Significance
Microbial communities have many applications, but current experimental strategies to investigate their behavior are limited by the combinatorial complexity of interactions between species. Here, we introduce a platform to automatically construct and test synthetic communities of microbes from a set of input species at a scale of ∼100,000 communities per day. As a first demonstration, we discovered specific compositions of bacteria isolated from local soil that promote the growth of a model plant symbiont. More broadly, our platform can be adopted for the discovery of microbial consortia with many useful properties, such as suppression of pathogens or degradation of recalcitrant substrates for use in biofuel production or environmental remediation.
Keywords: synthetic ecology, microbial interactions, community assembly, high-throughput screening, droplet microfluidics
Abstract
Microbial communities have numerous potential applications in biotechnology, agriculture, and medicine. Nevertheless, the limited accuracy with which we can predict interspecies interactions and environmental dependencies hinders efforts to rationally engineer beneficial consortia. Empirical screening is a complementary approach wherein synthetic communities are combinatorially constructed and assayed in high throughput. However, assembling many combinations of microbes is logistically complex and difficult to achieve on a timescale commensurate with microbial growth. Here, we introduce the kChip, a droplets-based platform that performs rapid, massively parallel, bottom-up construction and screening of synthetic microbial communities. We first show that the kChip enables phenotypic characterization of microbes across environmental conditions. Next, in a screen of ∼100,000 multispecies communities comprising up to 19 soil isolates, we identified sets that promote the growth of the model plant symbiont Herbaspirillum frisingense in a manner robust to carbon source variation and the presence of additional species. Broadly, kChip screening can identify multispecies consortia possessing any optically assayable function, including facilitation of biocontrol agents, suppression of pathogens, degradation of recalcitrant substrates, and robustness of these functions to perturbation, with many applications across basic and applied microbial ecology.
Microbial communities exhibit emergent consortia-level functions that are vital to all ecosystems on Earth. These functions include photosynthetic and chemosynthetic primary production (1), regulation of greenhouse gases (2), recycling of organic materials (3), and symbiotic protection of hosts against infectious agents (4, 5). The functionality and robustness of natural microbial communities suggest that synthetic consortia may someday be leveraged broadly as biotechnological tools (6). Indeed, such consortia have already been deployed for bioproduction (7), bioremediation (8), and probiotic-mediated therapies for a wide range of hosts and diseases (9–12).
The complexity of microbial interactions and environmental dependencies (13–16) can lead to unpredictable behaviors even in apparently simple communities, posing a challenge to design of consortia. Addressing this challenge likely requires the integration of multiple approaches, including the reverse-engineering of natural communities (e.g., inference-based co-occurrence analysis) (17) and further development of forward-engineering strategies (e.g., metabolic flux-balance analysis) (18, 19). Complimentarily, screening experimentally constructed synthetic combinations of strains can identify consortia with desired properties or validate rational designs (20–22).
High-throughput phenotypic screening has found widespread use as a discovery strategy for novel gene targets (23) and drugs (24), but its adoption in microbial consortia discovery has been hindered by the logistical complexity of constructing strain combinations. Conventional liquid handling techniques and platforms (e.g., pipette-based construction of combinations in multiwell plates) may not be sufficient to adequately sample combinatorial space in a single experiment (25). For example, from a library of just n = 16 strains, generating all subsets of size k = {1, 2, . . ., 7} in a single medium would require ∼160,000 liquid handling steps and 275 96-well plates (without replicates). As these combinations could not be prepared in advance and would have to be assembled on the timescale of cell division (∼1 h), generating even 10% of these combinations would likely be logistically impractical. Because constructing each community requires a unique set of liquid transfers, these experiments are also difficult to automate robotically. Indeed, combinatorial studies conducted in liquid media typically construct <103 unique synthetic communities (20, 26–28). Some of the largest combinatorial studies (29) use the Burkholder agar assay instead, whereby an array of n microbial colonies is introduced to an agar gel inoculated with a second species, generating n × 1 combinations per agar plate. Single studies using this assay can generate ∼103–104 interactions but are typically restricted to binary compositions. Diffusion between colonies further places an upper bound on the density of the colony array and throughput of the screen.
Here, we present a platform we call the kChip that addresses the experimental scale and setup time requirements to assay microbial community function in high throughput. The kChip system enables parallel construction and quantitative screening of ∼105 synthetic microbial communities per day and requires no robotic liquid handling. The platform screens n-multichoose-k combinations, meaning each community is composed of k inputs (e.g., strains, media) randomly selected from a larger library of n, where both n and k are selected by the user. The kChip platform generalizes a high-density microwell array approach (30, 31) that groups and merges sets of nanoliter droplets that each carry input components, a strategy we previously demonstrated for pairwise combinatorial compound screening (32). Droplets self-assemble randomly into groupings of k = {1, 2, . . ., 7, 19} dictated by microwell geometries, greatly reducing the time and logistical complexity of combination assembly. As with other droplet microfluidic systems, the kChip platform is amenable to fluorescent and label-free optical assays and uses small assay volumes that conserve valuable inputs. Furthermore, the length scale of kChip microwells (∼100–1,000 μm) is a natural ecological scale for interaction-driven microbial community assembly (33).
Results and Discussion
The kChip Rapidly Constructs Massively Parallel Community Sets of Controlled Size.
To generate parallel synthetic communities from a library of n inputs, each kChip contains tens of thousands of microwells where each microwell randomly groups k inputs. Multiple kChips and values of k can be used in accordance with the desired size, number, and replication of combinatorial groupings.
Setup (∼30 min) requires three steps: (i) droplet generation and pooling (10 min), (ii) droplet loading and grouping (20 min), and (iii) droplet merging (10 s) (Fig. 1A). Before droplet generation, a “color code” (unique ratio of three fluorescent dyes) is mixed with each input such that droplet contents can be reidentified via imaging (32). Color-coded droplet sets are generated separately on the benchtop (∼20,000 1-nL droplets per 20 μL) and pooled together to form a droplet library. The droplet library is loaded onto the kChip in a single pipetting step (via a custom kChip-loading jig; SI Appendix, Fig. S1). Droplets self-assemble into random groupings of k droplets determined by the size, shape, and internal design features of the microwells (Fig. 1B and SI Appendix, Figs. S2 and S3). The kChip is imaged (2× magnification) to identify the contents of each microwell from the droplet color codes (SI Appendix, Fig. S4). The droplets in each microwell are subsequently merged to combine their contents via exposure to an alternating-current electric field (corona treater; Electro-Technic Products) (Fig. 1) to finally generate the set of parallel n-multichoose-k synthetic communities.
Fig. 1.
kChip enables massively parallel construction of microbial communities. (A) To run a kChip screen, 1-nL droplets are first produced. Each droplet contains a color code (a specific ratio of three fluorescent dyes) that maps to a corresponding input. After they have been pooled, droplets are loaded onto the kChip, where they randomly group into microwells (SI Appendix, Fig. S1). The microwells are designed to group precisely k droplets. The kChip is imaged to identify the contents of each microwell from the droplet color codes. Droplets are then merged within their respective microwells via exposure to an alternating-current electric field, generating parallel synthetic communities. Community phenotypes can be tracked via optical assays, including fluorescent protein expression and respiration-driven reduction of resazurin to the fluorescent product resorufin. (B) Example micrographs show grouping and merging of droplets for different microwell types, the designs for which are described in SI Appendix, Fig. S2. Microwells are densely packed on the kChip, with microwell density varying inversely with size (k). A single microwell type can be arrayed across a kChip (“Full kChip”). For the screening application reported in Figs. 3 and 4, we have generated a “k = {1:7;19} Chip” that includes different microwell types arranged in parallel.
Microwells that group different numbers of inputs can be combined on a kChip in any organizational pattern (SI Appendix, Fig. S2). Because microwell density on a kChip decreases as k increases (owing to the increase in microwell size) (Fig. 1B), the total number of assay points on a kChip depends on its microwell layout (∼60,000 if all microwells are k = 1, ∼13,000 if all microwells are k = 7). Our working kChip, used for screening applications described below, has microwells that group up to seven or 19 droplets (“k = {1:7;19}”) with roughly even representation of each microwell type by number to enable simultaneous construction and assessment of communities of different richnesses (Fig. 1B).
Growth of Labeled and Unlabeled Strains Can Be Profiled Across Environmental Conditions.
kChip screening allows for rapid functional profiling of fluorescently labeled and unlabeled strains across libraries of environmental conditions (e.g., antibiotics, natural products, carbon sources) with flexible temporal resolution (limited only by kChip scan time, <15 min at 2× magnification). To compare kChip performance with a conventional approach, we obtained carbon utilization profiles [i.e., growth curves for each strain across different single carbon sources in a minimal medium (SI Appendix)] for a panel of both droplet cultures and conventional 96-well plate cultures (SpectraMax plate reader). On the kChip, we pooled a library of microbe-containing droplets with a library of carbon source-containing droplets and loaded the droplets onto a k = 2 Chip (i.e., all microwells on the kChip grouped two droplets). From microwells that received one droplet from each library (∼1/2 of the total microwells, or ∼17,000 microwells), we profiled the growth of each strain on each carbon source.
To track growth, we used one of two assays: (i) measurement of a constitutively expressed fluorescent protein [green fluorescent protein (GFP) or yellow fluorescent protein (YFP)] (Fig. 2A) or (ii) reduction of resazurin dye to the fluorescent product resorufin by cellular metabolism (proportional to cell density), a label-less assay that can be used with unlabeled or genetically intractable strains (34) (Fig. 2B). We selected 10 fluorescently labeled strains (SI Appendix, Table S1), and first confirmed that glucose utilization was recapitulated on a k = 1 Chip (SI Appendix, Fig. S5). We then crossed this strain panel with 13 carbon sources (SI Appendix, Table S2). Carbon utilization profiles produced from GFP or YFP signal on a k = 2 Chip and 96-well plates correlated strongly (Pearson r = 0.868) (Fig. 2C and SI Appendix, Fig. S6) with on-chip consistency between technical replicates (R2 = 0.968; SI Appendix, Fig. S7). Similarly, growth measurements of unlabeled strains using the resazurin assay (k = 2 Chip) correlated strongly with optical density (OD600) measurements in 96-well plates (Pearson r = 0.969) for a panel of three strains profiled across four carbon sources (Fig. 2D and SI Appendix, Fig. S8).
Fig. 2.
Carbon utilization profiles of labeled and unlabeled strains were measured on k = 2 Chips. (A) Droplet libraries can be made from a library of fluorescently labeled strains (SI Appendix, Table S1) and a set of carbon sources (SI Appendix, Table S2). The ability of each strain to grow on each carbon source can be measured by monitoring microwells that receive one microbe-containing droplet and one carbon source-containing droplet. (B) To measure growth of unlabeled strains, the dye resazurin is added to carbon source inputs before droplet production (postmerge concentration of 40 μM). Resazurin is reduced to the fluorescent product resorufin in the presence of metabolically active cells. (C) We measured fluorescence for a panel of 10 fluorescent strains (starting OD600 = 0.02) across 15 conditions [13 carbon sources at 0.5% (wt/vol), one additional glucose replicate control, and one negative control (no carbon)] in k = 2 Chip microwells (21 °C, no shaking) as well as 200-μL cultures in 96-well plates (21 °C, 220 rpm). Heatmaps show the relative signal at 50 h, with interleaved columns corresponding to the kChip and 96-well plates (Pearson r = 0.868) (full time course is shown in SI Appendix, Fig. S6). (D) We measured the resazurin signal (fluorescence due to resorufin accumulation) for three strains (starting OD600 = 0.005) across four carbon source conditions in k = 2 Chip microwells (21 °C, no shaking) and compared those signals with OD600 measurements from 200-μL cultures in 96-well plates (21 °C, 220 rpm). Heatmaps show signal at 22 h (Pearson r = 0.969) (full time course is shown in SI Appendix, Fig. S8). In C and D, the relative signal for each row is obtained by normalizing to the maximum across all carbon sources and time points after background subtraction. Ec, Escherichia coli; GlcNAc, N-acetylglucosamine; Hf, Herbaspirillum frisingense; Pae, Pseudomonas aeruginosa; Pau, Pseudomonas aurantiaca; Pch, Pseudomonas chlororaphis; Pci, Pseudomonas citronellolis; Pf, Pseudomonas fluorescens; Pp, Pseudomonas putida; Ps, Pseudomonas syringae; Pv, Pseudomonas veronii; Rep, replicate.
kChip Screening Identifies Compositions That Robustly Promote Growth of Herbaspirillum frisingense.
One application of kChip synthetic community screening is the discovery of compositions that promote or suppress the growth of a strain of interest. Moreover, the robustness of the effects of these compositions across abiotic environments and the presence of additional environmental strains (“isolates”) can be simultaneously assessed. Discovering such communities might inform the composition of defined probiotic interventions.
In a pilot screen, we measured the yield of a GFP-expressing strain of Herbaspirillum frisingense (“Hf-GFP”) (35), a model plant symbiont for which growth is likely impacted by variable biotic and abiotic environments in agricultural settings. We isolated a diverse set of soil bacteria (“isolates”) (SI Appendix, Fig. S9 and Dataset S1) and measured Hf-GFP yield across communities composed of Hf-GFP and combinations of isolates. These communities were constructed across different carbon sources that included carbohydrate oligomers (sucrose, lactose, and raffinose) and their monomeric constituents (glucose, galactose, and fructose) (Fig. 3A). Hf-GFP grew in monoculture to various extents on each of these carbon sources, with no detectable growth on sucrose (SI Appendix, Fig. S10).
Fig. 3.
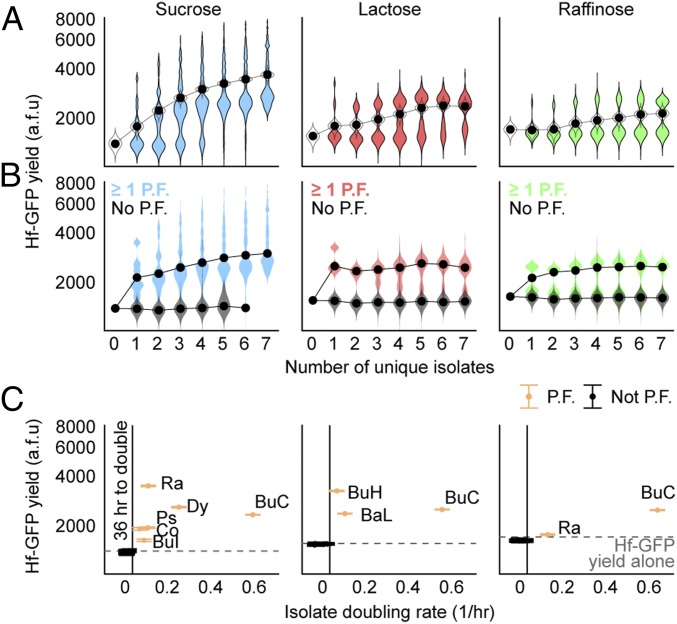
High-throughput kChip screening identifies H. frisingense-promoting compositions that are robust to carbon source and community context. (A) Screen schematic to identify Hf-GFP–promoting compositions. Assays are constructed whereby Hf-GFP represents half of the starting biomass (starting Hf-GFP OD600 = 0.02) and the other half is divided evenly among one to seven or 19 soil isolate inputs (starting total isolate OD600 = 0.02 if no control droplets are present). Each of these communities is constructed in one of six media that each contain a single carbon source. Each carbon source enables a different Hf-GFP monoculture yield (SI Appendix, Fig. S10). Droplets containing the same carbon source are pooled and loaded onto the same kChip (six kChips in total, 21 °C, no shaking). After droplet merging, Hf-GFP yield is measured (24 h, 48 h, and 72 h) in each community/carbon source environment. (B) Total number of assay points collected for different values of k (about evenly divided among the six kChips; Dataset S2). (C) Ranked Hf-GFP yield at 72 h for all constructed compositions. A median is represented when a composition is replicated more than one time (with a mean calculated in instances of two replicates), error bar = SEM, and dotted line = Hf-GFP yield in monoculture. (D) Effect of each s-sized composition on Hf-GFP was analyzed in two ways to identify the most facilitative and robust compositions. Here, the composition [BaL + Ra + Ps] is used as an example. (Top) First, all instances of [BaL + Ra + Ps] appearing in k = 3 microwells were identified across all carbon sources, and the median Hf-GFP yield for these was calculated (“Hf-GFP median yield”). (Bottom) Second, all instances of [BaL + Ra + Ps + one or more additional isolate] in k ≥ 4 microwells were identified across all carbon sources, and the 10th percentile of Hf-GFP yield for these was calculated (“Hf-GFP robustness”). The color of each data point indicates the carbon source. Gray dotted line = minimal viable Hf-GFP yield (1,500 GFP counts, or 1 SD above Hf-GFP monoculture yield in sucrose medium). (E) For compositions represented 30 or more times across all carbon sources (only k = 1, k = 2, and k = 3 compositions met this criterion; SI Appendix, Fig. S13), Hf-GFP median yield and Hf-GFP robustness were calculated as described in D. Dark blue points indicate a composition contains at least BuC. Dark green points indicate a composition contains at least [BaL + Ra]. The diagonal line is the x = y line. a.f.u., arbitrary fluorescence units; Av, Averyella dalhousiensis; Ch, Chryseobacterium sp.; Co, Collimonas sp.; Ew, Ewingella americana; Ps, Pseudomonas fluorescens.
We generated droplets that each contained the following: (i) Hf-GFP (starting OD600 = 0.02), (ii) one isolate (starting OD600 = 0.02, chosen among 14 isolates + one no-isolate control + one negative control) such that the synthetic communities contained the same initial [Hf-GFP]/[total isolate] ratio if no control droplets were present, and (iii) one carbon source. All droplets that received the same carbon source were loaded onto the same kChip such that droplet grouping produced combinations of k = 1, 2, . . ., 7, or 19 inputs with the carbon source type and concentration held constant. Overall, we produced ∼100,000 assay points (evenly divided among the carbon sources) (Fig. 3B and Dataset S2).
Most combinations of isolates affected Hf-GFP yield at 72 h, showing both suppressive (decrease in yield) and facilitative (increase in yield) effects (Fig. 3C and SI Appendix, Fig. S10). On carbon sources where Hf-GFP monocultures achieved high yield (glucose and galactose), the addition of other isolates almost always attenuate Hf-GFP growth. By contrast, facilitative compositions were common on carbon sources where Hf-GFP growth was low (fructose, raffinose, and lactose) and ubiquitous when undetectably low (sucrose).
To score the effect of each combination, we here differentiate between a “composition” as a given isolate subset of size s, for example, [A + B + C] (s = 3), and a “community” as a larger set of size k that contains the given composition and one or more additional isolates, for example, all communities [A + B + C + X + . . . + Y] (k ≥ 4) (Fig. 3D). For the majority of compositions, the facilitative effect did not persist across different carbon sources or community contexts. For example, the composition [Bacillus sp. I + Rahnella sp.] ([BaT + Ra]), measured at k = 2 on each kChip, facilitated Hf-GFP growth on fructose, sucrose, and raffinose, but suppressed its growth on the other carbon sources (SI Appendix, Fig. S11A). Similarly, the facilitation imparted by the composition [Enterobacter mori + Dyella sp.] ([En + Dy]) (measured at k = 2) in a medium containing galactose was not robust to community context [En + Dy + one or more additional unique isolates] (an s = 2 composition among all k ≥ 3 communities on the same kChip) (SI Appendix, Fig. S11B).
We sought to determine the facilitative compositions that were most robust to both carbon source and community context. We scored each composition in two ways (Fig. 3D). First, we computed median Hf-GFP yield at 72 h in coculture with just the composition across all carbon sources (“Hf-GFP median yield”). Second, we computed the 10th percentile of Hf-GFP yield in coculture with all communities containing the given composition to detect whether the composition’s effect on Hf-GFP was appreciably diminished by additional isolates across all carbon sources (“Hf-GFP robustness”). Based on analysis of variability in Hf-GFP monoculture data, we restricted the analysis to instances where a given composition was represented five or more times, on average, on a kChip (or ≥30 times in total) (SI Appendix, Fig. S12), which occurred for about half of s = k = 3 compositions (SI Appendix, Figs. S13 and S14).
We uncovered two isolate compositions that were strongly facilitative and robust to both carbon source and community context (Fig. 3E). While most facilitative compositions showed robustness to community context for a given carbon source (SI Appendix, Fig. S15), few showed robustness to both carbon source and community context. Interestingly, we identified that a single isolate, Burkholderia sp. I (BuC), or the isolate composition, [Bacillus sp. II + Rahnella sp.] ([BaL + Ra]), was consistently present among combinations that scored highly on both metrics. We validated these two compositions’ facilitative effects on Hf-GFP with the different carbon sources in 96-well plate coculture experiments (SI Appendix, Fig. S16).
Facilitation Increases with Community Richness and Is Driven by a Small Number of Strains.
Systematic screening data like those we generated here can reveal ecological trends that underpin particularly facilitative or robust compositions like BuC or [BaL + Ra]. Broadly, we found that Hf-GFP yield either increased or declined with community richness depending on its baseline growth on each carbon source in monoculture. In raffinose, lactose, and sucrose, the three carbon sources where it grew most poorly, Hf-GFP yield increased and then plateaued with community richness (Fig. 4A). We observed a similar but weaker trend in fructose, where Hf-GFP grew to a limited extent alone, and an isolate-agnostic suppressive effect in glucose and galactose, where Hf-GFP grew well in monoculture (SI Appendix, Fig. S17). We observe the same trend for k = 1–7 inputs and a roughly equivalent yield of Hf-GFP for seven and 19 inputs when considering all sampled community compositions (SI Appendix, Fig. S18).
Fig. 4.
Facilitation increases with community richness and is driven by a subset of strains. (A) In a medium containing sucrose, lactose, or raffinose, Hf-GFP yield increased with community richness. Colored distributions indicate median Hf-GFP yields for all unique compositions at a given k (i.e., all droplets in a given combination contain different strains). The black data point indicates the median of distribution. Outlined distributions represent medians of 100 bootstrap-resampled datasets at each k, whereby the Hf-GFP yield dataset for each k was resampled with replacement (with a resampling sample size equal to the actual sampling size), and a median of the resampled data was calculated each time. (B) Presence of one or more primary facilitator (P.F.) (SI Appendix, Fig. S19) was necessary and typically sufficient to enable Hf-GFP growth and drive a facilitative effect when additional isolates were present. In the case of raffinose, one of the two primary facilitators (Ra) facilitated Hf-GFP weakly, giving rise to a clear bimodal distribution. Colored distributions indicate Hf-GFP growth in communities possessing one or more primary facilitators. Gray distributions indicate Hf-GFP yield in communities with no primary facilitators (with distributions absent when there were no communities in the dataset consisting of all unique inputs and no primary facilitators). (C) Resazurin assay was conducted on a separate k = 2 Chip in parallel with the screen to measure the growth rate of each isolate (SI Appendix, Fig. S20). The subset of isolates that grew on a given carbon source (defined as one or more doubling of resorufin fluorescence by 36 h) corresponded to the subsets of isolates identified as primary facilitators. The number of data points in each distribution is given in Dataset S2. a.f.u., arbitrary fluorescence units; BuH, Burkholderia sp. II; BuI, Burkholderia sp. III; Co, Collimonas sp.; Ps, Pseudomonas fluorescens.
In coculture with a single isolate (k = 1 microwell), we identified “primary facilitator” strains that facilitated Hf-GFP growth on a given carbon source (SI Appendix, Fig. S19). We further found that the inclusion of one or more primary facilitator strains was necessary and almost always sufficient to facilitate Hf-GFP growth regardless of the number of other strains present (calculated from k > 1 microwells) (Fig. 4B). In conjunction with the screen, we also assayed growth rates of the isolates on the different carbon sources via the resazurin assay on a k = 2 Chip (SI Appendix, Fig. S20). For the raffinose, lactose, and sucrose conditions, we found that the subset of isolates that could grow (one or more doublings by 36 h) matched the subset of primary facilitators (Fig. 4C). We concluded that, for these conditions, growth of an isolate was necessary and sufficient for its ability to facilitate Hf-GFP growth. To investigate why facilitation increased with community richness beyond the presence of one primary facilitator, we first considered the k = 2 level. We found many instances where Hf-GFP yield in the presence of two isolates was greater than its yield with either isolate individually, particularly when the carbon source was sucrose (SI Appendix, Fig. S21). With a single primary facilitator present, we observed that the largest Hf-GFP yield increases were imparted by the addition of a second primary facilitator (SI Appendix, Fig. S22). We observed modest increases imparted by the addition of a non[primary facilitator] when a primary facilitator was present.
These data point to general design principles useful in constructing facilitative consortia. Based on carbon source utilization as a criterion for primary facilitation (Fig. 4C), we might expect that certain “core” compositions of isolates facilitate Hf-GFP across all carbon sources if at least one isolate within the core composition is able to grow on each carbon source. Further, based on the increase in Hf-GFP yield we see with community richness on carbon sources disfavored by Hf (Fig. 4A and SI Appendix, Figs. S21 and S22), we might expect improvements to the facilitative effect size or its robustness with the incorporation of specific isolates to the composition. Indeed, results from our two top-scoring compositions, BuC and [BaL + Ra], are consistent with these principles (SI Appendix, Fig. S23).
Conclusion
Droplet microfluidics have recently been applied in single-cell transcriptomics, drug discovery, and microbiology (30, 36). The kChip platform expands upon these applications to enable the rapid construction and high-throughput screening of beyond-pairwise combinations. Here, we have demonstrated that the kChip screening paradigm is compatible with diverse bacterial strains and media conditions, and supports various optical growth assays. Demonstrating the utility of kChip screening, we discovered and validated compositions that facilitate the model plant symbiont H. frisingense in a manner robust to carbon source and community context. We further extrapolated ecological trends in the data, derived principles from our large dataset about consortia design principles, demonstrated the applicability of these principles to top-scoring compositions identified in the screen, and explored the underlying ecology of our results (SI Appendix, Supplemental Discussion).
The kChip has numerous applications in elucidating microbial community ecology. For example, datasets might be leveraged to parameterize or assess mathematical models of growth or interactions as well as to determine how biotic metrics (e.g., species diversity) and abiotic factors (e.g., carbon substrates) drive metabolic decision making and interactions. Screens can also be used to suggest community design principles (37) and the environments that induce desirable interactions (38, 39).
Beyond fundamental ecological studies, kChip screens can generate short lists of “hit” microbial mixtures that are also robust to relevant biotic and abiotic perturbations and constitute attractive candidates for development into therapeutic probiotics (10–12, 40–43) (SI Appendix, Supplemental Discussion). More broadly, any optically detectable community-wide phenotype can be screened, providing access across many fields of application (e.g., the production of cryptic interaction-mediated metabolites for natural products discovery) (44).
Materials and Methods
Microbial Culture Input Preparation.
All bacterial cultures underwent an initial “starter phase,” whereby glycerol stocks of environmental isolates and fluorescently labeled strains were inoculated into 525 μL of lysogeny broth (LB) medium (2-mL-deep, 96-well plate via pin replicator) and 4 mL of LB (15-mL culture Falcon tube), respectively (30 °C, 220 rpm, 16 h). A subsequent “preculture phase” (30 °C, 220 rpm, 24 h) consisted of washing cells and resuspending them in M9 minimal medium (MM) with 0.5% (wt/vol) glucose at an initial OD600 of 0.01. The “experimental phase” consisted of washing and resuspending cells typically to a starting OD600 of 0.02 in MM (or ∼20 cells per droplet).
kChip Input Preprocessing.
For the screen described in Fig. 3, each droplet contained Hf-GFP, a single cocultured isolate, and a single carbon source. Every input received a “color code,” a unique ratio of three fluorescent dyes (1 μM or 10 μM) before generating droplets (SI Appendix, Fig. S4). Every input received 0.05% (wt/vol) bovine serum albumin to aid in retention of hydrophobic small molecules during droplet pooling and loading (32, 45).
Droplet Making and kChip Loading.
Droplets were produced on a Bio-Rad QX200 Droplet Generator in a fluorocarbon oil (3M Novec 7500). Droplets were pooled to prepare a total of 200 μL of droplet suspension (∼5 min) and injected into a custom-built kChip loading apparatus (SI Appendix, Fig. S1). Each microwell randomly sampled k droplets (∼5–10 min). The kChip was scanned at 2× magnification to identify the droplets in each microwell from their color codes (∼12–15 min) (SI Appendix, Fig. S4). Droplets were merged within their microwells via ∼10 s of exposure to an alternating-current electric field (4.5 MHz, 10,000–45,000 volts; Electro-Technic Products corona treater).
Fluorescence Microscopy.
All fluorescence microscopy was performed using a Nikon Ti-E inverted fluorescence microscope with fluorescence excitation by a Lumencor Sola light-emitting diode illuminator. Images were collected by means of a Hamamatsu ORCA-Flash 4.0 CMOS camera.
Supplementary Material
Acknowledgments
We thank Professor Otto Cordero and Professor Ahmad Khalil for early discussions about the use of droplet combinations to construct microbial communities. We also thank Prianca Tawde and Jameson Kief for assistance; members of the J.G. laboratory (MIT), Nadav Kashtan, and Mike Rothballer for samples; members of the laboratory of Otto Cordero (MIT), the S.K. laboratory (University of Illinois at Urbana–Champaign), the laboratory of Eric Alm (MIT), the J.G. laboratory (MIT), and the laboratory of Jim Collins (MIT, Broad Institute) for discussions; Alfonso Pérez-Escudero for reviewing the manuscript; and the Jupyter, numpy, scipy, scikit-image, scikit-learn, and pandas open-source development teams. This work was supported by National Science Foundation Graduate Research Fellowship Program [to J.K. (Fellow ID 2016220942) and A.K. (Fellow ID 2013164251)], an NIH grant [to C.M.A. (Grant F32CA236425)], the MIT Institute for Medical Engineering and Science Broshy Fellowship (to A.K.), a Career Award at the Scientific Interface from the Burroughs Wellcome Fund [to P.C.B. (Grant 1010240)], an MIT Deshpande Center Innovation Grant (to P.C.B.), a Scialog seed grant from the Gordon and Betty Moore Foundation and the Research Corporation for Science Advancement (to S.K. and P.C.B.), a grant from the Simons Foundation [to J.G. (Grant 542385)], a Bridge Project grant from the Dana Farber/Harvard Cancer Center and the Koch Institute for Integrative Cancer Research at MIT (to P.C.B.), a technology development seed grant from the Merkin Institute for Transformative Technologies in Healthcare at the Broad Institute (to P.C.B.), and a grant from the United States-Israel Binational Science Foundation [to J.F. (Grant 2017179)].
Footnotes
Conflict of interest statement: P.C.B. is an extramural faculty member of MIT’s Koch Institute for Integrative Cancer Research and a consultant to and equity holder in two companies in the microfluidics industry, 10X Genomics and General Automation Lab Technologies. The Broad Institute and MIT may seek to commercialize aspects of this work, and related applications for intellectual property have been filed.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900102116/-/DCSupplemental.
References
- 1.Hemingway J. D., et al. , Microbial oxidation of lithospheric organic carbon in rapidly eroding tropical mountain soils. Science 360, 209–212 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Bardgett R. D., Freeman C., Ostle N. J., Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2, 805–814 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Shi S., et al. , Plant roots alter microbial functional genes supporting root litter decomposition. Soil Biol. Biochem. 127, 90–99 (2018). [Google Scholar]
- 4.Buffie C. G., et al. , Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berendsen R. L., Pieterse C. M. J., Bakker P. A., The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Hays S. G., Patrick W. G., Ziesack M., Oxman N., Silver P. A., Better together: Engineering and application of microbial symbioses. Curr. Opin. Biotechnol. 36, 40–49 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Zhou K., Qiao K., Edgar S., Stephanopoulos G., Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 33, 377–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L., et al. , Removal of methyl parathion from artificial off-gas using a bioreactor containing a constructed microbial consortium. Environ. Sci. Technol. 42, 2136–2141 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Mendes R., et al. , Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Caballero S., et al. , Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe 21, 592–602.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawley T. D., et al. , Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8, e1002995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brugiroux S., et al. , Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2, 16215 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Mitri S., Foster K. R., The genotypic view of social interactions in microbial communities. Annu. Rev. Genet. 47, 247–273 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Ghoul M., Mitri S., The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Momeni B., Xie L., Shou W., Lotka-Volterra pairwise modeling fails to capture diverse pairwise microbial interactions. eLife 6, e25051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Gorostiaga A., Bajić D., Osborne M. L., Poyatos J. F., Sanchez A., High-order interactions dominate the functional landscape of microbial consortia. bioRxiv:10.1101/333534 (29 May 2018). [DOI] [PMC free article] [PubMed]
- 17.Tsai K.-N., Lin S.-H., Liu W.-C., Wang D., Inferring microbial interaction network from microbiome data using RMN algorithm. BMC Syst. Biol. 9, 54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harcombe W. R., et al. , Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 7, 1104–1115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnúsdóttir S., et al. , Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 35, 81–89 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Friedman J., Higgins L. M., Gore J., Community structure follows simple assembly rules in microbial microcosms. Nat. Ecol. Evol. 1, 109 (2017). [DOI] [PubMed] [Google Scholar]
- 21.de Vos M. G. J., Zagorski M., McNally A., Bollenbach T., Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proc. Natl. Acad. Sci. U.S.A. 114, 10666–10671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell T., Newman J. A., Silverman B. W., Turner S. L., Lilley A. K., The contribution of species richness and composition to bacterial services. Nature 436, 1157–1160 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Croston G. E., Functional cell-based uHTS in chemical genomic drug discovery. Trends Biotechnol. 20, 110–115 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Sundberg S. A., High-throughput and ultra-high-throughput screening: Solution- and cell-based approaches. Curr. Opin. Biotechnol. 11, 47–53 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Nai C., Meyer V., From axenic to mixed cultures: Technological advances accelerating a paradigm shift in microbiology. Trends Microbiol. 26, 538–554 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Foster K. R., Bell T., Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Mee M. T., Collins J. J., Church G. M., Wang H. H., Syntrophic exchange in synthetic microbial communities. Proc. Natl. Acad. Sci. U.S.A. 111, E2149–E2156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venturelli O. S., et al. , Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol. 14, e8157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordero O. X., et al. , Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 337, 1228–1231 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Kaminski T. S., Scheler O., Garstecki P., Droplet microfluidics for microbiology: Techniques, applications and challenges. Lab Chip 16, 2168–2187 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Cira N. J., Ho J. Y., Dueck M. E., Weibel D. B., A self-loading microfluidic device for determining the minimum inhibitory concentration of antibiotics. Lab Chip 12, 1052–1059 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Kulesa A., Kehe J., Hurtado J. E., Tawde P., Blainey P. C., Combinatorial drug discovery in nanoliter droplets. Proc. Natl. Acad. Sci. U.S.A. 115, 6685–6690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datta M. S., Sliwerska E., Gore J., Polz M. F., Cordero O. X., Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7, 11965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boedicker J. Q., Li L., Kline T. R., Ismagilov R. F., Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip 8, 1265–1272 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Straub D., Rothballer M., Hartmann A., Ludewig U., The genome of the endophytic bacterium H. frisingense GSF30(T) identifies diverse strategies in the Herbaspirillum genus to interact with plants. Front. Microbiol. 4, 168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terekhov S. S., et al. , Ultrahigh-throughput functional profiling of microbiota communities. Proc. Natl. Acad. Sci. U.S.A. 115, 9551–9556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johns N. I., Blazejewski T., Gomes A. L., Wang H. H., Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 31, 146–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hom E. F. Y., Murray A. W., Plant-fungal ecology. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science 345, 94–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W., Chow K.-H., Fleming E., Oh J., Selective colonization ability of human fecal microbes in different mouse gut environments. ISME J. 13, 805–823 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borody T. J., Khoruts A., Fecal microbiota transplantation and emerging applications. Nat. Rev. Gastroenterol. Hepatol. 9, 88–96 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Hu J., et al. , Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. MBio 7, e01790-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharyya P. N., Jha D. K., Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Benizri E., Baudoin E., Guckert A., Root colonization by inoculated plant growth-promoting rhizobacteria. Biocontrol Sci. Technol. 11, 557–574 (2001). [Google Scholar]
- 44.Seyedsayamdost M. R., Traxler M. F., Clardy J., Kolter R., Old meets new: Using interspecies interactions to detect secondary metabolite production in actinomycetes. Methods Enzymol. 517, 89–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skhiri Y., et al. , Dynamics of molecular transport by surfactants in emulsions. Soft Matter 8, 10618–10627 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.