Significance
Mutations in the metabolic enzyme isocitrate dehydrogenase are thought to contribute to cancer formation by inhibiting differentiation. The mechanisms involved in this block are not well understood, however. Here, we use a well-defined model system of differentiation of mesenchymal cells into muscle cells to outline how these mutations block the normal differentiation process through their effects on chromatin remodeling.
Keywords: isocitrate dehydrogenase, 2-hydroxyglutarate, differentiation, myogenesis, sarcoma
Abstract
Oncogenic IDH1/2 mutations produce 2-hydroxyglutarate (2HG), resulting in competitive inhibition of DNA and protein demethylation. IDH-mutant cancer cells show an inability to differentiate but whether 2HG accumulation is sufficient to perturb differentiation directed by lineage-specifying transcription factors is unknown. A MyoD-driven model was used to study the role of IDH mutations in the differentiation of mesenchymal cells. The presence of 2HG produced by oncogenic IDH2 blocks the ability of MyoD to drive differentiation into myotubes. DNA 5mC hypermethylation is dispensable while H3K9 hypermethylation is required for this differentiation block. IDH2-R172K mutation results in H3K9 hypermethylation and impaired accessibility at myogenic chromatin regions but does not result in genome-wide decrease in accessibility. The results demonstrate the ability of the oncometabolite 2HG to block transcription factor-mediated differentiation in a molecularly defined system.
The classic model of cellular differentiation centers on lineage-specifying transcription factors activating cognate targets and driving expression programs that mediate differentiation. More recently, it has been found that these transcription factors do not work in isolation. An increasing body of work has suggested that chromatin modifications and the pathways that regulate them can influence the ability of lineage-specifying transcription factors to activate their downstream pathways. Such a mechanism would allow differentiation to be influenced by environmental and metabolic conditions and controlled by signal transduction events beyond those that directly influence the activation of lineage-determining transcription factors.
One of the best studied models of in vitro cellular differentiation is that of the immortalized fibroblast cell line 10T1/2 (1). When 10T1/2 cells are treated with 5-azacytidine to induce DNA demethylation, they undergo differentiation into myotubes, adipocytes, or chondrocytes spontaneously. Expression profiling of 5-azacytidine–treated cells led to the identification of the lineage-specifying transcription factor MyoD (2, 3). When 10T1/2 cells were transfected with MyoD, the cells were reprogrammed to differentiate into myotubes in the absence of 5-azacytidine. This work demonstrated that DNA methylation can function to suppress the expression of lineage-specifying transcription factors. However, once such pioneering transcription factors were expressed, cellular differentiation was believed to be the inexorable consequence, as other transcription factors involved in lineage specification are activated by the pioneer factors and effect differentiation.
Recently, a high number of sarcomas have been found to harbor neomorphic mutations in the metabolic enzymes isocitrate dehydrogenase (IDH) 1 and 2, resulting in the production of 2-hydroxyglutarate (2HG) (4–7). The accumulation of 2HG in tumor cells leads to competitive inhibition of the alpha-ketoglutarate–dependent dioxygenases involved in DNA and histone demethylation (8–11). In hematopoietic tumors, IDH mutations were found to be mutually exclusive with loss-of-function mutations in the DNA demethylase, Tet2 (12, 13). By analogy, this suggested that 2HG might inhibit myogenic differentiation at the step of activation of lineage-specifying factors such as MyoD by blocking DNA demethylation. Furthermore, such a model would allow us to test whether once MyoD expression was induced, subsequent DNA and/or histone demethylation contributed to MyoD-directed cellular reprogramming and differentiation.
To address these questions, we have studied myogenic differentiation in 10T1/2 cells in the presence or absence of 2HG-producing IDH2 mutations. 2HG producing mutations blocked the ability of 5-azacytidine to induce differentiation into myotubes. Surprisingly, however, we found that 2HG or a 2HG-producing mutation also blocked the ability of MyoD to drive differentiation. This effect is not heritable and is dependent on hypermethylation of H3K9 as loss or inhibition of the H3K9 methyltransferase EHMT1 is sufficient to rescue the differentiation block. We show that oncogenic IDH2-R172K results in increased hypermethylation of H3K9, which, in turn, inhibits the gain in chromatin accessibility required for differentiation. This inhibition is not genome-wide but is seen preferentially at sites that gain accessibility after MyoD activation. These results provide mechanistic insights into how oncogenic IDH mutations contribute to suppressing differentiation.
Results
2HG-Producing IDH2 Mutations Block the Ability of MyoD to Drive Myotube Differentiation.
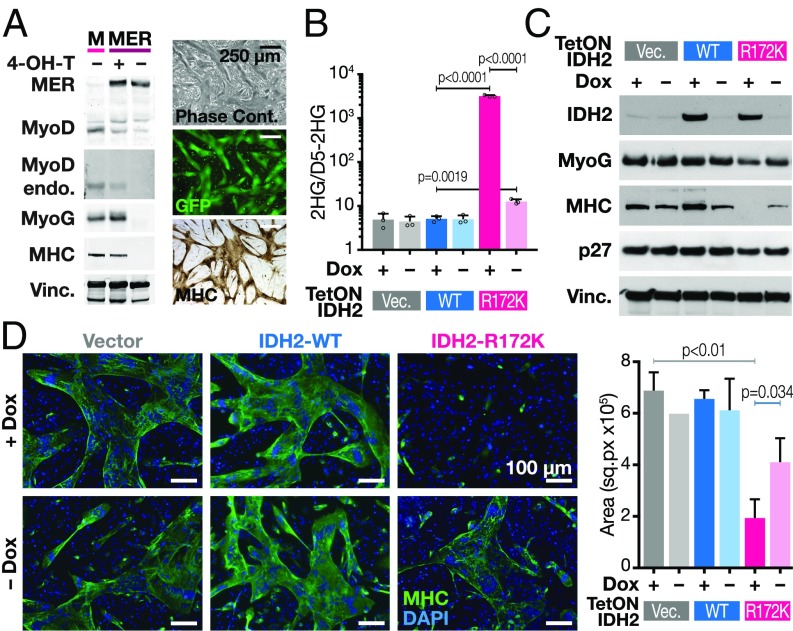
To dissect the effects of oncogenic IDH mutations on transcription factor-induced cellular differentiation, we sought a model where we could temporally control expression of both oncogenic IDH mutations and a lineage-specifying transcription factor that drives differentiation. 10T1/2 cells are mesenchymal progenitors capable of differentiating into myotubes, chondrocytes, or adipocytes upon exposure to a demethylating agent. Moreover, they readily differentiate into fused myotubes upon expression of either the canonical lineage-specifying transcription factor MyoD or 4-OH–tamoxifen-mediated activation of MyoD-ER (Fig. 1A) (14). Expression of ectopic MyoD leads to activation of the endogenous MyoD locus and the bHLH transcription factor myogenin within 12 h. Within 36–48 h, muscle-specific myosin heavy chain (MHC) expression is induced. At 48 h, MyoD expressing 10T1/2 cells fuse into 25–100 nuclei-containing myotubes that express MHC as seen by immunohistochemistry (Fig. 1A).
Fig. 1.
2HG-producing IDH2 mutations block the ability of MyoD to drive myotube differentiation. (A) 10T1/2 cells were transduced with wild-type MyoD (M) or MyoD-ER (MER) fusion cDNA-encoding lentiviral vectors and grown in the presence or absence of 4-OH-T. Western blots were probed with indicated antibodies (Top shows exogenous MyoD-ER and endogenous MyoD; second blot shows only endogenous MyoD; MyoG, myogenin; MHC, myosin heavy chain; Vinc, vinculin). Right are representative images of myotubes by phase contrast microscopy (Top), GFP (Middle), and immunohistochemistry for MHC (Bottom). (B) 10T1/2 cells were transduced with constructs containing doxycycline inducible (Tet-ON) vector, IDH2-WT, or IDH2-R172K in the presence or absence of doxycycline. 2HG levels shown were measured by GCMS and normalized to spiked-in D5-2HG. Data are mean ± SD of three biological replicates. P values were determined using unpaired Student’s t test. (C) Western blot of 10T1/2 cells transduced with doxycycline-inducible constructs as indicated in B. (D) Fluorescence microscopy of cells as described in C. MHC is in green; DAPI is in blue. Quantification of MHC stained areas is shown on Right. Data are mean ± SD of MHC signal from three representative areas. P values were determined using unpaired Student’s t test.
To determine whether IDH mutations are able to block MyoD-driven differentiation, we transduced 10T1/2 cells with lentiviral vectors encoding wild-type or R172K IDH2 cDNAs under the control of a doxycycline-dependent promoter. When grown in the presence of doxycycline, these cells produced 2HG at levels significantly higher than background (Fig. 1B). When 4-OH-tamoxifen (4-OH-T) was added to activate MyoD, cells transduced with IDH2-WT readily activated MHC expression and fused into large MHC-expressing myotubes (Fig. 1 C and D). In contrast, cells transduced with IDH2-R172K were incapable of either activating MHC expression or fusing into myotubes. Not all MyoD targets were equally affected in the IDH2-R172K setting. Early targets of MyoD like MyoG and p27 were still expressed, albeit at lower levels, while late targets like MHC failed to be induced.
The block in differentiation in the IDH2-R172K setting is not the result of an inability to exit the cell cycle, consistent with the MyoD-induced expression of p27. Reduction in EdU incorporation after 4-OH-T induction of MyoD still occurred in the IDH2-R172K setting (SI Appendix, Fig. S1A). Additionally, the CDK4/6 inhibitor Palbociclib was unable to rescue the differentiation block (SI Appendix, Fig. S1B) despite effectively inhibiting EdU incorporation. This argues that prevention of cell cycle exit is not the mechanism by which IDH2-R172K blocks MyoD-induced differentiation.
The above results suggest that the oncogenic IDH2-R172K mutation blocks the ability of the lineage transcription factor MyoD to drive differentiation by impairing a subset of the effects mediated by MyoD’s transcriptional targets.
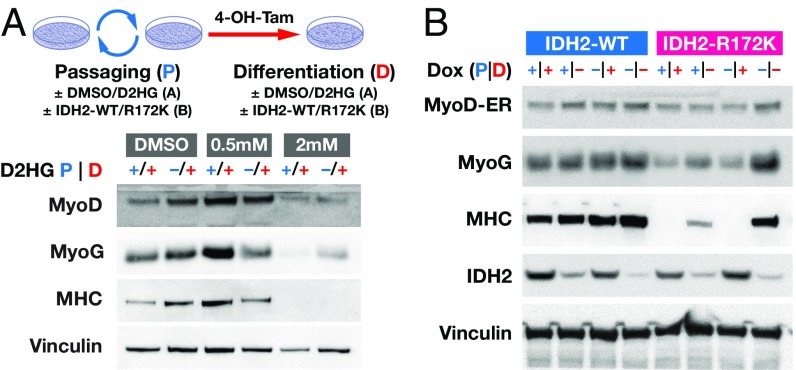
Continuous 2HG Production Is Required for the 2HG-Dependent Differentiation Block.
The inability of MyoD to drive differentiation in IDH2-R172K mutant cells may be the result of a repressed chromatin state that MyoD is incapable of accessing and that is inherited with cell division. Alternatively, oncogenic IDH mutations may actively impair events downstream of MyoD activation. To distinguish between these two possibilities MyoD-ER–expressing 10T1/2 cells were exposed to 2HG for four passages before MyoD activation and/or during the 24 h after 4-OH-T induction of MyoD. A clear differentiation block was seen when 2HG was present both during passaging and MyoD activation as evidenced by the lack of MHC expression (Fig. 2A). A comparable block was also observed when 2HG was present only following addition of 4-OH-T, i.e., only after MyoD is active and cells have exited the cell cycle. These results demonstrate that the product of mutant IDH1/2, 2HG, is capable of inhibiting the ability of MyoD to drive differentiation even after cells have induced MyoD and undergone cell cycle arrest.
Fig. 2.
Continuous 2HG production is required for the 2HG-dependent differentiation block. (A, Upper) Strategy for experiments shown in A and B; details indicated in the text. (A, Lower) 10T1/2 cells were passaged four times and differentiated for 2 days with 4-OH-T with 0.5 mM or 2 mM 2HG added throughout the experiment (+/+) or only during the differentiation step (−/+). Western blots were probed with antibodies indicated at Left. The line that appears in the reproduced panel quantitating MyoD represents the top edge of the membrane that was probed. The membrane was cut after transfer to allow the upper part of the western blot to be probed independently. (B) Western blot of 10T1/2 cells expressing doxycycline-inducible IDH2-WT or IDH2-R172K passaged and/or differentiated in the presence or absence of doxycycline.
To account for off-target or dosing effects of 2HG, we performed a complementary experiment to assess when inducible expression of control IDH2-WT or mutant IDH2-R172K was capable of inhibiting MyoD-dependent differentiation. IDH2-WT or IDH2-R172K expression was induced for 10 passages before differentiation and/or during the activation of MyoD in 10T1/2 cells harboring the respective inducible vectors and a MyoD-ER construct. Differentiation was assessed by expression of the early marker myogenin and the late marker MHC. As seen for the experiment with 2HG, activation of IDH2-R172K during the differentiation period alone was sufficient to block the ability of MyoD to induce differentiation (Fig. 2B). Moreover, the presence of IDH2-R172K during the 10 passages before MyoD activation was unable to block differentiation unless also present concurrently during MyoD activation. MHC expression was partially decreased in the condition in which IDH2-R172K had been expressed during passaging only. Although there was some leaky expression in the doxycycline system and low levels of 2HG are present in the IDH2-R172K lines even in the absence of doxycycline (Fig. 1B), these levels are not sufficient to block MyoD-induced differentiation. These two complementary results show that IDH2-R172K/2HG actively inhibit steps downstream of MyoD expression within the myogenic differentiation program and that IDH2-R172K mutations in this setting do not establish cellular memory of the undifferentiated state.
As the level of myogenin, an early MyoD target, was decreased when either mutant IDH2-R172K was induced or 2 mM exogenous 2HG was present, we investigated whether the reduced level of myogenin was still able to bind to its cognate targets by ChIP-qPCR. ChIP-Western blot confirmed selective immunoprecipitation of myogenin protein in nuclear extracts (SI Appendix, Fig. S2A). ChIP-qPCR showed that myogenin in the IDH2-WT condition was able to bind both to early and late myogenic target genes (SI Appendix, Fig. S2B). In IDH2-R172K mutant cells, myogenin was induced by MyoD expression to a significantly lower extent and bound to its cognate targets to a lower extent. These lower levels of myogenin in whole extracts were not a result of normal expression in a small fraction of cells in the lysate as immunofluorescence for myogenin confirmed consistent levels of cell-to-cell expression (SI Appendix, Fig. S2C). Furthermore, as previously reported (15), myogenin was excluded from the nucleus of fully differentiated myotubes.
Given the lower levels of myogenin seen in IDH2-R172K cells, we asked whether overexpression of myogenin could rescue the observed differentiation block. 10T1/2 cells expressing IDH2-WT or IDH2-R172K and inducible MyoD transgenes were further transduced with a control lentiviral vector or one expressing a myogenin cDNA. Overexpression of myogenin fully rescued the IDH2-R172K–mediated differentiation block (SI Appendix, Fig. S2D). While we cannot conclude that the cause of the IDH2-R172K–mediated differentiation block is an inability to reach a required myogenin threshold level of expression, myogenin overexpression can trigger a positive feedback loop sufficient to drive terminal differentiation.
C2C12 cells are mouse-derived myoblasts that constitutively express MyoD but differentiate into myotubes upon serum withdrawal or contact inhibition. While 10T1/2 cells represent an earlier multipotent progenitor capable of differentiating not only into myotubes but also into adipocytes and chondrocytes, C2C12 myoblasts can readily differentiate into myotubes but not into adipocytes or chondrocytes. To determine whether IDH mutations also impair the differentiation of C2C12 myoblasts, we transduced the same inducible IDH2-WT or IDH2-R172K constructs in these cells and allowed them to differentiate with serum withdrawal. IDH2-R172K did not impair the ability of C2C12 myoblasts to differentiate (SI Appendix, Fig. S2E). This is consistent with C2C12 myoblasts not requiring significant chromatin remodeling that could be inhibited by 2HG. These and the above results show that 10T1/2 and C2C12 cells expressing IDH2-R172K are permissive for differentiation if a sufficient constellation of transcription factors are ectopically introduced or is constitutively present.
2HG Blocks Differentiation by Impairing H3K9 Demethylation.
We next turned attention to delineating how oncogenic IDH mutations and 2HG block differentiation. To determine whether the inability to demethylate DNA 5mC residues is a mechanism by which 2HG blocks MyoD-mediated differentiation, we tested whether 5-azacytidine, a well-characterized DNA methyltransferase inhibitor, could rescue the 2HG-mediated differentiation block. As previously reported (1), treatment with 5-azacytidine is sufficient to induce differentiation of wild-type 10T1/2 cells. Treatment with 5-azacytidine of 10T1/2 cells expressing vector, IDH2-WT, or IDH2-R172K led to expression of MyoD in all three conditions although there was less induction in cells transduced with IDH2-R172K (SI Appendix, Fig. S3 A and B). However, while terminal differentiation to myotubes occurred in the vector and IDH2-WT cells, IDH2-R172K cells were blocked in their ability to differentiate. This result shows that DNA hypermethylation is not required for the IDH2-R172K–mediated differentiation block.
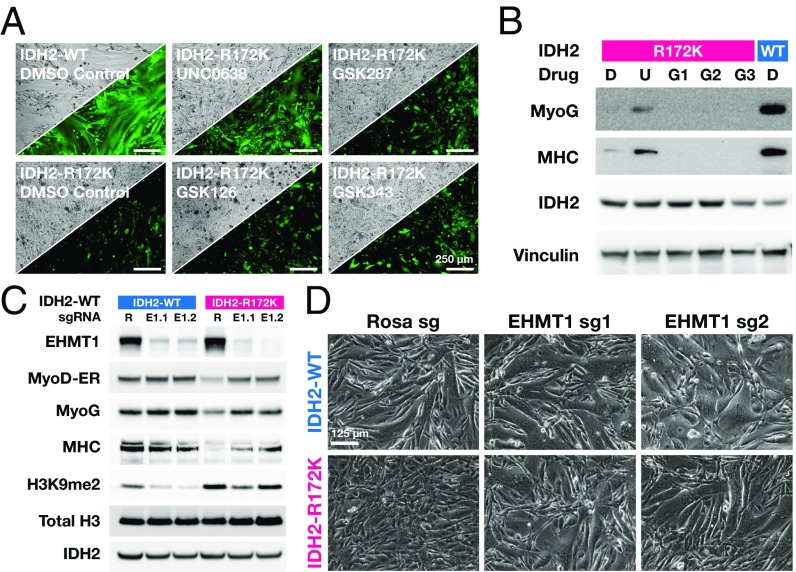
Whether histone lysine methylation marks require demethylation during MyoD-mediated differentiation is not known. By analogy to blocking DNA methylation with 5-azacytidine to induce 10T1/2 differentiation, we cultured 10T1/2 MyoD-ER cells expressing IDH2-WT or IDH2-R172K in the presence of inhibitors of several histone methyltransferases (UNC0638, inhibitor of EHMT1/2; GSK287, inhibitor of LSD1; GSK126 and GSK343, inhibitors of EZH2). At 48 h, the cells reached confluence and we exposed them to 4-OH-T to induce MyoD and trigger differentiation. Strikingly, the EHMT1/2 inhibitor, UNC0638, restored the ability of 10T1/2 cells expressing IDH2-R172K to form fused myotubes (Fig. 3A), an observation that correlated with increased expression of MHC by Western blot (Fig. 3B). In contrast, inhibition of LSD1 and EZH2 failed to restore MyoD-induced differentiation in 10T1/2 cells expressing IDH2-R172K.
Fig. 3.
2HG blocks differentiation by impairing H3K9 demethylation. (A) Phase contrast and GFP fluorescence microscopy of 10T1/2 MyoD-ER IDH2-WT (Upper Left) and IDH2-R172K (all others) transduced cells passaged with indicated inhibitors [DMSO (D), UNC0638 (U), GSK287 (G1), GSK126 (G2), GSK343 (G3)] and triggered to differentiate with 4-OH-T. (B) Western blot corresponding to cells indicated in A. (C and D) Western blot and phase contrast images of 10T1/2 MyoD-ER cells expressing IDH2-WT or IDH2-R172K and transduced with CRISPR Cas9/guide vectors targeting control Rosa26 (R) or EHMT1 using two different guides (E1.1 and E1.2). Blotted proteins indicated at C, Left).
We next investigated whether CRISPR-mediated deletion of the EHMT1/2 (GLP/G9a) H3K9 methyltransferase complex could phenocopy the effects seen with its purported inhibitor. MyoD-ER–transduced 10T1/2 cells expressing IDH2-WT or IDH2-R172K were transduced with control or EHMT1 targeting guide vectors, grown to near confluence, and MyoD-ER activated by 4-OH-T. CRISPR targeting led to nearly complete elimination of EHMT1 in the transduced population, and the expected decrease in dimethylated H3K9 was observed (Fig. 3C). The genetic knockout of EHMT1 led to restoration of MyoD-induced differentiation in 10T1/2 cells expressing IDH2-R172K as assayed by the induction of myogenin and MHC and myotube formation observed by phase-contrast microscopy (Fig. 3 C and D). Under these circumstances, addition of the EHMT1/2 inhibitor UNC0638 led to little further decrease in H3K9me2 levels or increased rescue of differentiation in this setting (SI ppendix, Fig. S3 C and D). These results show that in the context of MyoD-mediated differentiation, H3K9 hypermethylation is required for the ability of oncogenic IDH2-R172K to block differentiation.
The RNA N6-methyladenosine (m6A) demethylases FTO (Fat mass and obesity-associated protein) and ALKBH5 (AlkB homolog 5, RNA demethylase) are also potential targets of 2HG. To lower RNA N6-methyladenosine methylation, we targeted the methyltransferase Mettl3 responsible for placing the RNA methylation mark with two CRISPR guide vectors. Without RNA N6-methylation, differentiation was impaired in both the IDH2-WT and IDH2-R172K settings (SI Appendix, Fig. S3E). Thus, in the IDH2-R172K setting, it appears that 2HG-mediated inhibition of the m6A-specific demethylases FTO and ALKBH5 would favor differentiation. This result further reinforces the specificity of H3K9me2 as a mark whose removal is required for differentiation.
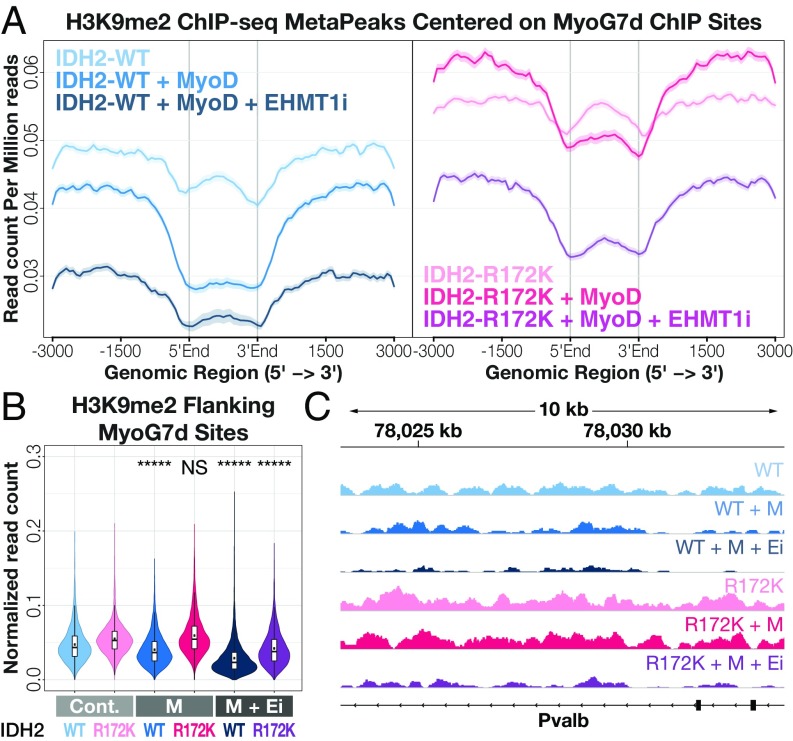
To better characterize the effects of IDH2-R172K mutations on global versus locus-specific H3K9 methylation, we performed ChIP-sequencing (seq) on IDH2-WT and IDH2-R172K samples before and after MyoD activation and in the presence or absence of UNC0638. Consistent with the whole cell lysate quantification (Fig. 3C), the overall levels of H3K9me2 and H3K9me3 were increased in IDH2-R172K cells compared with IDH2-WT cells (SI Appendix, Fig. S4), and treatment with the EHMT1/2 inhibitor UNC0638 led to global decreases in these marks. Across all genes, there was no appreciable difference in H3K9me2 or H3K9me3 levels upon activation of MyoD. When H3K9me2 and H3K9me3 ChIP-seq data were filtered for myogenic regions, there was a MyoD-dependent decrease in methylation within and flanking known MyoG-binding sites in IDH2-WT cells but not in IDH2-R172K cells (Fig. 4 and SI Appendix, Fig. S4). In fact, read counts for H3K9me2 in the flanks of MyoG-binding sites increased upon MyoD induction in IDH2-R172K cells. A decline in the H3K9me3 mark in flanking regions was also observed, albeit to a lesser extent (SI Appendix, Fig. S4). Importantly, while IDH2-R172K cells had much higher levels of H3K9me2 compared with WT cells upon MyoD activation, inhibition of EHMT1/2 led to a decrease in H3K9me2 to the range seen in IDH2-WT cells after MyoD activation (Fig. 4). This result reinforces the idea that MyoD activation results in removal of repressive H3K9me3 and H3K9me2 at myogenic regions that is, in turn, required for terminal differentiation, and this demethylation is inhibited by IDH2-R172K.
Fig. 4.
Oncogenic IDH mutations impair H3K9me2 demethylation at myogenic sites. (A) H3K9me2 MetaPeaks (mean read count per million mapped reads) for indicated samples centered around ChIP-seq peaks (delineated by gray vertical lines) for MyoG in 7-d myotubes; for clarity, IDH2-WT samples shown at Left and IDH2-R172K samples shown at Right. (B) Integrated normalized read counts for peak-flanking regions (±3 kb) shown in A; black dots indicate mean read count. *****P < 1 × 10e-50; NS: not significant with P > 0.5 based on Welch’s one-sided t test with expectation of decrease compared with control condition. Cont, no MyoD Control; Ei, EHMT1 inhibitor; M, MyoD. (C) Example H3K9me2 ChIP-seq tracks for indicated samples centered around the myogenic gene Pvalb.
Oncogenic IDH Mutations Impair a MyoD-Dependent Increase in Chromatin Accessibility at Myogenic Regions but Do Not Have a Global Effect on Chromatin Accessibility.
While the above results are consistent with H3K9me2/3 being the specific chromatin mark responsible for the IDH2-R172K effect, the effect of this impaired demethylation on the genomic landscape remains unknown. To determine whether impaired removal of H3K9me2/3 correlates with impaired accessibility at myogenic sites, we used Assay for Transposase-Accessible Chromatin (ATAC)-seq to interrogate the chromatin accessibility of IDH2-WT or IDH2-R172K 10T1/2 cells before or 24 h after activation of MyoD-ER, before myoblast fusion. The benefit of using ATAC-seq is that it is unbiased, genome-wide and chromatin mark agnostic, and can be done with relatively small numbers of cells (16). ATAC-seq profiles for biological triplicates of the four assayed conditions (IDH2-WT and IDH2-R172K with and without 4-OH-T) clustered separately by principal component analysis (SI Appendix, Fig. S5A). Consistent with their identity as fibroblast progenitors, 10T1/2 cells showed accessibility peaks at loci associated with adipocyte, chondrocyte, and myotube identity (SI Appendix, Fig. S6 A and B). Upon MyoD activation, adipocyte- and chondrocyte-specific peak counts decreased, while myogenic peak counts increased.
The IDH2-R172K mutation did not result in a global decrease in chromatin accessibility (Fig. 5A and SI Appendix, Fig. S5B). Rather, decreases in accessibility were seen in IDH2-R172K–expressing cells compared with IDH2-WT–expressing cells in the vicinity of genes that have been described to be essential for the process of myogenesis, such as the Linc-md1 noncoding RNA (Fig. 5A) (17). When gene set enrichment analysis was performed with ranked gene lists of loci with the highest difference in MyoD-induced increased accessibility between IDH2-WT and IDH2-R172K cells, the classic myogenesis gene sets scored highly (Fig. 5B), as did gene sets containing DNA-binding motifs for myogenic lineage transcription factors (SI Appendix, Fig. S5C). This decreased accessibility with the IDH2-R172K mutation was also seen surrounding known binding sites of MyoD and MyoG in differentiating myoblasts (C2C12 cells) (Fig. 5C and SI Appendix, Fig. S5D), and the strength of this difference increased with the stage of differentiation at which this ChIP data were obtained (MyoD 24 h, MyoG 60 h, MyoG 7 d). In contrast, areas surrounding CTCF (CCCTC-binding factor) binding sites were more accessible in IDH2-R172K mutant lines. Finally, when the H3K9me2 ChIP-seq data were filtered according to the regions with the highest differential accessibility change between IDH2-WT and IDH2-R172K, IDH2-WT cells showed a clear decrease in H3K9me2 upon MyoD activation (Fig. 5D) while IDH2-R172K cells showed an increase in H3K9me2. These data provide evidence that the IDH2-R172K mutation, and 2HG, lead to an impaired ability to open chromatin in myogenic regions in response to MyoD induction.
Fig. 5.
Oncogenic IDH mutations impair a MyoD-dependent increase in chromatin accessibility at myogenic regions but do not have a global effect on chromatin accessibility. (A) ATAC-seq signal tracks of IDH2-WT and IDH2-R172K cells before and after MyoD activation at a myogenic locus (Linc-md1). Differential accessibility peaks are highlighted. Also shown is the myogenin ChIP-seq track for C2C12 differentiated myotubes (MT) for orientation. (B) Gene set enrichment analysis plots for two myogenesis gene sets for genes with increased MyoD-dependent accessibility between IDH2-WT and IDH2-R172K. Vertical black line indicates position right of which genes are negatively correlated. (C) Superimposed differential accessibility (difference in MyoD-dependent accessibility change between IDH2-WT and IDH2-R172K samples) measured by differences in normalized read counts in regions flanking ChIP-seq peaks corresponding to MyoD or CTCF in C2C12 myoblasts and myogenin in differentiating C2C12 cells at 60 h or 7 d. All differential accessibility traces have nonoverlapping 99% confidence intervals with respect to CTCF and shuffled control. (D) H3K9me2 MetaPeaks (mean read count per million mapped reads) for indicated samples centered around differential ATAC peaks (peaks with 1.5× or higher increased MyoD-dependent accessibility in IDH2-WT but not in IDH2-R172K). (E) Integrated normalized read counts for peak-flanking regions (±3 kb) shown in D; black dots indicate mean read count. *****P < 1 × 10e-50; NS: not significant with P > 0.5 based on Welch’s one-sided t test with expectation of decrease compared with control condition.
Discussion
A block to differentiate is frequently observed in naturally occurring cancers. This has been interpreted to mean that during proliferation cells must first withdraw from the cell cycle before terminal differentiation. Since most driver oncogenes lead to persistent proliferation, this has long been believed to be the primary mechanism of differentiation resistance. Recurrent oncogenic mutations in genes involved in chromatin remodeling, particularly enriched in mesenchymal malignancies like sarcomas (18, 19), suggest that a requirement for oncogenesis is a redistribution of chromatin accessibility. These mutations, as seen also for IDH1/2, have been thought to contribute directly to a block to differentiate. However, IDH mutations rarely, if ever, occur alone and whether they can act alone to block differentiation has been unclear.
Here, we show that the oncogenic IDH2-R172K mutation does indeed block the ability of the canonical lineage-specifying transcription factor MyoD to drive transdifferentiation in a fibroblast progenitor cell line. The differentiation block mediated by 2HG is not inherited with cell division, and its effect is primarily mediated by the inability to demethylate H3K9me2. We also show that the repressive effect of 2HG is not genome-wide, but concentrated to regions that lose the H3K9me2/3 mark and acquire increased accessibility during the differentiation process.
Our findings regarding the heritability of the differentiation block have clinical consequences given the recent approval of mutant IDH2 inhibitors for the treatment of acute myelogenous leukemia (20–22). Were IDH2 mutations to impart a heritable change to tumor cells, later inhibition of the mutant enzyme would not be expected to result in differentiation. Our results suggest that mesenchymal cells harboring an IDH2 mutation, as reported in certain sarcomas (4, 5), are blocked at an early step of differentiation that requires a master lineage-specifying transcription factor such as MyoD. This is also in line with the observation that patients with IDH2 mutant AML can suffer life-threatening differentiation syndrome when treated with an IDH2 inhibitor, a black-box warning of the recently approved IDH2 inhibitor enasidenib (21, 22).
One unexpected finding of our studies is that inhibition of DNA methylation did not play a major role in the IDH2-R172K differentiation block, although it led to reactivation of the endogenous MyoD locus (SI Appendix, Fig. S3A). While this does not exclude a tumorigenic role for DNA 5mC hypermethylation in the biology of sarcomas or other IDH mutant solid tumors, it does point to the different biology of IDH mutations in different contexts. Tet1/2 mutations are mutually exclusive with IDH mutations in acute myelogenous leukemia (12). In contrast, Tet mutations are rarely, if ever, seen in sarcomas (4, 23). One possibility is that in these solid tumors the differentiation block is at the level of a near-committed precursor where differentiation requires histone rather than DNA demethylation. In keeping with the idea that these tumors represent blocks in differentiation at committed stages, H3K9me2 is a mark that is characteristic of facultative heterochromatin (24–26). Earlier work has shown H3K9 methylation positions facultative heterochromatin at the nuclear periphery (27). It may well be a mark whose active removal is required for the last stages of differentiation, even after cell cycle exit, as myogenic loci acquire nuclear positions critical for ongoing expression of genes required in the terminally differentiated cell.
While the removal of other chromatin marks has been found to be impaired in the context of 2HG or IDH mutations, none of these are required for the differentiation block seen in this myogenic model. However, the fact that we do not see a rescue of differentiation in the IDH2-R172K mutants with removal of other chromatin marks should not be interpreted as evidence that all of the 2HG effects are mediated through hypermethylation of H3K9. In fact, hypoxia has recently been shown to block differentiation of C2C12 myoblasts through the preferential inhibition of the H3K27 demethylase Kdm6a (28). Given the many enzymes inhibited by 2HG, there may be abnormalities in IDH2-R172K cells whose differentiation has been rescued in the absence of EHMT1/2. Notwithstanding, our findings are consistent with a model in which demethylation of H3K9me2 is triggered by the MyoD expression program and is required at myogenic genes for terminal differentiation. A limitation of our study is that we have not been able to determine the specific H3K9 demethylase(s) whose inhibition is responsible for the IDH2-R172K–mediated differentiation block. We were unable to reproducibly find H3K9-specific demethylases whose loss phenocopied the IDH-mutant effects. Genetic redundancy may be a plausible explanation, but it is worth noting that the inhibitory effect of 2HG may require the simultaneous inhibition of multiple demethylases and this may be part of the oncogenic mechanism of IDH mutations. Our results also suggest that inhibitors of H3K9 methyltransferase should be explored not only for IDH mutant tumors but also those driven by mutations in succinate dehydrogenase and fumarate dehydrogenase that accumulate metabolites that also block histone demethylation.
Finally, while IDH mutations lead to chromatin repression, little is known about whether this is a global or site-specific phenomenon, and if the latter, how certain regions escape repression. The observation that sites whose accessibility increases in a MyoD-dependent manner are preferentially affected by IDH2 mutations is consistent with MyoD-dependent targeting of demethylase activity to myogenic genes that acquire increased accessibility during differentiation. This is further supported by the preferential H3K9me2 demethylation seen flanking the myogenic sites by ChIP-seq. Of note, if global demethylation is forced with the EHMT1/2 inhibitor beyond a threshold level, differentiation can proceed, again favoring the model that 2HG blocks differentiation via H3K9 hypermethylation. However, chromatin accessibility is not globally reduced in the IDH2-R172K condition. For example, CTCF sites appear to show increased accessibility in IDH2-R172K cells. Alteration in CTCF-binding could impair the ability of insulator regions to regulate distant promoter–enhancer interactions (29). It is possible that this increased accessibility of some regions in the IDH2-R172K condition represents a shift in repressive marks to other regions of the genome. Further studies will be required to identify how IDH mutant cells adapt their chromatin to the effects of inhibition of alpha-ketoglutarate–consuming enzymes.
As noted, 10T1/2 cells are fibroblast-like progenitors capable of differentiating into adipocytes, chondrocytes, and myotubes. ATAC-seq profiles for genes of all three lineages were accessible in untreated 10T1/2 cells. Once MyoD was activated, however, myogenic genes became significantly more accessible while adipocyte and chondrocyte genes became less accessible. Interestingly, full myoblasts, as represented by C2C12 cells, were not inhibited in their ability to form myotubes by the IDH2-R172K mutation, suggesting that the demethylating activity recruited by MyoD is no longer required at myogenic loci of cells committed to myogenic differentiation.
In conclusion, our results show that the ongoing production of 2HG by cancer-associated IDH-mutant enzymes can block transcription factor mediated-differentiation through the dependence on removal of the facultative heterochromatic H3K9 methylation mark.
Materials and Methods
10T1/2 cells were obtained from American Type Culture Collection and cultured at 37 °C in 5% CO2 in high-glucose DMEM supplemented with 10% FBS. For a list of reagents used, see SI Appendix, Table S1. GC-MS analysis of 2HG was performed on 80% methanol lysed samples containing deuterated 2HG as an internal standard, dried, and derivatized and run on an Agilent 7890A GC with an HP-5MS column connected to a 5975C detector. CRISPR-Cas9 experiments were performed with lentiviral vectors expressing Cas9 in addition to indicated sgRNAs. ChIP-seq and ATAC-seq were performed as previously described (16, 30, 31). For a detailed description of methods, see SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the C.B.T. Laboratory for discussion and comments on the manuscript. J.-M.S. is a Hope Funds for Cancer Research 2017 Postdoctoral Fellow. This work made use of core facilities supplied in part by the Memorial Sloan Kettering Cancer Center Support Grant P30CA008748.
Footnotes
Conflict of interest statement: C.B.T. is a founder of Agios Pharmaceuticals and a member of its scientific advisory board. He also previously served on the board of directors of Merck and Charles River Laboratories.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov (accession no. GSE118440).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817662116/-/DCSupplemental.
References
- 1.Taylor S. M., Jones P. A., Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 17, 771–779 (1979). [DOI] [PubMed] [Google Scholar]
- 2.Davis R. L., Weintraub H., Lassar A. B., Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 (1987). [DOI] [PubMed] [Google Scholar]
- 3.Lassar A. B., Paterson B. M., Weintraub H., Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell 47, 649–656 (1986). [DOI] [PubMed] [Google Scholar]
- 4.Amary M. F., et al. , IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 224, 334–343 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Lu C., et al. , Induction of sarcomas by mutant IDH2. Genes Dev. 27, 1986–1998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang L., et al. , Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward P. S., et al. , The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury R., et al. , The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12, 463–469 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu W., et al. , Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koivunen P., et al. , Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483, 484–488 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C., et al. , IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaidzik V. I., et al. , TET2 mutations in acute myeloid leukemia (AML): Results from a comprehensive genetic and clinical analysis of the AML study group. J. Clin. Oncol. 30, 1350–1357 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Emadi A., et al. , Presence of isocitrate dehydrogenase mutations may predict clinical response to hypomethylating agents in patients with acute myeloid leukemia. Am. J. Hematol. 90, E77–E79 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Hollenberg S. M., Cheng P. F., Weintraub H., Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc. Natl. Acad. Sci. U.S.A. 90, 8028–8032 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferri P., et al. , Expression and subcellular localization of myogenic regulatory factors during the differentiation of skeletal muscle C2C12 myoblasts. J. Cell. Biochem. 108, 1302–1317 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesana M., et al. , A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride M. J., et al. , The SS18-SSX fusion oncoprotein hijacks BAF complex targeting and function to drive synovial sarcoma. Cancer Cell 33, 1128–1141.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadoch C., et al. , Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 49, 213–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waitkus M. S., Diplas B. H., Yan H., Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell 34, 186–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein E. M., et al. , Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130, 722–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amatangelo M. D., et al. , Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 130, 732–741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Totoki Y., et al. , Unique mutation portraits and frequent COL2A1 gene alteration in chondrosarcoma. Genome Res. 24, 1411–1420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker J. S., Nicetto D., Zaret K. S., H3K9me3-dependent heterochromatin: Barrier to cell fate changes. Trends Genet. 32, 29–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen B., Wu H., Shinkai Y., Irizarry R. A., Feinberg A. P., Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41, 246–250 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lienert F., et al. , Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet. 7, e1002090 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Towbin B. D., et al. , Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150, 934–947 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Chakraborty A. A., et al. , Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363, 1217–1222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flavahan W. A., et al. , Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C.-W., et al. , DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat. Med. 21, 335–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corces M. R., et al. , An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.