Abstract
The essential cellular process of ribosome biogenesis is at the nexus of various signalling pathways that coordinate protein synthesis with cellular growth and proliferation. The fact that numerous diseases are caused by defects in ribosome assembly underscores the importance of obtaining a detailed understanding of this pathway. Studies in yeast have provided a wealth of information about the fundamental principles of ribosome assembly, and although many features are conserved throughout eukaryotes, the larger size of human (pre‐)ribosomes, as well as the evolution of additional regulatory networks that can modulate ribosome assembly and function, have resulted in a more complex assembly pathway in humans. Notably, many ribosome biogenesis factors conserved from yeast appear to have subtly different or additional functions in humans. In addition, recent genome‐wide, RNAi‐based screens have identified a plethora of novel factors required for human ribosome biogenesis. In this review, we discuss key aspects of human ribosome production, highlighting differences to yeast, links to disease, as well as emerging concepts such as extra‐ribosomal functions of ribosomal proteins and ribosome heterogeneity.
Keywords: ribosomal protein, ribosome, ribosomopathy, RNA modification, RNA processing
Subject Categories: Protein Biosynthesis & Quality Control, RNA Biology, Structural Biology
Glossary
- cryo‐EM
cryo‐electron microscopy
- DBA
Diamond‐Blackfan anemia
- ETS
external transcribed spacer
- IRES
internal ribosome entry site
- ITS
internal transcribed spacer
- LSU
large ribosomal subunit
- Nm
ribose 2′‐O‐methylation
- NTP
nucleoside triphosphate
- RBF
ribosome biogenesis factor
- RNP
ribonucleoprotein complex
- RP
ribosomal protein
- snoRNP
small nucleolar ribonucleoprotein complex
- SSU
small ribosomal subunit
- UTR
untranslated region
- Ψ
pseudouridine
Ribosome production: going beyond the yeast model system
Production of proteins is an essential cellular process carried out by ribosomes, which are large ribonucleoprotein complexes (RNPs) composed of four ribosomal RNAs (rRNAs; 18S, 5S, 5.8S and 25S (yeast)/28S (humans)) and approximately 80 ribosomal proteins (RPs) (Ben‐Shem et al, 2010; Anger et al, 2013). Ribosome assembly requires the coordinated action of all three RNA polymerases and is one of the most energy‐consuming cellular processes, with approximately 7,500 new ribosomal subunits synthesised per minute in actively growing HeLa cells (Warner, 1999; Lewis & Tollervey, 2000). In eukaryotes, this process begins in the nucleolus with the RNA polymerase I‐mediated synthesis of three of the four rRNAs as a single precursor rRNA (pre‐rRNA) transcript. Recruitment of numerous RPs and trans‐acting ribosome biogenesis factors (RBFs) to this nascent transcript leads to formation of large, early pre‐ribosomal particles (90S). Concurrent with protein assembly, the pre‐rRNA transcript is extensively processed and numerous rRNA modifications take place. A central pre‐rRNA cleavage event gives rise to precursors of the small (SSU; 40S) and large (LSU; 60S) subunits. The pre‐40S and pre‐60S particles undergo extensive structural remodelling, involving the dynamic association and dissociation of numerous RBFs as well as the establishment of key ribosomal structures. After the immature subunits are independently exported to the cytoplasm, final maturation and quality control steps occur before they engage in translation.
Eukaryotic ribosome synthesis is best characterised in the model organism Saccharomyces cerevisiae (budding yeast; Woolford & Baserga, 2013). While the basic features of the pathway are conserved among all eukaryotes, the biogenesis pathway for human ribosomes has evolved to be considerably more complex due to the increased size of human ribosomes and pre‐ribosomes, and the evolution of regulatory networks that modulate ribosome assembly and function. Proteomic analyses of human nucleoli and several recent RNAi‐based screens (Andersen et al, 2002; Scherl et al, 2002; Wild et al, 2010; Tafforeau et al, 2013; Badertscher et al, 2015; Farley‐Barnes et al, 2018) have facilitated identification of human RBFs, and characterisation of some of these factors has already highlighted interesting differences between the assembly pathway of yeast and human ribosomes. Furthermore, these screens have revealed the extent to which ribosome biogenesis is coordinated with other cellular processes. The importance of uncovering such differences and gaining a comprehensive understanding of ribosome assembly in humans is underscored by the increasing number of genetic diseases (ribosomopathies) that have been found to be caused by mutations in genes encoding ribosomal proteins or RBFs (Narla & Ebert, 2010; Mills & Green, 2017). Furthermore, an expanding body of evidence demonstrates that ribosome assembly is regulated by several oncogenic signalling pathways, and defects in ribosome biogenesis have in turn been linked to the activation of tumour suppressors including p53 (Pelletier et al, 2018).
Here, we review the current knowledge of the ribosome biogenesis pathway in humans, focusing on aspects of the process that have been characterised in detail, and highlighting key differences to the pathway in yeast. We further highlight emerging topics such as ribosome heterogeneity, describe the regulation of ribosome assembly and discuss how defects in ribosome synthesis are linked to disease.
Identification and properties of human ribosome biogenesis factors
In yeast, a list of approximately 200 RBFs has been compiled over many years based on mass spectrometric analyses of isolated pre‐ribosomal complexes as well as examination of the effects of depletion of nucleolar proteins on ribosomal subunit production (Woolford & Baserga, 2013). While the inventory of yeast RBFs provides an excellent basis for analysing human ribosome assembly, more than 4,500 proteins have been detected in human nucleoli (Andersen et al, 2002; Scherl et al, 2002), implying that many additional factors may contribute to ribosome maturation in human cells. To identify novel factors required for ribosome assembly in human cells, several RNAi‐based, high‐throughput screens have recently been performed (Wild et al, 2010; Tafforeau et al, 2013; Badertscher et al, 2015; Farley‐Barnes et al, 2018). A pioneering microscopy‐based screen monitoring the mis‐localisation of pre‐ribosomal subunits confirmed the involvement of 153 proteins in human ribosome assembly (Wild et al, 2010), and this approach was subsequently extended to a genome‐wide level, revealing approximately 300 factors required for 40S maturation (Badertscher et al, 2015). Concurrently, 286 human RBFs, including 74 without yeast homologues, were identified in a screen for nucleolar proteins whose depletion affects pre‐rRNA processing (Tafforeau et al, 2013). Furthermore, 139 potential RBFs were recently discovered by a screen for factors affecting the number or morphology of nucleoli (Farley‐Barnes et al, 2018). These screens, together with numerous studies characterising individual or subsets of human RBFs, have not only confirmed the involvement of many of the homologues of yeast RBFs in human ribosome assembly, but also highlighted many additional proteins required for this process in human cells. A significant number of the factors identified have links to disease, emphasising the importance of correct ribosome production for cellular homeostasis. Some of the proteins found in these screens have by now been analysed in detail, but many of the newly discovered human RBFs still lack functional characterisation, and their precise roles in subunit assembly are still unknown. Although depletion of the factors identified in these screens affects ribosome biogenesis, it is unlikely that they are all bona fide components of pre‐ribosomal complexes, and many of the observed effects may rather point to mechanisms by which ribosome assembly can be regulated. For example, alongside the anticipated RPs and RBFs, proteins involved in transcription, pre‐mRNA splicing and translation were found to be required for pre‐40S maturation (Badertscher et al, 2015), probably indicating crosstalk between subunit assembly and other aspects of the gene expression pathway. Furthermore, various metabolic enzymes and signalling factors were identified as human RBFs (Badertscher et al, 2015; Farley‐Barnes et al, 2018), suggesting the co‐ordination of ribosome production with nutrient availability and cell growth; a model that is in line with the high energy demands of ribosome production. Depleting components of protein degradation machineries also affects pre‐40S maturation or nucleolar number (Badertscher et al, 2015; Farley‐Barnes et al, 2018), suggesting that maintaining the correct cellular levels of RPs and RBFs may be important for ongoing ribosome production. In the future, it will therefore be important to clearly differentiate between bona fide RBFs that are involved directly in subunit assembly, and proteins/pathways that modulate ribosome production.
The inventories of human RFBs highlight the diversity of proteins that contribute to ribosome maturation. As in yeast, some RBFs primarily mediate protein–protein interactions and act as structural scaffolds, while others are RNA‐binding proteins, and many possess catalytic activity. The spectrum of enzymatic RBFs includes many nucleases, modification enzymes and kinases (discussed in detail below), as well as a plethora of nucleoside triphosphate (NTP)‐dependent proteins such as GTPases, AAA‐ATPases and RNA helicases that mediate structural remodelling events. Six active GTPases (Nog1, Nog2, Nug1, Bms1, Lsg1, Efl1) are involved in yeast ribosome biogenesis, and although human homologues for each of these proteins have been found in screens for human RBFs, none of those proteins have been analysed in detail in human cells so far. Three yeast AAA‐ATPases (Rea1/Mdn1, Rix7, Drg1) are implicated in triggering the release of specific pre‐60S biogenesis factors (reviewed in Kressler et al, 2012), and human homologues of each of these proteins (MDN1, NVL2 and AFGH2, respectively) are known. In the case of MDN1, association with pre‐60S complexes and the PELP1‐TEX10‐WDR18 complex (Rix1 complex in yeast) suggest that the function of this enzyme is conserved from yeast to humans (Raman et al, 2016). Human NVL2 has been linked to the nuclear exosome and to telomerase (Her & Chung, 2012; Yoshikatsu et al, 2015), suggesting that it may be a multifunctional enzyme involved not only in ribosome assembly but also in other aspects of RNA biology. Although the roles of some RNA helicases are common to yeast and humans (e.g. yeast Dhr1 and human DHX37 both mediate release of the U3 snoRNA; Martin et al, 2013; Sardana et al, 2015; Choudhury et al, 2019), additional pre‐ribosomal functions have been described for several human RNA helicases. Examples include the requirement for DDX51 for release of the metazoan‐specific snoRNA U8 from pre‐LSU complexes (Srivastava et al, 2010), as well as the roles of DDX21 in coupling pre‐rRNA transcription and processing, and facilitating the access of late‐acting small nucleolar RNP (snoRNPs) to pre‐40S complexes (Calo et al, 2015; Sloan et al, 2015). The remodelling events catalysed by such enzymatic RBFs often serve as important checkpoints during ribosome assembly and thereby help maintain the directionality of the pathway.
While many RBFs associate with pre‐ribosomal particles independently, a hallmark of eukaryotic ribosome assembly is the modular assembly of pre‐ribosomal complexes. In both yeast and humans, the UTP‐A, UTP‐B and UTP‐C complexes, as well as the U3 snoRNP, the RCL1‐BMS1 heterodimer and the IMP3‐IMP4‐MPP10 and EMG1 complexes, are all recruited to the nascent pre‐rRNA transcript and form the core of the so‐called “SSU processome” (see Table 1 and references therein). Similarly, several sub‐complexes, such as the PeBoW (Nop7‐Erb1‐Ytm1 in yeast) and PELP1‐TEX10‐WDR18 (Rix1‐Ipi3‐Ipi1 in yeast) complexes, act during pre‐60S biogenesis. Despite the evolutionary conservation of these complexes, their composition varies between species, and several additional components have been identified in human cells (Table 1). DDX21 and DDX27, additional components of the human UTP‐B and PeBoW complexes, respectively (Kellner et al, 2015; Sloan et al, 2015), are both RNA helicases, possibly indicating the need for additional remodelling steps during the early stages of human pre‐ribosome assembly compared to yeast. Furthermore, several novel pre‐ribosomal sub‐complexes have been identified in human cells. For example, apoptosis‐antagonising transcription factor AATF, neuroguidin (NGDN) and NOL10 form a nucleolar sub‐complex (ANN) in which each of the components co‐stabilise the other proteins (Bammert et al, 2016). These proteins associate with early pre‐ribosomal complexes, and depletion of any of the ANN components leads to defects in early pre‐rRNA cleavage events. XND, another nucleolar complex composed of the G‐patch protein NF‐κB‐repressing factor (NKRF), the RNA helicase DHX15 and the 5′‐3′ exonuclease XRN2, is also involved in early aspects of human ribosome assembly (Memet et al, 2017). XRN2 is recruited by NKRF to pre‐ribosomal complexes, where it fulfils its various roles in pre‐rRNA processing and turnover of excised pre‐rRNA fragments. NKRF also stimulates the ATPase and unwinding activity of DHX15 (Memet et al, 2017), and it has been suggested that these proteins function together in a pre‐rRNA remodelling step that facilitates initial pre‐rRNA cleavage. The fact that DHX15 is required for processing of the 5′ external transcribed spacer (ETS) in human cells contrasts the roles of its yeast homologue Prp43, which is involved in release of snoRNAs from pre‐60S particles and facilitates cleavage of the 3′ end of the 18S rRNA in the cytoplasm (Bohnsack et al, 2009; Pertschy et al, 2009). Furthermore, the NF45‐NF90 heterodimer, which was initially characterised for its binding to the interleukin‐2 promoter, was found to be a component of human pre‐60S complexes (Wandrey et al, 2015). The interaction between NF45‐NF90 and pre‐60S particles was shown to be mediated by the double‐stranded RNA‐binding domains (dsRBDs) of NF90, implying a direct association with the rRNA. Although no clear defect in pre‐rRNA processing was observed upon depletion of either NF45 or NF90, lack of these factors causes changes in nucleolar morphology and the accumulation of pre‐60S complexes in the nucleoplasm, confirming the requirement for this complex for biogenesis of the LSU (Wandrey et al, 2015).
Table 1.
Composition of pre‐ribosomal sub‐complexes in yeast and humans
| Complex | Composition | Refs. | |
|---|---|---|---|
| Yeast | Human | ||
| UTP‐A | Utp4 | UTP4 (Cirhin) | Krogan et al (2004), Prieto and McStay (2007), Freed et al (2012) |
| Utp5 | UTP5 (WDR43) | ||
| Utp8 | – | ||
| Utp9 | – | ||
| Utp10 | UTP10 (BAP28) | ||
| Utp15 | UTP15 | ||
| Utp17 | UTP17 (WDR75) | ||
| – | NOL11 | ||
| UTP‐B | Utp1 | PWP2 | Krogan et al (2004), Sloan et al (2015) |
| Utp6 | UTP6 | ||
| Utp12 | UTP12 | ||
| Utp13 | TBL3 | ||
| Utp18 | UTP18 | ||
| Utp21 | WDR36 | ||
| – | DDX21 | ||
| – | NOP2 | ||
| UTP‐C | Rrp7 | RRP7A | Krogan et al (2004), Gérus et al (2010), Baudin‐Baillieu et al (1997) |
| Utp22 | NOL6 | ||
| Cka1 | CSNK2A1 | ||
| Cka2 | – | ||
| Ckb1 | CSNK2B | ||
| Ckb2 | – | ||
| Rrp36 | RRP36 | ||
| U3 snoRNP | U3 snoRNA | U3 snoRNA | Grandi et al (2002), Turner et al (2012) |
| Nop56 | NOP56 | ||
| Nop58 | NOP58 | ||
| Snu13 | 15.5K | ||
| Nop1 | Fibrillarin | ||
| Rrp9 | U3‐55K | ||
| IMP3‐IMP4 | Imp3 | IMP3 | Granneman et al (2003) |
| Imp4 | IMP4 | ||
| Mpp10 | MPHOSPH10 | ||
| RCL1‐BMS1 | Rcl1 | RCL1 | Wegierski et al (2001), Wang et al (2016) |
| Bms1 | BMS1 | ||
| EMG1 | Emg1 | EMG1 | Liu and Thiele (2001), Kühn et al (2009), Warda et al (2016) |
| Nop14 | NOP14 | ||
| Noc4 | NOC4L | ||
| UTP14A | |||
| PeBoW | Nop7 | PES1 | Holzel et al (2005), Rohrmoser et al (2007), Kellner et al (2015) |
| Erb1 | BOP1 | ||
| Ytm1 | WDR12 | ||
| DDX27 | |||
| Rix1 | Rix1 | PELP1 | Finkbeiner et al (2011a,b) |
| Ipi3 | TEX10 | ||
| Ipi1 | WDR18 | ||
| XND | NKRF | Memet et al (2017) | |
| XRN2 | |||
| DHX15 | |||
| ANN | AATF | Bammert et al (2016) | |
| NGDN | |||
| NOL10 | |||
| NF45‐NF90 | NF45 | Wandrey et al (2015) | |
| NF90 | |||
Specialisation of the pre‐rRNA processing pathway in higher eukaryotes
An aspect of human ribosome assembly that has been studied extensively and where several differences to yeast have emerged is the processing of the pre‐rRNA transcript. Maturation of the rRNAs requires removal of the external and internal transcribed spacers (ETS and ITS, respectively), which are up to five times longer in humans than yeast, by a combination of endonucleolytic and exonucleolytic processing (Henras et al, 2015; Tomecki et al, 2017). While removal of the 5′ ETS and cleavage in ITS1 largely occur co‐transcriptionally in yeast, the majority of human pre‐rRNA processing appears to occur post‐transcriptionally (Lazdins et al, 1997). Analogous endonucleolytic cleavage sites can be identified in yeast and human pre‐rRNAs. Several recent studies indicate that the corresponding endonucleases (Utp24/UTP24, Nob1/NOB1, RNase MRP, Las1/LAS1) are largely conserved (Figure 1), but there are notable exceptions (Schmitt & Clayton, 1993; Fatica et al, 2003; Preti et al, 2013; Sloan et al, 2013a; Gasse et al, 2015; Tomecki et al, 2015; Wells et al, 2016; Goldfarb & Cech, 2017). For example, in yeast, site B0 at the 3′ end of the 25S rRNA is cleaved by Rnt1, but depletion of the homologous RNase III enzyme does not support a role for this enzyme in the analogous 02 site cleavage in humans. The endonuclease(s) responsible for cleavage at A0 (yeast) and A0 (human) cleavage sites in the 5′ ETS remain unknown, but processing of the A0 site in humans has been linked to the putative PIN‐domain endonuclease UTP23, which unlike its degenerate yeast counterpart retains three of the four amino acids typically required for catalytic activity (Wells et al, 2017). Furthermore, an additional, metazoan‐specific cleavage site in the 5′ ETS, termed 01 or A’, has been identified (Lazdins et al, 1997). Studies so far suggest that depletion of any of the human orthologues of yeast pre‐rRNA processing enzymes does not prevent cleavage at this site. Therefore, it appears that an additional, hitherto unidentified endonuclease is involved in 5′ ETS processing in humans. The functional importance of this additional 5′ ETS processing event is not yet clear, but it has been suggested to serve as a quality control mechanism for the early stages of pre‐ribosome assembly (Wang & Pestov, 2011; Sloan et al, 2014). Serial endonucleolytic cleavages within the ETS and ITS release fragments that are degraded by exonucleases (Schillewaert et al, 2012; Sloan et al, 2013a, 2014; Memet et al, 2017; Kobyłecki et al, 2018), and it has been suggested that turnover of these fragments is important for recycling of bound RBFs. In some cases, mature ends of the rRNAs are directly formed by endonucleolytic cleavages (e.g. mature 5′ and 3′ ends of 18S rRNA cleaved by UTP24 and NOB1, respectively; Preti et al, 2013; Sloan et al, 2013a; Tomecki et al, 2015; Bai et al, 2016; Wells et al, 2016), but cleavage events can also occur within the spacer sequences and be followed by exonucleolytic processing. For example, in both yeast and humans, the 3′ end of the 5.8S rRNA is formed by the step‐wise action of several 3′‐5′ exonucleases (reviewed in Henras et al, 2015). Different from yeast, an additional exonuclease, ISG2OL2, is implicated in this process in human cells (Coute et al, 2008). Also, while the final trimming of the 3′ end of the 5.8S rRNA is mediated by Ngl2 in yeast, this step is performed by the multifunctional enzyme ERI1 in humans (Faber et al, 2002; Ansel et al, 2008). In striking contrast to human pre‐rRNA processing pathways, the central pre‐rRNA cleavage that separates precursors of the SSU and LSU rRNAs is in yeast followed directly by cleavage of the 3′‐end of the 18S rRNA by Nob1, while exonucleases play key roles in removing ITS1 sequences in human cells (Figure 1). Site 2 cleavage by human RNase MRP is followed by 3′‐5′ trimming by the sequential action of the nuclear exosome containing RRP6 and the poly(A)‐specific ribonuclease PARN (Sloan et al, 2013a; Goldfarb & Cech, 2017; Ishikawa et al, 2017; Montellese et al, 2017). Moreover, oligouridylated forms of 3′‐extended 18S rRNAs have been detected in the cytoplasm, implying that both terminal uridylyltransferases (TUTases) and additional 3′‐5′ exonucleases likely also contribute to ITS1 removal in humans (Montellese et al, 2017). Pre‐rRNA processing events can serve as checkpoints during ribosome assembly, and the utilisation of exo‐ as well as endonucleases for rRNA maturation likely provides mechanisms for coupling of pre‐rRNA processing with quality control and turnover of aberrant pre‐ribosomes. It remains unclear how the activity of processive exonucleases is regulated to ensure precise trimming of specific pre‐rRNA regions. However, it is tempting to speculate that the presence of proteins assembled on the pre‐rRNA or the establishment of pre‐rRNA secondary structures normally arrests the exonucleases at an appropriate position, while they may be poised to efficiently degrade incorrectly assembled pre‐ribosomes upon defects in pre‐ribosome assembly.
Figure 1. Pre‐rRNA processing in yeast and human cells.
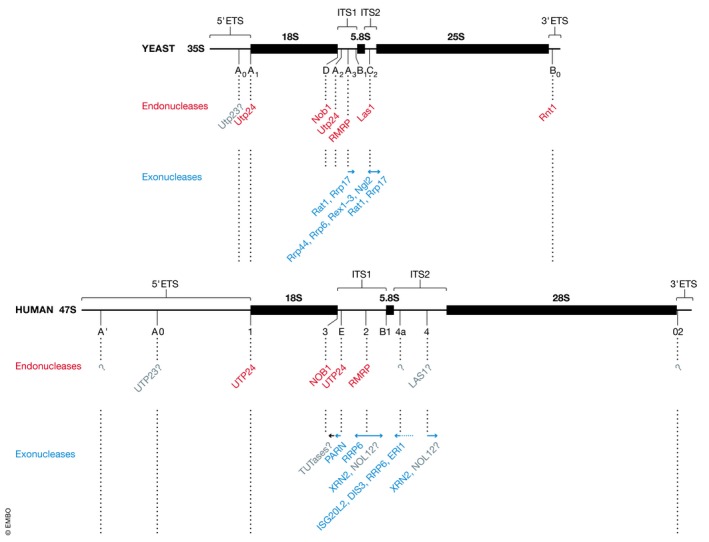
Schematic views of the primary pre‐rRNA transcripts from yeast (35S, upper panel) and human cells (47S, lower panel) are shown to scale. Mature 18S, 5.8S and 25S/28S rRNA sequences are indicated by black rectangles, external transcribed spacers (5′‐ETS and 3′‐ETS) and internal transcribed spacers (ITS1 and ITS2) are represented by black lines. Relative cleavage site positions are indicated, and sites are specified below the pre‐rRNA transcripts. Enzymes responsible for individual processing steps are indicated (Endonucleases—red; exonucleases—blue; putative enzymes—grey; unknown enzymes—?).
Analyses of the pre‐rRNA processing pathways in yeast and humans indicate that, in contrast to other aspects of ribosome assembly that occur in strict hierarchical order, alternative pre‐rRNA processing routes may be used to generate the mature rRNAs (reviewed in detail in Henras et al, 2015; Tomecki et al, 2017). In yeast, pre‐rRNA processing is initiated by removal of the 5′ ETS but subsequent cleavage in ITS1 can take place at either site A2 (approximately 85% of pre‐rRNAs) or at site A3 (approximately 15% of pre‐rRNAs; Schmitt & Clayton, 1993). The utilisation of alternative pre‐rRNA processing pathways appears more prominent in human cells, where step‐wise removal of the 5′ ETS and cleavage at either site E or site 2 in ITS1 can occur in different orders. This flexibility in the order of pre‐rRNA cleavages, together with differing kinetics of processing events, leads to variations in the steady‐state levels of pre‐rRNA intermediates between different organisms and cell types. The functional relevance of such alternative pre‐rRNA processing pathways remains unclear. While it is possible that having diverse mechanisms to generate the mature rRNAs simply ensures the robustness of ribosome production, the greater flexibility in cleavage order observed in higher eukaryotes may suggest that the alternative pre‐rRNA processing pathways serve as a means to regulate subunit assembly.
Comparison of the yeast and human rRNA modification machineries
A universally conserved feature of rRNAs is the presence of chemically modified residues in sequences that form the functional centres of the ribosome, such as the peptidyl transfer centre, decoding site and intersubunit interface (Decatur & Fournier, 2002; Polikanov et al, 2015; Sloan et al, 2017). Although the precise functions of many individual rRNA modifications remain unknown, they are collectively implicated in maintaining the stability of ribosome structure and regulating ribosome function. Mapping the sites of rRNA modifications in the yeast and human rRNAs has revealed 112 and 228 modified sites, respectively, and recent structural analysis of human ribosomes has enabled visualisation of many of these modifications in situ (Figure 2; Ofengand & Bakin, 1997; Piekna‐Przybylska et al, 2008; Carlile et al, 2014; Krogh et al, 2016; Taoka et al, 2016, 2018; Natchiar et al, 2017). The majority of modifications present in eukaryotic rRNAs are 2′‐O‐methylations of ribose moieties (Nm) and pseudouridylations (Ψ), which are introduced by box C/D and box H/ACA snoRNPs, respectively (Watkins & Bohnsack, 2012). In many cases, the snoRNAs responsible for guiding individual modifications have been computationally predicted (Lestrade & Weber, 2006; Gumienny et al, 2017), but most of these predictions still lack experimental confirmation in humans. The single non‐snoRNP‐dependent 2′‐O‐methylation in yeast 25S rRNA (Gm2922) is mediated by Sbp1 (Lapeyre & Purushothaman, 2004), but notably, depletion of its human homologue FTSJ3 causes defects in SSU maturation, suggesting that this enzyme has distinct or additional functions in human cells (Morello et al, 2011). Due to the extensive basepairing interactions formed between snoRNAs and their substrates, the majority of snoRNA‐guided modifications are expected to be installed during the early stages of ribosome biogenesis when the pre‐ribosome structure is likely to be more open, but whether the majority of human rRNA modifications are introduced co‐transcriptionally as in yeast remains to be determined. The total number of modified bases Nm and Ψ in the human rRNAs exceeds the number in yeast rRNAs by more than twofold (Piekna‐Przybylska et al, 2008; Taoka et al, 2016, 2018). The higher density of human rRNA modifications probably not only arises due to the larger size of human ribosomes, but also likely reflects a role for rRNA modifications in fine‐tuning gene expression and the greater need for such regulation in human cells (reviewed in Sloan et al, 2017; see also below).
Figure 2. Distribution of modifications in human rRNAs.
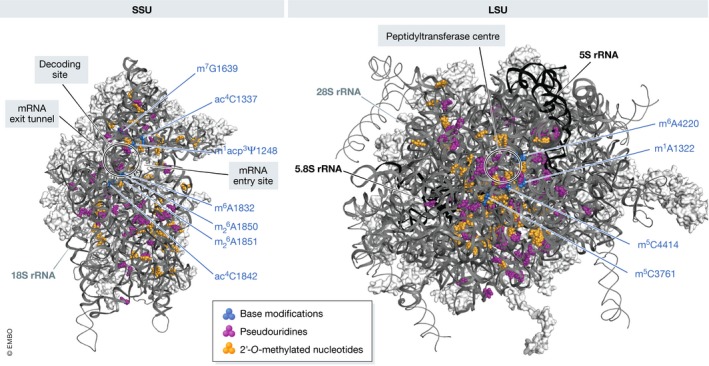
Tertiary structures of the human small ribosomal subunit (SSU) and large ribosomal subunit (LSU; PBD 4V6X) are shown, with rRNA sequences depicted as ribbons and ribosomal proteins in surface view. The type and nucleotide position of base modifications, as well as the positions of key functional regions of the ribosome, are indicated.
Of note, the number of identified human snoRNAs far exceeds the number of rRNA Nm and Ψ (Jorjani et al, 2016). Some human snoRNAs guide Nm and Ψ modifications in different RNA species such as U6 snRNA or likely mRNAs (Aw et al, 2016; Sharma et al, 2016; Bohnsack & Sloan, 2018), while others guide different types of modification (e.g. 18S‐ac4C1842 by SNORD13; Cavaille et al, 1996; Ito et al, 2014; Sharma et al, 2015; 2017b), and some, including U3 (SNORD3), U14A/B (SNORD14A/B), U17A/B (SNORA73A/B) and the metazoan‐specific snoRNAs U8 (SNORD118) and U22 (SNORD22), coordinate pre‐RNA folding by forming basepairing interactions with distant regions of the pre‐rRNA transcript (Savino & Gerbi, 1990; Peculis & Steitz, 1993; Tycowski et al, 1994; Mishra & Eliceiri, 1997; Dunbar & Baserga, 1998). Furthermore, extra‐ribosomal functions of some human snoRNAs in the regulation of alternative pre‐mRNA splicing have been uncovered; for example, HBII‐52 (SNORD115) regulates alternative splicing of the serotonin receptor 2C pre‐mRNA, while SNORD86 modulates expression of the snoRNP protein NOP56 by dictating usage of alternative splice sites (Kishore & Stamm, 2006; Lykke‐Andersen et al, 2018).
Other human rRNA base modifications and their cognate methyltransferases are largely conserved between yeast and humans, except for two N 3‐methyluridines (m3Us) at positions 25S‐U2634 and 25S‐U2843 of yeast 25S rRNA, neither of which are directly analogous to the single one at position 4,500 of the human 28S rRNA (Figure 2; Table 2) (Piekna‐Przybylska et al, 2008; Taoka et al, 2016, 2018). Furthermore, human 18S and 28S rRNAs both carry a single N 6‐methyladenosine (m6A) nucleotide not present in yeast, at positions A1832 and A4220, respectively; while the enzyme for A1832 modification remains still unknown, ZCCHC4 has recently been identified as the m6A methyltransferase responsible for the latter modification (Ma et al, 2019). N 6‐methylation of 28S‐A4420 increases base‐stacking with A4219 and G4222, thereby likely contributing to stability of this region of the 60S subunit. Consistent with this, lack of ZCCHC4 leads to decreased 60S subunit levels and a global reduction in protein synthesis (Ma et al, 2019).
Table 2.
The yeast and human rRNA modification enzymes
| Modification (human) | Modification (yeast) | Enzyme (human) | Enzyme (yeast) | References |
|---|---|---|---|---|
| 18S‐m1acp3Ψ1240 | 18S‐m1acp3Ψ1191 | SNORA14, EMG1, TSR3 | snR35, Emg1, Tsr3 | Wurm et al (2010), Meyer et al (2011, 2016), Warda et al (2016) |
| 18S‐ac4C1337 | 18S‐ac4C1280 | NAT10 | Kre33 | Ito et al (2014), Sharma et al (2015) |
| 18S‐m7G1639 | 18S‐m7G1575 | WBSCR22 | Bud23 | White et al (2008), Haag et al (2015) |
| 18S‐m6A1832 | – | Unknown | – | Piekna‐Przybylska et al (2008) |
| 18S‐ac4C1842 | 18S‐ac4C1773 | NAT10 | Kre33 | Ito et al (2014), Sharma et al (2015) |
| 18S‐m2 6A1850/1 | 18S‐m2 6A1781/2 | DIMT1L | Dim1 | Lafontaine et al (1998), Zorbas et al (2015) |
| 28S‐m1A1332 | 25S‐m1A645 | NML | Rrp8 | Peifer et al (2013), Waku et al (2016) |
| – | 25S‐m1A2142 | – | Bmt2 | Sharma et al (2013a) |
| 28S‐m5C3761 | 25S‐m5C2278 | NSUN5 | Rcm1 | Schosserer et al (2015) |
| – | 25S‐m3U2634 | – | Bmt5 | Sharma et al (2014) |
| 28S‐m6A4220 | – | ZCCHC4 | – | Ma et al (2019) |
| 28S‐m5C4414 | 25S‐m5C2870 | NSUN1 | Nop2 | Sharma et al (2013b), Bourgeois et al (2015) |
| – | 25S‐m3U2843 | – | Bmt6 | Sharma et al (2014) |
| 28S‐m3U4500 | – | Unknown | – | Piekna‐Przybylska et al (2008) |
| 28S‐Gm4469 | 25S‐Gm2922 | Unknown | Spb1 | Lapeyre and Purushothaman (2004) |
| Nm – (x112) | Nm ‐ (x55) | Fibrillarin in box C/D snoRNP | Nop1 in box C/D snoRNP | Kiss‐Laszlo et al (1996) |
| Ψ ‐ (x105) | Ψ ‐ (x46) | Dyskerin in box H/ACA snoRNP | Cbf5 in box H/ACA snoRNP | Ganot et al (1997) |
m1acp3Ψ—N 1‐methyl‐N 3‐aminocarboxypropylpseudouridine, ac4C—N 4‐acetylcytidine, m7G—N 7‐methylguanosine, m6A—N 6‐methyladenosine, m2 6A—N 6 N 6‐dimethyladenosine, m1A—N 1‐methyladenosine, m3U—N 3‐methyluridine, m5C—5‐methylcytosine, Nm—ribose 2′‐O‐methylation, Ψ—pseudouridine. Individual Nm and Ψ sites are not listed; instead, the total number present in the rRNAs is given.
Insights into pre‐ribosome export and cytoplasmic maturation steps
During their maturation, pre‐ribosomal subunits undergo extensive remodelling involving establishment of the rRNA folds present in mature ribosomes, incorporation of RPs and dynamic recruitment and dissociation of RBFs. The compartmentalisation of eukaryotic cells into nucleus and cytoplasm allows the physical separation of immature pre‐ribosomes and components of the translation machinery, and achieving export competence therefore represents a key milestone for pre‐ribosomal subunits. Pre‐ribosomal subunits are some of the largest RNPs that translocate through the permeability barrier of nuclear pore complexes, and their transport is facilitated by multiple nuclear transport factors and adaptors (reviewed in Sloan et al, 2016). As in yeast, the exportin CRM1 (XPO1) bound to RanGTP contributes to the nuclear export of both the pre‐40S and pre‐60S subunits in humans, and several putative adaptor proteins containing classical leucine‐rich nuclear export signals (NES; Guttler & Gorlich, 2011) have been identified. These include the LSU biogenesis factor NMD3 (Bai et al, 2013) and the SSU component RIOK2 (Zemp et al, 2009), both of which have been shown to bind to CRM1 in a RanGTP‐dependent manner. Upon RIOK2 depletion, the kinetics of pre‐SSU export are slowed, confirming a contribution of RIOK2 to pre‐SSU export (Zemp et al, 2009). However, the finding that RIOK2 is not essential for pre‐40S export suggests that additional export adaptors may exist in human cells and that there may be redundancy between them. Indeed, PDCD2L, which lacks a yeast homologue, was recently identified as a CRM1 adaptor for pre‐40S subunits in human cells (Landry‐Voyer et al, 2016). Furthermore, RAE1, the human homologue of yeast pre‐60S export factor Gle2, has been suggested to contribute not only to pre‐60S export, but also to transporting pre‐40S subunits to the cytoplasm (Wild et al, 2010). Moreover, while the Mex67‐Mtr2 heterodimer plays key roles in pre‐40S and pre‐60S export in yeast (Faza et al, 2012), its human counterpart NXF1‐NXT1 was not found in export‐competent pre‐ribosomal subunits purified from human cells (Wild et al, 2010), questioning whether these functions are conserved in metazoans. Finally, the well‐characterised pre‐microRNA‐ and tRNA‐export factor EXP5 (Leisegang et al, 2012) was identified as export adaptor for human pre‐60S complexes (Wild et al, 2010); while its pre‐ribosomal binding site remains to be determined, it is tempting to speculate that EXP5 may recognise a pre‐miRNA‐like hairpin structure within the pre‐rRNA.
Exciting insights into the composition and dynamics of late nuclear and cytoplasmic human pre‐40S complexes have recently been obtained by cryo‐electron microscopy (cryo‐EM; Larburu et al, 2016; Ameismeier et al, 2018). While these studies confirmed that the binding sites of most late‐acting pre‐40S biogenesis factors are largely conserved between yeast and humans, a striking compositional difference between yeast and human cytoplasmic pre‐40S complexes is the absence of DIMT1L (Dim1) in the latter. In yeast, N 6,N 6‐dimethylation of two adenosines close to the 3′ end of 18S rRNA by Dim1 occurs in the cytoplasm (Lafontaine et al, 1998), but the lack of DIMT1L in human cytoplasmic pre‐40S complexes is in line with observations that this event takes place prior to pre‐40S export in humans (Zorbas et al, 2015; Sloan et al, 2019). The significance of this temporal difference still remains to be uncovered.
A series of structural snapshots of different pre‐40S complexes has recently highlighted structural transitions that occur before nuclear export, and provided new information on how the formation of key ribosomal features is regulated (Ameismeier et al, 2018). These data have revealed that the export adaptor RRP12, which binds to the 3′ major domain of the 18S rRNA sequence, coordinates substantial rRNA rearrangements within the “head” and “beak” regions of nuclear pre‐40S subunits. Coupling maturation of the head region with nuclear export via RRP12 has therefore been suggested to serve as a quality control mechanism ensuring that only correctly assembled pre‐40S subunits reach the cytoplasm. Structural rearrangements that occur upon dissociation of RRP12 enable recruitment of RACK1, a protein that is not present in yeast pre‐40S particles (Larburu et al, 2016; Ameismeier et al, 2018). As RACK1 has previously been characterised as a translation initiation factor, its requirement for pre‐40S maturation (Larburu et al, 2016) raises the possibility that in human cells, the final stages of pre‐40S maturation are coupled to translation initiation. Maturation of the decoding centre, containing the ribosomal A and P sites, is one of the final pre‐40S biogenesis events to take place. This structure is formed by interconnections between helices h28, h44 and h45 of the 18S rRNA sequence, and the presence of various RBFs (such as TSR1, LTV1, RIO2K, PNO1) in pre‐40S particles stabilises h44 in an immature conformation. In this regard, PNO1 not only helps maintain the rRNA sequences that form the decoding centre in an immature state, but also forms close interactions with NOB1 that ensure that the catalytic centre of the endonuclease remains spatially separate from its target site at the 3′‐end of the 18S rRNA. It has been suggested that PNO1 release not only enables final assembly of the decoding centre, but also triggers a conformational change that allows NOB1 to contact the 3′‐end of the 18S rRNA, thereby enabling these two critical events to be coordinated (Ameismeier et al, 2018); a similar mechanism has also been proposed to operate in yeast.
Less is known about the late nuclear and cytoplasmic phases of human pre‐60S maturation; however, aspects that have been studied in detail are the assembly of the P‐stalk and the release mechanism of the pre‐60S maturation factor eIF6. In both yeast and humans, the RBF Mrt4/MRTO4 acts as a nuclear placeholder for the RP P0, which forms the base of the stalk structure of mature 60S subunits. Release of Mrt4/MRTO4 is triggered by the dual‐specificity phosphatase Yvh1/DUSP12 (Lo et al, 2010), allowing P0 recruitment and stalk assembly. Stalk assembly is a pre‐requisite for release of eIF6 (Tif6 in yeast), which plays an important role in preventing premature translation by impeding the association of pre‐60S particles with 40S subunits (Gartmann et al, 2010; Klinge et al, 2011). Detailed mechanistic insights into how this is achieved in human cells come from an elegant series of structural and biochemical analyses (Finch et al, 2011; Weis et al, 2015; Warren, 2018). SBDS, a protein mutated in Shwachman–Diamond syndrome (Table 3), initially binds to the ribosomal P‐site, the peptidyltransferase centre and the P‐stalk base, and proof‐reads the integrity of these features. Upon its recruitment to pre‐60S particles, the GTPase EFL1 competes with SBDS for binding to the P‐stalk and triggers a conformational change in SBDS that causes it to alternatively interact with h69 of the 28S rRNA. Rearrangement of EFL1 then leads to competition between EFL1 and eIF6 for binding to the sarcin–ricin loop of the 28S rRNA sequence. This not only induces release of eIF6 from the pre‐60S particle, but also activates GTP hydrolysis by EFL1, triggering concomitant dissociation of EFL1. Together, these structural studies provide insights into the mechanistic basis of the hierarchical release of RBFs during the final stages of subunit maturation. Furthermore, they highlight the extent to which the late stages of subunit assembly are closely coupled with quality control mechanisms to ensure the fidelity of the mature ribosomal subunits. Common themes that emerge are the roles of RBFs as placeholders for late‐associating ribosomal proteins, the proof‐reading of key ribosomal structures by RBFs and the positioning of RBFs to prevent premature subunit joining.
Table 3.
Inventory of genetic diseases caused by defects in ribosomal proteins or biogenesis factors
| Ribosomopathy | OMIM | Affected gene(s) | Mutation(s) | Clinical features | Key references |
|---|---|---|---|---|---|
| Diamond‐Blackfan anemia | 105650 | RPL5, RPL11, RPL27, RPL35A, RPS7, RPS10, RPS17, RPS19, RPS24, RPS26, RPS27, RPS28, GATA1, TSR2 | Various | Anaemia, microcephaly, hypertelorism, ptosis, micrognathia, cleft palate, short, webbed neck, malformed or absent thumbs, cataracts, glaucoma, strabismus | Boria et al (2010), Ellis and Gleizes (2011) |
| 5q‐myelodysplastic syndrome | 153550 | RPS14 | Deletion of ~1.5 Mb region of chr. 5 | Severe anaemia, thrombocytosis, dysmegakaryopoiesis | Ebert et al (2008) |
| Isolated congenital asplenia | 271400 | RPSA | pP199Sfs*25, pQ9*, p.T54N, p.L58F, p.R180W, p.R180G, p.R186C | Lack of spleen, immunodeficiency | Bolze et al (2013) |
| RPS23‐related ribosomopathy | 617412 | RPS23 | p.R67K | Microcephaly, hearing loss, simian palmar creases, epicanthic folds in the eyes, foetal finger pads, extra front teeth, low back hairline, facial asymmetry and high palate, intellectual disability, autism spectrum disorder | Paolini et al (2017) |
| Treacher Collins syndrome | 154500, 613717, 248390 | TCOF1, POLR1C, POLR1D | Various | Craniofacial defects; midface hypoplasia, micrognathia, microtia, conductive hearing loss and cleft palate | Valdez et al (2004), Dauwerse et al (2011), Vincent et al (2016) |
| Postaxial acrofacial dysostosis (POADS) | 263750 | DHODH | p.R135C, P.R346W, p.D392G, p.E52G, p.A357T, p.R326X, | Craniofacial defects (micrognathia, orofacial clefts, malar hypoplasia, cup‐shaped ears), postaxial limb deformities (lack of fourth/fifth rays of hands/feet; ulner/fibular hypoplasid) | Rainger et al (2012) |
| Roberts syndrome | 268300 | ESCO2 | p.E251fsX30, p.R169X, p.W539G, p.V84fsX7, p.K486fsX20, p.K369fsX34, p.R293fsX7, p.K138fsX10 | Prenatal growth retardation, craniofacial defects, e.g. cleft lip/palate or microcephaly, limb malformations | Vega et al (2005) |
| Scleroderma | 181750 | UTP14A | Gene hypermethylation – decreased expression | Hardened/thickened skin, ulcers/sores, swollen joints, fingers or toes, muscle weakness | Selmi et al (2012) |
| Dyskeratosis congenita | 305000 | DKC1, TERC, TERT, NOP10, NHP2, TINF2 | Various (prevalent Dyskerin p.A353V) | Mucocutaneous abnormalities, pulmonary fibrosis, bone marrow failure, immunodeficiency | Heiss et al (1998), Knight et al (2001), Vulliamy et al (2001, 2008) |
| Bowen‐Conradi syndrome | 211180 | EMG1 | p.D86G | Prenatal and postnatal growth retardation, microcephaly, prominent nose with an absent glabellar angle, micrognathia, joint abnormalities, camptodactyly, rocker‐bottom feet, severe psychomotor delay | Armistead et al (2009) |
| Cartilage‐hair hypoplasia | 250250 | RMRP | Various | Short stature, bone deformities, hair growth abnormalities | Ridanpää et al (2001) |
| North American Indian childhood cirrhosis | 604901 | UTP4, NOL11 | p.R565W | Biliary cirrhosis, portal hypertension | Freed and Baserga (2010), Freed et al (2012) |
| Shwachman–Diamond syndrome | 260400 | SBDS | Various | Exocrine pancreatic insufficiency, bone marrow dysfunction, leukaemia predisposition and skeletal abnormalities | Boocock et al (2003) |
| Alopecia, neurological and endocrinopathy syndrome (ANE) | 612079 | RBM28 | p.L351P | Alopecia, progressive neurological defects and endocrinopathy | Nousbeck et al (2008), Spiegel et al (2010) |
| Aplasia cutis congenita | 107600 | BMS1 | p.R930H | Skin defects (especially scalp vertex) | Marneros (2013) |
| Leukoencephalopathy, intercranial calcifications and cysts | 614561 | SNORD118 | Various | Leukoencephalopathy, intercranial calcifications and cysts | Jenkinson et al (2016) |
| Cancer‐prone bone marrow failure syndrome | 617052 | DNAJC21 | p.R173*, p. P32A, p.E265* | Bone marrow failure, short stature, microcephaly, decreased bone density, predisposition to acute myeloid leukaemia | Tummala et al (2016) |
| X‐linked intellectual disability, cerebellar hypoplasia and spondyloepiphyseal dysplasia | 300847 | RPL10 | p.A64V | Intellectual disability, cerebellar hypoplasia and spondyloepiphyseal dysplasia | Zanni et al (2015) |
Alongside the regulation of pre‐ribosome maturation events by spatial constraints or steric hinderance (e.g. as described above for 18S rRNA 3′‐end cleavage by NOB1), post‐translational modifications represent another mechanism that likely contributes to the regulation of subunit assembly. In both yeast and human cells, late‐stage pre‐40S particles contain several (putative) kinases (Rio1, Rio2 and Hrr25 in yeast; RIO1K, RIO2K, RIO3K and CK1δ/ε in humans). Phosphorylation of Ltv1/LTV1 and Enp1/ENP1 by Hrr25/CK1δ/ε appears to be a conserved event linked to dissociation of Ltv1/LTV1 from pre‐40S particles and to assembly of Rps3/RPS3 onto the beak structure (Schäfer et al, 2006; Zemp et al, 2014). While yeast pre‐40S particles contain two putative kinases of the RIO family (Rio1 and Rio2), human cells express three RIO kinases (RIOK1, RIOK2 and RIOK3) that are all required for pre‐40S maturation (Vanrobays et al, 2003; Zemp et al, 2009; Baumas et al, 2012; Widmann et al, 2012). Phosphorylation substrates of the RIO family kinases have long remained elusive, but human RIOK2 was recently demonstrated to phosphorylate DIMT1L in vitro (Sloan et al, 2019). Furthermore, both Rio1 and RIOK1 were recently found to orchestrate a network of interactions that may couple pre‐40S maturation with metabolism and cell proliferation (Iacovella et al, 2018).
Fundamental principles of ribosomal protein recruitment and assembly
Although RPs do not directly participate in peptide bond formation during protein synthesis, they play essential structural roles in ensuring correct rRNA folding to enable catalysis. As nascent RPs are synthesised on pre‐existing ribosomes, ribosomes contribute to their own production (Figure 3). The fundamental advantage of assembling these macromolecular complexes from numerous small RPs was recently demonstrated by mathematical modelling, which showed that this can be accomplished much more efficiently than through the expression of a minimal number of larger proteins (Reuveni et al, 2017). Most human RPs are expressed from single genes, whereas many yeast RPs exist in two isoforms. Approximately 50% of human RNA polymerase II transcription is dedicated to production of mRNAs encoding RPs (Warner, 1999), and the expression of these 80 RPs is carefully coordinated to ensure their equal availability for ribosome assembly. Most RP genes share a common promoter element to synchronise their transcription, as well as 5′‐terminal oligopyrimidine (TOP) sequences that allow RP mRNA translation to also be co‐regulated. Notable exceptions include the mRNAs encoding RPS27a and RPL40, which are expressed as ubiquitin‐fusion proteins (Webb et al, 1994). Most RPs are produced in excess, and as depletion of the CUL4 E3 ubiquitin ligase complex impairs both pre‐40S and pre‐60S maturation, it is likely that ubiquitination may target excess RPs for degradation (Lam et al, 2007 ; Badertscher et al, 2015). RPs are typically highly basic proteins, a property that renders them inherently prone to aggregation and degradation. Consequently, a common feature of yeast and human ribosome assembly is the employment of dedicated chaperones that bind (often co‐translationally) to nascent RPs to facilitate their nuclear import and recruitment to pre‐ribosomal complexes (reviewed in Pillet et al, 2017). While human homologues have been identified for several of the known yeast RP chaperones (Bcp1/BCCIP, Syo1/HEATR3, Rrb1/GRWD1, Sqt1/AMMP and Tsr2/TSR2), others (Acl4 and Yar1) do not appear to be conserved in metazoans (Schütz et al, 2014; Wyler et al, 2014; Calviño et al, 2015; Pausch et al, 2015; Pillet et al, 2015, 2017), implying the existence of additional mechanisms for facilitating recruitment of some human RPs. Notably, the ribosomal proteins RPL5 (uL18) and RPL11 (uL5) are recruited to pre‐ribosomes as a preformed sub‐complex together with the 5S rRNA. Pre‐5S rRNA is synthesised by RNA polymerase III, and maturation of its 3′‐end requires the REX1, REX2 and REX3 exonucleases as well as the presence of RPL5 (Figure 4A; van Hoof et al, 2000; Ciganda & Williams, 2011; Sloan et al, 2013b). In both yeast and humans, Rrs1/RRS1 and Rpf2/BXDC1 are required for integration of the 5S RNP into pre‐60S complexes, and the PICT1/GLTSCR2 tumour suppressor protein is an additional factor important for this step in human cells (Zhang et al, 2007; Sloan et al, 2013b).
Figure 3. Production, recruitment and functions of human ribosomal proteins.
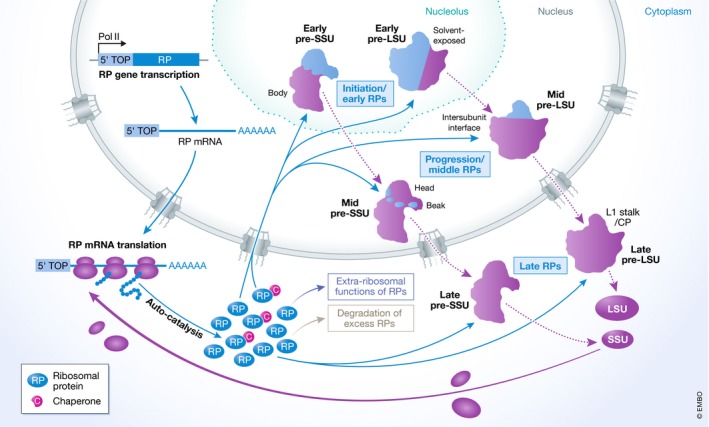
Schematic model showing the expression and assembly of ribosomal proteins (RP). Red arrows indicated sequential steps in the process, and dashed black lines indicate maturation of pre‐ribosomal particles. Pink circles containing a white “C” depict dedicated ribosomal protein chaperones. CP, central protuberance.
Figure 4. Assembly of the 5S RNP and its role in regulation of the tumour suppressor p53.
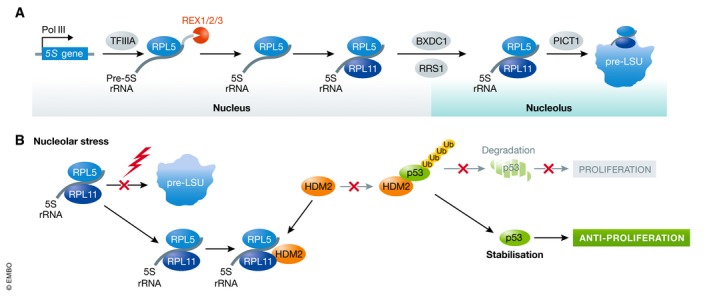
(A) Model of synthesis and assembly of the 5S RNP from 5S rRNA and ribosomal proteins RPL5 (uL18) and RPL11 (uL5), and 5S RNP integration into the pre‐LSU. (B) Upon nucleolar stress (e.g. in ribosomopathies), 5S RNP integration into the pre‐LSU is decreased and 5S RNP accumulates in the nucleoplasm, where it binds the E3 ubiquitin ligase HDM2. This impedes association of HDM2 with p53, promoting p53 stabilisation and inhibiting cell proliferation.
Many RPs are first weakly tethered to pre‐ribosomal complexes, where they chaperone correct rRNA folding and establishment of tertiary structures, in turn resulting in their stable incorporation into ribosomal complexes. A fundamental feature of ribosome assembly that is conserved not only throughout eukaryotes but also occurs during prokaryotic ribosome production (Chen & Williamson, 2013) is the hierarchical assembly of RPs, leading to the sequential assembly of particular subunit domains (Figure 3). Screens analysing pre‐rRNA processing defects arising from depletion of individual human RPs showed that the 5′‐, central and 3′‐minor domains of the 18S rRNA that form the “body” of the SSU assemble first, followed by the so‐called head and beak structures (O'Donohue et al, 2010). Similarly, the RPs that form the solvent‐exposed face of the LSU are positioned first, with those binding to the intersubunit interface and to the central protuberance recruited subsequently (Nicolas et al, 2016). In yeast, the 5S RNP is first tethered to pre‐60S complexes in an immature conformation, but undergoes a 180° rotation to establish its final position as the central protuberance of the LSU (Bassler et al, 2014; Leidig et al, 2014). Given the conservation of the temporal assembly of RPs, it is likely that a similar structural rearrangement also takes place during the late stages of human ribosome assembly, but this has not yet been confirmed. The universal nature of the hierarchical recruitment of RPs suggests that step‐wise assembly, stabilisation and compaction of different domains of the ribosomal subunits is an important mechanism that helps ensure the fidelity of the assembly pathway.
Extra‐ribosomal functions of human ribosomal proteins
A growing body of evidence indicates that several human RPs have additional functions beyond their roles as structural components of ribosomes (Figure 3). For example, extra‐ribosomal RPL26 (uL24) and RPL13 (eL13) have been found to modulate translation of selected mRNAs by binding to their 5′‐ and 3′‐UTRs, respectively (Takagi et al, 2005; Mukhopadhyay et al, 2008), and RPS3 (uS3) and RPS14 (uS11) have been suggested regulate transcription (Wan et al, 2007; Zhou et al, 2013). Furthermore, in addition to proposed roles for RPL22 (eL22) and RPL22L1 in the regulation of splicing of specific pre‐mRNAs (Zhang et al, 2017), several ribosomal proteins (e.g. RPS13 and RPL10a (uL16)) modulate (alternative) splicing of their own pre‐mRNAs (Malygin et al, 2007; Takei et al, 2016). However, the most prominent example of an extra‐ribosomal function of RPs is the role of the 5S RNP in a nucleolar stress response pathway that regulates the tumour suppressor p53 (Pelava et al, 2016). During homeostasis, the E3 ubiquitin ligase HDM2 (together with its partner HDMX) binds to p53, both inhibiting its transcriptional activity and leading to its ubiquitination‐induced proteasomal degradation, with a positive feedback loop in which p53 induces HDM2 expression ensuring that the levels of the tumour suppressor remain low in unstressed cells (Wu et al, 1993). Defects in ribosome assembly lead to increased levels of non‐ribosome‐associated 5S RNP in the nucleoplasm, where it binds to HDM2, inhibits p53 ubiquitination and thereby causes increased cellular p53 levels (Figure 4B) (Bursać et al, 2012; Donati et al, 2013; Sloan et al, 2013b). More specifically, it has been suggested that binding of the 5S RNP to HDM2 impedes its interaction with HDMX and that disruption of this interaction reduces ubiquitination of p53 by HDM2 (Li & Gu, 2011). This 5S RNP/HDM2 pathway of p53 regulation is implicated in the cellular response to many forms of stress, including hypoxia, nutrient stress, growth factor deprivation, UV/gamma radiation and oxidation stress. Furthermore, the tumour suppressor p14ARF, which is induced by oncogenic signals such as the expression of RAS or c‐MYC, is also implicated in this pathway, as it directly binds RPL11 (Dai et al, 2012) and as depletion of RPL5 or RPL11 suppresses the p53 accumulation normally observed in cells overexpressing p14ARF (Sloan et al, 2013b). Other ribosomal proteins, including RPS27a and RPL23, have also been shown to bind to HDM2, suggesting the presence of alternative mechanisms to regulate p53 levels in response to defects in ribosome assembly (Dai et al, 2004; Sun et al, 2011). In addition to p53 regulation, components of the 5S RNP have recently been shown to bind to the pro‐apoptotic tumour suppressor p73 and to prevent its interaction with HDM2, and RPL11 is implicated in regulating the transcriptional activity of c‐MYC (Riley & Lozano, 2012; Liao et al, 2014). The 5S RNP therefore constitutes a hub for coordinating ribosome assembly with multiple cell signalling pathways.
Regulation of ribosome production by cell signalling pathways
The pathway of ribosome assembly is closely coupled with cell cycle progression and modulated by various signalling pathways that control cell growth. Such co‐regulation of the ribosome synthesis pathway is unsurprising in light of the essential nature of protein synthesis and the high energy demand of their assembly pathway. Several oncogenes that drive cellular proliferation, such as c‐MYC, mTOR, ERK and AKT, and tumour suppressors that help maintain cellular homeostasis, including p53, ARF and PTEN, have been shown to regulate ribosome biogenesis. For example, a screen for c‐MYC target genes identified numerous RBFs and several RPs, and consistent with this, c‐MYC was found to regulate efficient rRNA maturation (Schlosser et al, 2003). Similarly, the mTOR‐containing mTORC1 complex, which connects cell growth to nutrient availability on various levels such as transcription, translation, protein degradation and membrane trafficking, promotes rDNA transcription by RNA polymerase I and 5S RNA synthesis by RNA polymerase III, and also regulates translation of most RP‐encoding mRNAs (Mayer & Grummt, 2006; Gentilella et al, 2015). Furthermore, mTORC‐dependent S6 kinase activation triggers phosphorylation of RPS6, and the expression of a large proportion of RBFs is altered in cells lacking S6 kinases (Chauvin et al, 2014). Notably, the mTORC2 complex component RICTOR and the L‐glutamine synthetase GLUL were recently identified in a screen for factors required for human ribosome biogenesis (Badertscher et al, 2015). As the effect of GLUL on 40S biogenesis was independent of mTORC1, intracellular glutamine synthesis may be important for efficient ribosome synthesis (Badertscher et al, 2015). Finally, in addition to the global up‐ and down‐regulation of ribosome biogenesis observed during tumorigenesis and senescence, respectively, the rate of ribosome biogenesis is also controlled by the circadian clock. Fluctuations in RP gene transcription and mRNA translation rates have been observed, and diurnal changes in mouse liver mass are accompanied by variations in the levels of newly synthesised rRNAs (Jouffe et al, 2013; Sinturel et al, 2017). A recent model integrating these two observations proposes that newly synthesised rRNAs not assembled with RPs are polyadenylated by PAPD5 and degraded by the nuclear exosome (Sinturel et al, 2017).
Ribosome heterogeneity
Given the precise molecular interactions and movements required of the ribosome during translation, the essential nature of many RBFs and the extensive monitoring that pre‐ribosomal subunits undergo during their maturation, it was long assumed that cells contain a uniform population of ribosomes. However, several lines of evidence suggest that this is not the case and different sources of ribosome heterogeneity have now been identified. On the rRNA level, alternative pre‐rRNA processing pathways generate two different (long and short) forms of the 5.8S rRNA. Recent quantitative analyses of rRNA modifications have further revealed that numerous Nm and Ψ sites are substoichiometrically modified, suggesting that subsets of ribosomes likely carry different combinations of modifications (Krogh et al, 2016; Sharma et al, 2017a). Variations in the extent or the sites of modification have been found in cancer cell lines of different origins, or depending on expression status of the tumour suppressor p53 (Krogh et al, 2016; Sharma et al, 2017a; Taoka et al, 2018). At the protein level, there is not only variation in the post‐translation modifications present on RPs in different contexts (Simsek & Barna, 2017), but tissue‐specific differences in RP mRNA levels have also been observed (Bortoluzzi et al, 2001), as well as differences in the RP composition of translating ribosomes (Shi et al, 2017). The existence of such ribosome heterogeneity raises the question of whether these compositional variations are merely biogenesis artefacts tolerated by (pre‐)ribosome quality control pathways, or whether they reflect active fine‐tuning of ribosome structure for specialised functions. Supporting the latter proposal, differences in rRNA 2′‐O‐methylation and pseudouridylation have been shown to influence the translational capacity of human ribosomes. For example, pathogenic mutations identified in the genetic disease dyskeratosis congenita cause changes in rRNA pseudouridylation that impair translation of the mRNAs encoding the tumour suppressors p53 and p27 and the anti‐apoptotic factors Bcl‐xL and XIAP, thereby promoting tumorigenesis (Yoon et al, 2006; Bellodi et al, 2010a,b). Similarly, changes in rRNA 2′‐O‐methylation have been observed to specifically affect translation of various IRES‐containing mRNAs (Erales et al, 2017). Moreover, human ribosomes containing RPS25 (eS25) or RPL10 (uL1) were observed to preferentially translate specific subsets of mRNAs involved in metabolism, cell cycle and development, indicating that ribosome heterogeneity can serve as a mechanism to fine‐tune translation (Shi et al, 2017). It is therefore likely that ribosome heterogeneity not only contributes to the adaptation of gene expression in different conditions, but also to development, disease and tissue identity.
Elucidating the molecular basis of ribosomopathies
The push for a comprehensive understanding of human ribosome assembly has primarily been driven by the recognition of a family of diseases, collectively termed ribosomopathies, that are caused by mutations in genes encoding RPs, RBFs or components of the rDNA transcription machinery (Table 3). The new inventories of human RBFs generated by RNAi‐based screens include numerous disease‐associated proteins and biomarkers, suggesting that the prevalence of disorders arising from ribosome dysfunction may be much greater than previously thought. While most ribosomopathies are relatively rare, the most prominent and best understood example is Diamond‐Blackfan anemia (DBA), which is characterised by bone marrow failure due to inhibition of hematopoietic stem cell differentiation along the erythroid lineage. DBA‐associated mutations have been identified in eleven different genes encoding RPs of both the small and large subunit (Table 3; Boria et al, 2010). Ribosomopathies manifest with a diverse set of phenotypes, but many share common characteristics or overlapping features such as haematopoietic dysfunction (e.g. DBA, 5q‐ syndrome, dyskeratosis congenita, Shwachman–Diamond syndrome) and/or skeletal (especially craniofacial) defects (e.g. Treacher Collins syndrome, Bowen‐Conradi syndrome and RPS23‐related ribosomopathy; Table 3).
Understanding how defects in a fundamental and ubiquitous cellular process such as ribosome synthesis can cause tissue‐specific human pathologies remains a major challenge, and several models to explain these phenomena have recently been proposed (Mills & Green, 2017; Emmott et al, 2019). As mutations in the genes encoding RPs or RBFs disrupt ribosome assembly, they lead to stabilisation of p53 via the 5S RNP/HDM2 pathway described above. Indeed, many of the tissue‐specific defects observed in ribosomopathies appear to be p53‐dependent, with symptoms of ribosomopathies such as Treacher Collins syndrome and 5q‐ syndrome rescuable by inhibition of p53 function (Jones et al, 2008; Barlow et al, 2010). Elevation of the cellular p53 level leads to cell cycle arrest and apoptosis, and it has been suggested that variations in the threshold or extent of p53 activation in different cell types may underlie tissue specificity in many ribosomopathies. An alternative proposal posits that variations in ribosome composition between different cell types and/or in different tissues (see above section on “ribosome heterogeneity”) lead to specialised translation; mutations that decrease the levels of specific RPs or impair the rRNA modification machinery may therefore reduce or alter production of proteins particularly required in certain cell types, thereby inducing the disease phenotype. Another possible molecular basis of the tissue‐specific effects observed in ribosomopathies may be alterations in translation caused by the high demand for ribosomes in fast‐proliferating tissues. Supporting this, DBA‐associated mutations have been found to reduce ribosome levels in haematopoietic cells such that the number of ribosomes becomes limiting, and translation of a specific subset of mRNAs is affected, which in turn impairs erythroid lineage commitment (Khajuria et al, 2018). The differential effects on mRNA translation caused by the lack of available ribosomes were suggested to be caused by inherent properties of specific mRNAs (e.g. highly structured 5′‐UTRs) that render these transcripts especially sensitive to reduced ribosome concentration. It is likely that differential p53 responses to ribosome assembly defects and alterations in the translation profile caused by limited ribosome production, and/or the assembly of specialised ribosomes, all contribute to different extents to ribosomopathy phenotypes. Nevertheless, it has also been suggested that some of the tissue‐specific effects observed may instead arise from varying abilities of different cell types to overcome such defects, for example via ribosome quality control pathways or recovery of RP levels by allelic compensation (Mills & Green, 2017).
Conclusions and outlook
For many years, the complex pathway of eukaryotic ribosome biogenesis was studied almost exclusively in the yeast model system, where the ease of genetic manipulation coupled with the possibility to isolate large amounts of pre‐ribosomal complexes for compositional and structural analysis has provided a wealth of knowledge about fundamental aspects of ribosome assembly. Motivated primarily by the importance of elucidating the molecular basis of ribosomopathies, analyses of the human ribosome assembly pathway have increased exponentially in recent years. While this work has confirmed that many features of the yeast and human ribosome assembly pathways are similar, the emerging differences provide important insights into particular aspects of the pathway that have adapted during evolution. Although a plethora of factors required for human ribosome biogenesis have now been uncovered, it is likely that future research will extend the inventory of human RBFs even further. Major challenges for the next years will be to determine which of the factors required for ribosome synthesis are directly associated with pre‐ribosomal complexes, and to dissect the individual roles of such proteins during subunit assembly. The recent cryo‐EM structures of yeast pre‐ribosomes have provided a vast amount of information on the temporal order, distribution and molecular functions of many RBFs. We eagerly anticipate a similar increase in structural analyses of human pre‐ribosomes, which should significantly advance our understanding of human ribosome assembly by enabling visualisation of novel RBFs in situ and revealing the dynamics of the ribosome assembly process. The recognition of ribosome biogenesis as a central hub coordinating protein synthesis and cell proliferation necessitates not only understanding of the assembly pathway itself, but also deeper insight into the complex regulatory networks modulating it. This should extend the scope of ribosome biogenesis research to a cellular and organismal level, and it will be exciting to discover the full extent of crosstalk between ribosome production and other aspects of cellular function.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We would like to thank Nicholas Watkins for interesting discussions on ribosomopathies and help with preparation of Table 3. This work was supported by the Deutsche Forschungsgemeinschaft (SFB1190) and the University Medical Centre Göttingen.
The EMBO Journal (2019) 38: e100278
See the Glossary for abbreviations used in this article.
Contributor Information
Katherine E Bohnsack, Email: katherine.bohnsack@med.uni-goettingen.de.
Markus T Bohnsack, Email: markus.bohnsack@med.uni-goettingen.de.
References
- Ameismeier M, Cheng J, Berninghausen O, Beckmann R (2018) Visualizing late states of human 40S ribosomal subunit maturation. Nature 558: 249–253 [DOI] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AKL, Lam YW, Steen H, Mann M, Lamond AI (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache J‐P, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R (2013) Structures of the human and Drosophila 80S ribosome. Nature 497: 80–85 [DOI] [PubMed] [Google Scholar]
- Ansel KM, Pastor WA, Rath N, Lapan AD, Glasmacher E, Wolf C, Smith LC, Papadopoulou N, Lamperti ED, Tahiliani M et al (2008) Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat Struct Mol Biol 15: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armistead J, Khatkar S, Meyer B, Mark BL, Patel N, Coghlan G, Lamont RE, Liu S, Wiechert J, Cattini PA et al (2009) Mutation of a gene essential for ribosome biogenesis, EMG1, causes bowen‐conradi syndrome. Am J Hum Genet 84: 728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, Boon KL, Tapsin S, Chan YS, Tan CP, Sim AYL et al (2016) In vivo mapping of eukaryotic RNA interactomes reveals principles of higher‐order organization and regulation. Mol Cell 62: 603–617 [DOI] [PubMed] [Google Scholar]
- Badertscher L, Wild T, Montellese C, Alexander LT, Bammert L, Sarazova M, Stebler M, Csucs G, Mayer TU, Zamboni N et al (2015) Genome‐wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Rep 13: 2879–2891 [DOI] [PubMed] [Google Scholar]
- Bai B, Moore HM, Laiho M (2013) CRM1 and its ribosome export adaptor NMD3 localize to the nucleolus and affect rRNA synthesis. Nucleus 4: 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai D, Zhang J, Li T, Hang R, Liu Y, Tian Y, Huang D, Qu L, Cao X, Ji J et al (2016) The ATPase hCINAP regulates 18S rRNA processing and is essential for embryogenesis and tumour growth. Nat Commun 7: 12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammert L, Jonas S, Ungricht R, Kutay U (2016) Human AATF/Che‐1 forms a nucleolar protein complex with NGDN and NOL10 required for 40S ribosomal subunit synthesis. Nucleic Acids Res 44: 9803–9820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JL, Drynan LF, Trim NL, Erber WN, Warren AJ, McKenzie AN (2010) New insights into 5q‐ syndrome as a ribosomopathy. Cell Cycle 9: 4286–4293 [DOI] [PubMed] [Google Scholar]
- Bassler J, Paternoga H, Holdermann I, Thoms M, Granneman S, Barrio‐Garcia C, Nyarko A, Stier G, Clark SA, Schraivogel D et al (2014) A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J Cell Biol 207: 481–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin‐Baillieu A, Tollervey D, Cullin C, Lacroute F (1997) Functional analysis of Rrp7p, an essential yeast protein involved in pre‐rRNA processing and ribosome assembly. Mol Cell Biol 17: 5023–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumas K, Soudet J, Caizergues‐Ferrer M, Faubladier M, Henry Y, Mougin A (2012) Human RioK3 is a novel component of cytoplasmic pre‐40S pre‐ribosomal particles. RNA Biol 9: 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Kopmar N, Ruggero D (2010a) Deregulation of oncogene‐induced senescence and p53 translational control in X‐linked dyskeratosis congenita. EMBO J 29: 1865–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, Ruggero D (2010b) Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res 70: 6026–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Shem A, Jenner L, Yusupova G, Yusupov M (2010) Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D (2009) Prp43 bound at different sites on the pre‐rRNA performs distinct functions in ribosome synthesis. Mol Cell 36: 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Sloan KE (2018) Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biol Chem 399: 1265–1276 [DOI] [PubMed] [Google Scholar]
- Bolze A, Mahlaoui N, Byun M, Turner B, Trede N, Ellis SR, Abhyankar A, Itan Y, Patin E, Brebner S et al (2013) Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science 340: 976–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boocock GRB, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM (2003) Mutations in SBDS are associated with Shwachman‐Diamond syndrome. Nat Genet 33: 97–101 [DOI] [PubMed] [Google Scholar]
- Boria I, Garelli E, Gazda HT, Aspesi A, Quarello P, Pavesi E, Ferrante D, Meerpohl JJ, Kartal M, Da Costa L et al (2010) The ribosomal basis of Diamond‐Blackfan Anemia: mutation and database update. Hum Mutat 31: 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoluzzi S, d'Alessi F, Romualdi C, Danieli GA (2001) Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics 17: 1152–1157 [DOI] [PubMed] [Google Scholar]
- Bourgeois G, Ney M, Gaspar I, Aigueperse C, Schaefer M, Kellner S, Helm M, Motorin Y (2015) Eukaryotic rRNA modification by yeast 5‐ methylcytosine‐methyltransferases and human proliferation‐associated antigen p120. PLoS One 10: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursać S, Brdovčak MC, Pfannkuchen M, Orsolić I, Golomb L, Zhu Y, Katz C, Daftuar L, Grabušić K, Vukelić I et al (2012) Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci USA 109: 20467–20472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J (2015) RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518: 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviño FR, Kharde S, Ori A, Hendricks A, Wild K, Kressler D, Bange G, Hurt E, Beck M, Sinning I (2015) Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA‐binding site. Nat Commun 6: 6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas‐Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J, Hadjiolov AA, Bachellerie JP (1996) Processing of mammalian rRNA precursors at the 3′ end of 18S rRNA. Identification of cis‐acting signals suggests the involvement of U13 small nucleolar RNA. Eur J Biochem 242: 206–213 [DOI] [PubMed] [Google Scholar]
- Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau‐Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O et al (2014) Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33: 474–483 [DOI] [PubMed] [Google Scholar]
- Chen SS, Williamson JR (2013) Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J Mol Biol 425: 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury P, Hackert P, Memet I, Sloan KE, Bohnsack MT (2019) The human RNA helicase DHX37 is required for release of the U3 snoRNP from pre‐ribosomal particles. RNA Biol 16: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciganda M, Williams N (2011) Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip Rev RNA 2: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coute Y, Kindbeiter K, Belin S, Dieckmann R, Duret L, Bezin L, Sanchez J‐C, Diaz J‐J (2008) ISG20L2, a novel vertebrate nucleolar exoribonuclease involved in ribosome biogenesis. Mol Cell Proteomics 7: 546–559 [DOI] [PubMed] [Google Scholar]
- Dai M‐S, Zeng SX, Jin Y, Sun X‐X, David L, Lu H (2004) Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 24: 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M‐S, Challagundla KB, Sun X‐X, Palam LR, Zeng SX, Wek RC, Lu H (2012) Physical and functional interaction between ribosomal protein L11 and the tumor suppressor ARF. J Biol Chem 287: 17120–17129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwerse JG, Dixon J, Seland S, Ruivenkamp CA, van Haeringen A, Hoefsloot LH, Peters DJ, Boers AC, Daumer‐Haas C, Maiwald R et al (2011) Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet 43: 20–22 [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ (2002) rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351 [DOI] [PubMed] [Google Scholar]
- Donati G, Peddigari S, Mercer CA, Thomas G (2013) 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2‐p53 checkpoint. Cell Rep 4: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar DA, Baserga SJ (1998) The U14 snoRNA is required for 2′‐O‐methylation of the pre‐18S rRNA in Xenopus oocytes. RNA 4: 195–204 [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR et al (2008) Identification of RPS14 as a 5q‐ syndrome gene by RNA interference screen. Nature 451: 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SR, Gleizes PE (2011) Diamond Blackfan anemia: ribosomal proteins going rogue. Semin Hematol 48: 89–96 [DOI] [PubMed] [Google Scholar]
- Emmott E, Jovanovic M, Slavov N (2019) Ribosome stoichiometry: from form to function. Trends Biochem Sci 44: 95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erales J, Marchand V, Panthu B, Gillot S, Belin S, Ghayad SE, Garcia M, Laforêts F, Marcel V, Baudin‐Baillieu A et al (2017) Evidence for rRNA 2′‐O‐methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci USA 114: 12934–12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber AW, Van Dijk M, Raué HA, Vos JC (2002) Ngl2p is a Ccr4p‐like RNA nuclease essential for the final step in 3′‐end processing of 5.8S rRNA in Saccharomyces cerevisiae . RNA 8: 1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley‐Barnes KI, McCann KL, Ogawa LM, Merkel J, Surovtseva YV, Baserga SJ (2018) Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep 22: 1923–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Oeffinger M, Dlakić M, Tollervey D (2003) Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol Cell Biol 23: 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG (2012) Role of Mex67‐Mtr2 in the nuclear export of 40S pre‐ribosomes. PLoS Genet 8: e1002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, Gonzalez Fernandez A, Simpson P, D'Santos CS, Arends MJ et al (2011) Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman‐Diamond syndrome. Genes Dev 25: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Muller S (2011a) The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J 30: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Raman N, Muller S (2011b) SUMO routes ribosome maturation. Nucleus 2: 527–532 [DOI] [PubMed] [Google Scholar]
- Freed EF, Baserga SJ (2010) The C‐terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Res 38: 4798–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EF, Prieto J‐L, McCann KL, McStay B, Baserga SJ (2012) NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre‐rRNA transcription and processing. PLoS Genet 8: e1002892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T (1997) Site‐specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Gartmann M, Blau M, Armache J‐P, Mielke T, Topf M, Beckmann R (2010) Mechanism of eIF6‐mediated inhibition of ribosomal subunit joining. J Biol Chem 285: 14848–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse L, Flemming D, Hurt E (2015) Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol Cell 60: 808–815 [DOI] [PubMed] [Google Scholar]
- Gentilella A, Kozma SC, Thomas G (2015) A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim Biophys Acta 1849: 812–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérus M, Bonnart C, Caizergues‐Ferrer M, Henry Y, Henras AK (2010) Evolutionarily conserved function of RRP36 in early cleavages of the pre‐rRNA and production of the 40S ribosomal subunit. Mol Cell Biol 30: 1130–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb KC, Cech TR (2017) Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev 31: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D et al (2002) 90S pre‐ribosomes include the 35S pre‐rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10: 105–115 [DOI] [PubMed] [Google Scholar]
- Granneman S, Gallagher JEG, Vogelzangs J, Horstman W, van Venrooij WJ, Baserga SJ, Pruijn GJM (2003) The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60‐80S ribonucleoprotein complexes. Nucleic Acids Res 31: 1877–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny R, Jedlinski DJ, Schmidt A, Gypas F, Martin G, Vina‐Vilaseca A, Zavolan M (2017) High‐throughput identification of C/D box snoRNA targets with CLIP and RiboMeth‐seq. Nucleic Acids Res 45: 2341–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttler T, Gorlich D (2011) Ran‐dependent nuclear export mediators: a structural perspective. EMBO J 30: 3457–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag S, Kretschmer J, Bohnsack MT (2015) WBSCR22/Merm1 is required for late nuclear pre‐ribosomal RNA processing and mediates N7‐methylation of G1639 in human 18S rRNA. RNA 21: 180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I (1998) X‐linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19: 32–38 [DOI] [PubMed] [Google Scholar]
- Henras AK, Plisson‐Chastang C, O'Donohue M‐F, Chakraborty A, Gleizes P‐E (2015) An overview of pre‐ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her J, Chung IK (2012) The AAA‐ATPase NVL2 is a telomerase component essential for holoenzyme assembly. Biochem Biophys Res Commun 417: 1086–1092 [DOI] [PubMed] [Google Scholar]
- Holzel M, Rohrmoser M, Schlee M, Grimm T, Harasim T, Malamoussi A, Gruber‐Eber A, Kremmer E, Hiddemann W, Bornkamm GW et al (2005) Mammalian WDR12 is a novel member of the Pes1‐Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol 170: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R (2000) Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 19: 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovella MG, Bremang M, Basha O, Giacò L, Carotenuto W, Golfieri C, Szakal B, Dal Maschio M, Infantino V, Beznoussenko GV et al (2018) Integrating Rio1 activities discloses its nutrient‐activated network in Saccharomyces cerevisiae . Nucleic Acids Res 46: 7586–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Yoshikawa H, Izumikawa K, Miura Y, Taoka M, Nobe Y, Yamauchi Y, Nakayama H, Simpson RJ, Isobe T et al (2017) Poly(A)‐specific ribonuclease regulates the processing of small‐subunit rRNAs in human cells. Nucleic Acids Res 45: 3437–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, Suzuki T (2014) Human NAT10 is an ATP‐dependent RNA acetyltransferase responsible for N4‐acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol Chem 289: 35724–35730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson EM, Rodero MP, Kasher PR, Uggenti C, Oojageer A, Goosey LC, Rose Y, Kershaw CJ, Urquhart JE, Williams SG et al (2016) Mutations in SNORD118 cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts. Nat Genet 48: 1185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, Glynn EF, Ellington L, Du C, Dixon J et al (2008) Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med 14: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, Zavolan M, Gruber AR (2016) An updated human snoRNAome. Nucleic Acids Res 44: 5068–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F (2013) The circadian clock coordinates ribosome biogenesis. PLoS Biol 11: e1001455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M, Rohrmoser M, Forne I, Voss K, Burger K, Muhl B, Gruber‐Eber A, Kremmer E, Imhof A, Eick D (2015) DEAD‐box helicase DDX27 regulates 3′ end formation of ribosomal 47S RNA and stably associates with the PeBoW‐complex. Exp Cell Res 334: 146–159 [DOI] [PubMed] [Google Scholar]
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR et al (2018) Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Stamm S (2006) The snoRNA HBII‐52 regulates alternative splicing of the serotonin receptor 2C. Science 311: 230–232 [DOI] [PubMed] [Google Scholar]
- Kiss‐Laszlo Z, Henry Y, Bachellerie JP, Caizergues‐Ferrer M, Kiss T (1996) Site‐specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts‐Hoffmann F, Leibundgut M, Arpagaus S, Ban N (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948 [DOI] [PubMed] [Google Scholar]
- Knight SW, Vulliamy TJ, Morgan B, Devriendt K, Mason PJ, Dokal I (2001) Identification of novel DKC1 mutations in patients with dyskeratosis congenita: implications for pathophysiology and diagnosis. Hum Genet 108: 299–303 [DOI] [PubMed] [Google Scholar]
- Kobyłecki K, Drążkowska K, Kuliński TM, Dziembowski A, Tomecki R (2018) Elimination of 01/A’‐A0 pre‐rRNA processing by‐product in human cells involves cooperative action of two nuclear exosome‐associated nucleases: RRP6 and DIS3. RNA 24: 1677–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bergler H, Bassler J (2012) The power of AAA‐ATPases on the road of pre‐60S ribosome maturation–molecular machines that strip pre‐ribosomal particles. Biochim Biophys Acta 1823: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP et al (2004) High‐definition macromolecular composition of yeast RNA‐processing complexes. Mol Cell 13: 225–239 [DOI] [PubMed] [Google Scholar]
- Krogh N, Jansson MD, Hafner SJ, Tehler D, Birkedal U, Christensen‐Dalsgaard M, Lund AH, Nielsen H (2016) Profiling of 2′‐O‐Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res 44: 7884–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn H, Hierlmeier T, Merl J, Jakob S, Aguissa‐Touré AH, Milkereit P, Tschochner H (2009) The Noc‐domain containing C‐terminus of Noc4p mediates both formation of the Noc4p‐Nop14p submodule and its incorporation into the SSU processome. PLoS One 4: e8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Preiss T, Tollervey D (1998) Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol Cell Biol 18: 2360–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry‐Voyer A‐M, Bilodeau S, Bergeron D, Dionne KL, Port SA, Rouleau C, Boisvert F‐M, Kehlenbach RH, Bachand F (2016) Human PDCD2L is an export substrate of CRM1 that associates with 40S ribosomal subunit precursors. Mol Cell Biol 36: MCB.00303‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B, Purushothaman SK (2004) Spb1p‐directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol Cell 16: 663–669 [DOI] [PubMed] [Google Scholar]
- Larburu N, Montellese C, O'Donohue M‐F, Kutay U, Gleizes P‐E, Plisson‐Chastang C (2016) Structure of a human pre‐40S particle points to a role for RACK1 in the final steps of 18S rRNA processing. Nucleic Acids Res 44: 8465–8478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazdins IB, Delannoy M, Sollner‐Webb B (1997) Analysis of nucleolar transcription and processing domains and pre‐rRNA movements by in situ hybridization. Chromosoma 105: 481–495 [DOI] [PubMed] [Google Scholar]
- Leidig C, Thoms M, Holdermann I, Bradatsch B, Berninghausen O, Bange G, Sinning I, Hurt E, Beckmann R (2014) 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat Commun 5: 3491 [DOI] [PubMed] [Google Scholar]
- Leisegang MS, Martin R, Ramírez AS, Bohnsack MT (2012) Exportin t and Exportin 5: tRNA and miRNA biogenesis ‐ and beyond. Biol Chem 393: 599–604 [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ (2006) snoRNA‐LBME‐db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34: D158–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Tollervey D (2000) Like attracts like: getting RNA processing together in the nucleus. Science 288: 1385–1389 [DOI] [PubMed] [Google Scholar]
- Li M, Gu W (2011) A critical role for noncoding 5S rRNA in regulating Mdmx stability. Mol Cell 43: 1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J‐M, Zhou X, Gatignol A, Lu H (2014) Ribosomal proteins L5 and L11 co‐operatively inactivate c‐Myc via RNA‐induced silencing complex. Oncogene 33: 4916–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PC, Thiele DJ (2001) Novel stress‐responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol Biol Cell 12: 3644–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW (2010) Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 39: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke‐Andersen S, Ardal BK, Hollensen AK, Damgaard CK, Jensen TH (2018) Box C/D snoRNP autoregulation by a cis‐Acting snoRNA in the NOP56 Pre‐mRNA. Mol Cell 72: 99–111.e5 [DOI] [PubMed] [Google Scholar]
- Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, Chen K, Lu Z, Chen H, Shi YG et al (2019) N 6‐Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol 15: 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malygin AA, Parakhnevitch NM, Ivanov AV, Eperon IC, Karpova GG (2007) Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res 35: 6414–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marneros AG (2013) BMS1 is mutated in aplasia cutis congenita. PLoS Genet 9: e1003573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Straub AU, Doebele C, Bohnsack MT (2013) DExD/H‐box RNA helicases in ribosome biogenesis. RNA Biol 10: 4–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I (2006) Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391 [DOI] [PubMed] [Google Scholar]
- Memet I, Doebele C, Sloan KE, Bohnsack MT (2017) The G‐patch protein NF‐kappaB‐repressing factor mediates the recruitment of the exonuclease XRN2 and activation of the RNA helicase DHX15 in human ribosome biogenesis. Nucleic Acids Res 45: 5359–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Wurm JP, Kötter P, Leisegang MS, Schilling V, Buchhaupt M, Held M, Bahr U, Karas M, Heckel A, Bohnsack MT, Wöhnert J et al (2011) The Bowen‐Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Ψ1191 in yeast 18S rRNA. Nucleic Acids Res 39: 1526–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Wurm JP, Sharma S, Immer C, Pogoryelov D, Kötter P, Lafontaine DLJ, Wöhnert J, Entian KD (2016) Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res 44: 4304–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Green R (2017) Ribosomopathies: There's strength in numbers. Science 358: 2755 [DOI] [PubMed] [Google Scholar]
- Mishra RK, Eliceiri GL (1997) Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc Natl Acad Sci USA 94: 4972–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montellese C, Montel‐Lehry N, Henras AK, Kutay U, Gleizes P‐E, O'Donohue M‐F (2017) Poly(A)‐specific ribonuclease is a nuclear ribosome biogenesis factor involved in human 18S rRNA maturation. Nucleic Acids Res 45: 6822–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello LG, Coltri PP, Quaresma AJC, Simabuco FM, Silva TCL, Singh G, Nickerson JA, Oliveira CC, Moore MJ, Zanchin NIT (2011) The human nucleolar protein FTSJ3 associates with NIP7 and functions in pre‐rRNA processing. PLoS One 6: e29174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Ray PS, Arif A, Brady AK, Kinter M, Fox PL (2008) DAPK‐ZIPK‐L13a axis constitutes a negative‐feedback module regulating inflammatory gene expression. Mol Cell 32: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A, Ebert BL (2010) Ribosomopathies: human disorders of ribosome dysfunction. Blood 115: 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, Klaholz BP (2017) Visualization of chemical modifications in the human 80S ribosome structure. Nature 551: 472–477 [DOI] [PubMed] [Google Scholar]
- Nicolas E, Parisot P, Pinto‐Monteiro C, de Walque R, De Vleeschouwer C, Lafontaine DLJ (2016) Involvement of human ribosomal proteins in nucleolar structure and p53‐dependent nucleolar stress. Nat Commun 7: 11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousbeck J, Spiegel R, Ishida‐Yamamoto A, Indelman M, Shani‐Adir A, Adir N, Lipkin E, Bercovici S, Geiger D, van Steensel MA et al (2008) Alopecia, neurological defects, and endocrinopathy syndrome caused by decreased expression of RBM28, a nucleolar protein associated with ribosome biogenesis. Am J Hum Genet 82: 1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donohue M‐F, Choesmel V, Faubladier M, Fichant G, Gleizes P‐E (2010) Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol 190: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand J, Bakin A (1997) Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J Mol Biol 266: 246–268 [DOI] [PubMed] [Google Scholar]
- Paolini NA, Attwood M, Sondalle SB, Vieira CMDS, van Adrichem AM, di Summa FM, O'Donohue M‐F, Gleizes P‐E, Rachuri S, Briggs JW et al (2017) A ribosomopathy reveals decoding defective ribosomes driving human dysmorphism. Am J Hum Genet 100: 506–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pausch P, Singh U, Ahmed YL, Pillet B, Murat G, Altegoer F, Stier G, Thoms M, Hurt E, Sinning I et al (2015) Co‐translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat Commun 6: 7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA (1993) Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 73: 1233–1245 [DOI] [PubMed] [Google Scholar]
- Peifer C, Sharma S, Watzinger P, Lamberth S, Kotter P, Entian KD (2013) Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res 41: 1151–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelava A, Schneider C, Watkins NJ (2016) The importance of ribosome production, and the 5S RNP‐MDM2 pathway, in health and disease. Biochem Soc Trans 44: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Thomas G, Volarević S (2018) Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 18: 51–63 [DOI] [PubMed] [Google Scholar]
- Pertschy B, Schneider C, Gnädig M, Schäfer T, Tollervey D, Hurt E (2009) RNA helicase Prp43 and its co‐factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 284: 35079–35091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna‐Przybylska D, Decatur WA, Fournier MJ (2008) The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res 36: D178–D183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillet B, García‐Gómez JJ, Pausch P, Falquet L, Bange G, de la Cruz J, Kressler D (2015) The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear Pre‐60S assembly site. PLoS Genet 11: e1005565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillet B, Mitterer V, Kressler D, Pertschy B (2017) Hold on to your friends: Dedicated chaperones of ribosomal proteins: Dedicated chaperones mediate the safe transfer of ribosomal proteins to their site of pre‐ribosome incorporation. BioEssays 39: 1–12 [DOI] [PubMed] [Google Scholar]
- Polikanov YS, Melnikov SV, Soll D, Steitz TA (2015) Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol 22: 342–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti M, O'Donohue M‐F, Montel‐Lehry N, Bortolin‐Cavaille M‐L, Choesmel V, Gleizes P‐E (2013) Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res 41: 4709–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J‐L, McStay B (2007) Recruitment of factors linking transcription and processing of pre‐rRNA to NOR chromatin is UBF‐dependent and occurs independent of transcription in human cells. Genes Dev 21: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainger J, Bengani H, Campbell L, Anderson E, Sokhi K, Lam W, Riess A, Ansari M, Smithson S, Lees M et al (2012) Miller (Genee‐Wiedemann) syndrome represents a clinically and biochemically distinct subgroup of postaxial acrofacial dysostosis associated with partial deficiency of DHODH. Hum Mol Genet 21: 3969–3983 [DOI] [PubMed] [Google Scholar]
- Raman N, Weir E, Müller S (2016) The AAA ATPase MDN1 Acts as a SUMO‐targeted regulator in mammalian pre‐ribosome remodeling. Mol Cell 64: 607–615 [DOI] [PubMed] [Google Scholar]
- Reuveni S, Ehrenberg M, Paulsson J (2017) Ribosomes are optimized for autocatalytic production. Nature 547: 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridanpää M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S et al (2001) Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage‐hair hypoplasia. Cell 104: 195–203 [DOI] [PubMed] [Google Scholar]
- Riley MF, Lozano G (2012) The many faces of MDM2 binding partners. Genes Cancer 3: 226–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrmoser M, Holzel M, Grimm T, Malamoussi A, Harasim T, Orban M, Pfisterer I, Gruber‐Eber A, Kremmer E, Eick D (2007) Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol 27: 3682–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R, Liu X, Granneman S, Zhu J, Gill M, Papoulas O, Marcotte EM, Tollervey D, Correll CC, Johnson AW (2015) The DEAH‐box helicase Dhr1 dissociates U3 from the pre‐rRNA to promote formation of the central pseudoknot. PLoS Biol 13: e1002083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R, Gerbi SA (1990) In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J 9: 2299–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T, Maco B, Petfalski E, Tollervey D, Böttcher B, Aebi U, Hurt E (2006) Hrr25‐dependent phosphorylation state regulates organization of the pre‐40S subunit. Nature 441: 651–655 [DOI] [PubMed] [Google Scholar]
- Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez J‐C, Greco A, Hochstrasser D, Diaz J‐J (2002) Functional proteomic analysis of human nucleolus. Mol Biol Cell 13: 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillewaert S, Wacheul L, Lhomme F, Lafontaine DLJ (2012) The evolutionarily conserved protein Las1 is required for pre‐rRNA processing at both ends of ITS2. Mol Cell Biol 32: 430–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D (2003) A role for c‐Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res 31: 6148–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA (1993) Nuclear RNase MRP is required for correct processing of pre‐5.8S rRNA in Saccharomyces cerevisiae . Mol Cell Biol 13: 7935–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle‐Perez A, Pircher A, Gerstl MP, Pfeifenberger S et al (2015) Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun 6: 6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz S, Fischer U, Altvater M, Nerurkar P, Peña C, Gerber M, Chang Y, Caesar S, Schubert OT, Schlenstedt G et al (2014) A RanGTP‐independent mechanism allows ribosomal protein nuclear import for ribosome assembly. Elife 3: e03473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmi C, Feghali‐Bostwick CA, Lleo A, Lombardi SA, De Santis M, Cavaciocchi F, Zammataro L, Mitchell MM, Lasalle JM, Medsger T Jr et al (2012) X chromosome gene methylation in peripheral lymphocytes from monozygotic twins discordant for scleroderma. Clin Exp Immunol 169: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Watzinger P, Kotter P, Entian KD (2013a) Identification of a novel methyltransferase, Bmt2, responsible for the N‐1‐methyl‐adenosine base modification of 25S rRNA in Saccharomyces cerevisiae . Nucleic Acids Res 41: 5428–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, Watzinger P, Kotter P, Entian KD (2013b) Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res 41: 9062–9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, Duttmann S, Watzinger P, Kotter P, Entian KD (2014) Identification of novel methyltransferases, Bmt5 and Bmt6, responsible for the m3U methylations of 25S rRNA in Saccharomyces cerevisiae . Nucleic Acids Res 42: 3246–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Langhendries J‐L, Watzinger P, Kotter P, Entian K‐D, Lafontaine DLJ (2015) Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res 43: 2242–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E, Sterne‐Weiler T, O'Hanlon D, Blencowe BJ (2016) Global mapping of human RNA‐RNA interactions. Mol Cell 62: 618–626 [DOI] [PubMed] [Google Scholar]
- Sharma S, Marchand V, Motorin Y, Lafontaine DLJ (2017a) Identification of sites of 2′‐O‐methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci Rep 7: 11490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, van Nues R, Watzinger P, Kotter P, Lafontaine DLJ, Granneman S, Entian K‐D (2017b) Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet 13: e1006804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Rost HL, Teruel MN, Barna M (2017) Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome‐wide. Mol Cell 67: 71–83.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Barna M (2017) An emerging role for the ribosome as a nexus for post‐translational modifications. Curr Opin Cell Biol 45: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinturel F, Gerber A, Mauvoisin D, Wang J, Gatfield D, Stubblefield JJ, Green CB, Gachon F, Schibler U (2017) Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169: 651–663.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJM, Watkins NJ (2013a) Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J Cell Biol 200: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Watkins NJ (2013b) The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 5: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Schneider C, Watkins NJ (2014) The roles of SSU processome components and surveillance factors in the initial processing of human ribosomal RNA. RNA 20: 540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Leisegang MS, Doebele C, Ramirez AS, Simm S, Safferthal C, Kretschmer J, Schorge T, Markoutsa S, Haag S et al (2015) The association of late‐acting snoRNPs with human pre‐ribosomal complexes requires the RNA helicase DDX21. Nucleic Acids Res 43: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Gleizes P‐E, Bohnsack MT (2016) Nucleocytoplasmic transport of RNAs and RNA‐Protein complexes. J Mol Biol 428: 2040–2059 [DOI] [PubMed] [Google Scholar]
- Sloan KE, Warda AS, Sharma S, Entian K‐D, Lafontaine DLJ, Bohnsack MT (2017) Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol 4: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Knox AA, Wells GR, Schneider C, Watkins NJ (2019) Interactions and activities of factors involved in the late stages of human 18S rRNA maturation. RNA Biol 16: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel R, Shalev SA, Adawi A, Sprecher E, Tenenbaum‐Rakover Y (2010) ANE syndrome caused by mutated RBM28 gene: a novel etiology of combined pituitary hormone deficiency. Eur J Endocrinol 162: 1021–1025 [DOI] [PubMed] [Google Scholar]
- Srivastava L, Lapik YR, Wang M, Pestov DG (2010) Mammalian DEAD box protein Ddx51 acts in 3′ end maturation of 28S rRNA by promoting the release of U8 snoRNA. Mol Cell Biol 30: 2947–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X‐X, DeVine T, Challagundla KB, Dai M‐S (2011) Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. J Biol Chem 286: 22730–22741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries J‐L, Mullineux S‐T, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DLJ (2013) The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre‐rRNA processing factors. Mol Cell 51: 539–551 [DOI] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123: 49–63 [DOI] [PubMed] [Google Scholar]
- Takei S, Togo‐Ohno M, Suzuki Y, Kuroyanagi H (2016) Evolutionarily conserved autoregulation of alternative pre‐mRNA splicing by ribosomal protein L10a. Nucleic Acids Res 44: 5585–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Yamauchi Y, Ishikawa H, Takahashi N, Nakayama H, Isobe T (2016) The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res 44: 8951–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Sato K, Ishikawa H, Izumikawa K, Yamauchi Y, Hirota K, Nakayama H, Takahashi N et al (2018) Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res 46: 9289–9298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Labno A, Drazkowska K, Cysewski D, Dziembowski A (2015) hUTP24 is essential for processing of the human rRNA precursor at site A1, but not at site A0. RNA Biol 12: 1010–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Sikorski PJ, Zakrzewska‐Placzek M (2017) Comparison of preribosomal RNA processing pathways in yeast, plant and human cells ‐ focus on coordinated action of endo‐ and exoribonucleases. FEBS Lett 591: 1801–1850 [DOI] [PubMed] [Google Scholar]
- Tummala H, Walne AJ, Williams M, Bockett N, Collopy L, Cardoso S, Ellison A, Wynn R, Leblanc T, Fitzgibbon J et al (2016) DNAJC21 mutations link a cancer‐prone bone marrow failure syndrome to corruption in 60S ribosome subunit maturation. Am J Hum Genet 99: 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Knox AA, Watkins NJ (2012) Nucleolar disruption leads to the spatial separation of key 18S rRNA processing factors. RNA Biol 9: 175–186 [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Steitz JA (1994) Requirement for intron‐encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science 266: 1558–1561 [DOI] [PubMed] [Google Scholar]
- Valdez BC, Henning D, So RB, Dixon J, Dixon MJ (2004) The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA 101: 10709–10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays E, Gelugne JP, Gleizes PE, Caizergues‐Ferrer M (2003) Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae . Mol Cell Biol 23: 2083–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K et al (2005) Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet 37: 468–470 [DOI] [PubMed] [Google Scholar]
- Vincent M, Geneviève D, Ostertag A, Marlin S, Lacombe D, Martin‐Coignard D, Coubes C, David A, Lyonnet S, Vilain C et al (2016) Treacher Collins syndrome: a clinical and molecular study based on a large series of patients. Genet Med 18: 49–56 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I (2001) The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413: 432–435 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Beswick R, Kirwan M, Marrone A, Digweed M, Walne A, Dokal I (2008) Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Natl Acad Sci USA 105: 8073–8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waku T, Nakajima Y, Yokoyama W, Nomura N, Kako K, Kobayashi A, Shimizu T, Fukamizu A (2016) NML‐mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53‐dependent manner. J Cell Sci 129: 2382–2393 [DOI] [PubMed] [Google Scholar]
- Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D et al (2007) Ribosomal protein S3: a KH domain subunit in NF‐kappaB complexes that mediates selective gene regulation. Cell 131: 927–939 [DOI] [PubMed] [Google Scholar]
- Wandrey F, Montellese C, Koos K, Badertscher L, Bammert L, Cook AG, Zemp I, Horvath P, Kutay U (2015) The NF45/NF90 heterodimer contributes to the biogenesis of 60S ribosomal subunits and influences nucleolar morphology. Mol Cell Biol 35: 3491–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Pestov DG (2011) 5′‐end surveillance by Xrn2 acts as a shared mechanism for mammalian pre‐rRNA maturation and decay. Nucleic Acids Res 39: 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhu Q, Huang L, Zhu Y, Chen J, Peng J, Lo LJ (2016) Interaction between Bms1 and Rcl1, two ribosome biogenesis factors, is evolutionally conserved in zebrafish and human. J Genet Genomics 43: 467–469 [DOI] [PubMed] [Google Scholar]
- Warda AS, Freytag B, Haag S, Sloan KE, Gorlich D, Bohnsack MT (2016) Effects of the Bowen‐Conradi syndrome mutation in EMG1 on its nuclear import, stability and nucleolar recruitment. Hum Mol Genet 25: 5353–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Warren AJ (2018) Molecular basis of the human ribosomopathy Shwachman‐Diamond syndrome. Adv Biol Regul 67: 109–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT (2012) The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414 [DOI] [PubMed] [Google Scholar]
- Webb GC, Baker RT, Coggan M, Board PG (1994) Localization of the human UBA52 ubiquitin fusion gene to chromosome band 19p13.1‐p12. Genomics 19: 567–569 [DOI] [PubMed] [Google Scholar]
- Wegierski T, Billy E, Nasr F, Filipowicz W (2001) Bms1p, a G‐domain‐containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA 7: 1254–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis F, Giudice E, Churcher M, Jin L, Hilcenko C, Wong CC, Traynor D, Kay RR, Warren AJ (2015) Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol 22: 914–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells GR, Weichmann F, Colvin D, Sloan KE, Kudla G, Tollervey D, Watkins NJ, Schneider C (2016) The PIN domain endonuclease Utp24 cleaves pre‐ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res 44: 5399–5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells GR, Weichmann F, Sloan KE, Colvin D, Watkins NJ, Schneider C (2017) The ribosome biogenesis factor yUtp23/hUTP23 coordinates key interactions in the yeast and human pre‐40S particle and hUTP23 contains an essential PIN domain. Nucleic Acids Res 45: 4796–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW (2008) Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre‐40S subunits. Mol Cell Biol 28: 3151–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann B, Wandrey F, Badertscher L, Wyler E, Pfannstiel J, Zemp I, Kutay U (2012) The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol Biol Cell 23: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U (2010) A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol 8: e1000522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Baserga SJ (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae . Genetics 195: 643–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ (1993) The p53‐mdm‐2 autoregulatory feedback loop. Genes Dev 7: 1126–1132 [DOI] [PubMed] [Google Scholar]
- Wurm JP, Meyer B, Bahr U, Held M, Frolow O, Kotter P, Engels JW, Heckel A, Karas M, Entian KD et al (2010) The ribosome assembly factor Nep1 responsible for Bowen‐Conradi syndrome is a pseudouridine‐N1‐specific methyltransferase. Nucleic Acids Res 38: 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler E, Wandrey F, Badertscher L, Montellese C, Alper D, Kutay U (2014) The beta‐isoform of the BRCA2 and CDKN1A(p21)‐interacting protein (BCCIP) stabilizes nuclear RPL23/uL14. FEBS Lett 588: 3685–3691 [DOI] [PubMed] [Google Scholar]
- Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D (2006) Impaired control of IRES‐mediated translation in X‐linked dyskeratosis congenita. Science 312: 902–906 [DOI] [PubMed] [Google Scholar]
- Yoshikatsu Y, Ishida Y, Sudo H, Yuasa K, Tsuji A, Nagahama M (2015) NVL2, a nucleolar AAA‐ATPase, is associated with the nuclear exosome and is involved in pre‐rRNA processing. Biochem Biophys Res Commun 464: 780–786 [DOI] [PubMed] [Google Scholar]
- Zanni G, Kalscheuer VM, Friedrich A, Barresi S, Alfieri P, Di Capua M, Haas SA, Piccini G, Karl T, Klauck SM et al (2015) A novel mutation in RPL10 (Ribosomal Protein L10) causes X‐Linked intellectual disability, cerebellar hypoplasia, and spondylo‐epiphyseal dysplasia. Hum Mutat 36: 1155–1158 [DOI] [PubMed] [Google Scholar]
- Zemp I, Wild T, O'Donohue M‐F, Wandrey F, Widmann B, Gleizes P‐E, Kutay U (2009) Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol 185: 1167–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Wandrey F, Rao S, Ashiono C, Wyler E, Montellese C, Kutay U (2014) CK1delta and CK1epsilon are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J Cell Sci 127: 1242–1253 [DOI] [PubMed] [Google Scholar]
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JLJ (2007) Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev 21: 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, O'Leary MN, Peri S, Wang M, Zha J, Melov S, Kappes DJ, Feng Q, Rhodes J, Amieux PS et al (2017) Ribosomal proteins Rpl22 and Rpl22l1 control morphogenesis by regulating Pre‐mRNA splicing. Cell Rep 18: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hao Q, Liao J‐M, Liao P, Lu H (2013) Ribosomal protein S14 negatively regulates c‐Myc activity. J Biol Chem 288: 21793–21801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorbas C, Nicolas E, Wacheul L, Huvelle E, Heurgue‐Hamard V, Lafontaine DLJ (2015) The human 18S rRNA base methyltransferases DIMT1L and WBSCR214‐TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol Biol Cell 26: 2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]


