Abstract
MicroRNAs (miRNAs) are widely expressed in human cells and closely associated with various types of cancer, including breast cancer. miR-876-5p has been indicated to participate in the tumorigenesis of certain types of cancer, such as hepatocellular carcinoma. Nevertheless, the roles of miR-876-5p in breast cancer remain unclear. In the present study, it was revealed that miR-876-5p expression levels were decreased in breast cancer cells compared with a normal cell line. miR-876-5p ectopic expression suppressed breast cancer cell proliferation and arrested progression of the cell cycle. In addition, miR-876-5p suppressed breast cancer cell migration and invasion. miR-876-5p was demonstrated to directly target transcription factor AP-2-α (TFAP2A) in breast cancer cells, and restoration of TFAP2A rescinded the suppressive role of miR-876-5p. In summary, the results from the present study provide evidence that miR-876-5p suppresses breast cancer progression by regulating cell proliferation, migration and invasion in a TFAP2A-dependent manner.
Keywords: microRNA, microRNA-876-5p, breast cancer, invasion, transcription factor AP-2-α
Introduction
Breast cancer has developed into the most common and aggressive cancer among women around the world (1). In 2012, breast cancer accounted for 25% of all cancer cases and 15% of all cancer deaths amongst females worldwide (1). Evidence has revealed that there are a number of risk factors that increase the incidence of breast cancer, such as obesity, alcohol abuse and genetic mutations (3). Despite numerous advances in breast cancer therapy, the mortality remains to increase (4). Currently, the major approaches for breast cancer treatment include surgery, radiotherapy, chemotherapy and hormonotherapy (5). The outcomes of patients with breast cancer remain poor. Thus, it is crucial to investigate the potential molecular mechanisms underlying breast cancer and develop novel therapeutic strategies.
MicroRNAs (miRNAs or miRs) are one group of small non-coding RNAs that are approximately 22 nucleotides in length (6). Like long non-coding RNAs, miRNAs are implicated in the development of multiple cancers, such as liver cancer (7,8). miRNAs may regulate gene expression through interacting with the 3′-untranslated region (3′-UTR) of target mRNAs (9). Evidence has demonstrated that miRNAs initiate or suppress cancer development by regulating cell proliferation, metastasis, apoptosis or differentiation (10). For example, miR-495 regulates the progression of gastric cancer via the mammalian target of rapamycin pathway (11). miR-150 suppresses thyroid cancer development by modulating Ras-related protein Rab-11A/WNT/β-Catenin signaling (12). miR-1179 inhibits the proliferation of non-small cell lung cancer cells through the protein kinase B pathway (13). In addition, miR-125b-5p has been reported to repress breast cancer cell growth and metastasis via targeting uncharacterized protein KIAA1522 (14). Thus, identification of key miRNAs involved in breast cancer will be important for understanding its potential underlying molecular mechanism.
miR-876-5p was previously been proven to inhibit metastasis of head and neck squamous cell carcinoma, liver cancer and lung cancer (15–17). However, its function in breast cancer development remains unclear. The aim of the present study was to analyse miR-876-5p expression in breast cancer and evaluate its effects on tumor cell malignant behaviors, whilst emphasizing the essential roles of the miR-876-5p/transcription factor AP-2-α (TFAP2A) axis in breast cancer.
Materials and methods
Cell culture and transfection
Breast cancer cell lines (MCF-7, MDA-MB-231 and T47D) and the normal breast epithelial cell line MCF-10A were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). These cell lines were cultured with RPMI 1640 medium containing 10% fetal bovine serum (both Invitrogen; Thermo Fisher Scientific, Inc.) in a humid cell incubator at 37°C. miR-876-5p inhibitors (5′-UGGUGAUUCACAAAGAAAUCCA-3′), miR-876-5p mimics (5′-UGGAUUUCUUUGUGAAUCACCA-3′) and negative controls (5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by GenePharma Biotech Corp., and transduced into cell lines at a concentration of 50 nM using Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For the rescue assay, the coding sequence of TFAP2A was cloned into pcDNA3 vector (Invitrogen; Thermo Fisher Scientific, Inc.) and transfected into cells (1 µg per well) using Lipofectamine™ 2000. Following 48 h, transfection efficiency was confirmed using reverse transcription-quantitative (RT-qPCR) as described below.
RNA extraction and RT-qPCR
RNAs were isolated from cultured cells using TRIzol® Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNAs were reverse transcribed using a PrimeScript RT reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol. qPCR analysis were performed using a SYBR Green I Master Mix kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. miRNA relative expression was normalized to U6 while mRNA expression was normalized to actin, cytoplasmic 1 (ACTB). Thermocycling conditions were as follows: Denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 sec and elongation at 60°C for 1 min. Gene expression was calculated according to the 2−ΔΔCq method (18). The primers were synthesized by Sangon Biotech Co., Ltd. The primer sequences were as follows: miR-876-5p: Forward, 5′-TGGATTTCTTTGTGAATCACCA-3′; reverse, 5′-AACGAGACGACGACAGAC-3′; U6: Forward, 5′-GCAAATTCGTGAAGCGTTCCATA-3′; reverse, 5′-AACGAGACGACGACAGAC-3′; TFAP2A: Forward, 5′-TGCTACACTGAGACTCCCGT-3′; and reverse, 5′-GAATGCCTGGAAATCGAGCG-3′; and ACTB: Forward 5′-CGGCGCCCTATAAAACCCA-3′ and reverse, 5′-GAGGCGTACAGGGATAGCAC-3′.
Cell proliferation assay
A total of 2,000 cells per well were seeded in the 96-well plate and cultured for 1, 2, 3 or 4 days at 37°C. Then 10 µl Cell Counting Kit (CCK) 8 solution (Dojindo Molecular Technologies) was added and incubated according the manufacturer's protocol. Finally, the absorbance at 450 nm was determined using a multimode microplate reader (Berthold Technologies GmbH & Co. KG).
Transwell assay
Cell migration and invasion was assessed using Transwell chambers (8 µm pore size; Corning Inc.). Briefly, 2×104 tumor cells per well were seeded into the upper chamber in serum-free RPMI 1640 medium (Invitrogen; Thermo Fischer Scientific, Inc.), and the lower chamber was filled with 600 µl complete RPMI 1640 medium with 10% FBS. Following incubation for 48 h at 37°C, the cells in the lower chamber were fixed with 4% paraformaldehyde for 1 h at 25°C and stained with 0.1% crystal violet for 30 min at 25°C. The cell number was counted using a light microscope under ×200 magnification. During invasion assay, Matrigel (Becton, Dickinson and Company) was used to pre-coat the upper chamber.
Western blotting
Breast cancer cells were lysed in cold radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Inc.), and protein concentration was determined using a Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). Protein (40 µg/lane) was separated via SDS-PAGE (10% gel) and transferred to a polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific, Inc.). The membrane was blocked using 5% non-fat milk in PBS (Thermo Fisher Scientific, Inc.) containing 0.1% Tween-20 (Sigma-Aldrich; Merck KGaA) at room temperature for 2 h. Subsequently, the PVDF membrane was incubated with anti-Cyclin D1 (1:1,000; cat. no. ab16663), anti-TFAP2A (1:1,000; cat. no. ab52222) and anti-GAPDH (1:1,000; cat. no. ab9485; all from Abcam) primary antibodies at room temperature for 2 h. Following washing with PBS for 10 min, the PVDF membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (1:5,000; cat. no. ab7090; Abcam) at room temperature for 1 h. Membranes were then washed with PBS for 10 min and the protein bands were visualized using the Pierce™ ECL Western Blotting Substrate kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Protein densitometry was performed using ImageJ Software (version 1.41; National Institutes of Health).
Dual-luciferase reporter assay
The TargetScan7 tool (http://www.targetscan.org/vert_71/) was used to predict the potential interaction between miR-876-5p and TFAP2A. The TFAP2A 3′-UTR sequence containing wild-type (WT) or mutant (MUT) binding site for TFAP2A was constructed into pGL3-reporter luciferase vector (Promega Corporation). For the luciferase reporter assay, tumor cells were co-transfected with TFAP2A reporter vector and miR-876-5p mimics or control using Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.). Following 48 h, luciferase activity was measured using a dual-luciferase reporter assay system (Promega Corporation) following the manufacturer's protocol. Luciferase activities were normalized to that of Renilla luciferase activity.
Statistical analysis
All experiments were repeated in triplicate and analyzed using SPSS software (version 22.0; IBM Corp.). The differences were determined via unpaired Student's t-test or a one-way analysis of variance followed by a Tukey's post-hoc test. Results were presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant result.
Results
miR-876-5p suppresses breast cancer cell proliferation
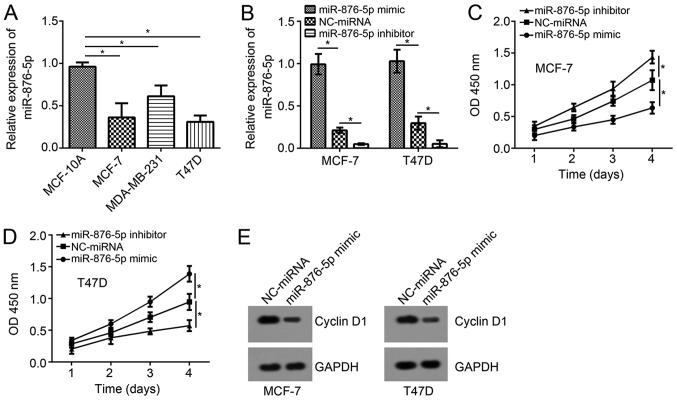
Expression patterns of miR-876-5p in breast cancer cells were analyzed using RT-qPCR. The results indicated that miR-876-5p expression levels are significantly downregulated in tumor cells when compared with MCF-10A cells (Fig. 1A). Among these three breast cancer cell lines, the expression levels of miR-876-5p were the lowest in MCF-7 and T47D cells. Thus, these two cell lines were selected for functional experiments. Using miR-876-5p mimics and inhibitors, the miR-876-5p expression levels were significantly upregulated and downregulated, respectively, in MCF-7 and T47D cells compared with NC-miRNA-transfected cells (Fig. 1B). According to the CCK8 assay, miR-876-5p upregulation suppressed the proliferation of breast cancer cells on day 4 compared to the NC-miRNA group while inhibition of miR-876-5p significantly promoted proliferation compared to the control (Fig. 1C and D). In addition, miR-876-5p upregulation inhibited the protein levels of Cyclin D1 in MCF-7 and T47D cells (Fig. 1E), indicating miR-876-5p prevents cell-cycle progression.
Figure 1.
miR-876-5p suppresses breast cancer cell proliferation. (A) RT-qPCR analysis demonstrated that miR-876-5p level was downregulated in breast cancer cell lines. (B) RT-qPCR analysis for miR-876-5p expression following transfection with miR-876-5p mimics, miR-876-5p inhibitors or negative controls. Cell Counting Kit-8 assays revealed that miR-876-5p mimics suppress proliferation of (C) MCF-7 and (D) T47D cells. (E) Western blot analysis indicated that miR-876-5p mimics suppress the expression of Cyclin D1 in MCF-7 and T47D cells. *P<0.05 with comparisons indicated by lines. miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; OD, optical density.
miR-876-5p inhibits migration and invasion of breast cancer cells
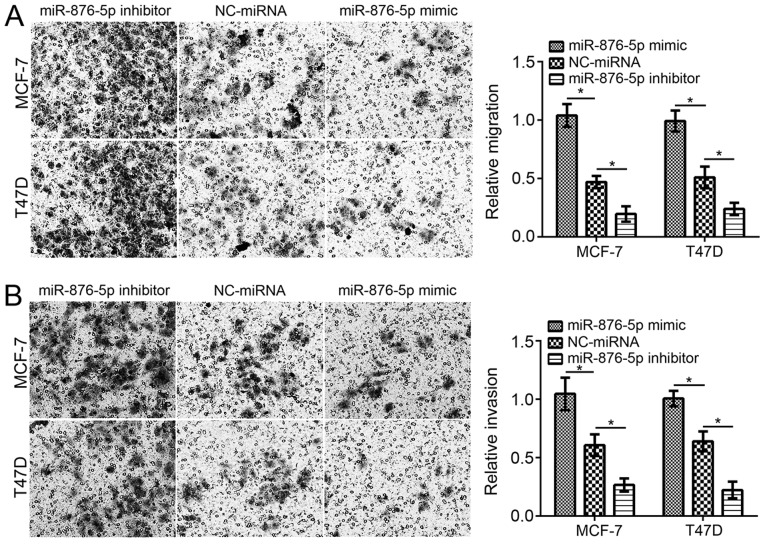
Furthermore, the effect of miR-876-5p on migration and invasion was evaluated using Transwell assays. Following miR-876-5p mimic transfection, the cell numbers of migration and invasion were significantly decreased compared with NC-miRNA-transfected cells (Fig. 2A and B). Inhibition of miR-876-5p significantly decreased the migration and invasion of MCF-7 and T47D cells compared with NC-miRNA-transfected cells.
Figure 2.
miR-876-5p inhibits the migration and invasion of breast cancer cells. miR-876-5p upregulation suppresses the (A) migration and (B) invasion of MCF-7 and T47D cells (magnification, ×200). *P<0.05 with comparisons indicated by lines. miR, microRNA; NC, negative control.
miR-876-5p targets TFAP2A in breast cancer cells
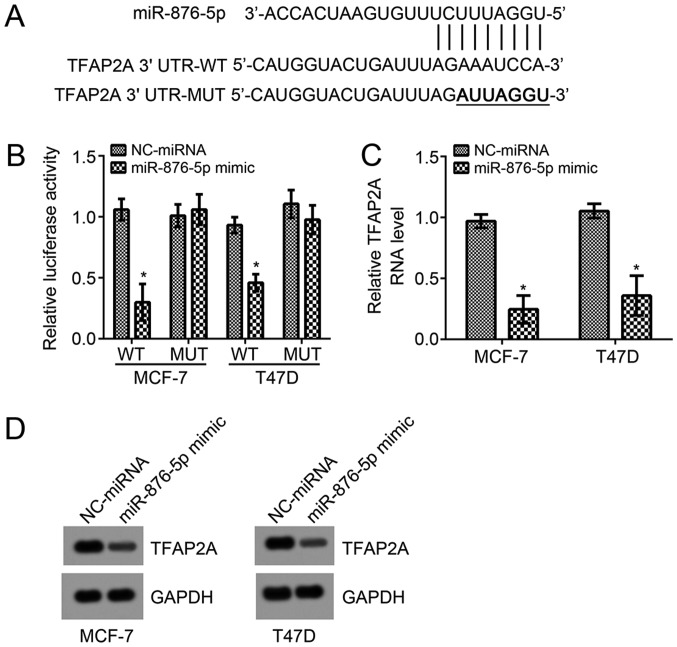
In order to investigate the underlying molecular mechanism of miR-876-5p, the TargetScan7 tool was used to predict the potential target gene of miR-876-5p. Among all candidates, TFAP2A ranked top. The potential binding site for miR-876-5p was identified in the 3′-UTR region of TFAP2A and it was mutated for the luciferase reporter assay (Fig. 3A). According to the luciferase reporter assay in MCF-7 and T47D cells, miR-876-5p mimic transfection significantly suppressed the activity of WT-reporter plasmid compared with NC-miRNA-transfected cells (Fig. 3B). Mutation of this site in the reporter plasmid rescinded the decrease in luciferase activity (Fig. 3B). Furthermore, miR-876-5p mimics significantly suppressed the mRNA levels compared with NC-miRNA-transfected cells, and markedly suppressed protein levels of TFAP2A in MCF-7 and T47D cells (Fig. 3C and D). These results demonstrated that miR-876-5p directly targets TFAP2A in breast cancer cells.
Figure 3.
miR-876-5p targets TFAP2A in breast cancer cells. (A) Predicted binding site for miR-876-5p in the 3′-UTR of TFAP2A by bioinformatics analysis. (B) Luciferase reporter assay revealed that miR-876-5p upregulation suppressed the activity of TFAP2A WT reporter but not the MUT one. (C) Reverse transcription-quantitative PCR and (D) western blot analysis revealed that TFAP2A mRNA and protein levels were suppressed by miR-876-5p mimics. *P<0.05 vs. NC-miRNA group. miR, microRNA; TFAP2A, transcription factor AP-2 α; WT, wild-type; MUT, mutant; NC, negative control; UTR, untranslated region.
Restoration of TFAP2A rescinds the effect of miR-876-5p mimics in MCF-7 cells
Finally, to validate whether miR-876-5p suppresses breast cancer progression through targeting TFAP2A, rescue assays were conducted. RT-qPCR analysis validated that co-transfection with pcDNA3-TFAP2A vector and miR-876-5p mimics significantly and successfully restored the expression of TFAP2A in MCF-7 cells compared with cells transfected with the miR-876-5p mimics alone (Fig. 4A). The CCK8 assay demonstrated that TFAP2A restoration markedly rescued the suppressed proliferation by miR-876-5p in MCF-7 cells (Fig. 4B). Furthermore, individual ectopic expression of TFAP2A further promoted MCF-7 cell proliferation. Similarly, TFAP2A restoration following co-transfection significantly rescued the migration and invasion abilities in MCF-7 cells compared with miR-876-5p mimics-transfected cells (Fig. 4C and D). Overall, these results demonstrated that miR-876-5p suppresses breast cancer progression by targeting TFAP2A.
Figure 4.
Restoration of TFAP2A rescinds the effect of miR-876-5p mimics in MCF-7 cells. (A) Relative expression levels of TFAP2A in transfected MCF-7 cells. (B) Cell Counting Kit-8 assay revealed that TFAP2A restoration rescued MCF-7 cell proliferation. Transwell assays indicated that TFAP2A restoration promoted the (C) migration and (D) invasion of MCF-7 cells (magnification, ×200). *P<0.05 with comparisons indicated by lines. TFAP2A, transcription factor AP-2-α; miR, microRNA; NC, negative control; OD, optical density; oe, overexpressed.
Discussion
Breast cancer remains to be a significant challenge for the health of women worldwide. However, the outcomes and survival rate urgently require improvement. A number of studies have indicated that miRNAs are involved in the progression of breast cancer (19) and may be good biomarkers for tumor diagnosis and prognosis (20). Furthermore, uncovering the functional mechanism of pivotal miRNAs involved in breast cancer will be helpful for developing novel therapeutic targets against this disease. The results from the present study indicated that miR-876-5p expression was decreased in breast cancer cell lines. miR-876-5p upregulation suppresses the proliferation, migration and invasion of tumor cells, which, to the best of our knowledge, indicates its anti-cancer role in breast cancer for the first time.
Up until now, the roles of miR-876-5p in tumorigenesis have been poorly understood. Xu et al (21) reported that elevated levels of miR-876 promotes apoptosis of endothelial cells. Bao et al (17) demonstrated that miR-876-5p inhibits metastasis in lung cancer through targeting bone morphogenetic protein 4. Sang et al (22) revealed that miR-876-5p targets the melanoma-associated antigen family to suppress the development of esophageal squamous cell carcinoma. Furthermore, previous studies have proved that miR-876-5p also prevents cell invasion in liver cancer (16,21). In addition, Dong et al (15) proved that miR-876-5p impeded the migration of head and neck squamous cancer cells via inhibiting vimentin. Currently, the way in which miR-876-5p regulates breast cancer remains unknown. In the present study, it was revealed that miR-876-5p expression levels were significantly decreased in breast cancer cell lines. A CCK8 assay demonstrated that miR-876-5p upregulation attenuates tumor cell growth. Furthermore, a Transwell assay demonstrated that miR-876-5p mimic transfection impaired the migration and invasion ability in breast cancer cell lines. The results from the present study indicate that miR-876-5p suppresses breast cancer progression. However, the expression levels of miR-876-5p should be investigated in the future, and analyzing the association between miR-876-5p and the clinical severity of breast cancer may further validate whether miR-876-5p could be a therapeutic target. Furthermore, whether miR-876-5p could serve as a diagnostic or prognostic marker for patients with breast cancer needs to be determined, alongside whether miR-876-5p regulates breast cancer cell apoptosis.
A number of studies have demonstrated that miRNAs exert roles through targeting the complementary sequence in the 3′UTR region of mRNAs (12,16). For example, miR-363-3p promotes glioma cell proliferation and metastasis through targeting pyruvate dehydrogenase B (22). miR-214 contributes to osteosarcoma progression via targeting tumor necrosis factor receptor-associated factor 3 (23). miR-543 enhances metastasis of gastric cancer cells by targeting speckle-type POZ protein (24). Furthermore, miR-1258 upregulation suppresses the growth of colon cancer cells and arrests cell-cycle progression through targeting transcription factor E2F8 (25). Through bioinformatics analyses, it was revealed that TFAP2A may be targeted by miR-876-5p. The luciferase reporter assay demonstrated the direct interaction between miR-876-5p and TFAP2A in breast cancer cells. Furthermore, it was also revealed that TFAP2A expression could be suppressed by miR-876-5p, suggesting that TFAP2A may be a downstream signal of miR-876-5p.
TFAP2A is a transcription factor involved in many different types of cancer (26). For example, Shi et al (27) revealed that TFAP2A promotes nasopharyngeal carcinoma cell proliferation and inhibits apoptosis. Notably, studies have indicated that TFAP2A promotes breast cancer progression (28,29). In the present study, it was also revealed that TFAP2A overexpression promotes the proliferation, migration and invasion of breast cancer cells. Furthermore, restoration of TFAP2A in miR-876-5p-upregulated breast cancer cells could abolish the suppressive effects of miR-876-5p mimics. Overall, the results from the present study demonstrated that miR-876-5p exerts roles by targeting TFAP2A.
To the best of our knowledge, the present study demonstrates for the first time that miR-876-5p suppresses breast cancer progression through attenuating proliferation, migration and invasion by targeting TFAP2A directly. Findings suggested that miR-876-5p may be a promising therapeutic target for breast cancer treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JX initiated and designed the present study, analyzed and interpreted the results. JZ, JW and JS performed various experiments. JX wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Zhao L, Zheng XY. MicroRNA-490 inhibits tumorigenesis and progression in breast cancer. Onco Targets Ther. 2016;9:4505–4516. doi: 10.2147/OTT.S100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudolph A, Chang-Claude J, Schmidt MK. Gene-environment interaction and risk of breast cancer. Br J Cancer. 2016;114:125–133. doi: 10.1038/bjc.2015.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 5.Liu RZ, Garcia E, Glubrecht DD, Poon HY, Mackey JR, Godbout R. CRABP1 is associated with a poor prognosis in breast cancer: Adding to the complexity of breast cancer cell response to retinoic acid. Mol Cancer. 2015;14:129. doi: 10.1186/s12943-015-0380-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Y, Chang C, Liu J, Qiang J. Targeting of GIT1 by miR-149* in breast cancer suppresses cell proliferation and metastasis in vitro and tumor growth in vivo. Onco Targets Ther. 2017;10:5873–5882. doi: 10.2147/OTT.S144280. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, Du Y, Gao G, Tian Y, He L, Fan Z. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15:1–19. doi: 10.1093/bib/bbs075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song B, Lin HX, Dong LL, Ma JJ, Jiang ZG. MicroRNA-338 inhibits proliferation, migration, and invasion of gastric cancer cells by the Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:1290–1296. doi: 10.26355/eurrev_201803_14470. [DOI] [PubMed] [Google Scholar]
- 11.Li N, Han M, Zhou N, Tang Y, Tang XS. MicroRNA-495 confers increased sensitivity to chemotherapeutic agents in gastric cancer via the mammalian target of rapamycin (mTOR) signaling pathway by interacting with human epidermal growth factor receptor 2 (ERBB2) Med Sci Monit. 2018;24:5960–5972. doi: 10.12659/MSM.909458. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Bai D, Sun H, Wang X, Lou H, Zhang J, Wang X, Jiang L. MiR-150 inhibits cell growth in vitro and in vivo by restraining the RAB11A/WNT/β-catenin pathway in thyroid cancer. Med Sci Monit. 2017;23:4885–4894. doi: 10.12659/MSM.906997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L, Dai Z, Zhang S, Zhang H, Liu C, Ma X, Liu D, Zan Y, Yin X. MicroRNA-1179 suppresses cell growth and invasion by targeting sperm-associated antigen 5-mediated Akt signaling in human non-small cell lung cancer. Biochem Biophys Res Commun. 2018;504:164–170. doi: 10.1016/j.bbrc.2018.08.149. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Wang Y, Fan H, Zhang Z, Li N. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem Biophys Res Commun. 2018;504:277–282. doi: 10.1016/j.bbrc.2018.08.172. [DOI] [PubMed] [Google Scholar]
- 15.Dong Y, Zheng Y, Wang C, Ding X, Du Y, Liu L, Zhang W, Zhang W, Zhong Y, Wu Y, Song X. MiR-876-5p modulates head and neck squamous cell carcinoma metastasis and invasion by targeting vimentin. Cancer Cell Int. 2018;18:121. doi: 10.1186/s12935-018-0619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Xie Y, Li X, Lin J, Zhang S, Li Z, Huo L, Gong R. MiR-876-5p acts as an inhibitor in hepatocellular carcinoma progression by targeting DNMT3A. Pathol Res Pract. 2018;214:1024–1030. doi: 10.1016/j.prp.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Bao L, Lv L, Feng J, Chen Y, Wang X, Han S, Zhao H. MiR-876-5p suppresses epithelial-mesenchymal transition of lung cancer by directly down-regulating bone morphogenetic protein 4. J Biosci. 2017;42:671–681. doi: 10.1007/s12038-017-9722-5. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Nagasawa S, Sedukhina AS, Nakagawa Y, Maeda I, Kubota M, Ohnuma S, Tsugawa K, Ohta T, Roche-Molina M, Bernal JA, et al. LSD1 overexpression is associated with poor prognosis in basal-like breast cancer, and sensitivity to PARP inhibition. PLoS One. 2015;10:e0118002. doi: 10.1371/journal.pone.0118002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Yin W, Lin Y, Yin K, Zhou L, Du Y, Yan T, Lu J. Downregulated circulating microRNAs after surgery: Potential noninvasive biomarkers for diagnosis and prognosis of early breast cancer. Cell Death Discov. 2018;4:21. doi: 10.1038/s41420-018-0089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Q, Zhu Q, Zhou Z, Wang Y, Liu X, Yin G, Tong X, Tu K. MicroRNA-876-5p inhibits epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by targeting BCL6 corepressor like 1. Biomed Pharmacother. 2018;103:645–652. doi: 10.1016/j.biopha.2018.04.037. [DOI] [PubMed] [Google Scholar]
- 22.Xu DX, Guo JJ, Zhu GY, Wu HJ, Zhang QS, Cui T. MiR-363-3p modulates cell growth and invasion in glioma by directly targeting pyruvate dehydrogenase B. Eur Rev Med Pharmacol Sci. 2018;22:5230–5239. doi: 10.26355/eurrev_201808_15721. [DOI] [PubMed] [Google Scholar]
- 23.Rehei AL, Zhang L, Fu YX, Mu WB, Yang DS, Liu Y, Zhou SJ, Younusi A. MicroRNA-214 functions as an oncogene in human osteosarcoma by targeting TRAF3. Eur Rev Med Pharmacol Sci. 2018;22:5156–5164. doi: 10.26355/eurrev_201808_15711. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Wang F, Wang X, He Z, Zhu X. miRNA-543 promotes cell migration and invasion by targeting SPOP in gastric cancer. Onco Targets Ther. 2018;11:5075–5082. doi: 10.2147/OTT.S161316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Li J, Huang Y, Peng W, Qian W, Gu J, Wang Q, Hu T, Ji D, Ji B, et al. Upregulated miR-1258 regulates cell cycle and inhibits cell proliferation by directly targeting E2F8 in CRC. Cell Prolif. 2018;51:e12505. doi: 10.1111/cpr.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan J, Zhang N, Zheng Y, Chen YD, Liu J, Yang M. LncRNA GAS5 indel genetic polymorphism contributes to glioma risk through interfering binding of transcriptional factor TFAP2A. DNA Cell Biol. 2018;37:750–757. doi: 10.1089/dna.2018.4215. [DOI] [PubMed] [Google Scholar]
- 27.Shi D, Xie F, Zhang Y, Tian Y, Chen W, Fu L, Wang J, Guo W, Kang T, Huang W, Deng W. TFAP2A regulates nasopharyngeal carcinoma growth and survival by targeting HIF-1alpha signaling pathway. Cancer Prev Res (Phila.) 2014;7:266–277. doi: 10.1158/1940-6207.CAPR-13-0271. [DOI] [PubMed] [Google Scholar]
- 28.Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V, Winkler R, Boniver J, Delvenne P, Begon DY. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10:R9. doi: 10.1186/bcr1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan F, He Q, Hu X, Li W, Wei K, Li L, Zhong Y, Ding X, Xiang S, Zhang J. Direct regulation of caspase3 by the transcription factor AP2α is involved in aspirininduced apoptosis in MDAMB453 breast cancer cells. Mol Med Rep. 2013;7:909–914. doi: 10.3892/mmr.2013.1257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.