Abstract
The class III peroxidase (POD) enzymes participate in plant development, hormone signaling, and stress responses. However, little is known about the POD family in cassava. Here, we identified 91 cassava POD genes (MePODs) and classified them into six subgroups using phylogenetic analysis. Conserved motif analysis demonstrated that all MePOD proteins have typical peroxidase domains, and gene structure analysis showed that MePOD genes have between one and nine exons. Duplication pattern analysis suggests that tandem duplication has played a role in MePOD gene expansion. Comprehensive transcriptomic analysis revealed that MePOD genes in cassava are involved in the drought response and postharvest physiological deterioration. Several MePODs underwent transcriptional changes after various stresses and related signaling treatments were applied. In sum, we characterized the POD family in cassava and uncovered the transcriptional control of POD genes in response to various stresses and postharvest physiological deterioration conditions. These results can be used to identify potential target genes for improving the stress tolerance of cassava crops.
Keywords: expression, genome-wide, identification of peroxidase genes, duplication pattern, stress, cassava
1. Introduction
Peroxidases (EC 1.11.1.X) form a large family of enzymes that are widely distributed in living organisms and catalyze the oxidoreduction reaction between hydrogen peroxide (H2O2) as an electron acceptor and diverse electron donors, such as auxin, phenolic compounds, or secondary metabolites [1,2]. According to their protein sequences and structure, peroxidases are classified as either non-heme peroxidases or heme peroxidases [3]. The majority of heme peroxidase members are divided into animal and non-animal groups [4]. On the basis of their amino acid sequences and catalytic properties, non-animal heme peroxidases are assigned to one of three large families: class I, II, or III [3,5]. The class III peroxidases (EC 1.11.1.7) are plant-specific oxidoreductases and have various abbreviations (POX, POD, Px, PER, and Prx) [2].
There are many multigenic class III peroxidasesin land plants, which is commonly secreted into the vacuole and cell wall [4,5,6,7]. The structures and weights of Prx proteins are highly conserved between paralogs and orthologs [1,3]. The class III plant peroxidases contain 10–12 conserved α-helices and two short β-strands [2,7,8,9,10], and they mainly participate in the peroxidative cycle and hydroxylic cycle to reduce the production of hydrogen peroxide and the formation of reactive oxygen species (ROS) [4,5,11,12]. Prx proteins are involved in a variety of physiological processes, such as the cross-linking of cell wall components, salt tolerance, defense against pathogen attack, the oxidation of toxic reductants, and the metabolism of phytohormones [2,3,13,14,15,16,17].
Some genetic evidence supports Prx proteins’ role in the plant response to biotic and abiotic stresses. Overexpression of AtPrx64 was able to enhance tolerance to aluminum stress in transgenic tobacco plants [18]. AtPrx3 was shown to positively regulate plant tolerance to drought and salt stresses in Arabidopsis [19]. Overexpression of the Catharanthus roseus genes CrPrx and CrPrx1 in tobacco led to enhanced chilling resistance and increased germination rates under dehydration and salt treatments, respectively [20]. Repressing the expression of Ep5C in tomato resulted in reduced susceptibility to bacterial speck caused by the pathogen Pseudomonas syringae pv tomato [21]. CaPO2 gene-silenced pepper plants were shown to be susceptible to infection by Xanthomonas campestris pv vesicatoria, whereas overexpression of CaPO2 in transgenic Arabidopsis thaliana conferred bacterial disease resistance [22]. Transgenic carrot plants overexpressing OsPrx114 exhibited enhanced resistance to necrotrophic fungal pathogens [23]. Together, these previous studies reveal the positive role of class III plant peroxidases in the response to biotic and abiotic stresses.
To date, the peroxidase (POD) family members have been characterized by whole-genome analyses in several plants, including 73 PODs in Arabidopsis [24,25,26], 138 PODs in rice [9], 93 PODs in Populus trichocarpa [27], 102 PODs in Medicago sativa [28], 119 PODs in maize [29], and 94 PODs in Pyrus bretschneideri [30]. However, there is less known about the POD family in cassava, a major tropical crop. Cassava is the third most valuable crop after maize and rice in Africa, Latin America, and Asia, supplying a carbohydrate source to 600 million people in tropical and subtropical regions [31]. Cassava can efficiently use water, heat, and light resources, and it is resistant to dehydration stress and lower-fertility soils [32,33]. Unfortunately, the potential of cassava as a food and industrial crop is restricted because its storage roots deteriorate within 72 h of its harvest [34]. ROS production is an early event that leads to the postharvest physiological deterioration (PPD) of cassava storage roots [35]. The mechanisms underlying cassava’s resistance to drought and sensitivity to PPD are not well understood. POD proteins function by reducing the production of H2O2 and formation of ROS, which are involved in various physiological processes. Systematic investigations of the cassava POD family would provide novel insights into the POD-mediated stress response and regulation of root deterioration.
2. Results
2.1. Genome-Wide Identification of PODs in Cassava
According to the 211 POD protein sequences from Arabidopsis and rice genome databases, 91 POD members were predicted from the cassava genome using BLAST and HMMER methods. After conserved domain detection was confirmed, these cassava POD proteins (MePODs) were named MePOD01 to MePOD91. The full length of these putative cassava POD proteins ranges from 153 (MePOD63) to 422 (MePOD46) amino acid residues, and their relative molecular weight varies from 16.64 (MePOD63) to 46.12 kDa (MePOD46), with isoelectric points ranging from 4.43 (MePOD26) to 9.63 (MePOD39) (Table S1).
2.2. Phylogenetic and Comparative Analyses of PODs in Cassava
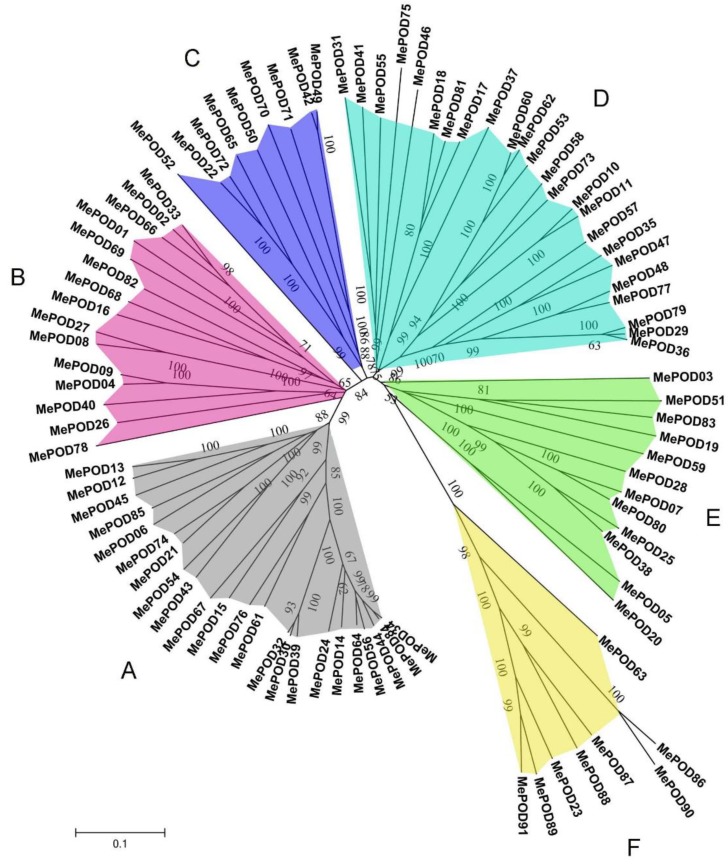
The homology and similarity of the POD genes in cassava were determined by performing multiple sequence alignments. Then, the radiation phylogenetic tree of the 91 POD genes was constructed using the neighbor-joining (NJ) method with a bootstrap value of 1000 using MEGA 5.1 (University College Dublin, Dublin, Ireland). The phylogenetic analyses indicated that MePOD genes can be divided into six subgroups on the basis of the observed genetic distance and bootstrap support (Figure 1). The large subgroups A and D consist of 23 and 24 MePOD members, respectively, whereas the small subgroups C and F contain 9 and 8 MePOD members, respectively. Subgroups B and E are composed of 15 and 12 MePODs members, respectively. These results show that a diversified POD family exists in cassava.
Figure 1.
Phylogenetic analyses of PODs from cassava. A total of 91 PODs from cassava were used to create the neighbor-joining (NJ) tree with 1000 bootstraps.
2.3. Conserved Motif and Gene Structure Analysis of POD Families in Cassava
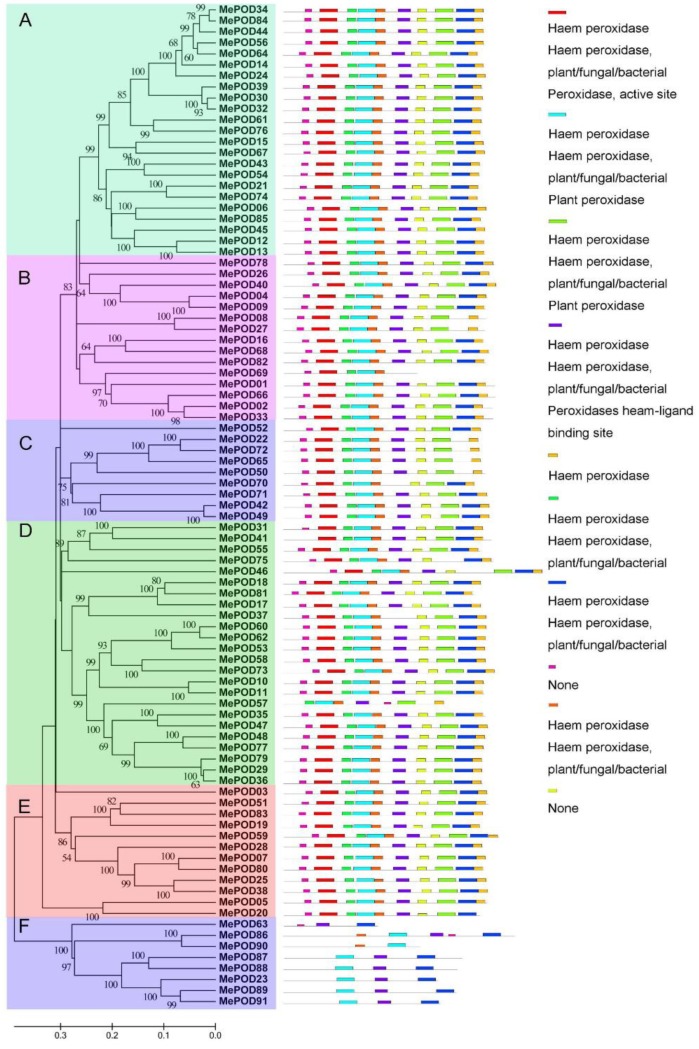
The structural features of MePODs were investigated by identifying 10 conserved motifs using the MEME database in accordance with the phylogenetic relationship. Then, the conserved motifs were submitted in their entirety to the InterProScan database for annotation. Eight domains (domains 1, 2, 3, 4, 5, 6, 7, and 9) were noted as POD protein motifs, which are an essential feature of the peroxidase family. On the basis of the motif analyses, 83 MePODs were assigned to one of five subgroups (A–E). Each of these 83 MePODs contains at least nine POD motifs, except for MePOD57 (in subgroup D) and MePOD69 (in subgroup B), which have seven and five motifs, respectively. The presence of these motifs suggests that the identified proteins are characteristic of the POD family (Figure 2). Subgroup F is distinct from the others: its members contain domains 2, 4, 5, 7, and 8. These results indicate that the proteins assigned to the same subfamilies share similar POD motif characteristics, further supporting their phylogenetic classification as PODs in cassava.
Figure 2.
The motif analyses of POD family members in cassava according to their evolutionary relationship. The POD motifs were identified by the MEME database. The 10 different colors of the boxes on the right represent diverse conserved motifs, while the gray lines indicate non-conserved sequences.
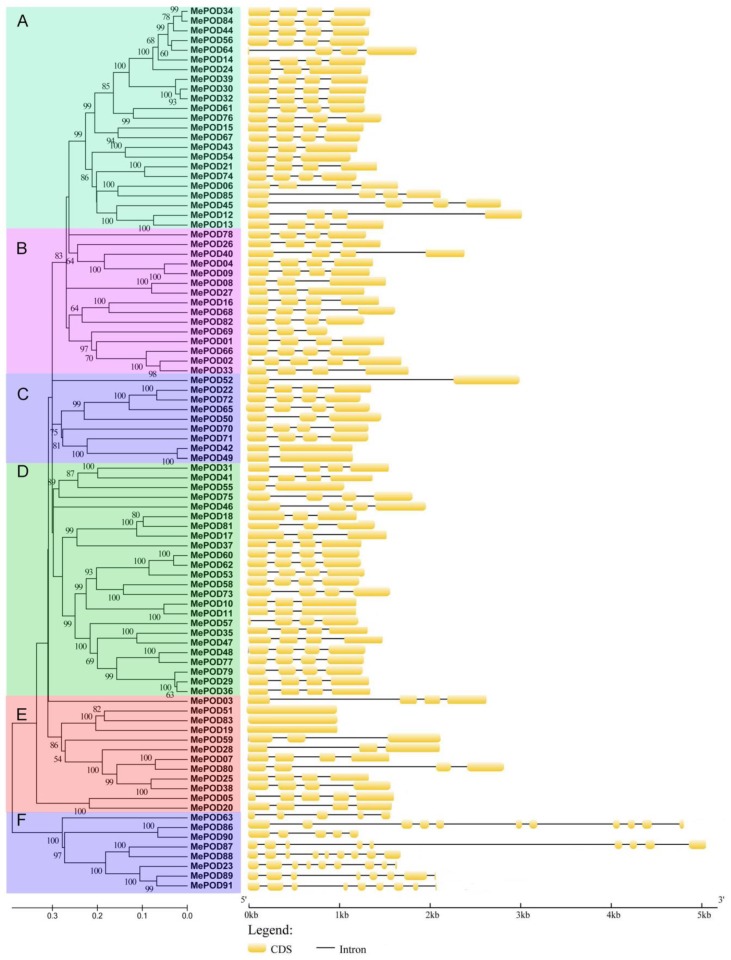
Next, the exon–intron structures of cassava POD genes were analyzed. Subgroup F is exon-rich, with five to nine exons, whereas other subfamilies have fewer (between one and four) exons, except for MePOD02 (in subgroup B), which has five exons (Figure 3). High proportions of POD genes contain four exons in subgroups A, B, C, and D, with four-exon POD genes forming 84%, 67%, 57%, and 76% of the genes in these subgroups, respectively, whereas only 50% of the subgroup E genes have four exons. Generally, POD genes in the same subgroup show similar exon–intron features, providing further evidence of their phylogenetic relationship.
Figure 3.
The exon–intron organization analyses of cassava PODs on the basis of the phylogenetic relationship. The exon–intron distribution was established using the GSDS database. The yellow boxes and the black lines represent exons and introns, respectively.
2.4. Analyses of Chromosomal Distribution and Duplication Events of the Cassava POD Genes
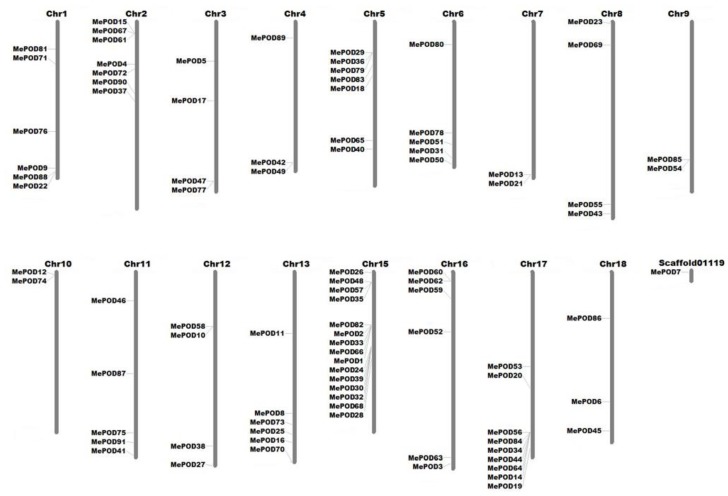
The locations of the cassava POD genes were determined by analyzing their chromosomal distribution (Figure 4). The 91 MePODs were mapped to chr1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 15, 16, 17, and 18, and scaffold01119. The 24 POD genes in subgroup D were distributed among chr1, 2, 3, 5, 6, 8, 11, 12, 13, 15, 16, and 17, making subgroup D the most widely distributed subgroup. Subgroup F contains eight POD genes, which are located on chr1, 2, 4, 8, 11, 16, and 18; thus, of the six subgroups identified, subgroup F is dispersed among the fewest chromosomes. The 23 members of subgroup A are found on chr1, 2, 7, 8, 9, 10, 15, 17, and 18, among which chromosomes 7, 9, and 10 only contain subgroup A genes. Generally, the cassava POD genes are widely distributed among chromosomes.
Figure 4.
Chromosome distribution analyses of POD gene subgroups in cassava. The chromosomal information of 91 MePODs was collected from the Phytozome 12.0 cassava database, and the genes were then mapped to 17 chromosomes and one scaffold. MapInspect software (Wageningen University, Wageningen, Netherlands) was used to draw this figure.
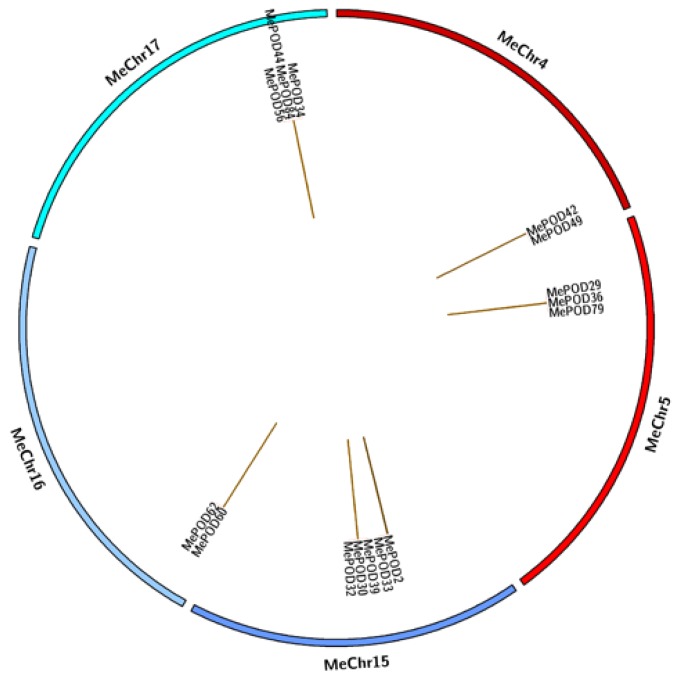
To further investigate the expansion of POD genes in cassava, we aligned the total nucleotide sequences of the 91 MePOD genes to identify duplication events. We identified 15 events involving 16 paralogs (MePOD2/MePOD33, MePOD29/36/79, MePOD30/32/39, MePOD34/44/56/84, MePOD42/MePOD49, MePOD60/MePOD62), suggesting that tandem duplication played a significant role in POD family expansion in the cassava genome (Figure 5).
Figure 5.
Analyses of POD gene duplication in cassava. The program Circos (Canada’s Michael Smith Genome Sciences Center, Vancouver, Canada) was used to draw different chromosomes in a circular distribution. The brown connection lines represent tandem duplication events of POD genes in cassava.
Next, we calculated nonsynonymous (Ka) and synonymous (Ks) ratios to understand the modes of evolutionary selection for the duplicated MePOD genes. We found that the Ka/Ks ratios of the paralogous genes are between 0.05 and 0.29, indicating that these genes underwent purifying selection during evolution (Table S2).
2.5. Expression Profiles of POD Genes in Different Tissues of Two Cassava Genotypes
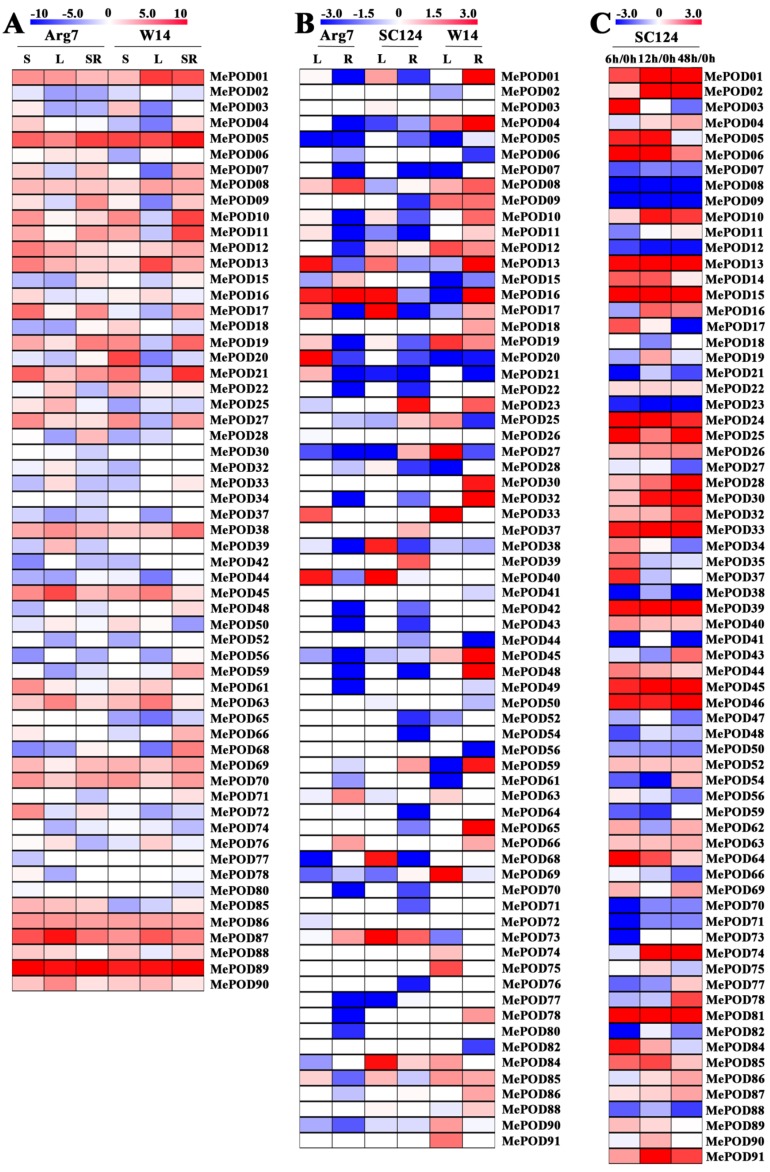
The expression levels of MePOD genes in different tissues were investigated by performing RNA-Seq analysis on the storage roots, stems, and leaves of a cultivated variety (Arg7) and wild subspecies (W14). The resulting expression data covered 59 and 56 MePOD genes in the transcriptome dataset of Arg7 and W14, respectively (Figure 6A; Table S3). Of these genes in Arg7, 15 (25%), 9 (15%), and 8 (14%) MePODs had high transcriptional levels (log2-based > 4) in stems, leaves, and storage roots, respectively. The number of MePODs with high expression (log2-based > 4) in the stems, leaves, and storage roots of W14 was 9 (16%), 7 (13%), and 10 (18%), respectively. Notably, MePOD5 in subgroup E and MePOD89 in subgroup F were strongly expressed (log2-based fold change > 4) in the three diverse tissues of Arg7 and W14. These POD genes may play a molecular role in the development and function of different cassava tissues.
Figure 6.
Transcriptomic analysis of cassava POD genes. (A) Expression of MePODs in the stem (S), leaf (L), and storage root (SR) of W14 and Arg7. The log2-based FPKM value was applied to build the heat map using Mev4.9.0 software (CCCB, Boston, USA). (B) Expression of MePODs in the leaf (L) and root (R) of Arg7, SC124, and W14 after drought treatment relative to under normal conditions. Log2-based fold changes (L/control; R/control) were applied to build the heat map using Mev4.9.0 software. (C) Expression of MePODs in the storage root at 6, 12, and 48 h relative to 0 h after harvest. Log2-based fold changes were applied to build the heat map using Mev4.9.0 software.
2.6. Expression Profiles of POD Genes After Drought Treatment
To study the possible role of MePODs in the cassava response to drought stress, water was withheld from a wild subspecies (W14) and two cultivated varieties (Arg7 and SC124) for 12 days. The leaves and roots of these samples were then collected to perform RNA-Seq. Of the transcriptome data, the expression data were obtained for 71 out of the 91 cassava POD genes (Figure 6B; Table S4). After drought treatment, 6 (8%) and 5 (7%) MePOD genes in Arg7 were transcriptionally upregulated (log2-based fold change > 1), whereas 7 (10%) and 29 (41%) were down-regulated (log2 based fold change < −1) in the leaves and roots, respectively. After SC124 was subjected to drought stress, 9 (13%) and 4 (6%) MePOD genes were upregulated (log2-based fold change > 1) but 6 (8%) and 29 (41%) were downregulated (log2-based fold change <−1) in the leaves and roots, respectively. After the W14 subspecies was exposed to drought, 13 (18%) and 21 (30%) MePOD genes were induced (log2-based fold change > 1), whereas 11 (15%) and 9 (13%) were depressed (log2-based fold change < −1) in the leaves and roots, respectively. MePOD13 (subgroup A) and MePOD16 (subgroup B) were upregulated (log2-based fold change > 1) by drought stress in the leaves of all three genotypes. The above data reveal that more MePOD genes were upregulated in response to drought treatment in W14 than in Arg7 and SC124.
2.7. Expression Profiles of Cassava PODs During PPD
Transcriptome analyses were conducted at different postharvest periods of the storage roots of SC124 to examine the possible function of MePOD genes during PPD (Figure 6C; Table S5). Transcriptional data were obtained for 71 of the 91 MePODs. Of these MePODs, 38 genes were induced (log2-based fold change > 1) at a minimum of one time point. Notably, MePOD-1, -6, -13, -15, -24, -25, -33, -39, -45, -46, -81, and -91 were continuously upregulated (log2-based fold change > 1) at all points. Conversely, MePOD-7, -8, -9, -12, -23, -38, -50, -70, -71, and -88 were downregulated (log2-based fold change < −1) at all tested times. These results suggest the possibility that MePODs play a role during PPD in cassava.
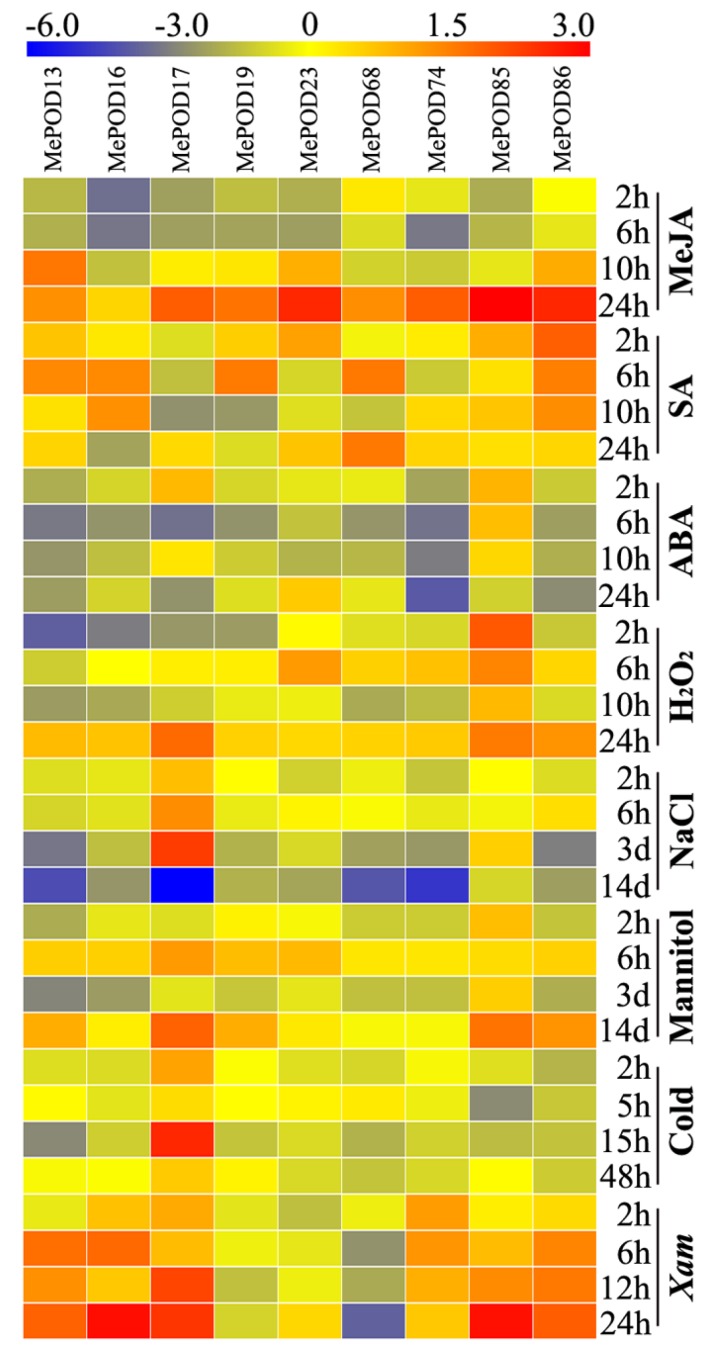
2.8. Expression Analysis of MePOD Genes in Response to Various Abiotic and Biotic Stresses and Related Signals
To test the transcription of MePOD genes upon exposure to methyl jasmonate (MeJA), salicylic acid (SA), abscisic acid (ABA), H2O2, salt, osmotic stress (by mannitol treatment), cold stress, and Xanthomonas axonopodis pv manihotis (Xam), nine genes (MePOD-13, -16, -17, -19, -23, -68, -74, -85, -86) that were induced by drought stress in at least two tissues in all three varieties were selected for quantitative real-time PCR (qRT-PCR) analysis (Figure 7; Table S6). We define upregulation as: log2-based fold change > 1. With MeJA treatment, the expression of eight genes (MePOD-13, -17, -19, -23, -68, -74, -85, and -86) was induced, with particularly high levels expressed 24 h after treatment. ABA treatment resulted in increased transcript levels of MePOD-17 and -85. With SA treatment, the expression levels of MePOD-13, -16, -19, -23, -68, -85, and -86 were amplified. H2O2 treatment led to the induction of MePOD-13, -17, -23, and -85. Salt treatment induced the expression of MePOD17 after two and six hours, and three days of treatment, but the gene was repressed after 14 days of treatment. Under osmotic stress, MePOD-13, -17, -19, -23, -85, and -86 were upregulated, among which MePOD-85 was induced throughout the entire treatment time. In response to cold treatment, MePOD17 was upregulated at 2 and 15 h. Exposure to the pathogen Xam led to the upregulated expression of six POD genes (MePOD-13, -16, -17, -74, -85, and -86) at a minimum of two time points. Together, these results demonstrate that the POD genes of cassava respond to multiple stresses and related signals (Figure 7).
Figure 7.
Expression profiles of cassava POD genes in the leaves of Arg7 after exposure to MeJA, SA, ABA, H2O2, salt, osmotic stress (mannitol treatment), cold stress, and Xam. Log2-based qRT-PCR fold changes were used to build the heat map with Mev4.9.0 software. The changes in color represent the relative gene expression level.
3. Discussion
Given the significant role of PODs in various physiological processes, including responses to biotic and abiotic stresses, it was necessary to scientifically investigate the potential functions of POD genes in cassava, which is an important crop. In this study, we identified 91 PODs in the cassava genome (Figure 1); thus, cassava has more POD members than Arabidopsis but fewer than rice, Populus trichocarpa, Medicago sativa, maize, and Pyrus bretschneideri [9,24,25,26,27,28,29,30]. We found that 92% (84/91) of the MePODs have a molecular mass in the range of 30 to 45 kDa, which is in accordance with previous studies [2,7]. Most of the POD genes (89/91) in cassava harbor more than one exon (Figure 3), which is similar to the proportion of single-exon POD genes in Pyrus bretschneideri and Zea mays (PbPRX (90/94) and ZmPRX (89/107), respectively) [29,30]. The similarities in gene structure and motif composition among the members in each MePOD subgroup support the phylogenetic classification presented here.
The expansion of a gene family primarily occurs via three kinds of modes: segmental duplication of multiple genes, tandem duplication of individual genes, and whole-genome duplication [36,37]. To analyze the duplication modes of the POD genes in cassava, we first identified the chromosomal locations of the MePOD genes. Chromosomal mapping revealed that these genes are widely distributed among 17 chromosomes and one scaffold (cassava has 18 chromosomes in total) (Figure 4), which is in accordance with the wide chromosomal distribution of PODs in Arabidopsis, rice, Populus trichocarpa, maize, and Pyrus bretschneideri [9,24,27,29,30]. Secondly, 16 paralogous POD genes were characterized in the cassava genome, indicating that tandem duplication contributed to MePOD expansion. Accumulated evidence has demonstrated that duplication events have been important for gene expansion in the POD family. A total of 37 PRX genes in Populus and 24 POD genes in maize were identified as tandem duplications, further supporting that tandem duplication has been a significant means of POD gene expansion [27,29]. Almost all these paralogous MePODs had low or no expression after drought treatment, but 63% (10 out of 16) from the postharvest transcriptome were expressed (Table S6), of which MePOD-2, -30, -32, -33, -39, and -44 were significantly upregulated and MePOD-34, -56, and -62 were repressed at some time point during the PPD process (Figure 6C). These results indicate that most of the MePOD genes resulting from tandem duplication-driven expansion are involved in the PPD process of cassava storage roots. Paralogous PRX genes were also found to be involved in other biological processes. In maize, paralogous genes ZmPRX-26, -42, and -75 were induced after NaCl, PEG, SA, or H2O2 treatment [29]. In Chinese pear, the expression of paralogous genes PbPRX-42 and -64 increased during fruit development [30].
The POD family is positively related to the reduced production of hydrogen peroxide and the decreased formation of reactive oxygen species, and the suppression of these species increases plant resistance to stresses [4,5,11,12,19,20]. In this study, the total number of POD genes responding to drought (log2-based fold change > 1) was greater in both the roots and leaves of W14 than that in Arg7 and SC124, suggesting the comprehensive activation of PODs in response to drought in W14 (Figure 6B). The wild ancestor W14 has been previously confirmed to be more resistant to drought than the two cultivars SC124 and Arg7 [38,39]. Accumulated evidence suggests that the overexpression of POD genes results in increased plant tolerance to drought and osmotic stresses [19,20,40,41]. The activity of POD enzyme was significantly enhanced under drought stress [42]. Consequently, we conclude that the high ratio of MePODs induced by drought in W14 might contribute to its strong drought tolerance.
Previous studies have suggested that ROS production results in the deterioration process in cassava during the postharvest period, and a reduction in ROS accumulation could delay the PPD process [35,43]. The POD family mainly participates in the peroxidative cycle and hydroxylic cycle, resulting in the reduced production of H2O2 and the decreased formation of ROS [4,5,11,12]. Some PRXs have been shown to change in expression during the fruit storage process [44,45]. The activity of POD enzyme significantly increased during cassava PPD process, suggesting their possible role during the postharvest period of cassava [35,44]. In this study, we found that 78% (71 out of 91) of PODs (log2-based fold change > 1) were upregulated in the storage roots of SC124 (Figure 6C). Interestingly, 13% (12 out of 91) of PODs (log2-based fold change > 1) were induced at all points. Collectively, these results indicate that MePOD genes are involved in the PPD process in cassava storage roots.
Previous research has indicated that PODs can extensively participate in plants’ responsse to biotic and abiotic stresses [18,19,20,21,22,23]. Here, we selected nine genes (MePOD-13, -16, -17, -19, -23, -68, -74, -85, and -86) to further examine their expression levels after various treatments (Figure 7; Table S6). These genes are located on different regions of chr7, 13, 3, 17, 8, 15, 10, 9, and 18, respectively (Figure 4). Phylogenetic analysis indicates that MePOD-16, -68, -74, and -85 belong to subgroup A; MePOD-17 belongs to subgroup D; and MePOD-13, -19, -23, and -86 belong to subgroup E (Figure 1). The results show that all nine of the analyzed MePODs were upregulated in response to at least two types of treatments. MePOD17 and MePOD85 (log2-based fold change > 1) were induced by six treatments (MeJA, salt, cold stress, osmotic stress, ABA, and Xam and MeJA, osmotic stress, SA, ABA, H2O2, and Xam, respectively); MePOD13 was upregulated by five treatments (MeJA, osmotic stress, ABA, SA, H2O2, and Xam); and MePOD23 and MePOD86 were upregulated by four treatments (MeJA, osmotic stress, SA, and H2O2 and MeJA, SA, H2O2, and Xam, respectively). Of these, MePOD13 and MePOD23 were induced after H2O2 treatment in cassava leaves (Figure 7) but exhibited the opposite trend of expression during the PPD process in storage roots (Figure 6C), suggesting their differential roles in diverse tissues. MePOD-13, -19, -23, -68, and -86 (belonging to subgroup E, except for MePOD68) were upregulated by MeJA and SA treatments but downregulated by ABA treatment (Figure 1 and Figure 7). The expression of some PODs has been induced by MeJA and SA treatments in other plant species [2,46]. The opposite direction of expression of these POD genes between MeJA and SA treatments and ABA treatment may be due to the antagonism between MeJA/SA and ABA [47,48]. Whereas ABA plays a prominent role in plants’ tolerance to drought stress [38], MeJA- and SA-mediated signaling pathways are also activated under drought stress [49,50]. The induction of these genes by MeJA, SA, and drought suggests their possible involvement in MeJA- and SA-mediated drought responses in cassava. The responses of POD genes to multiple treatments have been observed in other plants. In Arabidopsis, AtPrx33 and AtPrx34 were upregulated after H2O2 and flg22 treatments [51]. In maize, ZmPRX-26, -42, and -71 were induced by H2O2, salt, and PEG treatments [29]. Phylogenetic analysis of MePODs with AtPrx-33 and -34 and ZmPRX-26, -42, and -71 found that MePOD86 shares a close phylogenetic relationship with ZmPRX71 (Figure S1), suggesting their functional conservation in multiple treatments. Multiple stresses, such as cold, salt, or PEG, induced the activity of POD enzyme, demonstrating the response of POD genes to environmental stress [52,53,54]. These results suggest that MePODs participate in the response to multiple stresses or related signals and are candidate targets for the genetic improvement of cassava.
4. Materials and Methods
4.1. Plant Materials and Treatments
Three cassava genotypes, W14, SC124, and Arg7, were planted in the greenhouse of the Chinese Academy of Tropical Agricultural Science (Haikou, China). Their characteristics were described in our previous studies [38,55]. Stem segments containing three nodes were cut from eight-month-old cassava plants and planted in pots, as described in Hu’s study [51]. The transcripts of W14 and Arg7 MePOD genes in stems and leaves after being planted for 90 days and middle storage roots after being planted for 150 days were examined by RNA-Seq. After W14, SC124, and Arg7 were cultured for 90 days, they were subjected to drought stress by withholding water for 12 days, after which their leaves and roots were sampled to study the transcriptional responses by RNA-Seq. To examine the expression profiles of MePOD genes after the plants were exposed to stress and related signaling treatments, the 60-day-old Arg7 variety was treated with 100 μM MeJA for 0, 2, 6, 10, and 24 h; 300 mM NaCl for 0 h, 2 h, 6 h, 3 days, and 14 days; a low temperature (4 °C) for 0, 2, 5, 15, and 48 h; 100 μM SA for 0, 2, 6, 10, and 24 h; 200 mM mannitol (to induce osmotic stress) for 0 h, 2 h, 6 h, 3 days, and 14 days; 100 μM ABA for 0, 2, 6, 10, and 24 h; 10% H2O2 for 0, 2, 6, 10, and 24 h; or Xam for 0, 2, 6, 12, and 24 h. Ten-month-old cassava storage roots (CSR) were cut into 5-mm-thick slices and placed into Petri dishes containing wet filter paper for 0, 6, 12, and 48 h to study the expression changes in MePOD genes via RNA-Seq during CSR deterioration. All samples were frozen immediately in liquid nitrogen and stored at −80 °C for RNA-Seq and qRT-PCR.
4.2. Identification and Phylogenetic Analysis of PODs in Cassava
MePOD genes were identified in cassava on the basis of homology with 73 POD protein sequences from the Arabidopsis genome database (available online: http://www.arabidopsis.org/index.jsp) and 138 POD protein sequences from the rice genome database (available online: http://rice.plantbiology.msu.edu/index.shtml) [56,57]. The Hidden Markov Model-based search (HMMER: http://hmmer.wustl.edu/) profile of these confirmed POD proteins was constructed to search the cassava genome hub (available online: http://www.phytozome.net/cassava.php) [58]. Finally, all predicted POD protein sequences were further examined by CDD (available online: http://www.ncbi.nlm.nih.gov/cdd/) and PFAM (available online: http://pfam.sanger.ac.uk/) after being checked by BLAST analyses [59,60]. All the predicted cassava POD genes identified from HMMER and BLAST were confirmed only if they included the POD special domains examined by SMART (available online: http://smart.embl-heidelberg.de/) [61]. Multiple sequence alignment of all predicted MePOD protein sequences was performed with Clustal W in BioEdit software [62]. The phylogenetic tree of the full-length MePOD protein sequences was created using MEGA 5.0 (University College Dublin, Dublin, Ireland) with the neighbor-joining (NJ) method, and bootstrap analysis was conducted with 1000 replicates [63].
4.3. Protein Properties and Structure Analyses of PODs in Cassava
The ProtParam database (available online: http://web.expasy.org/protparam/) was used to predict the properties, including amino acid numbers, molecular weights (MW), and isoelectric points (pI), of all presumed POD proteins [64]. The motifs were analyzed using the MEME program (available online: http://meme-suite.org/tools/meme), in which the maximum number of motifs was set to 10, the optimum width of motifs was set to 15–50 amino acid residues, and the other settings were kept at default values [65]. Subsequently, these 10 motifs were annotated in InterProScan (available online: http://www.ebi.ac.uk/Tools/pfa/iprscan/) [66]. The gene structures of each MePOD were investigated using GSDS software (available online: http://gsds.cbi.pku.edu.cn/) using each MePOD’s genomic DNA sequence and its corresponding CDS sequence, which were retrieved from the cassava genome database [67].
4.4. Chromosomal Location and Duplication Pattern Analyses
According to the results of BLASTN in the Phytozome 12.0 cassava database, MePOD genes were mapped to different chromosomes. On the basis of the calculated value of nucleotide sequence similarity and the phylogenetic relationship of cassava POD genes, paralogous genes were identified. The gene duplication pattern of paralogous MePOD genes was determined by the following two criteria: (1) the identity of the aligned region was >90% and (2) the alignment covered >90% of the longer gene. Circos software (Canada’s Michael Smith Genome Sciences Center, Vancouver, Canada) was used to draw the duplication events of MePOD genes [68,69]. The values of nonsynonymous substitution (Ka) and synonymous substitution (Ks) were calculated suing DnaSP 5.0 software [70]. Ka/Ks rate > 1 indicates positive evolution, Ka/Ks rate = 1 indicates neutral evolution, and Ka/Ks rate < 1 indicates negative evolution [71].
4.5. Transcriptome Analyses of PODs in Cassava
RNA-Seq was used to determine the expression of cassava MePOD genes. Total RNA was isolated from frozen stems, leaves, roots, and storage roots using the plant RNeasy extraction kit (TIANGEN, Beijing, China) and quantified with a NanoDrop 2000c (Thermo Scientific Inc., Waltham, MA, USA). Total RNA (3 µg) of each sample was used to construct the cDNA library according to the Illumina instructions and then sequenced using an Illumina GA II (Illumina Inc., San Diego, USA). The original data processing and analysis methods were described in our previous study [72].
4.6. Quantitative Real-Time PCR Analyses
Leaf samples of the Arg7 variety subjected to MeJA, SA, ABA, NaCl, low temperature, mannitol, H2O2, or Xam treatments were collected to perform qRT-PCR analysis. Total RNA (1 µg) from each sample was used to synthesize the first-strand cDNA using oligo-dT primer by SuperScript reverse transcriptase (Takara, Dalian, China). The cDNA product was diluted to 50 ng·μL−1, and 1 μL was used for qRT-PCR. The qRT-PCR reaction mixtures (20 μL) contained 0.6 μL of each gene-specific primer (300 nmol·μL−1), 10 μL of 2× FastFire qPCR PreMix (Tiangen, Beijing, China), and 7.8 μL of RNase-free water. The qRT-qPCR thermal cycling included cDNA denaturation at 95 °C for 1 min, with 40 cycles of 95 °C for 5 s and 60 °C for 15 s in the Mx3005P Real-Time PCR System (Agilent Inc., Palo Alto, CA, USA) with the SYBR green method. The β-tubulin gene of cassava was chosen as an internal control. All qRT-qPCR experiments were performed in triplicate, and the gene-specific primers used in expression analysis are listed in Table S7. The data obtained from the qRT-qPCR were analyzed with Tukey’s post-hoc ANOVA in SPSS 22.0 (SPSS Inc., Chicago, USA) (P < 0.05) after fold treatment with the 2−△△Ct method.
5. Conclusions
In this study, we identified 91 PODs from the cassava genome and studied their basic classification, protein motif, gene structure, chromosomal distribution, and duplication pattern. Comprehensive transcriptional level analyses revealed the involvement of MePODs in biotic and abiotic stress responses, hormone responses, and storage root deterioration. Several MePOD genes (MePOD-13, -17, -85, and -86) were found to be transcriptionally upregulated after multiple different treatments, suggesting that these genes are good candidates to target for cassava improvement. These findings increase our understanding of POD-mediated stress and hormone responses and storage root deterioration in cassava, laying a foundation for the genetic improvement of cassava.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/11/2730/s1. Table S1. Characteristics of PODs in cassava. Table S2. The Ka/Ks ratios of duplicated POD genes in cassava. Table S3. The expression profiles (log2-based values) of the cassava POD genes in different tissues. Table S4. The expression profiles (log2-based fold changes) of the cassava POD genes after drought treatment. Table S5. The expression profiles (log2-based fold changes) of the cassava POD genes after harvest. Table S6. The expression profiles (log2-based values) of the cassava POD genes after various stress treatments. Table S7. Primers used in qRT-PCR analysis. Figure S1. Phylogenetic analyses of 91 MePODs, 2 AtPRXs, 3 OsPRXs, and 3 ZmPRXs.
Author Contributions
W.H., X.D., and H.Y. designed the research; C.W., X.D., Z.D., and Y.Y. performed the research; X.D., W.T., C.W., and Y.W. analyzed the data; X.D. and C.W. wrote the paper; W.H., Z.D., and H.Y. reviewed the paper. All authors have read and approved the final manuscript.
Funding
This research was funded by the Natural Science Foundation of Hainan Province (318MS092), the National Natural Science Foundation of China (31771859), the Central Public-Interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630052016005, 1630052016006, 1630052017021, 1630012019009, 1630052019023), the Central Public-Interest Scientific Institution Basal Research Fund for Innovative Research Team Program of CATAS (17CXTD-28, 1630052017017), and the earmarked fund for Modern Agro-industry Technology Research System (CARS-11).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Passardi F., Cosio C., Penel C., Dunand C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005;24:255–265. doi: 10.1007/s00299-005-0972-6. [DOI] [PubMed] [Google Scholar]
- 2.Almagro L., Gómez Ros L.V., Belchi-Navarro S., Bru R., Ros Barceló A., Pedreno M.A. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2009;60:377–390. doi: 10.1093/jxb/ern277. [DOI] [PubMed] [Google Scholar]
- 3.Welinder K.G. Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 1992;2:388–393. doi: 10.1016/0959-440X(92)90230-5. [DOI] [Google Scholar]
- 4.Mathé C., Barre A., Jourda C., Dunand C. Evolution and expression of class III peroxidases. Arch. Biochem. Biophys. 2010;500:58–65. doi: 10.1016/j.abb.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Hiraga S., Sasaki K., Ito H., Ohashi Y., Matsui H. A large family of Class III plant peroxidases. Plant Cell Physiol. 2001;42:462–468. doi: 10.1093/pcp/pce061. [DOI] [PubMed] [Google Scholar]
- 6.Zheng X.H., Van Huystee R.B. Oxidation of tyrosine by peroxidase isozymes derived from peanut suspension culture medium and by isolated cell walls. Plant Cell Tiss. Org. 1991;25:35–43. doi: 10.1007/BF00033910. [DOI] [Google Scholar]
- 7.Barceló A.R., Ros L.V., Carrasco A.E. Looking for syringyl peroxidases. Trends Plant Sci. 2007;12:486–491. doi: 10.1016/j.tplants.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Henriksen A., Mirza O., Indiani C., Teilum K., Smulevich G., Welinder K.G., Gajhede M. Structure of soybean seed coat peroxidase: A plant peroxidase with unusual stability and haem-apoprotein interactions. Protein Sci. 2001;10:108–115. doi: 10.1110/ps.37301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Passardi F., Longet D., Penel C., Dunand C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry. 2004;65:1879–1893. doi: 10.1016/j.phytochem.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Cosio C., Ranocha P., Francoz E., Burlat V., Zheng Y., Perry S.E., Ripoll J.J., Yanofsky M., Dunand C. The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 2016;213:250–263. doi: 10.1111/nph.14127. [DOI] [PubMed] [Google Scholar]
- 11.Passardi F., Penel C., Dunand C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004;9:534–540. doi: 10.1016/j.tplants.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Ostergaard L., Teilum K., Mirza O., Mattsson O., Petersen M., Welinder K.G., Mundy J., Gajhede M., Henriksen A. Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 2000;44:231–243. doi: 10.1023/A:1006442618860. [DOI] [PubMed] [Google Scholar]
- 13.Hiraga S., Yamamoto K., Ito H., Sasaki K., Matsui H., Honma M., Nagamura Y., Sasaki T., Ohashi Y. Diverse expression pro¢les of 21 rice peroxidase genes. Febs Lett. 2000;471:245–250. doi: 10.1016/S0014-5793(00)01409-5. [DOI] [PubMed] [Google Scholar]
- 14.Chen D., Ding Y., Guo W., Zhang T. Molecular cloning and characterization of a flower-specific class III peroxidase gene in G. Hirsutum. Mol. Biol. Rep. 2009;36:461–469. doi: 10.1007/s11033-007-9202-3. [DOI] [PubMed] [Google Scholar]
- 15.González A.M., Marcel T.C., Kohutova Z., Stam P., van der Linden C.G., Niks R.E. Peroxidase profiling reveals genetic linkage between peroxidase gene clusters and basal host and non-host esistance to rusts and mildew in barley. PLoS ONE. 2010;5:e10495. doi: 10.1371/journal.pone.0010495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin J., Hewezi T., Baum T.J. Arabidopsis peroxidase AtPRX53 influences cell elongation and susceptibility to Heterodera schachtii. Plant Signal. Behav. 2011;6:1778–1786. doi: 10.4161/psb.6.11.17684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigeto J., Tsutsumi Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016;209:1395–1402. doi: 10.1111/nph.13738. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y., Yang Z., How J., Xu H., Chen L., Li K. Overexpression of a peroxidase gene (AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Mol. Biol. 2017;95:157–168. doi: 10.1007/s11103-017-0644-2. [DOI] [PubMed] [Google Scholar]
- 19.Llorente F., López-Cobollo R.M., Catalá R., Martínez-Zapater J.M., Salinas J. A novel cold-inducible gene from Arabidopsis, RCI3, encodes a peroxidase that constitutes a component for stress tolerance. Plant J. 2002;32:13–24. doi: 10.1046/j.1365-313X.2002.01398.x. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S., Jaggi M., Sinha A.K. Ectopic overexpression of vacuolar and apoplastic Catharanthus roseus peroxidases confers differential tolerance to salt and dehydration stress in transgenic tobacco. Protoplasma. 2012;249:423–432. doi: 10.1007/s00709-011-0294-1. [DOI] [PubMed] [Google Scholar]
- 21.Coego A., Ramirez V., Ellul P., Mayda E., Vera P. The H2O2-regulated Ep5C gene encodes a peroxidase required for bacterial speck susceptibility in tomato. Plant J. 2005;42:283–293. doi: 10.1111/j.1365-313X.2005.02372.x. [DOI] [PubMed] [Google Scholar]
- 22.Choi H.W., Kim Y.J., Lee S.C., Hong J.K., Hwang B.K. Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 2007;145:890–904. doi: 10.1104/pp.107.103325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wally O., Punja Z.K. Enhanced disease resistance in transgenic carrot (Daucus carota L.) plants over-expressing a rice cationic peroxidase. Planta. 2010;232:1229–1239. doi: 10.1007/s00425-010-1252-4. [DOI] [PubMed] [Google Scholar]
- 24.Tognolli M., Penel C., Greppin H., Simon P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene. 2002;288:129–138. doi: 10.1016/S0378-1119(02)00465-1. [DOI] [PubMed] [Google Scholar]
- 25.Valério L., De Meyer M., Penel C., Dunand C. Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry. 2004;65:1331–1342. doi: 10.1016/j.phytochem.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Duroux L., Welinder K.G. The peroxidase gene family in plants: A phylogenetic overview. J. Mol. Evol. 2003;57:397–407. doi: 10.1007/s00239-003-2489-3. [DOI] [PubMed] [Google Scholar]
- 27.Ren L.L., Liu Y.J., Liu H.J., Qian T.T., Qi L.W., Wang X.R., Zeng Q.Y. Subcellular relocalization and positive selection play key roles in the retention of duplicate genes of populus Class III peroxidase family. Plant Cell. 2014;26:2404–2419. doi: 10.1105/tpc.114.124750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behr M., Legay S., Hausman J.F., Guerriero G. Analysis of cell wall-related genes in organs of Medicago sativa L. under different abiotic stresses. Int. J. Mol. Sci. 2015;16:16104–16124. doi: 10.3390/ijms160716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Wang Q., Zhao Y., Han G., Zhu S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subgroup involved in abiotic stress response. Gene. 2015;566:95–108. doi: 10.1016/j.gene.2015.04.041. [DOI] [PubMed] [Google Scholar]
- 30.Cao Y., Han Y., Meng D., Li D., Jin Q., Lin Y., Cai Y. Structural, evolutionary, and functional analysis of the Class III peroxidase gene family in Chinese pear (Pyrus bretschneideri) Front Plant Sci. 2016;7:1874. doi: 10.3389/fpls.2016.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveira E.J., Santana F.A., Oliveira L.A., Santos V.S. Genetic parameters and prediction of genotypic values for root quality traits in cassava using REML/BLUP. Genet Mol. Res. 2014;13:6683–6700. doi: 10.4238/2014.August.28.13. [DOI] [PubMed] [Google Scholar]
- 32.International Cassava Genetic Map Consortium High-resolution linkage map and chromosome-scale genome assembly for cassava (Manihot esculenta Crantz) from 10 populations. G3 (BethesdaMd.) 2014;5:133–144. doi: 10.1534/g3.114.015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maran J.P., Sivakumar V., Thirugnanasambandham K., Sridhar R. Degradation behavior of biocomposites based on cassava starch buried under indoor soil conditions. Carbohydr. Polym. 2014;30:20–28. doi: 10.1016/j.carbpol.2013.08.080. [DOI] [PubMed] [Google Scholar]
- 34.Vanderschuren H., Nyaboga E., Poon J.S., Baerenfaller K., Grossmann J., Hirsch-Hoffmann M., Kirchgessner N., Nanni P., Gruissem W. Large-Scale proteomics of the cassava storage root and identification of a target gene to reduce postharvest deterioration. Plant Cell. 2014;26:1913–1924. doi: 10.1105/tpc.114.123927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu W., Kong H., Guo Y., Zhang Y., Ding Z., Tie W., Yan Y., Huang Q., Peng M., Shi H., et al. Comparative physiological and transcriptomic analyses reveal the actions of melatonin in the delay of postharvest physiological deterioration of cassava. Front Plant Sci. 2016;7:736. doi: 10.3389/fpls.2016.00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freeling M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu Rev. Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- 37.Cao J., Shi F. Evolution of the RALF gene family in plants: Gene duplication and selection patterns. Evol. Bioinf. 2012;8:271–292. doi: 10.4137/EBO.S9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu W., Wei Y., Xia Z., Yan Y., Hou X., Zou M., Lu C., Wang W., Peng M. Genome-wide identification and expression analysis of the NAC transcription factor family in cassava. PLoS ONE. 2015;10:e0136993. doi: 10.1371/journal.pone.0136993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shang S., Wu C., Huang C., Tie W., Yan Y., Ding Z., Xia Z., Wang W., Peng M., Tian L., et al. Genome-wide analysis of the GRF family reveals their involvement in abiotic stress response in cassava. Genes. 2018;9:110. doi: 10.3390/genes9020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo X.H., Jiang J., Wang B.C., Li H.Y., Wang Y.C., Yang C.P., Liu G.F. ThPOD3, a truncated polypeptide from Tamarix hispida, conferred drought tolerance in Escherichia coli. Mol. Biol. Rep. 2010;37:1183–1190. doi: 10.1007/s11033-009-9484-8. [DOI] [PubMed] [Google Scholar]
- 41.Choi H.W., Hwang B.K. The pepper extracellular peroxidase CaPO2 is required for salt, drought and oxidative stress tolerance as well as resistance to fungal pathogens. Planta. 2012;235:1369–1382. doi: 10.1007/s00425-011-1580-z. [DOI] [PubMed] [Google Scholar]
- 42.Hu W., Huang C., Deng X., Zhou S., Chen L., Li Y., Wang C., Ma Z., Yuan Q., Wang Y., et al. Ta ASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Env. 2013;36:1449–1464. doi: 10.1111/pce.12074. [DOI] [PubMed] [Google Scholar]
- 43.Iyer S., Mattinson D.S., Fellman J.K. Study of the early events leading to cassava root postharvest deterioration. Trop. Plant Biol. 2010;3:151–165. doi: 10.1007/s12042-010-9052-3. [DOI] [Google Scholar]
- 44.Park M.H. Sucrose delays senescence and preserves functional compounds in Asparagus officinalis L. Biochem. Biophys. Res. Commun. 2016;480:241–247. doi: 10.1016/j.bbrc.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 45.Xi Y., Jiao W., Cao J., Jiang W. Effects of chlorogenic acid on capacity of free radicals scavenging and proteomic changes in postharvest fruit of nectarine. PLoS ONE. 2017;12:e0182494. doi: 10.1371/journal.pone.0182494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J.E., Liu K.K., Li D.W., Zhang Y.L., Zhao Q., Gong Z.H. A novel peroxidase CanPOD gene of pepper is involved in defense responses to Phytophtora capsici infection as well as abiotic stress tolerance. Int. J. Mol. Sci. 2013;14:3158–3177. doi: 10.3390/ijms14023158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moons A. Antagonistic effects of Abscisic Acid and Jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell. 1997;9:2243–2259. doi: 10.1105/tpc.9.12.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasuda M., Ishikawa A., Jikumaru Y., Seki M., Nakashita H. Antagonistic interaction between systemic acquired resistance and the Abscisic Acid-mediated abiotic stress response in Arabidopsis. Plant Cell. 2008;20:1678–1692. doi: 10.1105/tpc.107.054296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H.L., Deng H.P., Sun Z.Y., Zhao L.J., Han L., Ju G.S., Qian Y.Q. Effect of methyl jasmonate on physiological indexes of chrysanthemum cuttage under natural drought stress. For. Res. 2010;23:733–737. doi: 10.1080/00949651003724790. [DOI] [Google Scholar]
- 50.Pandey S., Chakraborty D. Salicylic acid and drought stress response: Biochemical to molecular crosstalk: Stress responses in plants. Stress Responses Plants. 2015;3:247–265. doi: 10.1007/978-3-319-13368-3_10. [DOI] [Google Scholar]
- 51.Daudi A., Cheng Z., O’Brien J.A., Mammarella N., Khan S., Ausubel F.M., Bolwell G.P. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012;24:275–287. doi: 10.1105/tpc.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Q., Wang Y., Li B., Chang J., Chen M., Li K., Yang G., He G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015;15:268. doi: 10.1186/s12870-015-0644-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meng D., Yu X., Ma L., Hu J., Liang Y., Liu X., Yin H., Liu H., He X., Li D. Transcriptomic Response of Chinese Yew (Taxus chinensis) to cold stress. Front. Plant Sci. 2017;8:868. doi: 10.3389/fpls.2017.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu L., Ding Z., Han B., Hu W., Li Y., Zhang J. Physiological investigation and transcriptome analysis of polyethylene glycol (PEG)-induced dehydration stress in cassava. Int. J. Mol. Sci. 2016;17:283. doi: 10.3390/ijms17030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu W., Yang H., Yan Y., Wei Y., Tie W., Ding Z., Zuo J., Peng M., Li K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016;6:22783. doi: 10.1038/srep22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawahara Y., de la Bastide M., Hamilton J.P., Kanamori H., McCombie W.R., Ouyang S., Schwartz D.C., Tanaka T., Wu J., Zhou S., et al. Improvement of the Oryza sativa nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:461. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potter S.C., Luciani A., Eddy S.R., Park Y., Lopez R., Finn R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marchler-Bauer A., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., et al. CDD: Specific functional annotation with the conserved domain database. Nucleic Acids Res. 2008;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finn R.D., Tate J., Mistry J., Coggill P.C., Sammut S.J., Hotz H.R., Ceric G., Forslund K., Eddy S.R., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2007;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Chris Ponting C.P., Bork P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 63.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown P., Baxter L., Hickman R., Beynon J., Moore J.D., Ott S. MEME-LaB: Motif analysis in clusters. Bioinformatics. 2013;29:1696–1697. doi: 10.1093/bioinformatics/btt248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McDowall J., Hunter S. InterPro protein classification. Methods Mol. Biol. 2011;694:37–47. doi: 10.1007/978-1-60761-977-2_3. [DOI] [PubMed] [Google Scholar]
- 67.Hu B., Jin J.P., Guo A.Y., Zhang H., Luo J.C., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang M., Yue H., Feng K., Deng P., Song W., Nie X. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.) BMC Genom. 2016;17:668. doi: 10.1186/s12864-016-2993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin X., Zhu L., Yao Q., Meng X., Ding G., Wang D., Xie Q., Tong Z., Tao C., Yu L., et al. Expression profiling of mitogen-activated protein kinase genes reveals their evolutionary and functional diversity in different rubber tree (Hevea brasiliensis) cultivars. Genes. 2017;8:261. doi: 10.3390/genes8100261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 71.Liang Y., Xiong Z., Zheng J., Xu D., Zhu Z., Xiang J., Gan J., Raboanatahiry N., Yin Y., Li M. Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus. Sci. Rep. 2016;6:24265. doi: 10.1038/srep24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu C., Hu W., Yan Y., Tie W., Ding Z., Guo J., He G. The late embryogenesis abundant protein family in cassava (Manihot esculenta crantz): Genome-wide characterization and expression during abiotic stress. Molecules. 2018;23:1196. doi: 10.3390/molecules23051196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.