Abstract
Cancer stem cells have been strongly linked to resistance and relapse in many malignancies. However, purifying them from within the bulk tumor has been challenging, so their precise genetic and functional characteristics are not well defined. The side population assay exploits the ability of some cells to efflux Hoechst dye via ATP-binding cassette transporters. Stem cells have increased expression of these transporters and this assay has been shown to enrich for stem cells in various tissues and cancers. This study identifies the side population within a zebrafish model of acute lymphoblastic leukemia and correlates the frequency of side population cells with the frequency of leukemia stem cells (more precisely referred to as leukemia-propagating cells within our transplantation model). In addition, the side population within the leukemia evolves with serial transplantation, increasing in tandem with leukemia-propagating cell frequency over subsequent generations. Sorted side population cells from these tumors are enriched for leukemia-propagating cells and have enhanced engraftment compared to sorted non-side population cells when transplanted into syngeneic recipients. RNA-sequencing analysis of sorted side population cells compared to non-side population cells identified a shared expression profile within the side population and pathway analysis yielded Wnt-signaling as the most overrepresented. Gene set enrichment analysis showed that stem cell differentiation and canonical Wnt-signaling were significantly upregulated in the side population. Overall, these results demonstrate that the side population in zebrafish acute lymphoblastic leukemia significantly enriches for leukemia-propagating cells and identifies the Wnt pathway as a likely genetic driver of leukemia stem cell fate.
Introduction
Although arising from a single cell initially, over time cancer cells acquire a series of genetic and epigenetic changes. At the time of diagnosis, the cancer consists of a heterogeneous collection of cells with differences in gene expression, proliferative capacity, morphology, and metastatic potential. Defined by their capacity to repopulate a tumor from a single cell, cancer stem cells (CSC) also evolve over time, leading to therapy resistance and relapse.1 Increased tumor heterogeneity and CSC have been associated with resistance and relapse for many tumor types, including acute myelogenous leukemia and acute lymphoblastic leukemia (ALL).2–5
ALL is the most common pediatric cancer with approximately 3,000 new cases each year.6 It can be derived from either T cells or B cells, with B-cell ALL (B-ALL) typically having much more positive treatment outcomes. T-cell ALL (T-ALL) is a more aggressive malignancy, resulting from the transformation of immature T cells.7,8 While the cure rate for pediatric patients has improved with more intensive treatment regimens, prognosis is bleak for the 20% of children and 60% of adult patients with refractory and relapsed T-ALL.9 Relapses and refractory disease are attributed to resistance to chemotherapy in the small sub-population of leukemia stem cells (LSC) with the capacity to regrow the tumor from a single cell.1,10 Thus, these LSC present an obvious target for preventing relapsed disease. However, there are no definitive markers to identify an LSC within the bulk ALL, and therefore our understanding of the molecular drivers of LSC fate in this disease is limited.
One promising method for studying CSC is the side population (SP) assay. This assay exploits the stem cell’s inherent ability to efflux the fluorescent dye Hoechst 33342 through an increased expression of certain ATP-binding cassette (ABC) transporters. This method was originally developed by Goodell et al., who discovered that normal bone marrow cells incubated with Hoechst 33342 could be analyzed by dual-wavelength flow cytometry to reveal a small subpopulation of cells that did not accumulate the dye.11 These cells were found to be highly enriched for hematopoietic stem and progenitor cells.11 The method has subsequently been applied to identify putative stem and progenitor cells in many other normal tissues12–15 as well as human cancers,16–18 including T-ALL.19
In this study, we characterize the SP cells within tumors generated in a robust model of zebrafish ALL that is morphologically and genetically similar to the human disease.20–22 Tumors are generated by co-injecting single cell zebrafish embryos with plasmids containing the zebrafish lymphocyte-specific rag2 promoter driving expression of a biofluorescent protein and the murine oncogene Myc.20 Most leukemias spread quickly beyond the thymus to invade the entire body. These tumors are transplantable by intraperitoneal injection into syngeneic and/or irradiated recipients. Through limiting-dilution transplantations, the frequency at which cells from a specific tumor engraft can be calculated using Extreme Limiting Dilution Analysis (ELDA) software.23 These cells are often classified as LSC but are more precisely designated leukemia-propagating cells (LPC). This approach allowed us to correlate the LPC frequency in relation to the frequency of SP cells, and then use the sorted side population as a proxy for LSC to identify transcriptional changes by RNA-sequencing.
Methods
Animals
All zebrafish experiments were performed as approved by the University of Chicago Institutional Animal Care and Use Committee using clonal golden 2 (CG2) syngeneic fish, which were a gift from Dr. Sergei Revskoy.24
Generation of mosaic transgenic fish that develop acute lymphoblastic leukemia
Rag2-GFP25 and rag2-Myc26 sequences were inserted into a pDEST-Tol2-pA2 backbone (gift from Dr. Christian Mosimann) to add inverted terminal repeats of minimal recognized tol2 sequence flanking the promoter/gene sequence. Co-injection of these vectors with M3 Tol2 transposase mRNA into single-cell embryos increases the efficiency of genomic integration27 and produces GFP+ ALL tumors as described previously.28,29 Fish are screened from 21-42 days post-fertilization (dpf) for green thymi to identify tumors for further experiments.
Cell transplantation and limiting-dilution analysis
Limiting-dilution transplantation of leukemia cells was performed as described previously.30 Briefly, fish were euthanized in a high dose (~0.1%) of Tricaine MS-222 (Western Chemical Inc.) and tumors harvested in 10% fetal bovine serum/90% 0.9X phosphate-buffered saline and heparin salt (300 U/mL) (Sigma), filtered through a 40 μm filter, and diluted into limiting cell doses along with carrier peripheral blood cells from a wild-type fish. Five microliters of the different cell dilutions were injected into the peritoneal cavity of anesthetized CG2 recipient fish. Engraftment was screened for weekly, starting at 2 weeks, and continued for 1 month after the final positive engraftment or until all members of the transplant group were positive or dead. Engraftment rates to calculate the LPC frequency were analyzed using ELDA software.31
Side population assay and fluorescent activated cell sorting
SP staining and analyses were performed as described previously.28 Briefly, tumors were harvested as described for cell transplantations, then incubated with 15 μg/mL Hoechst 33342 (Life Technologies) at 28°C for 2 h protected from light. The inhibitor sample also included 250 μM verapamil (Sigma). It should be noted that in our hands, the Hoechst dye was not stable at 4°C for more than 1-2 months, so fresh stock solution was made monthly for optimal staining. Cells were then washed and analyzed by flow cytometry on an LSR Fortessa X-20 instrument with a UV laser and Hoechst blue and Hoechst red bandpass filters (450/50 and 670/30, respectively). For fluorescent activated cell sorting, a BD AriaFusion instrument was used and cells were sorted with a 100 μm nozzle tip into 10% fetal bovine serum/90% 0.9X phosphate-buffered saline.
Quantitative polymerase chain reaction
RNA from each tumor was reverse transcribed in a 20 μL volume with random primers using the Maxima First Strand cDNA Synthesis Kit for reverse transcriptase-quantitative polymerase chain reaction (qPCR) (Fermentas) as described in the manufacturer’s instructions. qPCR was performed in 10 μL reactions that included 1-4 μL diluted cDNA, primers and SsoAdvanced SYBR Green Supermix supplemented with ROX (Biorad). Data were collected from triplicate reactions according to recommended settings (Biorad) on a 7900HT instrument with 384-well format (Applied Biosystems), and analyzed by the delta-delta CT method.32 Expression of ef1a was used as a reference gene.
RNA-sequencing and analysis
The RNA-sequencing and analysis are described in the Online Supplementary Methods.
EdU proliferation assay
Proliferation assays were performed with a Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Invitrogen) according to the manufacturer’s instructions with incubation adjusted to 2 h at 28°C.
Statistical analysis
Linear correlations were assessed by R2 and the Pearson correlation coefficient, and non-parametric correlations were assessed by the Spearman rank correlation coefficient, with two-tailed P-values used in both cases. LPC frequencies, confidence intervals, and pairwise test P-values were all determined by ELDA software.31 The statistical significance of data from qPCR experiments was determined by multiple t-tests.
Results
The frequency of side population cells correlates with leukemia-propagating cell frequency in zebrafish acute lymphoblastic leukemia
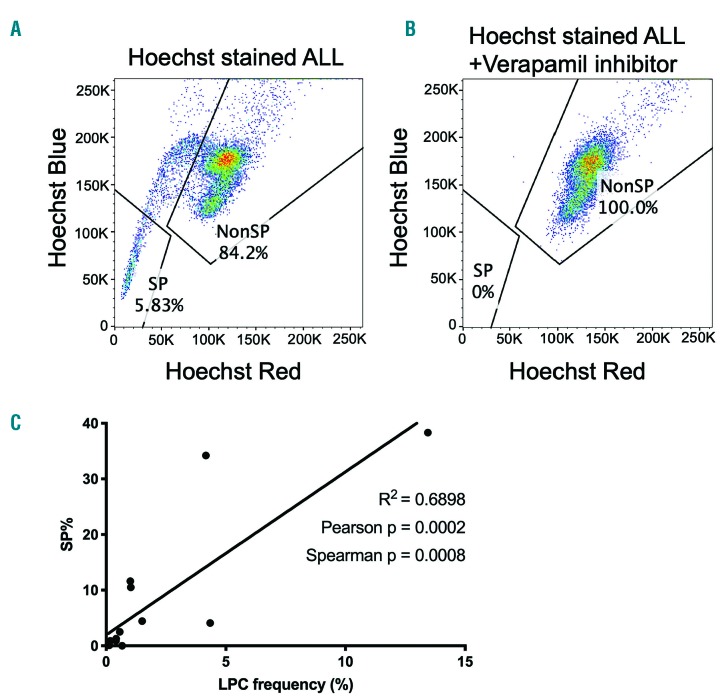
To evaluate the SP assay as a potential method for studying LPC in our zebrafish model, we harvested primary or transplanted tumors and measured the frequency of SP cells by flow cytometry as previously described.28 All zebrafish leukemias were generated in the clonal CG2 zebrafish line to facilitate syngeneic transplantation studies. In the 57 leukemias tested, SP frequency varied between 0 – 64.6%, with a mean of 14.84% and a median of 7.8% (Figure 1). Of the tumors tested, 87.7% (50 of 57) had a detectable SP. In order to determine LPC frequencies, tumor cells were transplanted at limiting dilutions into multiple syngeneic, sub-lethally irradiated CG2 recipient fish and evaluated for tumor engraftment for 2-4 months (Online Supplementary Table S1). Similarly to the wide range of SP frequencies, we also observed a wide range of LPC frequencies (0.09 – 13.4%), and these parameters were positively correlated (R2 = 0.6898) (Figure 1C). This suggests that the SP assay enriches for LPC in zebrafish ALL, as has been shown in other models and tissues.
Figure 1.
Frequency of side population cells in zebrafish acute lymphoblastic leukemia correlates with leukemia-propagating cell frequency. (A, B) Side population (SP) assay performed on a zebrafish acute lymphoblastic leukemia (ALL) tumor without (A) or with (B) verapamil inhibitor to block dye efflux. Live, singlet, tumor cells are shown separated by their emission of Hoechst blue (450/50 nm bandpass filter) and Hoechst red (670/30 nm bandpass filter). Data are shown for one representative tumor of 57 tested. (C) Correlation of the percentage of SP cells versus the frequency of leukemia-propagating cells (LPC).
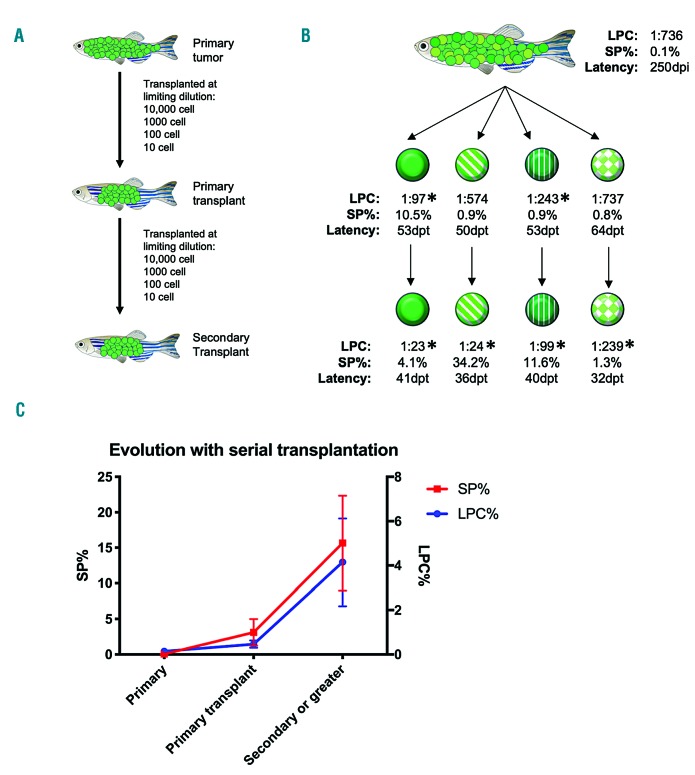
Leukemia-propagating cell frequency and side population frequency increase in tandem in serially transplanted tumors
From our correlative data, we noted that the tumors with the lowest SP% and LPC% were most often primary tumors, and those with the highest SP% and LPC% had been transplanted multiple times. Since Blackburn et al. had previously shown that the LPC frequency in this model undergoes clonal evolution,33 we were curious to know whether the SP% was evolving similarly. To test this, we transplanted primary tumors at limiting dilution, and measured the SP% on the day of transplantation. We then performed serial limiting-dilution transplantation of the tumors that grew out from these primary transplants, along with the SP assay for each subsequent passage (Figure 2) (Online Supplementary Table S2). Indeed, large increases in LPC frequency often corresponded with increases in SP%, and when these data were pooled with those from other lineages there was a general trend of increase in both parameters after multiple transplants (Figure 2C). Additionally, the latency and proliferation rate (measured by EdU incorporation) were independent of LPC frequency and SP% (data not shown), confirming previously reported data.33 These results provide further evidence for the connection between LPC frequency and the SP assay.
Figure 2.
Side population size undergoes clonal evolution in tandem with leukemia-propagating cell frequency. (A) Graphic depicting the workflow of serial limiting-dilution transplants. (B) Representative lineage of a primary tumor with leukemia-propagating cell (LPC) frequency, percent side population (SP), and latency to >50% tumor burden. Dpi: days post-injection; dpt: days post-transplant. The statistical significance of the increase in LPC% from the parent tumor was determined by pairwise tests using Extreme Limiting Dilution Analysis software *P<0.01. (C) Pooled data from multiple lineages showing percent SP (left y-axis, red) and LPC frequency (right y-axis, blue) (n=14).
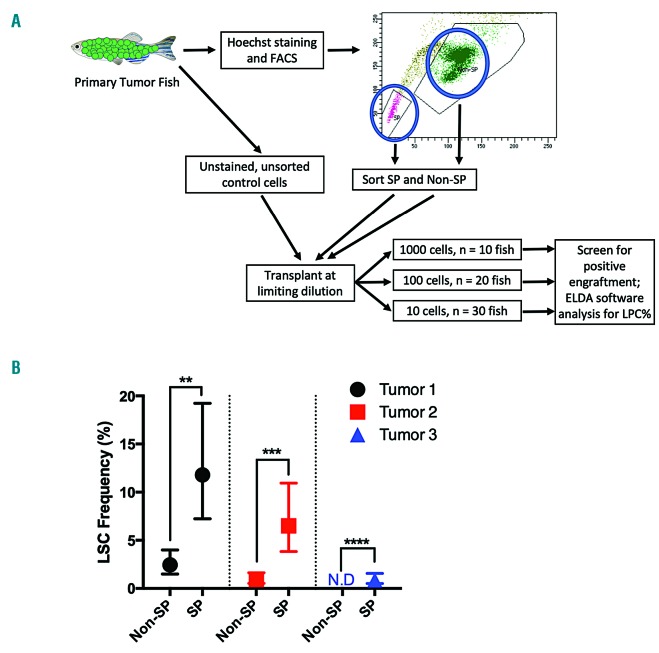
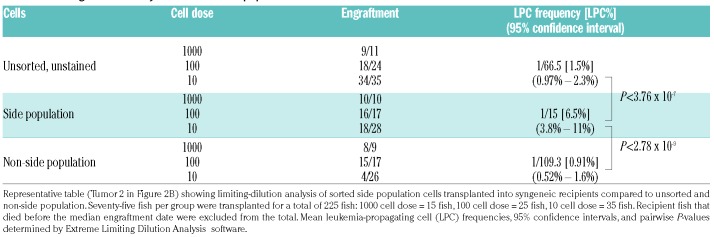
Sorted side population cells have increased leukemia-propagating cell frequency
To measure the ability of the SP to enrich for LPC in our model more directly, we sorted SP and non-SP cells and transplanted them at limiting dilutions along with unsorted, unstained control cells into recipient fish to determine the LPC frequency of each group (Figure 3A, Table 1 and Online Supplementary Table S3). In three independent experiments, there was a very significant increase in LPC frequency in the SP groups as compared to the non-SP groups (Figure 3B). These experiments showed both an enrichment of LPC activity in the SP groups and a depletion within the non-SP groups, including one tumor whose non-SP did not have engraftment of a single LPC. These results, in combination with the previous correlative data, strongly demonstrate that the SP in this zebrafish ALL model is enriched for LPC.
Figure 3.
Sorted side population cells have increased leukemia-propagating cell frequency. (A) Graphic depicting the workflow of determining leukemia-propagating cell (LPC) frequency in the side population (SP) and non-SP. FACS: fluorescent activating cell sorting; ELDA: Extreme Limiting Dilution Assay. (B) Three independent SP limiting-dilution analysis experiments. Data are shown as mean LPC frequency with 95% confidence intervals. The statistical significance of differences was determined by pairwise tests using ELDA software: **P<10−5, ***P<10−8, ****P<10−11 (n = 60 fish per experimental group, 360 total).
Table 1.
Limiting-dilution analysis of sorted side population cells.
Zebrafish acute lymphoblastic leukemia side population cells have a common expression profile with increased expression of stem cell- and Wnt pathway-associated genes
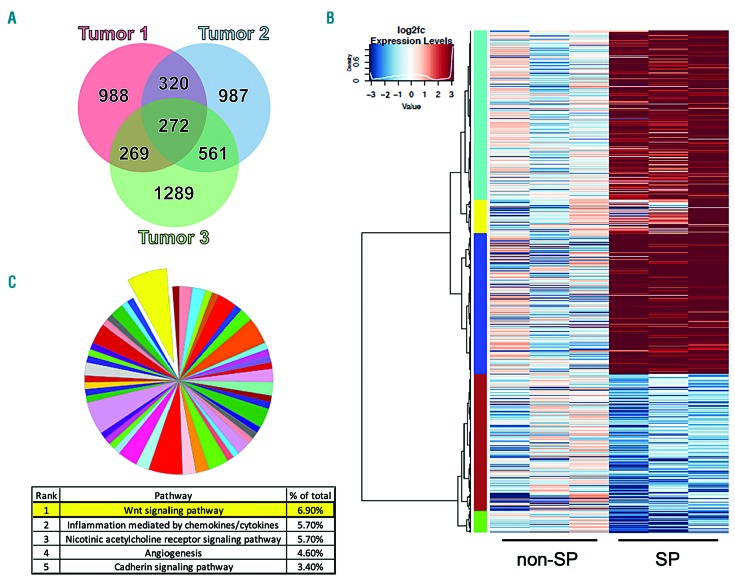
Once we had validated the SP assay as a method to enrich for LPC in our model, we did bulk RNA sequencing on sorted SP and non-SP cells from three distinct tumors (Figure 4). This produced 272 genes that were differentially expressed in the SP of each individual tumor (Figure 4A and Online Supplementary Table S4) and 761 genes whose mean expression across all three tumors (Figure 4B and Online Supplementary Table S5) was differentially expressed between SP and non-SP cells. Overall, the common expression profile showed a bias towards genes with increased expression in the SP, without a strong shared pattern of expression in the non-SP. This is not surprising as despite having been derived from genetically identical fish, individual tumors in this model are quite heterogeneous and have varied progression and functional parameters. PANTHER pathway analysis34 of the two differentially expressed gene sets identified Wnt signaling as the most overrepresented pathway (Figure 4C and Online Supplementary Table S6). The Wnt pathway has been implicated in regulating stem cells in normal and malignant tissues,35 shown to be necessary for self-renewal of LSC in acute myelogenous leukemia,36 and also been proposed to play a similar role in ALL.37,38
Figure 4.
Sorted side population cells from different tumors share similar expression profiles and increased Wnt signaling pathway gene expression. (A) Venn diagram representing the overlap of differentially expressed genes in the side population (SP) versus non-SP for three tumors: 272 genes were differentially regulated between the SP and non-SP in all three tumors. (B) Heatmap demonstrating differentially expressed genes between SP and non-SP cells. Cluster analysis performed using gene analysis set from mean expression across all three tumors (761 genes, listed in Online Supplementary Table S5). Expression shown as log2 fold change. (C) PANTHER pathway analysis of 272 differentially expressed genes highlighting Wnt signaling as the most overrepresented pathway.
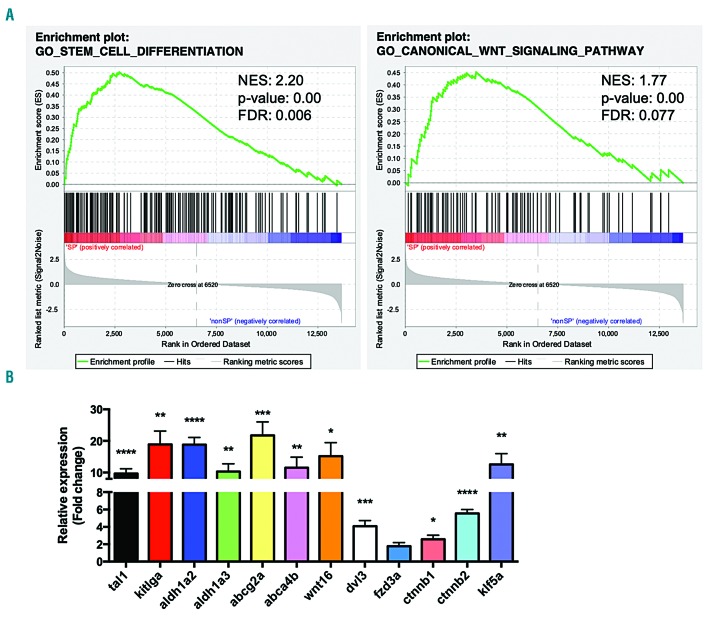
We further analyzed the entire RNA-sequencing dataset with gene set enrichment analysis (GSEA) using the Gene Ontology: Biological Processes gene set. This analysis identified 464 significantly upregulated pathways (Online Supplementary Table S7) including canonical Wnt signaling, and stem cell differentiation as the highest ranked gene set (Figure 5A). The majority of the 185 significantly downregulated pathways related to metabolic processes, including cell cycle transitions, protein translation and processing, and oxidative phosphorylation (Online Supplementary Table S7), suggesting that the SP includes an increased number of quiescent cells, another trait characteristic of stem cells.39,40 We validated expression of several stem cell genes and a panel of Wnt pathway genes by qPCR in four tumors sorted for SP and non-SP, which showed a significant increase in all but one of the tested genes (Figure 5B). Overall, the results described in this study demonstrate that the SP assay greatly enriches for LPC in zebrafish ALL and provides more evidence for the importance of the Wnt signaling pathway in LSC.
Figure 5.
Sorted side population cell gene expression is enriched for stem cell differentiation and canonical Wnt signaling. (A) Gene set enrichment analysis (GSEA) with the Gene Ontology (GO): biological processes gene sets for stem cell differentiation and canonical Wnt signaling. NES: normalized enrichment score; FDR: false discovery rate. (B) Quantitative polymerase chain reaction validation of selected stem/Wnt-associated genes identified in the side population (SP) versus non-SP differentially expressed genes and GSEA. Normalized to ef1a. Statistical significance determined by multiple t-tests: *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001 (n = 4).
Discussion
Although the importance of CSC to tumor maintenance, resistance, and relapse has long been proposed,41 the challenge of isolating and targeting these cells has been difficult to overcome. This is largely due to the simple fact that CSC are not equivalent to normal stem cells, and as such many of the markers used to isolate stem cells in specific tissues fail or underperform at isolating CSC. There is evidence that those tissue stem cell markers that do efficiently enrich for CSC in specific cancers, such as glioblastoma42 and acute myelogenous leukemia,43,44 are derived from transformed stem or early progenitor cells, although this point is still controversial. In malignancies without established CSC markers, such as ALL, the CSC can only be identified retrospectively, after it has demonstrated its ability to regrow a heterogeneous tumor, at which point the original cell is lost within the bulk tumor. This study establishes that the SP assay is a promising method for enriching LPC from ALL tumors, bypassing the lack of a reliable surface marker for isolation. This assay yields only an enrichment: not every LPC is found in the SP, nor are the SP cells exclusively LPC. However, this study demonstrates that the SP can be studied as a proxy for LSC to better understand their molecular drivers and, eventually, to identify better markers.
The Rag2-Myc zebrafish model has been extensively studied in the context of T-ALL, and because the tumors derive from the thymus, it was thought to produce ALL exclusively of T-cell origin. However, a recent study from Langenau’s laboratory has provided evidence for the existence of tumors of B-cell origin produced with this method, in addition to T-cell tumors.45 The origin of these B-ALL remains unclear, as they could be derived from early lymphoid progenitors located in the thymus, extra-thymic B-cell progenitors that home to the thymus after transformation, or possibly from B-cell precursors that line the thymus at later stages of development.46 Regardless, these tumors present a new opportunity to study B-ALL in zebrafish, a tumor type with few animal models.47 Another closely related model utilizing the human MYC oncogene was also recently demonstrated by Frazer’s team to produce both pre-B-ALL and T-ALL.48 Analysis of our own RNA-sequencing data showed a clear difference in expression of putative B- and T-cell genes between individual tumors, suggesting that ALL of both B- and T-cell origin are included in our dataset (Online Supplementary Figure S1). Our data indicate that SP cells from both T- and B-ALL share a common genetic signature and imply a likely role for Wnt/β-catenin signaling in stem cell pathways across a wide array of ALL. Further studies are needed to evaluate the impact of this signature on functional LSC and in human leukemia.
The identification of the Wnt signaling pathway associated with LSC has been reported previously, further validating our model system. Along with the observed enrichment of putative stem cell genes, the increased expression of Wnt pathway members in the zebrafish ALL SP is further evidence of the overlap between LSC and the SP. The Wnt pathway has been tied to normal stem cell and CSC function in many tissues and cancers35 including acute myelogenous leukemia36 and T-ALL.38 In a mouse T-ALL model, Giambra et al. found that leukemias with conditional loss of β-catenin1 had greatly impaired leukemia-propagating ability, but loss of β-catenin1 after the tumor had already been established did not affect disease progression. This coincides with our data highlighting a role for Wnt signaling in ALL specific to LSC. Our results also highlight a potential target for therapeutic intervention in Wnt16. While complete inhibition of the Wnt pathway could produce significant off-target effects, targeting an individual ligand isoform with a specific role in LSC may provide clinical benefit without substantial side effects. Wnt16 was significantly increased in the SP of all four tumors measured, and has been previously associated with transformation in a subset of ALL.49 While additional work will still be needed to validate Wnt16 as a potential therapeutic target, this and other targets identified through this study will be pursued to examine new treatments for ALL.
Supplementary Material
Acknowledgments
The authors would like to thank David Langenau and Sowmya Iyer for their help with the gene set enrichment analysis. We also thank Wilfredo Marin, Paula Herbst and Isabel Scharf for excellent fish care and technical support.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/7/1388
References
- 1.Adorno-Cruz V, Kibria G, Liu X, et al. Cancer stem cells: targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res. 2015;75(6):924–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson K, Lutz C, van Delft FW, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. [DOI] [PubMed] [Google Scholar]
- 3.Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322(5906):1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Notta F, Mullighan CG, Wang JC, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469(7330):362–367. [DOI] [PubMed] [Google Scholar]
- 6.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541–1552. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg JM, Silverman LB, Levy DE, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21(19):3616–3622. [DOI] [PubMed] [Google Scholar]
- 8.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371 (9617):1030–1043. [DOI] [PubMed] [Google Scholar]
- 9.Van Vlierberghe P, Ambesi-Impiombato A, De Keersmaecker K, et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122(1):74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355(12):1253–1261. [DOI] [PubMed] [Google Scholar]
- 11.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183(4):1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asakura A, Seale P, Girgis-Gabardo A, Rudnicki MA. Myogenic specification of side population cells in skeletal muscle. J Cell Biol. 2002;159(1):123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summer R, Kotton DN, Sun X, Ma B, Fitzsimmons K, Fine A. Side population cells and Bcrp1 expression in lung. Am J Physiol Lung Cell Mol Physiol. 2003;285(1):L97–104. [DOI] [PubMed] [Google Scholar]
- 14.Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245(1):42–56. [DOI] [PubMed] [Google Scholar]
- 15.Shimano K, Satake M, Okaya A, et al. Hepatic oval cells have the side population phenotype defined by expression of ATP-binding cassette transporter ABCG2/BCRP1. Am J Pathol. 2003;163(1):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Han Y, Cheng K, Zhang G, Wang X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47(6):587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Teng JS. Increased multi-drug resistance and reduced apoptosis in osteosarcoma side population cells are crucial factors for tumor recurrence. Exp Ther Med. 2016;12(1):81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi S, Zheng J, Zhu H, Yang L, Xiao X. Identification of neuroblastoma stem cells by characterization of side population cells in the human neuroblastoma SK-N-SH cell line. J Pediatr Surg. 2010;45(12):2305–2311. [DOI] [PubMed] [Google Scholar]
- 19.Chiarini F, Grimaldi C, Ricci F, et al. Activity of the novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor NVP-BEZ235 against T-cell acute lymphoblastic leukemia. Cancer Res. 2010;70(20):8097–8107. [DOI] [PubMed] [Google Scholar]
- 20.Langenau DM, Traver D, Ferrando AA, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299(5608):887–890. [DOI] [PubMed] [Google Scholar]
- 21.Rudner LA, Brown KH, Dobrinski KP, et al. Shared acquired genomic changes in zebrafish and human T-ALL. Oncogene. 2011;30(41):4289–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackburn JS, Liu S, Raiser DM, et al. Notch signaling expands a pre-malignant pool of T-cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia. 2012;26(9):2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1-2):70–78. [DOI] [PubMed] [Google Scholar]
- 24.Mizgireuv IV, Revskoy SY. Transplantable tumor lines generated in clonal zebrafish. Cancer Res. 2006;66(6):3120–3125. [DOI] [PubMed] [Google Scholar]
- 25.Jessen JR, Jessen TN, Vogel SS, Lin S. Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons. Genesis. 2001;29 (4):156–162. [DOI] [PubMed] [Google Scholar]
- 26.Langenau DM, Traver D, Ferrando AA, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299(5608):887–890. [DOI] [PubMed] [Google Scholar]
- 27.M KK, Esther F, Clemens G, et al. The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgene-sis constructs. Dev Dyn. 2007;236(11):3088–3099. [DOI] [PubMed] [Google Scholar]
- 28.Pruitt MM, Marin W, Waarts MR, de Jong JLO. Isolation of the side population in myc-induced T-cell acute lymphoblastic leukemia in zebrafish. J Vis Exp. 2017;(123). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith AC, Raimondi AR, Salthouse CD, et al. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 2010;115(16): 3296–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackburn JS, Liu S, Langenau DM. Quantifying the frequency of tumor-propagating cells using limiting dilution cell transplantation in syngeneic zebrafish. J Vis Exp. 2011;(53):e2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1):70–78. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 33.Blackburn JS, Liu S, Wilder JL, et al. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell. 2014;25(3):366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mi H, Huang X, Muruganujan A, et al. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wend P, Holland JD, Ziebold U, Birchmeier W. Wnt signaling in stem and cancer stem cells. Semin Cell Dev Biol. 2010;21(8):855–863. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belmonte M, Hoofd C, Weng AP, Giambra V. Targeting leukemia stem cells: which pathways drive self-renewal activity in T-cell acute lymphoblastic leukemia? Curr Oncol. 2016;23(1):34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giambra V, Jenkins CE, Lam SH, et al. Leukemia stem cells in T-ALL require active Hif1a and Wnt signaling. Blood. 2015;125 (25):3917–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Bhatia R. Stem cell quiescence. Clin Cancer Res. 2011;17(15):4936–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shyh-Chang N, Ng H-H. The metabolic programming of stem cells. Genes Dev. 2017;31(4):336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makino S. The role of tumor stem-cells in regrowth of the tumor following drastic applications. Acta Unio Int Contra Cancrum. 1959;15(Suppl 1):196–198. [PubMed] [Google Scholar]
- 42.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7)730–737. [DOI] [PubMed] [Google Scholar]
- 44.Pollyea DA, Gutman JA, Gore L, Smith CA, Jordan CT. Targeting acute myeloid leukemia stem cells: a review and principles for the development of clinical trials. Haematologica. 2014;99(8):1277–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia EG, Iyer S, Garcia SP, et al. Cell of origin dictates aggression and stem cell number in acute lymphoblastic leukemia. Leukemia. 2018;32(8):1860–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Li Y-S, Shinton SA, et al. Zebrafish B cell development without a pre-B cell stage, revealed by CD79 fluorescence reporter transgenes. J Immunol. 2017;199(5):1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baeten JT, Jong JLOd. Genetic models of leukemia in zebrafish. Front Cell Dev Biol. 2018;6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borga C, Park G, Foster C, et al. Simultaneous B and T cell acute lymphoblastic leukemias in zebrafish driven by transgenic MYC: implications for oncogene-sis and lymphopoiesis. Leukemia. 2019;33 (2):333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazieres J, You L, He B, et al. Inhibition of Wnt16 in human acute lymphoblastoid leukemia cells containing the t(1;19) translocation induces apoptosis. Oncogene. 2005;24(34):5396–5400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.