Abstract
Mutations of the β-catenin gene are common in various cancer types. MicroRNA (miR)-24 suppresses gene expression during the cell cycle. However, the effects of miR-24 on the cell viability and autophagy of glioma cells, and how these biological processes are regulated by β-catenin are largely unclear. The current study aimed to investigate the role of β-catenin in regulating the effects of miR-24 on the cell viability and autophagy of glioma cells. The expression levels of microtubule-associated proteins 1A/1B light chain 3B (LC3B) and Beclin1 were detected by immunohistochemistry and western blotting. Glioma C6 cells were transfected with miR-24 mimics, miR-24 inhibitors and negative control miRNAs. C6 cells transfected with miR-24 mimics or negative control miRNAs were treated with the β-catenin inhibitor, XAV-939. An MTT assay was utilized to evaluate the viability of C6 cells. The expression of miR-24 and mRNA expression of autophagy related 4a cysteine peptidase (ATG4A) were detected by quantitative polymerase chain reaction analysis. The protein expression of LC3B and Beclin1 decreased significantly in glioma tissue and glioma C6 cells compared with normal brain tissue. Compared with the negative control group, C6 cells transfected with miR-24 mimics exhibited significantly higher cell viability at 24 and 48 h, and those transfected with miR-24 inhibitors exhibited significantly lower cell viability at 48 h. XAV-939 decreased the stimulatory effects of miR-24 mimics on the viability of C6 cells. The expression of miR-24 significantly decreased and ATG4A mRNA significantly increased in C6 cells transfected with XAV-939 compared with those transfected with the negative control miRNA. XAV-939 attenuated the miR-24-induced decrease of the protein expression of LC3B and Beclin1, and decreased the stimulatory effects of miR-24 mimics on cell viability. In addition, XAV-939 attenuated the miR-24-induced decrease of autophagy marker expression by attenuating miR-24 expression and increasing ATG4A mRNA expression in glioma C6 cells. To the best of our knowledge, the present study is the first to demonstrate whether β-catenin regulates the intracellular effects of miR-24 on the viability and autophagy of glioma cells. The results also provide some mechanistic basis to the pharmaceutical targeting of WNT signaling in high grade glial tumors.
Keywords: β-catenin, microRNA-24, cell viability, autophagy, glioma C6 cells
Introduction
Gliomas comprise ~30% of brain and central nervous system tumors and 80% of all malignant brain tumors (1). The prognosis for patients with high-grade gliomas is generally poor, particularly in older patients. Notably, the median overall survival for grade IV glioblastoma is ~15 months (2). Catenins are a family of proteins found in complexes with the cell adhesion molecule, cadherin, in animal cells (3). The first two catenins that were identified were α-catenin and β-catenin. α-catenin can bind to β-catenin and actin. β-catenin binds the cytoplasmic domain of numerous cadherins (4). β-catenin is a dual function protein as it is involved in the coordination and regulation of cell-cell adhesion and gene transcription (5). The β-catenin gene is a proto-oncogene and mutations in the gene are commonly found in a variety of cancers, including primary hepatocellular carcinoma, colorectal cancer, skin cancer, prostate cancer and glioblastoma (6–10).
miR-24 is conserved in various species, and is clustered with miR-23 and miR-27 on human chromosome 9 and 19 (11). miR-24 was reported to suppress the expression of genes that are crucial for cell cycle control in hematopoietic differentiation, including E2F2 and myc (12). miR-24 also promoted the differentiation of keratinocytes by repressing actin-cytoskeleton regulators, including PAK4, Tsk5 and Rho GTPase-activating protein 19 (13). miR-24 was revealed to reduce the mRNA and protein levels of human activin receptor type-1B by targeting the 3′-untranslated region of the mRNA (14). Tripartate motif-containing protein 11, a direct target of miR-24-3p, was reported to promote cell proliferation and inhibit apoptosis in colon cancer (15). Additionally, overexpression of miR-24-3p in the small cell lung cancer cell line H446/EP led to a reduction of the autophagy related 4a cysteine peptidase (ATG4A) protein level, allowing small cell lung cancer cells to re-sensitize to the combination of chemotherapeutic etoposide (VP16) and cisplatin (DDP) (16). Therefore, to examine the direct function of miR-24, the mRNA expression of ATG4A, and protein expression of Beclin1 and microtubule-associated proteins 1A/1B light chain 3B (LC3B) were measured in the current study. Beclin1 and LC3B are essential proteins associated with autophagy (17). The effects of miR-24 on cell viability and autophagy of glioma cells, and how these biological processes are regulated by β-catenin remain unclear. Therefore, the role of β-catenin in regulating the effects of miR-24 on cell viability and autophagy of glioma cells was also examined.
Materials and methods
Cells, animals and reagents
Rat glioma C6 cells were purchased from iCell Bioscience, Inc. (Shanghai, China). Three male Sprague Dawley rats (weight range, 180 to 220 g; 6 weeks old) were purchased from JSJ Laboratory, Inc. (Shanghai, China). Three Sprague Dawley rats bearing C6 glioma were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). The animals were housed altogether in cages at ambient temperature under a 12-h light/dark cycle with free access to standard pelleted food and water. The project and number of animals were approved by the Animal Ethics Committee of Shanghai Changhai Hospital (Shanghai, China).
Dulbecco's modified Eagle medium (DMEM), fetal bovine serum, trypsin, penicillin and streptomycin antibiotics were purchased from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA). MTT cell proliferation assay kit was purchased from Aladdin Shanghai Biochemical Technology Co., Ltd. (Shanghai, China). Primary antibodies against GAPDH (ab181602) and LC3B (ab48394) were from Abcam (Cambridge, MA, USA); primary antibodies against Beclin1 (3495) and β-catenin (8480) were from Cell Signaling Technology, Inc. (Danvers, MA, USA); goat anti-rabbit antibody (65–6120) for western blotting and TRIzol™ reagent were from Invitrogen (Thermo Fisher Scientific, Inc.). TUNEL apoptosis detection kit was from Shanghai Yeasen Biotechnology Co., Ltd. (Shanghai, China). QuantScript RT kit and miRcute miRNA qPCR Detection kit were from Tiangen Biotech Co., Ltd. (Beijing, China). Specific primers for miR-24 and U6 were purchased from Sangon Biotech Co., Ltd. (Shanghai, China).
The equipment used was as follows: Cell incubator (Thermo Fisher Scientific, Inc.); microplate reader (Shanghai Kehua Bio-engineering Co., Ltd., Shanghai, China); light microscope (Olympus Corporation, Tokyo, Japan); table-type refrigerated centrifuge (USTC Zonkia Inc., Hefei, China); electric thermostatic drying oven (Huyue Inc., Shangyu, China); electrophoresis system (Beijing Liuyi Biotechnology Co., Ltd., Beijing, China); T-100 PCR machine (Bio-Rad Laboratories, Inc., Hercules, CA, USA); and StepOnePlus™ Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Transfection of miRNA
Following the culturing of rat glioma C6 cells at 37°C for 24 h, the complete DMEM medium was replaced with DMEM medium without serum and antibiotics. A mixture of Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) and miRNAs was prepared, including negative control miRNA (5-UUC UCC GAA CGU GUC ACG UTT-3), miR-24-3p mimics (5-UGG CUC AGU UCA GCA GGA ACA G-3) and miR-24-3p inhibitors (5-CUG UUC CUG CUG AAC UGA GCC A-3). Cells that were transfected with miRNA negative control served as controls. A total of 125 µl OPTI-MEM (Thermo Fisher Scientific, Inc.) was utilized to dilute RNA. A total 125 µl OPTI-MEM was also used to dilute 10 µl Lipofectamine 2000 and incubated at room temperature for 5 min. The prepared miRNA mixture and Lipofectamine 2000 mixture were then mixed, incubated at room temperature for 20 min, and added into antibiotic-free DMEM culture medium with glioma C6 cells. The final concentration of miR-24 inhibitors used was 100 nM and the final concentration of miR-24 mimics was 50 nM. The culture medium was changed to the complete DMEM medium after transfection of 4–8 h. The cells were typically cultured at 37°C for 48 h until harvested for subsequent experiments.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Glioma C6 cells were treated with miR-24-3p mimics, XAV-939 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) or a combination of XAV-939 and miR-24-3p mimics for 48 h. XAV-939 is a well-established Tankyrase inhibitor of the Wnt/β-catenin signaling cascade (18). For the combination group, the C6 cells were first treated with 50 nM miR-24-3p mimics then with 1 µM XAV-939 until harvested for the next procedure. Total RNA of C6 cells was extracted and purified by TRIzol, according to the manufacturer's protocol. A QuantScript RT kit (Tiangen Biotech Co., Ltd.) was utilized for reverse transcription at 37°C for 60 min. Each reaction contained 1 µl random hexamer primers (0.2 µg/µl) and 40 U M-MuLV Reverse Transcriptase (20 U/µl). The specific primer for the detection of miR-24 was 5-GGC TCA GTT CAG CAG GAA CA-3, whereas the patented reverse primer was supplied by the kit. he primer for ATG4A mRNA was forward (F): 5-AAC TGT GAC TGA GCC GAT TG-3′; reverse (R): 5-GTC TTT CAG GGA TGA CTT GGT G-3′. The primer for U6 was F: 5-CTC GCT TCG GCA GCA CA-3; R: 5-AAC GCT TCA CGA ATT TGC GT-3. miRcute miRNA qPCR Detection kit (Tiangen Biotech Co., Ltd.) was used for qPCR analysis. PCR conditions were as follows: Pre-denaturing at 94°C for 2 min, 40 cycles of denaturing at 94°C for 20 sec, and annealing and polymerization at 60°C for 34 sec, according to the supplied manual. PCR was performed in a StepOnePlus™ Real-Time PCR system. The relative expression of miR-24 and ATG4A was determined with the 2−ΔΔCq method (19), using U6 as the reference gene.
Immunohistochemistry (IHC)
The rats were euthanized by intraperitoneal injection with 40 mg/body weight kg sodium pentobarbital, following the guidelines by the Animal Ethics Committee of Shanghai Changhai Hospital. Normal brain tissue and glioma tissue from C6 glioma-bearing rats were surgically isolated and fixed in 10% neutral buffered formalin (Thermo Fisher Scientific, Inc.) at room temperature for 2 h, embedded in paraffin, and cut into 5 µm sections. The slides were then dewaxed and rehydrated. Following antigen retrieval, slides were blocked with bovine serum albumin, and incubated at 4°C overnight with anti-LC3B (1:400) and anti-Beclin1 (1:130) antibodies (Abcam). Next the slides were washed with PBS and incubated in the dark at 37°C with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibodies (1:50; OriGene Technologies, Inc., Beijing, China) for 30 min. Slides were observed under light microscope.
Western blotting analysis
The expression levels of LC3B and Beclin1 proteins were detected by western blotting. Cellular proteins of glioma C6 cells in different treatment groups were extracted with self-prepared radioimmunoprecipitation assay buffer (25 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100). Total protein concentration was determined by the BCA assay. The total proteins (~30 µg per lane) were separated by electrophoresis (120 V) on a 10% SDS-PAGE. The separated proteins were then electrophoretically (100 V for 120 min) transferred to polyvinylidene fluoride membranes. Following the blocking of the membranes with 5% non-fat milk powder for 1 h at room temperature, the membranes were incubated with anti-LC3B (1:1,000), anti-Beclin1 (1:1,000) and anti-GAPDH (1:5,000) antibodies at 4°C overnight. Following incubation, the membranes were washed three times with a solution of Tris-buffered saline with Tween-20. The membranes were then incubated for 1 h at room temperature with goat anti-rabbit secondary antibody labeled with HRP (1:3,000). They were washed and incubated for a short time period in enhanced chemiluminescence (ECL) solution (Chemiluminescent Western Blot Detection Kit, Thermo Fisher Scientific, Inc.). The films were exposed in a dark room. The densitometry was determined suing ImageJ bundled with 64-bit Java 1.6.0_24 (National Institutes of Health).
MTT assay
The cell viability of glioma C6 cells in different treatment groups was detected by MTT assay. Following culturing, 20 µl MTT solution (5 mg/ml) was added to each well in the 96 well plates and the cells were cultured at 37°C for 4 h. Cell supernatants were removed and discarded, and 150 µl dimethyl sulfoxide was added to each well. The plates were then agitated for 15 min to dissolve the purple formazan crystals and the absorbance of each sample was detected at 570 nm using an ELISA microplate reader. Relative cell viability was calculated by dividing the absorbance of the experimental groups by the absorbance of the control group.
Statistical analysis
Statistical analysis was performed and figures were created using GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Data are presented as the mean ± standard error of the mean and each experiment was performed in triplicate. Comparison between two groups was performed using Student's t tests and nonparametric tests. Differences between ≥3 groups were compared by one-way analysis of variance followed by the Bonferroni post hoc test. Differences between two groups were compared by unpaired t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
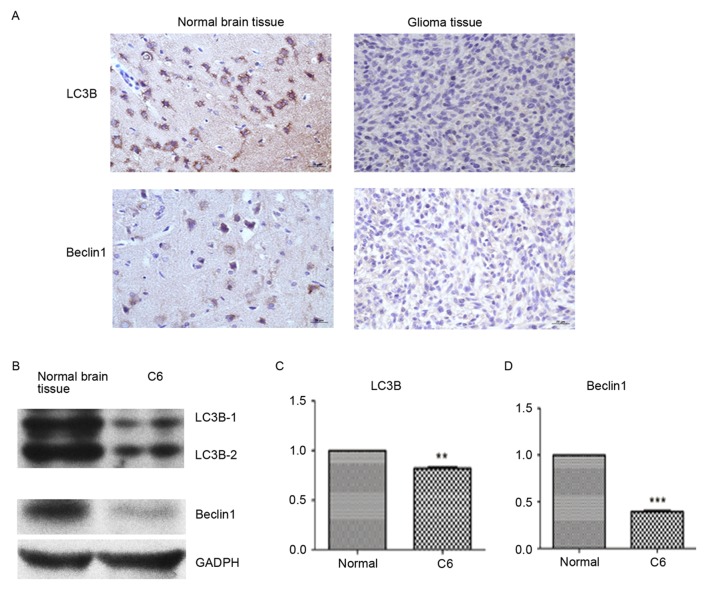
LC3B and Beclin1 protein expression decreases in glioma tissue and glioma C6 cells
The expression of LC3B and Beclin1 was detected by colorimetric IHC and western blotting. Compared with normal brain tissue, the expression of LC3B and Beclin1 was visually markedly decreased in glioma tissue (Fig. 1A). Similarly, the protein expression of LC3B and Beclin1 was detected by western blotting and determined by densitometry to be markedly decreased compared with normal brain tissue (Fig. 1B). LC3B (P<0.01) and Beclin1 (P<0.001) protein expression levels were significantly lower in glioma tissue compared to normal brain tissue (Fig. 1C and D).
Figure 1.
LC3B and Beclin1 protein expression decreases in glioma tissue and glioma C6 cells. The expression of LC3B and Beclin1 was detected by immunofluorescence and western blot analysis. (A) Immunohistochemistry results of normal brain tissue and glioma tissue. (B) Representative western blotting results of normal brain tissue and glioma C6 cells. Statistical analyses of (C) LC3B and (D) Beclin1 protein expression. Data are presented as the mean ± standard error of the mean; n=3/group. *P<0.01 and ***P<0.001 vs. normal brain tissue. LC3B, microtubule-associated proteins 1A/1B light chain 3B.
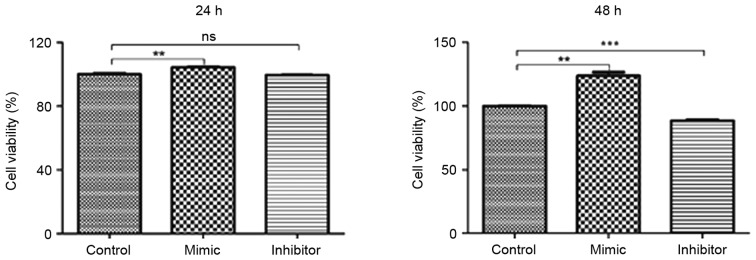
Glioma C6 cells transfected with miR-24 mimics exhibit greater cell viability
Glioma C6 cells were transfected with miR-24 mimic, miR-24 inhibitor and the negative control. An MTT assay was utilized to evaluate the viability of C6 cells. Compared with the negative control group, C6 cells transfected with miR-24 mimics exhibited significantly greater cell viability at 24 and 48 h (both P<0.01; Fig. 2). C6 cells transfected with miR-24 inhibitor exhibited significantly decreased cell viability at 48 h compared with the negative control group (P<0.001). No significant differences were identified in cell viability between the miR-24 inhibitor and negative control groups at 24 h.
Figure 2.
Glioma C6 cells transfected with miR-24 mimics exhibit greater cell viability. Glioma C6 cells were transfected with miR-24 mimics, miR-24 inhibitors and the negative control miRNA. MTT assays were utilized to evaluate the viability of C6 cells at 24 and 48 h. Data are presented as the mean ± standard error of the mean; n=3/group. **P<0.01 and ***P<0.001. ns, not significant; miR, microRNA.
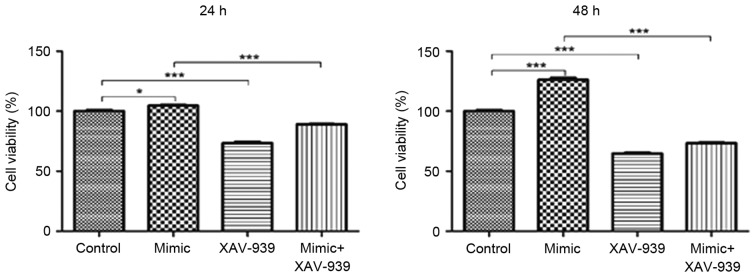
XAV-939 attenuates the miR-24 mimic-induced increase on the viability of glioma C6 cells
Glioma C6 cells transfected with miR-24 mimics or the negative control miRNA were treated with the β-catenin inhibitor, XAV-939. MTT assays were utilized to evaluate the viability of C6 cells. The viability of C6 cells was significantly decreased following the treatment with β-catenin inhibitor XAV-939 at 24 and 48 h compared with the negative control group (P<0.001, Fig. 3). In addition, the viability of C6 cells transfected with miR-24 mimics and treated with XAV-939 was significantly decreased at 24 and 48 h compared with C6 cells that were transfected with miR-24 mimics and untreated (P<0.001).
Figure 3.
β-catenin inhibitor XAV-939 decreases the stimulatory effects of miR-24 mimics on the viability of glioma C6 cells. Glioma C6 cells were transfected with negative control or miR-24 mimics and treated with β-catenin inhibitor, XAV-939. MTT assays were utilized to evaluate the viability of C6 cells at 24 and 48 h. Data are presented as the mean ± standard error of the mean; n=3/group. *P<0.05 and ***P<0.001. miR, microRNA.
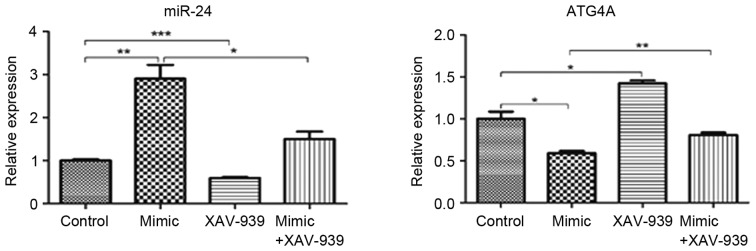
Expression of miR-24 decreases and ATG4A increases following treatment with XAV-939 in glioma C6 cells transfected with miR-24
Glioma C6 cells transfected with miR-24 mimics or negative control miRNA were treated with β-catenin inhibitor XAV-939. The mRNA expression levels of miR-24 and ATG4A were detected by RT-qPCR analysis. The expression of miR-24 significantly decreased in C6 cells transfected with miR-24 mimics and treated with XAV-939 compared with C6 cells transfected with miR-24 mimics (P<0.05; Fig. 4). Similarly, in C6 cells transfected with the negative control miRNA, miR-24 expression significantly decreased following XAV-939 treatment (P<0.001). In addition, ATG4A expression significantly increased following XAV-939 treatment either in C6 cells transfected with miR-24 mimics (P<0.01) or the negative control miRNA (P<0.05).
Figure 4.
miR-24 expression decreases and ATG4A increases following treatment with β-catenin inhibitor XAV-939 in glioma C6 cells transfected with miR-24. Glioma C6 cells transfected with miR-24 mimics, the negative control miRNA or either miRNA and the β-catenin inhibitor, XAV-939. The expression of miR-24 and mRNA expression of ATG4A were detected by reverse transcription-quantitative polymerase chain reaction analysis. Data are presented as the mean ± standard error of the mean; n=3/group. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; ATG4A, autophagy related 4a cysteine peptidase.
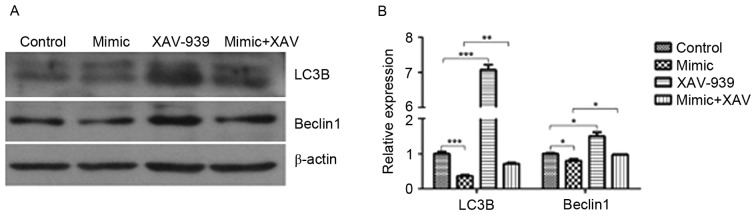
XAV-939 attenuates the miR-24-induced decrease in the protein expression of LC3B and Beclin1
The protein expression of LC3B and Beclin1 in glioma C6 cells was detected by western blotting. miR-24 induced a significant decrease in the protein expression of LC3B (P<0.001) and Beclin1 (P<0.05) compared with the cells transfected with negative control miRNA (Fig. 5). The protein expression of LC3B and Beclin1 in negative control miRNA and XAV-939-treated C6 cells was significantly increased compared with those treated with the negative control miRNA (P<0.001 and P<0.05, respectively). The protein expression of LC3B and Beclin1 in miR-24 mimic and XAV-939-treated C6 cells was significantly increased compared with those treated with miR-24 mimics alone (P<0.01 and P<0.05, respectively).
Figure 5.
XAV-939 attenuates the miR-24-induced decrease in the protein expression of LC3B and Beclin1. Glioma C6 cells were transfected with miR-24 mimics or negative control miRNA and treated with the β-catenin inhibitor, XAV-939. (A) Protein expression of LC3B and Beclin1 in glioma C6 cells was detected by western blotting. (B) Statistical analysis LC3B and Beclin1 expression in glioma C6 cells as assessed by western blotting. Data are presented as the mean ± standard error of the mean; n=3/group. *P<0.05, **P<0.01 and ***P<0.001. XAV, XAV-939; LC3B, microtubule-associated proteins 1A/1B light chain 3B; miR, microRNA.
Discussion
It was demonstrated in the present study that the β-catenin inhibitor, XAV-939, attenuates the miR-24 mimic-induced increase on the viability of glioma C6 cells. XAV-939 attenuated the miR-24-induced decrease in autophagy markers by decreasing miR-24 expression and increasing ATG4A expression in glioma C6 cells. This indicates that β-catenin promotes an increase in miR-24-induced increase cell viability in glioma C6 cells. It is likely that there is a β-catenin binding site at the promoter region of miR-24 or miR-24 stimulates cell proliferation via the β-catenin signaling pathway. The β-catenin gene is a proto-oncogene and its mutations are commonly found in a variety of cancers, including primary hepatocellular carcinoma, colorectal cancer, skin cancer, prostate cancer and glioblastoma (6–8,20,21). miR-96 was previously demonstrated to contribute to glioma tumor progression by activating the Wnt/β-catenin signaling pathway. Additionally, the miR-96/HMG box-containing protein 1/Wnt/β-catenin regulatory circuitry promoted the proliferation of glioma cells (22). It was also revealed that miR-603 promoted glioma cell growth via the Wnt/β-catenin signaling pathway by inhibiting Wnt inhibitory factor 1 and β-catenin-interacting protein 1 (23).
The ATG4A gene is the target gene of miR-24 (16). The results of the current study revealed that β-catenin increased miR-24 expression, thereby inhibiting autophagy in gliomas. β-catenin may bind to the promoter region of miR-24, and thus increase the expression and function of miR-24. In addition, it is possible that the β-catenin signaling pathway is involved in the downstream effects of miR-24 on autophagy. TGF-β1-induced autophagy was demonstrated to link β-catenin and Smad signaling to promote epithelial-mesenchymal transition in mouse tubular epithelial cells through the pY654-β-catenin/p-Smad2/integrin-linked protein kinase signaling pathway (20). Tissue kallikrein was reported to promote cell survival rate and β-catenin degradation in serum-starved SH-SY5Y cells via increasing autophagy (24). Additionally, the overexpression of progranulin inhibited TNF-α-induced inflammation in keratinocytes by positively mediating autophagy through the Wnt/β-catenin signaling pathway (25). More studies are required to elucidate the detailed role of β-catenin in the miR-24-mediated inhibition of autophagy. To the best of our knowledge, this is the first to demonstrate that β-catenin regulates the intracellular effects of miR-24 on the viability and autophagy of glioma cells. The present results also provide a direct mechanistic basis to support the growing pharmaceutical interest in targeting WNT signaling in high grade brain tumors (26).
Acknowledgements
Not applicable.
Funding
The present study received financial support from the National Science Foundation of China (grant no. ISIS: 81502163).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZY and DD conceived the idea and designed the study. HC and QL conducted the experiments, analysed the data and drafted the manuscript. CC, YD, YL and WM participated in the data acquisition and analysis. All authors read and approved the final version of this manuscript.
Ethics approval and consent to participate
The present study was approved by the Animal Ethics Committee of Shanghai Changhai Hospital (Shanghai, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205:613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Bleeker FE, Molenaar RJ, Leenstra S. Recent advances in the molecular understanding of glioblastoma. J Neurooncol. 2012;108:11–27. doi: 10.1007/s11060-011-0793-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weis WI, Nelson WJ. Re-solving the cadherin-catenin-actin conundrum. J Biol Chem. 2006;281:35593–35597. doi: 10.1074/jbc.R600027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saldanha G, Ghura V, Potter L, Fletcher A. Nuclear beta-catenin in basal cell carcinoma correlates with increased proliferation. Br J Dermatol. 2004;151:157–164. doi: 10.1111/j.1365-2133.2004.06048.x. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao H, Guo L, Chen L, Qiao G, Meng X, Xu B, Ye W. MSX1 inhibits cell migration and invasion through regulating the Wnt/β-catenin pathway in glioblastoma. Tumour Biol. 2016;37:1097–1104. doi: 10.1007/s13277-015-3892-2. [DOI] [PubMed] [Google Scholar]
- 7.Singh T, Katiyar SK. Green tea polyphenol, (−)-epigallocatechin-3-gallate, induces toxicity in human skin cancer cells by targeting β-catenin signaling. Toxicol Appl Pharmacol. 2013;273:418–424. doi: 10.1016/j.taap.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong HJ, Jang GB, Lee HY, Park SR, Kim JY, Nam JS, Hong IS. The Wnt/β-catenin signaling/Id2 cascade mediates the effects of hypoxia on the hierarchy of colorectal-cancer stem cells. Sci Rep. 2016;6:22966. doi: 10.1038/srep22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei A, Fan B, Zhao Y, Zhang H, Wang L, Yu X, Yuan Q, Yang D, Wang S. ST6Gal-I overexpression facilitates prostate cancer progression via the PI3K/Akt/GSK-3β/β-catenin signaling pathway. Oncotarget. 2016;7:65374–65388. doi: 10.18632/oncotarget.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL, et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut. 2016;65:1522–1534. doi: 10.1136/gutjnl-2015-310625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Jr, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, et al. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to ‘seedless’ 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amelio I, Lena AM, Viticchiè G, Shalom-Feuerstein R, Terrinoni A, Dinsdale D, Russo G, Fortunato C, Bonanno E, Spagnoli LG, et al. miR-24 triggers epidermal differentiation by controlling actin adhesion and cell migration. J Cell Biol. 2012;199:347–363. doi: 10.1083/jcb.201203134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Huang Z, Xue H, Jin C, Ju XL, Han JD, Chen YG. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- 15.Yin Y, Zhong J, Li SW, Li JZ, Zhou M, Chen Y, Sang Y, Liu L. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget. 2016;7:86755–86765. doi: 10.18632/oncotarget.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan B, Chen Y, Song H, Xu Y, Wang R, Chen L. Mir-24-3p downregulation contributes to VP16-DDP resistance in small-cell lung cancer by targeting ATG4A. Oncotarget. 2015;6:317–331. doi: 10.18632/oncotarget.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherra SJ, III, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, Chu CT. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Cui Y, Ma W, Lei F, Li Q, Su Y, Lin X, Lin C, Zhang X, Ye L, Wu S, et al. Prostate tumour overexpressed-1 promotes tumourigenicity in human breast cancer via activation of Wnt/β-catenin signalling. J Pathol. 2016;239:297–308. doi: 10.1002/path.4725. [DOI] [PubMed] [Google Scholar]
- 21.Wu S, Wang S, Zheng S, Verhaak R, Koul D, Yung WK. MSK1-mediated β-catenin phosphorylation confers resistance to PI3K/mTOR inhibitors in glioblastoma. Mol Cancer Ther. 2016;15:1656–1668. doi: 10.1158/1535-7163.MCT-15-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan Z, Wang J, Wang C, Jiao Y, Qi W, Che S. miR-96/HBP1/Wnt/β-catenin regulatory circuitry promotes glioma growth. FEBS Lett. 2014;588:3038–3046. doi: 10.1016/j.febslet.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 23.Guo M, Zhang X, Wang G, Sun J, Jiang Z, Khadarian K, Yu S, Zhao Y, Xie C, Zhang K, et al. miR-603 promotes glioma cell growth via Wnt/β-catenin pathway by inhibiting WIF1 and CTNNBIP1. Cancer Lett. 2015;360:76–86. doi: 10.1016/j.canlet.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Cui M, Lu Z, Yang Q, Dong Q. Tissue kallikrein promotes survival and β-catenin degradation in SH-SY5Y cells under nutrient stress conditions via autophagy. Mol Med Rep. 2016;13:1389–1394. doi: 10.3892/mmr.2015.4664. [DOI] [PubMed] [Google Scholar]
- 25.Tian R, Li Y, Yao X. PGRN suppresses inflammation and promotes autophagy in keratinocytes through the Wnt/β-catenin signaling pathway. Inflammation. 2016;39:1387–1394. doi: 10.1007/s10753-016-0370-y. [DOI] [PubMed] [Google Scholar]
- 26.McCord M, Mukouyama YS, Gilbert MR, Jackson S. Targeting WNT signaling for multifaceted glioblastoma therapy. Front Cell Neurosci. 2017;11:318. doi: 10.3389/fncel.2017.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.