Significance
Transport of specific mRNAs to defined sites in the cytoplasm allows local protein production and contributes to cell polarity, embryogenesis, and neuronal function. These localized mRNAs contain signals (i.e., zipcodes) that help direct them to their destination site. Zipcodes are recognized by RNA-binding proteins that, with the help of molecular motor proteins and supplementary factors, mediate mRNA trafficking. To identify all proteins assembling with a localized mRNA, we advanced a proximity labeling method, BioID, by tethering a biotin ligase to the 3′ UTR of mRNA encoding the conserved β-actin protein. We demonstrate that this method allows the identification of functionally important proteins required for mRNA localization.
Keywords: RNA-BioID, mRNA localization, RNA-binding protein, FUBP3
Abstract
The molecular function and fate of mRNAs are controlled by RNA-binding proteins (RBPs). Identification of the interacting proteome of a specific mRNA in vivo remains very challenging, however. Based on the widely used technique of RNA tagging with MS2 aptamers for RNA visualization, we developed a RNA proximity biotinylation (RNA-BioID) technique by tethering biotin ligase (BirA*) via MS2 coat protein at the 3′ UTR of endogenous MS2-tagged β-actin mRNA in mouse embryonic fibroblasts. We demonstrate the dynamics of the β-actin mRNA interactome by characterizing its changes on serum-induced localization of the mRNA. Apart from the previously known interactors, we identified more than 60 additional β-actin–associated RBPs by RNA-BioID. Among these, the KH domain-containing protein FUBP3/MARTA2 has been shown to be required for β-actin mRNA localization. We found that FUBP3 binds to the 3′ UTR of β-actin mRNA and is essential for β-actin mRNA localization, but does not interact with the characterized β-actin zipcode element. RNA-BioID provides a tool for identifying new mRNA interactors and studying the dynamic view of the interacting proteome of endogenous mRNAs in space and time.
The spatial distribution of mRNAs contributes to the compartmentalized organization of the cell and is required for maintaining cellular asymmetry, proper embryonic development, and neuronal function (1). Localized mRNAs contain cis-acting sequences, termed zipcodes or localization elements, that constitute binding sites for RNA-binding proteins (RBPs) (1). Together with these RBPs, localized mRNAs form transport complexes containing molecular motors, such as kinesin, dynein, and myosin (2, 3). These ribonucleoprotein complexes (RNPs) usually include accessory factors, such as helicases, translational repressors, RNA stability factors, and ribosomal proteins (3). Thus, mRNPs as functional units not only contain the information for an encoded polypeptide, but also determine the precise spatiotemporal regulation of the polypeptide’s translation and stability, thereby facilitating proper subcellular localization of the translation product (4).
One of the best-studied localized mRNAs is β-actin, which encodes the β isoform of the cytoskeleton protein actin (5). β-Actin mRNA is localized to the protrusion of migrating fibroblasts (6), where its local translation critically contributes to the migrating behavior of this cell type (7–11). In the developing mouse (12) and Xenopus (13, 14) neurons, β-actin mRNA is transported to the growth cone during axonal extension, and its deposition and local translation are highly regulated by external cues. In addition, translation of this mRNA in dendritic spines is involved in reshaping the postsynaptic site of synapses (14). A well-defined localization element is present in the proximal region of the β-actin 3′-untranslated region (UTR) (15). This cis-acting element is recognized and bound by the zipcode-binding protein ZBP1 (16), the founding member of the conserved VICKZ RBP family (17). ZBP1 (also called IGF2BP1 or IMP1) interacts with the β-actin zipcode via the third and fourth KH (hnRNP K homology) domains (16) and is required for RNA localization in fibroblasts and neurons (18). It has also been suggested that IGF2BP1 controls the translation of β-actin mRNA by blocking the assembly of ribosomes at the start codon (11). IGF2BP1 appears to act as a key RBP in β-actin mRNA distribution, but other proteins, including IGF2BP2 (19), RACK1 (20), KHSRP/FUBP2 (21), KHDRBS1/SAM68 (22), FMR1 (23), and HuR (24), also have been suggested to be involved in β-actin mRNA localization, although their molecular function is less clear.
To fully understand the mechanism(s) of mRNA localization, it is important to identify and study the mRNA-binding factors. Major technological advances, such as cross-linking and immunoprecipitation (CLIP) combined with next-generation sequencing, have allowed the identification of RNAs bound to specific RBPs (25) and the system-wide identification of RBPs bound to polyA RNA (26, 27). However, the major techniques for determining which proteins associate with a specific RNA include affinity purification of modified or tagged RNAs together with their bound proteins, along with coimmunoprecipitation (co-IP) of RNP components with the aid of known RBPs (28). In addition, affinity capturing of specific RNPs with hybridizing antisense probes or via integrated aptamers has been successful (29–31). A limitation of these techniques is the potential loss of low-affinity binders during purification, which so far has been addressed by in vivo UV cross-linking before cell lysis (25, 26). However, cross-linking enhances only the recovery of RBPs directly contacting nucleobases and thus does not overcome the loss of other physiologically important RNA interactors (e.g., motor or adapter proteins). These limitations could be overcome by in vivo labeling of proteins while they are associated with the target RNA.
Proximity-dependent biotin identification, or BioID (32–34), has been successfully used to detect subunits of large or dynamic protein complexes, such as the nuclear pore complex (32) and centrosome (34). In BioID, a protein of interest is fused to a mutant version of the Escherichia coli biotin ligase BirA (BirA*) that generates AMP biotin (“activated biotin”), which reacts with accessible lysine residues in its vicinity (33). After cell lysis, biotinylated proteins can be isolated via streptavidin affinity purification and identified using standard mass spectrometry techniques. Recently, BioID has also been applied to identify proteins associated with the genomic RNA of Zika virus (35).
In this study, we used BioID to characterize the proteome of endogenous β-actin mRNPs. We found that tethering of BirA* to an endogenous transcript not only allows identification of its associated proteins, but also can be used to probe the environment of this mRNA. We identified FUBP3/MARTA2, an RBP from the conserved FUBP family of proteins (36–38), which was previously shown to mediate dendritic targeting of MAP2 mRNA in neurons (39, 40). We found that FUBP3 binds to and facilitates localization of β-actin mRNA to the fibroblast leading edge. FUBP3 does not bind to the zipcode or IGF2BP1, but mediates β-actin RNA localization by binding to a distal site in its 3′ UTR. Therefore, the RNA-BioID approach allows the identification of novel functional mRNA interactors within the cell with high confidence.
Results
Tethering Biotin Ligases to the 3′ UTR of β-Actin mRNA.
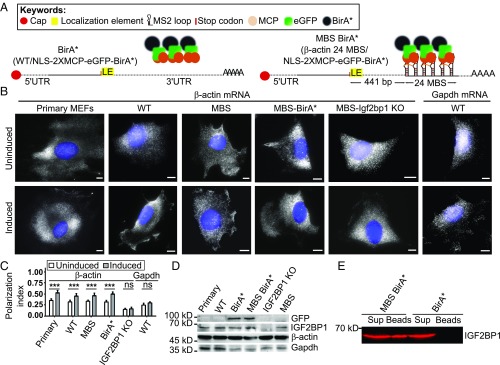
To tether BirA* to the 3′ UTR of β-actin mRNA (Fig. 1A), we stably expressed a fusion of the nuclear localized signal (NLS), MS2 coat protein (MCP) (41), GFP, and BirA* (MCP-GFP-BirA*) in immortalized mouse embryonic fibroblasts (MEFs) from transgenic β-actin-24 MBS mice (Fig. 1 A, Right) (8). These mice have both β-actin gene copies replaced by β-actin with 24 MS2 binding sites (MBS) in their distal 3′-UTR. In parallel, NLS-MCP-GFP-BirA* was stably expressed in WT (wildtype) MEFs with untagged β-actin mRNA, to generate a control cell line to eliminate background biotinylation due to the presence of constitutive expression of BirA* (Fig. 1 A, second left panel). Both constructs contain two copies of the MCP protein leading to a maximum of 24 GFP and 24 BirA* that can potentially bind to an mRNA. Since biotinylation or the expression of the MCP-GFP-BirA* might affect localization of the β-actin mRNA, we checked for the proper targeting of β-actin mRNA to the leading edge of the cell by single molecule fluorescent in situ hybridization (smFISH) (42) and analyzed RNA localization by polarization index calculation (9) (Fig. 1 B and C and SI Appendix, Fig. S1 A–F). The distribution of mRNAs within cells was assessed using probes against the β-actin ORF (for primary and immortalized MEFs) and β-actin–MBS (for the genetically modified immortalized MEFs: β-actin–MBS, or β-actin–MBS IGF2BP1 KO) (10). To account for random distribution of an mRNA within the cell, we used probes against Gapdh as a control. Gapdh mRNA is a highly abundant and uniformly distributed mRNA. To induce β-actin mRNA localization, cells were serum starved for 24 h followed by stimulation with serum addition for 1 h. The median of the polarization index of β-actin mRNA distribution was significantly lower in immortalized (WT) or genetically modified immortalized MEFs compared with primary MEFs (Fig. 1C). Stimulation of polarization by serum was observed for all of the cell types used in a similar manner (Fig. 1 C, gray bars). Also, as shown before (10) knockout of IGF2BP1 reduces significantly β-actin–MBS mRNA polarization (Fig. 1C). We observed that β-actin and Igf2bp1 mRNA or protein levels were not affected (Fig. 1D and SI Appendix, Fig. S2 A and B). Altogether these results suggest that biotinylation and/or the expression of the MCP-GFP-BirA* does not affect regulation of β-actin mRNA in MEFs. Furthermore, cells with similar expression levels of MCP-GFP-BirA* were sorted by FACS (fluorescence activated cell sorting). As shown before (43), we also found no differences in the biotinylation efficiency at labeling conditions of 50 µM to 300 µM of biotin for 6–48 h. For optimal biotinylation, we decided to perform proximity labeling by addition of 50 µM biotin to the medium for 24 h. To test if proximity labeling can identify known β-actin mRNA-associated proteins, we affinity purified biotinylated proteins followed by Western blot detection of IGF2BP1 (mouse ZBP1). IGF2BP1 was biotinylated in MEFs expressing β-actin–MBS/MCP-GFP-BirA* but not in those expressing only GFP-BirA* (Fig. 1E), which demonstrates that our tool can successfully biotinylate zipcode-interacting proteins. To differentiate between endogenously biotinylated proteins and RNA-dependently biotinylated proteins, we performed streptavidin pulldown in cells expressing β-actin–MBS/MCP-GFP-BirA* and in cells expressing only MCP-GFP and observed biotinylation of numerous additional proteins (SI Appendix, Fig. S3). We expected that MCP-GFP-BirA* represents a major fraction of these biotinylated proteins and therefore aimed at depleting the fusion protein from the lysate by GFP pulldown before streptavidin affinity purification. To our surprise, most of the biotinylated proteins were enriched in the GFP pulldown fraction (SI Appendix, Fig. S3), likely due to copurification of MCP-GFP-BirA*, β-actin mRNA, and biotinylated proteins via binding to the mRNA or the fusion protein. RNA degradation with RNase A (SI Appendix, Fig. S4) shifted a large part of the biotinylated proteins into the streptavidin fraction (SI Appendix, Fig. S3), supporting the idea that most of the biotinylated proteins are associated with β-actin mRNA. Additional treatment with high salt and 0.5% SDS further optimized the streptavidin affinity purification and decreased the background binding of the magnetic beads used in this purification (SI Appendix, Fig. S3).
Fig. 1.
RNA BioID to detect proteins interacting with localized β-actin RNA. (A) Schematic of the β-actin–MBS/GFP-BirA*. (Left) Control construct (BirA*) used to detect background biotinylation due to overexpression of the NLS-MCP-GFP-BirA* construct. Control cells expressing only NLS-2xMCP-eGFP-BirA* lack the MBS cassette in the β-actin mRNA. (Right) Construct used to detect β-actin mRNA-associated proteins (β-actin–MBS-BirA*). A 24xMS2 aptamer array (24MBS) was integrated in the 3′ UTR of the endogenous β-actin gene 441 bp downstream of the stop codon. BirA* is targeted to 24MBS by its fusion to a MS2 coat protein dimer (2xMCP). (B) Representative β-actin smFISH images of (from left to right) primary MEFs, immortalized MEFs (WT), β-actin–MBS, β-actin–MBS BirA*, and β-actin–MBS Igf2bp1 KO MEFs, as well as Gapdh smFISH images in immortalized (WT) MEFs (rightmost images). These and similar images were used to calculate the polarization index (C) of mRNA localization under serum-uninduced (Top) and serum-induced (Bottom) conditions. β-Actin mRNA was detected by probes against the β-actin ORF or MBS region, and Gapdh mRNA was detected by probes against its ORF (gray). (Scale bar: 10 µm.) (C) Bar graphs of the polarization index for Gapdh mRNA and β-actin mRNA in different MEFs [from left to right: primary, immortalized (WT), β-actin–MBS, β-actin–MBS BirA*, β-actin–MBS Igf2bp1 KO]. The polarization index was calculated in a total 100 of cells from three biological replicates. The line represents the median values. ***P < 0.005; not significant (ns), P > 0.05. (D) Protein levels of endogenous β-ACTIN, IGF2BP1, and heterologous MCP-GFP-BirA* detected by anti-GFP antibody. Quantification of Western blot analysis is provided in SI Appendix, Fig. S2. (E) Biotinylation of IGF2BP1 depends on MBS sites in β-actin. Following RNase A treatment, biotinylated proteins were affinity-purified with streptavidin-coated beads from cells expressing 2xMCP-eGFP-BirA* in the presence (β-actin-24MBS) or absence (β-actin) of MBS. The presence of IGF2BP1 was probed by a specific antibody in bead fractions (Beads) and supernatant (Sup).
β-Actin mRNA Interactors Under Serum-Induced and Uninduced Conditions.
β-Actin mRNA localization to the lamellipodia of chicken and mouse fibroblasts is known to increase after serum induction (6, 44). It also has been shown that cells enter a quiescent phase of the cell cycle during serum starvation (6), involving an overall reduction in actin stress fibers or focal adhesions (44). Since efficient biotinylation requires at least 6 h of incubation with biotin, we next applied smFISH to verify the persistence of β-actin mRNA localization during our labeling period. As has been shown previously (5), MEFs induced β-actin mRNA localization after serum addition (Fig. 1 B and C), and the fraction of MEFs with β-actin localized to lamellipodia increased within 1 h but then remained constant over the next 6 h.
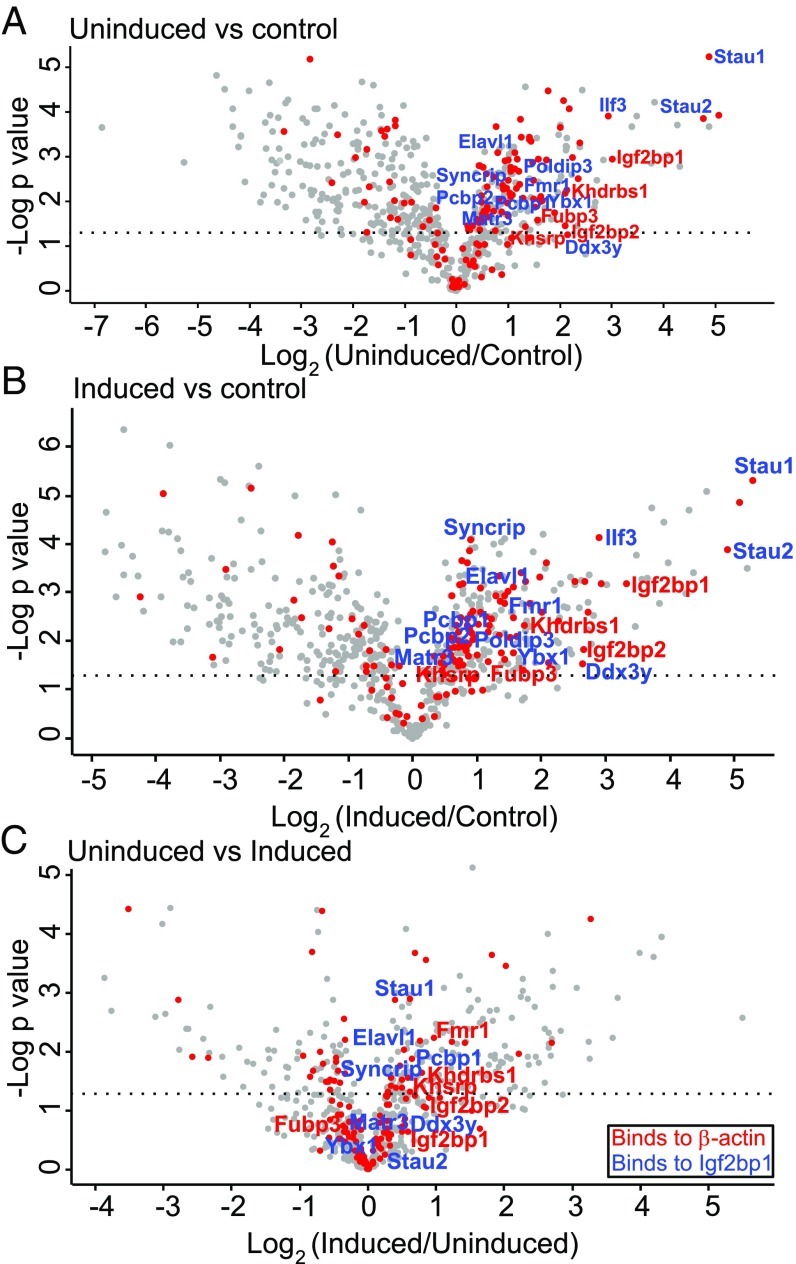
To identify and compare the β-actin–associated proteomes in uninduced and serum-induced MEFs, we performed RNA-BioID under both conditions (three replicate experiments each). Unspecific as well as endogenous biotinylation was assessed by performing BioID in MEFs expressing MCP-GFP-BirA* in the absence of MS2 aptamers in β-actin mRNA. Affinity-captured biotinylated proteins were identified and quantified by mass spectrometry using label-free quantification. Principal component analysis of the datasets revealed that the different conditions cluster apart from each other in dimensions 1 and 2 (explaining 33.8% and 15.5% of the variance, respectively), while the replicates with the same conditions cluster together, demonstrating biological reproducibility (SI Appendix, Fig. S5). Calculating the Spearman correlation between all sample types and replicates (SI Appendix, Fig. S6) supports the high reproducibility between biological replicates (correlation ≥0.97). In addition, it showed better correlation between uninduced and induced samples (average 0.95) compared with controls. In total, we found 169 (or 156) significantly enriched proteins in induced (or uninduced) MEFs compared with control cells (SI Appendix, Figs. S7 and S8A). Of these, 47 were enriched only under induced conditions (SI Appendix, Table S5). To assess the differential enrichment of the proteins under each condition, a Tukey post hoc test was performed after ANOVA, and significance was set to an adjusted P value of 0.05 following Benjamini–Hochberg multiple correction testing (Materials and Methods). Large fractions of the enriched proteins under induced conditions (30%) or uninduced conditions (34%) over control represent RBPs (Fig. 2, red solid circles); among these are RBPs (IGF2BP1, IGF2BP2, KHSRP, KHDRBS1, FMR1, HuR, RACK1, named in red) already known to control specific aspects of β-actin mRNA physiology. Other enriched RBPs have been associated with the localization of mRNAs in other cell types or organisms, including STAU1 and STAU2 (45–47), SYNCRIP (48), and FUBP3 (38). Furthermore, 85 proteins were significantly more enriched under serum-induced conditions than under uninduced conditions (SI Appendix, Fig. S8). However, the majority of the aforementioned RBPs (including IGF2BP1) become biotinylated under both induced and uninduced conditions, indicating that they are associated with β-actin mRNA under both conditions (Fig. 2C).
Fig. 2.
Enrichment of biotinylated proteins in control MEFs, or MEFs expressing β-actin–MBS-BirA* under serum-induced or uninduced conditions. Volcano plot representation of biotinylated proteins in uninduced MEFs compared with control MEFs (A), serum-induced MEFs compared with control MEFs (B), and serum-induced MEFs compared with uninduced MEFs (C). In the volcano plots, the x-axis represents log2 fold change in protein abundance and the y-axis represents the −log10 P value. Red circles are known RBPs identified by Gene Ontology (GO) molecular function analysis. Proteins in red represent known β-actin mRNA interactors, and proteins in blue are RBPs known to bind to IGF2BP1. The dotted line indicates P = 0.05.
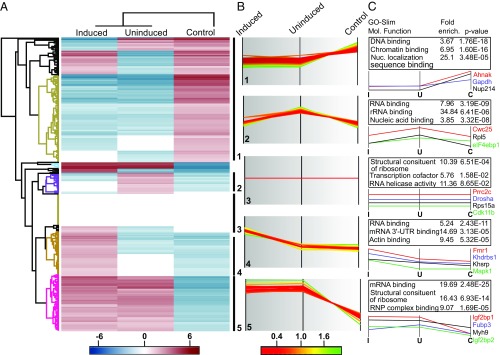
A cluster analysis (Fig. 3) reveals at least five different patterns of biotinylated proteins in induced, noninduced, and control MEFs (Fig. 3 B and C). In control MEFs, we see enrichment of mainly nuclear proteins (cluster 1). This is expected, since the unbound MCP-GFP-BirA* is enriched in the nucleus due to an N-terminal nuclear localization sequence (8). Cluster 1 also contains abundant cytoplasmic proteins, including glycerol aldehyde phosphate dehydrogenase (GAPDH). Cluster 3 represents proteins found equally in MEFs under all conditions and contains ribosomal proteins, among others. Proteins allocated to the other three clusters (clusters 2, 4, and 5) are overrepresented in the biotinylated proteome of MEFs expressing β-actin–MBS/GFP-BirA*. Of specific interest are clusters 4 and 5. In cluster 4, with proteins that are more biotinylated under serum-induced conditions, we find RBPs, including FMR1 and KHSRP, that have been reported to function in β-actin mRNA localization or to bind to IGF2BP1.
Fig. 3.
Cluster analysis of biotinylated proteins in control MEFs or MEFs expressing β-actin–MBS-BirA* under serum-induced and uninduced conditions. (A) Hierarchical clustering of biotinylated proteins in serum-induced and uninduced β-actin–MBS-BirA* MEFs and control MEFs (lacking β-actin–MBS). Enrichment is indicated in red; depletion, in blue. Various clusters of protein groups are highlighted in the dendrogram. (B) Profile plots of five selected clusters showing distinct enrichment patterns of biotinylated proteins: 1, strongly enriched in control MEFs; 2, enriched in β-actin–MBS-BirA* MEFs under uninduced conditions; 3, similar enrichment in all MEFs; 4, enriched in β-actin–MBS-BirA* MEFs under serum-induced conditions; and 5, enriched in β-actin–MBS-BirA* MEFs under serum-induced and uninduced conditions compared with control MEFs. Color-coding shows the degree of enrichment in each specific cluster: green, more enriched; red, less enriched). (C) Functional analysis of protein annotation terms results in multiple categories that are enriched in the selected clusters. GO-slim molecular function terms, the corresponding enrichment factors, and P values are shown in the table. Selected examples of proteins found in each cluster are shown below the tables.
Another group of proteins that are enriched in this cluster comprises proteins of the actin cytoskeleton (e.g., Filamin B, Cofilin-1, Myh9, Tpm4, Plastin-3). Their enrichment likely reflects deposition of the β-actin mRNA in the actin-rich cortical environment of the leading edge of MEFs. Finally, cluster 5 contains proteins found in β-actin–MBS MEFs under both induced and uninduced conditions but not in control MEFs. This cluster shows enrichment for proteins involved in mRNA-binding, RNP constituents, and ribosomal proteins. Since this cluster contains the RBP IGF2BP1, we hypothesized that other proteins in this cluster, such as FUBP3, are likely candidates for β-actin mRNA regulatory factors.
FUBP3 Is a Component of the β-Actin mRNP.
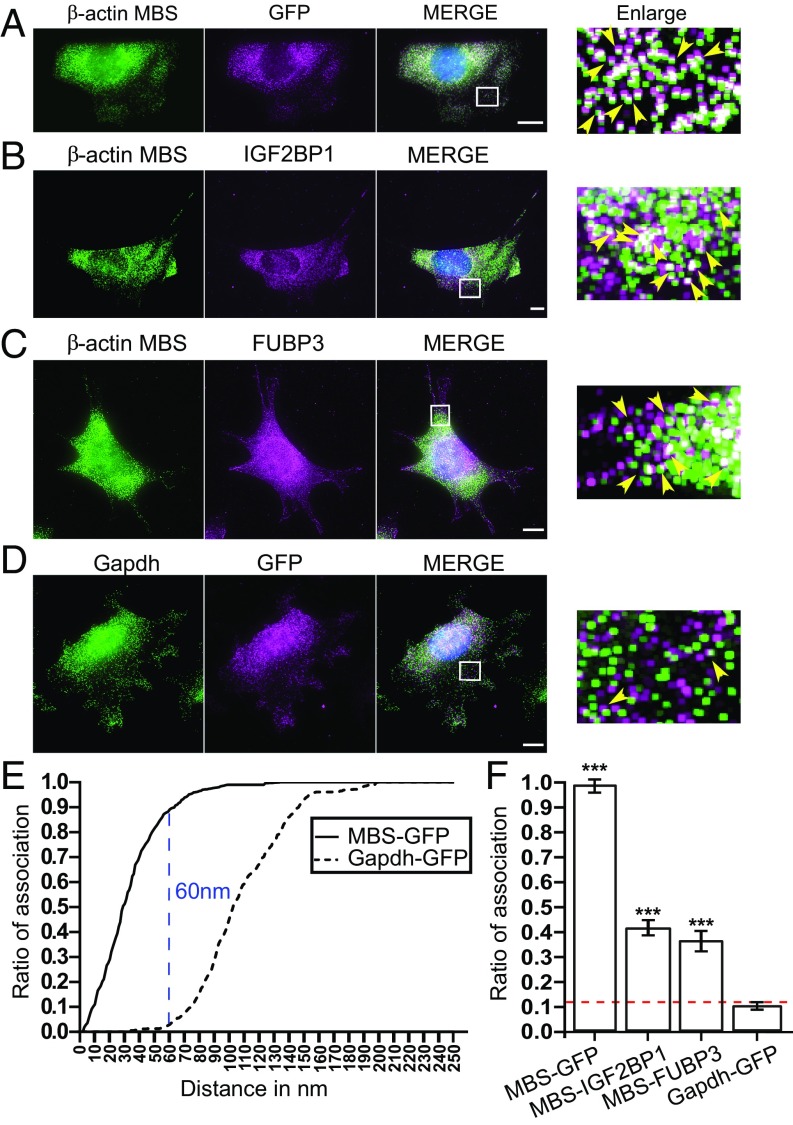
To confirm the association of the identified proteins and MS2-tagged β-actin mRNA, we combined single-molecule fluorescence in situ hybridization with immunofluorescence (smFISH-IF) using Cy3-labeled probes against either the ORF or the MBS of β-actin mRNA and antibodies against GFP, FUBP3, or IGF2BP1 in WT or β-actin–MBS MEFs (Fig. 4 A–C). While ORF probes were used to detect β-actin mRNA in WT MEFs, MBS probes against the MS2 loop sequences were used to detect the β-actin mRNA in β-actin–MBS MEFs. The association between β-actin mRNA and the proteins was determined by super-registration microscopy (47). In brief, we corrected the images for chromatic aberration and mechanical shifts in Cy3 and Cy5 channels using broad spectra fluorescent microsphere beads (SI Appendix, Fig. S9) and found that colocalization of smFISH and IF signals did not occur by chance within the cell using a positive control (MBS-GFP; Fig. 4A) and a negative control (Gapdh-GFP; Fig. 4D) for RNA–protein interaction. We calculated the association between the RNA and protein molecules as a function of their distances apart for positive and negative controls (Fig. 4E). For the positive control, 91% of the observed distances from the labeled probes to the MBS and from the antibodies to the GFP were within 60 nm (the optimal distance). In contrast, only 10% of the observed associations in the negative control (using Gapdh probes and MCP-GFP) were within 60 nm (Fig. 4 E and F). When combining smFISH of Gapdh with IF against MCP-GFP, fewer overlapping events were observed at a distance of <150 nm compared with MBS-GFP (Fig. 4 A, D, and E). At greater distances (>150 nm), the fluorescence signals in both channels were more likely to overlap by chance and thus are considered a random event. We found that at the optimal distance of 60 nm, β-actin mRNA was associated with IGF2BP1 and FUBP3 in MEFs. The RNA–protein associations were 37% for IGF2BP1 with β-actin and 29% for FUBP3 with β-actin in MEFs (Fig. 4 B, C, and F). These associations were significantly higher than the nonspecific interaction between Gapdh and MCP-GFP (10%), suggesting the physical contact between the molecules.
Fig. 4.
Association analysis of IGF2BP1 and FUBP3 with β-actin–MBS mRNA by super-registration microscopy. (A–D) Representative smFISH-IF images of MEFs expressing β-actin–MBS and MCP-GFP. Shown are MEFs stained for β-actin mRNA (MBS FISH probes, Cy3; green) and MCP-GFP (A), IGF2BP1 (B), and FUBP3 (C). Immunofluorescence staining is shown in magenta. MCP-GFP served as a positive control to determine the optimum distance between mRNA and protein. D represents staining of Gapdh mRNA (probes from Biosearch, Cy3; Green) together with MCP-GFP and served as a negative control to determine the distance for association between two signals occurring by chance. A 1-pixel dilated, enlarged version is shown on the right side of each panel (47). (E) Association curves between an mRNA (black, β-actin–MBS; dotted, Gapdh) and MCP-GFP protein. The curve of association is calculated as the cumulative ratio of association for intermolecular distances (in the range of 0–250 nm) that were less than a given observed distance, as described previously (48). The blue line represents the distance where the mRNA–protein association for MCP-MBS and MCP-Gapdh is maximally separated, the optimal distance (OD = 60 nm). (F) Summary of association analysis of β-actin mRNA and indicated proteins by smFISH-IF and super-registration. The dotted red line indicates background association defined by MCP-Gapdh. The error bar represents SD. P > 0.05; *P < 0.05; ***P < 0.001, unpaired t test.
FUBP3 and IGF2BP1 Bind on Different Regions of β-Actin mRNA and Interact with Each Other in an RNA-Dependent Manner.
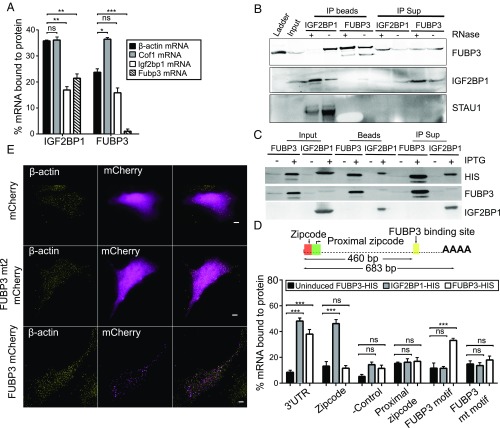
To validate the data demonstrating the RNA–protein association by super-registration microscopy, we performed co-IP of β-actin mRNA with FUBP3 and IGF2BP1 (Fig. 5A). Co-IP was tested with four mRNAs: β-actin, Cofilin1, Igf2bp1, and Fubp3 (Fig. 5A). IGF2BP1 bound to all the mRNAs tested, reflecting previous observations in HeLa cells, where almost 3% of the transcriptome was shown to bind to IGF2BP1 (49). Coprecipitation of β-actin with FUBP3 (23% of input bound to FUBP3) was similar to that with IGF2BP1 (37%). These values are consistent with the degree of RNA–protein association seen on colocalization (Fig. 4). In contrast, β-actin mRNA was not efficiently bound by the RBP VIGILIN, indicating that this mRNA does not associate with every RBP (SI Appendix, Fig. S10). The localized Cof1 mRNA (50) was bound by both FUBP3 and IGF2BP1 to a similar extent (48%).
Fig. 5.
FUBP3 binds to β-actin 3′ UTR. (A) Co-IP of selected mRNAs with IGF2BP1 and FUBP3. Bars represent percentage of input mRNA copurifying with the indicated protein. IGF2BP1 binds to several endogenous mRNAs, including Cofilin1, Igf2bp1, and Fubp3. FUBP3 binds to 23% of endogenous β-actin mRNA, while IGF2BP1 was associated with 37% of endogenous β-actin mRNA. Error bars represent mean ± SEM from three independent experiments. (B) Co-IP of STAU1, FUBP3, and IGF2BP1. Immunoprecipitation was performed from WT MEFs with either anti-FUBP3 or anti-IGF2BP1 antibodies in the presence and absence of RNase A. IGF2BP1 coprecipitates with FUBP3 only in the absence of RNase A, while binding of STAU1 to IGF2BP1 is RNA-independent. (C) Pulldown of His-tagged fusion proteins of IGF2BP1 and FUBP3 from bacterial lysates of E. coli grown under isopropyl β-d-1-thiogalactopyranoside (IPTG)-induced or IPTG-uninduced conditions. Magnetic beads were used to precipitate either IGF2BP1-HIS or FUBP3-HIS. (D, Top) Schematic representation of the 3′ UTR of β-actin mRNA. The 683-bp-long 3′ UTR contains the 54-nt zipcode sequence (after the stop codon), the proximal zipcode sequence (49 bp following the zipcode), and a potential FUBP3-binding sequence (460 bp downstream of the stop codon) with a consensus UAUG motif. (D, Bottom) Binding of in vitro transcribed RNA fragments of β-actin (complete 3′ UTR, zipcode, proximal zipcode, zipcode mutant, FUBP3-binding motif region, region with mutated FUBP3-binding motif) to IGF2BP1 or FUBP3. RNAs were added to E. coli lysates with or without (IPTG-uninduced) expressed His-tagged fusion protein. After affinity purification, bound RNAs were detected by quantitative RT-PCR. Bars represent percentage of input RNA. In contrast to IGF2BP1, FUBP3 shows little affinity for the zipcode sequence but binds to the 3′ UTR and a region containing the UAUG motif in the 3′ UTR. Error bars represent mean ± SEM from three independent experiments. Statistical significance of each dataset was determined using Student’s t test. *P < 0.05; ***P < 0.001; not significant (ns), P > 0.05. (E) RNA-binding domain KH2 is required for FUBP3 cytoplasmic granule formation. The conserved G-X-X-G motif of FUBP3 KH domains were individually mutated into G-D-D-G and WT and mutant proteins expressed in MEFs as mCherry fusion. Live cell imaging shows that WT FUBP3-mCherry forms cytoplasmic granules, whereas a KH2 mutant (FUBP3 mt2) is evenly distributed in the cytoplasm like the control mCherry protein. (Scale bars: 5 μm.)
To further substantiate our finding that FUBP3 can bind independently of IGF2BP1 to β-actin mRNA, we performed co-IP experiments with IGF2BP1 and FUBP3 (Fig. 5B) in the presence and absence of RNase A. The RBP STAU1 served as a positive control since it has been shown to bind to IGF2BP1 (51). Co-IP of IGF2BP1 and FUBP3 vanished on RNase treatment, indicating an RNA-dependent interaction between these two proteins. We conclude that FUBP3 does not bind to β-actin mRNA via IGF2BP1, but that both proteins may bind β-actin independently at different sites.
We next used recombinant histidine-tagged proteins (FUBP3-HIS and IGF2BP1-HIS) in pulldown assays (Fig. 5 C and D) to test binding to in vitro transcribed RNA fragments of β-actin mRNA. We selected the complete 643-bp-long β-actin 3′ UTR and the 54-nt localization zipcode element of β-actin. As negative control for IGF2BP1 binding, we used a mutant version of the zipcode region (16). In addition, we used a 49-nt region adjacent to the zipcode (proximal zipcode; ref. 16). A 79-nt region in the 3′ UTR at 460 nt downstream to the stop codon of β-actin mRNA, which spans a potential FUBP3-binding motif UAUG (52), along with a 75-nt fragment of the same region but carrying a deleted UAUG motif were used to specifically probe FUBP3 binding. The capturing assay was performed in total bacterial lysates to allow bacterial RBPs to compete for RNA binding. RNA captured by the His-tagged fusion proteins was detected by quantitative RT-PCR and normalized to the input (Fig. 5D). We found that IGF2BP1 and FUBP3 were bound to the 3′ UTR of β-actin mRNA, while neither could interact with the mutated zipcode or zipcode proximal region. Only FUBP3 was bound to the 79-nt region containing the UAUG motif on the 3′ UTR of β-actin mRNA, and the binding was abolished in absence of this motif (Fig. 5D). This is highly suggestive of direct binding of FUBP3 to the UAUG motif in the 3′ UTR of β-actin.
To identify the KH domain(s) of FUBP3 responsible for binding β-actin mRNA, we introduced mutations in the conserved KH domains of the protein. Each functionally important G-X-X-G motif in the four KH domains was changed to the inactive G-D-D-G (53), and individual mutant proteins were transiently expressed in MEFs as C-terminally tagged mCherry fusion protein. The G-D-D-G mutation in KH domain 2 resulted in loss of the cytoplasmic punctate signal seen in WT FUBP3, reminiscent of the punctate pattern observed for mRNPs (Fig. 5E). We conclude that KH2 in FUBP3 is important for its integration into RNP particles and likely constitutes the critical domain for RNA binding.
Loss of FUBP3 Affects β-Actin mRNA Localization.
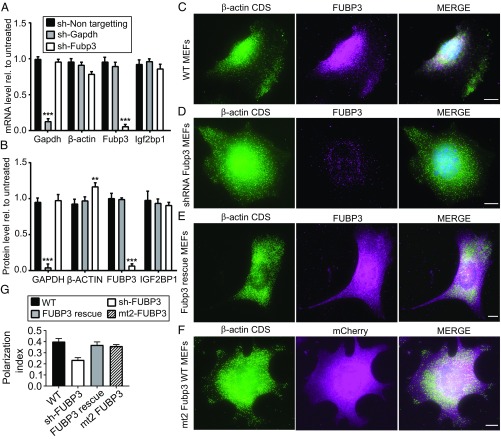
To validate that proteins identified by RNA-BioID are functionally significant for the mRNA used as bait, we performed shRNA-mediated knockdown experiments for FUBP3. The effectiveness of the knockdown was validated by quantitative RT-PCR and Western blot analysis (Fig. 6 A and B) using GAPDH as a control since it does not interact with β-actin mRNA, as shown by RNA BioID (Fig. 3C). FUBP3 knockdown only mildly reduced mRNA levels of β-actin or IGF2BP1 mRNAs (Fig. 6A). Similarly, IGF2BP1 protein levels did not significantly change on FUBP3 knockdown (Fig. 6B), ruling out an indirect effect of FUBP3 on β-actin mRNA by limiting IGF2BP1 levels. However, we observed a slight increase in β-ACTIN protein level, indicating that FUBP3 might coregulate β-actin mRNA translation or β-ACTIN protein stability.
Fig. 6.
Down-regulation of Fubp3 affects β-actin mRNA localization. (A) Western blot analysis to monitor shRNA-mediated knockdown of FUBP3 in nontargeted shRNA stably expressing in WT (second lane) compared with unmodified WT (lane 1) and WT stably expressing shRNA against Gapdh (lane 3) and stably expressing shRNA against FUBP3 (lane 4). Blot was reprobed against GAPDH and β-ACTIN and IGF2BP1 to access the changes in these proteins due to knockdown of FUBP3 or GAPDH. (B) Quantitative RT-PCR analysis of Gapdh, β-actin, Igf2bp1, and Fubp3 levels in knockdown cells. Corresponding mRNA levels in untreated cells were used for normalization. The statistical significance of each dataset was determined using Student’s t test. *P < 0.05; ***P < 0.001. (C) Western blot quantification of GAPDH, β-ACTIN, IGF2BP1, and FUBP3 protein levels in knockdown cells. Protein levels in untreated cells served as a normalization control. The statistical significance of each dataset was determined using Student's t test. *P < 0.05; ***P < 0.001. (D–G) Representative smFISH-IF images of immortalized MEFs before or after Fubp3 knockdown. (D–F) β-Actin mRNA (Cy3; green) and FUBP3 (magenta). Shown are MEFs before knockdown (WT) (D), MEFs after shRNA treatment (E), and shRNA-treated MEFs expressing a FUBP3 rescue construct (F) (Materials and Methods). (H) Representative smFISH-IF image of MEFs expressing the FUBP3 KH mutant mt2 (Fig. 5). (Scale bars: 10 µm) (I) Polarization index for β-actin mRNA in MEFs from experiments shown in D–G. The polarization index was calculated from a total of 65 cells from three biological replicates. Bars represent the median values. Statistical significance of each dataset was determined using Student’s t test. *P < 0.05; ***P < 0.001.
We assessed the effect of the FUBP3 knockdown or overexpression of mutant FUBP3 on β-actin mRNA localization by smFISH-IF (Fig. 6 C–F and SI Appendix, Fig. S11) and calculated the polarization index (Fig. 6G). In control cells (immortalized MEFs; Fig. 6C), FUBP3 and β-actin mRNA were expressed, and the polarization index of β-actin mRNA was 0.37 (Fig. 6G). In FUBP3 knockdown cells (Fig. 6D), almost no FUBP3 signal was detectable, and the β-actin mRNA polarization index dropped to 0.25 (Fig. 6G). To test whether the reduction in polarization is due to a loss of FUBP3, we expressed a knockdown-insensitive mCherry-tagged FUBP3 in these MEFs. Expression of this fusion protein was accessed by indirect immunofluorescence against mCherry, and β-actin mRNA was visualized by smFISH (Fig. 6E). The polarization index was determined using only MEFs positive for mCherry. Although full rescue was not observed, the polarization index was increased, to 0.33 (Fig. 6G). This indicates that FUBP3 is important for β-actin mRNA localization.
We also analyzed the effect on β-actin mRNA distribution when overexpressing a mCherry-tagged FUBP3mt2 mutant lacking a functional KH2 domain (Fig. 6F). As before, we selected MEFs with an mCherry signal for determination of the polarization index. We found a polarization index of 0.31 (Fig. 6G), which is not significantly different from that of β-actin mRNA in WT MEFs. These data suggest that although KH2 is important for the formation of FUBP3-containing RNP particle-like structures in the cytoplasm, it does not act as dominant negative mutation, probably because a mutant with this mutation does not compete with endogenous FUBP3.
Discussion
Proximity biotinylation has facilitated the characterization of dynamic protein complexes by in vivo labeling of interaction partners. Here we exploit this approach and demonstrate its utility for identifying functionally relevant RBPs of a specific mRNA, mammalian β-actin. This is achieved by combining MS2 tagging of the mRNA of choice and coexpression of a fusion protein of the MS2 coat protein (MCP) and the biotin ligase (BirA*).
The primary goal of RNA-based BioID is to identify novel RNA interactors. As seen in several proximity labeling (BioID or APEX-driven) approaches (43, 54, 55), the number of identified potential interactors for β-actin is far higher than the number of proteins identified by classical co-IP or coaffinity purification approaches. This might be due to proximity labeling’s greater sensitivity or its propensity to allow the capture of transient interactors (56). Although this can result in a skewed view of the actual components of a complex due to the rapid change in the composition of mRNP, it is beneficial to identify all mRNP components during the life stages of an mRNA. The most highly represented class of proteins was RBPs (Fig. 3 and SI Appendix, Fig. S8B), among them all RBPs previously associated with localization, translational control, or (de)stabilization of β-actin mRNA. Other RBPs, such as survival of motor neuron 1 (SMN1), which supports the association of IGF2BP1 with β-actin mRNA (57), were also found to be enriched in MEFs expressing β-actin–MBS compared with control MEFs, although with lower significance (P < 0.1).
We also analyzed our dataset for motor proteins involved in mRNA transport. Neither MYH10 (58) nor KIF11 (59), which have been suggested to work as β-actin mRNA transport motors, were found as biotinylated proteins. The only motor that we identified was MYH9, the heavy chain of an MYH10-related class II-A myosin, although it was not significantly enriched (P = 0.08). The lack of motor proteins is compatible with a recent observation that β-actin localization in fibroblasts works primarily by diffusion to and trapping in the microfilament-rich cortex (60). This is also corroborated by our finding that components of the actin-rich cell protrusion (Fig. 3, cluster 4) are heavily biotinylated in MEFs after serum-induced localization of β-actin.
Overall, our cluster analysis shows that the majority of previously identified β-actin RBPs behave similarly under the two test conditions (serum-induced and uninduced MEFs). This not only indicates that they interact with β-actin mRNA in MEFs even under steady-state conditions, but also makes it likely that other proteins, especially RBPs, found in this cluster might represent as-yet-unknown β-actin mRNA interactors. By choosing the far-upstream binding protein FUBP3 as a potential candidate, we demonstrate that this assumption holds true for at least this protein. Not only does FUBP3 bind to β-actin mRNA, but its knockdown also results in a similar decrease of β-actin localization to the leading edge as is seen with loss of IGF2BP1.
FUBP3 (also known as MARTA2) has been reported to bind to the 3′ UTR of the localized MAP2 mRNA in rat neurons (39) to regulate its dendritic targeting (40). Although the binding site of FUBP3 in MAP2 mRNA is not known, its preferred binding motif (UAUA/UAUG) was recently identified (52). This motif is present in the 3′ UTR of β-actin 460 nt downstream of the zipcode, and a 79-nt region containing this motif is bound by FUBP3. FUBP proteins might play a more substantial role in RNA localization, since homologs of a second member of the FUBP family, FUBP2, not only are reportedly involved in MAP2 or β-actin mRNA localization, but also are present among the biotinylated proteins that we identified. However, FUBP2 is mainly nuclear, and its role in β-actin mRNA localization might be indirect (61). In contrast, FUBP3 seems to have a direct function in localizing β-actin, as it binds to the 3′ UTR and its loss reduces β-actin mRNA localization independently of IGF2BP1. This independent function is supported by the observation that both proteins do not directly bind to each other but do bind to different regions of β-actin mRNA. A potential additional function could be translational regulation. Although less dramatic than seen for loss of IGF2BP1, knockdown of FUBP3 results in increased amounts of β-ACTIN protein, while β-actin mRNA levels are similar or even lower than those in untreated MEFs. This could be due to a loss of translational inhibition, as has been shown for IGF2BP1 (11).
Its role in β-actin and MAP2 mRNA localization suggests that FUBP3/MARTA2 is a component of several localizing mRNPs. Of note, RNA-BioID on β-actin mRNA has identified even more RBPs involved in the localization of other mRNAs, including SYNCRIP (48) and Staufen (45). Several of these RBPs (e.g., STAU1, STAU2) are highly enriched in our β-actin biotinylated proteome. This finding might reflect the participation of multiple RBPs in β-actin localization or regulation. It also shows that a common set of RBPs is used to control the fate of several different localized mRNAs in different cell types. Although RNA-BioID does not currently allow us to determine whether all these RBPs are constituents of the same β-actin mRNP, belong to different states of an mRNP, or belong to different populations, their identification allows us to address these questions to achieve a more detailed understanding of the common function of RBPs on diverse mRNAs.
Materials and Methods
RNA-BioID.
For RNA-BioID, cells were incubated with 50 µM biotin for at least 6 h. Following incubation, cells were washed twice with 1× PBS, lysed in IP lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 1 mM DTT, 1% Tween-20, and 1× protease inhibitor) and passed 10–12 times through a 21-gauge needle. The lysate was cleared by centrifugation at 12,000 × g for 10 min at 4 °C to remove cell debris. Protein from the supernatant (total cell lysate; 10 µg) was used to check for protein biotinylation. In the remaining lysate, NaCl was added to a final concentration of 500 mM. Then 200 µL of streptavidin magnetic bead suspension (GE Healthcare) were added, and the high salt lysate was incubated overnight at 4 °C with end-to-end rotation. The next day, the beads were collected (by keeping the beads on the magnetic stand for 2 min) and washed as described previously (43). The beads were washed twice for 5 min with 0.3 mL of wash buffer 1 (2% SDS), once with wash buffer 2 (0.1% wt/vol deoxycholate, 1% wt/vol Tween-20, 350 mM NaCl, 1 mM EDTA pH 8.0), once with wash buffer 3 (0.5% wt/vol deoxycholate, 0.5% wt/vol Tween-20, 1 mM EDTA, 250 mM LiCl, 10 mM Tris⋅HCl pH 7.4) and 50 mM Tris⋅HCl pH 7.5, once with wash buffer 4 (50 mM NaCl, 50 mM Tris⋅HCl pH 7.4), and finally twice with 500 µL of 50 mM ammonium bicarbonate. Then 20 µL of the beads were used for Western blot and silver staining, and 180 µL were subjected to mass spectrometry analysis. To release captured proteins from streptavidin beads for Western blot analysis, the beads were incubated in 2× Laemmli buffer containing 2 mM saturated biotin and 20 mM DTT for 10 min at 95 °C.
For biotinylation after serum induction, cells were starved for 24 h and then induced with 10% serum-containing medium containing 50 µM biotin for 6–24 h. Samples were processed for mass spectrometry analysis as described in SI Appendix, Materials and Methods.
Microscopy and Super-Registration Microscopy.
For live cell imaging, cells were imaged with a Zeiss Cell Observer wide-field fluorescence microscope, operated by ZEN software, illuminated with a xenon arc lamp, and detected with a CCD camera (Axiocam 506) with 100×/1.45 α-Plan fluor oil immersion objectives (Zeiss). Live cell imaging was done using a dual-band GFP/mCherry filter set (F56-319; AHF). For imaging of fixed cells, the microscope setup was the same as described by Eliscovich et al. (47).
Imaging Analysis.
Single-molecule localization was determined with FISH-QUANT (62), and super-registration analysis was performed as described by Eliscovich et al. (47) with existing software packages and custom algorithm programs written in MATLAB (MathWorks). For polarization index calculation, after taking the maximum projections from all of the Z-stacks, polarization and dispersion indices were measured as described previously (9) with an existing software package written in MATLAB.
smFISH-IF.
Immortalized WT MEFs or MEFs containing MS2-tagged β-actin but no MCP-GFP were seeded on a fibronectin-coated cover glass in a 12-well cell culture plate and grown for 24 h in serum-free medium, followed by the addition of serum-containing medium to the cells for 1–2 h. The protocol for smFISH-IF has been described previously (47). In brief, cells were washed three times with PBS, fixed for 10 min with 4% paraformaldehyde in PBS, washed three times in PBS and then quenched in 50 mM glycine, and finally permeabilized with 0.1% Triton X-100 (28314; Thermo Fisher Scientific) and 0.5% Ultrapure BSA (AM2616; Life Technologies) in 1× PBS-M for 10 min. After washing with PBS, cells were exposed to 10% (vol/vol) formamide, 2× SSC, and 0.5% Ultrapure BSA in RNase-free water for 1 h at room temperature, followed by incubation for 3 h at 37 °C with either 10-ng custom-labeled probes or 50-nM Stellaris RNA FISH probes (Biosearch Technologies) (SI Appendix, Table S4). Primary antibodies against GFP (GFP-1010; Aves Labs), IGF2BP1 (RN001M; MBL), or FUBP3 (Abcam) were diluted (SI Appendix, Table S3) in hybridization buffer containing 10% formamide, 1 mg/mL E. coli tRNA, 10% dextran sulfate, 20 mg/mL BSA, 2× SSC, 2 mM vanadyl ribonucleoside complex, and 10 U/mL SUPERase-In (Ambion) in RNase-free water. After incubation and quick washing, cells were further incubated twice with an Alexa Fluor 647-conjugated secondary antibody (Life Technologies) in 10% formamide and 2× SSC in RNase-free water for 20 min at 37 °C. After four washes in 2× SSC, DNA was counterstained with DAPI (0.1 μg/mL in 2× SSC; Sigma-Aldrich), and after a final wash, cells were mounted using ProLong Diamond Antifade Reagent (Life Technologies).
Data Availability.
Proteomic data supporting this study have been deposited in the PRIDE database, www.ebi.ac.uk/pride/archive/ (accession no. PXD010694).
Supplementary Material
Acknowledgments
We thank Jeff Chao (Friedrich Miescher Institute for Biomedical Research, Basel), Imre Gaspar (European Molecular Biology Laboratory, Heidelberg), Julién Bethune (Heidelberg University Biochemistry Center, Heidelberg), Dierk Niessing (University of Ulm), Michael Kiebler (University of Munich), Stefan Kindler (University of Hamburg), Stefan Hüttelmaier (University of Halle), and Ibrahim Muhammad Syed (Interfaculty Institute of Biochemistry, Tübingen), for plasmids, cell lines, antibodies, or spike RNA. We are grateful to Robert H. Singer for hosting J.M. during an imaging internship. We also thank Frank Essmann, Ruth Schmid (both at Interfaculty Institute of Biochemistry, Tübingen), and Silke Wahle (Proteome Center Tübingen) for technical support; Jeetayu Biswas (Albert Einstein College) for help with the polarization index scripts and the IGF2BP1 KO cell line; and Matthew Cheng (Interfaculty Institute of Biochemistry, Tübingen) for suggestions on the manuscript. The project was funded as a project of the Deutsche Forschungsgemeinschaft (DFG) Research Unit FOR2333 by a grant from the DFG (DFG JA696/11-1). C.E. was supported by an NIH grant (NS083085).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Proteomic data supporting this study have been deposited in the PRIDE archive, www.ebi.ac.uk/pride/archive/ (accession no. PXD010694).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820737116/-/DCSupplemental.
References
- 1.Martin K. C., Ephrussi A., mRNA localization: Gene expression in the spatial dimension. Cell 136, 719–730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eliscovich C., Buxbaum A. R., Katz Z. B., Singer R. H., mRNA on the move: The road to its biological destiny. J. Biol. Chem. 288, 20361–20368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchand V., Gaspar I., Ephrussi A., An intracellular transmission control protocol: Assembly and transport of ribonucleoprotein complexes. Curr. Opin. Cell Biol. 24, 202–210 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Dreyfuss G., Kim V. N., Kataoka N., Messenger RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3, 195–205 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Kislauskis E. H., Zhu X., Singer R. H., β-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 136, 1263–1270 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence J. B., Singer R. H., Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45, 407–415 (1986). [DOI] [PubMed] [Google Scholar]
- 7.Ross A. F., Oleynikov Y., Kislauskis E. H., Taneja K. L., Singer R. H., Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17, 2158–2165 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lionnet T., et al. , A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods 8, 165–170 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park H. Y., Trcek T., Wells A. L., Chao J. A., Singer R. H., An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Rep. 1, 179–184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz Z. B., et al. , β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 26, 1885–1890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hüttelmaier S., et al. , Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–515 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Bassell G. J., et al. , Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 18, 251–265 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao J., Sasaki Y., Wen Z., Bassell G. J., Zheng J. Q., An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat. Neurosci. 9, 1265–1273 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Turner-Bridger B., et al. , Single-molecule analysis of endogenous β-actin mRNA trafficking reveals a mechanism for compartmentalized mRNA localization in axons. Proc. Natl. Acad. Sci. U.S.A. 115, E9697–E9706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kislauskis E. H., Zhu X., Singer R. H., Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 127, 441–451 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao J. A., et al. , ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 24, 148–158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yisraeli J. K., VICKZ proteins: A multi-talented family of regulatory RNA-binding proteins. Biol. Cell 97, 87–96 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Yoon Y. J., et al. , Glutamate-induced RNA localization and translation in neurons. Proc. Natl. Acad. Sci. U.S.A. 113, E6877–E6886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wächter K., Köhn M., Stöhr N., Hüttelmaier S., Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Biol. Chem. 394, 1077–1090 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Ceci M., et al. , RACK1 is a ribosome scaffold protein for β-actin mRNA/ZBP1 complex. PLoS One 7, e35034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan F., Hüttelmaier S., Singer R. H., Gu W., ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol. Cell. Biol. 27, 8340–8351 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh M., Haga I., Li Q.-H., Fujisawa J., Identification of cellular mRNA targets for RNA-binding protein Sam68. Nucleic Acids Res. 30, 5452–5464 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rackham O., Brown C. M., Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. EMBO J. 23, 3346–3355 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dormoy-Raclet V., et al. , The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the beta-actin mRNA in a U-rich element-dependent manner. Mol. Cell. Biol. 27, 5365–5380 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee F. C. Y., Ule J., Advances in CLIP technologies for studies of protein-RNA interactions. Mol. Cell 69, 354–369 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Hafner M., et al. , Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castello A., et al. , Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Zielinski J., et al. , In vivo identification of ribonucleoprotein-RNA interactions. Proc. Natl. Acad. Sci. U.S.A. 103, 1557–1562 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogell B., et al. , Specific RNP capture with antisense LNA/DNA mixmers. RNA 23, 1290–1302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaspar I., Wippich F., Ephrussi A., Enzymatic production of single-molecule FISH and RNA capture probes. RNA 23, 1582–1591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slobodin B., Gerst J. E., A novel mRNA affinity purification technique for the identification of interacting proteins and transcripts in ribonucleoprotein complexes. RNA 16, 2277–2290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D. I., et al. , Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U.S.A. 111, E2453–E2461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roux K. J., Kim D. I., Burke B., May D. G., BioID: A screen for protein-protein interactions. Curr. Protoc. Protein Sci. 91, 19.23.1–19.23.15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Firat-Karalar E. N., Stearns T., Probing mammalian centrosome structure using BioID proximity-dependent biotinylation. Methods Cell Biol. 129, 153–170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramanathan M., et al. , RNA-protein interaction detection in living cells. Nat. Methods 15, 207–212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung H.-J., et al. , FBPs are calibrated molecular tools to adjust gene expression. Mol. Cell. Biol. 26, 6584–6597 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinn L. M., FUBP/KH domain proteins in transcription: Back to the future. Transcription 8, 185–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zivraj K. H., et al. , The RNA-binding protein MARTA2 regulates dendritic targeting of MAP2 mRNAs in rat neurons. J. Neurochem. 124, 670–684 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Blichenberg A., et al. , Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J. Neurosci. 19, 8818–8829 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rehbein M., Kindler S., Horke S., Richter D., Two trans-acting rat-brain proteins, MARTA1 and MARTA2, interact specifically with the dendritic targeting element in MAP2 mRNAs. Brain Res. Mol. Brain Res. 79, 192–201 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Peabody D. S., The RNA binding site of bacteriophage MS2 coat protein. EMBO J. 12, 595–600 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Femino A. M., Fay F. S., Fogarty K., Singer R. H., Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyagi S., Alsmadi O., Imaging native β-actin mRNA in motile fibroblasts. Biophys. J. 87, 4153–4162 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heraud-Farlow J. E., Kiebler M. A., The multifunctional Staufen proteins: Conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 37, 470–479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balasanyan V., Arnold D. B., Actin and myosin-dependent localization of mRNA to dendrites. PLoS One 9, e92349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eliscovich C., Shenoy S. M., Singer R. H., Imaging mRNA and protein interactions within neurons. Proc. Natl. Acad. Sci. U.S.A. 114, E1875–E1884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDermott S. M., Meignin C., Rappsilber J., Davis I., Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification. Biol. Open 1, 488–497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jønson L., et al. , Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell. Proteomics 6, 798–811 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Maizels Y., et al. , Localization of cofilin mRNA to the leading edge of migrating cells promotes directed cell migration. J. Cell Sci. 128, 1922–1933 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Weidensdorfer D., et al. , Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15, 104–115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dominguez D., et al. , Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell 70, 854–867.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hollingworth D., et al. , KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 40, 6873–6886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schopp I. M., et al. , Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 8, 15690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rhee H.-W., et al. , Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varnaitė R., MacNeill S. A., Meet the neighbors: Mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics 16, 2503–2518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fallini C., et al. , Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons. Dev. Neurobiol. 74, 319–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Latham V. M., Yu E. H., Tullio A. N., Adelstein R. S., Singer R. H., A Rho-dependent signaling pathway operating through myosin localizes beta-actin mRNA in fibroblasts. Curr. Biol. 11, 1010–1016 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Song T., et al. , Specific interaction of KIF11 with ZBP1 regulates the transport of β-actin mRNA and cell motility. J. Cell Sci. 128, 1001–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park H. Y., et al. , Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 343, 422–424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu W., Pan F., Zhang H., Bassell G. J., Singer R. H., A predominantly nuclear protein affecting cytoplasmic localization of β-actin mRNA in fibroblasts and neurons. J. Cell Biol. 156, 41–51 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mueller F., et al. , FISH-quant: Automatic counting of transcripts in 3D FISH images. Nat. Methods 10, 277–278 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic data supporting this study have been deposited in the PRIDE database, www.ebi.ac.uk/pride/archive/ (accession no. PXD010694).