Abstract
Objective
Leptin (Lep) plays a crucial role in controlling food intake and energy expenditure. Defective Lep/LepRb-signaling leads to fat accumulation, massive obesity, and the development of diabetes. We serendipitously noticed spontaneous development of obesity similar to LepR-deficient (db/db) mice in offspring from a C57BL/6J breeding and transmittance of the phenotype in a Mendelian manner. Candidate gene sequencing revealed a spontaneous point mutation in the LepRb gene. We investigated leptin responsiveness, leptin receptor signaling and metabolic phenotype of this novel LepRb mutant mouse variant.
Methods
Overexpression and functional tests of the mutant LepRb in 3T3 cells. Measurement of leptin responsiveness in hypothalamic nuclei, glucose tolerance, food uptake and energy expenditure in the mutant mice.
Results
The mutation results in the exchange of a glycine for serine (G506S) and introduces an alternative splice acceptor which, when used, encodes for a protein with a 40aa deletion that is retained in the cytoplasm. LepRb signaling was abrogated in the hypothalamic ventromedial nucleus (VMN) and dorsomedial nucleus (DMN), but only partially reduced in the hypothalamic arcuate nucleus (ARC) of LepRbG506S/G506S mice, most likely due to differential splicing in neurons located in the respective regions of the hypothalamus. Extensive metabolic characterization of these mice revealed interesting differences in the control of food intake, glucose tolerance, energy expenditure, and fat accumulation in LepRbG506S/G506S compared with LepRb-deficient db/db mice.
Conclusions
This study provides further insight into differences of the leptin responsiveness in VMN, DMN, and ARC and its metabolic consequences.
Keywords: Leptin receptor mutation, Ventromedial nucleus, Dorsomedial nucleus, Arcuate nucleus, Energy expenditure, Glucose tolerance
Highlights
-
•
A mutation in the leptin receptor gene results in alternative splicing and generates a cryptic protein that remains intracellular.
-
•
The mutation differentially affects leptin responsiveness of POMC neurons and SF1 neurons in the hypothalamus.
-
•
The mutation results in severe obesity but metabolic dysfunction distinct from LepR null mutants.
1. Introduction
Leptin is produced by adipose tissue, and its concentrations are proportional to the amount of body fat mass but also fluctuate in response to food intake [1]. Leptin reaches the brain via the bloodstream and binds to the leptin receptor expressed on neurons of the hypothalamus. Leptin binding induces the tetramerization of the long form of leptin receptor LepRb [2], and the resulting downstream signaling leads to the repression of food intake in a negative feedback loop [3], [4].
Leptin deficient (ob/ob) and leptin receptor deficient (db/db) mice are markedly obese from 4 weeks of age due to excessive food intake [5], [6], [7], [8]. The mutation in db/db mice results in an alternatively spliced leptin receptor RNA with a 106 bp deletion [9]. This deletion in the intracellular region of the long isoform of leptin receptor prevents signal transduction and therefore causes unresponsiveness to circulating leptin [10].
Leptin is mostly known for repressing appetite but it also controls other vital functions such as energy expenditure (EE), insulin sensitivity and glucose homeostasis [11], [12]. Indeed, ob/ob and db/db mice have increased body weight and show increased blood insulin, resulting from an attempt to compensate for the insulin resistance generated by the increased concentration of glucose in the blood. In addition, these mice also develop a fatty liver and hypercholesterolemia and are infertile [13], [14], [15]. Obesity and associated metabolic diseases are not entirely determined by increased food intake, since leptin-deficient mice, compared to lean mice, still accumulate fat and body weight when caloric uptake is restricted. This points to an important role of leptin in controlling energy expenditure independent of food intake [13].
The brain is the main target for the weight-reducing and neuroendocrine effects of leptin, as indicated by development of obesity in mice lacking LepRb specifically in neurons [16]. Conversely, reconstitution of LepRb in neurons of db/db mice reduces obesity [17]. Furthermore, leptin administration directly to the brain of both ob/ob and WT mice reduces food intake and body weigh t [18]. A particularly high expression of LepRb is observed in the arcuate nucleus (ARC), ventromedial nucleus (VMN), and dorsomedial nucleus (DMN) of the hypothalamus [19], [20]. In the ARC, neurons expressing LepRb can be further divided into anorexigenic proopiomelanocortin (POMC)-expressing neurons and orexigenic, agouti-related peptide (AgRP)-expressing neurons or neuropeptide Y (NPY) neurons [21], [22]. Under homeostatic conditions, leptin stimulates catabolic POMC neurons and POMC expression [21], [23] but inhibits AgRP neurons and AgRP expression [24], [25]. POMC neurons then release the catabolic neuropeptide α-melanocyte stimulating hormone (αMSH), which acts on melanocortin 3 and 4 receptors (MCR3/4) particularly in the paraventricular nucleus of the hypothalamus, to reduce food intake and increase energy expenditure via thermogenesis. AgRP serves as a potent endogenous antagonist (inverse agonist) of MCR3/4 [26]. NPY/AgRP neurons synapse onto POMC neurons to provide inhibitory action via release of NPY onto NPY receptor type 2 (Y2) and gamma-aminobutyric acid (GABA), further supporting a tightly regulated neuronal network (reviewed in [27], [28]).
In obesity models based on Lep/LepR-deficiency, hypothalamic POMC levels are downregulated, while AgRP and NPY are increased and administration of leptin in ob/ob mice reverses this effect [24], [29], [30]. Several studies have demonstrated that the various metabolic functions of leptin are controlled by different neurons. Leptin specifically activates SF1 neurons in the VMN to stimulate the sympathetic nervous system [31]. In addition, SF1 neurons are required for the control of energy expenditure [32], [33].
Other studies focusing on POMC neurons showed that even though they have little influence on the control of food intake, they are required for glucose homeostasis. Re-expression of LepRb selectively in POMC neurons in mice that were otherwise LepRb-deficient, normalized hyperglycemia but was insufficient to control food intake and the development of obesity [34], [35], [36], implying that POMC neurons are sufficient for glucose homeostasis. Similarly, selective re-expression of LepRb in AgRp neurons also normalized hyperglycemia in db/db mice, showing that AgRP neurons are sufficient for leptin's anti-diabetic effect [37]. Another hypothalamic region expressing high levels of leptin receptor is the DMN, which is responsible for leptin-mediated thermoregulation and thereby energy homeostasis [38].
Overall, it seems that there are subtle differences in the role of the various neurons expressing leptin receptor, but this does not exclude that some functions are redundantly shared by many neuron types.
We here describe a novel spontaneous point mutation of the LepRb gene in mice, leading to a truncated protein that predominantly affects LepRb signaling in the ventromedial and dorsomedial nucleus of the hypothalamus. Partial responsiveness to leptin in homozygous mutants resulted in differences in the control of energy expenditure and the severity of diabetes compared to db/db mice, although the development of obesity was comparable to full KO mice. These effects could be the result from the difference in LepRb expression and highlight the importance of the ARC in controlling energy homeostasis.
2. Material and methods
2.1. Mice
Mice were maintained in a temperature-controlled (21 ± 2 °C) room on a 12:12 h light/dark schedule with lights off at 1800 h. Food and water were provided ad libitum. The animals were fed standard chow diet (Diet 3436, Provimi Kliba AG, Kaiseraugst, Switzerland). C57BL/6 mice with the LepRbG506S mutation were maintained by crossing homozygous and heterozygous mice. Littermate controls were used in all experiments. C57BL/6 mice were obtained from the animal facility at ETH. Animals were kept in individually ventilated cages under specific pathogen-free conditions at the ETH Phenomics center. LepRb LoxTB [B6.129X1(FVB)-Leprtm1Jke/J, no. 019111; The Jackson Laboratory], and wildtype (WT) littermates were bred in the UZH LASC facility. LepRb LoxTB mice possess a transcriptional blocker cassette that prevents the transcription of the LepRb gene [35], [39]. db/db mice were purchased from Janvier Labs.
Prior to transfer to metabolic cages, mice were single-housed for adaptation for several days. All experiments were approved by the local Ethics Committees and were performed according to local guidelines and Swiss animal protection law.
2.2. Glucose and insulin tolerance test
Mice were fasted for 6 hours, and blood was sampled by tail nip for a baseline measurement. 2 g glucose/kg body weight or 0.5 IU insulin/kg body weight was then injected intraperitoneally (i.p.) and plasma glucose was measured after 15, 30, 60, 90, 120 and 180 minutes. Water was available ad libitum during the entire experiment.
2.3. Food intake
Metabolic and food intake measurements were performed in single-housed conditions in an automated monitoring system (PhenoMaster, TSE System). Before 24h-food intake measurements, mice were fasted for 2 hours, prior to dark onset.
2.4. Indirect calorimetry
The TSE PhenoMaster open circuit indirect calorimetry system for the determination of O2 consumption and CO2 production (TSE Systems; Bad Homburg, Germany) was used. Room air was passed through each cage at a flow rate of 0.41 L/min. Every 20 min, cage air was sampled from each individual cage and analyzed for O2 and CO2. From these values, energy expenditure (EE) and respiratory exchange ratio (RER) were calculated, based on the equations from Weir [40]. To account for differences in body weight and body mass composition, EE data were corrected for individual lean body mass (LBM in g) and fat mass (FM in g) using the following equation: LBM + 0.2FM, as recommended by Even and Nadkarni [41].
2.5. Measurement of body composition
CT scanning was performed using a La Theta LCT-100 (Hitachi Aloka Medical Ltd, Steinhausen, Switzerland). Mice were placed supine in the plexiglass holder with an inner diameter of 48 mm. The X-ray source tube voltage was set at 50 kV with 1 mA current. The entire body without head was evaluated for lean and fat mass. LaTheta LCT-100 software automatically distinguishes between visceral and subcutaneous fat; however, each image was examined and corrected if assigned incorrectly.
2.6. Analysis of plasma leptin, insulin, cholesterol and triglycerides
Blood was collected at sacrifice by heart puncture in K-EDTA tubes and was centrifuged at 10 000 rpm for 10 min and the plasma was stored at −80 °C until further processed. Insulin and leptin concentrations were determined in a 2-plex mesoscale assay (Mesoscale, MD, USA) following manufacturer's instructions. Cholesterol and triglycerides were measured with enzymatic photometric tests using the COBAS8000 autoanalyzer, both from Roche diagnostics (Rotkreuz, Switzerland).
2.7. Immunohistochemistry
2.7.1. Brain perfusion
Mice were fasted for 2h and at dark onset were injected with saline vs. leptin (i.p. 5 mg/kg, PeproTech, UK) and 45 min later perfused for 1.5 min with saline and 5 min with 2% paraformaldehyde in 0.1 M phosphate buffer (PB-PFA, pH 7.4). Brains were post-fixed for 2h in 2% PB-PFA and dehydrated overnight in 20% sucrose-0.1M PB. They were then frozen in hexane on dry ice for 3 min and stored at −80 °C. Frozen brains were sectioned serially at 25 μm through the ARC and VMN on a cryostat, sections were mounted onto superfrost plus slides (Life Technologies Europe, Zug, Switzerland) and stored in cryoprotectant (50% 0.02 M KPBS, 30% ethylene glycol, 20% glycerol) at −20 °C until staining for leptin-induced pSTAT3 and POMC immunocytochemistry (IHC) in the ARC.
2.7.2. pSTAT3 and POMC IHC
Brain slides were rinsed in 0.02 M potassium phosphate saline buffer (KPBS) and pre-treated for 20 min in KPBS containing 0.5% NaOH and 0.5% H2O2. After rinsing, brain slides were incubated in 0.3% glycine in KPBS, rinsed in KPBS and incubated for an additional 10 min in KPBS containing 0.03% SDS. Following rinses, slides were incubated in blocking solution containing 0.4% Triton X-100, 1% BSA and 4% normal goat serum (NGS) in KPBS for 1h followed by incubation at 4 °C for 48 h in modified blocking solution (0.4% Triton X-100, 1% BSA and 1% NGS) containing rabbit anti-pSTAT3 antibody (1:1000; Cell Signaling Technologies, BioConcept, Allschwil, Switzerland). After rinsing with KPBS, slides were incubated with secondary antibody (CY3 goat anti-rabbit, 1:250; Jackson Laboratories, Luzern, Switzerland) in KPBS for 2 h. After rinses in KPBS, slides were coverslipped with Vectashield hardset mounting medium (Vectorlabs, Servion, Switzerland).
In a second series, double-label pSTAT3-POMC IHC was carried out. The same pSTAT3 IHC protocol as above was followed. Sections were then incubated in blocking solution containing 0.4% Triton X-100, 1% BSA and 4% NGS in KPBS for 1h followed by incubation at 4 °C for 72 h in modified blocking solution (0.4% Triton X-100, 1% BSA and 1% NGS) containing rabbit anti-POMC antibody (1:1000; Phoenix Pharmaceuticals). Following rinses, sections were placed in Alexa-Fluor 488 goat anti-rabbit for 2 h (1:250; Life Technologies Europe, Zug, Switzerland), rinsed again and mounted using Vectashield hardset mounting medium [42].
2.7.3. Quantitative analysis of immunolabeled cells
Labeled cells were imaged using an L2 Imager upright microscope (Zeiss, Germany). Images of single and double labeled cells were counted using ImageJ (NIH) to allow for quantification. Three sections per brain were used for quantification. Slides were numerically coded to obscure the treatment group. To ensure similar imaging conditions for all images, the same microscope set-up and acquisition settings were used to acquire all images within the same experiment. For representative images, images were equally adjusted for brightness and contrast within the same experiment [39].
2.8. Isolation of ARC and RNA extraction
Mice were sacrificed by pentobarbital injection (100 mg/kg) 2 hours post starvation, and brains were snap frozen on dry ice. The brain were then mounted on a cryostat and cut in 250 μm sections through the ARC. ARC was punched out and stored at −80 °C until RNA extraction. Total RNA of cells or lysates was extracted using TRIzol (Life Technologies), followed by reverse transcription using GoScript Reverse Transcriptase (Promega) according to the manufacturer's instructions. LepR fragments were PCR amplified semi-quantitatively using oligonucleotides LepR(for)5′-gacttgcagatggtcacc-3′ and LepR(rev):5′-tgcacagaggtctgacac-3’.
2.9. Transduction of 3T3 cells
The wild-type LepRb coding region was amplified from brain cDNA and used to create LepRbG506S and LepRbΔ40aa prior to cloning into pMY vector carrying a GFP reporter. The plasmid was introduced into Phoenix cells, supernatant containing retrovirus was collected after 2 and 3 days and concentrated. Spin infection of 3T3 cells was performed at 37 °C for 45 minutes. GFP+ cells were sorted using a flow cytometry cell sorter (BD FACS ARIA IIIu). For double transductions, LepRbΔ40aa was transferred into the pMY vector carrying a dsRed2 reporter. Virus was produced as described and used to spin infect 3T3 cells that already overexpressed LepRbWT or LepRbG506S. GFP+dsRed2+ cells were FACS sorted.
2.10. Leptin stimulation of 3T3 cells
Cells were kept overnight in IMDM medium (ThermoFisher) with 100 U/mL Penicillin-Streptomycin (ThermoFisher) and 50 μM b-mercaptoethanol (ThermoFisher), without FBS. Various concentrations of leptin or BSA as a negative control were added for 30 minutes prior to trypsinization and collection of the cells for staining.
2.11. Staining and FACS analysis
Leptin receptor staining was performed using an α-leptin receptor antibody from R&D systems (polyclonal goat IgG, 1:300). Extracellular staining was performed in PBS + 2% FBS for 15 minutes. Intracellular staining was performed after 5 minutes fixation with 4% paraformaldehyde followed by 5 minutes permeabilization with 0.5% saponin in PBS + 2% FBS.
pSTAT3 staining (pY705, BD Biosciences, clone 4/P-STAT3) was performed diluting the antibody 1:5 in 2% FBS in PBS after 10 minutes fixation with 4% paraformaldehyde followed by permeabilization with 90% ice-cold methanol for 20 minutes.
Cells were acquired on BD FACS Canto II and analyzed with FlowJo software (Tree Star).
2.12. Western Blot
3T3 cells were lysed with RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM Na3VO4, 1% Triton X-100, supplemented with protease inhibitor [Sigma–Aldrich] and phosphatase inhibitor [Sigma–Aldrich]). Samples were then spun at 4 °C to remove all cell debris and protein concentrations were determined using the PierceTM BCA Protein Assay Kit (Thermo Scientific). 5 μg of proteins were fractionated by SDS-PAGE and transferred to a Polyvinylidene difluoride (PVDF) membrane using a transfer apparatus according to manufacturer's instructions (Bio-Rad). After blocking with 4% nonfat milk in TBST (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% Tween 20) for 45 min, the membrane was incubated with primary antibodies in 4% nonfat milk in TBST (Phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204), 1:1000, Cell Signaling Technology) at 4 °C for 16 h. After washing, membranes were incubated with horseradish peroxidase-conjugated anti-rabbit antibody in 4% nonfat milk in TBST (Goat Anti-Rabbit IgG(H + L), Mouse/Human ads-HRP, Biozol) at RT for 1 hour. Blots were washed with TBST three times and developed with the ECL system (Thermo Scientific) according to manufacturer's instructions. After acquisition with ChemiDoc MP Imaging System (Bio-Rad) using Image Lab program, membrane was stripped by incubating it two times 8 minutes with stripping buffer (1.5% glycine, 0.1% SDS, 1% Tween, pH 2.2), followed by two times with PBS and two times with TBST and finally blocked again for 45 minutes with 4% nonfat milk in TBST. Horseradish peroxidase-conjugated β-Actin antibody was incubated in 4% nonfat milk in TBST for 45 minutes at RT(AC-15; Sigma–Aldrich).
2.13. Statistics
GraphPad Prism 7 was used for all representations and statistical analysis. Mean and SD or mean and SEM are shown as described in the figure legend. One-Way ANOVA, two-way ANOVA, and Student's t-test were used for statistics. RMarkdown was used for ANCOVA analysis.
3. Results
3.1. A spontaneous point mutation in the leptin receptor gene is associated with the development of obesity in LepRbG506S/G506S similar to db/db mice
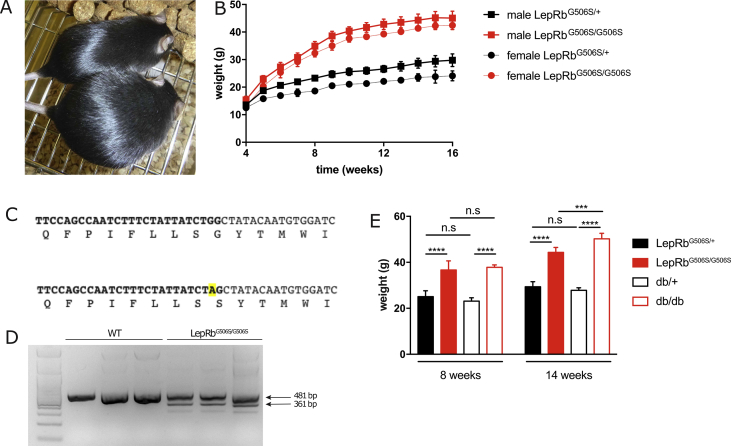
We serendipitously noticed that a breeding pair of C57BL/6 mice delivered offspring, some of which developed early onset obesity; this phenomenon was seen in both sexes (Figure 1A). When obese mice were bred with lean wild-type C57BL/6 mice and the F1 was interbred, transmission of the phenotype was Mendelian. Both males and females gained weight much faster compared to lean littermates and the difference was already discernible and statistically significant in 4 week-old males and 5 week-old females (Figure 1B). Genome sequencing revealed a single base pair mutation in the leptin receptor gene (G > A) at position 1516 from the ATG start codon (Figure 1C). This mutation causes an exchange of a glycine with a serine at position 506 (G506S) and homozygous mice were therefore dubbed LepRbG506S/G506S. In addition to the single amino acid exchange, the mutation also generated a new potential splice acceptor site, which would lead to an alternatively spliced mRNA and a protein with an internal deletion of 40 amino acids, including one of the two amino acids crucial for leptin binding. To address this possibility, we isolated the ARC from the hypothalamus, prepared cDNA, and semi-quantitively PCR amplified LepRb using primers that span the region including the potential splice product. In the ARC of homozygous LepRbG506S/G506S mice, besides the LepRb wild-type fragment, we identified a band corresponding in size and sequence to the product of the alternatively spliced RNA (LepRbΔ40aa, Figure 1D). The LepRbΔ40aa fragment was absent in WT mice. Therefore, we hypothesized that this mutation was responsible for the development of obesity in LepRbG506S/G506S mice. The increase in body weight of male LepRbG506S/G506S and db/db mice was comparable only until 9–10 weeks of age (Figure 1E). Thereafter, the increase in body weight was not as pronounced in LepRbG506S/G506S mice as it was for db/db.
Figure 1.
Spontaneous mutation in leptin receptor gene leads to development of obesity. (A) Picture of a three months old obese LepRbG506S/G506S and a lean LepRbG506S/+ mouse. (B) Body weight of male and female LepRbG506S/G506S and heterozygous LepRbG506S/+ littermates with age after weaning (n = 3–6/group). (C) Nucleotide and amino acid sequences from Q498 to I511 of wild-type (top) and mutant (bottom) leptin receptor. (D) ARC nuclei from wild-type and homozygous mutants (n = 3/group) were isolated and cDNA prepared. Semiquantitative PCR was performed using oligonucleotides flanking the G506S point mutation and allow identification of both the wild-type (481 bp) and alternatively spliced LepRb (361 bp) products. (E) Body weight of male LepRbG506S/G506S, db/db, and respective heterozygous controls (n = 6–11/group). Values in (B) and (E) show mean ± standard deviation. ***, P < 0.0021, ****, P < 0.0002.
3.2. A truncated LepRb due to alternative splicing is not transported to the cell surface and does not signal in response to leptin
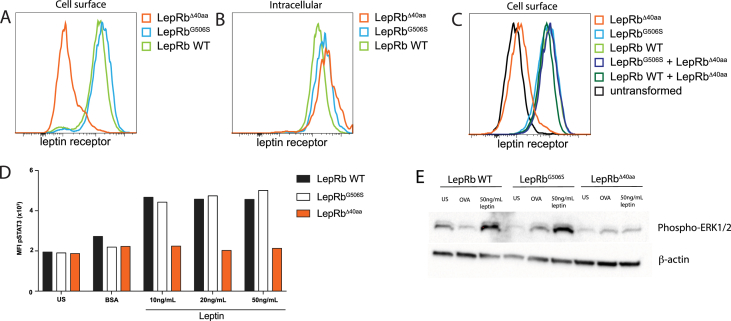
To investigate how the point mutation influences surface expression and leptin signaling, we generated fibroblast cell lines overexpressing the three different forms of leptin receptor. 3T3 cells were transduced with retroviruses expressing GFP together with the LepRb wild-type transcript (LepRbWT), a transcript with the point mutation (LepRbG506S) or a transcript with the deletion corresponding to the product of the alternatively spliced RNA (LepRbΔ40aa). GFP + cells were sorted and leptin receptor expression was measured by flow cytometry. Figure 2A shows that the leptin receptor WT and G506S mutant were comparably expressed. In contrast, the alternatively spliced form was not detected on the cell surface, while it could be detected by intracellular staining (Figure 2B), indicating that the protein is produced but cannot be transported to the cell surface. It is possible that cells expressing LepRb in obese LepRbG506S/G506S mice produce varying levels of the two alternatively spliced transcripts and therefore two protein forms: a LepRb with a point mutation and a truncated LepRb with a 40aa deletion. Since the leptin receptor needs to dimerize in order to be expressed on the cell surface [2], it was theoretically possible that the truncated LepRbΔ40aa protein acts in a dominant negative way by preventing dimerization with and/or cell surface translocation of the LepRbG506S with the point mutation. To investigate this, we generated 3T3 cells overexpressing both LepRbG506S (or LepRb WT) and LepRbΔ40aa simultaneously. Co-expression of dsRed and GFP reporters was used to select for double positive cells expressing both LepRb variants. Staining for the leptin receptor in double transformed cells indicated that the presence of LepRbΔ40aa was not sufficient to prevent transport of the full-length receptor to the surface (Figure 2C). It is important to note that this system has the limitation that the leptin receptors are most likely not expressed at levels comparable to what is observed in neurons of the hypothalamus. The overexpression could bypass the hypothesized dominant negative effect by producing enough of the longer form as homodimers which can be detected on the surface. Nevertheless, it indicates that if enough of the full-length form is present, it can be expressed on the cell surface.
Figure 2.
The alternatively spliced form of the leptin receptor is not expressed on the cell surface. (A) 3T3 cells were transduced with retroviral vectors encoding either LepRbWT-IRES-GFP, LepRbG506S -IRES-GFP, or LepRbΔ40aa-IRES-GFP. GFP + cells were sorted and expression of leptin receptor was analyzed by flowcytometry. (B) Intracellular staining for leptin receptor in transformed 3T3 cells as in (A). (C) Expression of leptin receptor on the surface of 3T3 cells transduced consecutively with LepRbWT-IRES-GFP and LepRbΔ40aa-IRES-dsRed2 or LepRbG506S-IRES-GFP and LepRbΔ40aa-IRES-dsRed2. (D) Induction of leptin signaling measured by phosphorylation of STAT3, 30 minutes after stimulation of transformed 3T3 cells with the indicated concentrations of leptin. US indicates unstimulated cells, BSA (0.5%) was used as unspecific stimulus. (E) Induction of leptin signaling measured by phosphorylation of ERK, 30 minutes after stimulation of transformed 3T3 cells. US indicates unstimulated cells, OVA (50 ng/mL) was used as unspecific stimulus.
The phenotype in LepRbG506S/G506S mice could be due either to alternative splicing of all leptin receptor gene transcripts, and therefore absence of cell surface expression, or to an inability of the single amino acid mutated form to bind leptin and transduce the signaling. To test the latter hypothesis, we starved 3T3 cells overexpressing the different receptor forms and then added leptin to measure the phosphorylation of STAT3 and ERK. Importantly, STAT3 and ERK activation were comparably strong in cells expressing either LepRbG506S or WT LepRb (Figure 2D,E), while cells expressing the alternatively spliced form LepRbΔ40aa failed to respond to leptin. We conclude that the point mutation is not responsible for defective leptin signaling in LepRbG506S/G506S mice.
3.3. Differential response to leptin in the ventromedial and arcuate nucleus of the hypothalamus of LepRbG506S/G506S mice
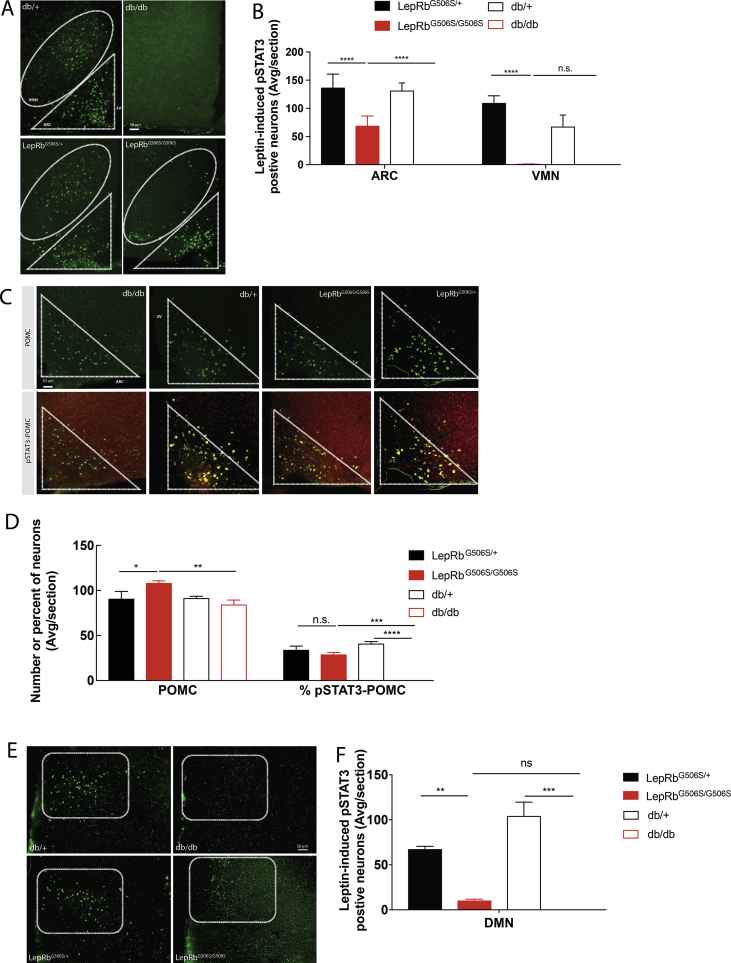
The metabolic activity of leptin is mainly determined in the hypothalamus by binding to the LepRb and triggering STAT3 activation in neurons of the arcuate nucleus and ventromedial nucleus of the hypothalamus [43], [44]. To monitor LepRb signaling in the brain, LepRbG506S/G506S, db/db, and WT control mice were fasted for 2 hours prior to injection of leptin and sacrificed 45 minutes later as previously published [45]. Expectedly, immunohistochemical staining of medio-basal hypothalamus sections showed abundant pSTAT3 positive neurons in both the VMN and ARC of WT mice (Figure 3A,B), which was undetectable in db/db mice, consistent with a complete unresponsiveness to leptin. Interestingly, LepRb signaling was also abrogated in the VMN of LepRbG506S/G506S mice, while the number of pSTAT3+ neurons in the ARC was only reduced to about one half of controls, pointing to an important difference between db/db and LepRbG506S/G506S mice in particular in respect to the activation of ARC neurons.
Figure 3.
STAT3 signaling is significantly impaired in the arcuate nucleus (ARC) and completely abrogated in the ventromedial nucleus (VMN) and dorsomedial nucleus (DMN) in the hypothalamus of LepRbG506S/G506Smice. (A and E) Immunohistochemistry of pSTAT3 on brain slices. (B) Quantification of pSTAT3-positive neurons in the ARC and in the VMN. Data of 2 separate experiments were pooled, in one experiment db/db were replaced by LepRloxTB/loxTB mice. (C) Co-staining of pSTAT3 and POMC on brain sections. (D) Quantification of POMC positive neurons and frequency of double positive neurons among POMC positive neurons. (F) Quantification of pSTAT3-positive neurons in the DMN. Mice were fasted for 2h prior to leptin injection and brains were removed 45 min later. Values represent mean with standard deviation (n = 3–6/group). *, P < 0.1234; **, P < 0.0332; ***, P < 0.0021, ****, P < 0.0002.
The ARC consists of at least two types of neurons, anorexigenic POMC and orexigenic AgRP, both of which are known to respond to leptin. Concomitant staining of pSTAT3 and POMC revealed a comparable number of pSTAT3+ POMC neurons in heterozygous LepRbG506S/+ and LepRbG506S/G506S mice (Figure 3C,D), suggesting that the decrease in leptin-induced pSTAT3 seen in the ARC is not due to a decrease in leptin signaling in POMC neurons. Another brain region exerting an important role in energy expenditure is the DMN. As for the VMN, DMN leptin-induced pSTAT3 was almost absent in LepRbG506S/G506S mice (Figure 3E,F).
3.4. Differences in glucose tolerance and subcutaneous fat accumulation in LepRbG506S/G506S and db/db mice
Since leptin signaling is abrogated in VMN but only partially affected in the ARC of LepRbG506S/G506S mice, we decided to further investigate the animals’ phenotype, in particular by focusing on metabolic differences compared to db/db.
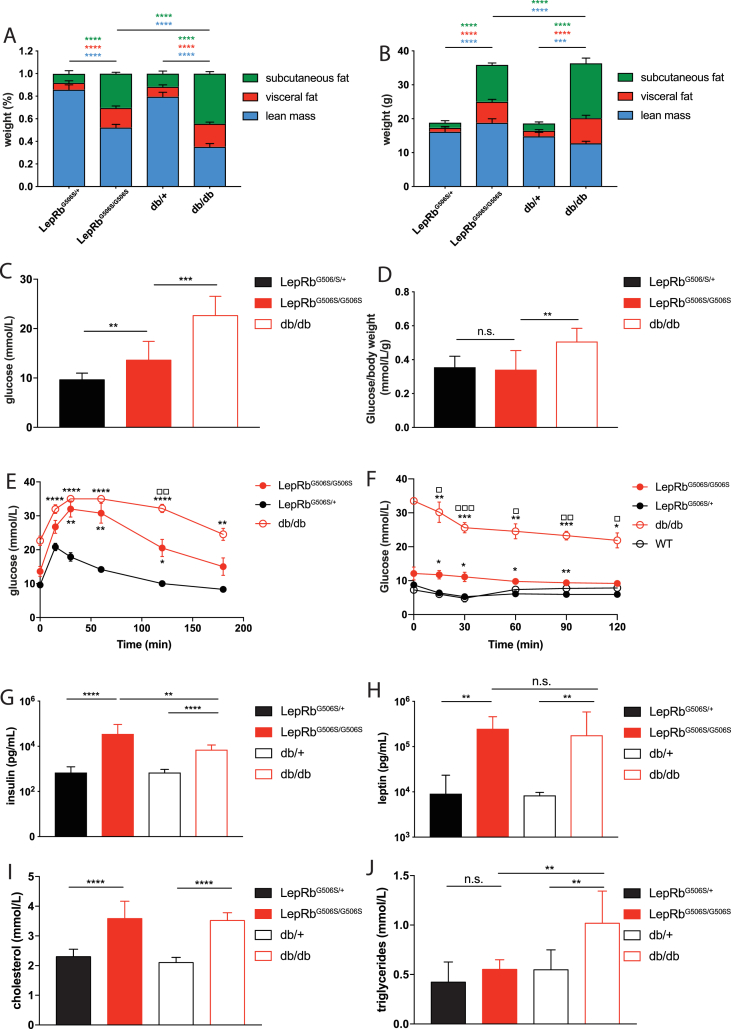
LepRbG506S/G506S, db/db and control mice were subjected to computed tomography (CT) to determine their body weight composition. Consistent with the obese phenotype, LepRbG506S/G506S mice had increased fat mass compared to littermates (Figure 4A,B). However, when compared to db/db mice, we found that even though the total weight was proportionally similar, LepRbG506S/G506S mice had more lean mass and less subcutaneous fat.
Figure 4.
Differences in metabolic parameters of 20 weeks old db/db and LepRbG506S/G506Smice. (A–B) Distribution of body mass in subcutaneous fat, visceral fat and lean mass represented as percentage of total body weight or gram of different tissues. (C) Fasting glucose levels. (D) Fasting glucose levels normalized to body weight. (E and F) Glucose and insulin tolerance test. Stars indicate comparison to controls, squares indicate comparison between db/db and LepRbG506S/G506S mice. (G–J) Fasting plasma levels of insulin (G), leptin (H), cholesterol (I) and triglycerides (J). (A–D) and (G–J) values represent mean and standard deviation, (E and F) values represent mean and standard error. C–E: n = 7–11; A, B, G, I, J: n = 6; F: n = 4–6; H: n = 3. *, P < 0.1234; **, P < 0.0332; ***, P < 0.0021, ****, P < 0.0002.
A glucose tolerance test showed that fasting plasma level of glucose in LepRbG506S/G506S mice was significantly lower compared to age matched db/db mice and only slightly elevated compared to WT littermates (Figure 4C). The difference between LepRbG506S/G506S and WT controls disappeared when plasma glucose was normalized to body weight (Figure 4D). LepRbG506S/G506S mice were glucose intolerant (Figure 4E); however, glucose levels came back to baseline faster than in db/db mice. It should be noted that blood glucose levels of db/db mice exceeded upper detection limit of the device at several time points indicating that peak level of db/db mice is higher than depicted. Despite differences in glucose metabolism, both LepRbG506S/G506S and db/db mice showed comparable insulin resistance during the ITT (Figure 4F).
In addition to plasma glucose, we also measured other parameters typically associated with obesity. As expected, plasma insulin was significantly increased in LepRbG506S/G506S mice compared to WT (Figure 4G). Interestingly, it was also increased in comparison to db/db mice. Both plasma leptin and cholesterol were significantly higher in LepRbG506S/G506S compared to WT mice and levels were comparable to those of db/db mice (Figure 4H,I). However, in contrast to db/db mice, triglycerides were not elevated in LepRbG506S/G506S mice (Figure 4J). Overall, these data show that while sharing many aspects of obesity, LepRbG506S/G506S mice are not as glucose intolerant as db/db and accumulate less fat.
3.5. Energy expenditure and food intake are differently regulated in LepRbG506S/G506S and db/db mice
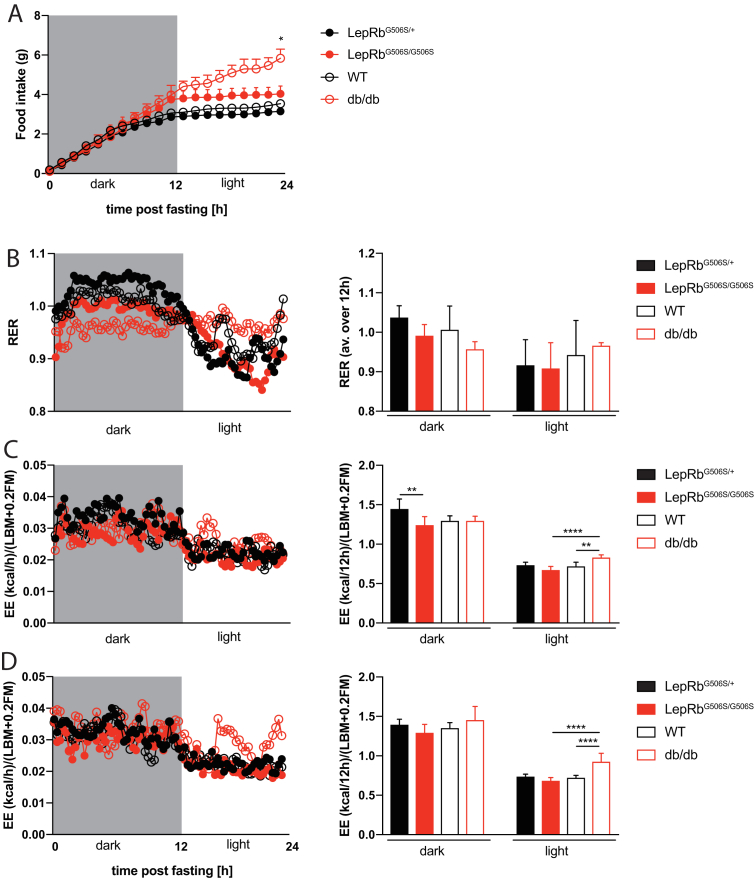
To investigate the metabolic state of LepRbG506S/G506S mice, they were put into metabolic cages and food intake, energy expenditure, oxygen consumption and CO2 production during day and night were determined and compared to db/db mice. As expected by the increased body weight, both db/db and LepRbG506S/G506S mice consumed more food than lean littermates (Figure 5A), but a difference was observed during the light phase, when db/db mice continued to eat, in contrast to the other groups. LepRbG506S/G506S mice ate until the very end of the dark phase while WT and heterozygous controls already reduced food consumption few hours before the end of the dark phase. Body weight of LepRbG506S/G506S and db/db mice was comparable at the time when this measurement was performed.
Figure 5.
Inhibition of food intake is compromised in LepRbG506S/G506Smice, but energy expenditure is comparable to lean littermates. (A) Food intake during 24 hours following 2 hours starvation, starting from 12 hours of dark. Values represent mean and standard error, stars indicate comparison to controls. (B) Respiratory exchange ratio and average of 12 hours dark or light. (C) Energy expenditure and sum of 12 hours dark or light. (D) Energy expenditure after 2 hours fasting and sum of 12 hours dark or light. (B–D) Dots represent mean; bars represent mean and standard deviation. n = 4 or 6/group. *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001.
The respiratory exchange ratio (RER, Figure 5B) also revealed a difference between LepRbG506S/G506S and db/db mice. While LepRbG506S/G506S and control groups switched from using carbohydrate oxidation (RER 1.0) during the dark phase to lipid oxidation (RER 0.7 if only fat is oxidized) during the light phase as an energy source, the RER of db/db mice did not differ markedly between day and night, which may be explained by continued food intake of db/db but not LepRbG506S/G506S mice during day time.
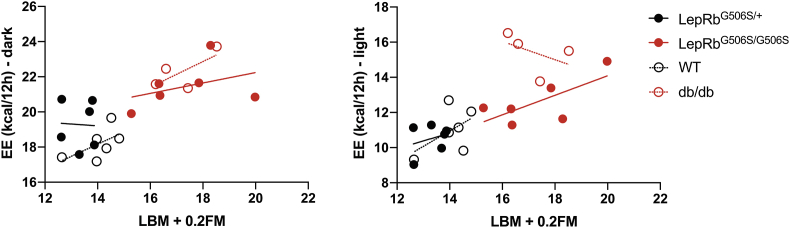
Finally, we compared energy expenditure to determine if this could explain the difference in fat mass observed between LepRbG506S/G506S mice and db/db. Indeed, during the light phase, LepRbG506S/G506S mice had an EE that was comparable to lean controls, whereas it was increased in db/db (Figure 5C). This difference became more obvious when looking at EE after a 2 hour fast (Figure 5D), when EE in LepRbG506S/G506S mice and control wild-type were comparably low while db/db's EE was elevated. ANCOVA analysis of EE and LBM+0.2FM, as recommended by Tschöp et al. [46], confirmed that the difference in EE is largely due to the mouse genotype and has very little correlation with LBM+0.2FM (Supplementary Figure 1), confirming the analysis of EE normalized to LBM+0.2FM.
Taken together, these results indicate that LepRbG506S/G506S and db/db mice differ in the control of energy expenditure. This mouse model might be useful to address the role of leptin signaling in different neurons without the need to generate conditional knockouts and targeted re-expression of the receptor in global knockouts.
4. Discussion
We discovered a novel spontaneous mutation in the leptin receptor gene that causes the development of obesity in homozygous mice. The DNA sequence mutation from G to A has two consequences: it generates an amino acid exchange (G506S), and it also creates an additional acceptor site for splicing that, when used, causes a 40 amino acid internal deletion in the receptor (LepRbΔ40aa). Since the position of the mutation is close to the leptin binding domain, the amino acid exchange could affect leptin binding and therefore signal transduction, irrespective of whether alternative splicing occurred or not. However, transfection of 3T3 cells with the full length LepRb transcript encoding the G506S point mutation proved that the exchange of this single amino acid did not compromise leptin signaling (i.e. STAT3 and ERK activation). In contrast, transfection of 3T3 cells with LepRbΔ40aa revealed that the alternatively spliced form of leptin receptor could not be expressed on the cell surface, indicating that abrogated or impaired cell surface expression, rather than impaired leptin binding due to the amino acid exchange, may be the underlying reason for the development of obesity in mice carrying this mutation. Since we found both spliced (LepRbΔ40aa) and unspliced transcripts with the G506S mutation in the medio-basal hypothalamus of homozygous LepRbG506S/G506S mice, we consider two likely scenarios. LepRbΔ40aa may exert a dominant negative effect on LepRbG506S, as LepRb needs to dimerize to allow translocation to the cell surface [2]. Association of LepRbΔ40aa with LepRbG506S monomers could prevent a functional dimerization of the receptor and therefore abrogate cell surface expression. Alternatively, the level of functionally intact LepRbG506S dimers on the cell surface could be too low to reach a threshold required for optimal signaling. The two possibilities are not mutually exclusive.
Leptin-induced pSTAT3 analysis revealed that leptin signaling was differentially affected in neurons localized in the DMN, VMN and ARC of the mutated mice. Abrogated signaling in response to exogenous leptin was observed in the VMN and in the DMN, suggesting that the LepRbΔ40aa product predominates in these neurons and prevents responsiveness to leptin in this region. On the other hand, leptin signaling was only halved in the ARC. Interestingly POMC neurons in the ARC of LepRbG506S/G506S mice showed normal pSTAT3 activation in response to leptin, suggesting that reduced signaling in the ARC may be caused by defective LepRb signaling in AgRP neurons. Unfortunately, staining of AgRP neurons in brain sections to verify this hypothesis is not possible. To reach a more definitive conclusion, selective depletion of AgRP neurons in LepRG506S/G506S mice will be required, for instance by crossing them to AgrpDTR mice [47], which still remains to be done. However AgrpDTR LepRG506S/G506S mice may have additional phenotypes caused by other functions of AgRP neurons independent of leptin.
We propose that the reduced number of pSTAT3+ cells found in the ARC is due to a difference in the efficiency of alternative splicing among different neuron populations, leading to efficient alternative splicing of the mutant LepRb transcript in SF1 neurons of the VMN and AgRP in the ARC but not in POMC neurons, possibly due to a different distribution of splicing factors. Since we hypothesized that the decrease in pSTAT3 signaling may be taking place in AgRP neurons and that no signal is present in VMN and DMN neurons, AgRP, VMN, and DMN neurons would be predicted to have similar splicing patterns generating the truncated LepRb protein that does not respond to leptin. This may be the underlying molecular reason for the differences in metabolic dysfunctions between LepRbG506S/G506S mice and db/db mice. The development of a massive obesity due to a defect in the inhibition of food intake is comparable in the two strains even though POMC signaling is maintained in the mutated mice. Interestingly, previous experiments showed that selective reconstitution of LepRb expression in AgRP neurons in db/db mice prevented uncontrolled food intake and obesity [37]. In contrast, selective depletion of leptin receptor in the VMN did not change food intake on normal chow diet [32], [33]. Similarly, depletion of LepRb from DMN neurons did not have any effect in food intake [38]. These data suggest that AgRP neurons in the ARC rather than VMN or DMN neurons play a pivotal role in the control of food intake. Furthermore, selective depletion or expression of the leptin receptor on POMC neurons does not change food intake [34], [35]. Thus, obesity and dysregulated food intake in LepRbG506S/G506S mice is most likely explained by defective signaling of the LepRbΔ40aa in AgRP neurons.
Even more interesting than the similarity in the development of obesity of db/db and LepRbG506S/G506S mice are the quantitative differences in glucose intolerance and fat accumulation. Fasting plasma glucose was lower in LepRbG506S/G506S compared to db/db mice, despite comparable body weight. Based on the altered leptin signaling observed in LepRbG506S/G506S mice, we speculate that improved glucose tolerance results from intact function of POMC neurons in the ARC. Although we cannot definitively conclude that this is due to a direct role of POMC neurons in the regulation of glucose homeostasis or rather by an indirect effect caused by the different body composition and slightly lower body weight of LepRbG506S/G506S mice compared to db/db mice, we favor the second hypothesis based on the data obtained. Normalization of fasting plasma glucose by body weight showed that LepRbG506S/G506S were comparable to lean littermates, while db/db mice had significantly higher plasma glucose, even when normalized. Notably, insulin resistance was comparable in LepRbG506S/G506S and db/db mice. This indicates that the improved control of glycemia in LepRbG506S/G506S compared to db/db mice occurs by an indirect mechanism such as reduced accumulation of adipose tissue in LepRbG506S/G506S mice. Most likely, the increased amount of food consumed by LepRbG506S/G506S mice is more efficiently converted into lean mass than in db/db mice.
It has been suggested previously that LepRb in the ARC, and in particular POMC neurons, play a major role controlling glucose homeostasis [35]. Melanocortin, which is produced by POMC neurons upon leptin stimulation, plays a crucial role in the regulation of glucose uptake by muscle cell. While food intake was unchanged, the amount of abdominal fat was reduced by treatment with α-MSH, and the response to insulin was potentiated, whereas melanocortin receptor antagonist mediated the opposite effect [48]. We also observed less subcutaneous fat in LepRbG506S/G506S mice compared to db/db. A plausible explanation is that the melanocortin system is at least partially intact in LepRbG506S/G506S mice, which is consistent with normal leptin-induced STAT3 activation observed in the POMC population. An intact melanocortin system may limit fat accumulation and ensure a better glycemic control without affecting food consumption. Indeed, mice expressing LepRb only on POMC neurons (LeprloxTB × POMC-cre mice) show obesity and food intake comparable to a full body LepRb KO, while energy expenditure, glycemia, and insulinemia were almost normal [35]. From 12 weeks of age LeprloxTB × POMC-cre mice start to display a difference in body weight compared to global LeprloxTB KO with global deletion, which is entirely due to less accumulation of fat mass. In addition, LepRb expression exclusively in POMC neurons reduced plasma cholesterol compared to global KO and restored triglycerides to levels of WT. Similarly, LepRbG506S/G506S mice with intact LepRb signaling in POMC neurons but defective LepRb in SF1 and AgRP neurons show improvement in glycemia, insulinemia, and accumulation of adipose tissue and plasma triglycerides compared to db/db mice. In addition, their energy expenditure is comparable to WT lean mice and, at day time, lower than in db/db. A possible explanation is that uncontrolled food intake during the day time accounts for increased expenditure in db/db mice, whereas the other groups rest during the day and therefore their energy expenditure is significantly lower and also lower than night time when they are more active [49].
These observations strongly support the hypothesis that alternative splicing of the LepRb occurs with different efficiencies in different neurons, thus abrogating the signal completely in some and leaving it almost intact in others. As a consequence, hyperphagic, obese mutant mice are able to control energy expenditure, reducing the accumulation of adipose tissue and ensuring improved glucose tolerance even by insulin resistance.
This natural mutation should be useful to separately and specifically study the role of leptin signaling in hypothalamic VMN and ARC and metabolic consequences.
Author contributions
FP, CLF, CS, ER performed and analyzed experiments. CLF, JK, TL, PN designed experiments and provided critical feedback. FP, MK designed study and experiments, wrote the manuscript, and obtained funding.
Acknowledgements
We thank Prof. Arnold von Eckhardstein, Clinical Chemistry, University Hospital Zürich, for measurement of cholesterol and triglycerides. This study was supported with funds from Swiss National Science Foundation (SNF 31003A_175458) to TL and the Swiss Federal Institute of Technology Zurich (ETH-28 18-2) to MK
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.04.010.
Conflict of interest
The authors declare no competing financial interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Figs1.
Supplementary Figure 1: Analysis of covariance of energy expenditure and LBM+0.2BM. ANCOVA of 12 hours energy expenditure (during dark and light respectively) and LBM+0.2FM. n = 4 or 6/group.
References
- 1.Matkovic V., Ilich J.Z., Badenhop N.E., Skugor M., Clairmont A., Klisovic D. Gain in body fat is inversely related to the nocturnal rise in serum leptin level in young females. Journal of Clinical Endocrinology & Metabolism. 1997;82:1368–1372. doi: 10.1210/jcem.82.5.3917. [DOI] [PubMed] [Google Scholar]
- 2.Moharana K., Zabeau L., Peelman F., Ringler P., Stahlberg H., Tavernier J. Structural and mechanistic paradigm of leptin receptor activation revealed by complexes with wild-type and antagonist leptins. Structure. 2014;22:866–877. doi: 10.1016/j.str.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Halaas J.L., Gajiwala K.S., Maffei M., Cohen S.L., Chait B.T., Rabinowitz D. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 4.Pelleymounter M.A., Cullen M.J., Baker M.B., Hecht R., Winters D., Boone T. Effects of the obese gene product on body weight regulation in ob/ob mice. Science. 1995;269:540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- 5.Ingalls A.M., Dickie M.M., Snell G.D. Obese, a new mutation in the house mouse. Journal of Heredity. 1950;41:317–318. doi: 10.1093/oxfordjournals.jhered.a106073. [DOI] [PubMed] [Google Scholar]
- 6.Tartaglia L.A., Dembski M., Weng X., Deng N., Culpepper J., Devos R. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Charlat O., Tartaglia L.A., Woolf E.A., Weng X., Ellis S.J. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 8.Hummel K.P., Dickie M.M., Coleman D.L. Diabetes, a new mutation in the mouse. Science. 1966;153:1127–1128. doi: 10.1126/science.153.3740.1127. [DOI] [PubMed] [Google Scholar]
- 9.Lee G.H., Proenca R., Montez J.M., Carroll K.M., Darvishzadeh J.G., Lee J.I. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 10.Baumann H., Morella K.K., White D.W., Dembski M., Bailon P.S., Kim H. The full-length leptin receptor has signaling capabilities of interleukin 6-type cytokine receptors. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8374–8378. doi: 10.1073/pnas.93.16.8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman D.L. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia. 1978;14:141–148. doi: 10.1007/BF00429772. [DOI] [PubMed] [Google Scholar]
- 12.Campfield L.A. Central mechanisms responsible for the actions of OB protein (leptin) on food intake, metabolism and body energy storage. Frontiers of Hormone Research. 2000;26:12–20. doi: 10.1159/000061019. [DOI] [PubMed] [Google Scholar]
- 13.Yen T.T., Allan J.A., Yu P.L., Acton M.A., Pearson D.V. Triacylglycerol contents and in vivo lipogenesis of ob/ob, db/db and Avy/a mice. Biochimica et Biophysica Acta. 1976;441:213–220. doi: 10.1016/0005-2760(76)90164-8. [DOI] [PubMed] [Google Scholar]
- 14.Koteish A., Diehl A.M. Animal models of steatosis. Seminars in Liver Disease. 2001;21:89–104. doi: 10.1055/s-2001-12932. [DOI] [PubMed] [Google Scholar]
- 15.Chehab F.F., Lim M.E., Lu R. Correction of the sterility defect in homozygous obese female mice by treatment with the human recombinant leptin. Nature Genetics. 1996;12:318–320. doi: 10.1038/ng0396-318. [DOI] [PubMed] [Google Scholar]
- 16.Cohen P., Zhao C., Cai X., Montez J.M., Rohani S.C., Feinstein P. Selective deletion of leptin receptor in neurons leads to obesity. Journal of Clinical Investigation. 2001;108:1113–1121. doi: 10.1172/JCI13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowalski T.J., Liu S.M., Leibel R.L., Chua S.C., Jr. Transgenic complementation of leptin-receptor deficiency. I. Rescue of the obesity/diabetes phenotype of LEPR-null mice expressing a LEPR-B transgene. Diabetes. 2001;50:425–435. doi: 10.2337/diabetes.50.2.425. [DOI] [PubMed] [Google Scholar]
- 18.Campfield L.A., Smith F.J., Guisez Y., Devos R., Burn P. Recombinant mouse OB protein: evidence for a peripheral signal linking adiposity and central neural networks. Science. 1995;269:546–549. doi: 10.1126/science.7624778. [DOI] [PubMed] [Google Scholar]
- 19.Elmquist J.K., Bjorbaek C., Ahima R.S., Flier J.S., Saper C.B. Distributions of leptin receptor mRNA isoforms in the rat brain. Journal of Comparative Neurology. 1998;395:535–547. [PubMed] [Google Scholar]
- 20.Fei H., Okano H.J., Li C., Lee G.H., Zhao C., Darnell R. Anatomic localization of alternatively spliced leptin receptors (Ob-R) in mouse brain and other tissues. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7001–7005. doi: 10.1073/pnas.94.13.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowley M.A., Smart J.L., Rubinstein M., Cerdan M.G., Diano S., Horvath T.L. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411:480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 22.Elias C.F., Aschkenasi C., Lee C., Kelly J., Ahima R.S., Bjorbaek C. Leptin differentially regulates NPY and POMC neurons projecting to the lateral hypothalamic area. Neuron. 1999;23:775–786. doi: 10.1016/s0896-6273(01)80035-0. [DOI] [PubMed] [Google Scholar]
- 23.Cheung C.C., Clifton D.K., Steiner R.A. Proopiomelanocortin neurons are direct targets for leptin in the hypothalamus. Endocrinology. 1997;138:4489–4492. doi: 10.1210/endo.138.10.5570. [DOI] [PubMed] [Google Scholar]
- 24.Wilson B.D., Bagnol D., Kaelin C.B., Ollmann M.M., Gantz I., Watson S.J. Physiological and anatomical circuitry between Agouti-related protein and leptin signaling. Endocrinology. 1999;140:2387–2397. doi: 10.1210/endo.140.5.6728. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz M.W., Baskin D.G., Bukowski T.R., Kuijper J.L., Foster D., Lasser G. Specificity of leptin action on elevated blood glucose levels and hypothalamic neuropeptide Y gene expression in ob/ob mice. Diabetes. 1996;45:531–535. doi: 10.2337/diab.45.4.531. [DOI] [PubMed] [Google Scholar]
- 26.Cone R.D. Anatomy and regulation of the central melanocortin system. Nature Neuroscience. 2005;8:571–578. doi: 10.1038/nn1455. [DOI] [PubMed] [Google Scholar]
- 27.Wu Q., Palmiter R.D. GABAergic signaling by AgRP neurons prevents anorexia via a melanocortin-independent mechanism. European Journal of Pharmacology. 2011;660:21–27. doi: 10.1016/j.ejphar.2010.10.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morton G.J., Cummings D.E., Baskin D.G., Barsh G.S., Schwartz M.W. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 29.Thornton J.E., Cheung C.C., Clifton D.K., Steiner R.A. Regulation of hypothalamic proopiomelanocortin mRNA by leptin in ob/ob mice. Endocrinology. 1997;138:5063–5066. doi: 10.1210/endo.138.11.5651. [DOI] [PubMed] [Google Scholar]
- 30.Wilding J.P., Gilbey S.G., Bailey C.J., Batt R.A., Williams G., Ghatei M.A. Increased neuropeptide-Y messenger ribonucleic acid (mRNA) and decreased neurotensin mRNA in the hypothalamus of the obese (ob/ob) mouse. Endocrinology. 1993;132:1939–1944. doi: 10.1210/endo.132.5.7682936. [DOI] [PubMed] [Google Scholar]
- 31.Satoh N., Ogawa Y., Katsuura G., Numata Y., Tsuji T., Hayase M. Sympathetic activation of leptin via the ventromedial hypothalamus: leptin-induced increase in catecholamine secretion. Diabetes. 1999;48:1787–1793. doi: 10.2337/diabetes.48.9.1787. [DOI] [PubMed] [Google Scholar]
- 32.Bingham N.C., Anderson K.K., Reuter A.L., Stallings N.R., Parker K.L. Selective loss of leptin receptors in the ventromedial hypothalamic nucleus results in increased adiposity and a metabolic syndrome. Endocrinology. 2008;149:2138–2148. doi: 10.1210/en.2007-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhillon H., Zigman J.M., Ye C., Lee C.E., McGovern R.A., Tang V. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron. 2006;49:191–203. doi: 10.1016/j.neuron.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 34.Balthasar N., Coppari R., McMinn J., Liu S.M., Lee C.E., Tang V. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Berglund E.D., Vianna C.R., Donato J., Jr., Kim M.H., Chuang J.C., Lee C.E. Direct leptin action on POMC neurons regulates glucose homeostasis and hepatic insulin sensitivity in mice. Journal of Clinical Investigation. 2012;122:1000–1009. doi: 10.1172/JCI59816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huo L., Gamber K., Greeley S., Silva J., Huntoon N., Leng X.H. Leptin-dependent control of glucose balance and locomotor activity by POMC neurons. Cell Metabolism. 2009;9:537–547. doi: 10.1016/j.cmet.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goncalves G.H., Li W., Garcia A.V., Figueiredo M.S., Bjorbaek C. Hypothalamic agouti-related peptide neurons and the central melanocortin system are crucial mediators of leptin's antidiabetic actions. Cell Reports. 2014;7:1093–1103. doi: 10.1016/j.celrep.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rezai-Zadeh K., Yu S., Jiang Y., Laque A., Schwartzenburg C., Morrison C.D. Leptin receptor neurons in the dorsomedial hypothalamus are key regulators of energy expenditure and body weight, but not food intake. Molecular Metabolism. 2014;3:681–693. doi: 10.1016/j.molmet.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lutz T.A., Coester B., Whiting L., Dunn-Meynell A.A., Boyle C.N., Bouret S.G. Amylin selectively signals onto POMC neurons in the arcuate nucleus of the hypothalamus. Diabetes. 2018;67:805–817. doi: 10.2337/db17-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weir J.B. New methods for calculating metabolic rate with special reference to protein metabolism. Journal of Physiology. 1949;109:1–9. doi: 10.1113/jphysiol.1949.sp004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Even P.C., Nadkarni N.A. Indirect calorimetry in laboratory mice and rats: principles, practical considerations, interpretation and perspectives. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2012;303:R459–R476. doi: 10.1152/ajpregu.00137.2012. [DOI] [PubMed] [Google Scholar]
- 42.Johnson M.D., Bouret S.G., Dunn-Meynell A.A., Boyle C.N., Lutz T.A., Levin B.E. Early postnatal amylin treatment enhances hypothalamic leptin signaling and neural development in the selectively bred diet-induced obese rat. American Journal of Physiology Regulatory Integrative and Comparative Physiology. 2016 doi: 10.1152/ajpregu.00326.2016. ajpregu 00326 02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banks A.S., Davis S.M., Bates S.H., Myers M.G., Jr. Activation of downstream signals by the long form of the leptin receptor. Journal of Biological Chemistry. 2000;275:14563–14572. doi: 10.1074/jbc.275.19.14563. [DOI] [PubMed] [Google Scholar]
- 44.White D.W., Kuropatwinski K.K., Devos R., Baumann H., Tartaglia L.A. Leptin receptor (OB-R) signaling. Cytoplasmic domain mutational analysis and evidence for receptor homo-oligomerization. Journal of Biological Chemistry. 1997;272:4065–4071. doi: 10.1074/jbc.272.7.4065. [DOI] [PubMed] [Google Scholar]
- 45.Le Foll C., Johnson M.D., Dunn-Meynell A.A., Boyle C.N., Lutz T.A., Levin B.E. Amylin-induced central IL-6 production enhances ventromedial hypothalamic leptin signaling. Diabetes. 2015;64:1621–1631. doi: 10.2337/db14-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tschop M.H., Speakman J.R., Arch J.R., Auwerx J., Bruning J.C., Chan L. A guide to analysis of mouse energy metabolism. Nature Methods. 2011;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luquet S., Perez F.A., Hnasko T.S., Palmiter R.D. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310:683–685. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- 48.Obici S., Feng Z., Tan J., Liu L., Karkanias G., Rossetti L. Central melanocortin receptors regulate insulin action. Journal of Clinical Investigation. 2001;108:1079–1085. doi: 10.1172/JCI12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butler A.A., Kozak L.P. A recurring problem with the analysis of energy expenditure in genetic models expressing lean and obese phenotypes. Diabetes. 2010;59:323–329. doi: 10.2337/db09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]