Significance
Cancer of the pancreas is highly heterogeneous. Improvement of diagnosis and prognosis requires the discovery of new biomarkers and new methods of classification. In this study, we identify TMEM16A calcium-activated chloride channel as a potential key biomarker for pancreatic cancer. We demonstrate that, through the modulation of chloride homeostasis and the initiation of a calcium signaling pathway, TMEM16A is an important regulator of ligand-induced EGFR signaling in pancreatic cancer cells. Taken together, our findings establish that EGF-induced TMEM16A-dependent calcium pathways constitute a gene set sufficient for classification of pancreatic cancer, and provide inroads for molecular characterization of different subtypes of pancreatic cancer.
Keywords: TMEM16A, calcium-activated chloride channel, pancreatic cancer, EGFR, store-operated calcium entry
Abstract
Pancreatic cancer typically spreads rapidly and has poor survival rates. Here, we report that the calcium-activated chloride channel TMEM16A is a biomarker for pancreatic cancer with a poor prognosis. TMEM16A is up-regulated in 75% of cases of pancreatic cancer and high levels of TMEM16A expression are correlated with low patient survival probability. TMEM16A up-regulation is associated with the ligand-dependent EGFR signaling pathway. In vitro, TMEM16A is required for EGF-induced store-operated calcium entry essential for pancreatic cancer cell migration. TMEM16A also has a profound impact on phosphoproteome remodeling upon EGF stimulation. Moreover, molecular actors identified in this TMEM16A-dependent EGFR-induced calcium signaling pathway form a gene set that makes it possible not only to distinguish neuro-endocrine tumors from other forms of pancreatic cancer, but also to subdivide the latter into three clusters with distinct genetic profiles that could reflect their molecular underpinning.
Pancreatic cancer is one of the most aggressive types of cancer, with a 5-y survival rate close to 6–8% and a strong ability to rapidly metastasize even before the detection of primary tumors (1). Pancreatic cancer is projected to become the second leading cause of cancer death by 2030, so there is an urgent need for new tools for its diagnosis and treatment (2). While it is well established that a core of four common mutations (KRAS, TP53, SMAD4, CDKN2) is at the origin of pancreatic cancer development, the heterogeneity of pancreatic cancer remains an important challenge to overcome for improving diagnosis and treatment (1). Therefore, it is important to identify biomarkers that not only can predict the prognosis of patients with pancreatic cancer and provide classification of pancreatic tumors, but also help with our understanding of their molecular characteristics and development of treatments.
TMEM16A, also known as ANO1, DOG1, and ORAOV2, is a calcium-activated chloride channel (CaCC) (3–5) with physiological functions in epithelial tissues, exocrine glands, dorsal root ganglion neurons, and smooth muscles (6–8). TMEM16A has also been detected in multiple cancers, including breast cancer, head and neck squamous cell carcinomas (HNSCC), gastrointestinal squamous tumors (GIST), lung cancer, pancreatic cancer, and ovarian cancer (9, 10). TMEM16A expression is associated either with tumor growth and cancer cell proliferation or cancer cell migration. In breast cancer and HNSCC, TMEM16A is associated with the EGF receptor (EGFR) signaling pathway. TMEM16A expression and its interaction with EGFR promote cancer cell proliferation by constitutive phosphorylation and activation of EGFR and its downstream signaling pathways (Akt, ERK, and CamK) (11, 12). However, it is still unclear how TMEM16A regulates EGFR signaling pathways and whether different types of cancer may involve different forms of EGFR regulation by TMEM16A.
In pancreatic cancer, the EGFR signaling pathway is involved in both cancer initiation and the development of metastasis (13, 14). Unlike breast cancer and HNSCC that are associated with mutations rendering EGFR constitutively active, pancreatic cancer involves activation of the EGFR signaling pathway by extracellular ligands such as EGF, TGF-α, or Epiregulin (15–18). It is therefore important to determine whether TMEM16A regulates EGFR signaling in pancreatic cancer cells in a manner that depends on ligand activation of EGFR.
In this study, we show that TMEM16A is a biomarker for pancreatic cancer with poor prognosis. TMEM16A is up-regulated in more than 75% of pancreatic cancers and high levels of TMEM16A expression are correlated with low probability of patient survival. We then show that this up-regulation of TMEM16A in pancreatic cancer tissues is positively correlated with an up-regulation of genes encoding molecular components of the EGFR signaling pathway. We tested whether TMEM16A regulation depends on EGFR activation in a pancreatic cancer cell line, and found that EGFR ligands promote cell migration by stimulating TMEM16A activity and TMEM16A-dependent store-operated calcium entry (SOCE), thereby unraveling a role of TMEM16A as initiator of SOCE. We further show that TMEM16A expression is required for the EGF-induced remodeling of phosphoproteome, leading to increased phosphorylation of proteins associated with EGFR signaling pathways and cell motility. Finally, building on the classification of pancreatic cancer in different molecular subtypes (19), we show that a group of genes associated with the EGF-induced TMEM16A-dependent Ca2+ signaling pathway was sufficient to distinguish neuro-endocrine tumors and classify the remaining pancreatic cancers into three subtypes, each with a particular molecular signature that can provide a better understanding of these cancers and help with the development of more specific therapeutics.
Results
TMEM16A Is Overexpressed in Pancreatic Ductal Adenocarcinoma and Influences the Overall Survival.
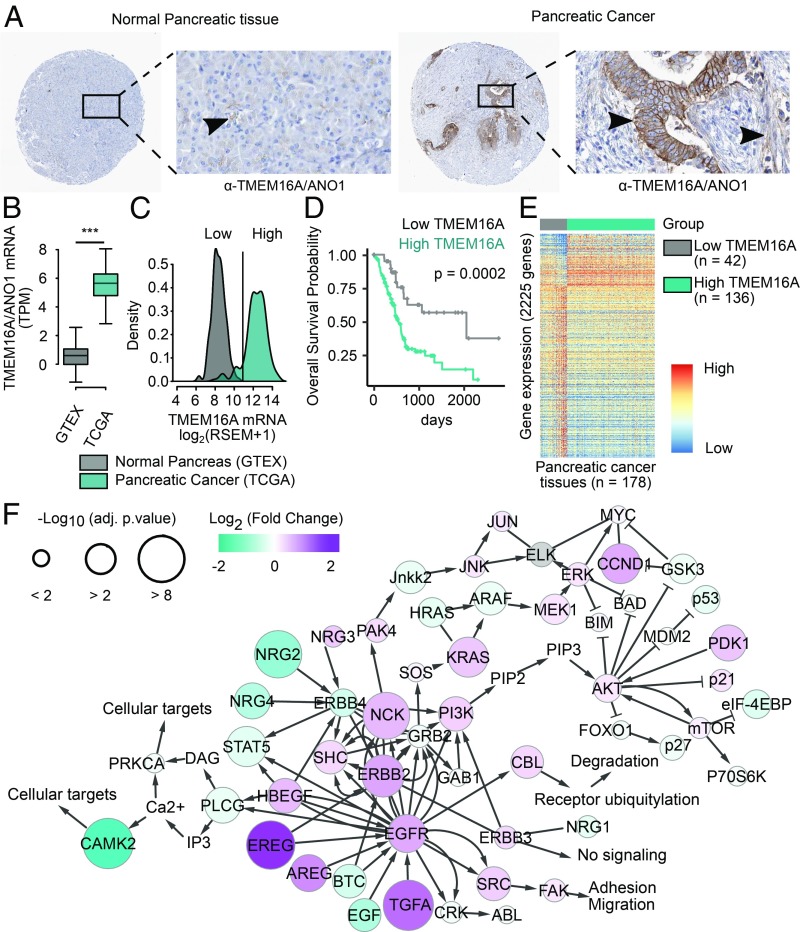
To investigate whether TMEM16A exhibits abnormal expression in human pancreatic cancer samples, we scrutinized available databases for signs of up-regulation of TMEM16A protein and mRNA in pancreatic cancer, and then looked for a correlation between TMEM16A expression level and overall survival of patients. Examination of data provided by the Human Protein Atlas (http://www.proteinatlas.org/) revealed greater expression of TMEM16A protein in 10 pancreatic cancer tissues compared with 5 normal pancreas (Fig. 1A and SI Appendix, Fig. S1A). Whereas lTMEM16A expression is low in the lumen of acini in normal pancreas, there is a strong TMEM16A staining in ductal-like epithelial cells and moderate TMEM16A staining in the surrounding mesenchymal-like cells in pancreatic cancer tissues (Fig. 1A). Comparison of normalized RNA-sequencing data of 178 pancreatic cancer from the The Cancer Genome Atlas (TCGA)-Pancreatic Ductal Adenocarcinoma Database (PAAD) dataset (19) with 168 normal pancreas from the Genotype Tissue Expression (GTEx) dataset (20, 21) revealed an increase of TMEM16A mRNA abundance by 60% (Fig. 1B). There is also up-regulation of TMEM16A mRNA in pancreatic cancer compared with normal pancreas in 7 of 12 microarrays available in the National Center for Biotechnology Information Gene Expression Omnibus (SI Appendix, Fig. S1B).
Fig. 1.
TMEM16A/ANO1 mRNA and protein are up-regulated in pancreatic cancer and associated to an enrichment of genes involved in EGFR signaling pathways. (A) Immunohistochemical staining of TMEM16A protein in normal and cancerous pancreatic tissues (data obtained from the Human Protein Atlas http://www.proteinatlas.org/). (A width, 1 mm; A, Insets width, 300 µm.) (B) RNA-sequencing measurement of TMEM16A/ANO1 mRNA expression expressed as log2 transcript per million (TPM) in normal pancreas and in pancreatic cancer (data obtained from GTEx and TCGA consortium, respectively). ***P < 0.001. (C) Density plot of TMEM16A/ANO1 mRNA expression in normal and cancerous pancreatic tissues. Pancreatic cancer samples with a normalized log2 value of TMEM16A/ANO1 mRNA expression superior to 11 were defined as high TMEM16A. (D) Overall survival probability curve for patients with pancreatic tumors with low or high expression of TMEM16A. (E) Heatmap of DEGs between pancreatic cancer with low or high TMEM16A expression (42 and 136 samples, respectively). (F) Representation of the ErbB pathway on which genes (nodes) are mapped with log2(FC) intensity (color) and negative log10 of adjusted P value (size) obtained from the differential expression analysis between pancreatic cancer with low or high TMEM16A expression.
By subdividing pancreatic cancer samples into those that up-regulate TMEM16A expression (“high TMEM16A”) and those with TMEM16A expression within the range of normal expression in normal pancreas (“low TMEM16A”) (Fig. 1C), we looked for correlation with poor prognosis. The overall survival of patients with pancreatic tumors noted as high TMEM16A is significantly reduced compared with the overall survival of patients with pancreatic tumors noted as low TMEM16A (Fig. 1D). Thus, TMEM16A is up-regulated in the majority of pancreatic cancer and is associated with poor survival.
TMEM16A overexpression in multiple cancer types is associated with amplification of the genomic region encompassing the TMEM16A gene (9, 10). Our mining of the TCGA-related whole-genome sequencing data revealed that the frequency of increased copy number of the TMEM16A gene is very low (∼10%) in pancreatic cancer compared with breast cancer (BRCA, ∼40%), HNSCC (HNSC ∼40%), and GIST (STAD, ∼30%) (SI Appendix, Fig. S1C), suggesting that mechanisms not involving gene amplification contribute to the up-regulation of TMEM16A in pancreatic cancer.
Up-Regulation of TMEM16A in Pancreatic Cancer Is Associated with Up-Regulation of Genes in Pancreatic Cancer Signaling and EGFR Signaling Pathways Including EGFR Ligands.
A differential expression analysis (DEA) between high TMEM16A and low TMEM16A pancreatic tumors of the TCGA-PAAD revealed that 8,743 genes of a total of 19,550 genes are differentially expressed between these two groups; 525 genes are up-regulated by more than 50%, while 1,700 genes are down-regulated by more than 50% in the high TMEM16A group of pancreatic tumors (Fig. 1E). Gene Set Enrichment Analysis (GSEA) revealed that TMEM16A up-regulation in pancreatic cancer is correlated with several signaling pathways, including the pancreatic adenocarcinoma and EGF/EGFR signaling pathways (Dataset S1). By mapping the EGFR signaling pathway with the log2 fold-change [log2(FC)] and the negative log10 of the adjusted P value [−log10(adj. p.value)] obtained by DEA, we found that several EGFR ligands showed significant association with the TMEM16A expression level (Fig. 1F). These results are in agreement with previous studies demonstrating the association of TMEM16A expression with the EGFR signaling pathway in breast cancer and HNSCC (11, 12, 22). Given that the EGFR signaling pathway in pancreatic cancer relies on the EGFR ligands (13, 14) and promotes pancreatic ductal adenocarcinoma (PDAC) development by regulating acinar-ductal metaplasia, cancer cell migration, and the occurrence of metastasis (15, 17, 18, 23), our finding suggests that TMEM16A could play an essential role in ligand-induced EGFR signaling pathway in pancreatic cancer.
EGF Induces a Transient Calcium Response and Activates CaCC Channels.
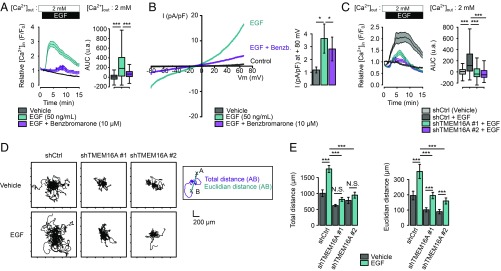
Previous investigations of other cancer types have examined the contribution of TMEM16A to the EGFR signaling pathway (11, 12). However, it is unknown if TMEM16A could be modulated by EGFR ligands and how TMEM16A modulates the EGFR signaling pathway. Here, we asked how EGFR ligands might affect TMEM16A channel activity in the pancreatic cancer cell line AsPC-1 with endogenous expression of both TMEM16A and EGFR. Because TMEM16A is a Ca2+-activated chloride channel, we used patch-clamp electrophysiology and calcium imaging to measure the chloride current and intracellular Ca2+ concentration, respectively. A dose-dependent acute application of EGF induced a transient increase of intracellular Ca2+ concentration and an outward-rectifiying chloride current, which were both inhibited by the TMEM16A blockers benzbromarone (10 μM), 1PBC (10 µM), and niclosamide (20 µM) (Fig. 2 A and B and SI Appendix, Fig. S2 A and B), implicating TMEM16A channel activity in the EGF-induced modulation of Ca2+ homeostasis as well as chloride conductance. Modifying the chloride electrochemical gradient by substituting chloride with gluconate ions prevents EGF-induced Ca2+ response, suggesting the requirement of chloride influx for this response (SI Appendix, Fig. S2C).
Fig. 2.
EGF stimulates TMEM16A-dependent chloride current and a TMEM16A-dependent Ca2+ influx. (A) Relative Ca2+ influx (F/F0) in AsPC-1 cells treated with EGF (50 ng/mL) alone or in combination with benzbromarone (10 µM). Boxplots represent the corresponding integrative fluorescence signal observed from three to five independent experiments regrouping 196–288 cells (t test: ***P < 0.001). (B) Representative chloride currents recorded from voltage ramps from −80 to +80 mV in whole-cell patch-clamp of AsPC-1 cells before and after the perfusion of a solution containing EGF (50 ng/mL) followed by benzbromarone (10 µM). Histograms represent the corresponding current amplitude. Values are mean ± SEM (9–13 cells; Mann–Whitney: *P < 0.05). (C) Relative Ca2+ influx (F/F0) in control (shCtrl) and TMEM16A-silenced (shTMEM16A #1, shTMEM16A #2) AsPC-1 cells treated with EGF (50 ng/mL). Boxplots represent the corresponding integrated fluorescence signal observed from five to eight independent experiments regrouping 198–345 cells per condition (t test: ***P < 0.001). (D) EGF stimulates AsPC-1 cell migration in a TMEM16A manner. Representative 12-h time lapse tracking plots from control (shCtrl) and TMEM16A-silenced (shTMEM16A #1, shTMEM16A #2) AsPC-1 cells cultured in presence or absence of EGF (50 ng/mL). (E) Corresponding histograms quantifying motility parameters (total length and the Euclidean distance traveled per cell). Values are mean ± SEM recapitulating the tracking of 45–156 cells (from two to three independent experiments; t test: ***P < 0.001, NS: nonsignificant).
Erlotinib, an EGFR inhibitor, suppressed the EGF-induced Ca2+ response (SI Appendix, Fig. S2D). Among various EGFR ligands and other growth factors we tested, only EGF and TGF-α generated an increase in the intracellular Ca2+ concentration (SI Appendix, Fig. S2E). Taken together, these results suggest that this EGF-induced TMEM16A-dependent Ca2+ response is mediated by EGFR. The shRNA knockdown of TMEM16A expression in AsPC-1 reduced the benzbromarone-sensitive chloride current (SI Appendix, Fig. S3A), while having moderate or no effect on the total and plasma membrane localized EGFR expression (SI Appendix, Fig. S3 B and C), although few complexes of EGFR and TMEM16A could be detected with a proximity ligation assay (SI Appendix, Fig. S3D). Notably, the molecular silencing of TMEM16A does not affect EGF endocytosis (SI Appendix, Fig. S3E). Both TMEM16A-deficient cell lines had a reduced EGF-induced intracellular Ca2+ response compared with the control cell line (Fig. 2C), thus confirming the contribution of TMEM16A to the EGF-induced Ca2+ response.
In agreement with a previous report (24), silencing of TMEM16A reduced AsPC-1 cell motility as well as the persistence of migration (i.e., Euclidian distance) (Fig. 2 D and E). EGF strongly stimulated both cell motility and the persistence of migration of AsPC-1 control cells. In shTMEM16A cell lines, EGF increased the persistence of migration but not the cell motility, and the extent of increase in persistence was greatly reduced compared with control AsPC-1 cell line. Altogether, these data indicate that TMEM16A regulates EGF-independent migration as well as the promotion of cancer cell migration by EGF, likely through Ca2+ signaling.
EGF Induces SOCE That Is Dependent on TMEM16A Channel Activity.
EGF can induce transient Ca2+ responses, which are required for EGFR internalization (25, 26). The EGF-induced Ca2+ response results from IP3R-mediated Ca2+ release from internal store due to the generation of IP3 by activated phospholipase-C (PLC). TMEM16A interacts with IP3R type 1 and can be activated by an IP3R-mediated release of Ca2+ (27). Moreover, TMEM16A can also be activated by Ca2+ influx through TRPC6 or ORAI1-mediated SOCE (8, 28).
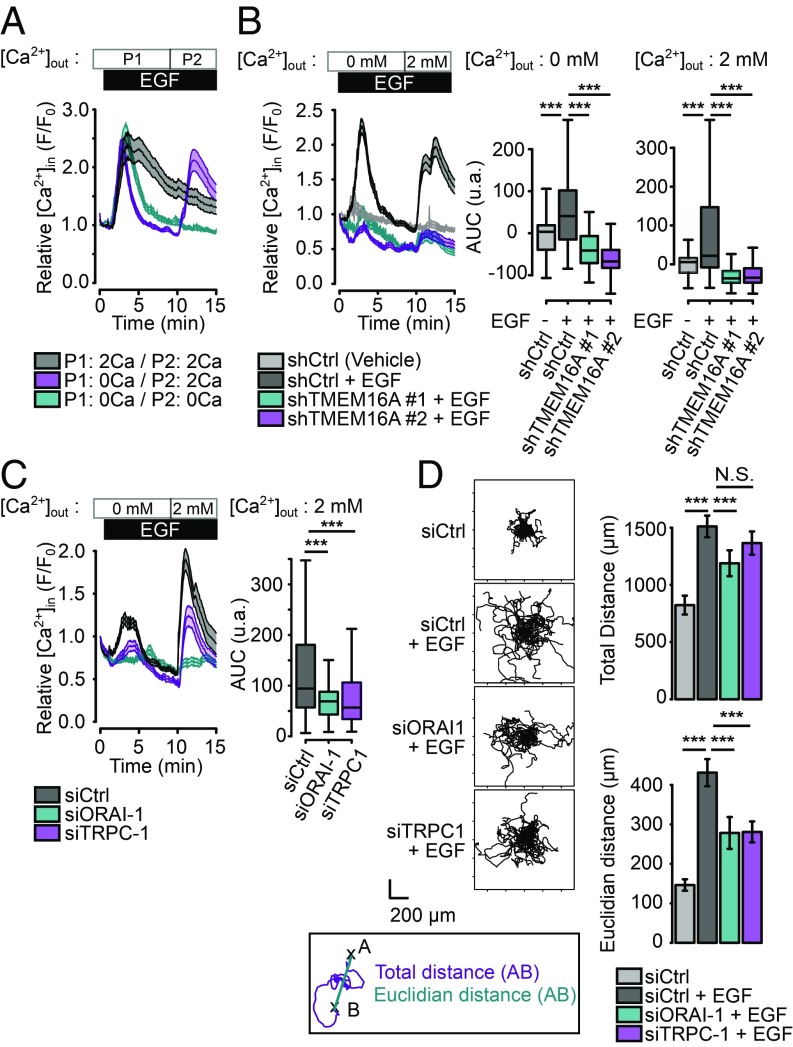
To determine the source of Ca2+ mobilization by EGF, we performed Ca2+ imaging in the presence or absence of extracellular Ca2+. Removal of extracellular Ca2+ led to shortening of the EGF-induced Ca2+ responses without affecting the amplitude (Fig. 3A), suggesting that the EGF-induced Ca2+ response in AsPC-1 requires both Ca2+ release from internal store and Ca2+ influx. We next tested whether this Ca2+ influx resulted from SOCE. First, EGF was applied in the absence of extracellular Ca2+ to monitor the intracellular Ca2+ release, and then extracellular Ca2+ level was raised so we could record the Ca2+ influx (Fig. 3A). Surprisingly, both components of the EGF-induced Ca2+ response were significantly reduced in the absence of TMEM16A (Fig. 3B), suggesting that TMEM16A activity regulates the Ca2+ release from internal store as well. Additionally, TMEM16A expression does not regulate store Ca2+ content as evidenced by ionomycin-induced Ca2+ release (SI Appendix, Fig. S4A). Interestingly, we observed that Thapsigargin-evoked SOCE or ATP-induced P2Y receptor-dependent intracellular Ca2+ release are moderately or insignificantly reduced in absence of the TMEM16A (SI Appendix, Fig. S4 C and D), suggesting that TMEM16A does not regulate IP3-independent SOCE and other PLC-coupled IP3-dependent SOCE. Thus, the contribution of TMEM16A to EGF-induced Ca2+ signaling appears to be particular related to the engagement of TMEM16A with EGFR.
Fig. 3.
EGF induces a SOCE involving ORAI-1 and TRPC1 channels. (A) Relative Ca2+ influx (F/F0) in AsPC-1 cells treated with EGF (50 ng/mL) in a Ca2+-free buffer (blue) for 15 min, in a 2 mM Ca2+ buffer (gray) for 15 min or in Ca2+-free buffer for 10 min followed by a 2 mM Ca2+ buffer for 5 min (magenta). Data are mean ± SEM from two to four independent experiments regrouping 95–155 cells per condition (t test: ***P < 0.001). (B and C) Relative Ca2+ influx (F/F0) in control (shCtrl) and TMEM16A-silenced (shTMEM16A #1, shTMEM16A #2) AsPC-1 cells and in control (siCtrl) or AsPC-1 silenced for ORAI-1 (siORAI1), TRPC-1 (siTRPC1) treated with EGF (50 ng/mL) in a Ca2+-free buffer for 10 min and a 2 mM Ca2+ buffer for 5 min. Boxplots represent the corresponding integrative fluorescence signal observed from three to five independent experiments regrouping 110–284 cells per condition (t test: ***P < 0.001). (D) EGF-induced cell migration is dependent of ORAI-1 and TRPC1 channels. (Left) Representative 12-h time lapse tracking plots from control (siCtrl) treated or not with EGF and AsPC-1 silenced for ORAI-1 (siORAI1), TRPC-1 (siTRPC1) treated with EGF. (Right) Corresponding histograms quantifying motility parameters (total length and the Euclidian distance traveled per cell). Values are mean ± SEM recapitulating the tracking of 23–25 cells (from two to three independent experiments). (Mann–Whitney test: *P < 0.05, **P < 0.01, ***P < 0.001, N.S. P > 0.05.)
SOCE is usually described by the aggregation of STIM1 into punctae following the depletion of intracellular Ca2+ stores, and their association with ORAI proteins at the plasma membrane to form functional Ca2+ channels that mediate Ca2+ influx (29, 30). A variety of ion channels in the ORAI or TRPC families have been implicated in SOCE (29, 31, 32). We observed that both the intracellular Ca2+ release and the SOCE are inhibited by 2-APB, an inhibitor of ORAI, TRPC, and IP3R channels, while application of the selective ORAI inhibitor, GSK7975-A, only inhibits the SOCE, suggesting that ORAI channels contribute to the SOCE and IP3R may mediate the release of intracellular Ca2+ following EGF application (SI Appendix, Fig. S4D). An RT-PCR screen of AsPC-1 cells revealed expression of IP3R, ORAI, STIM, and most of the TRPC channels except for TRPC7 (SI Appendix, Fig. S4E). Transfection of AsPC-1 cells with siRNA targeting ORAI-1 or TRPC-1, two channels particularly well associated with SOCE, decreased the expression of both proteins as verified by immunofluorescence (SI Appendix, Fig. S4F). As expected, knockdown of ORAI-1 or TRPC-1 had moderate effect on Ca2+ release from the internal store while causing significant reduction of the Ca2+ influx from the extracellular solution (Fig. 3C). We further tested whether cell migration is affected by siRNA mediated silencing of SOCE channels. Silencing ORAI-1 or TRPC-1 had a moderate effect on the overall cell motility and a stronger effect on the persistence of migration (Fig. 3D).
Taken together, these findings reveal a role for TMEM16A, in facilitating EGF-induced Ca2+ release from internal store and the subsequent Ca2+ entry, with significant impact on pancreatic cancer cell migration.
TMEM16A Regulates EGF-Induced EGFR Phosphorylation.
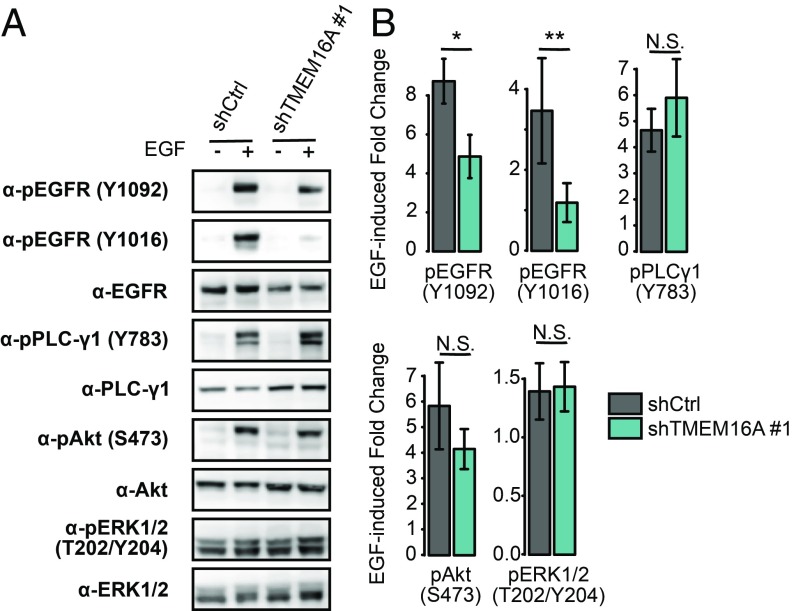
Having discovered that TMEM16A plays a role in regulating EGF-induced Ca2+ signaling, we wondered how TMEM16A may affect the EGFR signaling pathway. Given that inhibition or silencing of TMEM16A in breast cancer and HNSCC cell lines reduces EGFR constitutive phosphorylation as well as the Akt and ERK signaling pathways (12, 33), we first tested how silencing TMEM16A may affect EGFR phosphorylation. EGFR was only weakly phosphorylated in AsPC-1 cells without exposure to EGF (Fig. 4). Acute EGF stimulation of AsPC-1 cells induced a strong phosphorylation of tyrosine residues Y1016 and Y1092 of EGFR, which were significantly reduced in shTMEM16A cell lines (Fig. 4). Interestingly, this TMEM16A-dependent EGFR phosphorylation appears to be upstream of the SOCE as evidenced by the absence of effects on the phosphorylation of Y1092 of EGFR following the depletion of extracellular Ca2+ or the addition of the SOCE inhibitor 2-APB (SI Appendix, Fig. S5A). EGF also strongly stimulated the phosphorylation of PLC-γ1 (Y783) and Akt (S473), but these effects were not significantly affected by the silencing of TMEM16A (Fig. 4). Neither EGF nor TMEM16A silencing affected ERK phosphorylation (T202/Y204) (Fig. 4). We also observed that EGF increases the abundance of phosphorylated tyrosine residues while not affecting the abundance of phosphorylated serine residues (SI Appendix, Fig. S5 B and C). Interestingly, the absence of extracellular Ca2+ does not affected phosphorylated tyrosine and serine residues, whereas 2-APB reduces the abundance of phosphorylated serine and tyrosine residues (SI Appendix, Fig. S5 B and C). Thus, the EGF-induced and TMEM16A-dependent effects on EGFR signaling pathway in pancreatic cancer cells differ from the TMEM16A-dependent mechanisms found in other cancers. Moreover, TMEM16A appears to regulate EGFR activation independently of Ca2+ signaling.
Fig. 4.
TMEM16A modulates EGF-induced EGFR phosphorylation but does not affect PLC-γ1, Akt or ERK activation. Total protein extracts from control (shCtrl) and TMEM16A-silenced (shTMEM16A #1) AsPC-1 cells in the presence or absence of EGF (50 ng/mL) for 30 min were separated on SDS/PAGE and immunoblotted with different antibodies. (A) Representative Western blot obtained with antiphosphorylated EGFR (Y992, Y1048), anti-EGFR; antiphosphorylated PLC-γ1 (Y783), anti–PLC-γ1; antiphosphorylated Akt (S473), anti-Akt; anti-phosphorylated ERK1/2(T202/Y204), and anti-ERK1/2 antibodies. (B) Corresponding histograms representing the ratio of phosphorylated over total proteins normalized with GAPDH expression. Data are mean ± SEM from five to nine independent experiments (Mann–Whitney: *P < 0.05, **P < 0.01, N.S. P > 0.05).
TMEM16A Regulates EGF-Induced Phosphoproteome Remodeling.
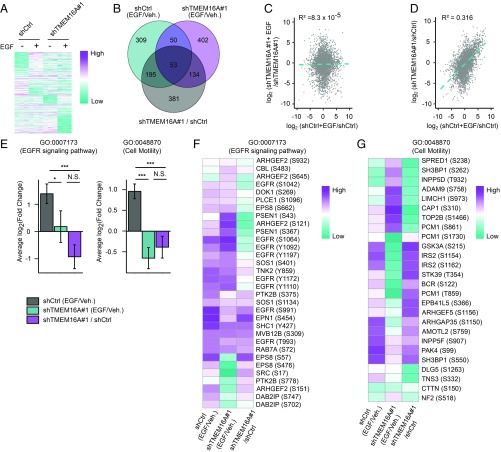
To elucidate the TMEM16A contribution to ligand-induced EGFR signaling in AsPC-1, we performed mass spectrometry for unbiased global phosphoproteome assessment of control and TMEM16A-deficient AsPC-1 cell lines in the presence or absence of EGF stimulation. After normalization and hybrid imputation, 5,608 unique phosphorylated residues belonging to 2,281 proteins were observed in all conditions. We calculated the EGF-induced fold-change [log2(FC)] and the corresponding P value of the phosphorylation level of each of these residues, and found significant differences in the pattern of phosphorylation generated by EGF treatment in AsPC-1 cells with or without TMEM16A (Fig. 5A and Dataset S2). Interestingly, silencing of TMEM16A alone partially mimics the EGF-induced phosphorylation observed in control cells.
Fig. 5.
TMEM16A regulates EGF-induced EGFR signaling pathways. Phosphoproteomics of control (shCtrl) and TMEM16A-silenced (shTMEM16A #1) AsPC-1 cells in the presence or absence of EGF (50 ng/mL) for 30 min show a TMEM16A-dependent regulation of EGF-induced EGFR signaling. Log2 values were calculated from data obtained in three independent experiments. (A) Heatmap of phosphorylated peptides differentially phosphorylated in EGF-treated cells. (B) Venn diagram representing the overlap of significantly altered phosphorylated peptides when comparing control cells treated by EGF with control cells treated by vehicle (shCtrl+EGF/shCtrl), TMEM16A-deficient cells treated with EGF and TMEM16A-deficient cells treated with vehicle (shTMEM16A+EGF/shTMEM16) and when comparing TMEM16A-deficient cells and control cells (shTMEM16A/shCtrl). (C and D) Correlation plot showing the log2(FC) induced by EGF in TMEM16A-deficient cells and the log2(FC) of TMEM16A-deficient cells over control cells as a function of the log2(FC) induced by EGF in control cells. (E) Histograms representing the mean ± SEM of the log2(FC) of phosphorylated peptides associated with EGFR signaling pathway (GO:0007173) or cell motility (GO:0048870) and significantly altered when comparing control cells stimulated or not with EGF (shCtrl+EGF/shCtrl), shTMEM16A#1 cells stimulated or not with EGF (shTMEM16A#1+EGF/shTMEM16A#1), and shTMEM16A#1 and control cells (shTMEM16A#1/shCtrl). *P < 0.05, ***P < 0.001, N.S. P > 0.05. (F and G) Heatmap of the log2 (FC) intensity of phosphorylated peptides associated with EGFR signaling pathway (GO:0007173) and cell motility (GO:0048870) between shCtrl stimulated or not with EGF and between shTMEM16A#1 stimulated or not with EGF and between shTMEM16A#1 and shCtrl.
EGF treatment significantly altered phosphorylation of 607 peptides in control and 639 peptides in shTMEM16A cells, respectively. However, only 17% (103) of these peptides are altered in both conditions (Fig. 5B). Interestingly, 40% (248) of those peptides altered by EGF in control cells are also affected by TMEM16A silencing alone (Fig. 5B). These observations still hold true when we separately examined peptides that were significantly up-regulated or down-phosphorylated upon EGF treatment (SI Appendix, Fig. S5 D and E). In contrast to the weak correlation (R2 = 8.1 × 10−5) between EGF-induced fold-change in shTMEM16A cells and the EGF-induced fold-change in control cells, there is a stronger correlation (R2 = 0.36) between the EGF-induced fold-change in control cells and the fold-change caused by silencing TMEM16A (Fig. 5 C and D). These results suggest that silencing TMEM16A partially mimics the EGF-induced remodeling of the phosphoproteome. As a result, EGF stimulation of control or shTMEM16A AsPC-1 cells will have dramatically different effects.
Examination of 10 different phosphorylated sites on EGFR protein revealed that EGF-induced phosphorylation of these sites was not fully abrogated by TMEM16A silencing, indicating that TMEM16A modulates the pattern of phosphorylation induced by EGF rather than EGFR activation (SI Appendix, Fig. S5F). Unlike Western blot using phosphospecific antibodies (Fig. 4), we could not detect alteration of the EGF-induced phosphorylation of EGFR Y1092 by TMEM16A silencing (SI Appendix, Fig. S5F), nor could we detect EGF-induced phosphorylation of EGFR Y1016 or Akt, likely indicative of technical limitations of our phosphoproteome approach. In agreement with results obtained in Western blot (Fig. 4), the EGF-induced phosphorylation of ERK and PLC-γ1 were not significantly altered by TMEM16A silencing (SI Appendix, Fig. S5 G and H). Thus, whereas mass spectrometry revealed massive remodeling of the phosphoproteome of AsPC-1 upon EGF addition, Western blot with phosphospecific antibodies may be a more sensitive assay for a particular site of phosphorylation.
TMEM16A Supports EGF-Induced Phosphorylation of Proteins Associated with EGFR Signaling and Cell Motility.
Monitoring changes of the phosphorylation status of proteins associated with a signaling pathway or a biological process could help us better understand how TMEM16A contributes to EGF signaling. To analyze the EGF-induced phosphorylation changes of biological processes, such as the EGFR signaling pathway [gene ontology (GO):0007173] and cell motility (GO:0048870), we generated lists of peptides that showed significant increase or decrease in phosphorylation induced by EGF treatment in control and TMEM16A-deficient AsPC-1 cell lines, as well as lists of peptides that showed significant increase or decrease in phosphorylation resulting from shTMEM16A knockdown compared with control cell lines. With these lists, we calculated the global average of the log2(FC) induced by either EGF or shTMEM16A for peptides associated with either the EGFR signaling pathway or cell motility.
In a control AsPC-1 cell line, EGF globally induced a strong up-phosphorylation of peptides associated with EGFR signaling pathway and cell motility (Fig. 5 E–G). Interestingly, in the shTMEM16A cell line, EGF did not change the phosphorylation of peptides associated with the EGFR signaling pathway; however, it decreased the phosphorylation of those peptides associated with cell motility. In addition, silencing of TMEM16A induced a global reduction of phosphorylation of both groups of peptides in cells with or without exposure to EGF, in contrast to the EGF-induced increase of phosphorylation observed in control cells.
Altogether, phosphoproteome analyses of AsPC-1 with or without TMEM16A indicate that, whereas EGF normally strongly promotes the phosphorylation of various sites of proteins associated with EGFR signaling pathway or cell motility, the absence of TMEM16A expression causes a reduction of basal phosphorylation of these peptides as well as EGF-induced de-phosphorylation of these sites. These results are in agreement with what we have found regarding the role of TMEM16A on the EGFR signaling pathway and on the EGF-induced cell motility.
TMEM16A as a Biomarker of Pancreatic Cancer.
We have identified TMEM16A as a central player in EGF/TGF-α–induced EGFR-dependent Ca2+ signaling involving ORAI-1, TRPC-1, and likely STIM-1 and IP3R Ca2+ channels. This Ca2+ signaling pathway regulates EGF-induced cancer cell migration and could potentially be involved in the spreading of pancreatic cancer cells. Because high TMEM16A expression is correlated with poor prognosis, we reasoned that this group of proteins could: (i) serve as a signature of aggressive pancreatic cancer, and (ii) provide a classification of pancreatic cancer into different subsets that may increase our understanding of molecular processes involved in each subset of pancreatic cancer.
Analyses of genomic or transcriptomic data have led to the classification of pancreatic ductal adenocarcinoma into two to four different subtypes (19, 34–36). Here, we applied our analysis to all samples of pancreatic tumors made available by TCGA, including nonadenocarcinoma pancreatic tumors to avoid restriction of our analysis to a particular cancer type (Dataset S3).
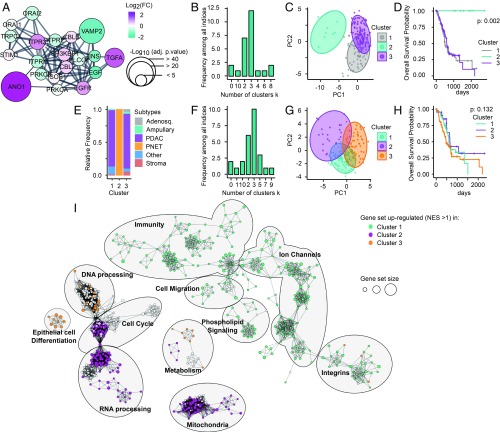
We enriched our group of selected proteins by adding 10 of their closest interactors (confidence = 0.8) by using the String App from Cytoscape to provide a more robust gene set (Fig. 6A). From the TCGA-PAAD dataset, we isolated only the transcriptomic measure of the expression of those genes involved in the EGF-induced TMEM16A-dependent signaling pathway. We determined three as the optimal number of clusters in our restricted dataset using NbClust package (Fig. 6B) and performed principal component analysis (PCA) (Fig. 6C).
Fig. 6.
Classification of pancreatic cancer samples into four clusters using EGF-induced TMEM16A-dependent Ca2+ signaling genetic signature. Genes involved in the EGF-induced TMEM16A-dependent Ca2+ signaling are selected to serve as a signature to classify patients in various clusters. (TMEM16A, EGF, TGFA, EGFR, ORAI1, TRPC1, STIM1, ITPR1, ITPR2, ITPR3). (A) Enrichment of the gene set by the addition of 10 of their closet interactors (confidence = 0.8) using StringApp from Cytoscape mapped with log2(FC) intensity (color) and negative log10 of adjusted P value (size) obtained from the differential expression analysis between pancreatic cancer with low or high TMEM16A expression. (B) Determination of the optimal number of clusters using “NbClust” package on the transcriptomic data obtained from the TCGA-PAAD dataset and restricted to our gene set defined in A. (C) PCA representation of pancreatic cancer samples from TCGA-PAAD dataset restricted to our gene set mRNA expression and mapped with the unsupervised K-means clusterization of these samples into three clusters. (D) Kaplan–Meier survival of patients according their classification into three clusters defined in B. (E) Histogram representing the distribution of TCGA-PAAD samples according the histopathological subtypes defined (19) and in function of clusters defined in C. (F) Determination of the optimal number of clusters using “NbClust” package on the transcriptomic data obtained from the TCGA-PAAD dataset removed from PNET-associated samples and restricted to our gene set defined in A. (G) PCA representation of pancreatic cancer samples from TCGA-PAAD dataset removed from PNET-associated samples restricted to our gene set mRNA expression and mapped with the unsupervised K-means clusterization of these samples into three clusters. (H) Kaplan–Meier survival of patients according their classification into three clusters defined in F. (I) Using the TCGA-PAAD dataset cleared from PNET-associated samples, the ranked list of DEGs for each cluster (obtained by DEA comparing each cluster against two others) was submitted to the GSEA and then visualized as network with the EnrichmentMap app of Cytoscape (FDR P < 0.01). Nodes represent each gene set/GO functions. Node size represents the number of genes in each gene set/GO. One color was applied for each cluster to gene set/GO that conditionally have a normalized enrichment score (NES) value > 1 in this cluster and a NES value < 1 in two others.
The Kaplan–Meier overall survival analysis of these different clusters revealed that the groups of genes we chose is sufficient to isolate a subset of surviving patients and to differentiate two other clusters based on their overall survival (Fig. 6D). According to the histopathological classification (19), samples in cluster 2 are exclusively pancreatic neuroendocrine tumors (PNET). The other clusters regroup a mix of various pancreatic cancer types (Fig. 6E and Dataset S3).
Thus, this group of genes is sufficient for differentiating PNET from other pancreatic cancers. To test the possibility that PNET as a particular group of pancreatic cancer may have obscured other subsets of pancreatic cancer, we removed these samples from our dataset and determined the optimal number of clusters. Surprisingly, we found that three is still the optimal number of clusters from the restricted dataset without PNET (Fig. 6 F and G). However, clustering of this restricted dataset into three clusters does not provide significant differences on the overall survival of the different groups (Fig. 6H). To understand the molecular signature of these clusters, we performed a DEA of each of these clusters against each of the other two clusters, followed by a GSEA on the ranked list of differentially expressed genes (DEG). We graphically represented significant GO functions [false-discovery rate (FDR) < 0.01)] using the EnrichmentMap app of Cytoscape (Fig. 6I).
Interestingly, we observed that samples from cluster 1 are enriched in GO functions associated with immunity, ion channels, cell migration, phospholipid signaling, and integrins, suggesting that pancreatic cancer belonging to this cluster could be potentially more aggressive or more prone to immune cell infiltration (Fig. 6I). The enrichment of these samples in GO functions associated with ion channels could suggest an important involvement of this particular group of proteins in pancreatic cancer development and provides a new reservoir of therapeutic target to validate. Samples belonging to cluster 2 are enriched in GO functions associated with cell cycle, RNA processing, metabolism, and mitochondria, suggesting that these pancreatic tumors may have undergone metabolic changes associated with mitochondrial dysfunction (Fig. 6I). Samples from cluster 3 are enriched in GO functions associated with DNA processing, epithelial cell differentiation, and metabolism, suggesting that these tumors may have gone through epithelial differentiation and accumulated aberrant mutations (Fig. 6I).
Discussion
Pancreatic cancer is heterogeneous; classification via biomarkers helps in better understanding of the etiology and possible treatment of different subtypes of pancreatic cancer. In the TCGA-PAAD dataset, the mRNA encoding TMEM16A is up-regulated in 75% of pancreatic cancer compared with the expression level observed in normal pancreas analyzed by the GTEx consortium. This up-regulation is also observed at the protein level in the collection of tissues of the Human Protein Atlas. At the clinical level, up-regulation of TMEM16A is associated with a poor prognosis, underscoring the significance of TMEM16A involvement in pancreatic cancer. Our findings, taken together with previous observations of up-regulation of TMEM16A in different cancers, such as breast cancer, HNSCC, or GIST, supports the notion that TMEM16A facilitates oncogenesis (9). Whereas our finding of a correlation between TMEM16A expression and the overrepresentation of genes associated with the EGFR signaling pathway is in agreement with previous reports describing the role of TMEM16A on the EGFR signaling pathway in breast cancer and HNSCC cell lines (11, 12), we found that EGFR ligands are up-regulated in pancreatic tumors with up-regulation of TMEM16A. Consistent with the ligand-dependent EGFR signaling in pancreatic cancer (13, 14, 17), our findings of EGFR ligand up-regulation as well as EGF-induced and TMEM16A-dependent EGFR signaling highlights the unique aspect of TMEM16A contribution to ligand-dependent EGFR signaling in pancreatic cancer.
While the contribution of TMEM16A to EGFR activation and associated-signaling pathways has been investigated in other cancers (11, 12), the nature of the molecular mechanism associating TMEM16A to EGFR remains an open question. Here, our investigation using a pancreatic cancer cell line, AsPC-1, has revealed that ligand-induced activation of the EGFR signaling pathway induces TMEM16A-dependent chloride current, as well as a Ca2+ response due to SOCE. Thus, the regulation of chloride and calcium homeostasis by TMEM16A appears to be a critical parameter during ligand-induced EGFR signaling in pancreatic cancer cells. We found that ORAI-1 and TRPC-1 are involved in the Ca2+ influx and that IP3R probably controls the Ca2+ release from internal store. Pharmacological inhibition of TMEM16A channel activity or molecular silencing of TMEM16A reduces both Ca2+ responses. Additionally, using a proximity ligation assay, we observed an interaction between TMEM16A and EGFR in AsPC-1.
Involvement of TMEM16A in Ca2+ release from internal store in pancreatic cancer cells is unexpected. Previous studies have identified TMEM16A as a downstream effector that can be activated by Ca2+ either through release from the internal store or influx of Ca2+ from extracellular media (8, 27, 28, 37, 38). This has led to the suggestion that TMEM16A is activated by IP3R-mediated Ca2+ release from the internal store, and then SOCE is initiated by the depletion of intracellular Ca2+ stores to sustain TMEM16A activity by refilling Ca2+ stores to sustain IP3R-mediated Ca2+ release (37). Our finding that TMEM16A activity itself regulates Ca2+ release from the internal store thus unveils a role for TMEM16A by positioning it upstream of the activation of IP3R as an initiator of the SOCE. In addition, our findings that TMEM16A does not contribute to IP3-independent or P2Y-related IP3-dependent SOCE suggest that the role of TMEM16A on EGF-induced SOCE could be explained by its proximity with EGFR.
Phospholipids could be a link between TMEM16A activity and this upstream role of TMEM16A in EGFR signaling. EGFR activation has been associated with PLC activation, PIP2 cleavage, and IP3 release to drive Ca2+ release from the internal store and promote EGFR endocytosis (26, 39). EGFR phosphorylation is also sensitive to the presence of PIP2 (40). Recent studies reveal that TMEM16A-mediated chloride homeostasis enhances PIP2 clustering at the plasma membrane (41). PIP2 could also modulate TMEM16A activity by preventing its rundown or desensitization (41–44). Given these findings, we hypothesize that TMEM16A interacts with EGFR and promotes EGFR localization into PIP2-rich microdomains at the plasma membrane, thereby facilitating the local release of IP3 in response to EGFR-induced PLC activation to induce Ca2+ release from internal store and SOCE. This proposed mechanism will create a positive feedback loop, sustaining TMEM16A activity, PIP2 clustering and IP3 release, to boost the Ca2+ signaling and chloride current. Future investigations will be required to validate this hypothesis.
We found that TMEM16A involvement in pancreatic cancer differs from the known role of TMEM16A in other cancers. In breast cancer and HNSCC cells, TMEM16A regulates EGFR phosphorylation and the activation of its downstream signaling pathways, such as Akt, ERK, and CamKII, and TMEM16A and EGFR expression were positively correlated (11, 12). Here, by Western blot and phosphoproteomics analysis, we found that silencing of TMEM16A modifies the pattern of EGF-induced phosphorylation of EGFR without affecting Akt or ERK phosphorylation. At the global level, TMEM16A expression strongly promotes the phosphorylation of proteins associated with EGF-induced EGFR signaling pathways and cell motility. In addition, we found that silencing TMEM16A does not significantly affect EGFR expression or its trafficking to the plasma membrane. Thus, notwithstanding the contribution of TMEM16A to EGFR activation to many cancer types, the downstream effects appear to be cancer-dependent, underscoring the importance of investigating the unusual ligand-induced and TMEM16A-dependent EGFR-signaling in pancreatic cancer. Interestingly, we found that TMEM16A-dependent EGFR phosphorylation is independent of the SOCE while SOCE inhibition could modify the global pattern of phosphorylation induced by EGF. This indicates that TMEM16A-dependent Ca2+ signaling does not support EGFR activation but is important for mediating EGF-induced remodeling of the phosphoproteome. To which extent EGF-induced SOCE contributes to the remodeling phosphoproteome remains to be investigated. Previous reports indicate that TMEM16A regulates EGFR activation through its chloride channel activity. Here, we ruled out the possibility of a Ca2+-dependent feedback regulation initiated by TMEM16A chloride channel activity. Future investigations will be required to elucidate the underlying molecular mechanism by which TMEM16A regulates EGFR activation.
We also observed that silencing of TMEM16A led to remodeling of the phosphoproteome of AsPC-1 that is reminiscent of EGF-induced phosphorylations. This finding could be indicative of a repressive role of TMEM16A that prevents targets of the EGFR signaling pathway from being phosphorylated in the absence of EGFR ligands. By maintaining a low level of phosphorylation of these targets in the absence of EGFR ligands, TMEM16A activity preserves the amplitude of the cellular response to EGF and the associated genetic reprogramming so as to enhance the signal above the background.
Recent genomic investigations have proposed several classifications of pancreatic cancer (19, 34–36). These classifications greatly improve our understanding of pancreatic cancer development and evolution, but they focus primarily on PDAC, the major form of pancreatic cancer. Here, in an attempt to examine the clinical relevance of the TMEM16A-dependent EGFR-induced Ca2+ signaling pathway characterized in vitro in AsPC-1, we focused on molecules identified in this pathway as a specific gene set, and we examined transcriptional data available for this gene set to classify patients into several groups. Unexpectedly, this small gene set was sufficient to distinguish neuro-endocrine tumors from other pancreatic cancers and to identify three clusters in the remaining pancreatic tumors with distinct genetic profiles that could reflect their molecular underpinning. Further characterizations of pancreatic cancers belonging to each of these clusters will: (i) help us better understand the contribution of the TMEM16A-dependent EGFR-induced Ca2+ signaling in each cluster, (ii) validate and improve this classification, (iii) improve our understanding of the inherent variability and heterogeneity of pancreatic cancers, and (iv) facilitate the development of therapeutic strategies with specific targeting of pancreatic cancers based on the molecular signature of the cluster they belong.
In summary, our study reveals a role of TMEM16A in EGFR-related signaling pathways of pancreatic cancer cells through the regulation of both chloride and calcium homeostasis. This signaling pathway that depends on both EGF ligands and TMEM16A activity provides a tool to improve the clinical classification of pancreatic tumors.
Materials and Methods
Reagents and Cell Culture.
All materials, chemicals, drugs, and antibodies used in this paper are listed in Dataset S4. The AsPC-1 cell line was purchased from ATCC (CRL-1682) and cultured as recommended by the manufacturer. Briefly, cells were maintained in a RPMI medium 1640 supplemented with FBS 10% and Penicillin/Streptomycin.
shRNA and siRNA.
Lentiviral particles were obtained from the University of California, San Francisco Viracore using pLKO.1 plasmids purchased from GE Dharmacon. On day 1, AsPC-1 cells were plated in a six-well plate at a density of 20,000 cells per well in complete medium. On day 2, medium was removed, and cells were incubated in complete medium containing 8 µg/mL of hexadimethrin bromide (Millipore Sigma) and transduced at a multiplicity of infection of 5. Clones RHS6848 (nontarget shRNA) and TRCN0000040265 and TRCN0000040263 (shTMEM16A targeted) were used for transduction. On day 4, puromycin (0.5 mg/mL) was added in fresh medium to start selection of transduced cells. siRNA targeting ORAI-1 or TRPC-1 were obtained from GE Dharmacon. On day 1, AsPC-1 cells were plated in a six-well plate at a density of 20,000 cells per well in complete medium. On day 2, medium was removed, and cells were tranfected with 50 nM of siRNA of interest in complete medium using JetPrime siRNA transfection protocol (Polyplus). On day 3, transfected AsPC-1 were trypsinized and plated either on coverslips or on 35-mm dishes.
Electrophysiology/Patch-Clamp.
Cells were trypsinized and plated on coverslips for 2 h. Then, coverslips were transferred to a recording chamber on a Nikon-TE2000 Inverted Scope (Nikon Instruments). The external solution was a physiological saline solution (PSS): 150 mM NaCl, 4 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, 10 mM d-glucose (pH adjusted at 7.3 with HCl, 295 mOsm/L). Patch borosilicate pipettes (Sutter Instrument) were pulled from a Sutter P-97 puller with resistances of 3–5 MΩ for whole-cell recordings. Internal recording solution was: 5 mM NaCl, 140 mM KCl, 1 mM MgCl2, 10 mM Hepes, 10 mM d-glucose (pH adjusted to 7.2 using HCl, 290 mOsm/L) completed with either a combination of CaCl2 0.85 mM and EGTA 1 mM or a combination of CaCl2 0.035 mM and EGTA 0.1 mM to achieve a final intracellular calcium concentration of 1 or 0.1 µM respectively. All experiments were performed at room temperature (22–24 °C). EGF and benzbromarone were added to the external solution, which were then administered at the vicinity of the cell selected for recording via a VC3-8xP pressurized perfusion system (ALA Science). Data were acquired using a Multiclamp 700B amplifier controlled by Clampex 10.2 via Digidata 1440A (Axon Instruments). A ramp protocol ranging from −100 to +100 mV in 2 s was applied from a holding of 0 mV. All data were analyzed using pClamp10 (Molecular Devices) and R.
Calcium Imaging.
Forty-eight hours before the experiment, subconfluent cells were trypsinized and plated in 35-mm plastic dishes. A day before experiment, media was replaced by a serum-free media and subject to an overnight (16–20 h) incubation. On the day of the experiment, cells were washed with PBS 1× and then loaded with 5 µM calcium-sensitive dye Fluo-8 (AAT Bioquest) at 37 °C for 35 min in a PSS containing: 150 mM NaCl, 4 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, and 10 mM d-glucose. Cells were then washed twice in PSS and incubate in PSS solution for 3 min before acquisition. For conditions requiring the depletion of extracellular calcium (PSS 0Ca), calcium chloride was omitted in the PSS preparation and EGTA 0.1 mM was added to chelate the residual calcium. For all solutions, pH was adjusted to 7.3 (295 mOsm/L). For drugs testing or changing the solution, an equal volume of a 2× concentrated compound of interest (prepared in PSS) was added to the dish. Fluorescence was acquired using a Nikon TE2000 inverted scope equipped with a thermostated chamber at 37 °C (Okolab), a CoolSnap HQ2 camera (Photometrics), a combination of dichroic cube, excitation and emission filters at 488 and 515 nm (SemRock), and a XCite Lamp (Excelitas Tech). Images were acquired every 3 s for 15 min. Fluorescence intensity of individual cells was obtained by defining a region-of-interest for each individual cell, substracting the background fluorescence and normalizing to the fluorescent intensity measured in the first image. All image processing was performed using ImageJ Software (NIH).
Time-Lapse Imaging.
Forty-eight hours before the experiment, subconfluent cells were trypsinized and plated at low confluence (5,000 cells per well) in a 12-well plate. On the day of experiments, media was changed 1 h the beginning of experiments. Time-lapse series were recorded using a Nikon TE2000 inverted microscope equipped with a thermostated chamber at 37 °C (Okolab), a CoolSnap HQ2 camera (Photometrics). Acquisition was performed over a 12-h period with an image every 5 min and two images per field. Cells were imaged every 10 min. For tracking cells, 12 individual cells in each field were manually tracked using the MTrack2 plugin of ImageJ/Fiji. For measuring cell migration, total and Euclidian distance traveled from each cells were measured.
Phosphoproteomics.
Forty-eight hours before the experiment, subconfluent cells were trypsinized and plated in 150-mm dishes (one for each condition). The day before the experiment, media was replaced by a serum-free media and subjected to overnight (16–20 h) incubation. On the day of the experiment, cells were treated in the presence or absence of EGF (50 ng/mL) for 30 min. Then, cells were washed in cold PBS and lysed in a solution containing 8 M urea, 0.1 M Tris pH 8.5, 40 mM 2-CAA, 10 mM TCEP, and 1× HALT protease/phosphate inhibitors. Collected lysates were immediately frozen on dry ice and stored in −80 °C while waiting for next steps. Lysates were sonicated, digested overnight with trypsin 1:50 (wt/wt) enzyme-to-substrate ratio, desalted and enriched for phosphopeptides using Fe3+-IMAC columns, dried down and stored at −80 °C. Peptides were reconstituted and injected (1 µg of samples) onto the instrument (LC-MS/MS). Raw data were then processed by MaxQuant software using Phospho (STY) settings and match-between-runs. Data were then filtered, normalized and submitted to hybrid imputation before analyzing them. Log2(FC) and corresponding P values for each peptides were generated.
Statistics.
All graphs and statistical analysis were generated using R software (CRAN project). Significant differences were determined with Mann–Whitney nonparametric test for small samples. For large samples, Shapiro and Bartlett tests were used to determine the normality and homogeneity of the sample. Student’s t test and ANOVA were used. In all cases, data represent mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, N.S. P > 0.05.
Supplementary Material
Acknowledgments
We thank Drs. Chin Fen Teo, Tina Han, and Sami Tuomivaara for helpful discussions and for editing the article. This study is supported in part by NIH Grants R01NS069229 (to L.Y.J.), and DP2 OD022552 and K08 CA184116 (to A.P.W.); and by a grant from the Philippe Foundation (to D.C.). Y.N.J. and L.Y.J. are Howard Hughes Medical Institute investigators.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900703116/-/DCSupplemental.
References
- 1.Collisson E. A., Maitra A., Pancreatic cancer genomics 2.0: Profiling metastases. Cancer Cell 31, 309–310 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Rahib L., et al. , Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74, 2913–2921 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Caputo A., et al. , TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 322, 590–594 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Schroeder B. C., Cheng T., Jan Y. N., Jan L. Y., Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 134, 1019–1029 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y. D., et al. , TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455, 1210–1215 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Huang F., et al. , Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc. Natl. Acad. Sci. U.S.A. 106, 21413–21418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedemonte N., Galietta L. J. V., Structure and function of TMEM16 proteins (anoctamins). Physiol. Rev. 94, 419–459 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Wang Q., Leo M. D., Narayanan D., Kuruvilla K. P., Jaggar J. H., Local coupling of TRPC6 to ANO1/TMEM16A channels in smooth muscle cells amplifies vasoconstriction in cerebral arteries. Am. J. Physiol. Cell Physiol. 310, C1001–C1009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bill A., Alex Gaither L., The mechanistic role of the calcium-activated chloride channel ANO1 in tumor growth and signaling. Adv. Exp. Med. Biol. 966, 1–14 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Wang H., et al. , Cell-specific mechanisms of TMEM16A Ca2+-activated chloride channel in cancer. Mol. Cancer 16, 152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bill A., et al. , ANO1/TMEM16A interacts with EGFR and correlates with sensitivity to EGFR-targeting therapy in head and neck cancer. Oncotarget 6, 9173–9188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britschgi A., et al. , Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc. Natl. Acad. Sci. U.S.A. 110, E1026–E1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ardito C. M., et al. , EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 22, 304–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navas C., et al. , EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell 22, 318–330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpenter B. L., et al. , Integrin α6β4 promotes autocrine epidermal growth factor receptor (EGFR) signaling to stimulate migration and invasion toward hepatocyte growth factor (HGF). J. Biol. Chem. 290, 27228–27238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang M., et al. , EGFR-dependent pancreatic carcinoma cell metastasis through Rap1 activation. Oncogene 31, 2783–2793 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., et al. , Genetic ablation of smoothened in pancreatic fibroblasts increases acinar-ductal metaplasia. Genes Dev. 30, 1943–1955 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stock A.-M., et al. , Induction of pancreatic cancer cell migration by an autocrine epidermal growth factor receptor activation. Exp. Cell Res. 326, 307–314 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network , Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carithers L. J., Moore H. M., The Genotype-Tissue Expression (GTEx) project. Biopreserv. Biobank. 13, 307–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamazon E. R., et al. ; GTEx Consortium , Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 50, 956–967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulkarni S., et al. , TMEM16A/ANO1 suppression improves response to antibody-mediated targeted therapy of EGFR and HER2/ERBB2. Genes Chromosomes Cancer 56, 460–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., et al. , Expression of amphiregulin predicts poor outcome in patients with pancreatic ductal adenocarcinoma. Diagn. Pathol. 11, 60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauter D. R. P., Novak I., Pedersen S. F., Larsen E. H., Hoffmann E. K., ANO1 (TMEM16A) in pancreatic ductal adenocarcinoma (PDAC). Pflugers Arch. 467, 1495–1508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caldieri G., et al. , Reticulon 3-dependent ER-PM contact sites control EGFR nonclathrin endocytosis. Science 356, 617–624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delos Santos R. C., et al. , Selective regulation of clathrin-mediated epidermal growth factor receptor signaling and endocytosis by phospholipase C and calcium. Mol. Biol. Cell 28, 2802–2818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin X., et al. , Activation of the Cl- channel ANO1 by localized calcium signals in nociceptive sensory neurons requires coupling with the IP3 receptor. Sci. Signal. 6, ra73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Concepcion A. R., et al. , Store-operated Ca2+ entry regulates Ca2+-activated chloride channels and eccrine sweat gland function. J. Clin. Invest. 126, 4303–4318 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambudkar I. S., de Souza L. B., Ong H. L., TRPC1, Orai1, and STIM1 in SOCE: Friends in tight spaces. Cell Calcium 63, 33–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan P. G., Rao A., Store-operated calcium entry: Mechanisms and modulation. Biochem. Biophys. Res. Commun. 460, 40–49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakriya M., Lewis R. S., Store-operated calcium channels. Physiol. Rev. 95, 1383–1436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salido G. M., Sage S. O., Rosado J. A., TRPC channels and store-operated Ca(2+) entry. Biochim. Biophys. Acta 1793, 223–230 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Duvvuri U., et al. , TMEM16A induces MAPK and contributes directly to tumorigenesis and cancer progression. Cancer Res. 72, 3270–3281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey P., et al. ; Australian Pancreatic Cancer Genome Initiative , Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Collisson E. A., et al. , Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moffitt R. A., et al. , Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47, 1168–1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma A., Ramena G., Yin Y., Premkumar L., Elble R. C., CLCA2 is a positive regulator of store-operated calcium entry and TMEM16A. PLoS One 13, e0196512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y., Birnbaumer L., Singh B. B., TRPC1 regulates calcium-activated chloride channels in salivary gland cells. J. Cell. Physiol. 230, 2848–2856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Rheenen J., et al. , EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 179, 1247–1259 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michailidis I. E., et al. , Phosphatidylinositol-4,5-bisphosphate regulates epidermal growth factor receptor activation. Pflugers Arch. 461, 387–397 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He M., et al. , Cytoplasmic Cl− couples membrane remodeling to epithelial morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 114, E11161–E11169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang S., et al. , Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Jesús-Pérez J. J., et al. , Phosphatidylinositol 4,5-bisphosphate, cholesterol, and fatty acids modulate the calcium-activated chloride channel TMEM16A (ANO1). Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 299–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye W., et al. , Phosphatidylinositol-(4, 5)-bisphosphate regulates calcium gating of small-conductance cation channel TMEM16F. Proc. Natl. Acad. Sci. U.S.A. 115, E1667–E1674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.