Abstract
ERG-deletions occur recurrently in acute lymphoblastic leukemia, especially in the DUX4-rearranged subtype. The ERG-deletion was shown to positively impact prognosis of patients with IKZF1-deletion and its presence precludes assignment into IKZF1plus group, a novel high-risk category on AIEOP-BFM ALL trials. We analyzed the impact of different methods on ERG-deletion detection rate, evaluated ERG-deletion as a potential marker for DUX4-rearranged leukemia, studied its associations with molecular and clinical characteristics within this leukemia subtype, and analyzed its clonality. Using single-nucleotide-polymorphism array, genomic polymerase chain reaction (PCR) and amplicon-sequencing we found ERG-deletion in 34% (16 of 47), 66% (33 of 50) and 78% (39 of 50) of DUX4-rearranged leukemia, respectively. False negativity of ERG-deletion by single-nucleotide-polymorphism array caused IKZF1plus misclassification in 5 patients. No ERG-deletion was found outside the DUX4-rearranged cases. Within DUX4-rearranged leukemia, the ERG-deletion was associated with higher total number of copy-number aberrations, and, importantly, the ERG-deletion positivity by PCR was associated with better outcome [5-year event-free survival (EFS), ERG-deletion-positive 93% vs. ERG-deletion-negative 68%, P=0.022; 5-year overall survival (OS), ERG-deletion-positive 97% vs. ERG-deletion-negative 75%, P=0.029]. Ultra-deep amplicon-sequencing revealed distinct co-existing ERG-deletions in 22 of 24 patients. In conclusion, our data demonstrate inadequate sensitivity of single-nucleotide-polymorphism array for ERG-deletion detection, unacceptable for proper IKZF1plus classification. Even using more sensitive methods (PCR/amplicon-sequencing) for its detection, ERG-deletion is absent in 22-34% of DUX4-rearranged leukemia and does not represent an adequately sensitive marker of this leukemia subtype. Importantly, the ERG-deletion potentially stratifies the DUX4-rearranged leukemia into biologically/clinically distinct subsets. Frequent polyclonal pattern of ERG-deletions shows that late origin of this lesion is more common than has been previously described.
Introduction
ERG (ETS transcription factor) gene deletions (ERGdel) can be found in 3-7% of pediatric B-cell precursor (BCP) acute lymphoblastic leukemia (ALL).1–3 It occurs almost exclusively in B-other ALL, a heterogeneous subset comprising 20-25% of pediatric BCP-ALL, defined by the absence of routinely tested (cyto)genetic classifying lesions. Two parallel European studies demonstrated that ERGdel frequently co-occurs with deletion of the IKZF1 gene (IKZF1del) and attenuates its negative prognostic impact.1,3 Subsequent study further investigating the prognostic impact of IKZF1del in a context of additional gene copy number changes identified a genetic pattern associated with poor outcome in children treated according to the AIEOP-BFM ALL 2000 protocol.4 This “IKZF1plus” genetic pattern is defined by co-occurrence of the IKZF1del with deletion of CDKN2A, CDKN2B, PAX5 and/or PAR1 region in the absence of ERGdel and will be used to refine risk stratification in the upcoming AIEOP-BFM ALL 2017 trial. The analysis of ERGdel thus becomes a part of the routine diagnostic algorithm in this large international clinical trial.
Several methods with different coverage and sensitivity can be used to detect ERGdel. Single nucleotide polymorphism (SNP) array can reveal any type of deletion (of minimal length varying with SNP array density) present in a major clone, representing more than 20-30% of the analyzed sample. Multiplex ligation-dependent probe amplification (MLPA) has similar sensitivity while the coverage depends on the design of the probe in the kits used. Genomic PCR has at least ten times higher sensitivity compared to SNP array/MLPA, while it only detects the most common deletion types which were considered in the PCR primer design. Importantly, the PCR approach was used in all the three aforementioned studies defining the prognostic impact of ERGdel in the context of IKZF1 del1,3 and IKZF1plus.4 Since ERGdel frequently occurs at subclonal levels that SNP arrays and MLPA fail to detect due to lower sensitivity,1 the genomic PCR should be used for diagnostics in accordance with these studies and the established definition of IKZF1plus. However, the genomic PCR-based ERGdel detection method used in those studies1,4,5 suffers from at least two disadvantages: 1) due to variable length of amplified region and uneven level of ERGdel-positive clones, the PCR products must be sequence-verified; and 2) strict precautions must be adopted when using Sanger sequencing in order to avoid carry-over contamination of samples, and sequence analysis can be complicated in samples with more distinct PCR products.
Based on the frequent subclonality of ERGdel and its instability between diagnosis and relapse, the ERGdel is considered a passenger genetic lesion.1,3 The early ALL gene expression profiling studies showed that it is specific to a subset of B-other ALL with a unique gene expression signature, likely representing a novel biological subtype.6,7 This novel ALL subtype was confirmed and further characterized by several recent studies, which also revealed its common genetic background, i.e. rearrangements of the DUX4 gene (DUX4r).7–9 In DUX4r-ALL, the expression of DUX4 (physiologically silent in somatic tissues) is activated by juxtaposition under the control of ectopic regulatory element, most frequently the IGH gene enhancer. The ERG gene was identified among direct DUX4 targets in DUX4r-ALL. It has been demonstrated that DUX4 deregulates ERG gene transcription in a complex manner; it induces expression of non-canonical ERG transcripts, including ERGalt which inhibits wild-type ERG and promotes leukemogenesis in mice. It is possible that DUX4 also renders the ERG gene prone to deletions via inducing (epi)chromatin changes.9 The DUX4r-ALL subtype has only recently been defined and for the moment there are no simple tools for DUX4r detection and DUX4r-ALL classification; thus, there are no data on its prognostic impact. A single American study reported favorable outcome of DUX4r-ALL suggesting that the favorable prognostic impact of ERGdel observed earlier is indeed inherent to this ALL subtype.9 However, ERGdel can only be found in a subset of DUX4r-ALL and its prognostic impact within this subgroup has not been explicitly addressed.
Here we analyzed the presence of ERGdel by SNP array, genomic PCR followed by Sanger sequencing, and by a newly-designed deep amplicon sequencing procedure (AmpliSeq). We aimed to determine to what extent the different sensitivity of methods impacts ERGdel detection and classification into IKZF1plus category. Next, we wanted to assess whether ERGdel can be detected in a significantly higher proportion of DUX4r-positive patients using a potentially more sensitive method and to what extent positivity could serve as a surrogate marker for the DUX4r-ALL classification. Moreover, we wanted to elucidate whether ERGdel-positive DUX4r-ALL differs from ERGdel-negative DUX4r-ALL, and whether the possible differences depend on the method used to detect ERGdel. Finally, use of AmpliSeq enabled us to study in detail also the repertoire and clonality of ERGdel in order to better understand its origin during leukemia clone evolution.
Methods
Patients and samples
The study analyzed diagnostic and remission bone marrow or peripheral blood samples from 118 children (aged 1-18 years) diagnosed with B-other ALL (negative for ETV6/RUNX1, TCF3/PBX1, BCR/ABL1, KMT2A-rearrangements, hyperdiploidy, hypodiploidy) and treated in the Czech Republic between August 1998 and July 2017 according to the BFM ALL protocols. Patients were selected according to the availability of the biological material and/or of already existing genomic data. A retrospective part of the cohort (treated August 1998-July 2010; n=30) was enriched for patients presenting with immunophenotypic features shown to be associated with ERGdel (CD2-positivity, immunophenotypic switch).10 The remaining 88 patients represent 84% of all consecutively diagnosed and prospectively analyzed B-other patients treated according to the AIEOP BFM ALL 2009 protocol (consecutive sub-cohort; August 2010 – July 2017). The study was approved by the Institutional Review Board of the University Hospital Motol and informed consent was obtained in accordance with the Declaration of Helsinki.
Genomic polymerase chain reaction to detect ERG (ETS transcription factor) deletions (ERGdel)
The presence of ERGdel was analyzed by multiplex PCR, as described previously.3 Two more primers corresponding to additional centromeric breakpoint sites1 were added: 5’-GCGGCTACTTGTTGGTCCAAGAA-3’ and 5’-CTATCCTGAACATTGCTGCCAG-3’. PCR products were analyzed on agarose gel; positive samples were sequenced by Sanger method.
Single nucleotide polymorphism array
Copy number aberrations (CNA) and regions of uniparental disomy (UPD) were analyzed in 104 patients using HumanOmni Express BeadChip (Illumina, San Diego, CA, USA) or CytoScan HD arrays (Affymetrix, Santa Clara, CA, USA). For six patients, the results from the analysis on GeneChip Mapping 250K Nsp and Sty arrays (Affymetrix, Santa Clara, CA, USA) were available from our previous study.10
DUX4r-acute lymphoblastic leukemia classification, analysis of DUX4, RAG1 and RAG2 expression
DUX4r-ALL was classified by supervised hierarchical clustering of patients based on expression of DUX4r-ALL signature genes (the top 150 up-regulated and the top 150 down-regulated genes in DUX4r-ALL compared to non-DUX4r-ALL9). Gene expression profiling was performed by whole transcriptome sequencing (RNAseq) and/or on microarrays, as described previously.11,12 Expression of DUX4, RAG1 and RAG2 was analyzed using data from RNAseq. RAG1/RAG2 reads were aligned and counted using hg19 reference genome.13 DUX4 reads were mapped to the DUX4 reference sequence and counted as described previously.9 Read counts were normalized using library size factor computed using R package Deseq2.14
Analysis of ERGalt expression
Expression of “ERGalt a” and “ERGalt b”10 was analyzed using RNAseq data. Reads containing the sequences specific for these ERGalt transcripts were counted and normalized by library size factors.
Amplicon sequencing
Libraries for the amplicon sequencing were prepared by one round multiplex PCR using FastStart™ High Fidelity PCR System (Roche, Basel, Switzerland). PCR primers used to amplify ERGdel spanning region and 1-2 control amplicons are listed in Online Supplementary Table S1 and their schematic position is shown in Figure 1. Sequencing was performed on an Ion Torrent PGM sequencer (Life Technologies, Carlsbad, CA, USA) using 400 bp chemistry according to the manufacturer’s instructions (Life Technologies). Reads were successively mapped to a custom reference using the Burrows-Wheeler Alignment (bwa) tool. First, reads were mapped to a custom reference comprising reference sequences for both control amplicons and reference sequences surrounding the five ERG 3’ breakpoint site clusters. Next, from all reads partially mapped on ERG 3’ breakpoint site clusters, the unmapped parts were exported and mapped against the reference sequence surrounding the common ERG 5’ breakpoint site. The unmapped parts of reads in between segments mapped to 3’and 5’ breakpoint site references were considered inserted non-templated nucleotides (N-segment). Data analysis was the same as that used for V-(D)-J rearrangements of immunoglobulin/T-cell receptor genes. Identified ERGdel alleles were defined by the position of last non-deleted 5’ nucleotide, N-segment (inserted non-tem-plated nucleotides), type of utilized 3’ breakpoint site cluster, and position of the first non-deleted 3’ nucleotide. Sequencing setting (target coverage, choice of control amplicons) and the coverage achieved are shown in Online Supplementary Table S2.
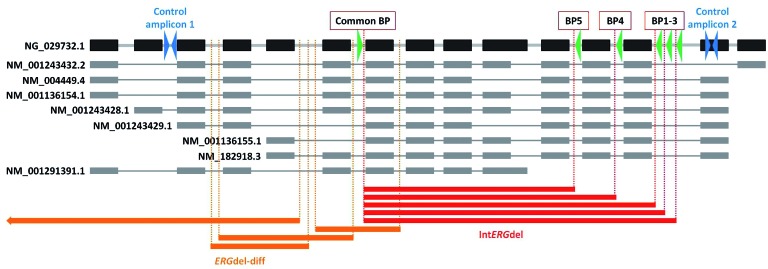
Figure 1.
Schematic representation of the ERG gene, its transcript variants, and different types of ERGdel found in the present study. Black (gray) boxes represent exons of the gene (individual transcript variants). Accession numbers for reference sequences from NCBI Reference Sequence Database are shown. The most common types of the deletion (IntERGdel) are shown in red, other types (ERGdel-diff) are shown in orange. Positions of primers used for the amplification of IntERGdel spanning region (green triangles) and 2 control amplicons (blue triangles) are shown. BP: breakpoint site.
Statistical analysis
The Mann-Whitney U test was used to compare numerical parameters in DUX4r-ALL stratified by ERGdel. The two-tailed Fisher exact probability test was used to compare frequencies. The Kaplan-Meier method was used to estimate survival rates, differences were compared with the two-sided log-rank test. Event-free survival (EFS) was defined as the time from diagnosis to the date of last follow up in complete remission or to the first event. Events were resistance to therapy (non-response), relapse, secondary neoplasm, or death from any cause. Failure to achieve remission due to early death or non-response was considered as events at time zero. Patients lost to follow up were censored at the time of their withdrawal.
Further details of the methods used are available in the Online Supplementary Appendix.
Results
Frequency of ERGdel in DUX4r- and non-DUX4r-acute lymphoblastic leukemia: performance of different ERGdel screening methods
We studied the presence of ERGdel in a cohort of 118 B-other ALL patients of whom 50 and 68 were assigned into the DUX4r or the non-DUX4r ALL subgroups, respectively, based on the presence of DUX4r-specific gene expression signature9 and DUX4 gene rearrangements (see Online Supplementary Results).
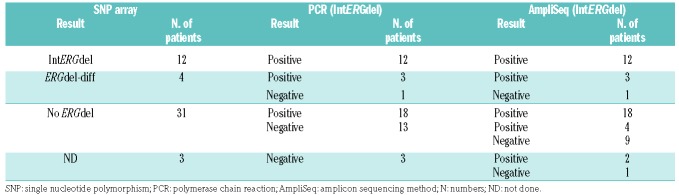
Using SNP array, we found ERGdel in 16 of 47 (34%) DUX4r-ALL patients. In 12 of 16 positive patients, the SNP array findings corresponded to the most frequent type of ERGdel, targeted by PCR/AmpliSeq (IntERGdel), while a different type of ERGdel was found in four of 16 positive patients (ERGdel-diff) (Table 1 and Figure 1). Using PCR, we found ERGdel in 33 of 50 (66%) DUX4r-ALL. As expected, all 12 patients with IntERGdel detected by SNP array were positive by PCR; surprisingly, also three of four patients with ERGdel-diff detected by SNP array were positive by PCR, demonstrating co-existence of various deletion types, each in a different proportion of cells. Importantly, PCR revealed IntERGdel in 18 of 31 patients negative by SNP array. Using AmpliSeq, ERGdel was detected in 39 of 50 (78%) of patients with DUX4r-ALL, including all 33 PCR-positive and an additional six of 17 PCR-negative patients.
Table 1.
Results of ERGdel screening by three different methods in 50 DUX4r-acute lymphoblastic leukemia.
In non-DUX4r-ALL, all 63 and 68 patients tested by SNP array and PCR, respectively, were ERGdel-negative. No ERGdel was found in nine non-DUX4r patients tested by AmpliSeq at higher coverage setting.
Prognostic impact of DUX4r and ERGdel
Neither the whole cohort (n=118) nor its consecutively analyzed part (n=88) showed a significant difference in EFS or OS between the DUX4r-ALL patients and the non-DUX4r B-other ALL (Online Supplementary Figure S5). Similarly, despite the fact that, according to all the analyses, patients with ERGdel fared better than patients lacking the deletion, the difference between ERGdel-positive and ERGdel-negative patients within the whole B-other ALL cohort did not reach a statistical significance, whatever method for the ERGdel detection was used (SNP array, PCR, AmpliSeq or combination of all) (Online Supplementary Figure S6).
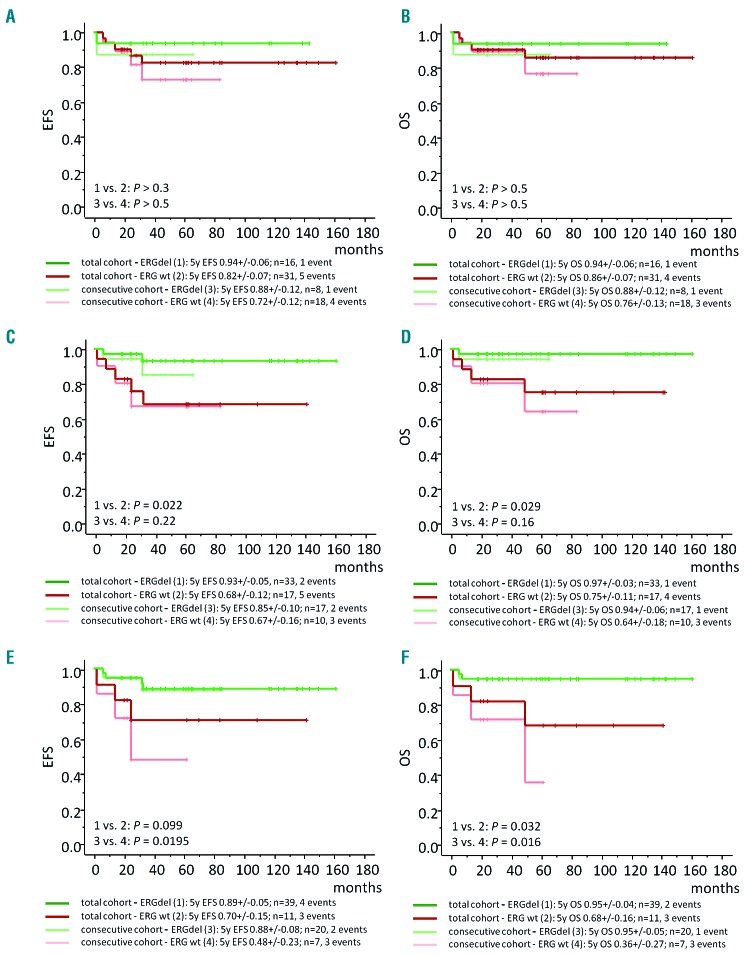
When the prognostic impact of the ERGdel was analyzed only within DUX4r-ALL, its favorable effect on outcome was again statistically non-significant when SNP array results were taken into account (combining both IntERGdel and ERGdel-diff patients; P>0.3). When PCR or AmpliSeq results were used, the positive prognostic impact of ERGdel was statistically significant in the whole DUX4r-ALL cohort (n=50) (PCR: EFS, P=0.022; OS, P=0.029. AmpliSeq: EFS, P=0.099; OS, P=0.032) and for AmpliSeq also in its consecutive part (n=27) (EFS, P=0.020; OS, P=0.016) (Figure 2).
Figure 2.
Impact of ERGdel on survival within acute lymphoblastic leukemia (DUX4r-ALL). Five-year event-free survival (EFS) (A, C, E) and overall survival (OS) (B, D, F) of DUX4r-ALL patients stratified according to the presence of ERGdel as defined by single nucleotide polymorphism array (A and B), genomic polymerase chain reaction (C and D), and amplicon sequencing (AmpliSeq) (E and F). n: number.
Expression of DUX4, RAG1 and RAG2 and ERGalt
Similarly to other recurrent deletions in BCP-ALL, ERGdel is thought to represent a result of illegitimate RAG1/RAG2-mediated V-(D)-J recombination. This theory is strongly supported by the presence of sequence motifs highly homologous to recombination signal sequences in close proximity to ERGdel breakpoints.3 It has also been suggested that DUX4 may facilitate ERGdel by increasing accessibility of ERG gene locus.9 We analyzed expression levels of DUX4, RAG1 and RAG2 in 44 DUX4r-ALL with available RNAseq data and did not find any difference between patients stratified by ERGdel presence based on results of any of the detection methods used (data not shown).
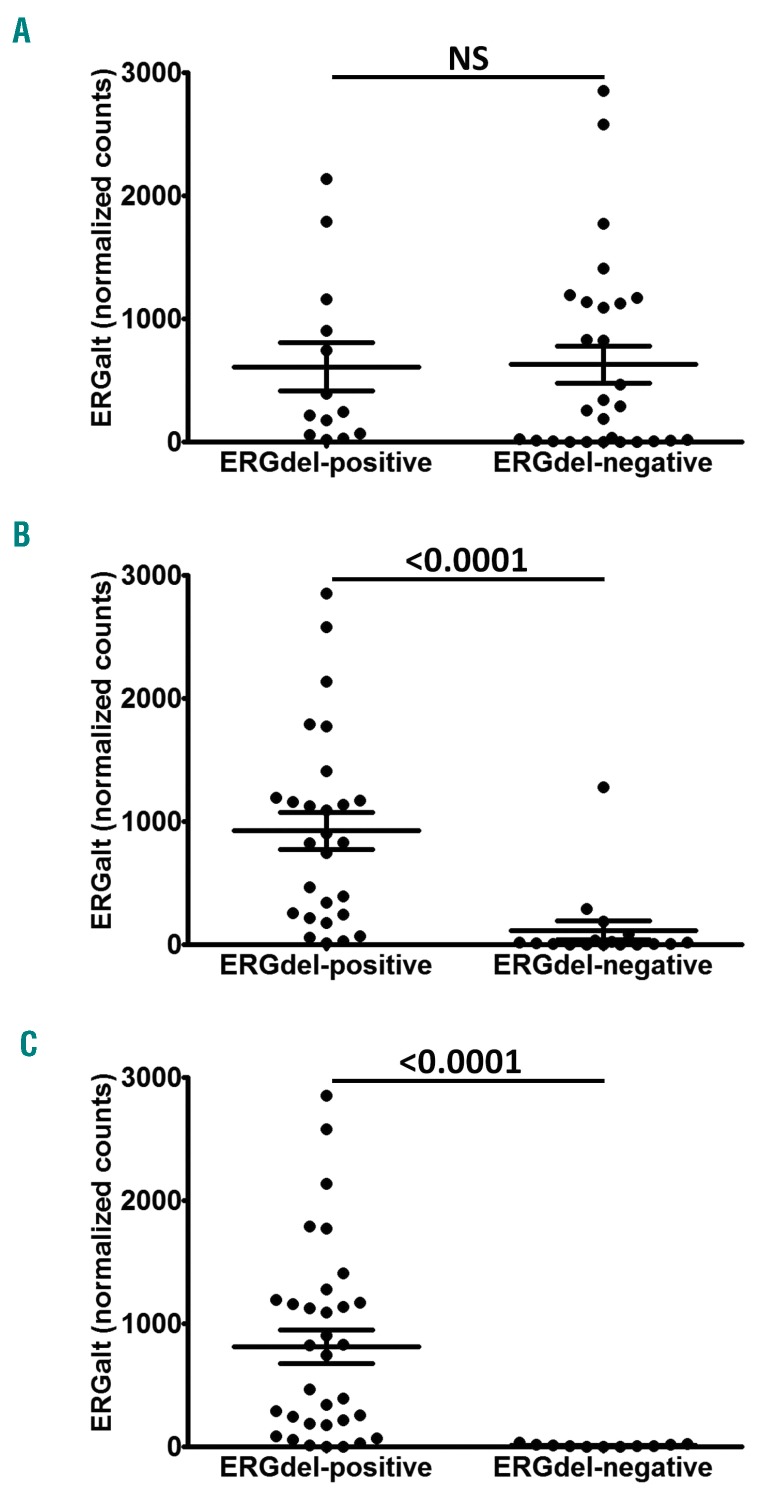
We analyzed the association between ERGdel and expression of ERGalt transcripts. In total, 41 of 44 DUX4r ALL cases expressed ERGalt. The expression levels varied substantially, and in some DUX4r ALL cases, the levels were very low and undistinguishable from those found in non-DUX4r ALL (Online Supplementary Figure S4). Interestingly, the expression of ERGalt was significantly higher in patients with versus patients without ERGdel detected by PCR/AmpliSeq, but not between patients with versus patients without ERGdel detected by SNP array (Figure 3A-C). There was no significant correlation of DUX4 and ERGalt expression levels in DUX4r cases (data not shown).
Figure 3.
ERGalt expression in acute lymphoblastic leukemia (DUX4r-ALL) stratified by ERGdel. ERGalt expression (y-axis: normalized read counts from RNAseq data) is shown for DUX4r-ALL patients stratified by ERGdel according to results of single nucleotide polymorphism array (A), polymerase chain reaction (B) and amplicon sequencing (C). P-values from Mann-Whitney U test are shown. NS: difference not statistically significant.
Frequency of copy number aberrations in DUX4r-ALL stratified by ERGdel
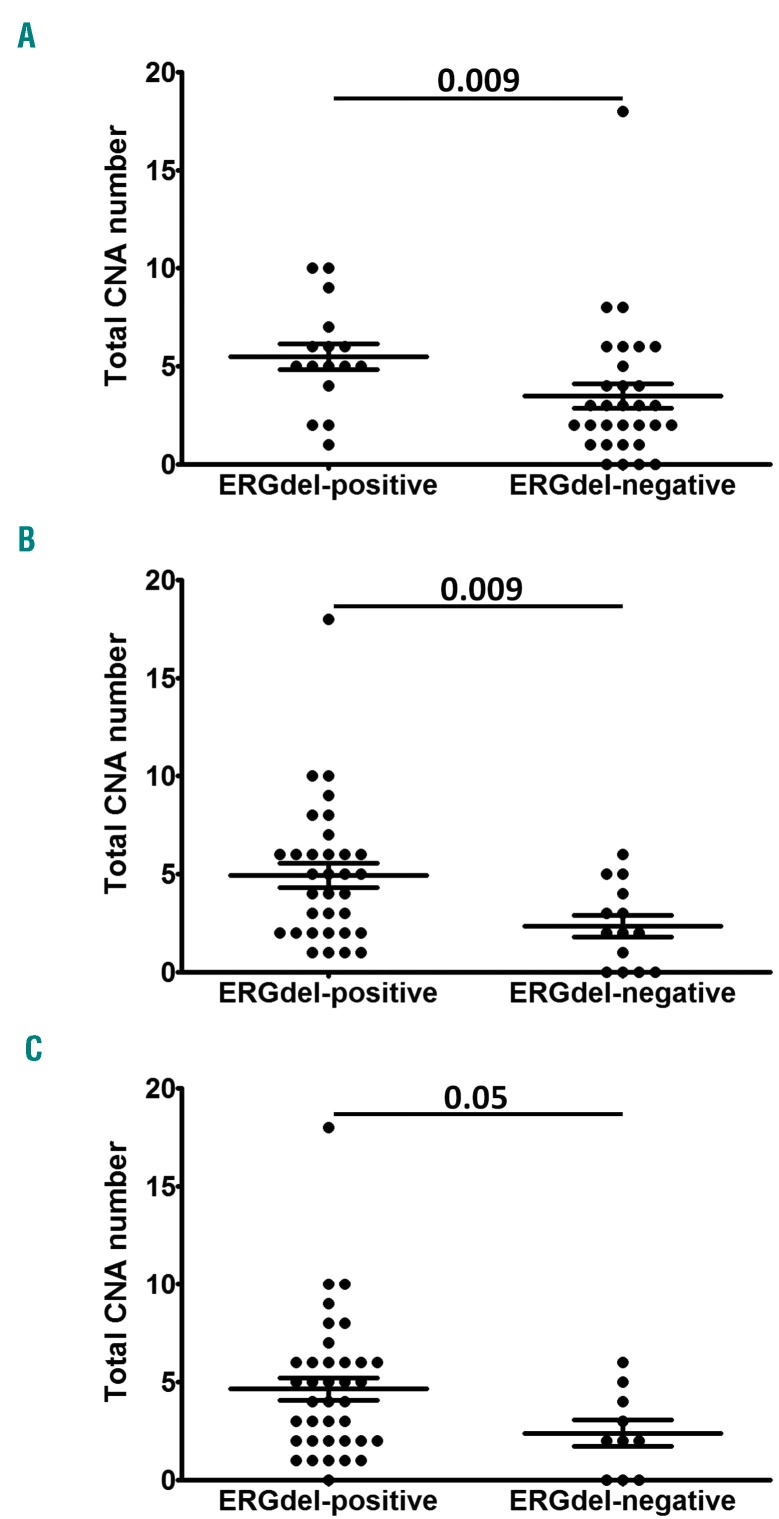
We analyzed the total number of copy number aberrations (CNA) in 47 DUX4r-ALLs with available SNP array data. We found a significantly higher number of CNA in patients positive compared to negative for ERGdel, independently of the ERGdel screening method used (Figure 4). The most common CNA were deletions (del) of the CDKN2A/B, IKZF1 and PAX5 genes (found in 43%, 23% and 20% of DUX4r-ALL patients, respectively) (Online Supplementary Table S2). Deletions of CDKN2A/B were evenly distributed among ERGdel-positive and ERGdel-negative patients. The frequency of IKZF1del was identical in the two groups when the ERGdel was determined by SNP array; however, when PCR results were used for the ERGdel assessment, the ERGdel-positive patients had a significantly higher percentage of IKZF1del compared to ERGdel-negative patients (33% vs. 0% of IKZF1del-positivity, respectively; P=0.02). Frequency of PAX5del was lower in ERGdel-positive compared to negative patients but the statistical significance was reached only on ERGdel stratification according to SNP array (0% vs. 29% PAX5del-positive patients; P=0.04). Simultaneous deletion of IKZF1 and CDKN2A/B or PAX5 (no PAR1 was found in DUX4r-ALL) was found in six DUX4r-ALL patients. All were ERGdel-positive using PCR screening which prevented their classification as IKZF1plus-ALL. Notably, SNP array revealed ERGdel in only one of the six patients.
Figure 4.
Total copy number aberrations (CNA) in acute lymphoblastic leukemia (DUX4r-ALL) stratified by ERGdel. The total number of CNA are shown for DUX4r-ALL patients stratified by ERGdel according to results of single nucleotide polymorphism array (A), polymerase chain reaction (B) and amplicon sequencing (C). P-values from Mann-Whitney U test are shown.
AmpliSeq: a novel method for ERGdel detection and analysis of ERGdel repertoire
To overcome the disadvantages of the “PCR + Sanger” method, we designed deep amplicon sequencing system (AmpliSeq) allowing us to easily distinguish between different ERGdel alleles co-occurring in one sample, as well as to filter out sequences of non-specific products and / or primers-dimers which complicate analysis of Sanger sequencing. During the PCR step of the AmpliSeq approach, individual samples were labeled with unique barcodes which enabled us to multiplex them and avoid carry-over contamination. To amplify the ERGdel spanning region, the same gene specific primers were used as for the PCR, targeting the most frequent type of ERGdel utilizing a common 5’ breakpoint site and one of the five different 3’ breakpoint site clusters. In the AmpliSeq, 1-2 ERG germline region(s) (control amplicons) were co-amplified within the same PCR reaction (Figure 1). This approach enabled us to quantify resulting amplicon libraries and achieve comparable sequencing depth (and thus sensitivity) across all patients, including potentially ERGdel-negative cases without any detectable or quantifiable ERGdel PCR product after PCR. The sensitivity of AmpliSeq was tested on a dilution series of positive control; using 1×106 coverage we achieved sensitivity of 0.01%. Two different levels of coverage were used in the AmpliSeq experiments. Fifteen PCR-positive patients were sequenced with standard coverage (2×105 reads per sample), and 18 PCR-positive DUX4r-ALL patients and 26 PCR-negative patients (17 from the DUX4r cohort and 9 from the non-DUX4r B-others) were sequenced with higher target coverage (1×106 reads per sample). The number of ERGdel reads in all but one patient was high (109-106); in the remaining single patient (UPN-004), who was PCR-negative, we found only 13 ERGdel reads. To assess sensitivity of AmpliSeq used at lower coverage, we performed random down-sampling of total reads to 5×104. ERGdel was reliably detected (supported by > 150 reads) in all AmpliSeq-positive patients except for Patient UPN-004, where only four ERGdel reads were found after down-sampling.
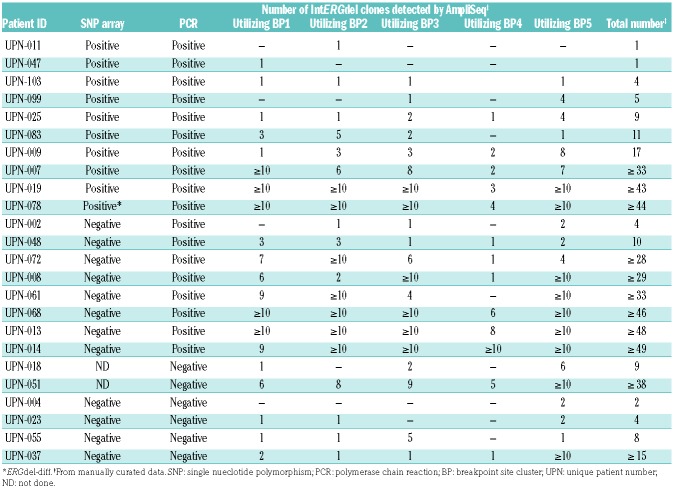
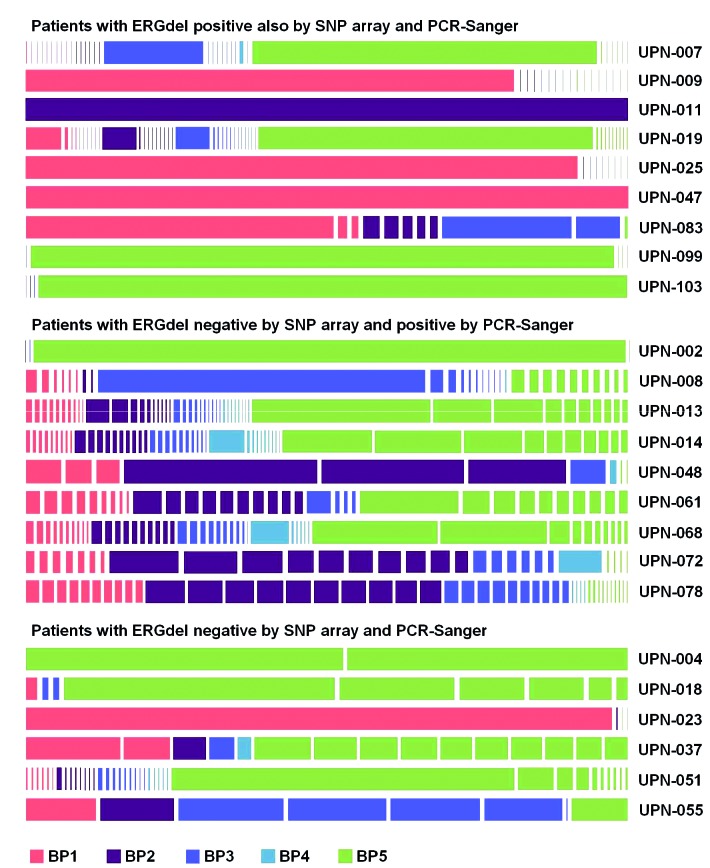
We have previously described a co-occurrence of multiple distinct ERGdel subclones at leukemia diagnosis.3 AmpliSeq provided a much deeper insight into this phenomenon. We studied IntERGdel repertoire in 24 patients sequenced at the higher coverage setting. Strikingly, we found two or more distinct IntERGdel subclones in 22 of 24 patients; in 14 of these, a polyclonal IntERGdel pattern (10-50 co-occurring subclones) was found (Table 2, Figure 5, and Online Supplementary Table S3). Notably, the polyclonal IntERGdel pattern was present in 50% of patients with ERGdel detected by SNP array, where the ERGdel could be incorrectly assumed to represent a “clonal lesion” acquired by a leukemia-founding cell and inherited by all its progeny (Figure 6). On the contrary, only the two patients with a single IntERGdel clone identified by AmpliSeq (and detectable by SNP array) might represent such a clonal lesion; however, we are unable to distinguish the genuine clonal from a “pseudoclonal” lesion, acquired later in leukemogenesis in progeny of a leukemia-founding cell and present in a major, dominant clone at diagnosis. Moreover, additional co-existing ERGdel(s) could have remained undetected in these two patients due to small size and/or inefficient sensitivity of AmpliSeq. Altogether, AmpliSeq revealed the rarity of the clonal/pseudoclonal pattern and a striking prevalence of the polyclonal pattern of IntERGdel.
Table 2.
IntERGdel repertoire in 24 patients positive for ERGdel by amplicon sequencing (AmpliSeq) at higher coverage setting.
Figure 5.
Schematic representation of the IntERGdel repertoire in 24 patients with ERGdel-positivity by amplicon sequencing. Distinct, manually curated IntERGdel clones are shown as bars colored according to the 3’ breakpoint site cluster (BP) used. Width of bars corresponds to the relative size (number of reads) of individual clones. SNP: single nucleotide polymorphism; PCR: polymerase chain reaction; UPN: unique patient number.
Figure 6.
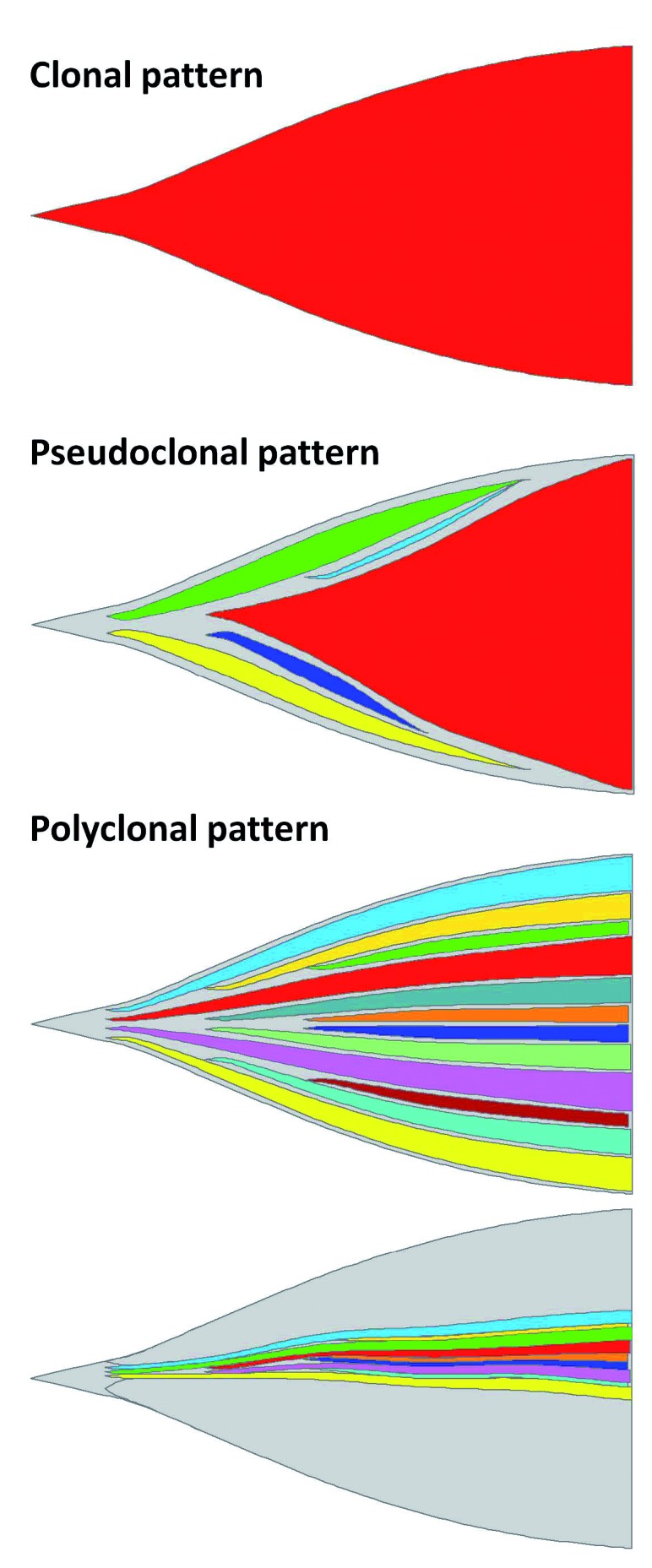
ERGdel patterns. Three different patterns of ERGdel are depicted. (Sub)clones with distinct ERGdel are represented by different colors; cells without ERGdel are shown in gray. Gain of ERGdel in leukemia-founding cell resulting in clonal pattern is probably extremely rare, if present at all. In acute lymphoblastic leukemia with a pseudoclonal pattern, only a single ERGdel gained by progeny of leukemia-founding cell is detected. However, an additional distinct subclonal ERGdel may co-exist at the level below sensitivity of the detection methods. Our data suggest that the most frequent ERGdel pattern is polyclonal, where multiple distinct ERGdel subclones co-exist, and based on the total proportion of ERGdel-positive cells, the ERGdel can be detected by single nucleotide polymorphism array/polymerase chain reaction (PCR) or by PCR only. Patterns were visualized using fishplot package for R.
Discussion
Our study demonstrates that the choice of detection method has a huge impact on the proportion of ERGdel-positive ALL patients that are identified. Only half of the patients with ERGdel identified by PCR were found positive also by SNP array. Thus, a switch to less sensitive methods (SNP array, array CGH, MLPA) could result in a significant deviation from original studies demonstrating the prognostic impact of ERGdel and such methods cannot be considered equivalent to the genomic PCR.
The IKZF1plus deletion pattern was recently identified to be a strong prognostic marker and it will be used to stratify patients with positive minimal residual disease (MRD) at the end of induction into a high-risk treatment arm on an upcoming AIEOP-BFM ALL trial.4 Importantly, we show here that, according to the false negative results of ERGdel screening by SNP array, five patients would be misclassified as IKZF1plus and assigned to the high-risk treatment and/or stem cell transplantation.
While SNP array provides a wide range of diagnostic data (analysis of hyper/hypo-diploidy, analysis of deletions involved in IKZF1plus pattern), the genomic PCR for the ERGdel detection represents an “extra” method and, moreover, it suffers from several disadvantages (as mentioned above) which may discourage diagnostic centers from introducing it. Here we present a novel method, AmpliSeq, which is comparably sensitive to original genomic PCR and overcomes the disadvantages of Sanger sequencing of PCR products. Massive parallel sequencing (MPS) becomes a standard technology, and is used in many routine diagnostic laboratories. The list of its applications is still growing, from whole genome/exome/transcriptome sequencing to custom panels including, e.g. screening of immunoreceptor genes [immunoglobulin (IG) and T-cell receptor (TR)] rearrangements used for the identification of targets for MRD monitoring. AmpliSeq for ERGdel detection can be easily coupled with such screening of IG/TR rearrangements or with other MPS applications. Moreover, as the analysis of IG/TR and ERGdel can use analogous algorithms, development of common analytical tools for these data can be expected in the near future, which would further facilitate the diagnostic use of ERGdel AmpliSeq.
Although we have sequenced a proportion of patients with very high coverage to be able to analyze the ERGdel repertoire in depth, the coverage used for diagnostic purposes can be substantially reduced without a significant increase in false negativity. Even with the reduced coverage, our data show slightly higher sensitivity of AmpliSeq over the PCR. However, the discordance between PCR and AmpliSeq is significantly lower compared to the discordance between SNP array and PCR, and, importantly, our data suggest that biological and clinical characteristics of the PCR-defined and AmpliSeq-defined ERGdel groups are very similar.
Previous studies have shown the rarity of ERGdel in non-B-other BCP-ALL. In the present study we did not find any ERGdel-positive patient within non-DUX4r B-other ALL; thus, ERGdel was 100% specific for the DUX4r-ALL subtype. However, even with the use of AmpliSeq at high coverage, approximately 1 in 5 to 1 in 4 of DUX4r-ALL remained ERGdel negative, confirming that ERGdel cannot serve as a marker for DUX4r-ALL classification. Similarly, we confirmed that nor do ERGalt transcripts represent a reliable surrogate marker for DUX4r ALL; they can be present in non-DUX4r ALL and, moreover, they are absent or expressed at low levels in a proportion of DUX4r ALL, and are undistinguishable from non-DUX4r ALL. Interestingly, higher levels of ERGalt were associated with the presence of PCR-defined ERGdel within DUX4r ALL in the present study. Since the ERGdel-negative DUX4r ALL cases in particular had zero or low levels of ERGalt, even simultaneous detection of ERGdel and ERGalt does not allow the DUX4 ALL subtype to be reliably identified.
Our study revealed additional biological and clinical differences between ERGdel positive and negative patients within the DUX4r-ALL. We found significantly more CNA in ERGdel-positive patients. Although we did not find any difference in expression of DUX4 and RAG1/2 at mRNA level, we believe that a higher CNA number may still reflect a higher rate of illegitimate V-(D)-J recombination in ERGdel-positive patients. Moreover, our data show significantly better outcome of ERGdel-positive compared to ERGdel-negative DUX4r-ALL patients defined by PCR-based techniques. Although this finding needs to be validated by further studies, it suggests that ERGdel might have additional value for outcome prediction than has been described so far.
A potential driving role for ERGdel in leukemogenesis has remained controversial until recently. Originally, it was thought that the ERGdel resulted in the expression of aberrant ERG protein with a potential dominant negative impact over wild-type ERG.6 However, our previous study demonstrated that this protein is not expressed from the ERGdel allele15 and Zhang et al. have shown recently that it is indeed encoded by ERGalt transcript.9 These findings further strengthen the likely passenger role of ERGdel. Our current study shows that ERGdel is independently acquired in multiple members of leukemic cell populations and could just represent collateral DNA damage resulting from continuous DUX4-induced exposure of the ERG gene locus. Our interpretation of ERGdel clonality presumes that this deletion is predominantly monoclonal at the single cell level. Biallelic ERG deletion in bulk leukemic samples has only rarely been reported so far. Moreover, as the aberrant ERG protein encoded by the intact ERG allele has a driver role in DUX4r-ALL,9 the biallelic ERGdel would be biologically disadvantageous. This is strongly supported by findings from in vitro study where silencing of ERG in a DUX4r-ALL NALM-6 cell line with monoallelic ERGdel led to cell death.16 Even though, in the absence of a single cell analysis, we cannot exclude the possibility that one dominant ERGdel (detectable by SNP array) is accompanied in small subclones by another ERGdel present on the second allele (e.g. in patients UPN-025, UPN-099) (Figure 4), taken together, our data clearly demonstrate the predominance of the polyclonal ERGdel pattern and the late origin of this lesion during leukemogenesis in the majority of cases.
To conclude, the ERG gene deletion represents a unique aberration among all the other recurrent genetic changes that have been described in ALL so far. We show here that it is predominantly polyclonal, most likely a passenger aberration whose presence, however, potentially stratifies DUX4r-ALL into two subsets that differ in their genomic profile and outcome. Since it is frequently present at subclonal levels below the sensitivity of SNP array/aCGH/MLPA, only genomic PCR or AmpliSeq should be used for ERGdel screening in order to appropriately define this subgroup and assess the IKZF1plus genotype. The methods used to detect ERGdel differ significantly in sensitivity and this should also be taken into consideration when comparing and interpreting the findings of individual studies.
Supplementary Material
Acknowledgments
This study was supported by grants from the Czech Science Foundation (GJ15-06049Y), Charles University (Primus/MED/28) and the Czech Health Research Council (NV15-30626A), by the Ministry of Education, Youth and Sports (NPU I nr. LO1604 and LM2015091) and by the project (Ministry of Health, Czech Republic) for conceptual development of research organization 00064203 (University Hospital Motol, Prague, Czech Republic).
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/7/1407
References
- 1.Clappier E, Auclerc MF, Rapion J, et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia. 2014;28(1):70–77. [DOI] [PubMed] [Google Scholar]
- 2.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. [DOI] [PubMed] [Google Scholar]
- 3.Zaliova M, Zimmermannova O, Dorge P, et al. ERG deletion is associated with CD2 and attenuates the negative impact of IKZF1 deletion in childhood acute lymphoblastic leukemia. Leukemia. 2014;28(1):182–185. [DOI] [PubMed] [Google Scholar]
- 4.Stanulla M, Dagdan E, Zaliova M, et al. IKZF1(plus) Defines a New Minimal Residual Disease-Dependent Very-Poor Prognostic Profile in Pediatric B-Cell Precursor Acute Lymphoblastic Leukemia. J Clin Oncol. 2018;36(12):1240–1249. [DOI] [PubMed] [Google Scholar]
- 5.Mullighan CG, Miller CBS, Radtke X, et al. ERG Deletions Define a Novel Subtype of B-Progenitor Acute Lymphoblastic Leukemia. Blood (ASH Annual Meeting Abstracts) 2007;110(11):691. [Google Scholar]
- 6.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–143. [DOI] [PubMed] [Google Scholar]
- 7.Lilljebjorn H, Henningsson R, Hyrenius-Wittsten A, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasuda T, Tsuzuki S, Kawazu M, et al. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet. 2016;48(5):569–574. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, McCastlain K, Yoshihara H, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016;48(12):1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slamova L, Starkova J, Fronkova E, et al. CD2-positive B-cell precursor acute lymphoblastic leukemia with an early switch to the monocytic lineage. Leukemia. 2014;28(3): 609–620. [DOI] [PubMed] [Google Scholar]
- 11.Zaliova M, Hovorkova L, Vaskova M, Hrusak O, Stary J, Zuna J. Slower early response to treatment and distinct expression profile of childhood high hyperdiploid acute lymphoblastic leukaemia with DNA index < 1.16. Genes Chromosomes Cancer. 2016;55(9):727–737. [DOI] [PubMed] [Google Scholar]
- 12.Zaliova M, Kotrova M, Bresolin S, et al. ETV6/RUNX1-like acute lymphoblastic leukemia: A novel B-cell precursor leukemia subtype associated with the CD27/CD44 immunophenotype. Genes Chromosomes Cancer. 2017;56(8):608–616. [DOI] [PubMed] [Google Scholar]
- 13.Anders S, McCarthy DJ, Chen Y, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8(9):1765–1786. [DOI] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potuckova E, Zuna J, Hovorkova L, et al. Intragenic ERG Deletions Do Not Explain the Biology of ERG-Related Acute Lymphoblastic Leukemia. PLoS One. 2016;11(8):e0160385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuzuki S, Taguchi O, Seto M. Promotion and maintenance of leukemia by ERG. Blood. 2011;117(14):3858–3868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.