Abstract
DNA double‐strand break repair by homologous recombination employs long‐range resection of the 5′ DNA ends at the break points. In Saccharomyces cerevisiae, this process can be performed by the RecQ helicase Sgs1 and the helicase–nuclease Dna2. Though functional interplay between them has been shown, it remains unclear whether and how these proteins cooperate on the molecular level. Here, we resolved the dynamics of DNA unwinding by Sgs1 at the single‐molecule level and investigated Sgs1 regulation by Dna2, the single‐stranded DNA‐binding protein RPA, and the Top3‐Rmi1 complex. We found that Dna2 modulates the velocity of Sgs1, indicating that during end resection both proteins form a functional complex and couple their activities. Sgs1 drives DNA unwinding and feeds single‐stranded DNA to Dna2 for degradation. RPA was found to regulate the processivity and the affinity of Sgs1 to the DNA fork, while Top3‐Rmi1 modulated the velocity of Sgs1. We hypothesize that the differential regulation of Sgs1 activity by its protein partners is important to support diverse cellular functions of Sgs1 during the maintenance of genome stability.
Keywords: DNA repair, Dna2, homologous recombination, RecQ helicases, single molecule
Subject Categories: DNA Replication, Repair & Recombination
Introduction
The genome of eukaryotic cells is constantly damaged by environmental factors, by‐products of the cellular metabolism, and transactions of the DNA metabolism. Damages appear in a variety of forms, such as base lesions, cross‐links between DNA strands or between DNA and proteins, and DNA single‐ and double‐strand breaks (DSBs) (Mehta & Haber, 2014). To avoid genome instability (Symington, 2014), cells use a number of intricate mechanisms to repair DNA lesions. DSBs are usually repaired by either of two main mechanisms—non‐homologous end joining (NHEJ) and homologous recombination (HR) (Mao et al, 2008; Brandsma & Gent, 2012; Aparicio et al, 2014).
HR in vegetative cells mostly uses genetic information stored in the sister chromatids in order to allow largely error‐free DSB repair (Mehta & Haber, 2014). This process is initiated by the resection of the 5′ DNA end at the break point, such that a 3′ overhang is created, which is immediately coated by the single‐stranded DNA (ssDNA)‐binding protein, replication protein A (RPA). In Saccharomyces cerevisiae (budding yeast), the long‐range DNA end resection is driven either by the exonuclease Exo1 or by the helicase Sgs1 together with the helicase–nuclease Dna2, which function in a synergistic manner (Cejka, 2015). In human cells, this conserved pathway is catalyzed by the Sgs1 homologs BLM or WRN together with human DNA2 (Nimonkar et al, 2011; Sturzenegger et al, 2014; Thangavel et al, 2015; Daley et al, 2017). Sgs1 is a processive 3′–5′ helicase of the RecQ family (Cejka & Kowalczykowski, 2010; Sarlós et al, 2012). In contrast, Dna2 possesses a highly processive and strictly unidirectional 5′–3′ motor activity (Levikova et al, 2013; Pinto et al, 2016), which likely functions as a ssDNA translocase in resection to facilitate the degradation of DNA unwound by Sgs1 (Levikova et al, 2017; Miller et al, 2017). DNA degradation during 5′ DNA end resection is accomplished by the barrel‐shaped nuclease domain of Dna2 containing a central tunnel that encircles the ssDNA strand. The nuclease domain travels along DNA ahead of the helicase motor (Zhou et al, 2015). The action of the Sgs1 and Dna2 motors with opposite polarities provides an intriguing similarity to the RecBCD complex that powers DNA end resection in E. coli. RecBCD uses the anti‐parallel helicase activities of the RecD and RecB subunits for DNA unwinding and the nuclease activity of RecB for DNA degradation (Spies et al, 2007). When reconstituting the DNA end resection reactions with the yeast proteins in vitro, a synergistic activity of Sgs1 and Dna2 was observed (Cejka et al, 2010a; Niu et al, 2010). However, the underlying mechanism of this stimulation remained undefined. Both proteins fulfill a number of additional cellular functions, which suggests a much more flexible and dynamic situation compared to the stable RecBCD complex carrying out a single functionality. For example, Sgs1 is also involved later in the HR pathway, including the dissolution of double Holliday junctions leading to non‐crossovers as well as in the regulation of aberrant HR (Oh et al, 2007; Daley et al, 2014b). Dna2 is engaged in Okazaki fragment maturation (Bae et al, 2001a), in processing of replication intermediates upon stress (Hu et al, 2012; Thangavel et al, 2015; Ölmezer et al, 2016), and in checkpoint signaling (Kumar & Burgers, 2013).
Previous biochemical studies have revealed that Sgs1, RPA, and Dna2 represent the minimal group of proteins that is able to reconstitute DNA end resection (Cejka et al, 2010a; Niu et al, 2010). On the protein level, Sgs1 and RPA are known to interact via the large Rfa1 subunit that binds to the N‐terminal acidic region of Sgs1, which is located next to the helicase domain (Hegnauer et al, 2012). DNA unwinding by Sgs1 does not require RPA but was found to be stimulated by this protein (Cejka et al, 2010a). In contrast, cognate RPA is essential for both the nuclease and the motor functions of Dna2 (Bae et al, 2001b, 2003). In mice, both proteins interact through the N‐terminal domain of the Rfa1 subunit and the alpha1 and OB folds of the Dna2 N terminus (Zhou et al, 2015). RPA prevents 3′ end degradation by Dna2 and instead promotes 5′ end degradation, enforcing thus the correct polarity of DNA end resection (Cejka et al, 2010a; Niu et al, 2010).
Although functional synergies and physical protein–protein interactions have been identified, it remains unclear how Sgs1 and Dna2 cooperate at the molecular level, and which functional steps are affected by these interactions. In order to gain insight into these processes, we studied DNA unwinding by Sgs1 using magnetic tweezers. This technique allowed us to monitor the DNA processing by Sgs1 and its physiological interaction partners—including RPA, Dna2, and the Top3‐Rmi1 complex—in real time on the single‐molecule level. Our data reveal how the DNA unwinding activity of Sgs1 is differentially and dynamically modulated by its cognate partners. This indicates that Sgs1 and Dna2 form together with RPA a functional complex during the DNA end resection reaction. Overall, our study helps explain how Sgs1 activities can be fine‐tuned to achieve diverse cellular functions.
Results
DNA unwinding by Sgs1
To study DNA unwinding by Sgs1 and its interaction partners, we employed a magnetic tweezers assay (Levikova et al, 2013; Pinto et al, 2016). A 6.6 kbp dsDNA molecule was bound at one end to a magnetic bead and on the other end to the surface of the fluidic cell of the magnetic tweezers setup (Fig 1A). A short 38 nucleotide (nt) gap with a 40‐nt 5′ flap about 0.5 kbp away from the surface attachment supported the initiation of DNA unwinding by Sgs1 and Dna2. A pair of magnets above the fluidic cell was used to apply defined forces of 15–25 pN onto the magnetic bead and therefore to stretch the DNA. Video microscopy was used to track the bead position and thus to monitor changes in the DNA length.
Figure 1. DNA duplex processing by Sgs1.
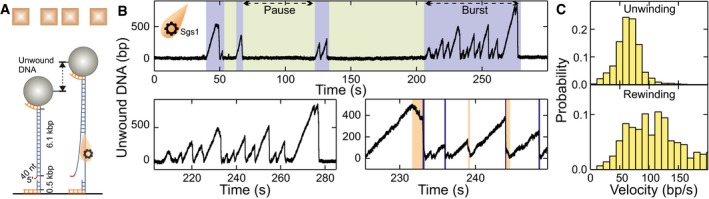
- Sketch of the employed magnetic tweezers assay and DNA processing by Sgs1.
- Observed dsDNA processing patterns of Sgs1, including an overview over consecutive bursts and pauses (upper panel) as well as detailed views into individual bursts containing multiple unwinding events (lower panels). A typical unwinding event of Sgs1 starts with slow, gradual unwinding of the dsDNA followed by DNA rehybridization that can be almost instant (76% of events, see blue sections in lower right panel) or contain slow rewinding sections (24% of events, see orange sections in lower right panel).
- Histograms of the observed unwinding and rewinding velocities for Sgs1. The mean unwinding rate was 65 ± 2 bp/s (N = 899). The mean rewinding rate was 115 ± 6 bp/s (N = 287). We attribute the large rewinding rate in the absence of RPA to an increased error in inferring the rewinding rate from the short rewinding sections.
When Sgs1 in the presence of ATP was added into the fluidic cell, DNA unwinding was observed as a gradual increase in the DNA length due to a larger extension of ssDNA compared to dsDNA at the applied forces (Smith et al, 1996; Fig 1B and Appendix Fig S1). Consistently, no DNA lengthening, i.e., no unwinding, was observed when omitting ATP or protein in the reaction (Appendix Fig S2). The unwinding rates followed a Gaussian‐like distribution with a mean of 65 ± 2 bp/s (Fig 1C, upper panel), indicating within error a unique unwinding rate for Sgs1. DNA unwinding was frequently terminated by an abrupt DNA length decrease (blue sections in Fig 1B, lower panel) that reflects rezipping, i.e., renaturation of the DNA duplex. Occasionally, the DNA rezipping contained short sections of a slow DNA length decrease (24% of the DNA closing events, see orange sections in Fig 1B, lower panel). Since the rate of these sections was approximately constant and was comparable to the magnitude of the unwinding rate (Fig 1C, lower panel), we attribute these sections to helicase‐driven DNA rewinding. We believe that in these cases, Sgs1 translocated on the opposite ssDNA strand away from the Y‐junction, and thus limited the rehybridization to the ssDNA translocation rate of the helicase. The single unwinding–rezipping events typically occurred in bursts comprising several individual events followed by long pauses (Fig 1B, upper panel). This indicated that a burst was likely initiated by the binding of a single Sgs1 unit (a molecule or a complex), which subsequently originated all events of the burst until the protein finally dissociated. Alternatively, more than one protein unit could bind to form an active unwinding complex. The observed behavior was similar to that seen for BLM (Wang et al, 2015), WRN (Lee et al, 2018), and the likely BLM homolog from Arabidopsis thaliana, AtRecQ2 (Klaue et al, 2013). For AtRecQ2, the transition between unwinding and rezipping most likely involves strand switching (Klaue et al, 2013). As a result, the enzyme is bound in a more loosely state since it lacks the DNA junction in its wake (Klaue et al, 2013). The lowered affinity of the helicase to ssDNA causes then predominantly fast rezipping, during which the enzyme is pushed by the rehybridizing junction. New DNA unwinding needs to be reinitiated by an additional strand switch. Due to the functional similarities between Sgs1, BLM, and AtRecQ2 (Oh et al, 2007; De Muyt et al, 2012; Klaue et al, 2013), we suggest that Sgs1 also undergoes cycles of strand switches during repetitive DNA unwinding–rezipping events, suggesting that this is a conserved characteristic of RecQ helicases.
DNA unwinding by Sgs1 in the presence of RPA
To systematically probe the influence of protein partners on the behavior of Sgs1, we first studied its DNA unwinding capacity in the presence of RPA (Fig 2A). Compared to Sgs1 alone, the unwinding–rezipping events appeared significantly altered when the reaction was supplemented with only 20 nM of the single‐stranded DNA‐binding protein (Fig 2B and C). No fast DNA rezipping events were observed; i.e., gradual DNA unwinding was exclusively followed by gradual DNA rewinding. Similar to Sgs1 alone, events occurred in a repetitive, burst‐like manner. The bursts were similarly separated by pauses (Fig 2B, upper panel), indicating that a single unwinding complex was likely driving the reaction.
Figure 2. DNA duplex processing by Sgs1 in the presence of RPA.
- Sketch of DNA processing by Sgs1 in the presence of RPA.
- Observed dsDNA processing pattern of Sgs1 in the presence of RPA including an overview over successive bursts and pauses (upper panel) and detailed views of single bursts (lower panels). Unwinding events are always followed by slow rewinding events (orange sections).
- Histograms of the observed unwinding and rewinding velocities for Sgs1 in the presence of RPA (brown bars). The mean unwinding rate was 51 ± 3 bp/s (N = 2,139). The mean rewinding rate was 66 ± 3 bp/s (N = 2,091). White bars show for comparison the distributions of unwinding and rewinding velocities for Sgs1 alone (taken from Fig 1C).
- Force dependence of the Sgs1 unwinding (black squares) (N = 2,139) and rewinding rates (black diamonds) (N = 2,091) in the presence of RPA (errors given as standard deviations). Red circles represent the force‐dependent DNA opening and closure rates measured for RPA alone (taken from Kemmerich et al, 2016a. A fit to these data is shown as a solid red line.).
The observed slow rewinding could result from either a limited velocity of Sgs1 translocating along the RPA‐coated ssDNA strand away from the junction or due to the limited rate at which RPA dissociated from the junction ends. To reveal whether active translocation or passive dissociation caused the slow rewinding, we carefully characterized the unwinding and rewinding velocities in a force range between 10 and 35 pN. No force dependence was detected for neither of the two processes (Fig 2D). The mean rates for unwinding and rewinding were 51 ± 3 bp/s and 66 ± 3 bp/s, i.e., rather similar (Fig 2C). This is in contrast to the expected rates at which RPA gets dissociated by a rezipping junction. Previous measurements found a strong exponential force dependence for such an RPA dissociation (see red curve in Fig 2D) (Kemmerich et al, 2016a). Therefore, DNA unwinding as well as rewinding is an active process that is driven by the Sgs1 helicase rather than the association and dissociation of RPA.
While the mean DNA unwinding velocity by Sgs1 was marginally reduced in the presence of RPA compared to in its absence, the rate distribution markedly differed. In particular, the presence of RPA caused a strong skew of the distribution with a maximum at 30–40 bp/s (Fig 2C) in contrast to the Gaussian‐distributed rates in the absence of RPA. A similarly skewed distribution was observed for DNA rewinding (Fig 2C). The shift of the rates toward lower values could be due to RPA acting as a roadblock in the way of Sgs1. However, this should only affect rewinding as RPA does not bind dsDNA. We therefore attribute the skewed rate distributions to a direct interaction between Sgs1 and RPA, in which the RPA‐bound form of Sgs1 has a slower and more variable unwinding/translocation rate. This conclusion is supported by the observation that a Sgs1 fragment (641–1,215) containing only the helicase core, but no specific RPA binding site (Bennett et al, 1998; Hegnauer et al, 2012), did not exhibit a shift of the unwinding velocities toward lower values in the presence of RPA (Appendix Fig S3). A similar skew was also observed at elevated RPA concentrations (Fig EV1A), suggesting that Sgs1 populations that differ in the number of bound RPA molecules are not the primary reason for the observed rate distributions. Overall, our conclusions are consistent with previous work, which demonstrated RPA binding region on Sgs1 (Hegnauer et al, 2012) that could modulate Sgs1 behavior.
Figure EV1. Skewness of the velocity distributions for unwinding and rewinding for the different protein combinations.
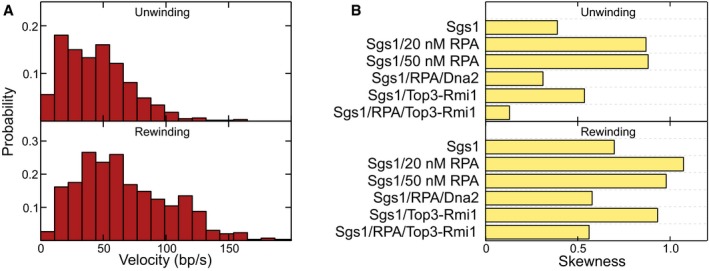
- Histograms of the observed unwinding and rewinding velocities for Sgs1 in the presence of 50 nM RPA. The mean unwinding rate was 46 ± 3 bp/s (N = 597). The mean rewinding rate was 68 ± 3 bp/s (N = 594).
- The skewness was calculated for the data shown in the velocity histograms in the main text.
DNA processing by the combined activity of Sgs1 and Dna2
Next, we set out to investigate the complete minimal DNA end resection reaction that included Sgs1, RPA, and Dna2 (Fig 3A). First, we tested DNA unwinding by Dna2 in the presence of RPA but in the absence of Sgs1. The Dna2 unwinding activity requires a 5′ ssDNA flap as present in our DNA substrate (Fig 1A). For wt Dna2, the nucleolytic degradation of such a flap effectively inhibits DNA unwinding by the Dna2 motor (Levikova et al, 2013) (Appendix Fig S4A). As expected, no DNA unwinding was detected with the helicase‐dead Dna2 K1080E (Appendix Fig S4B). In contrast, processive DNA unwinding was observed with the nuclease‐dead Dna2 E675A mutant (Fig 3B, inset and Appendix Fig S4C) (Levikova et al, 2013). Since the nuclease domain encircles the 5′‐terminated ssDNA strand (Zhou et al, 2015), dissociation or strand switching of Dna2 should be unlikely. In agreement with this model, the observed DNA unwinding by Dna2 was completely unidirectional (Levikova et al, 2013; Pinto et al, 2016); i.e., no DNA rewinding was observed. The unwinding rates by Dna2 were highly variable between the individual enzyme molecules ranging from 15 to 160 bp/s (Appendix Fig S4D) (Levikova et al, 2013).
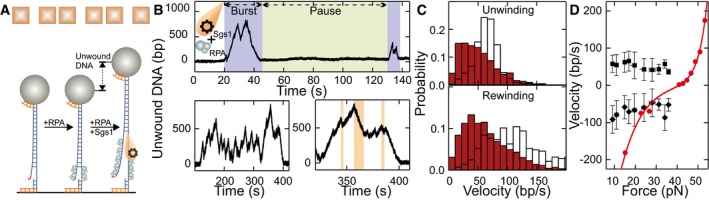
Figure 3. DNA duplex processing by the combined activity of Sgs1, Dna2, and RPA.
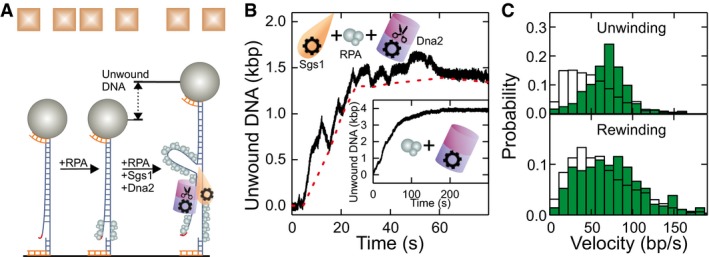
- Sketch of DNA processing by the Sgs1‐Dna2‐RPA complex.
- Typical dsDNA processing event. The dashed red line connects the minima at the end of the rewinding sections (see text for details). The inset shows a DNA unwinding event in the presence of nuclease‐dead Dna2 E675A and RPA.
- Histograms of the observed unwinding and rewinding rates (green bars) with mean values of 69 ± 3 bp/s (N = 365) and 74 ± 5 bp/s (N = 282), respectively. White bars show for comparison the unwinding and rewinding velocities of Sgs1 in the presence of RPA (taken from Fig 2C).
After assaying the activity of Dna2 alone, we studied the minimal reconstituted DNA end resection reaction in the presence of Sgs1, wt Dna2, and RPA in our magnetic tweezers setup. Notably, in contrast to wt Dna2 in the absence of Sgs1, significant DNA unwinding over kpb distances was observed (Figs 3B and EV2A). The trajectories exhibited a combination of the unwinding patterns of both helicases when investigated individually. Particularly, the typical Sgs1 patterns of alternating sections of DNA unwinding and rewinding were detected. In contrast to reactions with Sgs1/RPA alone, we observed a gradual increase in a baseline DNA unwinding; i.e., Sgs1 was not capable to fully rewind the full‐length DNA anymore in the presence of Dna2 (Fig 3B). This indicated that Dna2 was progressively moving along the 5′ DNA end, which was limiting the translocation of Sgs1 along this strand when moving backward. The approximate translocation/unwinding/DNA degradation distance is then obtained by connecting the lower turning points of Sgs1 in the trajectories (see dashed lines in Figs 3B and EV2A).
Figure EV2. DNA duplex processing by the DNA resection complex containing Sgs1, RPA, and mutated Dna2.
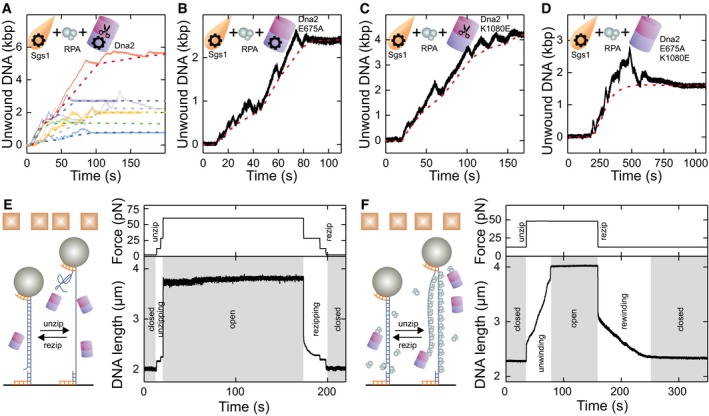
- Different dsDNA processing events in the presence of Sgs1, Dna2, and RPA. Solid lines show the measured DNA unwinding, while dashed lines reveal the estimated Dna2 position.
- dsDNA processing event in the presence of Sgs1, RPA, and the nuclease‐dead mutant Dna2 E675A.
- dsDNA processing event in the presence of Sgs1, RPA, and the helicase‐dead mutant Dna2 K1080E.
- dsDNA processing event in the presence of Sgs1, RPA, and the double helicase‐ and nuclease‐dead mutant Dna2 E675A K1080E.
- Mechanical unzipping of dsDNA in the presence of Dna2 E675A K1080E does not lead to inhibited DNA rezipping. dsDNA was mechanically unzipped in the presence of Dna2 E675A K1080E and kept open for > 100 s. Subsequently rezipping was induced by stepwise reduction of the applied force. The DNA length resumed to the original level, indicating rezipping over the entire molecule length.
- Mechanical unzipping of dsDNA in the presence of RPA and Dna2 E675A K1080E does not lead to inhibited DNA rezipping. dsDNA was mechanically unwound in the presence of RPA and Dna2 E675A K1080E. After keeping it in the unwound form for ˜70 s, the force was lowered to allow DNA rewinding against the bound RPA. The DNA length resumed to the original level, indicating that the rewinding occurred over the entire molecule length.
The limited DNA rewinding by Sgs1 may be due to Dna2 physically blocking DNA reannealing or due to DNA degradation. To clarify the underlying mechanism, we next examined the nuclease‐dead Dna2 E675A, the helicase‐dead Dna2 K1080E, and the double‐dead mutant (i.e., nuclease‐ and helicase‐dead Dna2) in combination with Sgs1 and RPA (Fig EV2B–D). Surprisingly, all three Dna2 variants promoted similar progressive DNA unwinding overlaid by short Sgs1 unwinding–rewinding cycles as observed with wt Dna2. For the nuclease‐dead variant, progressive ssDNA translocation by the helicase motor on the 5′ strand could block DNA rewinding by Sgs1. When the excess enzyme (wt or mutant) was removed from the flow cell and the complex was additionally challenged with 3 M NaCl after an unwinding reaction, the DNA remained unwound (i.e., no rezipping occurred, see Appendix Fig S5). This supports our conclusion that Dna2 can form an irreversible roadblock for Sgs1 translocating backward on the opposite, 5′‐terminated ssDNA strand. In this model, the nuclease domain of Dna2 encircles ssDNA, which prevents its dissociation even at high salt, in agreement with biochemical experiments and structural data (Kao et al, 2004; Zhou et al, 2015). For the helicase‐dead mutant, progressive degradation of the 5′ DNA end by its nuclease domain would prevent DNA rewinding by Sgs1 and thus also explain the progressive DNA unwinding. While the partial catalytic activity of the single mutants could explain a directional motion of Dna2 along the 5′ ssDNA end, progressive unwinding was surprisingly also seen with the catalytically dead double mutant (helicase and nuclease‐dead). One possibility to explain this observation is an external Dna2 binding to the unwound 5′ ssDNA at a random position, which would block rewinding. This mode of DNA binding would be distinct from the threading of Dna2 onto ssDNA as inferred from previous studies (Kao et al, 2004; Zhou et al, 2015). Nevertheless, to exclude this possibility, we mechanically unwound DNA in the presence of the Dna2 double‐dead mutant (Fig EV2E and F). When allowing DNA rezipping at lower forces, the DNA became always fully rewound; i.e., the Dna2 could not serve as a block to rewinding without Sgs1. We therefore hypothesize that even in the absence of catalytic activity, Dna2 can act as a ratchet‐like roadblock whose directionality and movement are facilitated by the physical interaction with Sgs1 (see Discussion) (Cejka et al, 2010a). The dispensability of the helicase and/or the nuclease activity for promoting progressive unwinding is corroborated by the observation that the unwinding processivity was not altered between helicase‐dead Dna2 in isolation and the different Dna2 variants in combination with Sgs1 (Appendix Fig S6).
When inspecting the rates of the Sgs1 unwinding and rewinding cycles in reactions containing wt Dna2, we found a Gaussian‐like distribution for both unwinding and rewinding, with significantly reduced skews compared to Sgs1 in the presence of RPA (Figs 3C and EV1B). This suggests that Sgs1 may directly interact with Dna2 both during unwinding and rewinding such that it can alleviate the inhibitory activity of RPA on the Sgs1 unwinding velocity. Since rewinding included only slow events, we hypothesize that RPA is still interacting with Sgs1, such that a ternary complex of the three different proteins is formed. Formation of a ternary complex is also supported by the observation that RPA is essential for the progressive unwinding (Appendix Fig S7). Overall, these data show that when both proteins act simultaneously during the end resection reaction, they unwind/translocate along DNA at different velocities. The modulation of the Sgs1 velocity by Dna2 supports the notion that the activities of the two enzymes are physically and functionally coupled.
DNA unwinding by Sgs1 together with Top3‐Rmi1
It is well established that Top3‐Rmi1 forms a complex with Sgs1 via interaction at the far end of the N terminus of Sgs1 (Fricke et al, 2001). Top3‐Rmi1 cooperates with Sgs1 in DNA end resection (Daley et al, 2014a; Fasching et al, 2015) as well as in the dissolution of double Holliday junctions (Fasching et al, 2015; Kaur et al, 2015). Generally, Top3‐Rmi1 is known to stimulate the rates of Sgs1 unwinding and DNA resection in biochemical assays (Cejka et al, 2010a); however, the underlying mechanism remained unclear. In order to study how the complex formation with Top3‐Rmi1 affects the activity of Sgs1, we conducted a set of experiments using our magnetic tweezers assay.
We first tested the activity of Top3‐Rmi1 alone on our flapped DNA substrate (Fig EV3A). Surprisingly, we observed stepwise increases in the DNA length of ~8 nm (Fig EV3B and C). This activity required the presence of a single‐stranded region on the DNA substrate. We attributed these shifts to the ssDNA cleavage activity of the type 1A topoisomerase Top3 (Dekker et al, 2002). Upon DNA cleavage, Top3 adopts an open form and remains covalently attached to 5′ end of cut strand (Bocquet et al, 2014; Mills et al, 2018). This can result in the formation of an elongated Top3‐DNA chain (Fig EV3A).
Figure EV3. Top3‐Rmi1 cleavage activity of ssDNA.
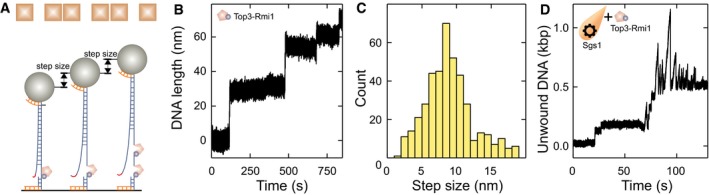
- Scheme illustrating the Top3‐Rmi1 ssDNA cleavage activity observed in the magnetic tweezers assay.
- Stepwise DNA length increase seen in the presence of Top3‐Rmi1.
- Histogram of the size of the stepwise length increase having a mean of 8.6 ± 0.2 nm (N = 433).
- dsDNA processing events observed for Sgs1 in the presence of Top3‐Rmi1. Stepwise DNA elongation due to ssDNA cleavage by Top3‐Rmi is observed together with dsDNA unwinding–rezipping bursts by Sgs1.
When testing Sgs1 and Top3‐Rmi1 together (Fig 4A), we observed the stepwise length increases due to the likely formation of the Top3‐DNA bridges, as well as the typical sawtooth‐like pattern from DNA unwinding by Sgs1 (Fig EV3D). Sgs1 unwinding could be clearly discriminated from Top3‐Rmi1 bridge formation and occurred in a burst‐like manner. Individual unwinding events comprised a gradual unwinding and typically an abrupt rezipping of all unwound DNA as seen for Sgs1 alone (Fig 4B). The presence of Top3‐Rmi1 reduced the fraction of partial rewinding events from 24 to 5%. The unwinding velocities were Gaussian‐like‐distributed and were more than 30% faster for the Sgs1 with Top3‐Rmi1 as compared to Sgs1 alone (86 ± 3 bp/s instead of 65 ± 2 bp/s, see Fig 4C).
Figure 4. DNA duplex processing by Sgs1 in the presence of Top3‐Rmi1.
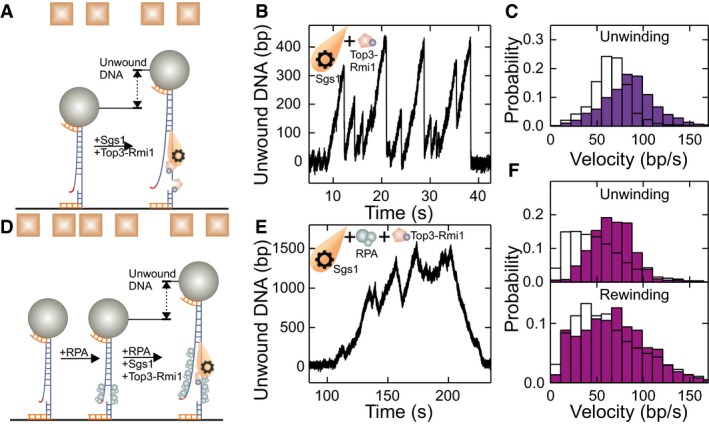
- Sketch of DNA processing by Sgs1 in the presence of Top3‐Rmi1.
- dsDNA processing events observed for Sgs1 in the presence of Top3‐Rmi1. Typical events consist of periods with slow gradual unwinding typically followed by instant DNA rezipping as seen also for Sgs1 alone.
- Histogram of the unwinding rate of Sgs1 in the presence of Top3‐Rmi1 (violet bars) with a mean rate of 86 ± 3 bp/s (N = 1,101). For comparison, the distribution of unwinding rates for Sgs1 alone is depicted with white bars (taken from Fig 1C).
- Sketch of DNA processing by Sgs1 in the presence of Top3‐Rmi1 and RPA.
- dsDNA processing by Sgs1 in the presence of Top3‐Rmi1 and RPA. In contrast to the absence of RPA, DNA closure is seen as a slow rewinding.
- Histograms of unwinding and rewinding rates of Sgs1 in the presence of Top3‐Rmi1 and RPA (purple bars). The mean rates for unwinding and rewinding are 66 ± 2 bp/s (N = 502) and 70 ± 4 bp/s (N = 369), respectively. For comparison, the velocity distributions for Sgs1 in the presence of RPA are shown as white bars (taken from Fig 2C).
When studying DNA unwinding by Sgs1, the Top3‐Rmi1 complex, and RPA (Fig 4D), no sudden length increases due to the formation of Top3‐DNA bridges were observed. This indicates that RPA protects ssDNA from the cleavage activity of Top3‐Rmi1. Gradual DNA unwinding was followed by gradual DNA rewinding as also seen for Sgs1 and RPA alone (Fig 4E). Taken together, the unwinding and rewinding velocities in the presence of Top3‐Rmi1 were higher for Sgs1 with RPA by 29% (66 ± 2 bp/s instead of 51 ± 3 bp/s) and 6% (70 ± 4 bp/s instead of 66 ± 3 bp/s), respectively. This suggests that Top3‐Rmi1 generally accelerates the motion of Sgs1 on DNA. Most importantly, the distribution of the unwinding and rewinding velocities was Gaussian‐like and significantly less skewed compared to Sgs1 and RPA alone (Figs 4F and EV1B). This is indicative of likely complex formation between Sgs1 and Top3‐Rmi1, which is similar to the reactions with Sgs1 and Dna2, for which an unskewed velocity distribution was also obtained.
Top3‐Rmi1 is known to stimulate the activity of Sgs1 in particular at elevated salt concentrations (Cejka et al, 2010a). To test this in our experiments, we challenged DNA unwinding by Sgs1 with “high‐salt” conditions (100 mM NaCl, 5 mM Mg2+), which is close to physiological ionic strength. Independently of the presence of Top3‐Rmi1, no DNA unwinding by Sgs1 was observed in the absence of RPA (Appendix Fig S8). When supplementing Sgs1 with RPA, similar unwinding–rewinding events as found for our standard reaction condition were observed (Fig EV4A). At 20 nM RPA, the velocity distribution of Sgs1 was however little skewed (Fig EV4B, gray bars). Complexes between RecQ helicases and RPA (Brosh et al, 2000; Machwe et al, 2011) are known to be rather stable. However, electrostatic interactions can become screened at elevated salt concentrations (Perez‐Jimenez et al, 2004), which can effectively increase the dissociation constant of the interaction. In agreement with this hypothesis, the Sgs1 unwinding velocity was found to be highly skewed in the presence of 200 nM RPA (Fig EV4B, dark blue bars). It also did not exhibit any significant force dependence (Fig EV4C). Addition of Top3‐Rmi1 to the reaction (Fig EV4D) reduced the skew in the velocity distribution, indicating likely a complex formation with Sgs1. Furthermore, the mean velocity in the presence of Top3‐Rmi1 was increased by 31% (77 ± 4 bp/s instead of 59 ± 2 bp/s). Given that RPA is an abundant protein, these results indicate that the observed velocity modulations by addition of RPA and Top3‐Rmi1 are relevant at physiological salt concentrations and that Top3‐Rmi1 serves as a general accelerator of the Sgs1 motor activity.
Figure EV4. DNA duplex processing by Sgs1 together with RPA and Top3‐Rmi1 at high‐salt conditions.
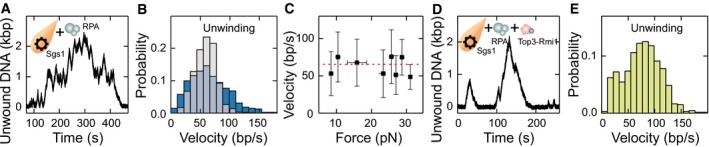
- dsDNA processing events observed for Sgs1 in the presence of RPA at high‐salt conditions.
- Histograms of unwinding velocity of Sgs1 in the presence of RPA at high‐salt conditions. Shown are the velocities in the presence of 20 nM RPA (gray) and 200 nM RPA (dark blue). Mean unwinding velocities are v = 59 ± 2 bp/s (N = 860) and v = 65 ± 3 bp/s (N = 1,095) for 20 and 200 nM RPA, respectively.
- Force dependence of the Sgs1 unwinding (black squares) (N = 1,095) velocities in the presence of 200 nM RPA. Red dashed lines represent mean values of the velocities, and error bars represent the standard deviation of the mean.
- dsDNA processing events observed for Sgs1 in the presence of RPA and Top3‐Rmi1 at high‐salt conditions.
- Histogram of the unwinding velocity of Sgs1 in the presence of RPA and Top3‐Rmi1. The mean unwinding velocity was v = 77 ± 4 bp/s (N = 678).
RPA promotes recruitment and processivity of DNA unwinding by Sgs1
So far, we analyzed the DNA unwinding and rewinding velocities by Sgs1. However, to an overall DNA unwinding, observed in vivo or in a test tube, other parameters of the whole reaction—including the rate of recruitment to the DNA template, the burst duration, and the processivity of unidirectional unwinding and rewinding—play an important role.
When analyzing the pause durations between individual bursts (see Figs 1B and 2B top panels), which are the inverse of the recruitment rate (Figs 5A and EV5A), we found longer pauses at high salt compared to standard reaction conditions. Overall, we did not see significant differences between the absence or the presence of the cofactors RPA and Top3‐Rmi1. Thus, at standard reaction conditions neither RPA nor Top3‐Rmi1 seem to contribute to the recruitment of Sgs1. At high‐salt conditions, RPA was essential for activity (Appendix Fig S9), i.e., recruitment, while Top3‐Rmi1 had only little influence. The influence of cofactors was different when analyzing the mean duration of the full unwinding bursts comprising many individual unwinding events (Figs 1B and 2B top panel). In the absence of RPA, burst durations were only 32 s on average but could exceed 200 s in the presence of RPA (Figs 5A and EV5A). Top3‐Rmi1 had little influence on the burst duration. Thus, RPA is a major determinant for the affinity of Sgs1 to the unwinding fork in agreement with the formation of a Sgs1‐RPA complex (Cejka et al, 2010a; Hegnauer et al, 2012; Kennedy et al, 2013).
Figure 5. Directionality, processivity and initiation of DNA processing events for the different protein combinations.
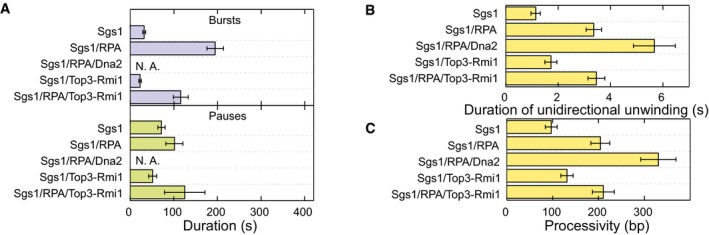
- Mean duration of bursts (violet bars) (N > 40) and pauses (green bars) (N > 40) for the different enzyme combinations. Error bars represent standard error of the mean.
- Mean duration of a unidirectional unwinding event (N > 350). Error bars represent standard error of the mean.
- Mean processivity of unidirectional DNA unwinding (N > 350). Error bars represent standard error of the mean.
Figure EV5. Directionality, processivity, and initiation of DNA processing events for the different protein combinations at high‐salt conditions.
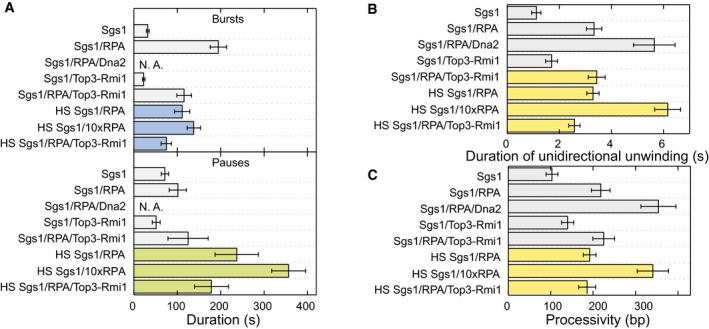
- Mean duration of bursts (upper panel, violet bars) (N > 40) and pauses (lower panel, green bars) (N > 40) for the different enzyme combinations at high‐salt conditions. Error bars represent standard error of the mean.
- Mean duration of unidirectional unwinding (N > 650). Error bars represent standard error of the mean.
- Mean processivity of unidirectional unwinding by the enzyme(s) (N > 650). Error bars represent standard error of the mean.
Finally, we analyzed the processivity of DNA unwinding by Sgs1. This can be either presented as the time during which Sgs1 is unidirectional unwinding without direction reversal (Fig 5B) or as a number of base pairs that are unwound during this time interval (Fig 5C). The addition of RPA to the reaction had the most significant influence, which increased the duration of continuous unwinding threefold to sixfold and the processivity twofold to 3.5‐fold. Top3‐Rmi1 increased the processivity more moderately and only at normal reaction conditions. Altogether, these data reveal that RPA modulates the recruitment at high salt, the affinity to the unwinding fork, and the processivity of DNA unwinding by Sgs1.
Discussion
In this study, we characterized how DNA unwinding by the yeast RecQ helicase Sgs1 is modulated by protein cofactors that cooperate with it during DNA end resection. We showed that a single Sgs1 protein complex can drive processive DNA unwinding over hundreds of base pairs (Fig 1). DNA unwinding by Sgs1 was found to be highly dynamic, involving many repetitive unwinding events separated by either rapid DNA rezipping or slower DNA rewinding. Sgs1 shares this highly dynamic activity pattern with other RecQ family helicases from prokaryotes (Harami et al, 2017; Bagchi et al, 2018) and eukaryotes (Klaue et al, 2013; Wang et al, 2015; Lee et al, 2018). It is thought that the switching between unwinding and rewinding involves repeated strand switching events to allow direction reversals of the helicase (Klaue et al, 2013). In the absence of any cofactor, the unwinding velocity of Sgs1 had a narrow distribution. In the presence of RPA, the distribution became rather broad and strongly skewed. Furthermore, only slow DNA rewinding due to active translocation by Sgs1 rather than fast rezipping was observed in the presence of RPA. We attributed this changed behavior to the likely formation of a Sgs1‐RPA complex. The broad distribution of the Sgs1 velocities with RPA may be due to multiple binding states of RPA that modulate the Sgs1 behavior in a different manner (Hegnauer et al, 2012).
When reconstituting the DNA end resection reaction by combining Sgs1, RPA, and Dna2, we observed that Sgs1 continued its dynamic DNA unwinding–rewinding activity including frequent direction reversals. In addition, the presence of Dna2 promoted a progressive overall unwinding; i.e., rewinding events did not succeed to close the full DNA duplex, but rather terminated away from the original flap position at a distance that increased with time (Figs 3 and EV2). This observed behavior is in agreement with the stringent unidirectional DNA unwinding of Dna2 (Levikova et al, 2013; Pinto et al, 2016) (see inset in Fig 3), combined with progressive degradation of the DNA 5′ end. Thus, the unwinding activity of Sgs1 and the unwinding/degradation activity of Dna2 occur at different velocities. Interestingly, when analyzing the Sgs1 velocities during DNA end resection, we observed that the distributions lost the pronounced skew observed in the presence of RPA. This indicates that the unwinding activity of Sgs1 is to some degree coupled to Dna2 and that Sgs1 directly interacts with Dna2. This is in agreement with previous biochemical data that revealed that Sgs1 and Dna2 can directly physically interact with each other (Cejka et al, 2010a). The direct interaction appears to alleviate the inhibitory effect of RPA on the unwinding velocity, either by disrupting Sgs1‐RPA contacts or by allosteric means. Since Dna2 requires RPA for correct loading onto the 5′ end (Zhou et al, 2015), and since the rewinding events of Sgs1 were exclusively slow during end resection, we hypothesize that RPA is still part of the formed DNA end resection machinery, forming a ternary complex. The stoichiometry of this complex remains to be determined. We hypothesize that it contains a single Dna2 (Zhou et al, 2015), a monomer or a dimer of Sgs1 (Cejka et al, 2010a), and a low number of RPA molecules. The latter may also make contacts to the unwound ssDNA and may be dynamically exchanged, e.g., be loaded or removed to or from ssDNA.
Since only Sgs1 can unwind dsDNA, and the average speed of the Sgs1 motor is about two times higher compared to that of Dna2 (Levikova et al, 2013), Sgs1 is the main factor for DNA unwinding, while Dna2 is trailing behind on the 5′ ssDNA end. Dna2 movement along the unwound ssDNA is powered by its motor activity (Levikova et al, 2017; Miller et al, 2017). Thus, a loop has to form in front of Dna2 (Fig 6). A similar loop formation has been found for the prokaryotic RecBCD complex (Dillingham et al, 2003), indicating that similar mechanisms can also exist in eukaryotic cells. A loop forming ahead of Dna2 would allow the binding of RPA to the unwound DNA behind Sgs1, which was shown to specifically promote degradation of the 5′‐terminated strand by Dna2 (Cejka et al, 2010a; Niu et al, 2010). A loop forming ahead of Dna2 thus explains how the regulatory function of RPA can be achieved. Next, Sgs1 occasionally switches strands and actively rewinds DNA, thus backtracking toward the slower moving Dna2 molecule. When Sgs1 encounters Dna2, it switches back again to DNA unwinding. What can be the reason for such a switching behavior? We hypothesize that the strand switching activity serves to limit the DNA unwinding by Sgs1, such that the ssDNA loop is not extensively long and prone to unscheduled cleavage. Furthermore, Dna2 is sensitive to obstacles on DNA such as secondary structures or protein blocks, which stall DNA degradation (Balakrishnan et al, 2010; Levikova et al, 2013). Sgs1, moving periodically toward Dna2, might help resolve these structures. Finally, Sgs1 has a function to promote the dissolution of double Holliday junctions in the late stage of the canonical DSB repair pathway (Cejka et al, 2010b), which is separate from its role in DNA end resection. To do so, it needs to migrate, in conjunction with Top3‐Rmi1, the two Holliday junctions toward each other. However, it was not apparent how the direction of the junction migration by Sgs1 is determined, as only convergent migration (i.e., the migration of the junctions toward each other) dissolves the entangled chromosomes (Cejka et al, 2010b). It has been speculated that chromatin may serve as a barrier to block junction migration in the “wrong” direction. This, coupled with the random switching of Sgs1 movement, would result in a “random walk” mechanism of junction migration, which would ultimately lead to convergence of the both junctions. Thus, the switching behavior of Sgs1 may be also relevant for processes separate from DNA end resection.
Figure 6. Model for DNA end resection by the Sgs1‐RPA‐Dna2 complex.
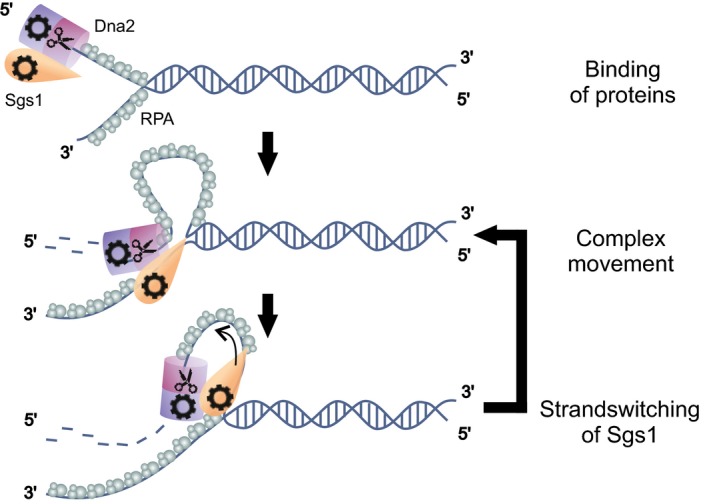
Sgs1‐Dna2 complex binds first to a 5′ DNA overhang assisted by RPA. At the DNA junction, both helicases start to move unidirectionally on either of the strands. Due to the faster movement of Sgs1, this helicase powers the unwinding of the DNA duplex and causes the formation of a ssDNA loop in front of Dna2. Upon an occasional strand switch of Sgs1, the protein rewinds the DNA and moves toward Dna2. This leads to shortening of the loop. Upon encounter of Dna2, Sgs1 switches back to the original strand to power further unwinding.
We observed that even the helicase‐ and nuclease‐dead double mutant of Dna2 could promote a progressive DNA unwinding in the presence of Sgs1. This behavior was not seen when DNA was unwound in the absence of Sgs1 (Fig EV2E and F); i.e., it must have been promoted by a direct interaction between Sgs1 and the double mutant of Dna2. We propose that Dna2, even without any catalytic activity, functions as a ratchet on ssDNA; i.e., it would be easily movable along ssDNA in the 5′–3′ direction but barely displaceable by Sgs1 backward in the 3′–5′ direction. Notably, an electrostatic ratchet mechanism has been suggested for the nuclease domain to allow progressive degradation of the 5′ DNA end (Zhang et al, 2011; Zhou et al, 2015). To allow progressive unwinding, a passive ratchet would still require an external energetic bias or an external force. We hypothesize that this could be provided from the coupling to Sgs1 and the limited conformational entropy of the forming RPA‐bound loop. We note that the hypothesized ratchet mechanism warrants further future investigations.
We also determined the effect of the known interaction partner Top3‐Rmi1 on DNA unwinding by Sgs1. Most importantly, we found that in the presence of RPA, the histograms of the Sgs1 unwinding and rewinding velocities exhibited significantly reduced skews (Fig 4F) compared to the presence of RPA only. Similarly, the interaction with Top3‐Rmi1 appeared to alleviate the inhibitory effect of RPA on the unwinding velocity. Additionally, we observed that the velocity of Sgs1 was increased in the presence of Top3‐Rmi1 regardless of RPA (Fig 4C and F) or the ionic strength (Fig EV4E). An increased unwinding/rewinding velocity thus explains the mechanism underlying the previously observed stimulation of Sgs1 by Top3‐Rmi1 (Cejka et al, 2010a). Since in the presence of RPA, DNA rewinding by Sgs1 in conjunction with the Top3‐Rmi1 complex was always slow, we hypothesize that Sgs1 is simultaneously interacting with RPA and Top3‐Rmi1. Notably, we observed that under the applied force, Top3‐Rmi1 can be trapped on ssDNA in the so‐called open‐gate configuration (Fig EV3A), which seems to be a general property of type IA topoisomerases (Mills et al, 2018). This conformational trapping may however be less relevant in vivo since it is abolished by the presence of RPA, which limits the access of Top3‐Rmi1 to ssDNA. The physiological relevance of this ssDNA cleavage activity of Top3‐Rmi1 may thus be limited.
As Top3‐Rmi1, also RPA appears to be an important regulator of Sgs1 activity. It is essential for the recruitment of Sgs1 at high ionic strength, which is similar to E. coli RecQ, whose initiation is supported by SSB (Mills et al, 2017). Furthermore, RPA slows the rewinding of DNA by Sgs1 (Fig 2B and C) and increases the processivity of Sgs1 for unidirectional unwinding (Fig 5C). Finally, RPA increases the duration of unwinding bursts; i.e., it stabilizes the interaction of Sgs1 with the DNA substrate (Fig 5A). Altogether, these results indicate that RPA ensures that DNA is kept in a partially unwound state for longer periods of time, which may promote the end resection process.
Altogether, our data show that the various Sgs1 protein partners lead to surprisingly diverse modulations of the Sgs1 activity. We believe that these modulations allow fine‐tuning of DNA unwinding by Sgs1, which helps it to tackle its diverse functions to promote genome stability.
Materials and Methods
Recombinant proteins
Sgs1, RPA, and Dna2 from Saccharomyces cerevisiae and their mutants were expressed and purified as described previously (Kantake et al, 2003; Cejka et al, 2010a; Levikova et al, 2013). In short, Sgs1 and Sgs1 (641–1,215) were expressed using pFB‐MBP‐Sgs1‐His vector and the Bac‐to‐Bac baculovirus expression system in Sf9 cells. The proteins were first bound to amylose resin (New England Biolabs). Afterward, the proteins were treated with PreScission protease to remove the MBP‐tag. Next, Sgs1 and Sgs1 (641–1,215) were bound to Bio‐Rex70 resin and Ni2+‐NTA Agarose, eluted, and dialyzed (Cejka & Kowalczykowski, 2010). Yeast RPA was expressed in Escherichia coli from p11d‐scRPA vector (a kind gift from M. Wold, University of Iowa) and purified as described for the human recombinant RPA (Anand et al, 2018). Wild‐type Dna2 and its mutants were expressed from altered pGAL:DNA2 vector containing N‐terminal Flag and HA tags as well as C‐terminal His6 tag, in S. cerevisiae strain WDH668 (Solinger et al, 2001). Dna2 was purified by affinity chromatography using Ni2+‐NTA Agarose (Qiagen) and M2 anti‐FLAG affinity resin (Sigma), washed, and eluted with buffer containing 3xFLAG Peptide (Sigma) (Levikova et al, 2013).
DNA substrate
The DNA construct for the magnetic tweezers experiments containing the 40‐nt flap (see Fig 1A) was prepared as previously described (Luzzietti et al, 2012; Levikova et al, 2013). The main DNA fragment of 6.6 kbp in length was excised from plasmid pNLrep (Luzzietti et al, 2011) using the restriction enzymes BamHI and BsrGI. It was simultaneously digested with the nicking enzyme Nt.BbvCI to produce a 63‐nt gap at an engineered site containing 5 consecutive, 15‐nt spaced Nt.BbvCI sites. The gap was located approximately 0.5 kbp from the BamHI‐cut end of the fragment. 63 nt of the gap was filled by hybridizing a 25‐nt DNA oligomer that carried an additional 40‐nt polythimidine tail on its 5′ end that served as the flap. In a subsequent ligation reaction, the oligomer was ligated at its 3′ end inside the gap. Furthermore, 600 bp DNA handles carrying either multiple biotin or digoxigenin modifications were attached at either end. The handle duplexes were produced by PCR in the presence of biotin and digoxigenin‐modified nucleotides and digested with BsrGI and BamHI, respectively.
Magnetic tweezers experiments
Single‐molecule experiments were carried out in a custom‐made magnetic tweezers setup (Klaue & Seidel, 2009; Huhle et al, 2015) at room temperature. Fluidic cells were prepared from two coverslips (Menzel, Braunschweig, Germany) and a parafilm (Bemis, Oshkosh, USA) spacer. The bottom coverslip was previously coated by spin coating using a 1% solution of polystyrene in toluene (Sigma‐Aldrich, St. Louis, USA). To allow specific DNA tethering in the fluidic cell, anti‐digoxigenin antibodies (Roche, Penzberg, Germany) were adsorbed to the polystyrene layer overnight from a 50 mg/ml anti‐digoxigenin in standard aqueous phosphate‐buffered saline (PBS) solution. Subsequently, the fluidic cell was incubated with 10 mg/ml bovine serum albumin (BSA, New England Biolabs, Ipswich, USA) to prevent non‐specific surface binding. 3 μm latex beads (Life Technologies, Darmstadt, Germany) serving as reference particles and 2.8 μm streptavidin‐coated magnetic beads (Life Technologies, Darmstadt, Germany) with prebound DNA molecules were flushed into the flow cell. The beads and the DNA were allowed to bind to the surface, and subsequently, unbound particles were removed by washing the chamber with phosphate‐buffered saline (PBS). Lowering the magnets allowed to stretch and to identify bead‐tethered DNA molecules. The measurements were then performed at 300 Hz using video microscopy, and real‐time GPU accelerated image analysis (Huhle et al, 2015). One measurement usually was performed with 15–25 molecules at a time. Magnetic forces were calibrated using fluctuation analysis (Daldrop et al, 2015). Unless stated otherwise, the measurements were performed in reaction buffer (25 mM Tris‐acetate pH 7.5, 2 mM magnesium acetate, 1 mM ATP, 1 mM DTT, 0.1 mg/ml BSA) using protein concentrations of 0.2 nM Sgs1, 20 nM yRPA, 0.4 nM Top3‐Rmi1, and 5 nM of Dna2 or its variants.
Data analysis
Analysis of the results was performed using custom‐written MATLAB program (Kemmerich et al, 2016b). Particularly, the unwinding and rewinding velocities were determined from fitting linear segments to periods of constant velocities of the recorded trajectories. For converting measured unwinding velocities in μm/s into unwinding rates in bp/s, a conversion factor was obtained from recording force–extension curves of bare DNA construct and RPA‐coated construct. Errors of obtained rates and times are given as standard error of the mean (S.E.M.) throughout.
Author contributions
PC and RS designed the study. KK, PC, and RS wrote the manuscript. KK, FF, and PD carried out the measurements and analyzed the data. ML and RA purified and characterized the employed proteins.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We greatly acknowledge stimulating discussions and experimental support by Dr. Martin Göse, Felix Kemmerich, Marius Rutkauskas, Dominik Kauert, and Pierre Aldag. This work was supported by a Consolidator grant of the European Research Council (724863) to R.S. and by the Swiss National Science Foundation (31003A_175444) and European Research Council grants (681630) to P.C.
The EMBO Journal (2019) 38: e101516
Contributor Information
Petr Cejka, Email: petr.cejka@irb.usi.ch.
Ralf Seidel, Email: ralf.seidel@physik.uni-leipzig.de.
References
- Anand R, Pinto C, Cejka P (2018) Chapter two ‐ methods to study DNA end resection I: recombinant protein purification In Mechanisms of DNA recombination and genome rearrangements: methods to study homologous recombination, Spies M, Malkova A. (eds), Vol. 600, pp 25–66. Cambridge, MA: Academic Press; [DOI] [PubMed] [Google Scholar]
- Aparicio T, Baer R, Gautier J (2014) DNA double‐strand break repair pathway choice and cancer. DNA Repair 19: 169–175 Cutting‐edge Perspectives in Genomic Maintenance [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SH, Bae KH, Kim JA, Seo YS (2001a) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412: 456 [DOI] [PubMed] [Google Scholar]
- Bae SH, Kim JA, Choi E, Lee KH, Kang HY, Kim HD, Kim JH, Bae KH, Cho Y, Park C et al (2001b) Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Res 29: 3069–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae KH, Kim HS, Bae SH, Kang HY, Brill S, Seo YS (2003) Bimodal interaction between replication‐protein A and Dna2 is critical for Dna2 function both in vivo and in vitro . Nucleic Acids Res 31: 3006–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi D, Manosas M, Zhang W, Manthei KA, Hodeib S, Ducos B, Keck JL, Croquette V (2018) Single molecule kinetics uncover roles for E. coli RecQ DNA helicase domains and interaction with SSB. Nucleic Acids Res 46: 8500–8515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan L, Polaczek P, Pokharel S, Campbell JL, Bambara RA (2010) Dna2 exhibits a unique strand end‐dependent helicase function. J Biol Chem 285: 38861–38868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Sharp JA, Wang JC (1998) Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae . J Biol Chem 273: 9644–9650 [DOI] [PubMed] [Google Scholar]
- Bocquet N, Bizard AH, Abdulrahman W, Larsen NB, Faty M, Cavadini S, Bunker RD, Kowalczykowski SC, Cejka P, Hickson ID et al (2014) Structural and mechanistic insight into holliday‐junction dissolution by topoisomerase IIIα and RMI1. Nat Struct Mol Biol 21: 261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma I, Gent DC (2012) Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr 3: 9–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA (2000) Replication protein A physically interacts with the bloom's syndrome protein and stimulates its helicase activity. J Biol Chem 275: 23500–23508 [DOI] [PubMed] [Google Scholar]
- Cejka P, Kowalczykowski SC (2010) The full‐length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J Biol Chem 285: 8290–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC (2010a) Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1‐Top3. Nat Struct Mol Biol 17: 1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda‐Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC (2010b) DNA end resection by Dna2‐Sgs1‐RPA and its stimulation by Top3‐Rmi1 and Mre11‐Rad50‐Xrs2. Nature 467: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P (2015) DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J Biol Chem 290: 22931–22938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daldrop P, Brutzer H, Huhle A, Kauert D, Seidel R (2015) Extending the range for force calibration in magnetic tweezers. Biophys J 108: 2550–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Chiba T, Xue X, Niu H, Sung P (2014a) Multifaceted role of the Topo IIIα‐RMI1‐RMI2 complex and DNA2 in the BLM‐dependent pathway of DNA break end resection. Nucleic Acids Res 42: 11083–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Gaines WA, Kwon Y, Sung P (2014b) Regulation of DNA pairing in homologous recombination. Cold Spring Harbor Perspect Biol 6: a017954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Jimenez‐Sainz J, Wang W, Miller AS, Xue X, Nguyen KA, Jensen RB, Sung P (2017) Enhancement of BLM‐DNA2‐mediated long‐range DNA end resection by CtIP. Cell Rep 21: 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M (2012) BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker NH, Rybenkov VV, Duguet M, Crisona NJ, Cozzarelli NR, Bensimon D, Croquette V (2002) The mechanism of type IA topoisomerases. Proc Natl Acad Sci USA 99: 12126–12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Spies M, Kowalczykowski SC (2003) RecBCD enzyme is a bipolar DNA helicase. Nature 423: 893 [DOI] [PubMed] [Google Scholar]
- Fasching C, Cejka P, Kowalczykowski S, Heyer WD (2015) Top3‐Rmi1 dissolve Rad51‐mediated D loops by a topoisomerase‐based mechanism. Mol Cell 57: 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Kaliraman V, Brill SJ (2001) Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J Biol Chem 276: 8848–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harami GM, Seol Y, In J, Ferencziová V, Martina M, Gyimesi M, Sarlós K, Kovács ZJ, Nagy NT, Sun Y et al (2017) Shuttling along DNA and directed processing of D‐loops by RecQ helicase support quality control of homologous recombination. Proc Natl Acad Sci USA 114: E466–E475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegnauer AM, Hustedt N, Shimada K, Pike BL, Vogel M, Amsler P, Rubin SM, van Leeuwen F, Guénolé A, van Attikum H et al (2012) An N‐terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J 31: 3768–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, Zhang M, Hu Y, Wang Q, Xu W et al (2012) The intra‐S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 149: 1221–1232 [DOI] [PubMed] [Google Scholar]
- Huhle A, Klaue D, Brutzer H, Daldrop P, Joo S, Otto O, Keyser UF, Seidel R (2015) Camera‐based three‐dimensional real‐time particle tracking at kHz rates and Ångström accuracy. Nat Commun 6: 5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantake N, Sugiyama T, Kolodner RD, Kowalczykowski SC (2003) The recombination‐deficient mutant RPA (rfa1‐t11) is displaced slowly from single‐stranded DNA by Rad51 protein. J Biol Chem 278: 23410–23417 [DOI] [PubMed] [Google Scholar]
- Kao HI, Campbell JL, Bambara RA (2004) Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J Biol Chem 279: 50840–50849 [DOI] [PubMed] [Google Scholar]
- Kaur H, De Muyt A, Lichten M (2015) Top3‐Rmi1 DNA single‐strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol Cell 57: 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerich FE, Daldrop P, Pinto C, Levikova M, Cejka P, Seidel R (2016a) Force regulated dynamics of RPA on a DNA fork. Nucleic Acids Res 44: 5837–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerich FE, Kasaciunaite K, Seidel R (2016b) Modular magnetic tweezers for single‐molecule characterizations of helicases. Methods 108: 4–13 DNA Helicases [DOI] [PubMed] [Google Scholar]
- Kennedy JA, Daughdrill GW, Schmidt KH (2013) A transient α‐helical molecular recognition element in the disordered N‐terminus of the Sgs1 helicase is critical for chromosome stability and binding of Top3/Rmi1. Nucleic Acids Res 41: 10215–10227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaue D, Seidel R (2009) Torsional stiffness of single superparamagnetic microspheres in an external magnetic field. Phys Rev Lett 102: 028302 [DOI] [PubMed] [Google Scholar]
- Klaue D, Kobbe D, Kemmerich F, Kozikowska A, Puchta H, Seidel R (2013) Fork sensing and strand switching control antagonistic activities of RecQ helicases. Nat Commun 4: 2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Burgers PM (2013) Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev 27: 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Shin S, Uhm H, Hong H, Kirk J, Hyun K, Kulikowicz T, Kim J, Ahn B, Bohr VA et al (2018) Multiple RPAs make WRN syndrome protein a superhelicase. Nucleic Acids Res 46: 4689–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levikova M, Klaue D, Seidel R, Cejka P (2013) Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc Natl Acad Sci USA 110: E1992–E2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levikova M, Pinto C, Cejka P (2017) The motor activity of DNA2 functions as an ssDNA translocase to promote DNA end resection. Genes Dev 31: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzietti N, Brutzer H, Klaue D, Schwarz FW, Staroske W, Clausing S, Seidel R (2011) Efficient preparation of internally modified single‐molecule constructs using nicking enzymes. Nucleic Acids Res 39: e15–e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzietti N, Knappe S, Richter I, Seidel R (2012) Nicking enzyme‐based internal labeling of DNA at multiple loci. Nat Protoc 7: 643 [DOI] [PubMed] [Google Scholar]
- Machwe A, Lozada E, Wold MS, Li GM, Orren DK (2011) Molecular cooperation between the Werner syndrome protein and replication protein A in relation to replication fork blockage. J Biol Chem 286: 3497–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bozzella M, Seluanov A, Gorbunova V (2008) Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 7: 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Haber JE (2014) Sources of DNA double‐strand breaks and models of recombinational DNA repair. Cold Spring Harbor Perspect Biol 6: a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AS, Daley JM, Pham NT, Niu H, Xue X, Ira G, Sung P (2017) A novel role of the Dna2 translocase function in DNA break resection. Genes Dev 31: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills M, Harami GM, Seol Y, Gyimesi M, Martina M, Kovács ZJ, Kovács M, Neuman KC (2017) RecQ helicase triggers a binding mode change in the SSB–DNA complex to efficiently initiate DNA unwinding. Nucleic Acids Res 45: 11878–11890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills M, Tse‐Dinh YC, Neuman KC (2018) Direct observation of topoisomerase IA gate dynamics. Nat Struct Mol Biol 25: 1111–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC (2011) BLM‐DNA2‐RPA‐MRN and EXO1‐BLM‐RPA‐MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L et al (2010) Mechanism of the ATP‐dependent DNA end‐resection machinery from Saccharomyces cerevisiae . Nature 467: 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PYH, Taylor AF, Smith GR, Hunter N (2007) BLM Ortholog, Sgs1, prevents aberrant crossing‐over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ölmezer G, Levikova M, Klein D, Falquet B, Fontana GA, Cejka P, Rass U (2016) Replication intermediates that escape Dna2 activity are processed by Holliday junction resolvase Yen1. Nat Commun 7: 13157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Jimenez R, Godoy‐Ruiz R, Ibarra‐Molero B, Sanchez‐Ruiz JM (2004) The efficiency of different salts to screen charge interactions in proteins: a Hofmeister effect? Biophys J 86: 2414–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto C, Kasaciunaite K, Seidel R, Cejka P (2016) Human DNA2 possesses a cryptic DNA unwinding activity that functionally integrates with BLM or WRN helicases. Elife 5: e18574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarlós K, Gyimesi M, Kovács M (2012) RecQ helicase translocates along single‐stranded DNA with a moderate processivity and tight mechanochemical coupling. Proc Natl Acad Sci USA 109: 9804–9809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Cui Y, Bustamante C (1996) Overstretching B‐DNA: the elastic response of individual double‐stranded and single‐stranded DNA molecules. Science 271: 795–799 [DOI] [PubMed] [Google Scholar]
- Solinger JA, Lutz G, Sugiyama T, Kowalczykowski SC, Heyer WD (2001) Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J Mol Biol 307: 1207–1221 [DOI] [PubMed] [Google Scholar]
- Spies M, Amitani I, Baskin RJ, Kowalczykowski SC (2007) RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell 131: 694–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzenegger A, Burdova K, Kanagaraj R, Levikova M, Pinto C, Cejka P, Janscak P (2014) DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long‐range DNA end resection in human cells. J Biol Chem 289: 27314–27326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS (2014) End resection at double‐strand breaks: mechanism and regulation. Cold Spring Harbor Perspect Biol 6: a016436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA et al (2015) DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol 208: 545–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Qin W, Li JH, Lu Y, Lu KY, Nong DG, Dou SX, Xu CH, Xi XG, Li M (2015) Unwinding forward and sliding back: an intermittent unwinding mode of the BLM helicase. Nucleic Acids Res 43: 3736–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, McCabe KA, Bell CE (2011) Crystal structures of λ exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc Natl Acad Sci USA 108: 11872–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Pourmal S, Pavletich NP (2015) Dna2 nuclease‐helicase structure, mechanism and regulation by Rpa. Elife 4: e09832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


