Significance
Most organisms experience daily light–dark changes and show rhythms of basic biological processes such that they occur at optimal times of the day. Rhythms are also observed in a multitude of marine organisms, but their molecular foundations are still largely unknown. Here, we report daily oscillations of gene expression and cell fluorescence in the diatom Phaeodactylum tricornutum, which persist in the absence of external timing cues. We demonstrate that the protein RITMO1, encoded by a bHLH-PAS gene, which is widely represented in algal genomes, regulates these rhythms. By demonstrating circadian regulation in the most species-rich algal group in the ocean, this study unveils critical features of diatom biology, thus advancing the field of diurnal and circadian rhythms in marine algae.
Keywords: diatom, circadian rhythms, bHLH-PAS, gene expression, cellular fluorescence
Abstract
Periodic light–dark cycles govern the timing of basic biological processes in organisms inhabiting land as well as the sea, where life evolved. Although prominent marine phytoplanktonic organisms such as diatoms show robust diel rhythms, the mechanisms regulating these processes are still obscure. By characterizing a Phaeodactylum tricornutum bHLH-PAS nuclear protein, hereby named RITMO1, we shed light on the regulation of the daily life of diatoms. Alteration of RITMO1 expression levels and timing by ectopic overexpression results in lines with deregulated diurnal gene expression profiles compared with the wild-type cells. Reduced gene expression oscillations are also observed in these lines in continuous darkness, showing that the regulation of rhythmicity by RITMO1 is not directly dependent on light inputs. We also describe strong diurnal rhythms of cellular fluorescence in wild-type cells, which persist in continuous light conditions, indicating the existence of an endogenous circadian clock in diatoms. The altered rhythmicity observed in RITMO1 overexpression lines in continuous light supports the involvement of this protein in circadian rhythm regulation. Phylogenetic analysis reveals a wide distribution of RITMO1-like proteins in the genomes of diatoms as well as in other marine algae, which may indicate a common function in these phototrophs. This study adds elements to our understanding of diatom biology and offers perspectives to elucidate timekeeping mechanisms in marine organisms belonging to a major, but under-investigated, branch of the tree of life.
The Earth’s rotation means that life evolved under a daily cycle of alternate light and dark periods. Most living organisms have developed rhythms of many fundamental biological processes, ranging from physiology to behavior, such that they occur at optimal times of the day, which can enhance fitness (1). These rhythms are the product of the coordinated action of signals from endogenous biological clocks, together with environmental and metabolic inputs (2–4). In most eukaryotes, biological rhythms are controlled by interconnected transcriptional-translational feedback loops involving transcription factors (TFs). Although this regulatory framework is conserved among eukaryotes, the regulators orchestrating these rhythms seem to have emerged several times through evolution (5, 6).
Robust diel rhythms in growth, gene expression, pigment synthesis, phototactic movements, and bioluminescence have been observed in a variety of phytoplanktonic organisms, including diatoms (7–10). Diatoms represent the most species-rich group of algae in the ocean and populate a wide range of aquatic environments (11). These algae of the Stramenopila lineage, derived by secondary endosymbiosis events (12), show peculiar genomic, metabolic, and cellular features and have an impressive capacity to deal with environmental changes (e.g., refs. 12–16). Genome-wide analyses have also shown that 25% of the diurnal transcriptome is influenced by light–dark cycles in the centric diatom Thalasiossira pseudonana (17). Additional studies in the pennate diatom Phaeodactylum tricornutum have highlighted strong synchronization of the cell cycle with day–night cycles and a strict temporal separation of transcriptional gene networks and metabolism (18–21), as observed in other algae (22, 23). However, the molecular mechanisms orchestrating these processes are still unknown in diatoms and many other phytoplanktonic organisms, except for the green alga Ostreococcus tauri, which possesses a plant-like circadian clock (24). Indeed, no clear homologs of the circadian clock components discovered in bacteria, fungi, animals, or plants have been found in the diatom genomes except for cryptochromes and casein kinases (13).
In this work, by characterizing gene transcription and cell fluorescence rhythms in P. tricornutum, we uncover the existence of circadian rhythms in diatoms and demonstrate the involvement of the bHLH-PAS protein bHLH1a, here named RITMO1, in the regulation of these processes. Notably, the bHLH-PAS domains feature in proteins involved in the regulation of rhythmic processes in animals (5). Phylogenetic analyses reveal a wide distribution of RITMO-like homologs in the genomes of diatoms and other marine algae. Thus, RITMO1 represents a key molecular entry point for the identification of the timekeeper components in diatoms and paves the way for a deeper understanding of marine rhythms and their evolutionary and ecological relevance.
Results
Transcriptome Profiling Identifies Potential Regulators of Diurnal Rhythms in P. tricornutum.
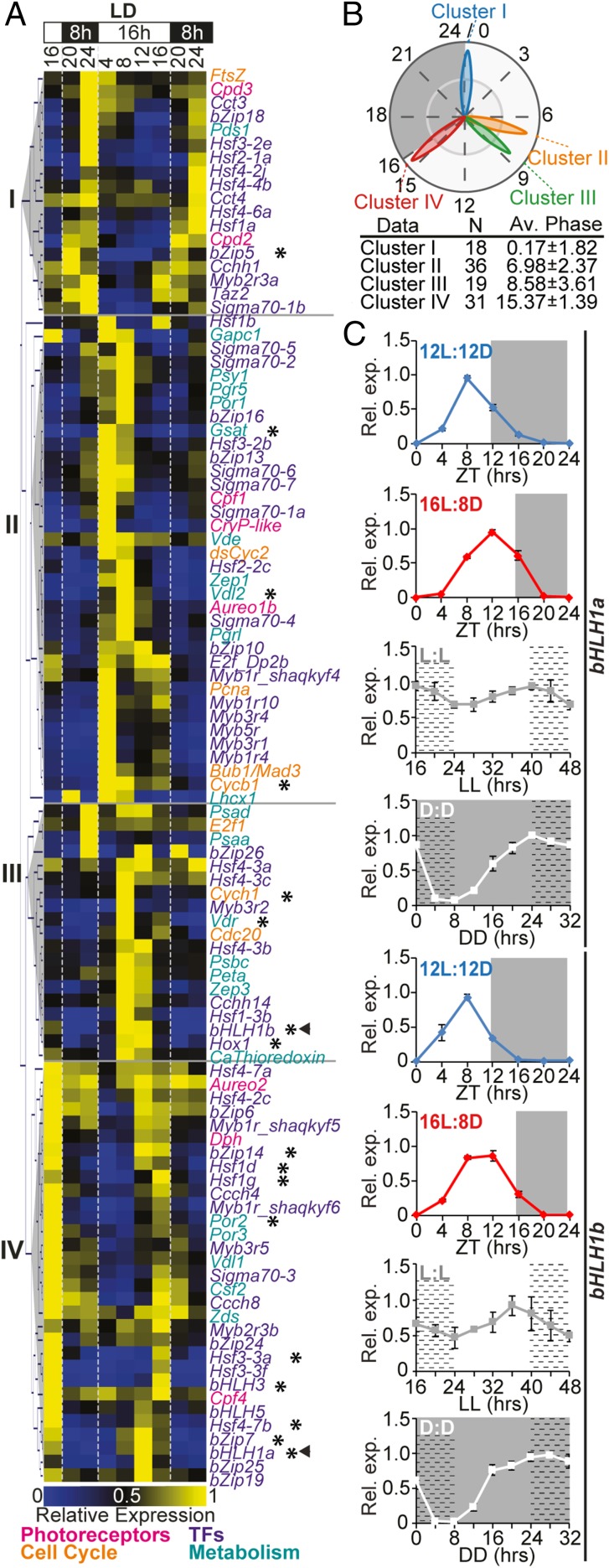
To identify potential regulators of cellular rhythmicity in P. tricornutum, a publicly available diurnal transcriptomic dataset (18) was analyzed. One hundred and four genes with robust diel oscillating expression were selected, including eight photoreceptors (19, 25–27), 66 TFs (28), and 30 potential output genes implicated in diel rhythmic processes (pigment synthesis, cell cycle regulation, and photosynthesis) (SI Appendix, Table S1). The transcriptional dynamics of the selected genes were further examined in a 16:8-h light:dark (L:D) photocycle for 32 h. Hierarchical clustering analysis revealed four main clusters of coexpressed genes, termed I–IV (Fig. 1 A and B). Cluster I phased at dawn, at around Zeitgeber Time 0 h (ZT: hours after illumination), suggesting a transcriptional anticipation of the light onset. This cluster comprised 18 genes including 14 TFs, mostly belonging to the Heat Shock Transcription Factor family (HSF), the two DNA repair enzymes CPD photolyases (CPD2 and CPD3), and one carotenoid synthesis enzyme (PDS1). Cluster II phased around ZT7 and encompassed 36 genes, including the dsCYC2 gene controlling the onset of cell division (19). Cluster II also contained 18 TFs, of which 8 were sigma factors putatively involved in the regulation of chloroplast transcription, three genes implicated in photoprotection (LHCX1, Vdl2, and Zep1), and the chlorophyll synthesis POR1 gene. Such active transcription of genes involved in chloroplast activity during the light period has been shown previously (18, 29). The blue light sensors Aurochrome1b and the cryptochromes CPF1 and CryP-like also belonged to cluster II and showed a strong expression following light onset in accordance with previous observations (26, 30). Cluster III phased around ZT9 and comprised 9 TFs and 10 metabolism-related genes. Finally, cluster IV phased just before dusk, at around ZT15, and included 23 TF genes, likely contributing to preparing cells for light-to-dark transition. Cluster IV also contained the CPF4 and the far-red light sensing phytochrome (DPH1), the peak expression of which at the end of the light period has been observed previously (26, 31).
Fig. 1.
Diurnal expression of rhythmic P. tricornutum genes. (A) Hierarchical clustering of gene expression profiles in 16L:8D, analyzed by nCounter analysis. Arrowheads highlight bHLH1a and bHLH1b; asterisks indicate the transcript profiles oscillating in D:D, shown in SI Appendix, Fig. S1. (B) Polar plot showing average phases of expression of the four clusters. N: number of genes within each cluster; Av. Phase: average phases ± SD. (C) bHLH1a and bHLH1b expression in 12L:12D, 16L:8D, L:L, and D:D (mean ± SEM, n = 3). In L:L and D:D, cells entrained under 16L:8D cycles were released to continuous light or dark at ZT16. White and gray regions represent light and dark periods; dashed regions represent subjective nights in L:L and D:D. The expression value of each gene is relative to its maximum expression.
To explore transcription dynamics in the absence of light inputs, the expression of these rhythmic genes was further analyzed in cells exposed to continuous darkness (D:D) for 30 h. The analysis of transcript profiles revealed that around 20% of the analyzed genes showed persistent oscillating expression in D:D, although some profiles displayed reduced amplitudes and/or shifted phases of expression compared with the L:D condition (SI Appendix, Fig. S1). In particular, we identified 18 genes, including putative TFs, pigment-related enzymes, and cell cycle-related genes, showing the highest amplitudes of expression in both L:D and D:D. Conversely, the other 80% of the analyzed expression profiles did not have persistent oscillating patterns in darkness and instead had strongly reduced amplitudes or altered expression timing compared with the L:D condition (SI Appendix, Fig. S2).
Rhythmic bHLH1a and bHLH1b Expression Is Adjusted in a Photoperiod-Dependent Manner and Persists in Continuous Light and Continuous Dark Conditions.
Our analysis identified two TFs—bHLH1a (Phatr3_J44962) belonging to cluster IV and bHLH1b (Phatr3_J44963) belonging to cluster III—which each have a Per-ARNT-Sim (PAS) domain in conjunction with a bHLH DNA-binding domain. Because bHLH-PAS proteins have been shown to be involved in the regulation of rhythmic processes in animals (5), the expression profiles of bHLH1a and bHLH1b were further examined in P. tricornutum cells grown under different photoperiods. bHLH1a expression peaked at ZT8 in the 12L:12D photoperiod and at ZT12 in the 16L:8D photoperiod, 4 h before the end of the light period in both cases, and then gradually decreased to below detection limits at ZT0 (Fig. 1C). Transcription of bHLH1b appeared to start earlier than that of bHLH1a. In cells entrained in 12L:12D cycles, bHLH1b expression peaked at ZT8, whereas it peaked between ZT8 and ZT12 in 16L:8D photoperiods (Fig. 1C). Rhythmic expression was maintained in continuous light (L:L), although amplitude was reduced and period elongated for both genes. In D:D (Fig. 1C), bHLH1a and bHLH1b showed an increase in relative transcript abundance during the subjective day that was comparable to that observed in L:D, but the decrease during the subjective night was less pronounced (Fig. 1C and SI Appendix, Fig. S1). Because iron metabolism is diurnally regulated in P. tricornutum and iron starvation strongly affects rhythmic gene expression (32), we further examined the bHLH1a and bHLH1b expression profiles in cells grown in iron replete and deplete conditions by using public transcriptome datasets (32). In this analysis, bHLH1a and bHLH1b showed similar expression patterns in both control and iron starvation conditions, with peak expression at ZT9 in cells grown in 12L:12D photocycles (SI Appendix, Fig. S3). Together, these results demonstrate the robust control of bHLH1a and bHLH1b expression timing. Of these proteins, we decided to further investigate the possible involvement of bHLH1a in the regulation of P. tricornutum rhythms.
Cellular Localization of bHLH1a Protein.
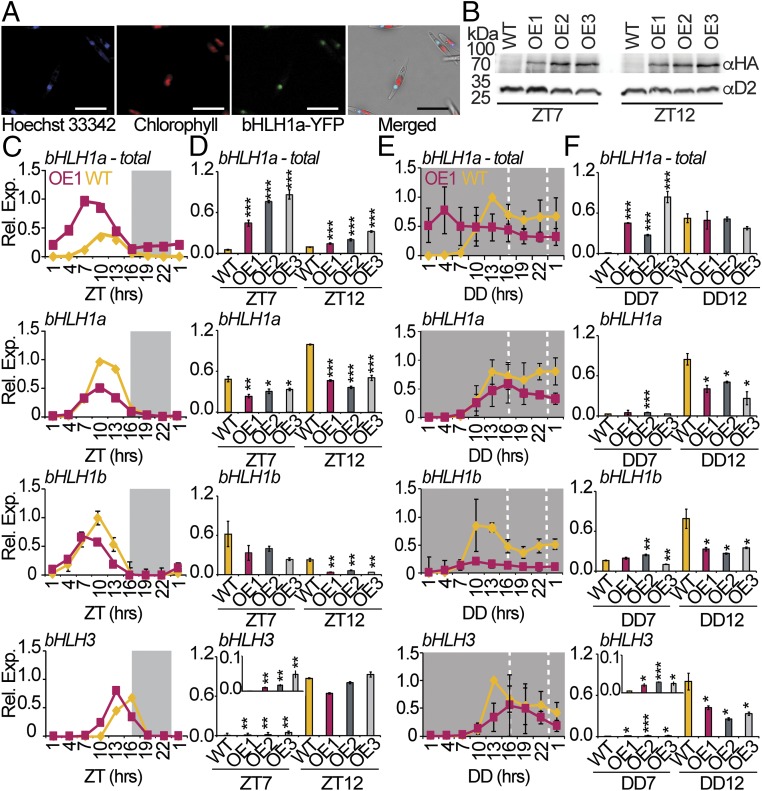
To examine bHLH1a cellular localization, we generated transgenic lines expressing bHLH1a fused to a YFP-tag at the C-terminal under the regulation of the Lhcf2p promoter (33). Confocal laser scanning microscopy of bHLH1a-YFP–expressing cells showed colocalized signals of YFP with the Hoechst 33342 (DNA tracker) stain in the nucleus (Fig. 2A). This confirmed the predicted in silico localization (SI Appendix) of bHLH1a in the nucleus of P. tricornutum, further supporting its possible role as a transcriptional regulator.
Fig. 2.
bHLH1a localizes in the nucleus, and its overexpression alters rhythmic diel gene expression. (A) Confocal fluorescence microscopy of P. tricornutum cells expressing the bHLH1a-YFP protein under the control of the Lhcf2p promoter. (Scale bar: 15 µm.) (B) Immunoblot analysis of bHLH1a-HA protein at ZT7 and ZT12 in 16L:8D entrained WT and OE lines, using the anti-HA and anti-D2 (loading control) antibodies. (C) qRT-PCR analysis of bHLH1a, bHLH1b, and bHLH3 in WT and OE1 cultures sampled in L:D every 3 h over 24 h (mean ± SEM, n = 2). (D) qRT-PCR analysis of the same genes at ZT7 and ZT12 in WT and OE1, OE2, and OE3 lines (mean ± SEM, n = 3). Inset in the bHLH3 graph magnifies the ZT7 time point. (E) nCounter analysis of the bHLH1a, bHLH1b, and bHLH3 over 24 h of continuous dark (D:D) in WT and OE1 lines (mean ± SEM, n = 3). (F) qRT-PCR analysis of the same genes at DD7 and DD12 in WT and OE1, OE2, and OE3 lines (mean ± SEM, n = 3). bHLH1a-total includes endogenous bHLH1a and bHLH1a-HA transgene transcripts; for all of the other genes, we refer to endogenous transcripts. The expression value of each gene is relative to its maximum expression. *P < 0.05, **P < 0.01, ***P < 0.001, t test.
bHLH1a Regulates Pace of Diel Gene Expression.
To explore possible functions of bHLH1a in diel rhythmicity, we generated cell lines expressing HA-tagged bHLH1a under the regulation of the Lhcf2p promoter, which drives gene expression 3 h after the light onset (33). Immunoblot analysis performed at ZT7 and ZT12 in a 16L:8D regime revealed three independent bHLH1a-HA overexpressing lines, hereafter named OE1, OE2, and OE3 (Fig. 2B). The ectopic protein was detected at ZT7 and ZT12 in these lines. Analysis of bHLH1a total transcripts, both endogenous (bHLH1a) and transgenic (bHLH1a-HA) mRNAs, also showed earlier expression in the OE lines compared with the wild type (WT) (Fig. 2 C and D).
To investigate the effect of bHLH1a deregulation on diel gene expression, WT and OE lines were grown in 16L:8D photocycles and sampled for RNA analysis. The expression of selected genes exhibiting a diurnal rhythmic expression pattern (Fig. 1) was analyzed in WT and OE1 sampled every 3 h over 24 h (Fig. 2C and SI Appendix, Fig. S4A). As described above, total bHLH1a transcript levels were higher in the OE line compared with the WT (Fig. 2 C and D), but a decrease in endogenous bHLH1a transcripts was also observed (Fig. 2 C and D). A similar pattern was also reported for the bHLH1b gene while bHLH3 showed earlier expression in OE1 compared with the WT (Fig. 2 C and D). In addition to the bHLH genes, altered expression patterns in OE1 were also found for other putative TF genes and cell cycle regulators (SI Appendix, Fig. S4A). To test a possible function of bHLH1a in the maintenance of diurnal rhythms in the absence of light inputs, the expression of genes found to be rhythmic in the WT in D:D (SI Appendix, Fig. S1) was analyzed also in OE1. Ten of 11 genes displayed reduced amplitudes and shifts in the phase of expression in OE1, compared with WT (Fig. 2E and SI Appendix, Fig. S4B). Analysis of bHLH1a, bHLH1b, and bHLH3 in OE2 and OE3 at ZT7 and ZT12 in L:D (Fig. 2D) and at DD7 and DD12 in D:D (Fig. 2F) suggested a similar deregulation of gene expression in independent transgenic lines.
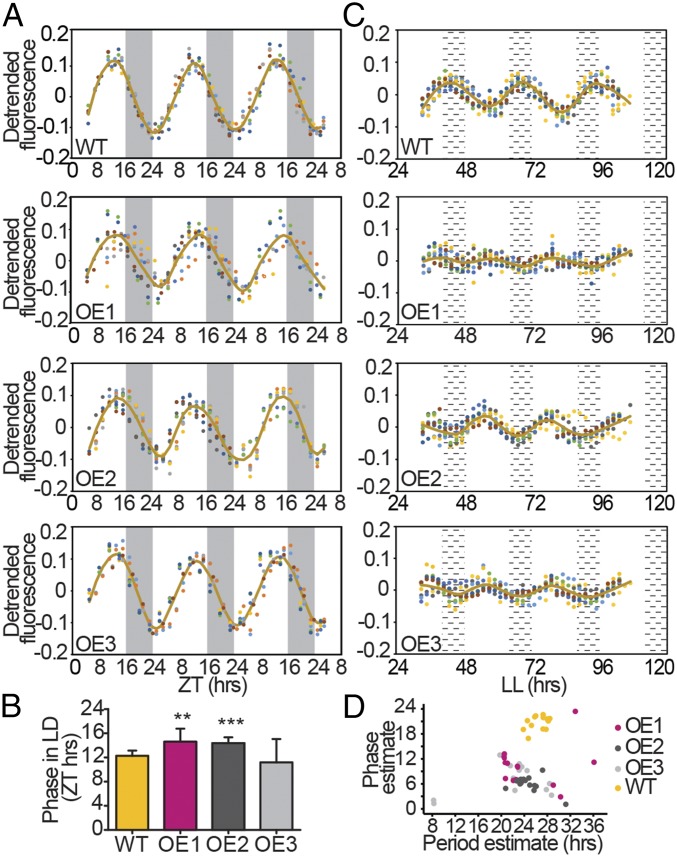
The Ectopic Overexpression of bHLH1a Results in Altered Circadian Rhythms.
We next questioned if, beside gene expression, other physiological rhythms were regulated by bHLH1a. With this aim, daily cellular fluorescence was analyzed using the flow cytometer channel FL3-A that estimates chlorophyll a cellular content (8, 29). Cellular fluorescence displayed strong oscillations in 16L:8D grown cultures with a periodicity of ∼24 h (Fig. 3A and SI Appendix, Table S2). Cell fluorescence in WT cultures increased during daytime to phase around ZT12 (Fig. 3A) and then started to decrease before night onset. Fluorescence progressively declined during the night period, reaching a trough in the early morning at ZT0 (Fig. 3A). In synchronized cells, the increase of FL3-A fluorescence is concomitant with the increase in the proportion of G2/M cells, and a decrease in the FL3-A relates to an increase in cell concentration (29) (SI Appendix, Fig. S5), likely reflecting chloroplast partitioning to daughter cells during cell division. Despite maintaining period rhythmicity in the cellular fluorescence dynamics, all three OE lines displayed less synchronized phenotypes, resulting in higher variability of waveforms among replicates compared with WT (Fig. 3A). Phase-time estimations using the FFT-NLLS method identified significant shifts of ∼2 h in the maximum fluorescence timing in OE1 and OE2 compared with the WT (Fig. 3 A and B and SI Appendix, Fig. S6 and Table S2). We then questioned if these oscillations were maintained in continuous blue light (L:L) as observed in other microalgae (24, 34). First, we found that under these conditions bHLH1a gene expression showed a stronger rhythmic pattern on the second subjective day (SI Appendix, Fig. S7), compared with the white L:L condition (Fig. 1C). WT cells also showed persistent fluorescence oscillations from LL33, with a period of ∼27 h. These rhythms lasted to at least the fifth day of continuous light (Fig. 3C), supporting the hypothesis that they are self-sustained by an endogenous clock. Patterns of cell fluorescence oscillations in L:L were strongly altered in all three OE lines compared with WT (Fig. 3 C and D and SI Appendix, Figs. S8 and S9) although similar growth rates were observed (SI Appendix, Fig. S10). Analysis via the Fast Fourier Transform Nonlinear Least Squares (FFT-NLLS) method showed that all lines maintained residual rhythmicity, but with a strong phase shift compared with the WT (Fig. 3 C and D) and a reduced amplitude, with the OE2 being the less affected for the latter one (Fig. 3D and SI Appendix, Figs. S8 and S9 and Table S3). Additionally, OE lines also presented shorter periods than the WT, although OE1 lines lacked a significant difference most likely due to the variability introduced by the replicates with high relative amplitude error (SI Appendix, Supplementary Methods, Fig. S8 C and D, and Table S3). Fluorescence oscillations were similar between WT and an independent transgenic line expressing only the antibiotic resistance cassette (SI Appendix, Fig. S11), excluding a generic alteration of rhythmicity due to transgenesis. Based on these results, we named bHLH1a “RITMO1” (Italian word for “rhythm”) after its role as regulator of diatom diel rhythms.
Fig. 3.
bHLH1a overexpression alters circadian rhythms of cellular fluorescence. (A) Diurnal oscillation of chlorophyll fluorescence (FL3-A parameter) in WT and OE lines entrained under 16L:8D over 3 d (n ≥ 8). (B) Phase-time calculation of the FL3-A value in WT and OE lines (mean ± SD, n ≥ 8, **P < 0.01, ***P < 0.001, t test). (C) Circadian oscillation of chlorophyll fluorescence in representative WT and OE lines under continuous blue light (L:L) over 4 subjective days (n = 8). (D) Plot of phase against period estimates in L:L (n = 15). Dots of different color indicate independent replicate cultures. White and gray regions represent light and dark periods; black dashed regions represent subjective nights. Brown lines in plots represent the fitted curves (lowess fit) of the average FL3-A.
RITMO1-Like Proteins Are Widely Represented in the Genome of Marine Algae.
The bHLH-PAS proteins were thought to be restricted to the animal (Opisthokonta) lineage (35) until various sequencing projects revealed bHLH-PAS family members in microalgae (diatoms and Nannochloropsis) (36). Diatom proteins show peculiar features including a single predicted PAS domain, whereas animal bHLH-PAS proteins have two and an N-terminal extension that is absent in their animal counterparts (Fig. 4A). Through searching transcriptomic and genomic databases from Archeaplastida, Cryptophyta, Stramenopila, Alveolata, and basal Opisthokonta organisms (SI Appendix, Table S4), we discovered about 90 bHLH-PAS proteins. With one exception, the identified proteins showed a single predicted PAS domain, short C-terminal extensions, and N-terminal regions of variable length, similar to the predicted structure of diatom bHLH-PAS (Fig. 4A). Notably, we identified the first Archaeplastida bHLH-PAS that possesses two PAS domains like the animal proteins in Galdieria sulphuraria (Rhodophyta).
Fig. 4.
bHLH-PAS protein family structure and phylogeny. (A) bHLH-PAS protein domain schematic architecture in eukaryotes. Dotted line indicates possible absence of the second PAS in some Opisthokonta species; gray patterns represent the variations in N-terminal and C-terminal length in different groups. (B) Maximum likelihood (ML) phylogenetic tree of the bHLH-PAS family (outgroup: the Opisthokonta clade) midpoint rooted. Numbers refer to bootstrap values of the basal nodes using ML (RAxML, 1,000 bootstraps) and Bayesian inference (MrBayes). Symbols indicate the position of P. tricornutum RITMO1 and bHLH1b (arrows) and T. pseudonana bHLH1 (square).
All of the identified sequences, including selected bHLH-PAS from Opisthokonta lineages, were used to perform a detailed phylogenetic analysis of the protein family using the bHLH and PAS domains. This analysis revealed that the majority of microalgal bHLH-PAS proteins fall into three separate clades, the first containing 9 TFs from diatoms and Ectocarpus siliculosus, the second comprising RITMO1 together with 35 proteins from diatoms and Alveolata (Dinoflagellata), and the third comprising 41 proteins from Alveolata (Ciliophora and Dinoflagellata) and diatoms, including bHLH1b (Fig. 4B). Interestingly, domain organization and branching positions of proteins from basal Opisthokonta (Monosiga brevicollis) and secondary endosymbiotic microalgae [Guillardia theta (Cryptophyta) and Nannochloropsis species (Stramenopila)] (Fig. 4B) support a possible common origin for this TF family from a heterotrophic ancestor (37) featuring single bHLH and PAS domains. However, the basal position and domain organization of the G. sulphuraria bHLH-PAS protein hint at a more complex scenario possibly involving endosymbiotic (horizontal) gene transfer, gene duplication/loss, and convergent evolution in the diversification of this family.
Discussion
This study shows that diatoms integrate light signals from the environment as well as from an endogenous circadian clock to tightly regulate diel cellular activities. It also unveils the P. tricornutum RITMO1 protein of the bHLH-PAS transcription factor family as the first regulator of circadian rhythms to be found in these algae. RITMO1 displays robust diel expression patterns, which are adjusted in a photoperiod-dependent manner and are unaffected by iron deficiency. As for other circadian clock-regulated genes in algae (24), the rhythmic expression of RITMO1 persists in cells exposed to continuous blue light, a dominant waveband in the ocean (27). However, RITMO1 transcription shows reduced oscillation amplitude in constant darkness, likely due to the lack of critical light time setting signals and alteration of metabolism due to the fact that photosynthesis is not dispensable in P. tricornutum. Thus, the RITMO1 expression timing is tightly regulated and likely subjected to multiple levels of control from different input regulators, conceivably blue light photoreceptors (27) and metabolism, as already observed for other clock genes in plants and animals (3, 4).
RITMO1 is involved in the transcriptional regulation of diel rhythms, as supported by consistent deregulation of transcript diurnal oscillations for several genes in RITMO1 OE lines in L:D, this pattern being accentuated in D:D. These results suggest, on one hand, that multiple light-driven processes participate in the regulation of daily gene expression, partially masking RITMO1’s contribution to this process in cyclic environments and, on the other hand, reveal a key role for RITMO1 in the maintenance of rhythms in the absence of light–dark inputs. Furthermore, the observed down-regulation of the endogenous RITMO1 transcription in OE lines compared with the WT may reflect negative feedback regulation of RITMO1 controlling its own transcription. This would be compatible with this gene being part of a transcriptional feedback loop operating over the daily cycle (2). We also report robust oscillations of diatom cellular fluorescence in L:D cycles, as observed for gene expression. These rhythms persisted for at least five subjective days in L:L, supporting the existence of a circadian clock regulating diatom physiology. The oscillations in L:L have a period slightly longer than 24 h under free running conditions, a common feature in circadian clock-regulated processes (1, 2). The deregulation of RITMO1 expression in transgenic lines shifts the phase of fluorescence rhythms in L:D compared with the WT. In constant blue light (L:L), all of the independent OE lines showed altered rhythmicity if compared with the WT and a transgenic control line, with a strongly altered phase, reduced amplitude, and the period also being remarkably perturbed. Although multiple factors may account for the slight phenotypic differences observed between OE lines (e.g., chromosomal position effects on the transgenes, deregulation of other genetic or epigenetic processes due to the overexpression), the consistent deregulation of rhythmicity is coherent with a role of RITMO1 in the regulation of circadian rhythms. Because all of the OE lines show a normal growth rate compared with WT, the observed altered rhythmicity is likely due to a defect in retrieving synchrony in chloroplast ontogeny and cell cycle progression, which are closely coupled in diatoms (20).
Together, our results support the hypothesis that RITMO1 is one component of the still-uncharacterized endogenous circadian clock in diatoms. RITMO1 contains bHLH and PAS protein domains that are also present in the CLOCK and BMAL proteins, components of the mammalian central circadian oscillator (2, 5, 38). Interestingly, previous studies have shown that the P. tricornutum animal-like blue light sensor Cpf1 can repress the transcriptional activity of these proteins in a heterologous mammalian cell system (25), suggesting at least partial conservation in the regulatory program generating rhythmicity in animals and diatoms. We have reported several other putative TFs showing altered diel expression patterns in RITMO1 OE cells (i.e., bHLH1b, bHLH3, bZIP7, HSF1g, HSF4.7b), which represent direct or indirect targets of RITMO1 activity and possible additional components of the network generating circadian rhythms. RITMO1 might also act downstream of signal transduction cascades activated by the diatom photoreceptors (27). Therefore, RITMO1 now represents a key molecular entry point for the identification of other diatom timekeeper components and the input–output pathways interacting with the clock.
Biological rhythms are still poorly understood at the molecular level in many phyla of marine algae such as Stramenopila. Our phylogenetic analysis revealed a wide distribution of RITMO1-like bHLH-PAS proteins in diatoms, as well as in other algae. The similarities in the timing of expression between RITMO1 and its homolog in the diatom T. pseudonana (TpHLH1) (17) suggest that these proteins may play a similar role in the regulation of cellular rhythmicity. Regulators such as RITMO1 may have played a critical role for diatom prominence in marine ecosystems by synchronizing cellular activities in optimal temporal programs and maximizing the diatoms’ ability to anticipate and adapt to cyclic environmental variations.
bHLH-PAS RITMO-like proteins might have independently acquired a function in rhythm regulation by convergent evolution (6). However, the existence of this function in an ancient heterotrophic marine ancestor that subsequently acquired plastids via endosymbiosis events (12) and before colonization of land cannot be excluded. Thus, further characterization of RITMO1-like proteins is expected to provide insights into the evolution of biological rhythms and their significance for life in the marine environment.
Methods
Culture Conditions.
WT P. tricornutum (Pt1 8.6) cells and transgenic lines were grown in f/2 Guillard media, as described in SI Appendix.
Gene Expression Analyses.
To select genes with rhythmic expression, we used data from (18, 39). Total RNA was extracted as described in ref. 31 and analyzed by qRT-PCR and nCounter analysis as described in SI Appendix. Proteins were analyzed as previously described (25).
Generation of the bHLH1a Transgenic Lines.
Diatom cell lines overexpressing bHLH1a-HA and bHLH1a-YFP were obtained by cotransformation of the Nourseothricin resistance plasmid (pNAT) together with the pDEST-C-HA-bHLH1a and the pDEST-C-YFP-bHLH1a plasmids, respectively. Details of vectors and transformation are provided in SI Appendix.
Microscopic Analysis.
Images were produced with the confocal laser scanning microscope Leica SP8× as described in SI Appendix.
Data Mining, Protein Sequence, and Phylogenetic Analysis.
Detailed information about data mining, protein sequence, and phylogenetic analysis is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank M. Jaubert, L. De Veylder, R. Dorrell, and P. Oliveri for critical suggestions; D. Petroutsos and G. Finazzi for support in monitoring cell physiology; G. Benvenuto for assistance in microscopy; and the Institut de Biologie Paris-Seine imaging core facility for help with flow cytometry. This work was funded by Human Frontier Science Program Grant RGY0082/2010; the Gordon and Betty Moore Foundation Grant GBMF 4966; the European Union H2020 EMBRIC Grant G.A. 654008; and the Fondation Bettencourt Schueller (A.F.). M.J.J.H. is currently an employee of the European Research Council Executive Agency. The views expressed are purely those of the writer and may not in any circumstances be regarded as stating an official position of the European Commission.
Footnotes
This article is a PNAS Direct Submission.
Data deposition: Code set information and raw nCounter data are available from the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo/ (Series GSE112268).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819660116/-/DCSupplemental.
References
- 1.Pittendrigh C. S., Temporal organization: Reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55, 16–54 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Bell-Pedersen D., et al. , Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenham K., McClung C. R., Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16, 598–610 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Reinke H., Asher G., Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 20, 227–241 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Dunlap J. C., Molecular bases for circadian clocks. Cell 96, 271–290 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Rosbash M., The implications of multiple circadian clock origins. PLoS Biol. 7, e62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moulager M., et al. , Light-dependent regulation of cell division in Ostreococcus: Evidence for a major transcriptional input. Plant Physiol. 144, 1360–1369 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ragni M., D’Alcala M. R., Circadian variability in the photobiology of Phaeodactylum tricornutum: Pigment content. J. Plankton Res. 29, 141–156 (2007). [Google Scholar]
- 9.Noordally Z. B., Millar A. J., Clocks in algae. Biochemistry 54, 171–183 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Poliner E., Cummings C., Newton L., Farré E. M., Identification of circadian rhythms in Nannochloropsis species using bioluminescence reporter lines. Plant J., 10.1111/tpj.14314 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Malviya S., et al. , Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. U.S.A. 113, E1516–E1525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moustafa A., et al. , Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324, 1724–1726 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Bowler C., et al. , The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Flori S., et al. , Plastid thylakoid architecture optimizes photosynthesis in diatoms. Nat. Commun. 8, 15885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen A. E., et al. , Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473, 203–207 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Bailleul B., et al. , Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Ashworth J., et al. , Genome-wide diel growth state transitions in the diatom Thalassiosira pseudonana. Proc. Natl. Acad. Sci. U.S.A. 110, 7518–7523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauton M. S., Winge P., Brembu T., Vadstein O., Bones A. M., Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol. 161, 1034–1048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huysman M. J., et al. , AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). Plant Cell 25, 215–228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huysman M. J., et al. , Genome-wide analysis of the diatom cell cycle unveils a novel type of cyclins involved in environmental signaling. Genome Biol. 11, R17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volpert A., Graff van Creveld S., Rosenwasser S., Vardi A., Diurnal fluctuations in chloroplast GSH redox state regulate susceptibility to oxidative stress and cell fate in a bloom-forming diatom. J. Phycol. 54, 329–341 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Zones J. M., Blaby I. K., Merchant S. S., Umen J. G., High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27, 2743–2769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poliner E., et al. , Transcriptional coordination of physiological responses in Nannochloropsis oceanica CCMP1779 under light/dark cycles. Plant J. 83, 1097–1113 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Corellou F., et al. , Clocks in the green lineage: Comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell 21, 3436–3449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coesel S., et al. , Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 10, 655–661 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortunato A. E., Annunziata R., Jaubert M., Bouly J. P., Falciatore A., Dealing with light: The widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J. Plant Physiol. 172, 42–54 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Jaubert M., Bouly J. P., Ribera d’Alcalà M., Falciatore A., Light sensing and responses in marine microalgae. Curr. Opin. Plant Biol. 37, 70–77 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Rayko E., Maumus F., Maheswari U., Jabbari K., Bowler C., Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 188, 52–66 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Hunsperger H. M., Ford C. J., Miller J. S., Cattolico R. A., Differential regulation of duplicate light-dependent protochlorophyllide oxidoreductases in the diatom Phaeodactylum tricornutum. PLoS One 11, e0158614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee A., et al. , Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes. Nucleic Acids Res. 44, 5957–5970 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortunato A. E., et al. , Diatom phytochromes reveal the existence of far-red-light-based sensing in the Ocean. Plant Cell 28, 616–628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith S. R., et al. , Transcriptional orchestration of the global cellular response of a model pennate diatom to diel light cycling under iron limitation. PLoS Genet. 12, e1006490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo M. T., Annunziata R., Sanges R., Ferrante M. I., Falciatore A., The upstream regulatory sequence of the light harvesting complex Lhcf2 gene of the marine diatom Phaeodactylum tricornutum enhances transcription in an orientation- and distance-independent fashion. Mar. Genomics 24, 69–79 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Braun R., Farré E. M., Schurr U., Matsubara S., Effects of light and circadian clock on growth and chlorophyll accumulation of Nannochloropsis gaditana. J. Phycol. 50, 515–525 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Yan J., Ma Z., Xu X., Guo A. Y., Evolution, functional divergence and conserved exon-intron structure of bHLH/PAS gene family. Mol. Genet. Genomics 289, 25–36 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Thiriet-Rupert S., et al. , Transcription factors in microalgae: Genome-wide prediction and comparative analysis. BMC Genomics 17, 282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brodie J., et al. , The algal revolution. Trends Plant Sci. 22, 726–738 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Gekakis N., et al. , Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Annunziata R., et al. , Data from “bHLH-PAS protein RITMO1 regulates diel biological rhythms in the marine diatom Phaeodactylum tricornutum.” Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112268. Deposited 23 March 2018. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.