Significance
We have demonstrated the design, prototyping, and implementation of a non-natural route to isoprenoid biosynthesis, termed the isoprenoid alcohol (IPA) pathway. Opposed to working within the confines of native metabolic pathways, our approach of utilizing all available biochemical reactions identified a synthetic route to isoprenoid compounds that is both different and provides a more energy-efficient pathway compared with native isoprenoid biosynthesis. By improving the overall pathway design, the IPA pathway has the potential to facilitate the synthesis of a diverse range of isoprenoid compounds at high product titers and yields.
Keywords: isoprenoids, de novo pathway design, synthetic biology, retrobiosynthesis, IPA pathway
Abstract
The more than 50,000 isoprenoids found in nature are all derived from the 5-carbon diphosphates isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). Natively, IPP and DMAPP are generated by the mevalonate (MVA) and 2-C-methyl-d-erythritol-4-phosphate (MEP) pathways, which have been engineered to produce compounds with numerous applications. However, as these pathways are inherently constrained by carbon, energy inefficiencies, and their roles in native metabolism, engineering for isoprenoid biosynthesis at high flux, titer, and yield remains a challenge. To overcome these limitations, here we develop an alternative synthetic pathway termed the isoprenoid alcohol (IPA) pathway that centers around the synthesis and subsequent phosphorylation of IPAs. We first established a lower IPA pathway for the conversion of IPAs to isoprenoid pyrophosphate intermediates that enabled the production of greater than 2 g/L geraniol from prenol as well as limonene, farnesol, diaponeurosporene, and lycopene. We then designed upper IPA pathways for the generation of (iso)prenol from central carbon metabolites with the development of a route to prenol enabling its synthesis at more than 2 g/L. Using prenol as the linking intermediate further facilitated an integrated IPA pathway that resulted in the production of nearly 0.6 g/L total monoterpenoids from glycerol as the sole carbon source. The IPA pathway provides an alternative route to isoprenoids that is more energy efficient than native pathways and can serve as a platform for targeting a repertoire of isoprenoid compounds with application as high-value pharmaceuticals, commodity chemicals, and fuels.
Metabolic engineering approaches that rely on exploiting native metabolism are inherently limited by the range of known biological pathways and by metabolic constraints shaped by evolution and a given fitness advantage rather than maximizing the synthesis of a desired compound. Therefore, attainable product titers, rates, and yields are often limited by intrinsic carbon and energy inefficiencies and negative cross talk between product-forming and growth-sustaining reactions (1). The biosynthesis of isoprenoids illustrates both the extraordinary diversity of biology and the inherent limitations of native metabolism. Isoprenoids, comprising more than 50,000 structures found in all kingdoms of life (2), play a wide range of ecological, physiological, and structural roles and have been exploited for applications ranging from high-value pharmaceuticals to commodity chemicals and fuels (2–5). In all known organisms, isoprenoids are derived from the 5-carbon diphosphates IPP and DMAPP. IPP and DMAPP are generated through either the MVA or the MEP pathway (SI Appendix, Fig. S1A) (2) with the recent discovery of a 5′-methylthioadenosine-isoprenoid pathway also providing a unique MEP shunt linking methionine salvage to isoprenoid biosynthesis (6). Although these pathways have been engineered for isoprenoid production in a variety of organisms, their inherent inefficiencies, complex chemistry, and intrinsic regulatory mechanisms (Table 1) present significant challenges that have limited product titers and yields to well below theoretical maxima (3, 5).
Table 1.
Comparison of native and synthetic pathways for the generation of isoprenoid precursors
| Attribute* | Native isoprenoid pathways | Isoprenoid alcohol pathway | |||
| MVA | MEP | Fig. 4A | SI Appendix, Fig. S10† | SI Appendix, Fig. S12‡ | |
| Starting central carbon metabolite(s) | Acetyl-CoA | Pyruvate; glyceraldehyde 3-phosphate | Acetyl-CoA | Acetyl-CoA; propionyl-CoA | Pyruvate |
| Number of steps§ | 6 | 7 | 8 | 8 (17) | 10 |
| Initial isoprenyl precursor produced | IPP | IPP and DMAPP | DMAPP | DMAPP (GPP) | DMAPP |
| Carbon efficiency¶ | 0.83 | 0.83 | 0.83 | 1.00 | 0.83 |
| ATP balance# | −3 ATP | −3 ATP (1 ATP; 1 CTP to CMP) | −2 ATP | −2 (−2) ATP | −2 ATP |
| Redox balance#,|| | −2 NAD(P)H | −3 NAD(P)H | −2 NAD(P)H | −3 (−7) NAD(P)H | −4 NAD(P)H |
| Notable pathway/enzyme required cofactors | CoA, ATP | TPP, CTP, ATP, [4Fe-4S] | CoA, ATP | CoA, ATP | CoA, ATP, [4Fe-4S] |
Carbon efficiency, ATP/redox balance, and number of steps determined for the generation of initial C5 precursor(s) (IPP or DMAPP) from starting central metabolite(s).
Attributes for route to C5 precursor and direct to C10 precursor (in parentheses when different) shown.
Attributes shown for route involving the dehydration of 2-hydroxyisovaleryl-CoA shown in SI Appendix, Fig. S12.
For IPA pathway designs, the number of steps calculated assuming two step reduction of CoA intermediates to associated alcohol.
Carbon molar basis.
Plus (+) or minus (−) refers to generation or consumption, respectively.
For ease of comparison, redox balance calculated assuming NAD(P)H as an electron donor.
Opposed to working within the constraints of native metabolism, the development of synthetic metabolic pathways can exploit the ever-expanding repertoire of biochemical reactions to both improve attenable product titers and yields as well as confer novel biological capabilities for utilizing promising feedstocks and the synthesis of non-natural products (7, 8). Here, we sought to develop de novo pathways for isoprenoid synthesis by exploiting enzymes and biochemical reactions beyond those involved in the MVA and MEP pathways. We utilized a retrobiosynthetic approach that draws inspiration from the notion of retrosynthesis in organic chemistry by defining a target molecule of interest and “walking” backward through known biochemical transformations to key precursor(s) (9, 10). This allowed us to identify target intermediate molecules with the potential to be readily generated from common central carbon intermediates and converted to isoprenoid precursors through a minimal number of steps, whereas also circumventing inherent inefficiencies and complexities of native pathways. Through this approach, we developed a non-natural isoprenoid alcohol (IPA) pathway, termed the IPA pathway, enabling the demonstration of isoprenoid synthesis from a single unrelated carbon source.
Results
Design of a De Novo Pathway for the Synthesis of Isoprenoid Precursors.
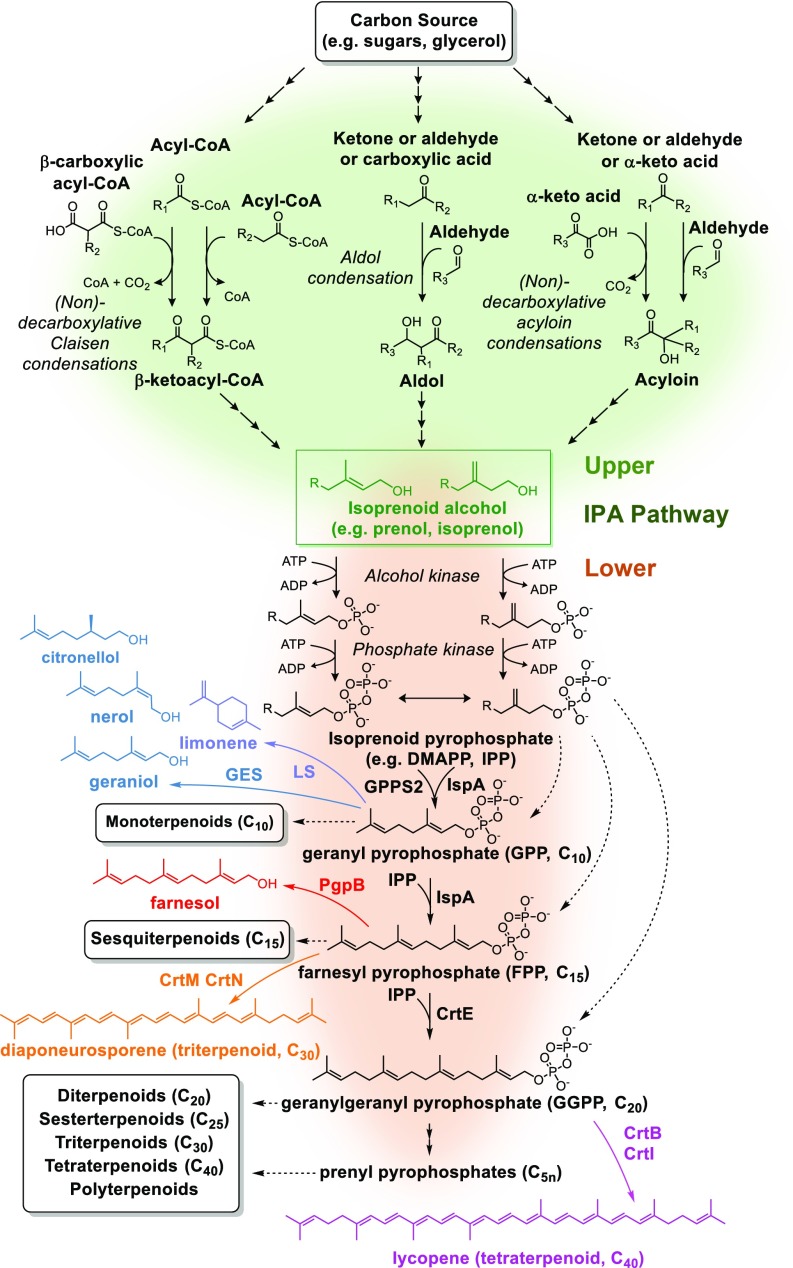
IPP, DMAPP, as well as longer-chain length isoprenoid precursors formed through iterative condensation of DMAPP with IPP units [e.g., geranyl pyrophosphate (GPP)/C10, farnesyl pyrophosphate (FPP)/C15, etc.], all share a common chemical structure with an aliphatic hydrocarbon chain (C5n) forming a phosphate ester with a pyrophosphate group (Fig. 1). Considering this general structure, a retrobiosynthetic analysis suggests that these isoprenoid precursors can be generated from the corresponding alcohols through the phosphorylation of the hydroxyl group followed by a second phosphorylation of the resulting phosphate esters (Fig. 1). When these sequential phosphorylation reactions are combined with pathways for the synthesis of IPAs from central carbon metabolites, a non-native route to isoprenoid biosynthesis is realized (Fig. 1). As IPAs are the key metabolic intermediates linking isoprenoid precursors to central carbon metabolites, we termed this design the IPA pathway. In this pathway architecture, an IPA(s) connects the upper IPA branch, generating C5n IPA(s) from central carbon intermediates and the lower IPA branch converting the C5n IPA(s) to various isoprenoid precursors (e.g., DMAPP, IPP, GPP, etc.) and, hence, facilitates the synthesis of isoprenoid compounds (Fig. 1). In principle, any C5n IPA could serve as the intermediate(s), however, targeting C5 (iso)prenol would enable the generation of all chain length pyrophosphate intermediates through the formation of DMAPP and IPP and subsequent iterative condensation with C5 units.
Fig. 1.
Retrobiosynthetic based design of the de novo IPA pathway for the generation of isoprenoid precursors from common C2 and C3 central carbon metabolites. Claisen, aldol, or acyloin condensation reactions combined with additional biochemical reactions could support the generation of IPAs from various central carbon metabolites (upper IPA pathway, green shading). These IPAs (green box) serve as the key intermediates linking the upper pathway to the lower IPA pathway (orange shading) in which diphosphorylation generates isoprenoid precursors from IPAs. Examples of individual pathways identified through this approach are shown in the SI Appendix, Figs. S10–S12. Representative isoprenoid products demonstrated in this study and enzymes utilized for their production are shown (geraniol, limonene, farsnesol, diaponeurosporene, and lycopene).
Identification and Characterization of Enzymes Supporting Lower IPA Pathway Designs.
Our design targeting (iso)prenol as the key intermediate(s) for generating universal isoprenoid precursors relies on the ability to phosphorylate either C5 IPA to the corresponding pyrophosphate intermediate (Fig. 1). Although (iso)prenol is not known to be the physiological substrate of any kinase, reports have confirmed the ability for various thermophilic isopentenyl phosphate kinases (IPKs) to phosphorylate both (iso)prenol as well as the monophosphate (e.g., DMAP) (11, 12), and a protein engineering strategy was recently utilized to construct a Thermoplasma acidophilum IPK variant (IPKTHA*) with improved prenol phosphorylation activity (11). In attempting to utilize IPKTHA* for catalyzing (iso)prenol kinase activity, we found it to have low soluble expression in Escherichia coli resulting in low levels of purified IPK and hence a potential for low functional activity to limit the utility of this enzyme.
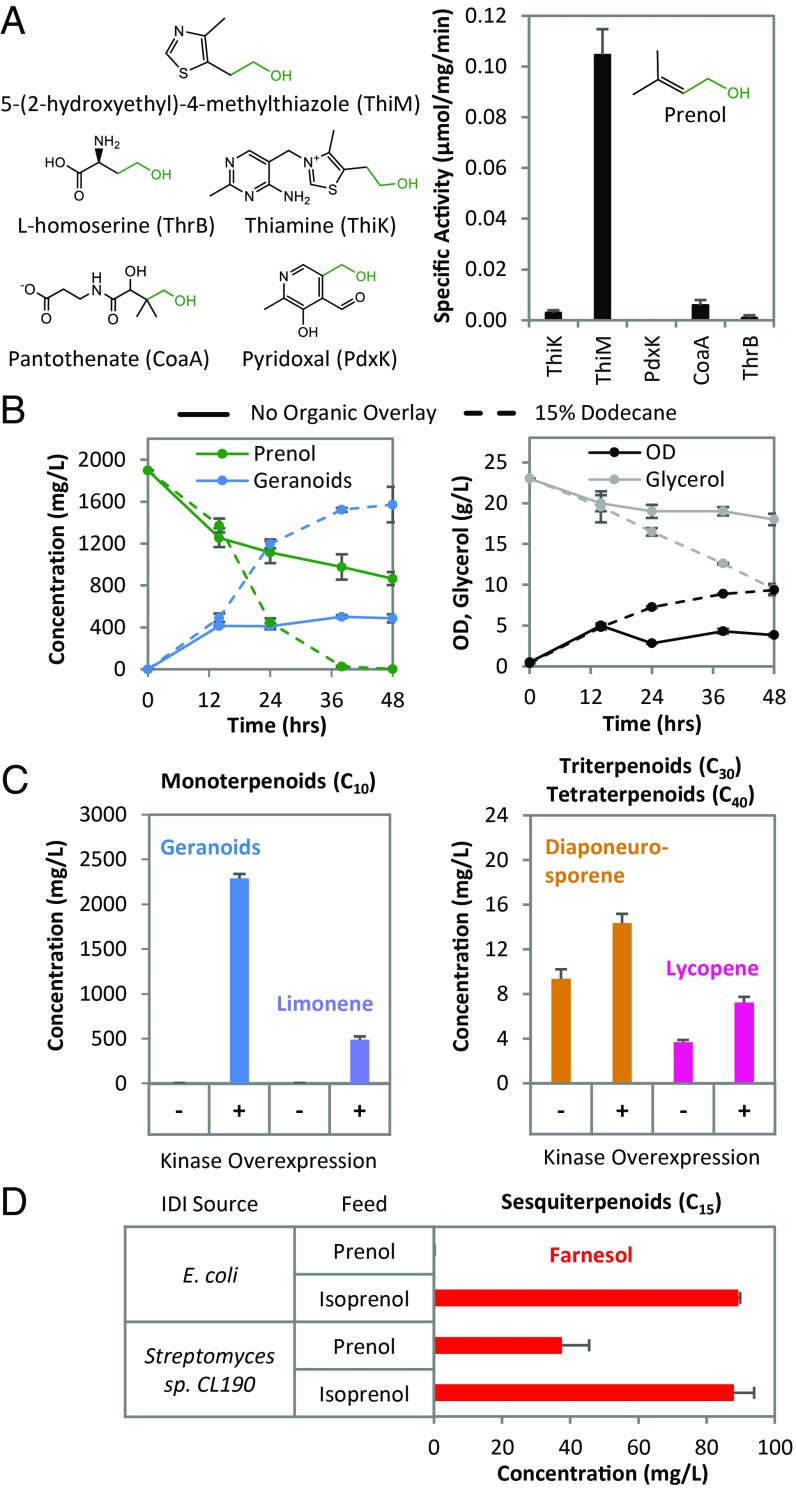
Our initial efforts to identify a better performing enzyme(s), through both sequence similarity search via HMMER (13) and mining for candidate enzyme sequences using our desired reaction(s) via Selenzyme (14), returned a vast number of potential enzymes difficult to narrow to the most promising subset (SI Appendix, Figs. S2 and S3). Given the low soluble expression of our template protein in E. coli and the relatively low level of (iso)prenol kinase activity to begin with (11, 12), parsing this set of enzymes for ideal candidates to test remains a challenge. As such, we utilized a more rational approach wherein the –CHx–CH2OH structure of (iso)prenol was used to identify potential E. coli enzymes catalyzing the phosphorylation of a primary alcohol with a double bond, methyl branch, or functional group at carbon 2 or 3 (Fig. 2A). Of the enzymes identified, purification and characterization of prenol kinase activity revealed that E. coli hydroxyethylthiazole kinase (ThiM) can catalyze our desired phosphorylation reaction (Fig. 2A).
Fig. 2.
Design and evaluation of the lower IPA pathway. (A) E. coli kinases (in parentheses) evaluated for prenol kinase activity based on native substrate chemical structure (green atoms and bonds depict the R–CH2OH group). (B) Geranoid (blue; total of geraniol, nerol, and citronellol) production, prenol consumption (green), OD (black), and glycerol consumption (gray) by JST06 (DE3) atoBCT5 ΔfadB harboring the pathway shown in Fig. 1 (with ThiM and IPKMTH as the alcohol and phosphate kinase, respectively) for conversion of prenol to geraniol in fermentations without (solid lines) and with (dotted lines) 15% dodecane organic overlay. Estimates on product loss due to volatilization described in SI Appendix, Fig. S7. (C) Synthesis of monoterpenoids, triterpenoids, and tetraterpenoids by JST06 (DE3) atoBCT5 ΔfadB harboring the corresponding pathways shown in Fig. 1 (with ThiM and IPKMTH) for the conversion of prenol to the specified product. For geranoid and limonene production, strains were grown in 25 mL flasks (stoppered with foam plugs) containing 10 mL medium [3 g/L prenol and 50 µM isopropyl β-d-1-thiogalactopyranoside (IPTG) included for geranoids; 2 g/L prenol and 100 µM IPTG for limonene] with a 15% dodecane overlay. For diaponeurosporene and lycopene production, strains were grown in 25 mL flasks (stoppered with foam plugs) containing 5 mL of medium (0.5 g/L prenol, 100 µM IPTG). (D) Synthesis of farnesol by strain JST06 (DE3) atoBCT5 ΔfadB harboring the pathway shown in Fig. 1 (with ThiM and IPKMTH) utilizing either E. coli or Streptomyces sp. CL190 Idi. Strains were grown as described above for diaponeurosporene and lycopene production.
To assess the lower IPA pathway for conversion of (iso)prenol to C10 isoprenoids, we combined the expression of ThiM with the IPK from Methanothermobacter thermautotrophicus (IPKMTH), which in addition to its native function of converting IP to IPP can also convert DMAP to DMAPP (kcat/Km = 7.9 × 105) (12). Furthermore, E. coli IPP isomerase (Idi) was overexpressed for the interconversion of DMAPP and IPP, and a truncated version of geranyl diphosphate synthase (GPPS2) from Abies grandis (15) was utilized for subsequent condensation of DMAPP and IPP to GPP (Fig. 1). Geraniol production was selected as a proxy for GPP generation with the expression of a truncated version of the geraniol synthase (GES) from Ocimum basilicum enabling conversion of GPP to geraniol (16). Using a biotransformation approach in which a growth and expression phase was followed by resuspension of the cells in minimal media containing 10 mM prenol, nearly 200 mg/L geranoids (C10 IPAs geraniol, nerol, and citronellol) were produced when ThiM was overexpressed in conjunction with other lower IPA pathway enzymes (SI Appendix, Fig. S4). The substrate and product flexibility of terpene synthases (2) as well as endogenous enzymes for generating or interconverting isoprenoid products (17) are likely contributing causes to the formation of three distinct monoterpenoids observed here. Although ThiM also appears capable of phosphorylating isoprenol as the same combination of enzymes resulted in ∼125 mg/L C10 IPAs when 10 mM isoprenol was used (SI Appendix, Fig. S4), the 60% higher geranoid production using prenol indicated this C5 alcohol was preferable. Furthermore, the use of ThiM in place of the aforementioned IPKTHA* resulted in more than a 15-fold increase in geranoids from prenol (SI Appendix, Fig. S4). Characterization of E. coli ThiM further confirmed the ability for this enzyme to catalyze the phosphorylation of prenol with the following kinetic parameters: Km = 5.8 ± 0.1 mM; kcat = 0.11 ± 0.01 s−1; kcat/KM = (1.9 ± 0.09) × 101 M−1 s−1 (SI Appendix, Fig. S5).
Fermentative Production of Isoprenoids through a Lower IPA Pathway.
To better assess the ability of the lower IPA pathway to support product synthesis, we evaluated isoprenoid production in fermentations with cells actively growing on a common carbon source (glycerol). For this, we utilized a two-plasmid system for the overexpression of lower IPA pathway enzymes in a genetic background, MG1655 derivative JST06(DE3), engineered for the synthesis of various products from glycerol (18). The ThiM-mediated lower IPA pathway enabled geranoid production from prenol (Fig. 2B). Since significant amounts of prenol remained unutilized in the medium after 48 h, we hypothesized and confirmed that product/geraniol toxicity was a major issue, including its intracellular accumulation (SI Appendix, Fig. S6). To alleviate these issues, we utilized a 15% dodecane overlay which enabled the complete utilization of ∼2 g/L prenol and significantly reduced the accumulation of intracellular geranoids (SI Appendix, Fig. S6B) yielding a total of ∼1.5 g/L geranoids (Fig. 2B). When 3 g/L of prenol was included, over 2 g/L of geranoids were produced compared with only ∼0.005 g/L without kinase overexpression (Fig. 2C), which demonstrates minimal generation of diphosphate intermediates through the native MEP pathway.
We also explored the use of various condensation and product synthesis enzymes (Fig. 1). Replacement of GES with a truncated version of the limonene synthase from Mentha spicata (19) enabled the production of nearly 0.5 g/L limonene (Fig. 2C). We next utilized alternative enzymes for the condensation of diphosphate intermediates in conjunction with appropriate termination enzymes (20, 21) to demonstrate the synthesis of longer-chain products, farnesol, a sesquiterpenoid (C15), as well as the C30 and C40 isoprenoids diaponeurosporene and lycopene, respectively (Fig. 2 C and D). Although the lower product titers for these longer-chain products could be explained, in part, due to the competition for FPP (e.g., ubiquinone and menaquinone biosynthesis) (2), the ratio of DMAPP to IPP generated via the phosphorylation of C5 IPAs may also play a key role, especially with the use of prenol which dictates that DMAPP is the initial C5 pyrophosphate generated. Indeed, when isoprenol, instead of prenol, was utilized as the feed for the lower IPA pathway, farnesol production was observed (Fig. 2D). As such, the utilization of E. coli Idi, which favors the formation of DMAPP over IPP (22), may limit the availability of IPP units required for longer-chain length isoprenoids. In agreement with this hypothesis, the use of an Idi from Streptomyces sp. strain CL190 (23), an enzyme that favors the formation of IPP over DMAPP (22, 23), led to the production of ∼40 mg/L farnesol from prenol compared with negligible amounts with the use of E. coli Idi (Fig. 2D). Interestingly, neither strategy significantly improved diaponeurosporene or lycopene production (SI Appendix, Fig. S8) indicating that additional constraints beyond the DMAPP to IPP ratio are potentially limiting C30 and C40 product synthesis from the IPA pathway.
Design and In Vivo Implementation of the Upper IPA Pathway.
Capitalizing on the ability of the engineered lower IPA pathway to support the synthesis of isoprenoid precursors in the context of the overall synthetic route requires upper IPA pathway designs for the efficient generation of IPAs. However, to date, the synthesis of iso(prenol) has only been accomplished via cleavage of the pyrophosphate group from the corresponding isoprenoid intermediate, essentially the reverse of the IPA pathway reactions, following engineering of native isoprenoid pathways (24, 25). To generate upper IPA routes to these C5 alcohols from C2 and C3 central carbon metabolites, enzymes catalyzing carbon–carbon bond forming Claisen, aldol, or acyloin condensation reactions were considered (Fig. 1). When combined with other biochemical reactions and various starting points from the central carbon metabolism, these condensation reactions could support an array of different routes to IPAs of varying chain lengths (SI Appendix, Figs. S9–S12). Although the design and prototyping of the lower IPA pathway indicated that prenol would be preferable as a linking metabolite, our initial designs for the upper IPA pathway also considered isoprenol and longer-chain length IPAs to determine all potential routes.
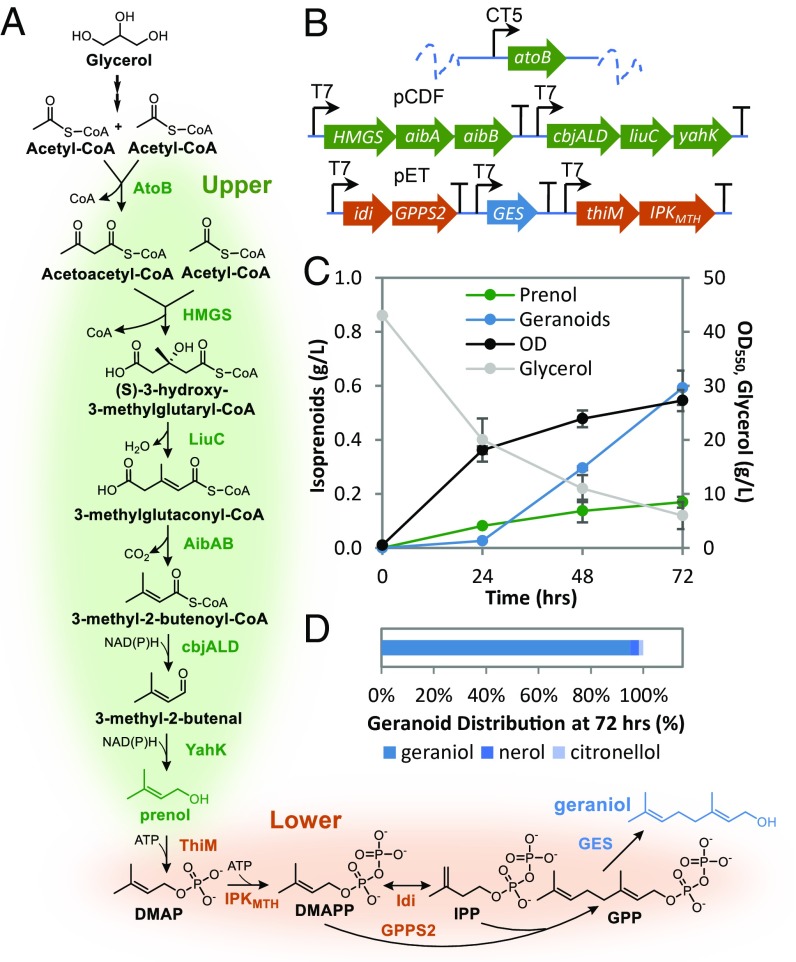
The potential and practical applications of select routes were assessed through various metrics, including both quantitative carbons, ATP, and redox balances, and qualitative factors, such as the complexity and availability of the specific biochemical reactions, in direct comparison with native isoprenoid pathways (Table 1). Although pathways based on nondecarboxylative Claisen condensation of acetyl-CoA and propionyl-CoA, and decarboxylative condensation of two molecules of pyruvate have promising carbon, ATP, and redox balances (Table 1), implementation of these pathways would be limited, in part, due to the need for challenging biochemical reactions for which no known enzyme has demonstrated the required substrate to product conversion (SI Appendix, Supplementary Text). Thus, we decided to focus on the synthesis of prenol from acetyl-CoA through Claisen condensation reactions. This upper IPA design exploits the condensation of two acetyl-CoA molecules, followed by the nondecarboxylative condensation of acetoacetyl-CoA and acetyl-CoA (Fig. 3A). Subsequent dehydration (26), decarboxylation (27), and reduction results in the generation of prenol (Fig. 3A). This route has the potential to operate with reduced ATP requirements compared with either the MVA or the MEP pathways, whereas supporting the same carbon efficiency from starting intermediates and utilizing well established biochemical reactions, albeit with a higher number of enzymatic steps (Table 1).
Fig. 3.
Engineering E. coli for prenol production via an upper IPA pathway. (A) Proposed biosynthetic pathway to prenol starting from acetyl-CoA. Enzymes overexpressed for various pathway steps shown. (B) Schematic of E. coli construct for prenol production. Host strain JC01 (MG1655 ΔldhA ΔpoxB Δpta ΔadhE ΔfrdA) (DE3) is a mixed-acid fermentation-deficient E. coli strain. Chromosomal expression of E. coli native atoB was regulated by cumate-inducible PCT5 promoter. All other enzymes were expressed from pCDF plasmid. Specifically, expression of HMGS, aibA, aibB was placed under the first IPTG-inducible PT7 promoter (P1) with cbjALD, liuC, and yahK under the second inducible PT7 promoter (P2). (C) Prenol production by JC01 (DE3) atoBCT5 harboring pCDF prenol plasmid. The strain was grown in LB-like 3-(N-morpholino)propanesulfonic acid (MOPS) medium with 20 g/L glycerol in 25 mL flasks (stoppered with foam plugs) with or without a 20% tributyl phosphate (+TBP) organic layer incubated at 37 °C and 200 rpm for 48 h. Estimates on product loss due to volatilization described in SI Appendix, Fig. S14.
Although enzymes catalyzing the condensation steps, dehydration (26) and 3-methylglutaconyl-CoA decarboxylation (27) of this upper IPA pathway design have been established, identification of specific enzyme(s) for reducing 3-methylcrotonyl-CoA to prenol was required (Fig. 3A). Leveraging our previous findings for the ability of the Clostridium beijerinckii acyl-CoA reductase (cbjALD) to reduce functionalized acyl-CoA intermediates with low activity for the reduction of acetyl-CoA (18) as well as alcohol dehydrogenases catalyzing the reversible oxidation of functionalized alcohols (28), we tested these enzymes for their ability to function in the context of 3-methylcrotonyl-CoA reduction. These included the aforementioned cbjALD for which detectable activity for the NADH-dependent reduction of 5 mM 3-methylcrotonyl-CoA was measured with a purified enzyme (0.008 ± 0.001 μmol/mg/min) as well as the E. coli aldehyde reductase YahK for which crude extracts of cells overexpressing this enzyme had a measured specific activity of 0.30 ± 0.03 μmol/mg/min for the NADP+-dependent oxidation of 1 mM prenol. These results provided initial feasibility for the overall reduction to prenol and established a full set of enzymes for the upper IPA pathway.
The functionality of this upper IPA pathway design was tested by cloning genes encoding Staphylococcus aureus HMGS, Myxococcus xanthus LiuC, AibA, and AibB, cbjALD, and E. coli YahK into pCDFDuet-1 under IPTG control (Fig. 3B). This allowed us to utilize our previously developed MG1655 derivative JC01 (MG1655 ΔldhA ΔpoxB Δpta ΔadhE ΔfrdA) (DE3) with chromosomal overexpression of the E. coli thiolase AtoB under cumate control (Fig. 3B) (28). Expression of this full pathway from acetyl-CoA to prenol along with the use of conditions previously demonstrated to enable the synthesis of products from the acetyl-CoA node at high titer with glycerol as the carbon source (28–30) enabled the production of prenol (Fig. 3C). Further evaluation of conditions (SI Appendix, Fig. S13) and the inclusion of an organic solvent overlay (20% TBP) increased overall prenol titer with the production of nearly 2 g/L in 48 h (Fig. 3C). This selective production of prenol (use of MVA or MEP pathways leads to a mixture of C5 isoprenoid alcohols) achieved titer an order of magnitude higher than those previously reported (24, 25, 31).
Integrated IPA Pathway for the Synthesis of Isoprenoids from Glycerol.
To evaluate an integrated IPA pathway for isoprenoid synthesis from a single, nonrelated carbon source (Fig. 4A), the pCDF vector expressing the genes encoding enzymes for prenol production from glycerol was combined with a pET vector expressing genes encoding all downstream pathway enzymes for geraniol production from prenol in a JST06(DE3) atoBCT5 ΔfadB background (Fig. 4B). Initial testing of the total integrated pathway with 20 g/L glycerol resulted in the production of over 90 mg/L geranoids, nearly fourfold higher than the concentration observed under the same conditions without the overexpression of ThiM and IPKMTH (SI Appendix, Fig. S15). This increase in geranoid production upon kinase overexpression was also accompanied by a significant decrease in prenol production, suggesting that prenol synthesized from glycerol was further converted to our desired product. Further evaluation of conditions (media volume, IPTG concentration, and dodecane overlay; SI Appendix, Fig. S16) and the use of 40 g/L glycerol resulted in significant increases to geranoid production with nearly 0.6 g/L geranoids (0.59 ± 0.06 g/L) produced after 72 h (Fig. 4C) compared with 0.013 ± 0.004 g/L with the control strain without the overexpression of ThiM and IPKMTH (SI Appendix, Fig. S17). Geraniol represented the major C10 product with only small amounts (<20 mg/L) of nerol and citronellol produced (Fig. 4D). Prenol production was also observed with increasing concentration throughout the time course (Fig. 4C). These results mark the demonstration of a synthetic isoprenoid pathway to produce isoprenoid compounds from a single unrelated carbon source.
Fig. 4.
Isoprenoid production from a single nonrelated carbon source (glycerol) through the IPA pathway. (A) Integrated IPA pathway for geraniol production. Enzymes overexpressed for various pathway steps shown. (B) Schematic of E. coli construct for geranoids production. Host strain JST06 (DE3) ΔfadB is a JC01 (DE3) derivative with additional deletions to native thioesterase genes. Chromosomal expression of E. coli native atoB was regulated by a cumate-inducible PCT5 promoter. All other enzymes were expressed from pCDF or pET plasmid as shown. (C) Monoterpenoids (blue; total of geraniol, nerol, and citronellol), prenol (green), and cell growth (gray) by JST06 (DE3) atoBCT5 ΔfadB harboring pCDF-P1-HMGS-aibA-aibB-P2-cbjALD-liuC-yahK and pET-P1-idi-trGPPS2-P2-GES-P3-ThiM-IPKMTH. The strain was grown in 5 mL LB-like MOPS medium with 40 g/L glycerol with a 15% dodecane organic in 25 mL flasks (stoppered with foam plugs) incubated at 30 °C and 200 rpm. Estimates on product loss due to volatilization described in SI Appendix, Fig. S7. (D) Distribution of monoterpenoids after 72 h.
Discussion
We have demonstrated the design, prototyping, and implementation of a non-natural pathway to isoprenoid biosynthesis, termed the IPA pathway (Fig. 1). The IPA pathway is based on the conversion of central carbon metabolites to C5n alcohol(s) (upper IPA), which are subsequently phosphorylated to the corresponding isoprenoid precursors (lower IPA). The identification of a native E. coli kinase (ThiM) capable of phosphorylating (iso)prenol facilitated the development of a lower IPA pathway capable of supporting isoprenoid pyrophosphate generation and enabling the synthesis of multiple isoprenoid products, including geraniol, limonene, farnesol, diaponeurosporene, and lycopene (Fig. 2). With prenol as the linking metabolite between the upper and the lower IPA pathways, we developed a route to this branched unsaturated alcohol from central carbon metabolites recruiting biochemical reactions from various organisms that enabled its selective production at nearly 2 g/L (Fig. 3). When the upper and lower branches were combined into an integrated IPA pathway, a full synthetic route to isoprenoids from the single unrelated carbon source glycerol was realized, resulting in the production of 0.6 g/L monoterpenoids (Fig. 4).
The IPA pathway design circumvents many of the stoichiometric and regulatory constraints associated with native isoprenoid pathways, which have evolved for the generation of biosynthetic intermediates required for cell growth opposed to efficient product synthesis (2). For example, whereas the MVA and MEP pathways both consume three net ATP equivalents per 5-carbon diphosphate generated from their respective starting central carbon metabolites (3), the IPA pathway requires two ATP molecules (Table 1). The IPA pathway variant demonstrated here is relatively easy to implement as it does not require complex chemistry or enzymes with intrinsic characteristics that limit their potential in vivo functionality under various conditions. Compared with the MVA pathway, the IPA pathway avoids the tight regulation associated with many pathway enzymes, such as 3-hydroxy-3-methyl-glutaryl-CoA reductase proposed to be regulated at the transcriptional, post-transcriptional, translational, and post-translational level, including inhibition by free CoA, HMG, NADPH, and NADP+ (3, 32).
We have also identified potential bottlenecks for which future engineering efforts may serve to improve product synthesis and overall pathway operation. Specific to longer-chain length isoprenoids, control of the DMAPP to IPP ratio appears to represent an important factor, especially when prenol is the linking IPA. For overall IPA pathway operation, the accumulation of prenol in the medium indicates that the lower pathway, specifically prenol kinase, represents a key bottleneck. In addition, the development of alternative upper IPA routes to IPAs has the potential to further improve pathway operation. For example, preliminary results for the route proceeding via pyruvate through an acetolactate synthase (SI Appendix, Fig. S12) suggest a potential for high titer and flux (33). Finally, although our demonstration of this synthetic isoprenoid pathway was implemented in E. coli, accessing the vast range of isoprenoid products requires the use of other microbial systems, such as yeast(s), which offer distinct advantages for the production of high value isoprenoids and oxygenated derivatives (5). Given its modularity and design directly linking to central carbon metabolism, expression of the IPA pathway could be explored in a range of micro-organisms to further unlock the potential of this route for isoprenoid synthesis.
Materials and Methods
Strains, Plasmids, and Genetic Methods.
Wild-type K12 E. coli strain MG1655 (34) was used as the host for all genetic modifications with gene knockouts and chromosomal expression constructs introduced by P1 phage transduction. Strains used in this study as well as details on plasmid construction and molecular biology techniques can be found in the SI Appendix.
Culture Medium and Cultivation Conditions.
The minimal medium designed by Neidhardt et al. (35), with 125 mM MOPS and Na2HPO4 in place of K2HPO4, supplemented with 20 g/L glycerol, 10 g/L tryptone, 5 g/L yeast extract, 100 μM FeSO4, 5 mM calcium pantothenate, 1.48 mM Na2HPO4, 5 mM (NH4)2SO4, and 30 mM NH4Cl was used for all fermentations unless otherwise stated. Calcium carbonate was included at 5.5% wt/vol for pH control. Fermentations were conducted in 25 mL Pyrex Erlenmeyer flasks (Corning, Inc., Corning, NY) filled with varying media volumes and foam plugs filling the necks. Individual flasks from the same inoculum were stopped at various times to determine cell growth and product synthesis profiles. Additional details, including an estimation of potential product losses due to volatilization, can be found in the SI Appendix.
Analytical Methods.
OD was measured at 550 nm in a Thermo Spectronic Genesys 20 (Thermo Scientific, Waltham, MA) and used as an estimate of cell mass (1 OD550 nm = 0.34 g dry weight/L). Concentrations of glycerol, ethanol, and organic acids were determined via HPLC using a Shimadzu Prominence SIL 20 system (Shimadzu Scientific Instruments, Inc., Columbia, MD) equipped with a refractive index detector and a HPX-87H organic acid column (Bio-Rad, Hercules, CA) with the following operating conditions: 0.3 mL/min flow rate, 30 mM H2SO4 mobile phase, column temperature 42 °C. Quantification and identification of additional compounds were carried out using an Agilent 7890B series custom gas chromatography system equipped with a 5977B inert plus mass selective detector turbo EI bundle (for identification), a flame ionization detector (for quantification), and an Agilent HP-5ms capillary column (0.25-mm internal diameter, 0.25-μm film thickness, 30-m length) as described in the SI Appendix.
Enzyme Assays.
Details on the preparation of cell extracts, protein purification, and specific enzyme assays are described in the SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by Bioactive Ingredients Corporation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest statement: The authors have filed a patent application (International Application No. PCT/US2017/022581). R.G. owns shares of Bioactive Ingredients Corporation.
This article is a PNAS Direct Submission. T.J.E. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821004116/-/DCSupplemental.
References
- 1.Chubukov V., Mukhopadhyay A., Petzold C. J., Keasling J. D., Martín H. G., Synthetic and systems biology for microbial production of commodity chemicals. NPJ Syst. Biol. Appl. 2, 16009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schrader J., Bohlmann J., Eds., Biotechnology of Isoprenoids (Springer International Publishing, 2015), vol. 148, pp. 1–475. [Google Scholar]
- 3.Ward V. C. A., Chatzivasileiou A. O., Stephanopoulos G., Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS Microbiol. Lett. 365, fny079 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Beller H. R., Lee T. S., Katz L., Natural products as biofuels and bio-based chemicals: Fatty acids and isoprenoids. Nat. Prod. Rep. 32, 1508–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Vickers C. E., Williams T. C., Peng B., Cherry J., Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr. Opin. Chem. Biol. 40, 47–56 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Erb T. J., et al. , A RubisCO-like protein links SAM metabolism with isoprenoid biosynthesis. Nat. Chem. Biol. 8, 926–932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel J. B., et al. , Computational protein design enables a novel one-carbon assimilation pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 3704–3709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwander T., Schada von Borzyskowski L., Burgener S., Cortina N. S., Erb T. J., A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354, 900–904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadadi N., Hatzimanikatis V., Design of computational retrobiosynthesis tools for the design of de novo synthetic pathways. Curr. Opin. Chem. Biol. 28, 99–104 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Bachmann B. O., Biosynthesis: Is it time to go retro? Nat. Chem. Biol. 6, 390–393 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Liu Y., Yan Z., Lu X., Xiao D., Jiang H., Improving the catalytic activity of isopentenyl phosphate kinase through protein coevolution analysis. Sci. Rep. 6, 24117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M., Poulter C. D., Characterization of thermophilic archaeal isopentenyl phosphate kinases. Biochemistry 49, 207–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn R. D., Clements J., Eddy S. R., HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbonell P., et al. , Selenzyme: Enzyme selection tool for pathway design. Bioinformatics 34, 2153–2154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke C., Croteau R., Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Arch. Biochem. Biophys. 405, 130–136 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Iijima Y., Gang D. R., Fridman E., Lewinsohn E., Pichersky E., Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol. 134, 370–379 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J., et al. , Engineering Escherichia coli for selective geraniol production with minimized endogenous dehydrogenation. J. Biotechnol. 169, 42–50 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Cheong S., Clomburg J. M., Gonzalez R., Energy- and carbon-efficient synthesis of functionalized small molecules in bacteria using non-decarboxylative Claisen condensation reactions. Nat. Biotechnol. 34, 556–561 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Alonso-Gutierrez J., et al. , Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab. Eng. 19, 33–41 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Wang C., Park J. E., Choi E. S., Kim S. W., Farnesol production in Escherichia coli through the construction of a farnesol biosynthesis pathway–Application of PgpB and YbjG phosphatases. Biotechnol. J. 11, 1291–1297 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Furubayashi M., et al. , A high-throughput colorimetric screening assay for terpene synthase activity based on substrate consumption. PLoS One 9, e93317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn F. M., Hurlburt A. P., Poulter C. D., Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J. Bacteriol. 181, 4499–4504 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneda K., Kuzuyama T., Takagi M., Hayakawa Y., Seto H., An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc. Natl. Acad. Sci. U.S.A. 98, 932–937 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George K. W., et al. , Metabolic engineering for the high-yield production of isoprenoid-based C-5 alcohols in E. coli. Sci. Rep. 5, 11128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y., et al. , Metabolic engineering of Escherichia coli for high-specificity production of isoprenol and prenol as next generation of biofuels. Biotechnol. Biofuels 6, 57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Luxenburger E., Müller R., An alternative isovaleryl CoA biosynthetic pathway involving a previously unknown 3-methylglutaconyl CoA decarboxylase. Angew. Chem. Int. Ed. Engl. 52, 1304–1308 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Bock T., et al. , AibA/AibB induces an intramolecular decarboxylation in isovalerate biosynthesis by Myxococcus xanthus. Angew. Chem. Int. Ed. Engl. 56, 9986–9989 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Clomburg J. M., et al. , Integrated engineering of β-oxidation reversal and ω-oxidation pathways for the synthesis of medium chain ω-functionalized carboxylic acids. Metab. Eng. 28, 202–212 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Clomburg J. M., Contreras S. C., Chou A., Siegel J. B., Gonzalez R., Combination of type II fatty acid biosynthesis enzymes and thiolases supports a functional β-oxidation reversal. Metab. Eng. 45, 11–19 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Kim S., Clomburg J. M., Gonzalez R., Synthesis of medium-chain length (C6-C10) fuels and chemicals via β-oxidation reversal in Escherichia coli. J. Ind. Microbiol. Biotechnol. 42, 465–475 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Zada B., et al. , Metabolic engineering of Escherichia coli for production of mixed isoprenoid alcohols and their derivatives. Biotechnol. Biofuels 11, 210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemmerlin A., Post-translational events and modifications regulating plant enzymes involved in isoprenoid precursor biosynthesis. Plant Sci. 203-204, 41–54 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Cheong S., Clomburg J. M., Gonzalez R., A synthetic pathway for the production of 2-hydroxyisovaleric acid in Escherichia coli. J. Ind. Microbiol. Biotechnol. 45, 579–588 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Kang Y. S., et al. , Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186, 8548 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neidhardt F. C., Bloch P. L., Smith D. F., Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.