Abstract
Colorectal cancer (CRC) is the third most common cancer diagnosed worldwide and represents the second most common cause of all cancer-related deaths in Korea. Although epidemiological data indicate a sharp increase in the incidence of CRC among individuals older than 50 years, more than 10% of CRCs occur before reaching 50. These are known as early-onset CRCs (EOCRCs) and are likely to be suggestive of hereditary predisposition. However, known familial CRC syndromes account for only 20% of genetic aberrations of EOCRC, and the remaining 80% are still in question. Therefore, we aimed to establish reproducible biological resources and contribute to expand the mutation database of EOCRC. Four cell lines derived from the original tumor mass of CRC patients diagnosed under age 30 years were established, and next-generation sequencing technique was used to identify the genetic features of EOCRC. We have identified one novel fusion gene, FAM174A-WWC1, and analyzed its functional role. The induction of FAM174A-WWC1 to normal fibroblast caused alternations in cellular morphology as well as intercellular expression of E-cadherin and N-cadherin. Moreover, WWC1 carrying the fused FAM174A domain not only abrogated the membrane expression of YAP1 but also significantly increased the levels of nucleic YAP1. As a result, the FAM174A-WWC1 expression increased the oncogenic capacity and invasiveness of normal fibroblasts, which suggests its role as a potential driver mutation of EOCRC.
Introduction
Colorectal cancer (CRC) is the third most common cancer diagnosed worldwide and is the second most common cause of cancer deaths in Korea [1], [2]. Although epidemiological data indicate the incidence of CRC sharply rises among individuals older than 50 years, more than 10% of CRCs occur before age 50. Such early-onset CRC (EOCRC) may suggest hereditary predisposition [3], [4], [5]. However, known familial CRC syndromes account for only 20% of genetic aberrations of EOCRC, and the remaining 80% are still poorly known [5]. Epidemiological factors such as obesity, diabetes mellitus, and diet have been proposed as potential risk factors but do not fully explain the development of EOCRCs [6], [7]. The clinicopathologic features of EOCRCs are known to be poorly differentiated, carrying mucinous and signet ring features, and are diagnosed at advanced stages, which partially reflect the aggressive behavior of EOCRC. Based on their distinct clinicopathological characteristics, previous studies suggested the creation of a unique molecular subgroup for classification of these tumors [8].
Structural chromosome rearrangements such as translocation, interstitial deletion, or chromosomal inversion may result in the formation of a chimeric gene from previously separate genes [9]. Such hybrid gene often generates a novel protein with functional capabilities distinct from those of the parental genes. Especially, the specificity of gene fusion in neoplastic tissues and their diverse functionalities during carcinogenesis provide critical clues for disease mechanisms underlying tumorigenesis, enabling tumor subclassification [10]. Since gene fusion is closely associated with specific tumor phenotype, which is derived from the unique chimeric domains, they represent ideal targets for anticancer treatment and risk stratification [11]. Although the effect of translocation involving coding and regulatory DNA sequences has been intensely studied in chronic myeloid leukemia [12], [13] and lung cancer [14], [15], its role in EOCRC has yet to be elucidated.
In this study, four cell lines derived from patients with EOCRC were established and analyzed via next-generation sequencing to identify molecular and genetic features. We also have identified one novel fusion gene, FAM174A_WWC1, and analyzed its functional role.
Materials and Methods
Establishment of Cell Lines and Cell Culture Condition
Cell lines were established from pathologically proven colorectal carcinomas. Detailed procedure was described previously [16]. The locations and stages of original tumors of newly established EOCRC cell lines are listed in Table 1.
Table 1.
Clinicopathologic Characteristics of Four EOCRC Cell Lines
|
In Vitro |
In Vivo |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Growth Pattern | Doubling Time (h) | Cell Morphology | Sex | Age | Family History | Degree of Differentiation | Stage | Pathology | Location | Recur. | |
| NCC-375 | Adherent | 35 | Fusiform | F | 24 | None | M/D | T3N2M1 | adc | Rectal cancer with liver and lung metastasis | Recur. |
| SNU-1460 | Adherent | 48 | Polygonal | M | 25 | None | M/D | T4aN0 | adc | S-colon cancer | NED |
| SNU-1826 | Adherent | 45 | Polygonal | M | 15 | GF (stomach ca.) | P/D | adc | Cecal cancer | NED | |
| SNU-2446 | Adherent/floating | 82 | Round/Polygonal | F | 21 | none | T4bN2 | Mucinous carcinoma | D-colon cancer with p-seeding | PD | |
GF, grandfather; adc, adenocarcinoma; M/D, moderately differentiated; P/D, poorly differentiated.
DNA Fingerprinting Analysis
Total DNA was isolated from cell pellet by using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's protocol. DNA was amplified using an AmpFlSTR identifiler PCR Amplification Kit (Applied Biosystems, Foster City, CA). Detailed procedure was described previously [17]. The STR profiles of established EOCRC cell lines are listed in Table 2.
Table 2.
STR Profiles of Four EOCRC Cell Lines
| Cell Name | D8S1179 | D21S11 | D7S820 | CSF1PO | D3S1358 | TH01 | D13S317 | D16S539 | D2S1338 | D19S433 | Vwa | TPOX | D18S51 | Amelogenin | D5S818 | FGA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCC-375 | 13,15 | 32.2,33.2 | 10,12 | 10,12 | 18 | 7 | 9,12 | 10 | 17,23 | 13,13.2 | 18 | 11 | 15 | X | 12,17 | 20,23 |
| SNU-1460 | 13 | 30.2,31 | 8,11 | 12 | 15,17 | 6,7 | 9,12 | 10 | 23,24 | 13,14.2 | 14,18 | 8,11 | 13 | X | 11 | 21.23 |
| SNU-1826 | 13 | 32,32.2 | 9,10 | 10 | 15 | 6 | 8,11 | 9,11 | 17 | 11.2,14 | 17,18 | 8 | 15 | X | 11 | 22 |
| SNU-2446 | 12 | 29,34 | 9,10 | 10,12 | 15,17 | 9 | 11 | 13 | 18,20 | 13 | 14 | 9,11 | 19 | X | 12 | 24 |
Growth Properties and Morphology In Vitro
Suspensions of 5×103 cells were seeded to 35-mm tissue culture dishes containing a culture medium. The number of cells was counted in triplicate at 24-hour intervals for 10 days using WST-1 assay. The doubling time of the cells was calculated from the growth phase. Mycoplasma contamination was tested by the 16S-rRNA-gene–based polymerase chain reaction (PCR) amplification method using e-Myco Mycoplasma PCR Detection Kit (Intron Biotechnology, Kyonggi, Korea).
Nucleic Acid Isolation and Complementary DNA Synthesis
Genomic DNA was extracted from the cell lines using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), and RNA was extracted using the TRIzol (Life Technologies, CA, USA) and RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. For cDNA synthesis, detailed procedure was described previously [17].
Whole Exome Sequencing Analysis
Total DNA was isolated from cell pellet using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Exomic sequences were enriched in samples using the Illumina Exome Enrichment protocol, and the captured libraries were sequenced using Illumina HiSeq 2000 Sequencers (Illumina, CA, USA). The reads were mapped to the human genome assembly UCSC hg19 using Burrows-Wheeler Alignment (BWA version 0.5.8, http://bio-bwa.sourceforge.net/). Single nucleotide polymorphisms and small insertions/deletions were called with SAMtools (version 0.1.7, http://samtools.sourceforge.net/). The annotated variants were filtered against the variations reported in dbSNP132, the 1000 Genomes project November 2010 edition, and the NHLBI Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) databases. Computational tools SIFT [18] and PolyPhen-2 [19] were used to predict the impact of missense mutations, while MaxEntScan [20] and NNSplice (http://www.fruitfly.org/seq_tools/splice.html) were used to predict the effect of splice variants.
RNA Sequencing Analysis
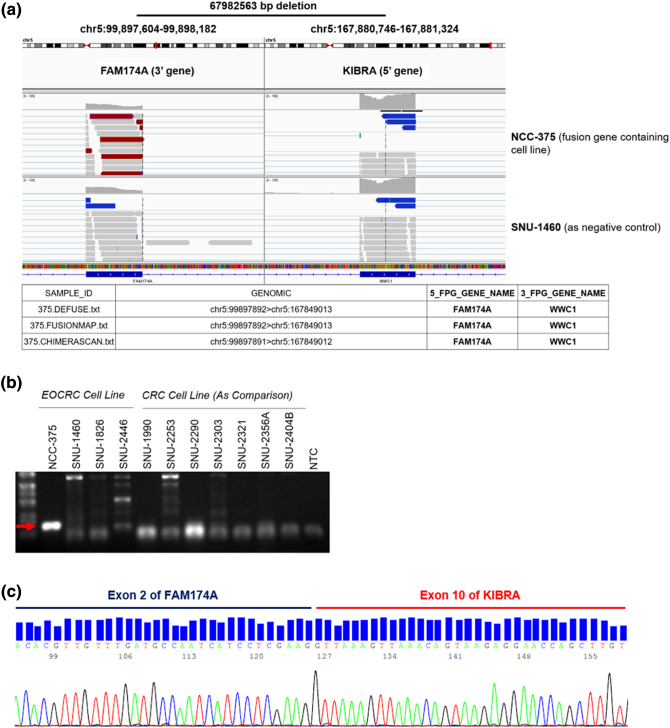
Total RNA was isolated from cell lysate using TRIzol (Qiagen, Hilden, Germany) and Qiagen RNeasy Kit (Qiagen, Hilden, Germany). Sequencing libraries were prepared using the Illumina TruSeq Stranded Total RNA Library Prep Kit. Fifty-one million reads were obtained from the cell lysates. Following base-calling and alignment with the Tuxedo Suite, the rejected reads were analyzed using FusionMap, ChimeraScan, and Defuse with default parameters for RNA and alignment to GRCh37.72. The output was filtered to include in-frame fusions, with at least one rescued read and two unique seed reads and excluded known and recurrent artifacts. Sequencing of RNA isolated from EOCRC cell lines was used to identify RNA sequences spanning two different regions in a chromosome. The in-frame fusion with the highest read score spanned the FAM174A gene on chromosome 5q21.1 and WWC1 on chromosome 5q34. The exon RPKM track displays RPKM calculated for each exon and where the library size normalization factor was considered as the number of reads aligned to exons alone. The fusion read spanning exon 2 in FAM174A and exon 10 in WWC1 was also confirmed with IGV.
Verification of FAM174A-WWC1 in Cancer Cell Line Sample
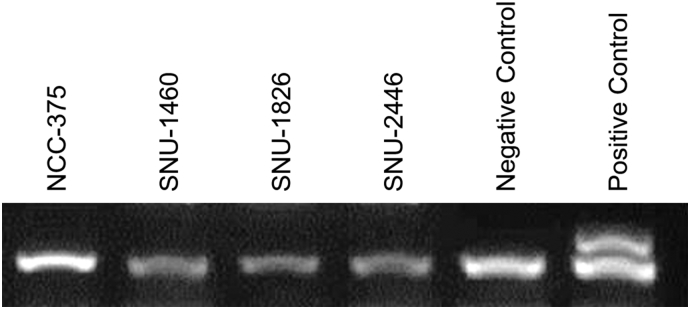
Based on previously described gene fusion isoforms involving the FAM174A and WWC1 genes, the oligonucleotide primers were designed to encompass breakpoint junctions of FAM174A-WWC1 using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primerblast) (FAM174A Forward: GTC AGG ACG GTC AGG ATG AGA, WWC1 Reverse: GCT TCC TCC AGT TCT CTC ACA A). The PCR cycling conditions were: 94°C for 5 minutes; followed by 35 cycles of 94°C for 30 seconds, 55°C for 45 seconds, and 72°C for 1 minutes; and a final extension of 72°C for 10 minutes using Intron PCR Kit (Intron). Using primers complimentary to the sequence in exon 2 of FAM174A and exon 10 of WWC1, reverse transcription followed by polymerase chain reaction (RT-PCR) resulted in a product with a sequence suggesting in-frame junction.
Cloning of FAM174A-WWC1
After confirming the FAM174A-WWC1 fusion gene by RT-PCR, cloning primers were designed to amplify the complete coding sequence of the FAM174A-WWC1 gene fusion. (FAM174A forward: CAC CAT GAA GGC CTC GCA GTG CTG C, WWC1 reverse: GAC GTC ATC TGC AGA GAG AGC TGG GAT). The PCR cycling conditions were: 94°C for 5 minutes, followed by 3 cycles of 94°C for 30 seconds and 72°C for 3 minutes, and a final extension of 72°C for 10 minutes using Q5 Hot Start High-Fidelity Taq (NEB). The amplified FAM174A-WWC1 sequence was cloned into a pENTR/SD/D/TOPO vector according to the manufacturer's recommendations. The fusion junction was verified by Sanger sequencing. The entry vector was then subcloned into a plenti6.2/V5-DEST lentiviral expression vector according to the manufacturer's instructions.
Immunocytochemistry
Cells were seeded on chambered cover glass with a desirable cell confluency. The chambered cover glass was designed to be hydrophilic, with no ECM component treated before seeding. Cells were washed three times with cold DPBS for 15 minutes, 72 hours after cell seeding. The cells were fixed and permeabilized with BD Cytofix/Cytoperm. Cells were washed with the washing solution and treated with DPBS containing 2% FBS for an hour for blocking. After cells were washed with cold DPBS, they were incubated with KIBRA and V5 Tag antibody diluted in 0.05% of PBST for 1.5 hours at room temperature. Subsequently, cells were washed with 0.05% of PBST, followed by incubation with Alexa 488 and Alexa 568 secondary antibodies diluted in 0.05% of PBST for 1 hour at room temperature, and an additional 30 minutes with 1× DAPI and Rhodamine-conjugated Phalloidin diluted in distilled water. The cells were washed three times with DPBS and imaged under an LSM800 Confocal Laser Scanning Microscope equipped with ZEN software to analyze the cells. The intensity of each channel was fixed to compare the target protein expression between the samples. Digital resolution, scan speed, and the average number of images were set to 1024 × 1024, 40 seconds per one channel, and 8, respectively. The images were focused at the very bottom of the fixed cells to investigate the protruding regions of the cell.
2D and 3D Migration Assay
The 2D Gap closure migration assay was performed using a CytoSelect 24-Well Wound Healing Assay kit according to the manufacturer's protocol. Briefly, the NIH3T3 Control and NIH3T3 FAM174A-WWC1 cells were diluted to 2 × 105 cells/ml in RPMI1640 treated with 10% of FBS and 1% penicillin streptomycin. Once the inserts provided in the assay kit were placed into each well of the 24-well tissue culture plate, 500 μl of previously diluted cell solution was dispensed to each side of the insert to obtain a total cell solution of 1 ml per each well. After 24 hours of incubation at 37°C in an atmosphere of 5% CO2 and 95% air, inserts were removed. Immediately, cells were washed with complete media twice. The wound closure was recorded under phase contrast microscope at 24-hour intervals for 5 days. The closed wound area was calculated with ImageJ version 1.8.0. The whole procedures were performed in triplicate, and the average closed wound area was analyzed using GraphPad Prism version 7.00 for Windows.
The 3D migration assay was performed using the AIM 3D cell culture chips according to the manufacturer's protocol. Briefly, cells were diluted to 2 × 105 cells/ml in RPMI1640 without FBS. The cell solution was mixed with a reduced growth factor basement membrane matrix with 1:1 ratio, and 10 μl of the mixture was filled into a gel inlet channel. After incubation for 30 minutes in a 37°C incubator, the media channels were hydrated with 120 μl of RPMI1640 mixed with 10% of FBS and 1% of penicillin streptomycin, followed by further incubation for 24 hours at 37°C in an atmosphere of 5% CO2 and 95% air. Visual inspection and images were obtained with a phase contrast microscope using the 4× objective with 24-hour intervals. Images were analyzed using the ImageJ version 1.8.0. The entire procedures were performed in triplicate, and the average closed wound area was analyzed using GraphPad Prism version 7.00 for Windows.
3D Spheroid Formation Assay and 3D Spheroid Cell Invasion Assay
The 3D spheroid formation assay was performed using Cultrex 96-Well 3D Spheroid BME Cell Invasion Assay kit according to the manufacturer's protocol. Briefly, the reduced growth factor basement membrane matrix was thawed on ice overnight in a 4°C refrigerator. HEK293 cell aggregates were dissociated into a single cell suspension with trypsin. Cells were diluted to 1 × 106 cells/ml in RPMI1640 combined with 10% of FBS and 1% penicillin streptomycin. The cell solution was mixed with the reduced growth factor basement membrane matrix, and 50 μl of the solution was dispensed per well of the 3D culture–containing 96-well spheroid formation plate (provided). The plate was centrifuged at 200×g for 3 minutes at room temperature in a swinging bucket rotor and incubated at 37°C in a tissue culture incubator for 72 hours to promote spheroid formation. After 24 hours, in ice, 50 μl of invasion matrix (provided) was added per well of the 96-well spheroid formation plate. The plate was centrifuged at 300×g at 4°C for 5 minutes in a swinging bucket rotor to eliminate bubbles and position spheroids within the invasion matrix towards the middle of the well. The plate was transferred to a tissue culture incubator set at 37°C for 1 hour to promote gel formation of the invasion matrix. The images were obtained after 72 hours using a phase contrast microscope equipped with the 4× objective.
The 3D spheroid cell invasion assay was conducted by replacing the reduced growth factor basement membrane matrix with 10× Spheroid Formation ECM (provided) to artificially induce the spheroid formation. We also substituted the 96-well spheroid formation plate for 96 hang-in-drop plate to retain cell aggregation. Once the spheroid was formed, it was transferred to a 96 U-bottom plate with 50 μl of invasion matrix (provided). After 1 hour, 100 μl of warm (37°C) cell culture medium was added containing chemoattractant. The spheroid in each well was photographed every 24 hours for 5 days under phase contrast microscope using the 4× objective. Images were analyzed using ImageJ version 1.8.0. All the procedures were performed in triplicate, and the intact form of the spheroid and the rate of disseminated cells were analyzed with circularity estimation using Plugin from GraphPad Prism version 7.00 for Windows.
Results
General Characteristics of the Cell Lines
On in vitro cultivation, three cell lines (NCC-375, SNU-1460, and SNU-1826) were cultured as monolayers of substrate-adherent cells, and one cell line (SNU-2446) formed floating and adherent aggregates. The majority of tumor cells displayed a polygonal shape and had exhibited round-to-oval nuclei with prominent single-to-double nucleoli (Figure 1). Each cell line was passaged at least three times prior to characteristic analysis. Population doubling times ranged from 35 to 82 hours. A summary of clinicopathologic information is provided in Table 1. All cell lines were confirmed to be free of bacterial and mycoplasma contamination (Supplementary Figure 1). Fifteen tetranucleotide repeat loci and the gender-determining marker amelogen were heterogeneously distributed in each cell line, without cross-contamination (Table 2).
Figure 1.
Phase-contrast microscopy of new established early onset colorectal cancer cell lines. On in vitro cultivation, three cell lines (NCC-375, SNU-1460, and SNU-1826) grew as monolayer of substrate-adherent cells, and one cell line (SNU-2446) formed floating and adherent aggregates. The majority of tumor cells displayed a polygonal shape and had round-to-oval nuclei with prominent single-to-double nucleoli.
Supplementary Figure 1.
Four newly established early-onset colorectal cancer cell lines were free of mycoplasma contamination.
Genomic Analyses
Due to the uncharacteristically aggressive clinical course and in the absence of the prototypical FAM174A-WWC1 fusion, the histological diagnosis was in question. To more comprehensively characterize the molecular basis of disease, all the cell lines were subjected to whole exome and transcriptome analysis. The cell lines were primarily derived from pathologically proven tumor tissue. In order to integrate whole exome sequencing data from EOCRC patients with a larger database, the top 100 mutated genes in COAD cohorts under age 35 were extracted from the TCGA-GDC data portal. Intersection between gene lists from TCGA-GDC and from EOCRC patients was sorted, and the mutations that affected the protein structure were screened. The PCLO gene was mutated in all the EOCRC samples and four COAD samples obtained from TCGA-GDC. Mutations in the PCLO gene are associated with depressive disorders [21], [22]. The relationship between PCLO gene and EOCRC requires further analysis. Summarized results are listed in Table 3, Table 4, Table 5, Table 6.
Table 3.
Shared Mutation in NCC-375 and TGCA-GDC Dataset
| GENE | CHR | POS | ID | REF | ALT | AA_CHANGE |
|---|---|---|---|---|---|---|
| MUC4 | 3 | 195,518,112 | rs142781032 | T | TGTCTCCTGCGTAACA | p.T113MLRRRP |
| FNDC1 | 6 | 159,660,779 | rs141435210 | GCCACCACCCGCCGCACGA | G | p.ATTRRTT1471A |
| PCLO | 7 | 82,581,488 | rs10630259 | T | TTCA | p.E2927VK |
| SYNE2 | 14 | 64,560,092 | rs2781377 | G | A | p.W4001* |
Table 4.
Shared Mutation in SNU-1460 and TGCA-GDC Dataset
| GENE | CHR | POS | ID | REF | ALT | AA_CHANGE |
|---|---|---|---|---|---|---|
| FNDC1 | 6 | 159,660,779 | rs141435210 | GCCACCACCCGCCGCACGA | G | p.ATTRRTT1471A |
| THSD7A | 7 | 11,871,469 | A | AGCAGCG | p.35RC | |
| PCLO | 7 | 82,581,488 | rs10630259 | T | TTCA | p.E2927VK |
| NCOR2 | 12 | 124,824,721 | rs61519723 | C | CGCCGCTGCT | p.G1846GAAA |
| NCOR2 | 12 | 124,887,058 | rs35831183 | G | GGCT | p.511S |
| ZFHX3 | 16 | 72,821,593 | rs374416547 | AGCCGCCGCC | A | p.GGG3525Del |
| ZFHX3 | 16 | 72,822,563 | T | TTGCTGCTGC | p.Q3204RSSK | |
| TP53 | 17 | 7,578,370 | C | T | n/a |
Table 5.
Shared Mutation in SNU-1826 and TGCA-GDC Dataset
| GENE | CHR | POS | ID | REF | ALT | AA_CHANGE |
|---|---|---|---|---|---|---|
| MUC4 | 3 | 195,518,118 | rs71180965 | - | TGCGTAACAGTCTCC | p.M111METVTQ |
| FNDC1 | 6 | 159,660,804 | rs3842694 | CCCGCCGCACGACCACCA | - | p.ATTRRTT1471A |
| THSD7A | 7 | 11,871,487 | . | GGCAGCGGCAGC | - | p.25_29del |
| PCLO | 7 | 82,581,493 | rs10694231 | - | ATC | p.D2926DD |
| FRMD4A | 10 | 13,698,938 | . | - | CGC | p.R884RG |
| NCOR2 | 12 | 124,824,739 | rs61519723 | - | GCCGCTGCT | p.S1834SSGS |
| NCOR2 | 12 | 124,887,095 | . | - | CTG | p.Q499QQ |
| ZFHX3 | 16 | 72,822,586 | . | - | CTG | p.Q2283QQ |
| ZFHX3 | 16 | 72,831,382 | . | TTGTTG | - | p.818_819del |
| RTTN | 18 | 67,863,854 | rs58913700 | - | TCC | p.G242GG |
Table 6.
Shared Mutation in SNU-2446 and TGCA-GDC Dataset
| GENE | CHR | POS | ID | REF | ALT | AA_CHANGE |
|---|---|---|---|---|---|---|
| TTN | 2 | 179,472,617 | . | C | A | p.E17633* |
| PCLO | 7 | 82,581,488 | rs10694231 | T | TTCA | p.E2927DE |
| WDFY4 | 10 | 50,165,232 | . | C | A | p.S2679* |
Verification of FAM174A-WWC1 in Patient Sample
The novel fusion gene, FAM174A-WWC1, was detected in NCC-375 cell line using three different fusion gene detection algorithms. The reads spanning the breakpoint of the fusion gene were visualized with an IGV program, which revealed a 67,892,563-bp deletion in chromosome 5 (Figure 2A). Oligonucleotide primers were designed to encompass breakpoint junctions of FAM174A and WWC1. Using primers complimentary to the sequence in exon 2 of FAM174A and exon 10 of WWC1, RT-PCR of the tumor derived cell line resulted in a product whose sequence verified the in-frame junction (Figure 2B). Amplicons were detected in other cell lines and confirmed as nonspecific PCR products by Sanger sequencing. The amplicon containing the target fusion gene was verified with direct Sanger sequencing which showed that the exon 2 of FAM174A was connected to the exon 10 of WWC1 (Figure 2C).
Figure 2.
Verification of FAM174A-WWC1 in Patient Sample. (A) The novel fusion gene, FAM174A-WWC1, was detected in NCC-375 cell line with three different fusion gene detection algorisms. The reads spanning the breakpoint of the fusion gene were visualized with IGV program. (B) Oligonucleotide primers were designed to encompass breakpoint junctions of FAM174A and WWC1. (C) The amplicon containing the target fusion gene was verified with direct Sanger sequencing which represented that the exon 2 of FAM174A is connected to the exon 10 of WWC1.
Cloning and In Vitro Assessment of FAM174A-WWC1
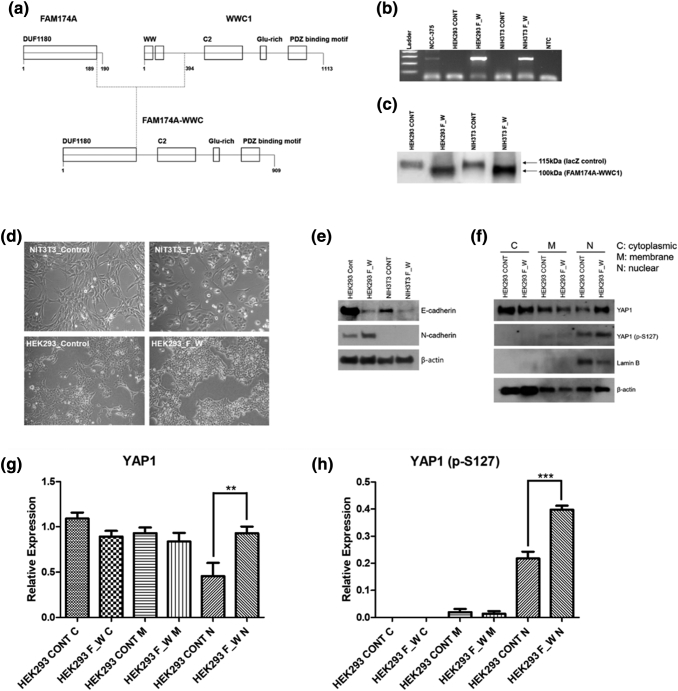
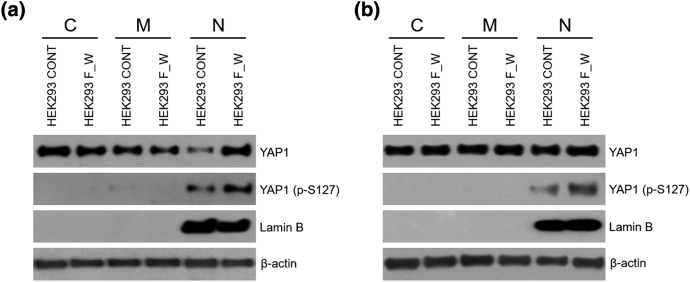
We employed RT-PCR to clone the entire coding sequence of the FAM174A-WWC1 fusion transcript using RNA isolated from the NCC-375 cell line. In silico analysis of the FAM174A-WWC1 coding sequence (http://web.expasy.org/compute_pi/) predicted the expression of a ∼123-kDa protein. Examination of the WWC1-encoding sequence revealed that the truncation of WWC1 resulted in the loss of its WW domains and preservation of its C2 and PDZ binding motif. The truncation of the FAM174A portion in FAM174A-WWC1 resulted in conserved exons 1-2 encoding the DUF1180 domain at which the function is unknown (Figure 3A). Murine fibroblast NIH3T3 cells and HEK293 cells were transduced with packaged lentivirus encoding FAM174A-WWC1. The expression of the fusion transcript was verified by RT-PCR with primers spanning the fusion breakpoint (Figure 3B). The expression of fusion protein was assessed by Western blotting using a V5-tag antibody, which demonstrated a protein at the predicted molecular weight of 100 kDa (Figure 3C). The induction of FAM174A-WWC1 caused morphological alternation in both HEK293 and NIH3T3 cells. The morphology of both cells was changed from stellate to round and polygonal (Figure 3D). Induction of FAM174A-WWC1 decreased the protein expression of E-cadherin and augmented N-cadherin in HEK293 cell lines as well. No basal N-cadherin was expressed in NIH3T3 cell lines, and only decreased E-cadherin was observed (Figure 3E). Subcellular fractional protein extraction indicated that the fusion of WWC1 with DUF1180 domain increased the levels of both total YAP1 and phosphorylated YAP1 in the nucleus (Figure 3F). The augmented levels of total YAP1 (Figure 3G) and phosphorylated YAP1 (Figure 3H) in the cytosolic and nuclear fractions were quantitatively analyzed with statistical significance.
Figure 3.
Cloning and in vitro assessment of FAM174A-WWC. (A) Mimetic diagram showing the structure of FAM174A-WWC1. The expression of the fusion transcript was verified by RT-PCR (B) with primers spanning the fusion breakpoint and by Western blotting (C) using an V5-tag antibody, demonstrating a protein at the predicted molecular weight of 100 kDa. (D) The induction of FAM174A-WWC1 caused morphologic alternation in both HEK293 and NIH3T3 cells. (E) Induced FAM174A-WWC1 decreased protein expression of E-cadherin and augmented N-cadherin in HEK293 cell lines as well. (F) Subcellular fractional protein extraction indicated that WWC1 fused with DUF1180 domain increased both total YAP1 and phosphorylated YAP1 level in nucleus. (G) Quantitative analysis of expression of YAP1 in cytoplasm, membrane, and nuclear. (H) Quantitative analysis of expression of YAP1 (p-S127) in cytoplasm, membrane, and nuclear.
Oncogenic Capacity of FAM174A-WWC1
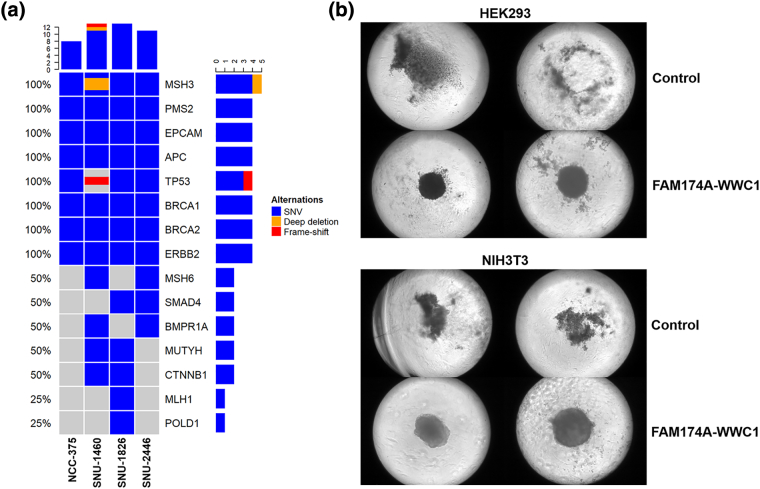
The SNV profiling of EOCRC patients implicated oncogenic potential of the fusion gene. The tumor expressing FAM174A-WWC1 carried the lowest number of oncogene mutations (Figure 4A). To further elucidate the tumorigenic capacity of FAM174A-WWC1, cells were grown under conditions of anoikis, as tumorspheres, to assay their tumor-initiating capability and stem-like properties. FAM174A-WWC1 expression significantly increased the tumorsphere propagation (Figure 4B).
Figure 4.
Oncogenic capacity of FAM174A-WWC1. (A) SNV profiling of the enrolled EOCRC patients. The tumor expressing FAM174A-WWC1 had lowest number of oncogene mutations. (B) 3D spheroid forming assay indicated FAM174A-WWC1 expression significantly increased tumorsphere propagation.
Metastatic Potential of FAM174A-WWC1
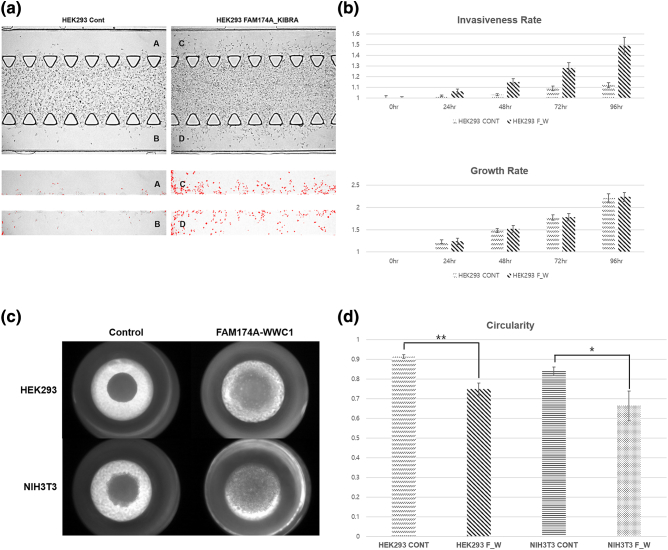
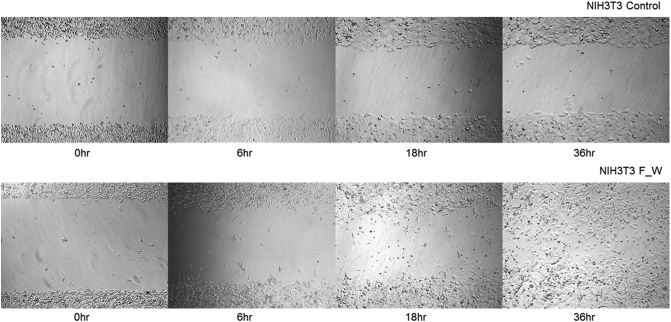
To determine the role of FAM174A-WWC1 in cell migration and invasion, cells were seeded into a narrow channel with a basement membrane matrix containing reduced growth factors, which mimics extravasation of tumor cells. Compared with controls, cells expressing FAM174A-WWC1 significantly increased the number of invaded cells (Figure 5, A and B). Increased ability to invade into a membrane matrix was re-confirmed with three-dimensional spheroid invasion assay. Cell were grown as 3D cyst-like forms and seeded with invasion matrix. Consistent with previous findings, the FAM174A-WWC1 expression promoted dissemination of cells into the surrounding matrigels (Figure 5, C and D). Accordingly, FAM174A-WWC1 expression also enhanced cell migration in two dimensions (Supplementary Figure 2).
Figure 5.
Metastatic potential of FAM174A-WWC1. (A, B) 3D chip invasion assay indicated that, compared with controls, cells with FAM174A-WWC1 significantly increased the number of invaded cells. Cell growth rate was rarely affected. (C, D) 3D spheroid cell invasion assay indicated that FAM174A-WWC1 expression promoted dissemination of cells into the surrounding invasion matrix.
Supplementary Figure 2.
Wound healing assay indicated that NIH3T3 cells with FAM174A-WWC1 fusion gene acquired increased migration ability.
Metastatic Potential of FAM174A-WWC1 in Focal Adhesion and ECM Receptor Pathway
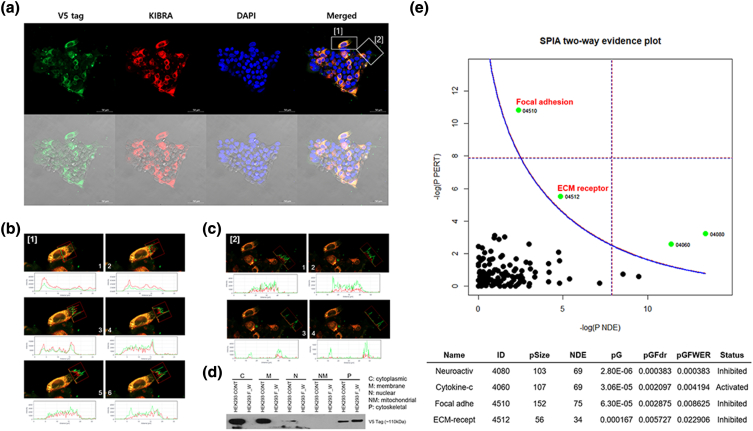
The intercellular localization of WWC1 and FAM174A-WWC1 was analyzed with immunocytochemistry. WWC1 and FAM174A-WWC1 were mostly colocalized in the cytoplasmic and cytoskeletal region. Nevertheless, the expression of fusion protein was mostly centered in the cytoskeletal region, which may indicate the role of fusion protein in focal adhesion and ECM binding (Figure 6A). Signal intensity analysis revealed the expression of both WWC1 and FAM174A-WWC1 in lamellipodia, with prominent FAM174A-WWC1 expression at the edge (Figure 6, B and C). The subcellular compartment of FAM174A-WWC1 was further investigated with fractional protein extraction. As in preceding, the fusion protein was mostly detected in the cytoskeletal fraction (Figure 6D). To identify genes and pathways affected by the fusion protein, RNA sequencing was performed. Consistent with the prior findings, focal adhesion and ECM receptor pathways were significantly inhibited by FAM174A-WWC1 fusion protein. Correctively, these findings suggest the potential role of fusion protein in movement (Figure 6E).
Figure 6.
Metastatic potential of FAM174A-WWC1 is involved in focal adhesion and ECM receptor pathway aberration. (A) WWC1 and FAM174A-WWC1 were mostly colocalized in cytoplasmic and cytoskeletal region. Nevertheless, the expression of fusion protein was mostly centered on cytoskeletal region. (B, C) Signal intensity analysis discovered that both WWC1 and FAM174A-WWC1 were expressed in lamellipodia, yet FAM174A-WWC1 expression is prominent at the edge. (D) Fractional protein extraction showed that the fusion protein was mostly detected in cytoskeletal fraction. (E) RNA sequencing indicated that focal adhesion and ECM receptor pathways are significantly inhibited when FAM174A-WWC1 fusion protein was induced.
Discussion
We present four cases of EOCRC. The cases did not harbor any of the fusion oncogenes reported previously in EOCRC; however, it was found to express a novel fusion gene involving FAM174A and WWC1 in one of the patients enrolled in this study. Sequence verification and cloning of the FAM174A-WWC1 fusion gene facilitated for functional characterization of the fusion protein. We have used HEK293, human embryonic kidney cells, and NIH3T3, mouse embryonic fibroblasts, which are normal cell lines to pinpoint the tumorigenic effect of the FAM174A-WWC1. We demonstrated that the expression of FAM174A-WWC1 in HEK293 and NIH3T3 cells was sufficient for cellular transformation.
Various fusion genes including BRAF, NTRK3, RAS, and RET have been reported in CRC [23], [24]; however, no study specifically elucidated gene fusions in EOCRC. WWC1 is associated with several other fusion genes. For instance, the interchromosomal in-frame fusion gene WWC1-ADRBK2 was found in BRCA-mutated breast cancer [25]. Yuri Choi et al. idenfitied an intrachromosomal in-frame fusion gene, BOD1-WWC1, in CRC patient tissue [26]. Multiple fusions involving the WWC1 gene may be triggered by the instability of chromosome 5q arm. Chromosome 5q deletions have been associated with biological and prognostic significance of myeloid malignancies [27] and triple-negative breast cancers [28]. The FAM174A-WWC1 gene fusion resulted from a 67,892,563-bp deletion in the 5q arm, which may also reflect the loss of chromosome 5q arm in EOCRC case (Figure 2A). Due to this recurrent instability, fusion genes may carry various breakpoints. Thus, we constructed five different primer sets targeting different exons in FAM174A and WWC1 based on the identified break junction, and no variable breakpoint existed other than exon 2 of FAM174A and exon 10 of WWC1 (data not shown).
Analysis of the FAM174A-WWC1 sequence revealed that the WWC1 portion of FAM174A-WWC1 maintains the entire C2 domain and PDZ bind motif of WWC1, and lacks the WW domain (Figure 3A). WWC1 has been suggested as a key regulator of the hippo pathway, regulating organ size, cell contact inhibition, as well as tissue regeneration and tumorigenesis. WWC1 activates both large tumor suppressor (LATS1/2) kinases and Yes-associated protein (YAP) via phosphorylation on the hydrophobic motif [29], [30]. Our results indicated that FAM174A-WWC1 not only lowered the membrane level of YAP1 but also significantly increased its level in the nucleus (Figure 3F), which demonstrated that the replacement of WW domains with DUF1180 domain is associated with nuclear localization of YAP1. Phosphorylation at Ser127 by LATS kinases promotes YAP translocation from the nucleus to the cytoplasm, where it is sequestered through association with 14-3-3 proteins [31]. The nucleic p-S127 YAP1 level was higher in HEK293 cells containing FAM174A-WWC1 (Figure 3F), which may imply that the fusion protein hinders the cytoplasmic localization of phosphorylated YAP1.
The SNV profiling of EOCRC patients suggested the oncogenic potential of the fusion gene. The tumor expressing FAM174A-WWC1 carried the lowest number of oncogene mutations (Figure 4A), which was concurrent with a previous study suggesting tumors harboring fusion genes tended to exhibit fewer oncogene mutations [23]. Nevertheless, cancer development in patients with FAM174A-WWC1 may be influenced by oncogene and tumor suppressor gene mutations involving APC, in addition to the fusion gene. To further determine whether FAM174A-WWC1 was involved in tumorigenic capacity, cells were grown under conditions of anoikis as tumorspheres to examine their tumor-initiating capability and stem-like properties since sphere-forming efficiency indicates tumorigenic potential [32]. FAM174A-WWC1 expression significantly increased the tumorsphere propagation (Figure 4B). It was reported that WW1/2 domains of WWC1 mediated the inhibition of tumorsphere formation [28]. The FAM174A-WWC1 not only lacks WW1/2 domains of WWC1 but also contains DUF1180 domain of FAM174A replacing WW domains, which may demonstrate that the DUF1180 domain is associated with accelerated tumor progression.
The patient harboring FAM174A-WWC1 developed multiple metastases to lungs and liver after surgery (Table 1). Two dimensional and 3D invasion assays were performed for clinicopathological analysis. Without exception, cells expressing FAM174A-WWC1 showed increased invasiveness and dissemination compared with controls (Figure 5 and Supplementary Figure 2). These data support a role for FAM174A-WWC1 in promoting metastatic dissemination, which was inconsistent with the role of naive WWC as a metastatic suppressor [28]. This finding may imply again that the replacement of WW domains with DUF1180 domain reverses the primary role of WWC1. Collectively, these data indicate that the expression of FAM174A-WWC1 augments the tumorigenic and metastatic capacity and, therefore, that its expression can confer significant advantages to EOCRC.
In conclusion, we identified and analyzed one novel fusion gene, FAM174A-WWC1, from a patient with EOCRC based on comprehensive and integrated clinicopathological analysis, NGS data, and in vitro assessments. We demonstrated the oncogenic and metastatic capacity of FAM174A-WWC1 through in vitro analysis. This fusion gene may represent a robust molecular and therapeutic target in EOCRC. Although cases of EOCRC carry an overall unfavorable prognosis, identification of young patients diagnosed with EOCRC may represent a population of high-risk cases, with tumors driven by novel genes such as FAM174A-WWC1. It would be interesting to determine the prevalence of FAM174A-WWC1 in archival samples of EOCRC and to correlate FAM174A-WWC1 status with clinical outcome.
The following are the supplementary data related to this article.
Supplementary Figure 3.
The nucleus level of YAP1 and phosphorylated YAP1 was increased in HEK293 cells when transducted with FAM174A-WWC1 fusion gene. The whole processes were repeated three times.
Conflicting Interests
The authors declare that there is no conflict of interest.
Funding
This research was supported by the Korean Cell Line Research Foundation, the Priority Research Centers Program (2009-0093820) and the Basic Science Research Program (2012R1A1A3010709) through the National Research Foundation (NRF) of Korea funded by the MSIP. The first author received a scholarship from the BK21-plus education program provided by the NRF.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Guarantor
None.
Acknowledgement
None.
Contributor Information
Seung-Yong Jeong, Email: syjeong@snu.ac.kr.
Ja-Lok Ku, Email: kujalok@snu.ac.kr.
References
- 1.Siegel RL, Miller KD. Jemal A (2016). Cancer statistics. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Jung KW, Won YJ, Kong HJ, Lee ES. Cancer statistics in Korea: incidence, mortality, survival. and prevalence in 2015 Cancer research and treatment : official journal of Korean Cancer Association. 2018;50:303–316. doi: 10.4143/crt.2018.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dozois EJ, Boardman LA, Suwanthanma W, Limburg PJ, Cima RR, Bakken JL, Vierkant RA, Aakre JA, Larson DW. Young-onset colorectal cancer in patients with no known genetic predisposition: can we increase early recognition and improve outcome? Medicine. 2008;87:259–263. doi: 10.1097/MD.0b013e3181881354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.You YN, Xing Y, Feig BW, Chang GJ, Cormier JN. Young-onset colorectal cancer: is it time to pay attention? Arch Intern Med. 2012;172:287–289. doi: 10.1001/archinternmed.2011.602. [DOI] [PubMed] [Google Scholar]
- 5.Ahnen DJ, Wade SW, Jones WF, Sifri R, Mendoza Silveiras J, Greenamyer J, Guiffre S, Axilbund J, Spiegel A, You YN. The increasing incidence of young-onset colorectal cancer: a call to action. Mayo Clin Proc. 2014;89:216–224. doi: 10.1016/j.mayocp.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Pan SY, DesMeules M. Energy intake, physical activity, energy balance, and cancer: epidemiologic evidence. Methods in molecular biology (Clifton, NJ) 2009;472:191–215. doi: 10.1007/978-1-60327-492-0_8. [DOI] [PubMed] [Google Scholar]
- 7.Yuhara H, Steinmaus C, Cohen SE, Corley DA, Tei Y, Buffler PA. Is diabetes mellitus an independent risk factor for colon cancer and rectal cancer? Am J Gastroenterol. 2011;106:1911–1921. doi: 10.1038/ajg.2011.301. quiz 1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavestro GM, Mannucci A, Zuppardo RA, Di Leo M, Stoffel E, Tonon G. Early onset sporadic colorectal cancer: worrisome trends and oncogenic features Digestive and liver disease : official journal of the Italian Society of Gastroenterology and the Italian Association for the. Study of the Liver. 2018;50:521–532. doi: 10.1016/j.dld.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Harewood L, Fraser P. The impact of chromosomal rearrangements on regulation of gene expression. Hum Mol Genet. 2014;23:R76–R82. doi: 10.1093/hmg/ddu278. [DOI] [PubMed] [Google Scholar]
- 10.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 11.Dai X, Theobard R, Cheng H, Xing M, Zhang J. Fusion genes: a promising tool combating against cancer. Biochimica et biophysica acta Reviews on cancer. 2018;1869:149–160. doi: 10.1016/j.bbcan.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 13.Rejlova K, Musilova A, Kramarzova KS, Zaliova M, Fiser K, Alberich-Jorda M, Trka J, Starkova J. Low HOX gene expression in PML-RARalpha-positive leukemia results from suppressed histone demethylation. Epigenetics. 2018;13:73–84. doi: 10.1080/15592294.2017.1413517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H. Identification of the transforming EML4-ALK fusion gene in non–small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 15.Rolfo C, Raez L. New targets bring hope in squamous cell lung cancer: neurotrophic tyrosine kinase gene fusions. Laboratory investigation; a journal of technical methods and pathology. 2017;97:1268–1270. doi: 10.1038/labinvest.2017.91. [DOI] [PubMed] [Google Scholar]
- 16.Ku JL, Shin YK, Kim DW, Kim KH, Choi JS, Hong SH, Jeon YK, Kim SH, Kim HS, Park JH. Establishment and characterization of 13 human colorectal carcinoma cell lines: mutations of genes and expressions of drug-sensitivity genes and cancer stem cell markers. Carcinogenesis. 2010;31:1003–1009. doi: 10.1093/carcin/bgq043. [DOI] [PubMed] [Google Scholar]
- 17.Kim SC, Hong CW, Jang SG, Kim YA, Yoo BC, Shin YK, Jeong SY, Ku JL, Park JG. Establishment and characterization of paired primary and peritoneal seeding human colorectal cancer cell lines: identification of genes that mediate metastatic potential. Translational oncology. 2018;11:1232–1243. doi: 10.1016/j.tranon.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng PC, Henikoff S. 2003. SIFT: predicting amino acid changes that affect protein function Nucleic acids research31; pp. 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adzhubei I, Jordan DM, Sunyaev SR. 2013. Predicting functional effect of human missense mutations using PolyPhen-2 Current protocols in human geneticsChapter 7, Unit7; p. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shamsani J, Kazakoff SH, Armean IM, McLaren W, Parsons MT, Thompson BA, O'Mara TA, Hunt SE, Waddell N, Spurdle AB. Oxford; England: 2018. A plugin for the Ensembl Variant Effect Predictor that uses MaxEntScan to predict variant spliceogenicity Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mbarek H, Milaneschi Y, Hottenga JJ, Ligthart L, de Geus EJC, Ehli EA, Willemsen G, Davies GE, Smit JH, Boomsma DI. Genome-wide significance for PCLO as a gene for major depressive disorder Twin research and human genetics : the official journal of the. International Society for Twin Studies. 2017;20:267–270. doi: 10.1017/thg.2017.30. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed MY, Chioza BA, Rajab A, Schmitz-Abe K, Al-Khayat A, Al-Turki S, Baple EL, Patton MA, Al-Memar AY, Hurles ME. Loss of PCLO function underlies pontocerebellar hypoplasia type III. Neurology. 2015;84:1745–1750. doi: 10.1212/WNL.0000000000001523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kloosterman WP, Coebergh van den Braak RRJ, Pieterse M, van Roosmalen MJ, Sieuwerts AM, Stangl C, Brunekreef R, Lalmahomed ZS, Ooft S, van Galen A. A systematic analysis of oncogenic gene fusions in primary colon cancer. Cancer Res. 2017;77:3814–3822. doi: 10.1158/0008-5472.CAN-16-3563. [DOI] [PubMed] [Google Scholar]
- 24.(2012). Comprehensive molecular characterization of human colon and rectal cancer Nature 487, 330–337. [DOI] [PMC free article] [PubMed]
- 25.Ha KC, Lalonde E, Li L, Cavallone L, Natrajan R, Lambros MB, Mitsopoulos C, Hakas J, Kozarewa I, Fenwick K. Identification of gene fusion transcripts by transcriptome sequencing in BRCA1-mutated breast cancers and cell lines. BMC Med Genomics. 2011;4:75. doi: 10.1186/1755-8794-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi Y, Kwon CH, Lee SJ, Park J, Shin JY, Park DY. Integrative analysis of oncogenic fusion genes and their functional impact in colorectal cancer. Br J Cancer. 2018;119:230–240. doi: 10.1038/s41416-018-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giagounidis AA, Germing U, Aul C. Biological and prognostic significance of chromosome 5q deletions in myeloid malignancies Clinical cancer research : an official journal of the American Association for. Cancer Res. 2006;12:5–10. doi: 10.1158/1078-0432.CCR-05-1437. [DOI] [PubMed] [Google Scholar]
- 28.Knight JF, Sung VYC, Kuzmin E, Couzens AL, de Verteuil DA, Ratcliffe CDH, Coelho PP, Johnson RM, Samavarchi-Tehrani P, Gruosso T. KIBRA (WWC1) is a metastasis suppressor gene affected by chromosome 5q loss in triple-negative breast cancer. Cell Rep. 2018;22:3191–3205. doi: 10.1016/j.celrep.2018.02.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu FX, Zhao B, Guan KL. Vol. 163. 2015. Hippo pathway in organ size control, tissue homeostasis, and cancer Cell; pp. 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park HW, Guan KL. Regulation of the Hippo pathway and implications for anticancer drug development. Trends Pharmacol Sci. 2013;34:581–589. doi: 10.1016/j.tips.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia (New York, NY) 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]