Cancers of the same histotype may show profound differences in clinical behavior because they can arise from cells having acquired multiple mutations, which presumably occur over many years.1 In contrast, it is well known that cancers with different morphologies and phenotypes, and those that originate in different organs, may be clonally related.2–4 Here we present a case exhibiting different lymphoma entities occurring in the same patient, exemplifying major intratumoral heterogeneity at the morphological, immunophenotypic, and genetic levels.
A 52-year old Caucasian woman presented with seizures in March 2007. A contrast-enhanced magnetic resonance image (MRI) scan of her brain demonstrated an irregular space-occupying lesion in the left occipital cortex. Due to the presence of a clinically symptomatic lesion, the patient started steroid treatment before the diagnostic biopsy could be performed, and the lesion was no longer visible in a subsequent MRI scan.
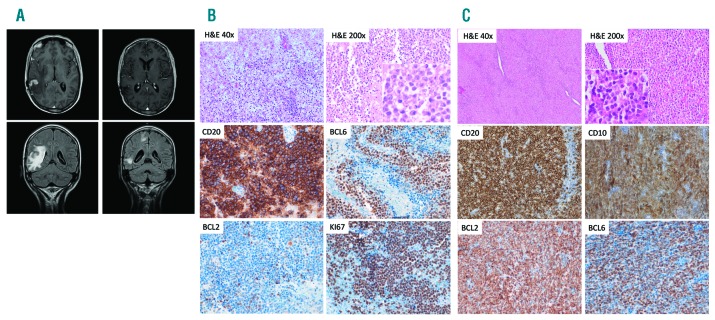
She returned in May 2008 with nausea, vomiting, instability and holocranial headache. Peripheral lymphadenopathies were not identified in a physical examination. An MRI scan of the brain revealed a 1 cm lesion in the white matter of the right parietal region, in addition to millimetric lesions in the supratentorial white matter (Figure 1A). A computed tomography scan and positron emission tomography/computed tomography did not reveal any lesions other than those in the brain.
Figure 1.
Radiological and pathological characteristics of the patient’s tumors. (A) Magnetic resonance imaging scan of the brain at diagnosis (2008) and after therapy. (B) Brain biopsy showing extensive infiltration of the parenchyma by primary central nervous system lymphoma. (C) Lymph node biopsy (1998), showing distinctive features of follicular lymphoma. Insets show cytological details.
Laboratory tests showed a normal hemogram without leukocytosis. All biochemical tests gave results within the normal range, except for a slightly elevated lactate dehydrogenase level (568 IU/L; normal 266-500 IU/L). Serological studies for hepatitis B virus, hepatitis C virus, Epstein-Barr virus, and human immunodeficiency virus all yielded negative results.
A cerebral biopsy was performed and showed extensive infiltration of the cerebral parenchyma by a lymphoproliferative process, with a diffuse and perivascular pattern and areas of necrosis, consisting of large atypical cells with a centroblastic or plasmablastic appearance (Figure 1B). Immunohistochemistry and in situ hybridization studies were performed using standard routine procedures. They revealed a germinal center B-cell phenotype that was positive for CD20, CD79a, BCL6, CD10, and negative for MUM1, BCL2, and p53, with a very high proliferative index (Ki67) (close to 100%). The in situ hybridization study for Epstein-Barr virus (EBER) gave a negative result. Fluorescence in situ hybridization studies for the t(8; 14) (IGH/BCL2) translocation and other MYC rearrangements also yielded negative results. Given these observations, a primary central nervous system lymphoma (PCNSL)5 was diagnosed.
Flow cytometry analysis of the cerebrospinal fluid identified 6 cells/μL, 2% of which were large CD22+ B-lymphocytes with heterogeneous expression of CD20, consistent with infiltration by PCNSL. A bone marrow biopsy was negative for tumor infiltration.
The patient completed five cycles of chemotherapy with rituximab, methotrexate, vincristine, plus procarbazine, combined with intrathecal liposomal cytarabine (cycles 1 and 3). Subsequently, whole-brain radiotherapy was given, with a total dose of 36 Gy. She finally received systemic cytarabine. An MRI scan of the brain was performed 3 months after the end of the treatment and positron emission tomography/computed tomography was carried out after 6 months. Both studies revealed a complete remission with cicatricial changes (Figure 1A). Follow-up MRI scans were performed every 6 months for 1 year and annually thereafter, without detecting relapse of the disease.
Remarkably, the patient’s clinical records indicated a previous medical history of nodal lymphoma, diagnosed in 1998 in another institution in an incidental inguinal lymph node. After obtaining approval from the institutional review board and written informed consent from the patient, a representative formalin-fixed, paraffin-embedded tissue block was obtained and the pathology, including histopathology and immunohistochemistry, was reviewed (Figure 1C). The lymph node biopsy showed complete effacement of the architecture by a lymphoproliferative process with a follicular growth pattern, composed of small lymphocytes with a centrocytic appearance, and focal marginal-zone differentiation. Tumor cells showed the immunohistochemical characteristics of follicle lymphoma (FL),5 being positive for B-cell markers, CD10, BCL2, and BCL6, and negative for CD5, CD23, IgD, Cyclin D1, and MNDA.6 The proliferative index (Ki67) was very low (2-5%). The case was classified at that time as localized stage I disease, and was treated with six cycles of cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) plus radiotherapy, which produced complete remission. The patient never manifested splenomegaly, peripheral blood, and/or bone marrow infiltration, either at diagnosis or during her follow up.
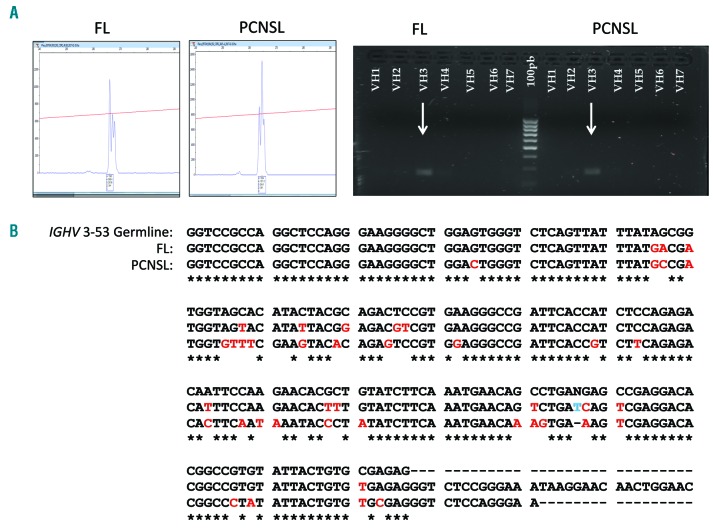
Even though the clinical presentation, chronology of the lesions, and histopathological features indicated that a clonal relationship between the two tumors in the present case was extremely unlikely, we performed a routine IGH clonality analysis (Biomed-2 protocol)7 using the IGH Vн-FR2-Jн and Vн-FR3-Jн primer sets. IGHV family usage was determined using single primers for the different Vн families, as previously described.8 The results demonstrated monoclonal peaks of almost identical size (265-266 bp) in both lesions. Both lymphomas also exhibited an identical Vн3 rearrangement (Figure 2A).
Figure 2.
IGH clonality analysis of the patient’s tumors. (A) Monoclonal Vн-FR2-Jн rearrangements and Vн3 usage. (B) Sanger sequencing of the rearranged regions showing hypermutated IGHV 3-53 sequences and ongoing mutations in both lymphomas: FL (91.9 % homology with the germline) and PCNSL (82.2% homology). Asterisks indicate identity with germline. FL: follicular lymphoma; PCNSL: primary central nervous system lymphoma.
Conventional Sanger sequencing of the rearranged Vн-FR2-Jн regions and comparison with the reference germline genes (NCBI IGBLAST database) demonstrated somatically hypermutated sequences that had originated from IGHV 3-53 germline genes in both lymphomas. The chronologically first tumor, FL, was 91.9% homologous with the germline and the second tumor, PCNSL, exhibited 82.2% homology. The two lesions shared 13 mutations, thereby demonstrating the existence of a common lymphoid precursor. In addition, we found four private mutations in the FL and 16 private mutations in the PCNSL. Figure 2B illustrates the comparisons of all these variants.
Taking into account these results and from a conceptual point of view, the brain tumor does not represent a true primary brain lymphoma, but a neoplasia clonally related to the previous FL. It is also noteworthy that this tumor was of germinal center type according to the Hans algorithm, whereas most true PCNSL are of activated B-cell type.5
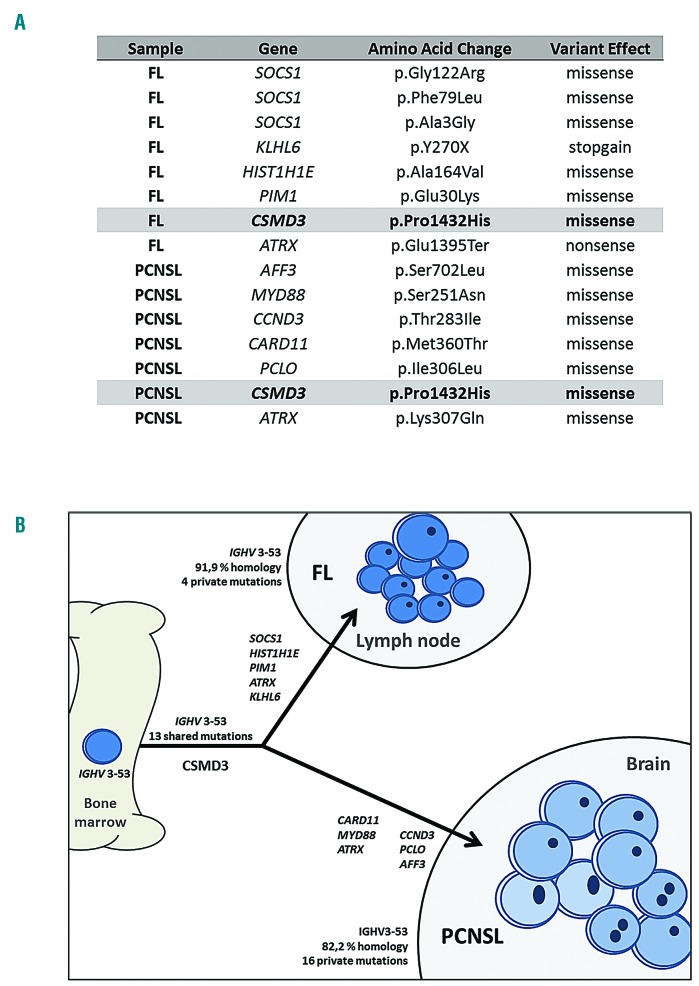
Once the clonal relationship between the tumors had been demonstrated by IGH analyses, the mutational profiles of the two tumors were further analyzed using next-generation sequencing with a targeted custom panel of 110 genes recurrently mutated in B-cell lymphomas (Online Supplementary Table S1).9,10 Libraries were prepared using the HaloPlex Target Enrichment protocol (Agilent, Santa Clara, CA, USA) from 30-50 ng of input DNA extracted from formalin-fixed, paraffin-embedded samples, and sequenced using Ion Torrent S5 (Thermo Fisher Scientific, NY, USA) technology. Variant calling was performed with software tools available in the instrument (Torrent Suite v4.0.2). Polymorphic variants were filtered using germline DNA extracted from normal bone marrow aspirate smears. Figure 3 and Online Supplementary Table S2 summarize the variants: the first tumor had mutations affecting six genes and the second tumor exhibited mutations affecting seven genes. Only one mutated gene was common to both tumors (CSMD3 c.4295C>A, P1432H). The brain lymphoma showed several mutations that are rather typical of PCNSL, including MYD88 and CARD11, and rare in FL.
Figure 3.
Mutational analysis of the tumors. (A) Summary of variants detected by next-generation sequencing in the two tumors. (B) Mutational relationship and branched pattern. FL: follicular lymphoma; PCNSL: primary central nervous system lymphoma.
This peculiar landscape, in which most of the mutations are private in each tumor, supports the hypothesis of a distant common precursor that underwent branched evolution, giving rise to two almost independent neoplasms. Nevertheless, they may be clonally related, since IGH gene rearrangement is a very early phenomenon in normal lymphoid maturation. Since the occurrence of BCL2 translocation is also considered an early event in most FL, we performed new cytogenetic analyses using fluorescence in situ hybridization for the translocation t(14;18)(IGH/BCL2) in both samples, with negative results. Supplementary fluorescence in situ hybridization analyses for BCL6 and MYC rearrangements also yielded negative results. BCL2 mutations were not identified in any sample. The next-generation sequencing panel covered the BCL2 promoter, exon 1 and 2 regions, including intronic sequences (Online Supplementary Table S1), and it could, therefore, detect any variant (via the somatic hypermutation machinery or variants affecting the antibody binding domain).11
The only shared variant affected CSMD3, which is a gene that is physiologically expressed mainly in the brain. This gene has been suggested to be a tumor suppressor in several tumor types, including oligodendrogliomas. Interestingly, among hematolymphoid diseases, mutations detected in CSMD3 have been described in PCNSL, suggesting that alterations in genes having a role in central nervous system development, which would have been acquired by a common precursor, may facilitate late lymphoma central nervous system tropism in PCNSL.12
The present case illustrates discordant lymphomas as an example of spatial heterogeneity and branched evolution,1 through the occurrence of two histologically distinct lymphoma types in different anatomical locations. Spatial heterogeneity is a phenomenon that can be driven by genomic instability, epigenetic events, interaction with the tumor microenvironment and stroma, and stochastic variations in cellular development and antitumor therapies. These mechanisms may lead to branched sub-clonal evolution from a common progenitor clone, resulting in spatial variation between different tumor sites and differences in histotypes and phenotypes.13 In contrast, tumor transformation is an example of temporal heterogeneity: progression of a low-grade to a high-grade lymphoma during the course of the disease,14 representing a model of linear clonal evolution. Transformation is a phenomenon usually associated with treatment resistance, clinical disease progression, and increased disease-specific mortality.3,15
Supplementary Material
Acknowledgments
We are indebted to Mar Lopez for her expertise and excellent technical assistance with the IGH clonality experiments.
Footnotes
Funding: this work was supported by grants from the Plan Nacional de I+D+I, co-financed by the ISCIII-Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER), PI12/1832, PI17/00020, CIBERONC - CB16/12/00291); Programas para Grupos de Investigación de la Comunidad Autónoma de Madrid (Biomedicina 2017); Fundación Carolina-Fundación BBVA; and the Spanish Association for Cancer Research (AECC). MSB currently holds a Miguel Servet II contract (CPII16/00024) from ISCIII-MINECO-AES-FEDER (Plan Estatal I+D+I 2013-2016).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Schürch CM, Federmann B, Quintanilla-Martinez L, Fend F. Tumor heterogeneity in lymphomas: a different breed. Pathobiology. 2018;85(1-2):130–145. [DOI] [PubMed] [Google Scholar]
- 2.Jiang Y, Redmond D, Nie K, et al. Deep sequencing reveals clonal evolution patterns and mutation events associated with relapse in B-cell lymphomas. Genome Biol. 2014;15(8):432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green MR, Gentles AJ, Nair RV, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121(9):1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landau DA, Carter SL, Getz G, Wu CJ. Clonal evolution in hematologic malignancies and therapeutic implications. Leukemia. 2014; 28(1):34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metcalf RA, Monabati A, Vyas M, et al. Myeloid cell nuclear differentiation antigen is expressed in a subset of marginal zone lymphomas and is useful in the differential diagnosis with follicular lymphoma. Hum Pathol. 2014;45(8):1730–1736. [DOI] [PubMed] [Google Scholar]
- 7.Evans PA, Pott C, Groenen PJ, et al. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia. 2007;21(2):207–214. [DOI] [PubMed] [Google Scholar]
- 8.McDonald TJ, Kuo L, Kuo FC. Determination of VH family usage in B-cell malignancies via the BIOMED-2 IGH PCR clonality assay. Am J Clin Pathol. 2017;147(6):549–556. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Grubor V, Love CL, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1398–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011; 43(9):830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkhard R, Bhagat G, Cogliatti SB, et al. BCL2 mutation spectrum in B-cell non-Hodgkin lymphomas and patterns associated with evolution of follicular lymphoma. Hematol Oncol. 2015;33(1):23–30. [DOI] [PubMed] [Google Scholar]
- 12.Vater I, Montesinos-Rongen M, Schlesner M, et al. The mutational pattern of primary lymphoma of the central nervous system determined by whole-exome sequencing. Leukemia. 2015;29(3):677–685. [DOI] [PubMed] [Google Scholar]
- 13.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012; 486(7403):395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agbay RL, Loghavi S, Medeiros LJ, Khoury JD. High-grade transformation of low-grade B-cell lymphoma: pathology and molecular pathogenesis. Am J Surg Pathol. 2016;40(1):e1–16. [DOI] [PubMed] [Google Scholar]
- 15.Kreso A, O’Brien CA, van Galen P, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339(6119):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.