Abstract
Targeting subunits of BAF/PBAF chromatin remodeling complexes has been proposed as an approach to exploit cancer vulnerabilities. Here we develop PROTAC degraders of the BAF ATPase subunits SMARCA2 and SMARCA4 using a bromodomain ligand and recruitment of the E3 ubiquitin ligase VHL. High-resolution ternary complex crystal structures and biophysical investigation guided rational and efficient optimization towards ACBI1, a potent and cooperative degrader of SMARCA2, SMARCA4 and PBRM1. ACBI1 induced antiproliferative effects and cell death caused by SMARCA2 depletion in SMARCA4 mutant cancer cells, and in acute myeloid leukemia cells dependent on SMARCA4 ATPase activity. These findings exemplify a successful biophysics- and structure-based PROTAC design approach to degrade high profile drug targets and pave the way towards new therapeutics for the treatment of tumors sensitive to the loss of BAF complex ATPases.
Introduction
Proteolysis targeting chimeras (PROTACs) are an emerging new class of drug molecules wherein a target-binding ligand linked covalently to an E3 ligase-binding ligand forms a target-PROTAC-ligase ternary complex, directing the ubiquitin proteasome system to degrade the target protein1–3. In contrast to classical small molecule drugs, PROTAC-driven degradation functions in a sub-stoichiometric nature thus requiring lower systemic exposures to achieve efficacy4, 5. PROTACs have been shown to display higher degrees of selectivity for protein degradation than the target ligand itself due to complementarity differences in the protein-protein-interaction interfaces of the formed ternary complexes6–9. In addition, PROTACs promise to expand the druggable proteome as degradation is not limited to the protein domain functionally responsible for the disease. In the case of challenging multidomain proteins, traditionally viewed as undruggable targets, the most ligandable domain can be targeted for degradation independent of its functionality or vulnerability to small molecule blockade10, 11.
The ATP-dependent activities of the BAF (SWI/SNF) chromatin remodeling complexes affect the positioning of nucleosomes on DNA and thereby many cellular processes related to chromatin structure, including transcription, DNA repair and decatenation of chromosomes during mitosis12, 13. The BAF complex is mutated in approximately 20% of human cancers and contains one of two mutually exclusive ATPases, SMARCA2 or SMARCA413–16. While SMARCA4 acts as a tumor suppressor in solid tumors, the role of SMARCA4 in acute myeloid leukemia (AML) is markedly different, such that it is required to maintain the oncogenic transcription program and drive proliferation17. Selective suppression of SMARCA2 activity has been proposed as a therapeutic concept for SMARCA4-mutated cancers18–20. A recent disclosure by Papillon et al. (published while this manuscript was under review) demonstrated that dual allosteric inhibitors of SMARCA2/4 ATPase activity show anti-proliferative effects in a SMARCA4 mutant xenograft model21.
Small molecule ligands targeting the bromodomains of SMARCA2 and SMARCA4 (SMARCA2BD/SMARCA4BD) have also been reported18, 22, 23. Although cells lacking SMARCA4 activity are vulnerable to the loss of SMARCA218, SMARCA2/4BD inhibitors have failed to phenocopy these anti-proliferative effects. Indeed, re-expression of SMARCA2 variants in cells, where the endogenous protein had been suppressed, showed that an intact bromodomain is not required to maintain proliferation24. SMARCA2/4BD inhibitors are thus precluded from use for the treatment of SMARCA4 mutant cancers but could provide attractive ligands for PROTAC conjugation. Small molecules binding to other bromodomains have been successfully converted into PROTACs by conjugating them with structures capable of binding to the E3 ligases VHL or cereblon5, 6, 10, 11, 25–27. In the case of the BET protein BRD4, this has been achieved through the use of structure-based drug design via ternary complex crystal structures7. We therefore reasoned that a PROTAC targeting the non-functional bromodomain of SMARCA2/4 should offer an opportunity to exploit the vulnerability of SMARCA2- or SMARCA4-dependent cancer cells for therapeutic purposes.
Here we show how structure-based PROTAC design enabled the identification of a potent degrader of SMARCA2 and SMARCA4 with anticancer activity. Biophysical analysis identified a prototype that forms cooperative ternary complexes, electing it as a lead for further investigation. Co-crystallization of ternary complexes guided rational design to yield an optimized chemical probe, ACBI1, in only two design steps. With this compound we demonstrate how depletion of the ATPases can lead to a reduction in other BAF/PBAF subunits within these stable complexes due to dissociation following SMARCA2/4 depletion. Furthermore, rapid and profound PROTAC-induced knockdown of SMARCA2/4 led to pronounced anti-proliferative effects and apoptosis across multiple cancer cell lines, substantiating the potential of targeted degradation of BAF complex ATPases as a viable cancer therapeutic strategy.
Results
Identification of a partial SMARCA2/4 degrader
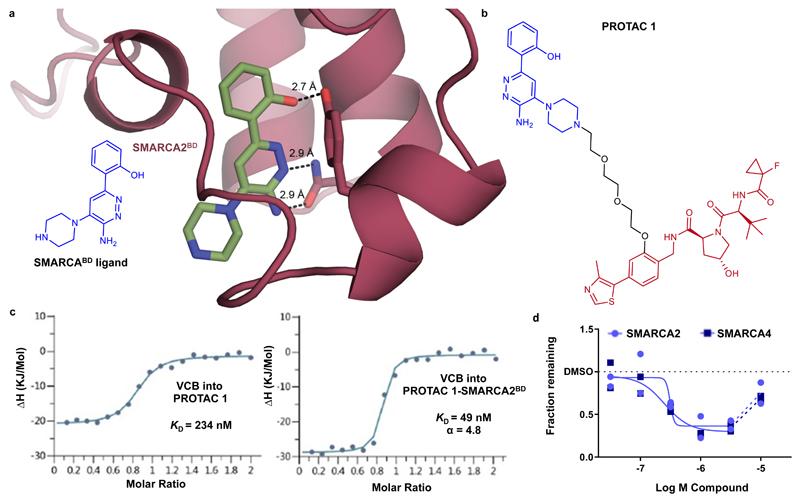
In order to identify a targeted SMARCA degrader we first looked to select a suitable binding ligand. Great effort has been applied to discover SMARCA bromodomain inhibitors, so we turned our attention to a series of 2-(6-aminopyridazin-3-yl)phenols reported to possess robust SMARCA2/4 binding22, 28, 29. To elucidate the binding mode of these ligands and guide PROTAC conjugation design, we solved the co-crystal structure of a piperazine substituted SMARCABD ligand (1) with SMARCA2BD (Fig. 1a, Supplementary Table 1, PDB ID: 6HAZ). The observed binding mode is characterized by an interaction deep in the acetyl-lysine binding site between the phenol of the ligand and Y1421 as well as a donor/acceptor interaction between the aminopyridazine core and N1464. The piperazine ring of the ligand protrudes out into the solvent, suggestive of a suitable vector for PROTAC linkage. To recruit the von Hippel–Lindau (VHL) E3 ubiquitin ligase, we chose a VHL ligand with high affinity and known binding mode, recently published by our group30. Previous data also suggested a phenolic linkage point would be tolerated by VHL and simultaneously permit incorporation of the affinity-improving fluorocyclopropyl amide group30–32. By combining these observations and utilizing polyethylene glycol-based linkers, a small set of PROTACs were designed and subsequently evaluated. Effective PROTACs have been shown to induce stabilizing protein-protein interactions of the E3-ligase with the target protein, leading to cooperative formation of a ternary complex in which the affinity of the PROTAC to both its binding partners is higher than when binding to each protein individually7, 32, 33. In isothermal titration calorimetry (ITC) assays, binding of PROTAC 1 (2) with the VHL-ElonginC-ElonginB (VCB) complex displays 4.8 fold greater affinity when already in complex with SMARCA2BD than it does alone, indicating that ternary complexes formed by this PROTAC are positively cooperative (Fig. 1b and 1c, Supplementary Fig. 1a and 1b). We also observed that PROTAC 1 was able to induce partial degradation of SMARCA2 and SMARCA4 in MV-4-11 cells (Fig. 1d). Interestingly, PROTAC 1 showed a >10 fold weaker binding affinity for the SMARCA bromodomain when compared to the SMARCABD ligand, as measured by SPR (Supplementary Table 2). This drop-off in binary affinity, perhaps due to an inhibitory conformation of the PROTAC for binding to the ATPase bromodomain, would be classically viewed as a step in the wrong direction. However, the cooperative nature, and so enhanced binding affinity of PROTAC 1 at forming the key ternary complex species, encouraged us to investigate this compound further.
Figure 1. Rational design and evaluation of a partial degrader of SMARCA2 and SMARCA4, PROTAC 1.
a) 2D chemical structure of a SMARCABD ligand and crystal structure of this ligand in complex with SMARCA2BD. The piperazine ring was selected as an exit vector for PROTAC linkage as it is directed into solvent away from the binding site. b) 2D chemical structure of PROTAC 1 c) Inverse ITC titrations of VCB into PROTAC 1 (left) and VCB into the preformed PROTAC 1–SMARCA2BD complex (right), n = 2. PROTAC 1 binds VCB with higher affinity when in complex with SMARCA2BD and is therefore cooperative, α = 4.8. d) Degradation of SMARCA2 and SMARCA4 in MV-4-11 cells following treatment with PROTAC 1, analysed via capillary electrophoresis (see online methods). For SMARCA2 and SMARCA4, maximal degradation is ~65 % and ~70%, and DC50 is 300 nM and 250 nM, respectively. Data represent means from two biologically independent experiments, ± S.E.M.
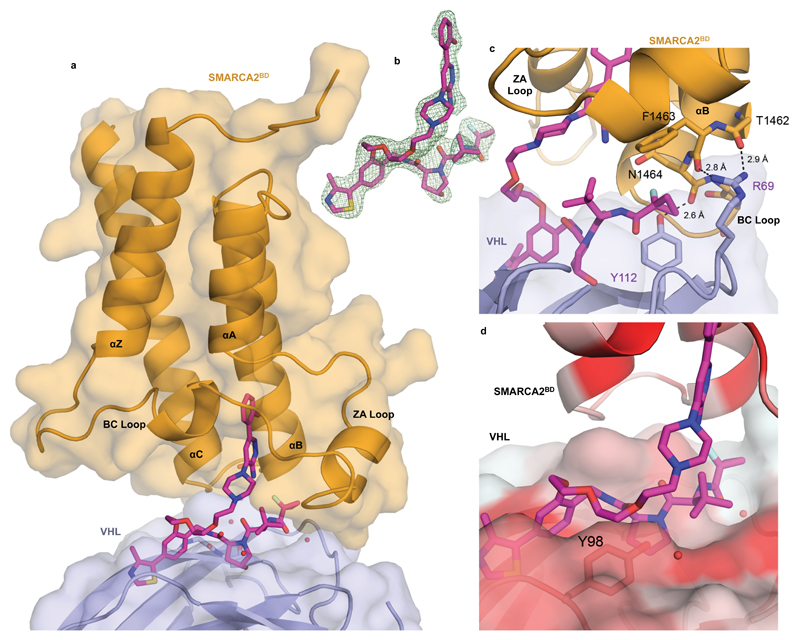
To rationalize the cooperative recognition and potentially enable structure-based PROTAC design we solved a high resolution co-crystal structure of the ternary complex of VCB: PROTAC 1: SMARCA2BD (2.25 Å, PDB ID: 6HAY, Fig. 2a, 2b, Supplementary Table 1 and Supplementary Fig. 1c and 2). The crystal structure contains two copies of the ternary complex in the asymmetric unit, with minor differences between them. The binding mode of PROTAC 1 with both SMARCA2BD and VHL in the ternary complex recapitulates the key elements of the respective binary structures for each isolated binder, with a minor rearrangement of the BC loop of SMARCA2BD when compared to the co-crystal structure of the SMARCABD ligand, to accommodate closer packing against VHL (Supplementary Fig. 1c). Of particular note, de novo protein-protein interactions were observed in the region of the BC loop of SMARCA2BD and the periphery of the VHL binding site (Fig. 2a). The interface appears to be stabilized by an electrostatic interaction between R69 of VHL and the carbonyl groups of residues T1462 and F1463 within the electronegative C-cap of the B helix of SMARCA2BD, as well as a hydrogen bond between Y112 of VHL and N1464 of SMARCA2BD (Fig. 2c). Close packing of these residues around the fluorocyclopropyl amide group is a key feature of this ternary complex crystal structure. Collapse of the flexible polyethylene glycol-based linker onto a lipophilic face created in part by Y98 of the VHL protein is also visible in the structure (Fig. 2d). In a Caco-2 permeability assay, we observed a very low A-B rate of 1.1 x 10-7 cm/s, a B-A rate of 20.7 x 10-6 cm/s, and therefore an efflux permeability ratio of 190 for PROTAC 1, indicating poor passive permeability (A-B) and a high degree of transporter-mediated efflux (see online methods). Taken together, our data suggest that PROTAC 1 forms moderately cooperative ternary complexes allowing for subsequent degradation and is likely limited by cellular permeability.
Figure 2. Ternary co-crystal structure of SMARCA2BD: PROTAC 1: VCB.
a) Overall co-crystal structure of VCB: PROTAC 1: SMARCA2BD in ribbon representation with bound PROTAC shown in stick with magenta carbons. b) Fo-Fc omit map (green meshes) of PROTAC 1 prior to ligand modelling (complex 1, interface of chains A/B) contoured at 3.0 σ with a carve radius of 2.2 Å. c) Key electrostatic protein-protein interactions between R69 of VHL and C-terminal backbone carbonyl groups of the B helix of SMARCA2BD d) Surface representation colored using normalized consensus hydrophobicity scale, red = hydrophobic, white = hydrophilic. Linker of PROTAC 1 seen to be collapsing on to a lipophilic face formed in part by Y98 of VHL.
Structure-based design of a SMARCA2/4 degrading PROTAC
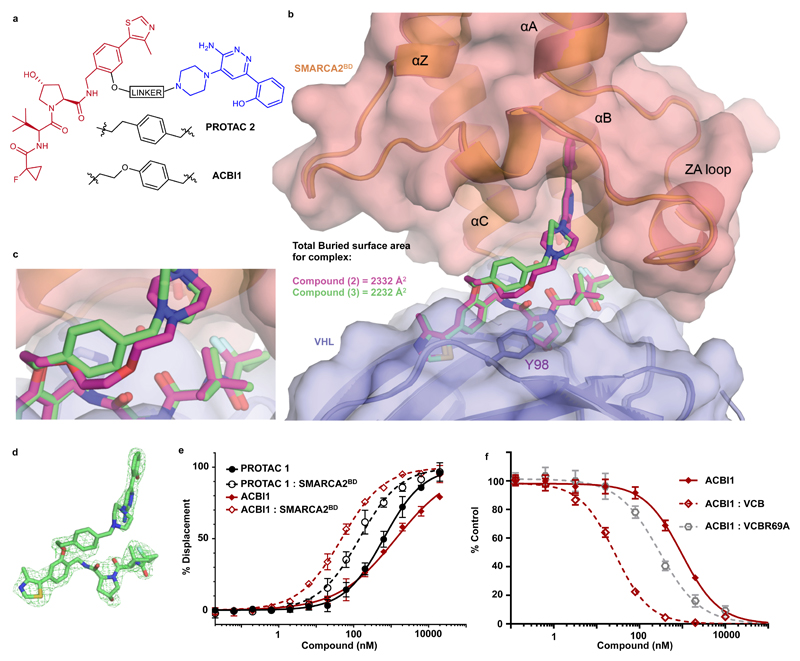
In order to avoid lengthy empirical exploration of structure-activity relationships based on PROTAC 1, we endeavoured to use the ternary complex co-crystal structure to efficiently guide us towards the design of a more optimal degrader. Inspection of the ternary complex structure of PROTAC 1 indicates that a large proportion of the de novo protein-protein interactions exist around the fluorocyclopropyl amide moiety (Fig. 2a and 2c). Further, this crystal structure also points towards all other hydrogen bond donor containing functionalities in PROTAC 1 being engaged in PROTAC-protein interactions. We therefore decided to leave these regions unchanged and optimized the linker interaction with the lipophilic face. To increase conformational restraint, and attempt to form pi-stacking interactions to Y98, we introduced a benzylic group attached to the piperazine of the SMARCA-binding motif. Aware that linker geometry could play an important role in defining the ternary complex arrangement, we used a 1,4-substitution of the newly introduced phenyl ring to mimic the linker conformation observed for PROTAC 1 in our ternary complex structure and significantly reduce the polarity of the linker. The resulting compound, PROTAC 2 (3), exhibited a more favourable molecular recognition for the SMARCABD within the ternary complex (Fig. 3a and Supplementary Table 2). Furthermore, an improvement in the Caco-2 permeability assay was also observed with A-B rate of 8.4 x 10-7 cm/s, a B-A rate of 7.6 x 10-6 cm/s and efflux ratio of 9.1. The ternary complex co-crystal structure of VCB: PROTAC 2: SMARCA2BD was solved at 2.35 Å resolution (PDB ID: 6HAX, Fig. 3b-d and Supplementary Table 3). This structure reveals a near identical ternary complex to that observed with PROTAC 1 with all de novo protein-protein and protein-PROTAC interactions maintained. As designed, the 1,4-substitution of the newly introduced phenyl ring in PROTAC 2 indeed recapitulates the geometry of the more flexible linker of PROTAC 1 (Fig. 3c), with the phenyl ring forming an ideal T-stack to Y98 of VHL. In addition, the co-crystal structure for VCB: PROTAC 2: SMARCA4BD was also solved (1.76 Å resolution, PDB ID: 6HR2, Supplementary Fig. 3 and Supplementary Table 3) showing negligible differences in complexes formed by the bromodomains of either of the two ATPases with PROTAC 2 and VCB.
Figure 3. Ternary co-crystal structure of SMARCA2BD: PROTAC 2: VCB and biophysical data validate a rational design strategy.
a) 2D chemical structures of PROTAC 2 and ACBI1 b) Overlays of ternary crystal structures of VCB: PROTAC 1 : SMARCA2BD (orange, PROTAC carbons in magenta) and VCB: PROTAC 2 : SMARCA2BD (2.35 Å, salmon, PROTAC carbons in green). Near identical ternary complexes are formed by both PROTACs; with the phenyl ring of PROTAC 2 in close proximity to Y98 of VHL. c) Overlays show that the constrained 1,4-disubstituted phenyl ring of PROTAC 2 (green carbons) accurately recapitulates the linker geometry observed previously for PROTAC 1 (magenta carbons). d) Fo-Fc omit map (green meshes) of PROTAC 2 prior to ligand modelling contoured at 3.0 σ with a carve radius of 2.2 Å. e) Fitted curves from Fluorescence Polarization competition assays measuring displacement of a VHL peptide by PROTACs in the presence or absence of SMARCA2BD. ACBI1 forms more cooperative and stable complexes compared with PROTAC 1. Curves are a best fit of means from n = 3 biologically independent experiments, ± S.E.M. f) Fitted curves for TR-FRET assays measuring displacement of a biotinylated SMARCA2BD probe by PROTAC alone, in complex with VCB or in complex with an R69A variant of VCB. A significant rightward shift when using VCBR69A vs VCB highlights the importance of this residue in ternary complex formation in solution. Curves are a best fit of means from n = 3 biologically independent experiments, ± S.D.
A series of competition biophysical assays were also established to further understand the formation of ternary complexes induced by PROTAC 1 and PROTAC 2. When using PROTAC 1, all assays produced data in good agreement with the ITC data obtained earlier for formation of VCB: PROTAC 1: SMARCA2BD complexes (Supplementary Tables 2 and 4). A fluorescence polarization (FP) assay was developed to measure displacement of a VHL-binding HIF1α peptide and to determine the stability and cooperativity of ternary complexes (Supplementary Table 2 and Supplementary Fig. 4)30, 34. The data show that cooperativity (α) and ternary Ki values for VCB: PROTAC: BAF unit complexes are significantly improved for PROTAC 2 as compared to PROTAC 1. Furthermore, the free energy change of the two-step complex formation process (ΔGcomplex), calculated as a sum of ΔG values derived from the binary interaction of PROTAC with SMARCA2BD (from SPR KD) and ternary complex formation of SMARCA2BD: PROTAC complex with VCB (from FP Ki) was found to be 1.4 kcal/mol lower for PROTAC 2 compared with PROTAC 1 (Supplementary Table 5). This shift is equivalent to a ten-fold increase in ternary complex stability as a result of the structure-based optimization. TR-FRET-based competition assays, measuring the displacement of a biotinylated SMARCA2 probe (4) in the presence or absence of VCB, were developed as an orthogonal approach to the FP assay. In TR-FRET competition assays cooperativity of PROTAC 2 is likewise improved relative to PROTAC 1 (Supplementary Table 4, Supplementary Fig. 5). Using the same biotinylated SMARCA2 probe, the cooperative nature of both compounds was confirmed in an AlphaLISA assay evaluating PROTAC-induced ternary complexes between full-length proteins within cell lysates (Supplementary Table 6 and Supplementary Fig. 6, see online methods for details).
Emboldened by our crystallographic and biophysical data, we sought to further optimize our degrader by maintaining the benzylic functionality on the piperazine which had produced improvements in PROTAC 2, while extending to the same linker length that had been employed in PROTAC 1, by introducing an oxygen atom between the VHL ligand and the phenyl ring of the linker (Fig. 3a). Based on this design rationale, ACBI1 (5) was prepared and also subjected to evaluation in our FP and TR-FRET biophysical assays (Fig. 3e and f, Supplementary Tables 2, 4 and 5 and Supplementary Fig. 5). In both the FP and TR-FRET assays, ACBI1 demonstrated a cooperativity (α) of approximately 30. Despite an unimpressive TR-FRET IC50 of 770 nM when measured against the SMARCA2BD alone, the ternary complex IC50 for ACBI1 in this assay was 26 nM. ACBI1 also demonstrated significant improvements in the Caco-2 permeability assay with A-B rate of 2.2 x 10-6 cm/s, a B-A rate of 3.8 x 10-6 cm/s and efflux ratio of 1.7.
The ternary complex crystal structures for PROTAC 1 and PROTAC 2 both suggest a key stabilising role of the electrostatic interaction between R69 of VHL and the electronegative C-terminal cap of the B helix of SMARCA2BD. To validate the specificity of the induced PPI in solution a modified VCB complex with an R69A mutation in VHL (VCBR69A) was utilized. As predicted by the ternary crystallographic data, a significant decrease in cooperativity is seen for all PROTACs when VCBR69A is used in the TR-FRET assay relative to wild-type (Fig. 3f, Supplementary Table 4, and Supplementary Fig. 5). Notably, irrespective of the initial cooperativity when using VCB, for all PROTACS the cooperativity values revert to ~3-4 when using VCBR69A, indicating a limit to the level of molecular recognition possible when this arginine residue is not present as an anchor point for these complexes. This data is also highly suggestive that an interaction between R69 of VHL and SMARCA2BD is similarly important for molecular recognition within the ternary complex formed by ACBI1, as it is for PROTAC 1 and PROTAC 2.
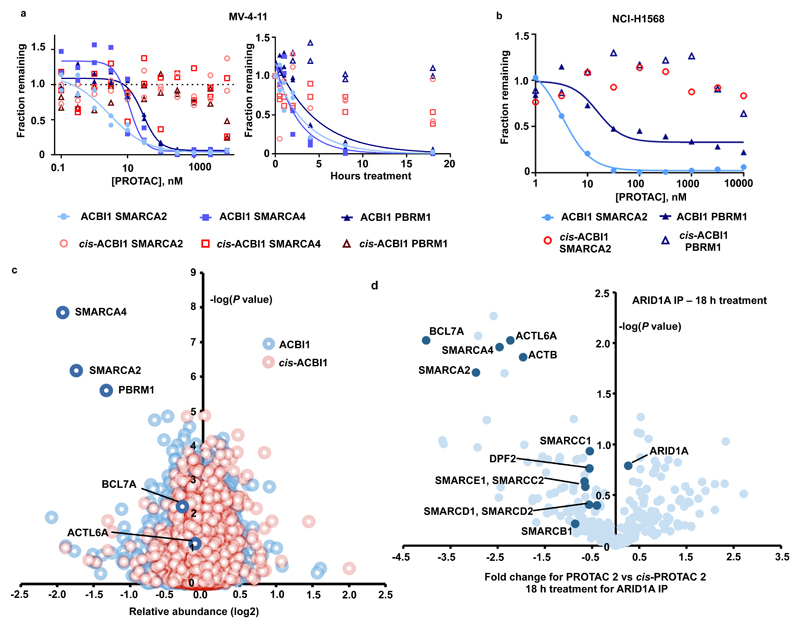
Finally, we evaluated the activity of our PROTACs in cellular assays (Fig. 4a and b and Supplementary Figures 10-11). Complete and potent degradation induced by ACBI1 was observed for SMARCA2 (DC50 of 6 nM) and SMARCA4 (DC50 of 11 nM) in MV-4-11 cells, with similar effects on SMARCA2 in SMARCA4-deficient NCI-H1568 cells. The SMARCABD ligand used in our PROTACs also binds to the 5th bromodomain of the PBAF complex member PBRM1, and ACBI1 is also seen to degrade that protein with a DC50 of 32 nM. As expected, degradation by PROTAC 2 was sensitive to competition with a VHL inhibitor (6) and was dependent on neddylation35 and proteasome activity (Supplementary Fig. 11). Using 1 µM ACBI1, a half maximal degradation occurred well within 2 h for SMARCA2 and SMARCA4 in MV-4-11 cells (Fig. 4a). In line with increased ternary complex stabilities and expected permeabilities based on evaluation in Caco-2 assays, ACBI1 is a more potent degrader than PROTAC 2, which in turn is more potent than PROTAC 1.
Figure 4. ACBI1 is a potent and selective degrader of SMARCA2, SMARCA4 and PBRM1.
a) Degradation of endogenous SMARCA2, SMARCA4 and PBRM1 in MV-4-11 cells treated for 18 h gave a DC50 of 6, 11 and 32 nM respectively (left). Time course of degradation in MV-4-11 in the presence of 1 µM of ACBI1 or 1 µM cis-ACBI1 (right). Two independent experiments. b) Degradation of endogenous SMARCA2 in NCI-H1568 cells treated for 18 h gave a DC50 of 3.3nM for SMARCA2 and 15.6 nM for PBRM1 Two independent experiments. c) Effects of ACBI1 (blue) and cis-ACBI1 (red) at 333 nM for 8 h on the proteome of MV-4-11 cells. Data plotted Log2 of the fold change versus DMSO control against –Log10 of the p value per protein from n = 3 independent experiments. All t-tests performed were two-tailed t-tests assuming equal variances d) SWI/SNF complexes were immuno-purified from MV-4-11 cell lysates following PROTAC treatment and abundance of subunits was determined by label free quantitation. Data plotted as fold change in abundance for PROTAC 2 vs cis-PROTAC 2 treatment against –Log10 of the p value per protein from n = 3 biologically independent experiments. Subunits of the SWI/SNF complex are highlighted. All t-tests performed were two-tailed t-tests assuming equal variances.
To understand the wider effects of our PROTACs on the cellular proteome, unbiased quantitative Tandem Mass Tag (TMT) labelling and mass spectrometry proteomics was performed with 333 nM of ACBI1 or cis-ACBI1 (7) and a treatment time of 8 h in MV-4-11 cells (Fig. 4c). The data corroborate a significant knockdown of SMARCA2, SMARCA4 and PBRM1 with minimal down-regulation of other proteins across the proteome. To further understand the effects of chemically induced ATPase degradation on BAF/PBAF complex integrity, we immunoprecipitated complexes from cell extracts using antibodies against the SMARCC2/BAF170 and ARID1A subunits and identified specifically enriched associating proteins by mass spectrometry. Treatment with PROTAC 2 led, as expected, to a less efficient recovery of SMARCA2, SMARCA4 and PBRM1 (Fig. 4d and Supplementary Fig. 12 and 13). The majority of accessory subunits remain associated, but it is clear that some core subunits such as ACTL6A, the BAF specific BCL proteins and the PBAF specific PHF10 protein are co-depleted. The TMT proteomics data with both PROTAC 2 and ACBI1 show no reduction in ACTL6A suggesting dissociation of this subunit from the complex following ATPase degradation (Fig. 4c and Supplementary Fig. 12a).
Degradation of SMARCA2/4 causes cancer cell death
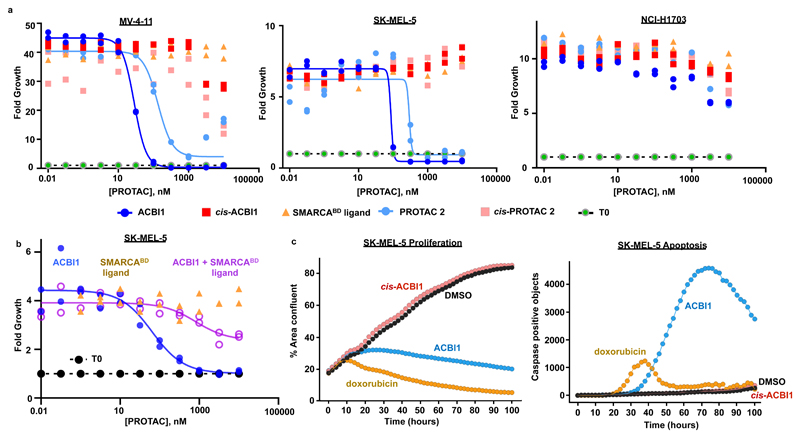
The ATPases of the BAF complex have been shown to be essential in certain cancer cell lines. SMARCA4 supports AML cell proliferation by maintaining an essential enhancer17, whereas in several solid cancer types, mutational loss of SMARCA4 leads to a dependence on active SMARCA218–20, 24. We therefore tested the effects of PROTAC-mediated loss of SMARCA2 and SMARCA4 on proliferation of a panel of cancer cell lines (Fig. 5a and Supplementary Fig. 14). We first tested the activity of ACBI1 on cells known to depend on an intact BAF complex: leukemia cell lines, such as MV-4-11, as well as the SMARCA4-deficient cells SK-MEL-5 and NCI-H1568. In all cell lines tested, ACBI1 exerted potent anti-proliferative effects. In MV-4-11 and SK-MEL-5 cells, its non-degrading distomer cis-ACBI1 remained inactive. ACBI1 demonstrated an anti-proliferative IC50 of 28 nM in MV-4-11 cells, broadly in line with its DC50 of 6 nM. The SMARCABD ligand, i.e. the bromodomain binder without the linker and VHL-binding part, had no antiproliferative effect in any cell line. Surprisingly, an antiproliferative effect was seen with cis-ACBI1 in NCI-H1568 cells, albeit at significantly higher concentration than ACBI1 (IC50 of 441 nM vs. 68 nM, Supplementary Fig. 14b). The reasons for this effect are presently unclear. A series of studies were next carried out to demonstrate that the effects of the PROTACs were due to on-target degradation of SMARCA2 and SMARCA4. Firstly, we combined high amounts of the SMARCABD ligand with titrations of ACBI1 to compete for bromodomain binding. As expected, competing amounts of SMARCABD ligand reduced ACBI1-induced degradation of SMARCA2 as well as the negative effects on cell proliferation (Fig.5b and Supplementary Fig. 14c and d). Secondly, a lung adenocarcinoma cell line deficient in both SMARCA2 and SMARCA4, NCI-H1703, was chosen to assess potential off-target activity of ACBI1. Both ACBI1 and cis-ACBI1 show no anti-proliferative effects in this SMARCA2/4 null cell line (Fig. 5a). These cells remain competent to degrade BAF complex members, as ectopically expressed SMARCA2 was efficiently eliminated in the presence of PROTAC 2, but not the non-VHL-binding version of this, cis-PROTAC 2 (8) (Supplementary Fig. 15a). SMARCA2 bearing an inactivating mutation (see online methods) in the bromodomain (SMARCA2 bromodead) was stable in the presence of PROTAC 2, confirming that degradation depends on the compound interacting with an intact SMARCA2 bromodomain (Supplementary Fig. 15a). In NCI-H1568 cells over-expressing PBRM1, ACBI1 showed similar anti-proliferative effect in comparison to a vector control, whereas the overexpression of SMARCA2 produced a significant rightward shift in anti-proliferative IC50 as well as attenuating maximal effect (Supplementary Fig. 15b-d). Taking into consideration that ACBI does not induce degradation of proteins other than SMARCA2, SMARCA4 and PBRM1, this supports insufficient amounts of SMARCA2 following ACBI1 treatment being the driver of the observed phenotype.
Figure 5. Effects on the proliferation and apoptosis of cancer cells in the presence of ACBI1.
a) Cell viability of an AML cell line, MV-4-11 (left), a SMARCA4 deficient melanoma cell line, SK-MEL-5 (centre) and a SMARCA2 and SMARCA4 deficient NSCLC cell line, NCI-H1703 (right), after 7 days in the presence of ACB.I1. IC50s for ACBI1 and cis-ACBI1 in MV-4-11 cells are 29 nM and 1.4 µM, and in SK-MEL-5 cells are 77 nM and > 10000 nM, respectively. Two independent experiments. b) Cell viability of a melanoma cell line, SK-MEL-5 after 3 days in the presence of dose titrations of ACBI1, the SMARCABD ligand or a dose titration of ACBI1 in the presence of 10 µM of SMARCABD ligand, representative of two biologically independent experiments. c) Real-time measurement of proliferation (left) and apoptosis (right) in SK-MEL-5 cells after treatment with doxorubicin (1 µM), ACBI1 (0.3 µM) and cis-ACBI1 (0.3 µM). Representative of two biologically independent experiments.
We then assessed the extent of cellular apoptosis induced by compound treatment. ACBI1-mediated loss of SMARCA2 led to increased caspase activity in SK-MEL-5, comparable to that seen with the chemotherapeutic drug doxorubicin (Fig. 5c). In SK-MEL-5 cells, the fraction of caspase-positive cells steeply increased after 40 h of treatment alongside the appearance of cleaved PARP, confirming the induction of apoptosis in these cells (Supplementary Fig. 15e). These results confirm that the vulnerability of cancer cells to SMARCA subunit loss demonstrated by genetic means can be recapitulated upon chemically induced degradation.
Discussion
In this study we describe how structure-based design guided us to improve the characteristics of a bifunctional degrader molecule. We showed that despite binding to the protein target of interest with weaker affinity than the analogous SMARCABD ligand, a prototype compound PROTAC 1 formed cooperative ternary complexes and was active as a degrader, and thus a promising lead for PROTAC optimization. Co-crystal structures identified the key induced interactions that were central to stabilize the ternary complex and delineated the conformation and interactions of the polyethylene glycol linker. This information saved considerable time and effort by guiding us away from changing regions of the PROTAC structure that could have been inhibitory to complex formation, a likely path if following an empirical approach. Rather, this data informed the introduction of a disubstituted phenyl ring to enhance conformational restraint and form novel protein-linker interactions, while simultaneously reducing polarity of the molecule in a targeted way to give PROTAC 2. A subsequent ternary co-crystal structure and allied biophysical studies with PROTAC 2 showed improvements in molecular recognition and free energy of ternary complex formation for this compound, validating the design hypotheses. In a further round of design the linker length was extended to that used in PROTAC 1 while retaining the benzyl moiety on the piperazine ring to yield ACBI1. These three PROTACs showed progressively more cooperative behavior in FP and TR-FRET ternary complex assays. In line with this and with improvements in permeability, these PROTACs induced degradation of their targets with increasing potency in cells. Our findings together qualify ACBI1 as a potent degrader of BAF complex ATPases in multiple cancer cell lines, and furnish a first-in-class chemical tool for acute and profound SMARCA2/4 knockdown.
We exemplify the efficiency of structure-based PROTAC design and further demonstrate how non-functional domains can provide suitable handles for targeted degradation10. Prioritizing and designing bifunctional molecules on the basis of their ability to induce stable complexes between the E3 ligase and the target is broadly applicable and may greatly reduce the empirical nature of PROTAC design that has dominated to date. Whilst cooperativity of ternary complexes is not essential for PROTAC-induced protein degradation8, 33, 36, we show that the identification of compounds that induce stable complexes can provide rational starting points for optimization, even if those molecules are suboptimal at inducing protein degradation or retaining the binary binding affinity of the constitutive target/ligase-binding units.
Unbiased whole cell proteomic analysis confirmed exquisitely selective ACBI1-mediated target protein knockdown and IP-MS experiments with its analogue PROTAC 2, showed that the majority of BAF/PBAF subunits remain associated following PROTAC treatment. Several BAF subunits including ACTB, ACTL6A, BCL7A and PHF10 were consistently co-depleted from immunopurified complexes with SMARCA2, SMARCA4 and PBRM1. Many of these proteins are known to interact with SMARCA2, SMARCA4 and PBRM1 and are likely to dissociate as a result of loss of contact with these subunits, supported by our data showing total protein levels of ACTL6A are not changed20, 37, 38. At the outset of our study, it was not known if degradation of individual subunits out of stable BAF/PBAF multi-protein complexes by PROTACs was possible. By addressing this we have demonstrated the potential for acute chemically mediated manipulation of BAF/PBAF composition, which is of value both in the design of future therapeutics and as research tools.
The dependence of certain cancer types on the BAF complex paralog ATPases SMARCA2 and SMARCA4 has been unequivocally shown both by RNA interference and CRISPR-based knockouts18–20, 24, 39, 40. However, chemically-induced knockdown targeting these BAF complex subunits has not been demonstrated so far. Our work shows that targeting the SMARCA2/4 bromodomains for protein degradation can exploit these identified cancer vulnerabilities and phenocopies the genetic knockdowns. The nature of the anti-proliferative effect of loss of BAF complex members has not been comprehensively investigated20. We show here that SMARCA2/4-degrading PROTACs can induce rapid apoptosis in a subset of SMARCA4-deficient cells. Additional studies are required to understand the differential rate in responses of a subset of cancer cells that have adapted to loss of SMARCA4 (e.g. NCI-H1568 vs. SK-MEL-5). Using our dual SMARCA2/4-degrading PROTACs, we were also able to show anti-proliferative effects in AML cells, consistent with earlier knockdown studies showing sensitivity of these cells to SMARCA4 depletion17. While mice lacking functional SMARCA2 develop normally, the loss of SMARCA4 results in embryonic death in mice39–42. Therefore, whilst it is likely that a selective degrader of SMARCA2 will not have dramatic on-target side effects, it remains to be seen whether SMARCA4 degradation can be therapeutically translated. In addition to targeting the BAF complex ATPases, ACBI1 also causes degradation of PBRM1 albeit with lower potency. However, overexpression of PBRM1 in NCI-H1568 cells failed to rescue the effects of ACBI1 on cell growth whereas SMARCA2 overexpression indeed attenuated the effect, suggesting that that loss of PBRM1 alone does not exert the anti-proliferative effects observed. Future studies are warranted toward deeper mechanistic understanding of the downstream effects of degrading BAF ATPase subunits in both cancerous and non-cancerous cells. We anticipate that progress in this direction will be greatly advanced by sharing the chemical tools that have emerged from our work with the wider community. Finally, we hope that structure-based design becomes a central approach in the emerging PROTAC field as it has in more traditional areas of drug discovery.
Online Methods
Chemical synthesis
Synthesis of compounds described in this paper and their intermediates are described in the Supplementary Note.
Cell lines and culture
Cell lines were obtained through ATCC, verified for identity by satellite repeat analysis and tested for mycoplasma contamination. Cells were grown in the medium as specified by the supplier unless described otherwise and not used beyond passage 25.
Caco-2 permeability assay
Caco-2 cells (1-2 × 105 cells/1 cm2) were seeded on filter inserts (Costar transwell polycarbonate or PET filters, 0.4 µm pore) and cultured in DMEM for 10 to 25 days. Compounds were dissolved in appropriate solvent (1-20 mM stock solutions were diluted with HTP-4 buffer (128.13 mM NaCl, 5.36 mM KCl, 1 mM MgSO4, 1.8 mM CaCl2, 4.17 mM NaHCO3, 1.19 mM Na2HPO4.7H2O, 0.41 mM NaH2PO4.H2O, 15 mM HEPES, 20 mM glucose, pH 7.2) containing 0.25% BSA to prepare the transport solutions (0.1 - 300 µM compound, final dimethyl sulfoxide (DMSO) concentration ≤0.5%). Transport solution (TL) was applied to the apical or basolateral donor side for measuring A-B or B-A permeability (3 filter replicates), respectively. The receiver side contained HTP-4 buffer supplemented with 0.25% BSA. Samples were collected at the start and end of experiment from the donor and at various time intervals for up to 2 h also from the receiver side for concentration measurement by HPLC-MS/MS. Sampled receiver volumes were replaced with fresh receiver solution.
Constructs, protein expression and purification of VCB and SMARCA2, SMARCA4 and PBRM1 bromodomains
Wild type and mutant versions of human proteins were used for all protein expression, as follows: VHL (UniProt accession number: P40337), ElonginC (Q15369), ElonginB (Q15370) and the bromodomains (BDs) of SMARCA2 (SMARCA2BD; P51531-2, residues 1373-1493 with additional N-terminal SM residues (cloning artefact)), SMARCA4 (SMARCA4BD; P51532, residues 1448-1569 with additional N-terminal SM residues (cloning artifact)), and the fifth bromodomain of PBRM1 (PBRM1BD5, Q86U86, residues 645-766). SMARCA4BD was provided by the Structural Genomics Consortium (SGC), Toronto43 and SMARCA2BD and PRM1BD5 were synthesised by GeneArt then subcloned into pDEST15 vectors (Invitrogen). The VCB complex was expressed and purified as described previously, with the modification that 0.3 mM IPTG was used for induction of expression7. Briefly, N terminally His6 tagged VHL (54-213), ElonginC (17-112) and ElonginB (1-104) were co expressed and the complex isolated by Ni affinity chromatography, the His6 tag was removed using TEV protease, and the complex further purified by anion exchange and size exclusion chromatography (SEC). The VCBR69A mutant, in which R69 of VHL (54-213) was mutated to alanine, was generated using a QuickChange II site directed mutagenesis kit (Agilent) according to the manufacturer’s instructions and expressed and purified as for VBC. Both were stored in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 150 mM sodium chloride and 1 mM tris(2-carboxyethyl)phosphine (TCEP) pH 7.5. SMARCA2BD, SMARCA4BD and PBRM1BD5 were expressed in Escherichia coli BL21(DE3) as N terminal GST fusion proteins with a TEV protease cleavage site. Expression and purification of these proteins has been described previously43, 44. Starter cultures were grown overnight at 37 °C in 10 mL of Luria-Bertani (LB) medium with ampicillin (100 μg/mL). The starter cultures were diluted (1:100) in Terrific Broth (TB) medium with ampicillin (100 μg/mL) and grown in a shaking incubator at 37 °C to an optical density (OD600) ~ 1 before the temperature was lowered for induction (to 23 °C for SMARCA2BD or 18 °C for SMARCA4BD and PRRM1BD5). Expression was induced using IPTG (final concentration 0.3 mM for SMARCA2BD, PBRM1BD5, 0.4 mM for SMARCA4BD) for 16 h at the specified temperatures. Cells were harvested by centrifugation and stored at -80 °C prior to purification. Cells were resuspended in Lysis buffer and lysed by sonication (20 pulses of 5 s for 4 min) using a Sonopuls HD 3080 (Bandelin, Berlin, Germany) or by homogenization using a Stansted Pressure Cell Homogenizer (Stansted Fluid Power). The Lysis buffers were as follows: for SMARCA2BD: 50 mM HEPES, 500 mM sodium chloride, 5% glycerol, 5 mM dithiothreitol (DTT); for SMARCA4BD: 25 mM HEPES, 0.3 M sodium chloride, 5% glycerol, 10 mM DTT, pH 7.5; for PBRM1BD5: 25 mM HEPES, 300 mM sodium chloride, 5% glycerol, 10 mM DTT, pH 7.8; in each case supplemented with complete protease inhibitors (Roche). Affinity purification was performed using Glutathione Sepharose 4B (GE Healthcare) in batch mode or on-column. Cleavage of the GST-tag was performed using TEV protease for 16 h at 4 °C, either on column, or in solution following elution of the GST tagged BDs with Lysis buffer containing 20 mM reduced L glutathione (Sigma Aldrich). For SMARCA2BD, prior to TEV protease cleavage the eluted GST-tagged BD was first dialysed into desalting buffer (20 mM HEPES, 250 mM sodium chloride, pH 7.0 + 0.5% glycerol). Where TEV cleavage was performed in solution, a second affinity (GST-trap) column purification step was carried out to remove the GST-tag and uncleaved GST-tagged protein. Proteins were further purified by SEC (HiLoad Superdex-75, 16/600) (GE Healthcare) and stored in Storage buffer (for SMARCA2BD: 20 mM HEPES, 150 mM sodium chloride, pH 7.5; for SMARCA4BD: 10 mM HEPES, 0.5 M NaCl, 5% glycerol, pH 7.5; for PBRM1BD5: 20 mM HEPES, 300 mM NaCl, 5 mM DTT, pH 7.8). For AlphaScreen assays, eluted GST-tagged BDs were purified directly by SEC in the respective storage buffers without TEV cleavage. All chromatography purification steps were performed either at room temperature or 4 °C using an ÄKTA FPLC purification system (GE Healthcare) or a plastic Econo-Pac column (Bio-Rad).
Isothermal titration calorimetry (ITC)
Titrations for cooperativity experiments were performed at 25 °C on a MicroCal PEAQ-ITC or ITC200 micro calorimeter (Malvern) in reverse mode (protein in syringe, ligand in cell) as described previously7. Briefly, PROTACs (ACBI1 or ACBI2) were diluted from a 10 mM DMSO stock solution to 20 μM in buffer consisting of 20 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane (BIS-TRIS) propane, 150 mM NaCl, 1 mM TCEP, pH 7.4 and a final DMSO concentration of 0.2%. Two sets of sequential titrations were performed; firstly, a titration of SMARCA2BD (200 μM, in the syringe) into the PROTAC solution (20 μM, in the cell), followed by a titration of VCB complex solution (168 μM in the syringe) into the PROTAC–SMARCA2BD complex solution retained in the cell. Binary experiments consisted of an analogous initial single injection of buffer (syringe) into the PROTAC solution (20 μM, in the cell), followed by a titration of VCB complex solution (168 μM in the syringe) into the diluted PROTAC solution retained in the cell. Data were fitted to a single-binding-site model to obtain the stoichiometry (n), the dissociation constant (KD) and the enthalpy of binding (ΔH) using MicroCal PEAQ-ITC Analysis Software (Malvern). Cooperativity values were calculated as the ratio of KD values determined for VCB binding to a PROTAC or to a PROTAC–SMARCA2BD complex (i.e. binary KD/ternary KD). The reported values are the mean ± S.D. from two independent measurements.
FP competition assays
To evaluate cooperativity of PROTAC binding to VCB in the absence or presence of saturating concentrations of each bromodomain, an existing fluorescence polarisation assay in competition format was utilized, with only minor modifications30, 34. Fluorescence polarisation experiments were performed at room temperature on a PHERAstar FS (BMG LABTECH) in 384-well plates (Corning 3820), with excitation at 485 nm and emission at 520 nm. The fluorescent peptide employed in these experiments was a 5,6 Carboxyfluorescein (FAM)-labelled HIF-1α peptide (FAM-DEALAHyp YIPMDDDFQLRSF, measured KD for VCB ~ 1-2 nM) (‘JC9’). The assay buffer was 100 mM BIS-TRIS, 100 mM NaCl, 1 mM DTT, pH 7, supplemented with compound/DMSO vehicle (final DMSO concentration 1%). The final assay volume per well was 15 µL. A direct titration of VCB against the fluorescent peptide was firstly performed using 5 nM of peptide and decreasing concentrations of VCB (15-point two fold serial dilution from 100 µM to 6.1 nM, triplicate wells). Next a competition experiment was performed using each PROTAC (14 point half-log-fold serial dilution from 50 µM to 0.016 nM) in the absence or presence of a saturating concentration of SMARCA2BD, SMARCA4BD or PBRM1BD5 (75 µM for SMARCA2BD 150 µM for SMARCA4BD and 100 µM for PBRM1BD5). The concentration of bromodomain was calculated such that it remained in molar excess relative to PROTAC, with the lowest concentration selected being greater than 10 fold the weakest PROTAC KD for that particular bromodomain (>90% fractional occupancy). For each independent experiment, titrations were performed using duplicate wells. IC50 values were calculated in Prism (Version 7.03, GraphPad). Curves were fitted using nonlinear regression (four parameter fit) following normalisation to percentage of maximum signal based on corresponding DMSO controls for each treatment type. KD values were back calculated from fitted IC50 values as described previously30, 34. Cooperativity values (α) for each PROTAC were calculated using the ratio:
The reported values are the mean ± S.E.M. from 3 independent measurements (duplicate wells).
TR-FRET competition assays
To evaluate cooperativity of PROTAC binding to SMARCA2BD, a TR-FRET assay was developed in a competition format using (4)28 (KD for SMARCA2BD ~ 400 nM) and His6-tagged SMARCA2BD. PROTAC binding to SMARCA2BD was then measured in the absence or presence of saturating concentrations of VCB.
All assays were performed at room temperature in a Proxiplate-384 PLUS, white (PerkinElmer) plate. Reagents were diluted in buffer, consisting of 50 mM HEPES, 50 mM sodium chloride, 2 mM DTT, 0.008% Brij, 0.01% BSA at pH 7.3, and were allowed to equilibrate to room temperature prior to addition to plates. The final concentrations in the assay were as follows: 40 nM SMARCA2BD, a concentration series of each PROTAC (10 µM to 1 nM, 11-point half-log-fold serial dilution), 16.6 nM biotinylated probe 5, 2.5 nM Lance Eu-W1024 labeled Streptavidin (Perkin Elmer Cat No AD0062, 50 µg, 500 µg/mL, 8.3 µM), 50nM ULight-anti His6 AB (Perkin Elmer TRF0105-M, 5 µM, 800 µg/mL) and either assay buffer or 5 µM VCB at a final DMSO concentration of 1%. SMARCA2BD, Streptavidin-Eu and U-Light AB were mixed in assay buffer (SMARCA2 solution) and kept at room temperature prior to use. 5 nL of the biotinylated probe solution was added to rows 1-23 (with the Labcyte Echo 55x) and 187.5 nL VCB (or assay buffer) was transferred with the Labcyte Echo 55x to rows 1-23. 15 µL of the SMARCA2 solution was subsequently added to rows 1-24. Plates were incubated at room temperature for 60 minutes and the signal was measured in a PerkinElmer Envision HTS Multilabel Reader using the TR-FRET LANCE Ultra specs from PerkinElmer (665 nm/615 nm × 10000).
Each plate contained 16 wells of a negative control (diluted DMSO instead of test compound with biotinylated probe) and 16 wells of a positive control (diluted DMSO instead of test compound without biotinylated probe). The average of the background control (positive control, without enzyme) was calculated and subtracted from the measured values for each plate. IC50 values were calculated and analyzed using a 4 parametric logistic model for each PROTAC in the absence or presence of saturating concentrations of VCB. Cooperativity values (α) for each PROTAC were calculated using the ratio:
Note; ‘VCB’ represents either VCB or VCBR69A. The reported values are the mean ± S.E.M. from 3 independent measurements (duplicate wells).
SMARCA2: (1) crystallography
The binary crystal complex structure of SMARCA2BD: (1) was obtained by ligand soaking into crystals of SMARCA2BD. Crystallisation of SMARCA2BD (14 mg/mL in 25 mM tris(hydroxymethyl)aminomethane (TRIS) pH 8, 150 mM sodium chloride) was carried out at 4 °C using the sitting drop method (0.2 μL of each of protein/reservoir solutions, using a small seed stock of disrupted crystals added to the protein immediately prior to drop setting). Crystals were obtained after approximately 14-21 days in reservoir solution consisting of 25% (w/v) PEG 6000, 0.1 M HEPES pH 6.1, 0.01 M zinc chloride, 0.01 M cobalt (III) hexamine chloride, 8% (v/v) ethylene glycol. For ligand soaking experiments, 1 μL of a 10 mM DMSO stock solution of (1) was dried and re-dissolved in 1 μL of reservoir solution, into which crystals of SMARCA2BD were soaked for 5 days, then flash cooled in liquid nitrogen using reservoir solution containing 25% ethylene glycol as a cryo-protectant. Data were collected at the SLS beam line PXII (Swiss Light Source, Paul Scherrer Institute, λ = 1.000 Å) using a PILATUS 6M detector. The crystals belonged to space group P 41 with unit cell parameters a = 42.5, b = 42.5, c = 164.2 Å and α, β, γ = 90° and contained two monomers per asymmetric unit. Images were processed with autoPROC incorporating STARANISO (Tickle, I.J., Flensburg, C., Keller, P., Paciorek, W., Sharff, A., Vonrhein, C., Bricogne, G. (2018). STARANISO. Cambridge, United Kingdom: Global Phasing Ltd.)45. The resolution limit was set to 1.33 Å using local(I/σI) ≥ 1.20. The structure was solved by molecular replacement using a SMARCA2BD crystal structure (PDB entry 4QY4; https://www.thesgc.org/structures/4qy4) as a search model. Subsequent iterative model building and refinement was done according to standard protocols using CCP446, COOT47 and autoBUSTER (Global Phasing Ltd.). The structure was refined to Rwork and Rfree values of 17.9% and 19.3%, respectively, with 100% of the residues in Ramachandran favoured regions as validated with MOLPROBITY48.
Ligand restraint and initial coordinate files for all structures were generated using GRADE (O. S. Smart, T. O. Womack, A. Sharff, C. Flensburg, P. Keller, W. Paciorek, C. Vonrhein, and G. Bricogne. (2011) grade, version 1.2.13. Cambridge, United Kingdom, Global Phasing Ltd., http://www.globalphasing.com) incorporating crystallographic geometry information from MOGUL49. Data collection and refinement statistics for all structures are described in Supplementary Table 1.
VCB: (2):SMARCA2BD ternary complex crystallography
VCB, (2) and SMARCA2BD were mixed in a 1:1:1 stoichiometric ratio in 20 mM HEPES, pH 7.5, 150 mM sodium chloride, 1 mM TCEP, 2% DMSO, incubated for 5 minutes at room temperature and concentrated to a final concentration of approximately 8 mg/mL. Drops were prepared by mixing 1 μL of the ternary complex with 1 μL of well solution and crystallised at 4°C using the hanging drop vapour diffusion method. Crystals were obtained in 18% (w/v) PEG 3350, 0.2 M sodium formate, 0.1 M HEPES, pH 7.75, and formed within 24 h but were fully grown after a few days. Harvested crystals were flash cooled in liquid nitrogen following gradual equilibration into cryo protectant solution consisting of 25% (v/v) ethylene glycol in 19% PEG 3350, 0.2 M sodium formate, 0.1 M HEPES, pH 7.75. Diffraction data were collected at Diamond Light Source beamline I04 1 (λ = 0.91587 Å) using a Pilatus 6M-F detector and processed using XDS50. The crystals belonged to space group P 21 21 21 with unit cell parameters a = 79.7, b = 117.3, c = 121.6 Å and α, β, γ = 90° and contained two copies of the ternary complex per asymmetric unit. The structure was solved by molecular replacement using PHASER with VCB coordinates derived from the VCB:MZ1:Brd4BD2 complex (PDB entry 5T35) and SMARCA2BD (PDB entry 4QY4)51 as search models. Subsequent iterative model building and refinement was done according to standard protocols using CCP446, COOT47 and autoBUSTER (Global Phasing Ltd.). The structure was refined to Rwork and Rfree values of 19.3% and 22.4%, respectively, with 97.9% of the residues in Ramachandran favoured regions as validated with MOLPROBITY48.
VCB:(3):SMARCA2BD ternary complex crystallography
The ternary complex of VCB: (3):SMARCA2BD (at approximately 14 mg/mL) was prepared and crystallised as for the VCB: (2):SMARCA2BD complex, in well solution comprising 13% (w/v) PEG 3350, 0.2 M sodium formate, 0.1 M HEPES, pH 8.0. Harvested crystals were flash cooled in liquid nitrogen following gradual equilibration into cryo protectant solution consisting of 25% (v/v) ethylene glycol in 20% PEG 3350, 0.2 M sodium formate, 0.1 M HEPES, pH 8.0. Diffraction data were collected at Diamond Light Source beamline I24 (λ = 0.97998 Å) using a Pilatus3 6M detector and processed using XDS50. The crystals belonged to space group P 21 21 21 with unit cell parameters a = 80.5, b = 116.2, c = 120.6 Å and α, β, γ = 90° and contained two copies of the ternary complex per asymmetric unit. The structure was solved by molecular replacement and built, refined and the geometry validated as for the VCB: (2):SMARCA2BD ternary complex, to final Rwork and Rfree values of 23.2% and 26.8%, respectively, with 97.6% of the residues in Ramachandran favoured regions as validated with MOLPROBITY48.
VCB:(3):SMARCA4BD ternary complex crystallography
The ternary complex of VCB:ACBI1:SMARCA4BD (at approximately 14 mg/mL) was prepared and crystallised in well solution comprising 20% (w/v) PEG 1500, 0.1 M MIB buffer, pH 6.0. Harvested crystals were flash cooled in liquid nitrogen following gradual equilibration into cryo protectant solution consisting of 25% (v/v) ethylene glycol in 25% PEG 1500, 0.1 M MES, pH 6.0. Diffraction data were collected at SLS beamline X10SA (λ = 1.000 Å) using a Pilatus3 6M detector and processed using AUTOPROC. The crystals belonged to space group P 1 with unit cell parameters a = 66.33, b = 61.68, c = 81.63 Å and α = 69.43, β =83.38, γ = 86.42° and contained two copies of the ternary complex per asymmetric unit. The structure was solved by molecular replacement and built, refined and the geometry validated as for the VCB: (2):SMARCA2BD ternary complex, to final Rwork and Rfree values of 21.6% and 23.6%, respectively, with 97.4 % of the residues in Ramachandran favoured regions as validated with MOLPROBITY9.
Transfection, lysis and protein detection for AlphaLISA assays
HEK293 cells in DMEM (Invitrogen) were seeded at 3-3.5 × 105 cells/well of 6 well plates and the medium was replaced on the day of transfection. The pIRESpuro3 vector (Clontech) containing an insert for C-terminal 3×FLAG-tagged wild-type SMARCA2 (Uniprot identifier: P51531-1) or SMARCA4 (P51532-1) were transiently transfected into HEK293 cells using lipofectamine 2000 (Invitrogen) and OptiMEM (Invitrogen). Untransfected cells and cells transfected with the empty vector were used as negative controls. After 48 h, cells were washed twice in cold PBS (Invitrogen) and lysed in hypotonic buffer containing 10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 50 units/mL benzonase nuclease (Sigma) and protease inhibitor cocktail (Roche) by Dounce homogenisation. Protein samples were cleared by centrifugation at 4 °C, at 1000 × g for 5 min and the supernatants stored at -80 °C. Protein concentration was determined by BCA assay (Pierce) and the absorbance at 562 nm measured by spectrophotometry (NanoDrop ND1000). Samples were separated by SDS-PAGE using 40 μg of protein per well of NuPAGE Novex 4-12% BIS-TRIS gels (Invitrogen) and transferred to 0.2 μm pore nitrocellulose membrane (Amersham) by wet transfer. Over-expression was confirmed by Western blot detection of mouse anti-FLAG (1:1000, Sigma #F1804), rabbit anti-SMARCA2 (1:1000, Sigma #HPA029981), rabbit anti-SMARCA4 (1:1000, abcam #49360) and rabbit anti-β-actin (1:25000, Cell Signaling Technology, #4970S) antibodies with goat anti-mouse or donkey anti-rabbit IRDye 800CW secondary antibodies (1:10000, LI-COR #925-32210 and #926-32213) using a ChemiDoc MP imaging system (Bio-Rad). Quantification of band intensity was performed in Image Studio Lite (LI-COR) and normalised to β-actin expression and the DMSO control.
Ex cell AlphaLISA competition assays
Protein lysates containing either FLAG-tagged SMARCA2 or SMARCA4, were utilized for evaluation of ternary complex formation with full-length protein. Three biological repeats were performed with protein from three separate transfections and assay wells were ran in duplicate. All assays were performed at room temperature in 384-well alphaplates (PerkinElmer) with a final assay volume of 30 μL. Before each 1 h incubation, plates were sealed with film (PerkinElmer), centrifuged at 100 × g for 30 s and mixed by shaking at 450 rpm for 2 min. All assay components were made up as 6× stocks in assay buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 0.1% w/v BSA, 0.02% w/v CHAPS, 0.2 μm filtered). A 12 point, half-log-fold dilution series was used for all PROTACs and un-tagged VCB. (4)28 (100 nM final) and excess untagged VCB (5 μM-50 μM final) or an equivalent volume of assay buffer were incubated with PROTAC (all 0.1 nM-31.62 μM final) and 15 μg protein lysate or equivalent volume of lysis buffer for 1 h. Anti-FLAG beads (10 μg/mL final, PerkinElmer) were added in low light conditions and incubated for 1 h in the dark. Anti-streptavidin acceptor beads (10 μg/mL final, PerkinElmer) were added in low light conditions and incubated for 1 h in the dark. Plates were read on a PHERAstar FS (BMG LABTECH) with an AlphaLISA optic module (BMG LABTECH, excitation 680 nm and emission 615 nm). Signal intensity was plotted against PROTAC concentration in GraphPad Prism 7. Data was normalised to the average DMSO signal intensity for binary (without VCB) and ternary (with VCB) values and the percentage signal plotted with an [inhibitor] vs. response variable slope four parameter fit with a bottom constraint equal to zero. The binary IC50 was divided by the ternary IC50 to generate the α value.
SPR binding studies
SPR experiments were performed on Biacore 8K or T200 instruments (GE Healthcare). Immobilization of target protein was carried out at 25 °C on a CM5 chip using amine coupling (EDC/NHS, GE Healthcare or XANTEC) in HBS-P+ running buffer, containing 2 mM TCEP, pH 7.4. Following activation of the surface with EDC/NHS (contact time 600 s, flow rate 10 μL/min), the target bromodomains (SMARCA2BD, SMARCA4BD prepared at 0.5 - 0.7 mg/mL or PBRM1BD5 at 0.005 mg/mL in coupling buffer consisting of 10 mM Na-Acetate pH 6.5, 0.005% Tween-20 and 50 µM PFI-324, were coupled to a density of 1000-20000 Response Units (RU) (SMARCA2BD and SMARCA4BD) or 100 2000 RU (PBRM1BD5). Surfaces were deactivated using 1 M ethanolamine. For VHL target protein, streptavidin (Sigma Aldrich) (prepared at 1 mg/mL in 10 mM sodium acetate coupling buffer, pH 5.0) was first immobilized by amine coupling to a density of 1000-10000 RU), after which biotinylated VCB complex (2.8 μM in running buffer) was streptavidin-coupled to a density of 1000-20000 RU. Biotinylated VCB was prepared as previously described7. The reference surface consisted of an EDC/NHS-treated surface deactivated with 1 M ethanolamine.
All interaction experiments were performed at 6 °C in running buffer containing 20 mM TRIS, 150 mM potassium chloride, 2 mM magnesium chloride, 2 mM TCEP, 0.005% TWEEN 20, 1% dimethyl sulfoxide; pH 8.3. Sensorgrams from reference surfaces and blank injections were subtracted from the raw data prior to data analysis using Biacore T200 or Biacore 8K evaluation software. Sensorgrams recorded at different compound concentrations in multi-cycle experiments were fitted using a 1:1 interaction model, with a term for mass-transport included.
Proliferation assays
1000 cells/well were seeded in 384 well plates. After overnight incubation, compounds were added to the cells at logarithmic dose series using the HP Digital Dispenser D300 (Tecan), normalising for added DMSO. 1 day and 8 days after seeding, cellular ATP content was measured using CellTiterGlo (Promega). Measurements after 8 days were divided by the measurement after 1 day (=i.e. the T0 plate) to derive fold proliferation.
For online analysis of cell growth, 80000 cells in 800 µL/well were seeded in clear 12-well plates (costar #3513). IncuCyte Caspase-3/7 Green Apoptosis Assay Reagent (1:1000, Essen BioScience #4440) was added and cells were incubated at 37 °C and 5% CO2 overnight. On the next day, compounds were added at the desired concentration using the hp digital dispenser D300 and plates were read in an Incucyte ZOOM. Every 2 h, phase object confluence (percent area) for proliferation and green object confluence (percent area) for apoptosis were measured. Values for apoptosis were normalised for the number of cells by dividing the percent area for apoptosis by the percent area for proliferation and multiplying by 1000.
Protein degradation assays
For capillary electrophoresis, 20000 cells in 300 µl/well were seeded in a 24 well plate and incubated at 37 °C overnight. Compounds were added from DMSO stock solution using a Digital Dispenser D300 (Tecan), normalising for added DMSO and cells were incubated at 37 °C for the indicated times. Medium was removed, cells washed with PBS and lysed in 80 µl lysis buffer (1% Triton, 350 mM KCl, 10 mM TRIS pH 7.4, phosphatase-protease inhibitor cocktail (Thermo Scientific #1861281), 10 mM DTT, Benzonase 0.5 µL/mL (Novagen #70746 10KU, 25 U/µL)) for 30 min on ice before insoluble debris was pelleted by centrifugation. SMARCA2 levels were determined on a WES capillary electrophoresis instrument (Proteinsimple) using rabbit anti-SMARCA2 antibody (1:25, Sigma #HPA029981), SMARCA4 antibody (CellSignaling #49360, 1:25), PBRM1 antibody (Bethyl #A301-591A-M, 1:40) and anti-GAPDH antibody (1:100, abcam #ab9485) for normalization.
For Western blots, 3-4 x 105 NCI-H1568 cells in 2 mL/well were seeded into 6 well plates and 5 x 106 Mv-4-11 cells in 10 mL/plate were seeded into 10 cm plates 24 h before treatment. For two biological replicates, cells were treated for the indicated times, washed with PBS and lysed with lysis buffer (1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 50 mM Tris pH 7.4, protease inhibitor cocktail (Roche), 50 units/mL benzonase nuclease (Sigma)). NCI-H1568 cells were lysed in 80 µL lysis buffer/well and Mv-4-11 cells were lysed in 250 µL/plate. Lysates were sonicated then cleared by centrifugation at 4 °C, at 15800 × g for 10 min and the supernatants stored at -20 °C. Protein concentration was determined by BCA assay (Pierce) and the absorbance at 562 nm measured by spectrophotometry (NanoDrop ND1000). Samples were separated by SDS-PAGE using 40 μg of protein per well of NuPAGE Novex 4-12% BIS-TRIS gels (Invitrogen) and transferred to 0.2 μm pore nitrocellulose membrane (Amersham) by wet transfer. Western blot images were obtained through detection of rabbit anti-SMARCA2 (1:1000, Sigma #HPA029981), rabbit anti-SMARCA4 (1:1000, abcam #ab108318), rabbit anti-PBRM1 (1:1000, Bethyl Laboratories #A301-591A-M) and rabbit anti-β-actin (1:25000, Cell Signaling Technology #4970S) antibodies with donkey anti-rabbit IRDye 800CW secondary antibody (1:10000, LI-COR #926-32213) using a ChemiDoc MP imaging system (Bio-Rad).
Specificity of antibody detection was determined by the absence of the protein band in cells that did not express the respective protein.
PARP cleavage
PARP cleavage was detected by Western blotting using an anti PARP antibody (cell signaling #9542).
Expression constructs for studies with NCI-H1703 and NCI-H1568 cell lines
The coding sequences of SMARCA2 were expressed from the retroviral plasmid LMH, a version of the vector LMN52 in which the neomycin resistance gene had be replaced by hygromycin resistance using the BClII and MluI sites for cloning. To inactivate the bromodomain, the conserved asparagine 1482 was mutated to alanine.
MS proteomics
Sample preparation
MV-4-11 cells in RPMI 1640 (Invitrogen) were seeded at 5 × 106 cells on a 100 mm plate 24 h before treatment. Cells were treated in triplicate by addition of test compound. After 8 h, the cells were centrifuged at 250 × g for 5 min and washed twice with 12 mL of cold PBS. Cells were lysed in 500 µL of 100 mM TRIS pH 8.0, 4% (w/v) SDS supplemented with protease inhibitor cocktail (Roche). The lysate was pulse sonicated briefly and then centrifuged at 15,000 × g for 10 min at 4 °C. Samples were quantified using a micro BCA protein assay kit (Thermo Fisher Scientific) and 200 μg of each sample was processed and digested using the filter aided sample preparation (FASP) method followed by alkylation with iodoacetamide and digestion with trypsin as previously described7. The samples were then desalted using a 7 mm, 3 mL C18 SPE cartridge column (Empore, 3M) and labelled with TMT 10plex Isobaric Label Reagent Set (Thermo Fisher Scientific) as per the manufacturer's instructions. After labelling, the peptides from the 9 samples were pooled together in equal proportion. The pooled sample was fractionated using high pH reverse-phase chromatography on an XBridge peptide BEH column (130 Å, 3.5 μm, 2.1 × 150 mm, Waters) on an Ultimate 3000 HPLC system (Thermo Scientific/Dionex). Buffers A (10 mM ammonium formate in water, pH 9) and B (10 mM ammonium formate in 90% acetonitrile, pH 9) were used over a linear gradient of 5% to 60% buffer B over 60 min at a flow rate of 200 μL/min. 80 fractions were collected before concatenation into 20 fractions based on the UV signal of each fraction. All the fractions were dried in a Genevac EZ-2 concentrator and resuspended in 1% formic acid for MS analysis.
nLC-MS/MS analysis
The fractions were analyzed sequentially on a Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific) coupled to an UltiMate 3000 RSLCnano UHPLC system (Thermo Scientific) and EasySpray column (75 μm × 50 cm, PepMap RSLC C18 column, 2 μm, 100 Å, Thermo Scientific). Buffers A (0.1% formic acid in water) and B (0.08% formic acid in 80% acetonitrile) were used over a linear gradient from 5% to 35% buffer B over 125 min at 300 nL/min. The column temperature was 50 °C. The mass spectrometer was operated in data dependent mode with a single MS survey scan from 335-1600 m/z followed by 15 sequential m/z dependent MS2 scans. The 15 most intense precursor ions were sequentially fragmented by higher energy collision dissociation (HCD). The MS1 isolation window was set to 0.7 m/z and the resolution set at 120,000. MS2 resolution was set at 60,000. The AGC targets for MS1 and MS2 were set at 3e6 ions and 1e5 ions, respectively. The normalized collision energy was set at 32%. The maximum ion injection times for MS1 and MS2 were set at 50 ms and 200 ms respectively. The data for Compounds 3 and 4 were obtained using a Q Exactive HF-X Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific) using an MS2 resolution of 45,000.
Peptide and protein identification
The raw MS data files for all 20 fractions were merged and searched against the Uniprot-sprot-Human-Canonical database by Maxquant software 1.6.0.16 for protein identification and TMT reporter ion quantitation. The Maxquant parameters were set as follows: enzyme used Trypsin/P; maximum number of missed cleavages equal to two; precursor mass tolerance equal to 10 p.p.m.; fragment mass tolerance equal to 20 p.p.m.; variable modifications: oxidation (M), acetyl (N-term), deamidation (NQ), Gln -> pyro-Glu (Q N-term); fixed modifications: carbamidomethyl (C). The data was filtered by applying a 1% false discovery rate followed by exclusion of proteins with less than two unique peptides. Quantified proteins were filtered if the absolute fold-change difference between the three DMSO replicates was ≥ 1.5.
Immunoprecipitation-Mass Spectrometry (IP-MS)
MV-4-11 cells in RPMI 1640 (Invitrogen) at ~1 × 106 cells/mL were treated with 0.1% DMSO, 1 µM ACBI2 or 1 µM cis-ACBI2 for 8 h and 18 h. Treatment was performed as three biological repeats and administered to pooled cells which were then seeded into separate flasks for each time point. At the indicated time points, cells were centrifuged at 250 × g for 5 min, washed with PBS and centrifuged again. Cell pellets were snap frozen and stored at -80 °C. Each IP 2 µg anti-SMARCA2 antibody (Santa Cruz, sc-17838x) or 1 µg anti-ARID1A antibody (SIGMA, HPA005456) was DMP-crosslinked to Dynabeads Protein G or A (Invitrogen, 10004D and 10002D, respectively). Mouse or rabbit IgG was used to establish specificity of the bait antibody as shown in the supplementary figure 7. Cells were lysed on ice in buffer containing 50 mM TRIS HCl pH 7.5, 150 mM NaCl, 0.5% C7BzO and protease inhibitors for 30 min and spun at 21,000 × g for 15 min at 4 °C. 750 µg of protein lysate, as determined by Bradford assay (Thermo Scientific), was incubated with crosslinked beads for 4 h at 4 °C. IPs were washed three times with 50 mM TRIS HCl pH 7.5, 750 mM NaCl and eluted with 7.5% SDS, 10 mM TCEP, 50 mM triethylammonium bicarbonate buffer pH 8.5 (TEAB) for 20 min at 65 °C. Alkylation was performed for 15 min with 20 mM iodoacetamide at 25 °C in the dark and proteins were cleaned up using the SP3 (Sera-Mag, VWR) procedure as described but using 300 µg of stock beads per sample53. Elution was performed in 50 mM TEAB and digestion with 1 µg Trypsin (Pierce) per sample overnight at 37 °C. Resulting peptides were cleaned up according to the SP3 protocol and eluted into 2% DMSO in water and vacuum dried. Liquid Chromatography–Mass Spectrometry (LC-MS): Data dependent acquisition was carried out on the Orbitrap Velos in CID mode. The top 15 precursors were selected for MS2 over the mass range of 335-1800 (m/z) at a resolution of 60,000. MS2 scans were acquired with a minimum charge state of 2, a normalized collision energy of 35%, an activation time of 10 ms. Data searching and analysis: The data were processed using MaxQuant,54 version 1.5.0.25 and the Andromeda search engine with the following parameters55: proteins and peptides were identified using the human UniProt database (downloaded on 07.01.2018) and peptides were searched with a fixed cysteine carbamidomethyl modification and variable methionine oxidation and N-terminal acetylation modifications and matched between runs. T-test differences between means for protein abundance of cells treated with ACBI2 or cis-ACBI2 were calculated using LFQ intensities and plotted vs. log 10 P values determined by two-tailed t-test (Perseus Software1.6.1.3). To test for specificity of IPs, t-test difference of IPs vs. the IgG controls were plotted vs. the log P value calculated using the means of LFQ intensities. Each experiment was performed in triplicate except the ARID1A-IP at 8 hr treatment time which was performed in quadruplicate.
Statistical methods
No statistical methods were used to predetermine sample size, experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. Number of replicates, mean value and error values are described in the respective figure legends and/or methods. Error bars are show for all data points with replicates as a measure of variation within the group. All t-tests performed were two-tailed t-tests assuming equal variances.
Supplementary Material
Acknowledgements
This project has received funding from Boehringer Ingelheim. A.C. was partially supported by a European Research Council (ERC) Starting Grant (grant agreement no. ERC-2012-StG-311460 DrugE3CRLs). Biophysics, drug discovery and proteomics/computing activities at Dundee were partially supported by Wellcome Trust strategic awards 100476/Z/12/Z, 094090/Z/10/Z and 097945/C/11/Z, respectively. We are thankful to D. Covini for synthesis support. G. Glendinning and S. Mayer for compound logistics. D. Haering and A. Weiss for help with cooperativity and protein degradation measurements. S. Winkler and M. Scharnweber for help with SPR measurements. K. Mayr for LogD measurements. S. Doebel, G. Flotzinger, G. Siszler and P. Werni for protein production and purification. P. Fyfe for support with the in-house X-ray facility. M. Gadd for the gift of the VHL R69A construct. D. Lamont for assistance in proteomics. V. Vetma for data analysis. The Diamond Light Source for beamtime (BAG proposal MX14980) and for beamline support at beamline IO4-1 and I24.
Footnotes
Data availability
Atomic coordinates and structure factors for new protein structures SMARCA2BD: (1), SMARCA2BD: (2):VCB, SMARCA2BD:compound (3):VCB and SMARCA4BD: (3):VCB have been deposited in the Protein Data Bank (PDB) under accession numbers 6HAZ, 6HAY, 6HAX and 6HR2 respectively. All data generated and analyzed during this study are included in this published article and its supplementary information or are available from the corresponding authors upon reasonable request.
Author Contributions
AC, MP and MK conceived the idea. AC, WF, MK, SS, MP, DM and PE directed the project. WF, SS and CD designed compounds. MK, AC, WF, TOH, CW, NT, NW, ED, HW, CD, BS, SW and MR designed experiments and interpreted data. ED, NT, JKO, OP and PG synthesized compounds. Crystallography was performed by MR, GBader and AWP. GBoehmelt helped in the design of a cell line used for testing PROTAC induced protein degradation. Protein production was performed and supervised by MR and AZ. TGerstberger supervised TR-FRET competition experiments, BS performed TR-FRET competition experiments. KR supervised SPR experiments, MG performed SPR experiments. ET and CW performed degradation studies supervised by RS and MK. TGmaschitz, JW, KE and CR performed over-expression studies, proliferation, degradation and apoptosis assays. MR and DZ developed and performed FP cooperativity assay and MR performed the ITC cooperativity assays. CW developed and performed the cell lysate AlphaLISA assay. NT and NW performed MS proteomics. MYW developed IP-MS protocols. JR and HA supervised in vitro DMPK experiments. WF and MK wrote the manuscript with input from all other authors.
Competing financial interests
A.C. is a scientific founder, director and shareholder of Amphista Therapeutics, a company that is developing targeted protein degradation therapeutic platforms.
References
- 1.Toure M, Crews CM. Small-molecule PROTACS: New approaches to protein degradation. Angewandte Chemie International Edition. 2016;55:1966–1973. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- 2.Collins I, Wang H, Caldwell JJ, Chopra R. Chemical approaches to targeted protein degradation through modulation of the ubiquitin–proteasome pathway. Biochemical Journal. 2017;474:1127–1147. doi: 10.1042/BCJ20160762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes Scott J, Ciulli A. Molecular recognition of ternary complexes: A new dimension in the structure-guided design of chemical degraders. Essays In Biochemistry. 2017;61:505–516. doi: 10.1042/EBC20170041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bondeson DP, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nature Chemical Biology. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter GE, et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zengerle M, Chan K-H, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chemical Biology. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadd MS, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nature Chemical Biology. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowak RP, et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nature Chemical Biology. 2018;706 doi: 10.1038/s41589-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bondeson DP, et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chemical Biology. 2018;25:78–87.e75. doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gechijian LN, et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nature Chemical Biology. 2018;14:405–412. doi: 10.1038/s41589-018-0010-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassi ZI, et al. Modulating PCAF/GCN5 Immune Cell Function through a PROTAC Approach. ACS Chemical Biology. 2018;13:2862–2867. doi: 10.1021/acschembio.8b00705. [DOI] [PubMed] [Google Scholar]
- 12.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Science Advances. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St Pierre R, Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev. 2017;42:56–67. doi: 10.1016/j.gde.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature Genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodges C, Kirkland JG, Crabtree GR. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harbor Perspectives in Medicine. 2016;6:a026930. doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman GR, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proceedings of the National Academy of Sciences. 2014;111:3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oike T, et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013;73:5508–5518. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 20.Wilson BG, et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol Cell Biol. 2014;34:1136–1144. doi: 10.1128/MCB.01372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papillon JPN, et al. Discovery of orally active inhibitors of brahma homolog (BRM)/SMARCA2 ATPase activity for the treatment of brahma related gene 1 (BRG1)/SMARCA4-mutant cancers. Journal of Medicinal Chemistry. 2018;61:10155–10172. doi: 10.1021/acs.jmedchem.8b01318. [DOI] [PubMed] [Google Scholar]
- 22.Sutherell CL, et al. Identification and development of 2,3-dihydropyrrolo[1,2-a]quinazolin-5(1H)-one inhibitors targeting bromodomains within the switch/sucrose nonfermenting complex. Journal of Medicinal Chemistry. 2016;59:5095–5101. doi: 10.1021/acs.jmedchem.5b01997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerstenberger BS, et al. Identification of a chemical probe for family VIII bromodomains through optimization of a fragment hit. Journal of Medicinal Chemistry. 2016;59:4800–4811. doi: 10.1021/acs.jmedchem.6b00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vangamudi B, et al. The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF-mutant cancers: Insights from cDNA rescue and PFI-3 inhibitor studies. Cancer Research. 2015;75:3865–3878. doi: 10.1158/0008-5472.CAN-14-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remillard D, et al. Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angewandte Chemie International Edition. 2017;56:5738–5743. doi: 10.1002/anie.201611281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoppi V, et al. Iterative Design and Optimization of Initially Inactive Proteolysis Targeting Chimeras (PROTACs) Identify VZ185 as a Potent, Fast, and Selective von Hippel–Lindau (VHL) Based Dual Degrader Probe of BRD9 and BRD7. Journal of Medicinal Chemistry. 2019;62:699–726. doi: 10.1021/acs.jmedchem.8b01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin C, et al. Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression. Journal of Medicinal Chemistry. 2018;61:6685–6704. doi: 10.1021/acs.jmedchem.8b00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albrecht BK, et al. WO2016138114 WIPO patent. 2016
- 29.Myrianthopoulos V, et al. Discovery and optimization of a selective ligand for the switch/sucrose nonfermenting-related bromodomains of polybromo protein-1 by the use of virtual screening and hydration analysis. Journal of Medicinal Chemistry. 2016;59:8787–8803. doi: 10.1021/acs.jmedchem.6b00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soares P, et al. Group-based optimization of potent and cell-active inhibitors of the von Hippel–Lindau (VHL) E3 ubiquitin ligase: Structure–activity relationships leading to the chemical probe (2S,4R)-1-((S)-2-(1-cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298) Journal of Medicinal Chemistry. 2017;61:599–618. doi: 10.1021/acs.jmedchem.7b00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckley DL, et al. HaloPROTACS: Use of small molecule PROTACs to induce degradation of halotag fusion proteins. ACS Chemical Biology. 2015;10:1831–1837. doi: 10.1021/acschembio.5b00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniaci C, et al. Homo-PROTACs: Bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nature Communications. 2017;8:830. doi: 10.1038/s41467-017-00954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan K-H, Zengerle M, Testa A, Ciulli A. Impact of target warhead and linkage vector on inducing protein degradation: Comparison of bromodomain and extra-terminal (BET) degraders derived from triazolodiazepine (JQ1) and tetrahydroquinoline (I-BET726) BET inhibitor scaffolds. Journal of Medicinal Chemistry. 2017;61:504–513. doi: 10.1021/acs.jmedchem.6b01912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Molle I, et al. Dissecting fragment-based lead discovery at the von Hippel-Lindau protein:Hypoxia inducible factor 1α protein-protein interface. Chemistry & Biology. 2012;19:1300–1312. doi: 10.1016/j.chembiol.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 36.Zorba A, et al. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proceedings of the National Academy of Sciences. 2018;115:E7285–E7292. doi: 10.1073/pnas.1803662115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao K, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 38.Qiu Z, Ghosh A. A calcium-dependent switch in a CREST-BRG1 complex regulates activity-dependent gene expression. Neuron. 2008;60:775–787. doi: 10.1016/j.neuron.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald ER, et al. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell. 2017;170:577–592.e510. doi: 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Tsherniak A, et al. Defining a cancer dependency map. Cell. 2017;170:564–576.e516. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyes JC, et al. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) The EMBO Journal. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Molecular Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 43.Fedorov O, et al. Selective targeting of the BRG/PB1 bromodomains impairs embryonic and trophoblast stem cell maintenance. Science Advances. 2015;1:e1500723. doi: 10.1126/sciadv.1500723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vonrhein C, et al. Data processing and analysis with the auto PROC toolbox. Acta Crystallographica Section D Biological Crystallography. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project, N. The CCP4 suite: Programs for protein crystallography. Acta Crystallographica Section D Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D Biological Crystallography. 2009;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruno IJ, et al. Retrieval of crystallographically-derived molecular geometry information. Journal of Chemical Information and Computer Sciences. 2004;44:2133–2144. doi: 10.1021/ci049780b. [DOI] [PubMed] [Google Scholar]
- 50.Kabsch W. Xds. Acta Crystallographica Section D Biological Crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 53.Hughes CS, et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Molecular Systems Biology. 2014;10:757–757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 55.Cox Jr, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. Journal of Proteome Research. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.