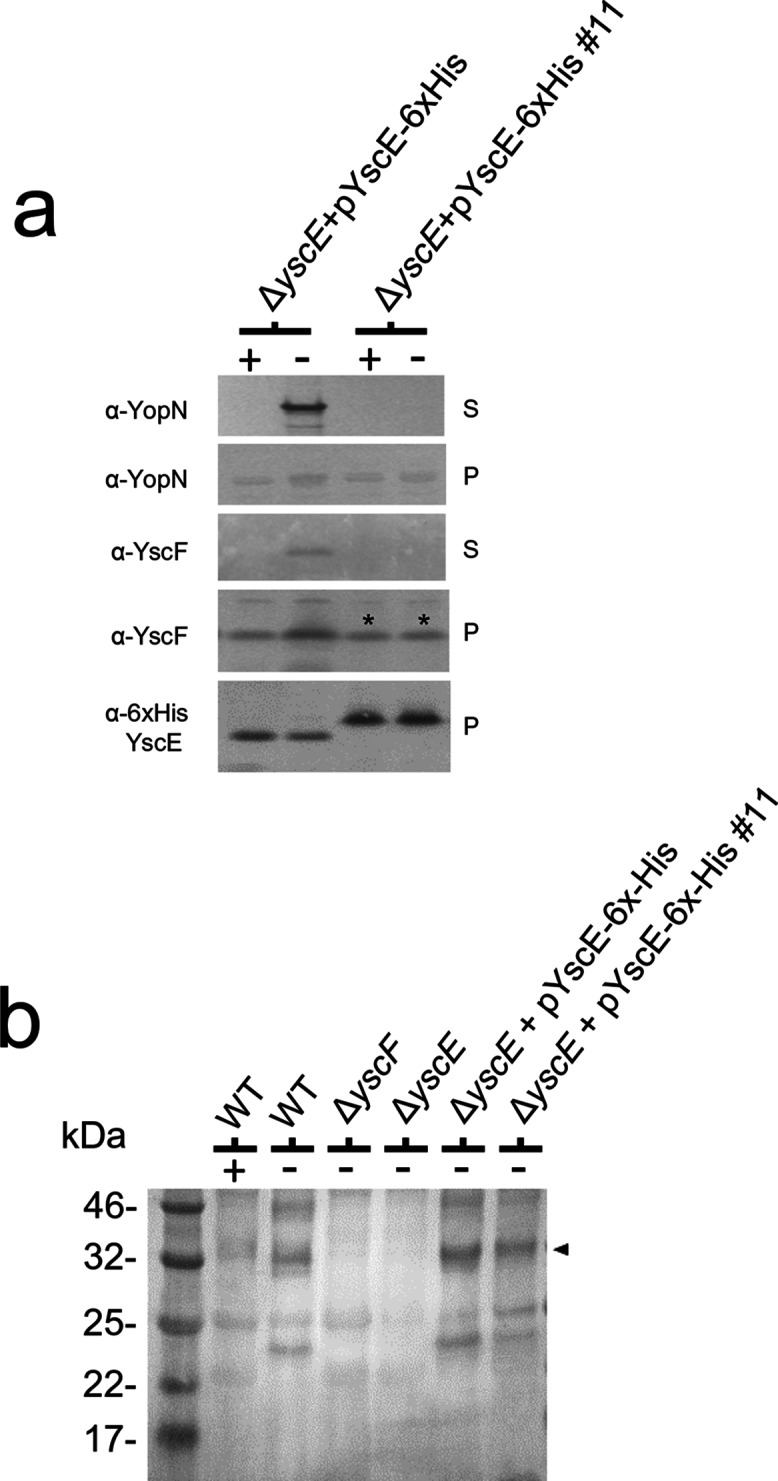
Expression of the YscE-6xHis (G35K G36H D37K) mutant in ΔyscE Y. pestis stabilizes YscF as part of a cytoplasmic YscEFG complex, but YscF is not secreted. (a) A Y. pestis ΔyscE mutant complemented with pYscE-6xHis or pYscE-6xHis (G35K G36H D37K) was grown in the presence (+) or absence (−) of 2.5 mM CaCl2, and the cell pellets and culture supernatants were analysed by SDS-PAGE and immunoblot analysis with antibodies specific for YopN, YscF and penta-His. The YscF stabilized by YscE-6xHis (G35K G36H D37K) is marked by an asterisk. (b) The presence of a cytoplasmic YscEFG complex was determined by BS3 crosslinking, SDS-PAGE and immunoblot analysis with antibodies specific for YscF. The location of the YscEFG complex in the parent (WT) and yscE deletion strains expressing YscE-6xHis or YscE-6xHis #11 is shown by an arrowhead.
YscF deleted for amino acids 2 to 15 is secreted and functional
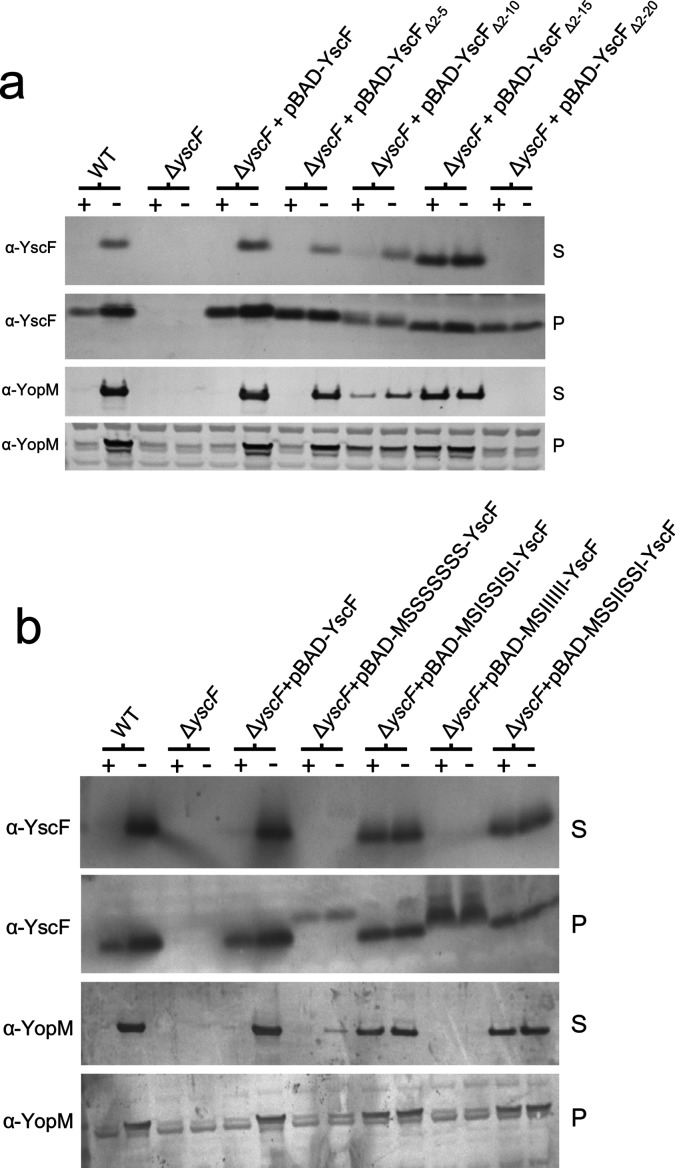
The targeting of secretion substrates to the T3S apparatus is dependent upon N-terminal amino acid signals and in many cases T3S chaperones [36]. Nevertheless, recent studies have demonstrated that the N-terminal 15 to 20 residues of YscF can be removed without significantly altering its secretion or post-secretion function, suggesting that YscF may not require typical N-terminal signals for its secretion [37, 38]. To explore this further, pBAD-YscF plasmids deleted for DNA sequences encoding YscF residues 2 to 5 (pBAD-YscFΔ2-5), 2 to 10 (pBAD-YscFΔ2-10), 2 to 15 (pBAD-YscFΔ2-15) and 2 to 20 (pBAD-YscFΔ2-20) were generated and moved into a Pla- ΔyscF Y. pestis strain. Providing plasmid pBAD-YscF or pBAD-YscFΔ2-5 restored the calcium-regulated expression and secretion of both YscF and YopM (Fig. 5a). Y. pestis (ΔyscF) expressing YscFΔ2-10 and YscFΔ2-15 also expressed and secreted the truncated YscF protein as well as YopM; however, these strains showed either a partial defect (YscFΔ2-10) or a complete defect (YscFΔ2-15) in the calcium-dependent regulation of the T3S process. In contrast, production of YscFΔ2-20 in ΔyscF Y. pestis failed to restore secretion of YscF or YopM, suggesting that YscFΔ2-20 was non-functional due to a defect in secretion or post-secretion function. Overall, these results confirm that the N-terminal region of YscF (residues 2 to 15) can be eliminated without significantly altering YscF secretion or the function of YscF in the T3S process; however, deletions in this region did disrupt a likely function of YscF in the regulation of the T3S process.
Fig. 5.
The N-terminal 2 to 15 residues of YscF are not required for stable YscF expression or secretion and synthetic amphipathic but not synthetic hydrophilic or hydrophobic N-terminal secretion signals function to promote YscF secretion. (a) Y. pestis KIM5-3001, an isogenic ΔyscF mutant, and the ΔyscF mutant carrying plasmids pBAD-YscFΔ2-5, pBAD-YscFΔ2-10, pBAD-YscFΔ2-15 and pBAD-YscFΔ2-20 were grown in the presence (+) or absence (−) of 2.5 mM CaCl2 and the cell pellets and culture supernatants were analysed by SDS-PAGE and immunoblot analysis with antibodies specific for YopN and YscF. (b) Y. pestis KIM5-3001, an isogenic ΔyscF mutant, and the ΔyscF mutant carrying plasmids pBAD-MSSSSSSS-YscF, pBAD-MSISSISI-YscF, pBAD-MSIIIIII-YscF and pBAD-MSSIISSI-YscF were similarly grown in the presence (+) or absence (−) of 2.5 mM CaCl2 and the cell pellets and culture supernatants were analysed by SDS-PAGE and immunoblot analysis with antibodies specific for YopM and YscF.
Replacement of the YscF N-terminal 8 residues with synthetic serine/isoleucine sequences
Lloyd et al. [39] demonstrated that synthetic amphipathic N-terminal amino acid sequences can function as N-terminal T3S signals for YopE; conversely, strictly hydrophobic or hydrophilic N-terminal sequences were very poor T3S signals. To determine whether N-terminal amino acid sequences that are compatible with the T3S process are required for YscF secretion, we replaced the DNA sequences of pBAD-YscF encoding residues 1 to 8 of YscF with amphipathic functional (MSISSISI or MSSIISSI), hydrophilic non-functional (MSSSSSSS) or hydrophobic non-functional (MSIIIIII) synthetic serine/isoleucine sequences. Plasmids encoding the hybrid proteins were moved into a ΔyscF Y. pestis strain and the expression and secretion of YscF and YopM were determined by immunoblot analysis (Fig. 5b). Y. pestis strains expressing wild-type YscF exhibited typical calcium-regulated expression and secretion of YscF and YopM. Providing plasmids that express YscF with the N-terminal 8 residues replaced with synthetic amphipathic serine/isoleucine sequences previously demonstrated to function as N-terminal T3S signals (MSISSISI and MSSIISSI) also facilitated YscF secretion and restored T3S system activity and YopM secretion; however, the secretion of these proteins was no longer regulated by calcium. Conversely, ΔyscF Y. pestis carrying plasmids that express YscF with non-functional N-terminal hydrophilic (MSSSSSSS) or hydrophobic (MSIIIIII) synthetic serine/isoleucine sequences expressed the hybrid YscF proteins; however, no significant amounts of YscF or YopM were secreted. These results indicate that functional amphipathic N-terminal T3S signals facilitate or at least do not interfere with YscF secretion; in contrast, hydrophobic or hydrophilic N-terminal sequences that have been shown to not function as T3S signals for YopE do not facilitate YscF secretion, or possibly interfere with or block YscF secretion.
The YscF N-terminal region carries a functional T3S signal
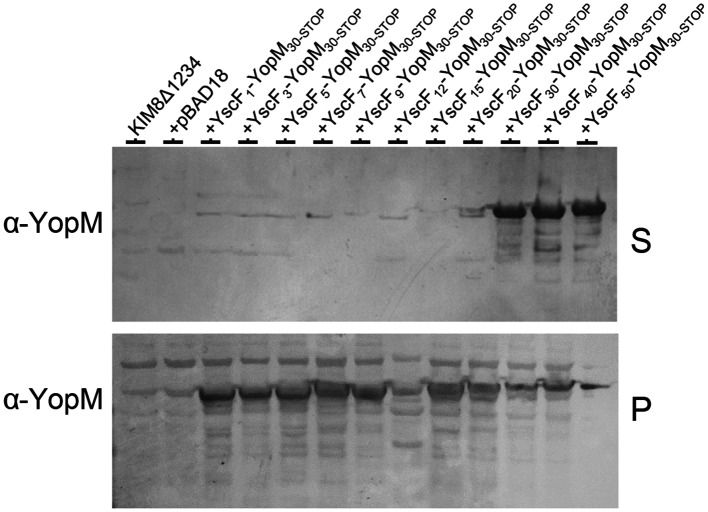
To determine whether the wild-type YscF N-terminal region carries a functional N-terminal T3S signal, we generated gene fusions encoding the N-terminal 1, 3, 5, 7, 9, 12, 15, 20, 30, 40 and 50 residues of YscF fused to the DNA sequences encoding YopM deleted for its N-terminal secretion signal (YopM30-STOP) [40]. YopM was chosen as a secretion reporter because it is a natural substrate of the Yersinia T3S system and has no T3S chaperone [41]. Plasmids encoding the YscF–YopM30-STOP gene fusions were moved into Y. pestis KIM8Δ1234 [42], a strain that expresses a functional T3S apparatus but is deleted for DNA sequences encoding all of the identified effector proteins including YopM. All of the YscF–YopM30-STOP fusion proteins were expressed under secretion-permissive conditions (−calcium); however, only the YscF30–YopM30-STOP, YscF40–YopM30-STOP and YscF50–YopM30-STOP hybrid proteins were secreted efficiently (Fig. 6). These results confirm that the N-terminal 30 residues of YscF contains a functional T3S signal capable of targeting YopM30-STOP to the T3S apparatus independently of any known T3S chaperone. Overall, the results presented indicate that YscF is targeted to the T3S apparatus via the combined action of an N-terminal secretion signal and the YscE/YscG chaperone.
Fig. 6.
Expression and secretion of YscF–YopM30-STOP hybrid proteins. Y. pestis KIM8Δ1234 carrying derivatives of plasmid pBAD18 encoding YscF–YopM30-STOP hybrid proteins was grown in the presence (+) or absence (−) of 2.5 mM CaCl2, and the cell pellets and culture supernatants were analysed by SDS-PAGE and immunoblot analysis with antibodies specific for YopM.
Discussion
The secretion and polymerization of T3S needle proteins is a highly regulated process that is essential for the assembly and function of all T3S systems [12]. In the Y. pestis Ysc T3S system and closely related T3S systems, a unique heterodimeric chaperone plays a critical role in this process [10, 23, 34, 43]. We examined the role of the YscE/YscG chaperone and YscF N-terminal signals in the secretion and post-secretion function of the Y. pestis needle protein YscF. Y. pestis strains deleted for yscE or yscG showed no stable YscF expression, no YscEFG complex assembly and no T3S activity, suggesting that YscF is unstable in the absence of a functional YscE/YscG chaperone (Fig. 1). These results replicate results obtained with P. aeruginosa pscE and pscG mutants, indicating that a role in substrate stabilization is likely conserved among the YscE/YscG family of chaperones (type V chaperones) [34]. Plasmid-based co-expression of PcrG and PcrF in a P. aeruginosa pscE deletion mutant restored PscF expression and T3S activity in the absence of PscE, indicating that the PscE component of the PscE/PscG chaperone is not absolutely essential for PscF expression or secretion [24]. Instead, PscE was designated as a co-chaperone for PscG. Interestingly, overexpression of YscG, as a functional MBP-YscG hybrid protein, in a ΔyscE ΔyscG mutant, restored stable cytosolic production of YscF in the absence of YscE; however, the YscF was not secreted and T3S activity was not restored, suggesting that YscE is required to target YscF for secretion in Y. pestis (Fig. 3). Furthermore, a YscE-6xHis (G35K G36H D37K) mutant was identified that functioned with YscG to stabilize cytosolic YscF but YscF was not targeted for secretion (Figs 4a and S1). The YscE-6xHis G35K G36H D37K mutant provides an additional line of evidence that YscE of Y. pestis, unlike PscE of P. aeruginosa, likely plays a more direct role in targeting YscF for secretion. BS3 crosslinking studies confirmed that the YscE-6xHis G35K G36H D37K mutant was present in a cytoplasmic YscEFG complex (Fig. 4b). In both cases, the stabilization of YscF in the absence of functional YscE suggests that YscF was bound by YscG, but the mutant chaperone–substrate complexes were not competent for delivery of YscF to the T3S apparatus. The G35K G36H D37K mutations mapped to the loop region that connects the two alpha helices of YscE and that is located on the surface of the YscEFG complex [10]. At this time, the role of YscE in the secretion of YscF is not clear. YscE may play a direct role in this process by mediating contact with the T3S apparatus. Alternatively, structural changes in YscG in the absence of functional YscE may interfere with the delivery of YscF to the T3S apparatus.
Surprisingly, the secretion of YscF was not dependent upon its N-terminal 15 residues being intact, as is normally the case with T3S substrates [44]. YscF proteins deleted for N-terminal residues 2 to 5, 2 to 10 or 2 to 15 were still secreted and capable of assembling a functional T3S apparatus (Fig. 5a). Deletion of yscF DNA sequences encoding residues 2 to 20 did not produce a functional T3S system due to a loss of YscF secretion or function. The lack of T3S activity associated with the YscFΔ2-20 mutant was surprising as Osei-Owusu et al. [37] had previously demonstrated that a Y. pestis YscFΔ2-20 mutant secreted YscF and expressed a functional T3S system. At this time, we have no explanation for these differences. Overall, our studies and studies by other laboratories agree that the N-terminal 15 residues of YscF are not essential for YscF secretion or the function of YscF in needle assembly and T3S.
Replacement of the N-terminal 8 residues of YscF with hydrophilic or hydrophobic synthetic serine/isoleucine sequences that do not function as T3S signals prevented YscF secretion and T3S activity (Fig. 5b), whereas amphipathic synthetic serine/isoleucine sequences that previously functioned as N-terminal T3S signals for YopE facilitated or did not interfere with YscF secretion (Fig. 5b) [39]. These findings suggest that YscF secretion requires a functional N-terminal secretion signal or that the presence of sequences incompatible with the T3S process can interfere with YscF secretion. We confirmed the presence of an N-terminal signal by demonstrating that the N-terminal 30 residues of YscF were able to function as a T3S signal when fused to a T3S substrate lacking its own T3S signal (YopM30-STOP) (Fig. 6). Overall, our results are consistent with YscF having an N-terminal secretion signal that functions with the YscE/YscG chaperone to target YscF to the T3S apparatus. Deletion of the N-terminal 15 residues of YscF likely disrupts a YscF N-terminal signal; however, the newly generated N-terminal sequences likely also function as a minimal N-terminal T3S signal. The definitive characteristics of an N-terminal T3S signal are not fully understood, but the general consensus is that N-terminal T3S signals are unstructured amphipathic amino acid sequences [39]. Importantly, the N-terminal 49 residues of YscF have been shown to be in an unstructured disordered state when YscF is complexed with the YscE/YscG chaperone [10]. Furthermore, this region of YscF has an overall amphipathic character, suggesting that N-terminally truncated YscF peptides would likely possess unstructured amphipathic N-terminal sequences that might be expected to function as minimal T3S signals. Alternatively, N-terminal sequences critical to YscF secretion may also be located upstream of YscF residue 15.
Although deletion of DNA sequences encoding YscF residues 2 to 10 and 2 to 15 did not significantly affect YscF secretion, Y. pestis strains expressing these truncated YscF proteins secreted YscF and Yops constitutively in the presence and absence of extracellular calcium (Fig. 5b). Similarly, replacement of the N-terminal 8 residues of YscF with amphipathic serine/isoleucine N-terminal sequences also disrupted the normal calcium-dependent regulation of the T3S process (Fig. 5b). Previous studies have suggested a role for the T3S needle in the regulation of the T3S process and for the calcium-dependent regulation of this process in the yersiniae [20, 21, 45]. Indeed, YscF point mutants I13A and D17A located in the N-terminal region of YscF have previously been implicated in the calcium-dependent regulation of the T3S process [21]. In contrast, Osei-Owusu et al. [37] found that YscF deleted for N-terminal residues 2 to 15 or 2 to 20 exhibited normal calcium-dependent regulation of Yop secretion. The different results obtained in these two studies may be due to differences in the level of YscF production or differences in the composition of the growth media. Further studies will be required to determine whether the YscF N-terminus has a specific role in the regulation of the T3S process.
Supplementary Data
Funding information
This research was supported by a grant from the National Institutes of Health (AI101823).
Acknowledgements
We thank Dr David Waugh for providing the purified YscEFG heterotrimeric complex. C. A. S. and K. R. contributed equally to this manuscript. The molecular graphics were performed with the UCSF Chimera package. Chimera was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Two supplementary tables and one supplementary figure are available with the online version of this article.
Abbreviations: HIB, heart infusion broth; MBP, maltose-binding protein; Pla, plasminogen activator; PCR, polymerase chain reaction; TBA, tryptose blood agar; TMH, thoroughly modified Higuchi’s; T3S, type III secretion; Ysc, Yersinia secretion.
Edited by: P. Langford and M. Whiteley
References
- 1.Perry RD, Fetherston JD. Yersinia pest is—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eidson M, Thilsted JP, Rollag OJ. Clinical, clinicopathologic, and pathologic features of plague in cats: 119 cases (1977–1988) J Am Vet Med Assoc. 1991;199:1191–1197. [PubMed] [Google Scholar]
- 3.Cobbs CG, Chansolme DH. Plague. Dermatol Clin. 2004;22:303–312. doi: 10.1016/j.det.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Galindo CL, Rosenzweig JA, Kirtley ML, Chopra AK. Pathogenesis of Y. enterocolitica and Y. pseudotuberculosis in Human Yersiniosis. J Pathog. 2011;2011:1–16. doi: 10.4061/2011/182051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plano GV, Schesser K. The Yersinia pestis type III secretion system: expression, assembly and role in the evasion of host defenses. Immunol Res. 2013;57:237–245. doi: 10.1007/s12026-013-8454-3. [DOI] [PubMed] [Google Scholar]
- 6.Kubori T, Sukhan A, Aizawa SI, Galán JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 8.Mueller CA, Broz P, Müller SA, Ringler P, Erne-Brand F, et al. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- 9.Haddix PL, Straley SC. Structure and regulation of the Yersinia pestis yscBCDEF operon. J Bacteriol. 1992;174:4820–4828. doi: 10.1128/jb.174.14.4820-4828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun P, Tropea JE, Austin BP, Cherry S, Waugh DS. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J Mol Biol. 2008;377:819–830. doi: 10.1016/j.jmb.2007.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day JB, Guller I, Plano GV. Yersinia pestis YscG protein is a Syc-like chaperone that directly binds yscE. Infect Immun. 2000;68:6466–6471. doi: 10.1128/IAI.68.11.6466-6471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewoody RS, Merritt PM, Marketon MM. Regulation of the Yersinia type III secretion system: traffic control. Front Cell Infect Microbiol. 2013;3:4. doi: 10.3389/fcimb.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edqvist PJ, Olsson J, Lavander M, Sundberg L, Forsberg A, et al. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol. 2003;185:2259–2266. doi: 10.1128/JB.185.7.2259-2266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood SE, Jin J, Lloyd SA. YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J Bacteriol. 2008;190:4252–4262. doi: 10.1128/JB.00328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day JB, Plano GV. A complex composed of SycN and YscB functions as a specific chaperone for YopN in Yersinia pestis . Mol Microbiol. 1998;30:777–788. doi: 10.1046/j.1365-2958.1998.01110.x. [DOI] [PubMed] [Google Scholar]
- 16.Ferracci F, Schubot FD, Waugh DS, Plano GV. Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol Microbiol. 2005;57:970–987. doi: 10.1111/j.1365-2958.2005.04738.x. [DOI] [PubMed] [Google Scholar]
- 17.Cheng LW, Kay O, Schneewind O. Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica . J Bacteriol. 2001;183:5293–5301. doi: 10.1128/JB.183.18.5293-5301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosqvist R, Magnusson KE, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosqvist R, Persson C, Håkansson S, Nordfeldt R, Wolf-Watz H. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contrib Microbiol Immunol. 1995;13:230–234. [PubMed] [Google Scholar]
- 20.Davis AJ, Mecsas J. Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, YscF, that specifically abrogate effector translocation into host cells. J Bacteriol. 2007;189:83–97. doi: 10.1128/JB.01396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torruellas J, Jackson MW, Pennock JW, Plano GV. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol Microbiol. 2005;57:1719–1733. doi: 10.1111/j.1365-2958.2005.04790.x. [DOI] [PubMed] [Google Scholar]
- 22.Quinaud M, Plé S, Job V, Contreras-Martel C, Simorre JP, et al. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc Natl Acad Sci USA. 2007;104:7803–7808. doi: 10.1073/pnas.0610098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee C, Kumar S, Chakraborty S, Tan YW, Leung KY, et al. Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila . PLoS One. 2011;6:e19208. doi: 10.1371/journal.pone.0019208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plé S, Job V, Dessen A, Attree I. Cochaperone interactions in export of the type III needle component PscF of Pseudomonas aeruginosa . J Bacteriol. 2010;192:3801–3808. doi: 10.1128/JB.00117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindler LE, Klempner MS, Straley SC. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect Immun. 1990;58:2569–2577. doi: 10.1128/iai.58.8.2569-2577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Une T, Brubaker RR. In vivo comparison of avirulent Vwa- and Pgm- or Pstr phenotypes of yersiniae. Infect Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goguen JD, Yother J, Straley SC. Genetic analysis of the low calcium response in Yersinia pestis mu d1(Ap lac) insertion mutants. J Bacteriol. 1984;160:842–848. doi: 10.1128/jb.160.3.842-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali SA, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. Biotechniques. 1995;18:746–750. [PubMed] [Google Scholar]
- 30.Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 31.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai Y, Matsushima Y, Sugimura T, Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plano GV, Straley SC. Mutations in yscC, yscD, and yscG prevent high-level expression and secretion of V antigen and Yops in Yersinia pestis . J Bacteriol. 1995;177:3843–3854. doi: 10.1128/jb.177.13.3843-3854.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinaud M, Chabert J, Faudry E, Neumann E, Lemaire D, et al. The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa . J Biol Chem. 2005;280:36293–36300. doi: 10.1074/jbc.M508089200. [DOI] [PubMed] [Google Scholar]
- 35.Phan J, Austin BP, Waugh DS. Crystal structure of the Yersinia type III secretion protein YscE. Protein Sci. 2005;14:2759–2763. doi: 10.1110/ps.051706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 37.Osei-Owusu P, Jessen Condry DL, Toosky M, Roughead W, Bradley DS, et al. The N terminus of type III secretion needle protein YscF from Yersinia pestis functions to modulate innate immune responses. Infect Immun. 2015;83:1507–1522. doi: 10.1128/IAI.02687-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allaoui A, Schulte R, Cornelis GR. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 39.Lloyd SA, Sjöström M, Andersson S, Wolf-Watz H. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol Microbiol. 2002;43:51–59. doi: 10.1046/j.1365-2958.2002.02738.x. [DOI] [PubMed] [Google Scholar]
- 40.Boland A, Sory MP, Iriarte M, Kerbourch C, Wattiau P, et al. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y.enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 41.Trülzsch K, Roggenkamp A, Aepfelbacher M, Wilharm G, Ruckdeschel K, et al. Analysis of chaperone-dependent Yop secretion/translocation and effector function using a mini-virulence plasmid of Yersinia enterocolitica . Int J Med Microbiol. 2003;293:167–177. doi: 10.1078/1438-4221-00251. [DOI] [PubMed] [Google Scholar]
- 42.Bartra SS, Jackson MW, Ross JA, Plano GV. Calcium-regulated type III secretion of Yop proteins by an Escherichia coli hha mutant carrying a Yersinia pestis pCD1 virulence plasmid. Infect Immun. 2006;74:1381–1386. doi: 10.1128/IAI.74.2.1381-1386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson DL, Stone CB, Mahony JB. Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J Bacteriol. 2008;190:2972–2980. doi: 10.1128/JB.01997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michiels T, Cornelis GR. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991;173:1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenjale R, Wilson J, Zenk SF, Saurya S, Picking WL, et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 46.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.