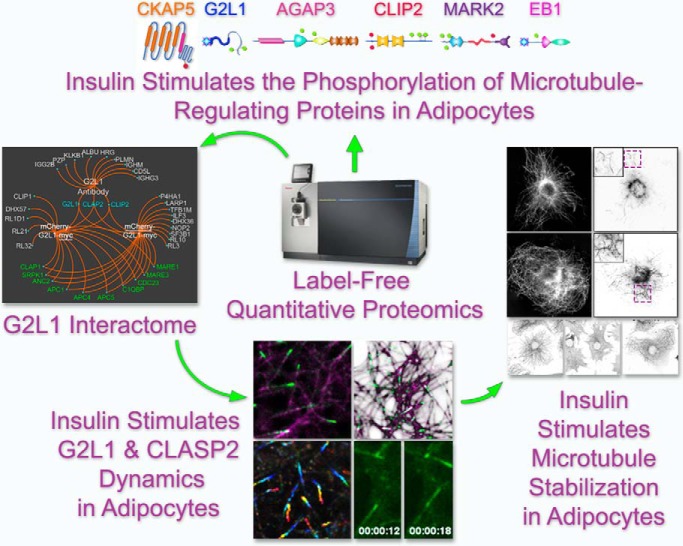
In adipocytes, our quantitative proteomics studies have revealed that insulin affects the phosphorylation of a new microtubule-associated protein network. Mapping microtubule plus-end interacting protein interactomes resulted in the live-cell based discovery that insulin has novel effects on microtubule plus-end protein and microtubule dynamics. In addition, we present microtubule stabilization as a new paradigm in insulin action. Taken together, our findings introduce new directions aimed at pursuing a better understanding of the relationship between insulin and the microtubule network.
Keywords: Affinity proteomics, Label-free quantification, Phosphorylation, Protein-Protein Interactions*, Quantification, CLASP2, G2L1, Insulin, Interactome, Microtubules
Graphical Abstract
Highlights
Insulin Affects the Phosphorylation of G2L1, MARK2, CLIP2, EB1, AGAP3, and CKAP5.
Insulin Increases CLASP2 +TIP Density and Decreases CLASP2 +TIP Velocity.
Insulin Stimulates CLASP2 and G2L1 Trailing Along Microtubules.
Insulin Stimulates α-Tubulin Acetylation at Lysine 40 and Microtubule Stabilization.
Abstract
Insulin-stimulated glucose uptake is known to involve microtubules, although the function of microtubules and the microtubule-regulating proteins involved in insulin action are poorly understood. CLASP2, a plus-end tracking microtubule-associated protein (+TIP) that controls microtubule dynamics, was recently implicated as the first +TIP associated with insulin-regulated glucose uptake. Here, using protein-specific targeted quantitative phosphoproteomics within 3T3-L1 adipocytes, we discovered that insulin regulates phosphorylation of the CLASP2 network members G2L1, MARK2, CLIP2, AGAP3, and CKAP5 as well as EB1, revealing the existence of a previously unknown microtubule-associated protein system that responds to insulin. To further investigate, G2L1 interactome studies within 3T3-L1 adipocytes revealed that G2L1 coimmunoprecipitates CLASP2 and CLIP2 as well as the master integrators of +TIP assembly, the end binding (EB) proteins. Live-cell total internal reflection fluorescence microscopy in adipocytes revealed G2L1 and CLASP2 colocalize on microtubule plus-ends. We found that although insulin increases the number of CLASP2-containing plus-ends, insulin treatment simultaneously decreases CLASP2-containing plus-end velocity. In addition, we discovered that insulin stimulates redistribution of CLASP2 and G2L1 from exclusive plus-end tracking to “trailing” behind the growing tip of the microtubule. Insulin treatment increases α-tubulin Lysine 40 acetylation, a mechanism that was observed to be regulated by a counterbalance between GSK3 and mTOR, and led to microtubule stabilization. Our studies introduce insulin-stimulated microtubule stabilization and plus-end trailing of +TIPs as new modes of insulin action and reveal the likelihood that a network of microtubule-associated proteins synergize to coordinate insulin-regulated microtubule dynamics.
Since the initial discovery of insulin-stimulated cytoskeletal remodeling over 25 years ago (1), the role of both actin and microtubule reorganization in insulin action and the proteins that regulate these processes have been the subject of investigation. On insulin stimulation, cell models of insulin target tissues including 3T3- L1 adipocytes as well as L6 and C2C12 myotubes (2–6) exhibit profound actin reorganization at the plasma membrane as a result of actin branching. Rac1 is a master regulator of actin dynamics and inhibiting either Rac1 or Rac1-effector proteins severely diminishes trafficking of the insulin responsive glucose transporter (Solute carrier family 2, facilitated glucose transporter member 4 or “GLUT4”1) and subsequent insulin-stimulated glucose uptake (7). Therefore, insulin not only relies on signal transduction through protein-protein communication, but also requires a specific cytoskeletal environment to properly enhance acute glucose uptake. Studies characterizing the proteins involved in insulin-controlled actin regulation have been more prevalent than those focused on proteins involved in regulating microtubule dynamics on insulin stimulation. As a result, much more progress has been made on elucidating the actin system under insulin control versus that of microtubules.
Microtubules possess the unique characteristic of “dynamic instability,” their growth and shrinkage serve as modes of functional regulation across a multitude of molecular platforms. Microtubule assembly and disassembly rates as well as duration of stability are controlled in part by “microtubule-associated proteins” (MAPs), a large family of proteins with several notable subgroups (reviewed in (8)). Although many of the MAPs localize at the base of the microtubule (the minus-end) or along the length of the microtubules (the lattice), the plus-end tracking proteins (+TIPs) are found at the growing tip of the microtubule (the plus-end). The +TIPs are a functionally diverse group of almost 20 proteins, and as such, their regulation and cooperativity have been explored at length (reviewed in (9)), although not in the context of insulin action. One exception is CLIP-associating protein 2 (CLASP2) (10), a +TIP that was found to be enriched in an unbiased proteomics screen for proteins that undergo insulin-stimulated phosphorylation (11). Just like the rest of the +TIPs, CLASP2 tracks the plus-end of microtubules, associates with an assortment of other +TIPs, and is involved in a variety of microtubule-regulated cellular events (12). Notably, CLASP2 has been linked to the “search and capture” behavior of microtubules (13), wherein microtubule tips are observed to seek out specific landing zones through CLASP2 binding of proteins localized at subcellular regions (14–18). For example, CLASP2 binding to microtubule capture sites on the cell cortex creates a delivery route for acetylcholine receptors to neuromuscular junctions (19) as well as exocytotic vesicles traveling to focal adhesions (20). These observations revealed that under certain cellular contexts, microtubules adopt specific patterns of reorganization that possess both temporal and spatial characteristics that synchronize with +TIP function.
Microtubules and actin routinely cooperate through proteins including the formins (21), the spectraplakins (22), and members of the growth-arrest-specific 2 (GAS2) family (23). One of the GAS2 family members, G2L1, was recently discovered in the CLASP2 interactome in 3T3-L1 adipocytes (24). G2L1 interacts with both actin and microtubules to coordinate actin and microtubule alignment (23, 25, 26), an event that is of potential interest within insulin action because the insulin-responsive GLUT4 storage vesicle has been proposed to switch tracks at an interface between microtubules and actin at the plasma membrane (27).
Early immunofluorescence studies coupled with biochemical techniques first discovered that insulin increases tubulin polymerization in 3T3-L1 adipocytes (28). A decade later, using live-cell total internal reflection fluorescence (TIRF) microscopy, it was demonstrated that insulin increases microtubule density and curvature in the 200 nm immediately proximal to the plasma membrane in 3T3-L1 adipocytes (29). The purpose of these dynamic microtubule events has never been established and the mechanisms underlying microtubule regulation in insulin action are still unknown. This report presents new findings that significantly expand the number of +TIPs affected by insulin, evidence for the existence of an undiscovered microtubule-associated protein system under insulin control. We found that insulin stimulation promotes a redistribution of CLASP2 and G2L1 from exclusive microtubule plus-end localization to “trailing” behind the growing tip of the microtubule along the microtubule lattice. We discovered that insulin acutely stimulates acetylation of a-tubulin at Lysine 40 and promotes microtubule stabilization, a phenomenon never before linked to acute insulin action. Our studies introduce a new insulin-responsive protein system as well as novel insulin-regulated microtubule and microtubule-associated protein dynamics, findings that significantly expand our understanding of the relationship between insulin and the microtubule network.
EXPERIMENTAL PROCEDURES
Cell Culture, Immunoprecipitation, and Western Blot Analysis
Mouse 3T3-L1 fibroblasts were differentiated into adipocytes exactly as previously described (24). For cell treatment experiments, cells were starved for four hours in serum free media containing 0.3% BSA and then either left untreated or stimulated with 100 nm insulin for 15 min (unless otherwise indicated) at 37 °C. Cells were lysed with 500 μl of lysis buffer containing 40 mm HEPES (pH 7.6), 120 mm NaCl, 0.3% CHAPS, 10 mm NaF, 10 mm β-glycerol phosphate, 1 mm EDTA (pH 8.0), 2 mm sodium orthovanadate, 17 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mm PMSF. Cell lysates were rotated at 4 °C for 20 min followed by centrifugation (14,000 RPM, 4 °C, 20 min), and the clarified supernatants were used for immunoprecipitation (IP). IPs and preparation for SDS-PAGE were performed exactly as previously described (24). The eluates were separated by 10% SDS-PAGE and the gels were either stained with Bio-Safe Coomassie G-250 Stain (Bio-Rad, Hercules, CA) or transferred to a nitrocellulose membrane for subsequent Western blotting. Western blots were performed exactly as previously described (24). Primary antibodies used: anti-G2L1 (cat. # H00010634-B01P, Novus Biologicals, Littleton, CO), anti-mCherry (cat. # NBP2–43720, Novus Biologicals), anti-myc (cat. # 2276, Cell Signaling Technologies, Danvers, MA), anti-EB1 (cat # E3406, Sigma-Aldrich, St. Louis, MO), anti-α-tubulin (cat. # T9026, Sigma-Aldrich), anti-acetylated Lysine 40 of α-tubulin (“AcK40”) (cat. # 5335, Cell Signaling Technologies), anti-MARK2 (cat. # NBP1–71890, Novus Biologicals), anti-CLIP2 (cat. # SAB1412760, Sigma Aldrich), anti-AGAP3 (cat. # SAB2700915, Sigma Aldrich), and anti-CKAP5 (cat. # 620401, Biolegend).
In-gel Digestion
Proteins were separated by SDS-PAGE and stained with Bio-Safe Coomassie G-250 Stain. For the interactome experiments, each lane of the SDS-PAGE gel was cut into seven slices. For the specific protein targeted phosphoproteomics experiments, the single band corresponding to the location of the individual protein on the SDS-PAGE gel was excised. The gel slices were subjected to trypsin digestion and the resulting peptides were purified by C18-based desalting exactly as previously described (24). The dried peptides were resuspended in 6 μl of 0.1% FA (v/v) followed by sonication for 2 min. 2.5 μl of the final sample was then analyzed by mass spectrometry.
Mass Spectrometry and Spectrum Count Data Processing
HPLC-ESI-MS/MS was performed in positive ion mode on a Thermo Scientific Orbitrap Fusion Lumos tribrid mass spectrometer fitted with an EASY-Spray Source (Thermo Scientific, San Jose, CA). NanoLC was performed without a trap column using a Thermo Scientific UltiMate 3000 RSLCnano System with an EASY Spray C18 LC column (Thermo Scientific, 50 cm × 75 μm inner diameter, packed with PepMap RSLC C18 material, 2 μm, cat. # ES803); loading phase for 15 min at 0.300 μl/min; mobile phase, linear gradient of 1–34% Buffer B in 119 min at 0.220 μl/min, followed by a step to 95% Buffer B over 4 min at 0.220 μl/min, hold 5 min at 0.250 μl/min, and then a step to 1% Buffer B over 5 min at 0.250 μl/min and a final hold for 10 min (total run 159 min); Buffer A = 0.1% FA/H2O; Buffer B = 0.1% FA in 80% ACN. All solvents were liquid chromatography mass spectrometry grade. Spectra were acquired using XCalibur, version 2.3 (Thermo Scientific). A “top speed” data-dependent MS/MS analysis was performed. Dynamic exclusion was enabled with a repeat count of 1, a repeat duration of 30 s, and an exclusion duration of 60 s. Tandem mass spectra were extracted from Xcalibur 'RAW' files and charge states were assigned using the ProteoWizard 3.0.1 msConvert script using the default parameters (30). The fragment mass spectra were then searched against the mouse SwissProt_2016_10 database (23,550 entries) using Mascot (Matrix Science, London, UK; version 2.4) using the default probability cut-off score. The search variables that were used were: 10 ppm mass tolerance for precursor ion masses and 0.5 Da for product ion masses; digestion with trypsin; a maximum of two missed tryptic cleavages; variable modifications of oxidation of methionine and phosphorylation of serine, threonine, and tyrosine. Cross-correlation of Mascot search results with X! Tandem was accomplished with Scaffold (version Scaffold_4.8.7; Proteome Software, Portland, OR). Probability assessment of peptide assignments and protein identifications were made using Scaffold. Only peptides with ≥ 95% probability were considered. Reported peptide FDR rates from Scaffold ranged from 0.1–0.2%. For the Spectrum Count Profiles, the Uniprot IDs are listed.
Label-free Quantitative Proteomics
Progenesis QI for proteomics software (version 2.4, Nonlinear Dynamics Ltd., Newcastle on Tyne, UK) was used to perform ion-intensity based label-free quantification. In brief, in an automated format, .raw files were imported and converted into two-dimensional maps (y axis = time, x axis = m/z) followed by selection of a reference run for alignment purposes. An aggregate data set containing all peak information from all samples was created from the aligned runs, which was then further narrowed down by selecting only +2, +3, and +4 charged ions for further analysis. The samples were then grouped in basal versus insulin. A peak list of fragment ion spectra from only the top eight most intense precursors of a feature was exported in Mascot generic file (.mgf) format and searched against the mouse SwissProt_2016_10 (23,550 entries) database using Mascot (Matrix Science, London, UK; version 2.4). The search variables that were used were: 10 ppm mass tolerance for precursor ion masses and 0.5 Da for product ion masses; digestion with trypsin; a maximum of two missed tryptic cleavages; variable modifications of oxidation of methionine and phosphorylation of serine, threonine, and tyrosine; 13C = 1. The resulting Mascot .xml file was then imported into Progenesis, allowing for peptide/protein assignment, whereas peptides with a Mascot Ion Score of <25 were not considered for further analysis. Precursor ion-abundance values for specific phosphopeptide ions were normalized to a selected series of standard peptide ions for the same target protein. The selected series of standard peptide ions had to conform to a set of rules previously established (32–34), i.e. (1) detected by HPLC-ESI-MS with high intensity among the peptides for the target protein; (2) no missed cleavage observed; (3) no methionine in the sequence to avoid variability because of methionine oxidization and no N-terminal Gln residues; (4) cannot be a nonphosphorylated version of a peptide that was detected as being phosphorylated. Each phosphopeptide ion's normalized abundance value was normalized by the mean value of the respective basal sample and then expressed as a fold change over basal ±S.E. When multiple peptide ions were present for a phosphopeptide ion, a representative peptide ion was chosen to reflect the effect of insulin. For the proof of principle dose curve test, no normalization was performed.
Generation of Viruses and Transduction of 3T3-L1 Adipocytes
Negative control null GFP adenovirus as well as N-terminal GFP plus C-terminal HA-tagged mouse CLASP2 adenovirus were described before (24). Negative control null mCherry adenovirus as well as N-terminal mCherry plus C-terminal myc-tagged mouse G2L1 adenovirus using G2L1 cDNA, cat. # MC204613 from Origene (Rockville, MD) were created by VECTOR BIOLABS (Malvern, PA). Viral infections were performed on 150 mm plates of 3T3-L1 adipocytes as previously described (24). Lentiviral transfer plasmid pCIG3 (Addgene #78264, a gift from Felicia Goodrum) was modified to express a puromycin resistance gene in place of GFP. mRuby2-Tubulin-6 (human Tubulin alpha 1b; Addgene #55914, a gift from Michael Davidson) was subcloned into this backbone by addition of the restriction sites KpnI and BamHI by PCR, to generate pLenti-mRuby2-Tubulin. To generate pLenti-iRFP670-Tubulin, tubulin was first subcloned out of the mRuby2-Tubulin-6 vector and into piRFP670-N1 (Addgene #45457, a gift from Vladislav Verkhusha). iRFP670-Tubulin was then subcloned out of the piRFP670-Tubulin vector and into the pLenti backbone to generate pLenti-iRFP670-Tubulin. Second generation lentiviral particles were generated by PEI transfection of 293T cells as previously described (35) with transfer plasmid, pMD2.G, and psPAX2 (Addgene #12259, #12260, gifts from Didier Trono). At 48 and 72 h posttransfection, 293T media containing lentiviral particles was collected and combined followed by centrifugation at 500 × g for 15 min. The supernatant was filtered and either immediately used or concentrated (Lenti Concentrator, cat. # TR30025, Origene), depending on the virus. The virus was added directly to adipocyte cultures with polybrene. The following day, cells were split onto Poly-d-Lysine (cat. # A-003-E, Sigma Aldrich) treated glass bottom dishes (cat. # P35GCol-1.5–14-C, MatTek Corporation), and the cells were imaged 24 h later.
Immunofluorescence and Live-Cell Imaging
For live-cell imaging experiments, cells were cultured on No. 1.5 coverslip bottom dishes (Mattek, Ashland, MA). Adipocytes were starved for one hour in serum free media containing 0.3% BSA and then either left untreated or stimulated with 100 nm insulin for the indicated duration at 37 °C. For immunofluorescence studies, 3T3-L1 adipocytes were split onto coverslips (cat. # 1254580, Fisher Scientific). The following day, the cells were starved for 4 h in serum free media containing 0.3% BSA followed by the indicated cell treatments. Cells were fixed with 100%, −20 °C methanol for 20 min followed by permeabilization with 1% Tween-20 and 2% PFA. The background signal was quenched with 0.1 m glycine followed by incubation in 1% BSA. Coverslips were incubated with anti-α-Tubulin and anti-AcK40 Tubulin, followed by incubation with 568 anti-mouse (cat. # A10037, Life Technologies) and 488 anti-rabbit (cat. # A11034, Life Technologies) secondary antibodies, and then mounted onto slides with ProLong Diamond (cat. # p36965, Fisher Scientific).
Microscopy and Image Analysis
Total Internal Reflection Fluorescence Microcopy was performed on a Nikon Eclipse Ti (Nikon Instruments Inc., Melville, NY) inverted microscope equipped with multiple laser lines with AOTF control, Nikon Perfect Focus System, and an environmental chamber supplying 5% CO2 and 37 °C temperature control. Samples were imaged with a 100× Apo TIRF 1.49 NA oil-immersion objective. TIRF excitation angle was determined by adjusting for maximum extinction. Image acquisition was performed using a Hamamatsu ORCA-Flash 4.0 V2 cMOS camera (Hamamatsu Photonics). Live images were acquired on a 2 s interval for the indicated total duration. Images were deconvolved in NIS Elements using five iterations of Richardson-Lucy deconvolution algorithm. Overall microtubule structure and acetylation were imaged using a Zeiss Axio Observer 7- ApoTome.2 inverted microscope with 63× oil-immersion objective and Axiocam 503 mono camera. Microscopy figures and videos were prepared using ImageJ/FIJI (36) and Adobe Photoshop. For the quantification of the time courses on the effect of insulin on CLASP2 and G2L1 trailing, the live cell images were analyzed using Imaris 9.2.1 with FIJI extension. Specifically, +TIP microtubule decoration lengths were measured at different time points at fixed intervals with respect to the length of the time course using a frame within the first minute. The manual filament tools of Imaris were used for measurement of +TIP trail lengths, which were identified by scrubbing the time slider. Measurement was done visually, starting at the moving end of the trail or trail break with the AutoPath feature of Imaris. Once all visible trails were measured on frame, a new filament object was created, and time was scrubbed to the next frame along the fixed interval specified earlier. After image analysis was completed, all filament objects were compared with the serum-starved length of the first frame using Imaris Vantage. +TIP length data was graphed using RStudio. For quantification of +TIP velocity, CLASP2 position was tracked during a 30-s imaging interval using the Manual Tracking FIJI plug-in. The density of +TIPs was quantified by manually counting motile CLASP2 signal on microtubules and normalizing to cell area. For quantification of tubulin stabilization after nocodazole treatment with or without insulin treatment, the length of the microtubule network was measured using Simple Neurite Tracer FIJI plug-in (37) and normalized to cell area. Quantification for microtubule acetylation was determined using ImageJ corrected total cell fluorescence (CTCF) which calculates the normalized fluorescence based on the formula (Integrated density - (Area of selected cell × Mean background fluorescence) (38).
Experimental Design and Statistical Rationale
Previous interactome experiments using the mean and standard deviations for the spectrum counts of proteins identified by SAINT analysis as significantly enriched over the negative control showed a sample size of four biological replicates per group provided more than sufficient power to detect differences between groups (24). Based on the high degree of reproducibility, we used a sample size of two biological replicates for the interactome experiments and followed those experiments with confirmatory reciprocal interactomes as well as IP and western blots to further validate potential protein partners. The varying statistical analyzes for the remainder of the other types of experiments are listed in the Figure Legends and were justifiable based on the data and basic experiment performed. For spectral counting measurements, modified peptides, semitryptic peptides, and shared peptides were included. Statistical analysis of +TIP density, +TIP velocity, and tubulin stabilization experiments was performed by confirming the distribution of the data by Shapiro-Wilk normality test and performing a paired or unpaired t test as indicated. Statistical analysis of CTCF and +TIP microtubule decoration lengths was performed by a paired or unpaired t test as indicated
RESULTS
Insulin Regulates the Phosphorylation of G2L1, MARK2, CLIP2, EB1, AGAP3, and CKAP5
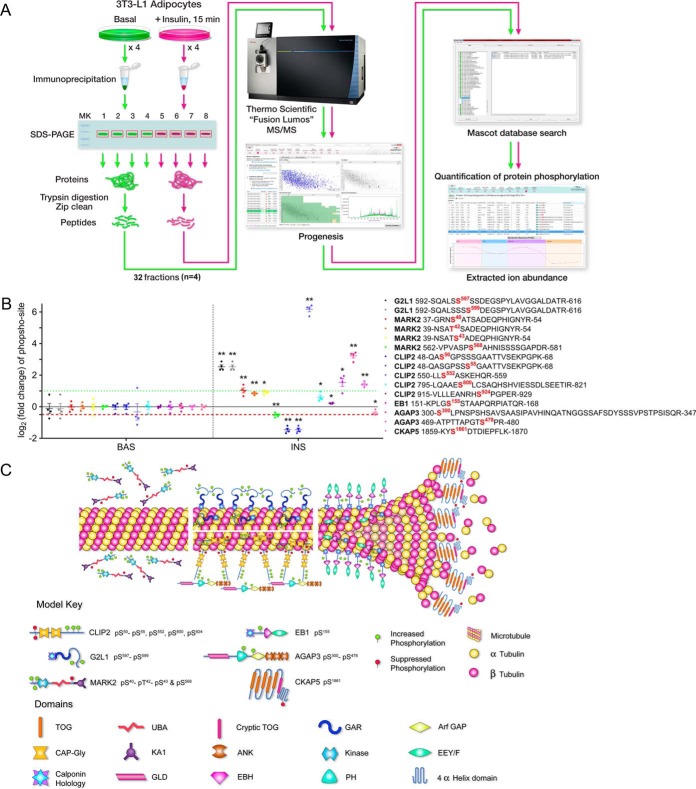
We recently reported characterization of a CLASP2 protein network in 3T3-L1 adipocytes (24), and because we previously detected CLASP2 in an unbiased proteomics screen for proteins that undergo insulin-stimulated phosphorylation (11), we hypothesized that the members of the CLASP2 network in 3T3-L1 adipocytes (24) also undergo insulin-regulated phosphorylation. We adapted our previously reported technique (32–34, 39) of label-free quantification of phosphorylation, where extracted ion abundances from parent ion phosphopeptides are normalized to parent ion peptides of the same protein, except we now take advantage of automated processing of the data using Progenesis (Fig. 1A). We validated the quantitative approach via Progenesis using immunoprecipitated CLASP2 digests (supplemental Fig. S1). On stimulating 3T3-L1 adipocytes with insulin, we discovered that the CLASP2 protein network members G2L1, CLIP2, AGAP3, MARK2, and CKAP5 all undergo insulin-regulated phosphorylation, as well as EB1, a fundamental +TIP with ties to CLASP2, G2L1, CLIP2, and CKAP5 (40) (Fig. 1B–1C, supplemental Fig. S2, and supplemental Tables S1–S6). These findings highlight a completely new series of proteins that are responsive to insulin and reveal the existence of a systematic microtubule-regulating protein response to insulin.
Fig. 1.
G2L1, MARK2, CLIP2, EB1, AGAP3, and CKAP5 undergo insulin-regulated phosphorylation. A, Serum-starved 3T3-L1 adipocytes were either left untreated or treated with insulin, lysed, and target proteins were immunoprecipitated as described under “Experimental procedures”. The IPs were separated by SDS-PAGE and the gel slice corresponding to the protein of interest was excised and subjected to trypsin digestion. The resulting tryptic digest was purified and subsequently analyzed by tandem mass spectrometry. Raw data processing for quantification was executed in Progeneis QI for Proteomics and peptide/protein identification was performed by database searching with Mascot. The resulting Mascot peptide and protein identifications were imported into Progenesis QI for Proteomics and quantification of changes in phosphopeptide abundance was performed via extracted ion abundance in Progenesis QI for Proteomics. MS/MS, tandem mass spectrometry. B, G2L1, MARK2, CLIP2, EB1, AGAP3, and CKAP5 phosphorylation was analyzed as described above and under Experimental Procedures (n = 4 per protein). Phosphopeptides are labeled with both the starting and final amino acid position of the phosphopeptide within the protein, along with the phospho-site (in red). The basal versus insulin data is separated by the vertical dashed black line. The horizontal red dashed line represents a 50% decrease in phosphorylation while the horizontal green dashed line represents a 2-fold increase in phosphorylation. *p ≤ 0.05; **p ≤ 0.01 insulin compared with basal; t test. BAS, basal. INS, insulin. C, The illustration depicts all identified phosphorylation sites affected by insulin, the location of the phosphorylation sites within each protein, the effect of insulin on phosphorylation (increased = green, suppressed = red), as well as the hypothetical localization of the microtubule-regulating proteins respective to the microtubule. TOG, tumor overexpressed gene; UBA, ubiquitin-associated; GAR, Gas2-related; GAP, GTPase-activating protein; KA1, kinase associated domain 1; ANK, ankyrin; EEY/F, EEY/F motif; GLD, GTP-binding protein-like domain; EBH, EB homology; PH, pleckstrin homology.
The G2L1 Interactomes
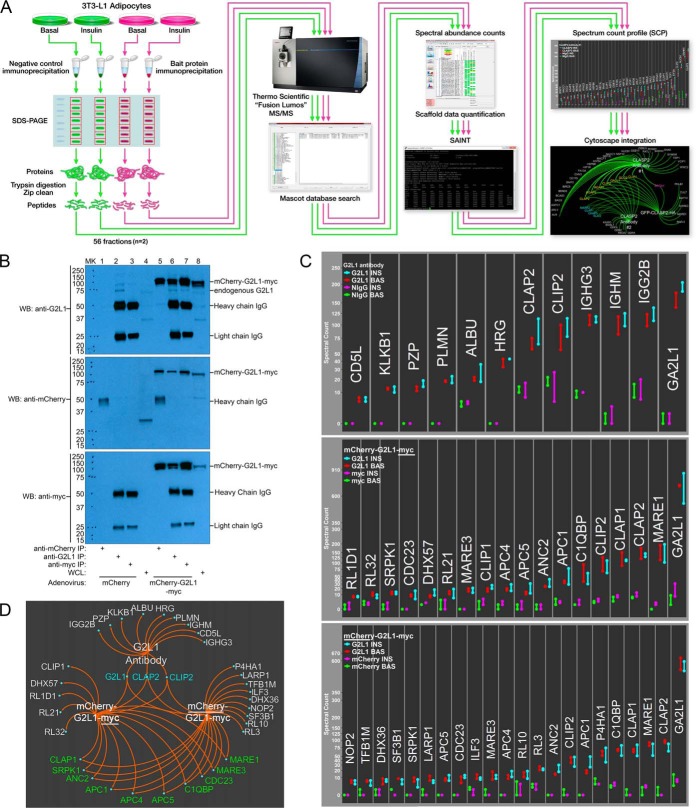
We previously characterized the CLASP2 protein network in 3T3-L1 adipocytes and discovered G2L1 was enriched in three alternative, cross-reference CLASP2 interactomes (24). To follow-up on a possible relationship between CLASP2 and G2L1 in 3T3-L1 adipocytes, we performed multiple G2L1 affinity purification coupled with mass spectrometry (AP-MS) experiments using our previously described label-free quantitative proteomics approach (24) (Fig. 2A). We performed basal versus insulin treatment interactome analysis of endogenous G2L1 IPs as well as overexpressed G2L1 immunoprecipitated with two alternative antibodies, one to a myc-epitope tag and the other to a mCherry fluorescent protein tag (western blots of all G2L1 protein IPs are shown in Fig. 2B). The resulting spectrum count data for the G2L1 IPs was then scored for enrichment over the various appropriate negative control IPs using the established bioinformatic tool Significance Analysis of Interactome (SAINT) as previously described (24, 41–43). On the SAINT Probability Score (“P-Score”) scale from 0 to 1, ≥0.85 was used to signify a protein as “SAINT qualified.” Protein SAINT scores, spectrum counts, distinct peptides, and percent protein coverage for the interactomes are included in the supplemental Tables S7–S15. We used our Spectrum Count Profile (“SCP”) to hierarchically summarize the raw spectral count data of all the SAINT-qualified proteins (Fig. 2C) followed by Cytoscape-based (44) integration to simultaneously visualize all three G2L1 SAINT-qualified interactomes (Fig. 2D). All three G2L1 interactomes (Uniprot ID for G2L1 is GA2L1) had SAINT-qualified enrichment of CLASP2 and CLIP2, discoveries which serve to reciprocally confirm our previous report that both CLASP2 and CLIP2 can co-IP G2L1 (24). G2L1 was identified during a proteome-wide screen for EB-binding proteins (26). In our overexpressed G2L1 interactomes, EB1 (Uniprot ID: MARE1) and to a lesser extent EB3 (Uniprot ID: MARE3) were both strongly enriched, with spectrum count numbers like those detected for CLASP2. CLASP2 at a predicted mass of ∼141kDa has the potential for a larger number of spectrum counts compared with EB1 at ∼30kDa, so the fact that EB1 and CLASP2 had comparable spectrum counts suggests a larger amount of EB1 associates with G2L1 versus CLASP2 (as well as CLIP2). Antibodies targeted to endogenous protein can disrupt protein-protein interactions and possess nonspecific cross-reactivity (45, 46), which may explain why EB1 was undetected in the endogenous G2L1 interactome. To further validate the association between EB1 and G2L1, we performed an endogenous EB1 interactome and confirmed the reciprocal co-IP of G2L1 with EB1 (supplemental Fig. S3). The overexpressed G2L1 interactomes possessed levels of CLASP1 like those detected for CLASP2, EB1, and CLIP2 and contained a lower amount of EB3 as compared with EB1. There were other similarities between the two overexpressed G2L1 interactomes, including the presence of five members (APC1, ANC2/APC2, APC4, APC5, and CDC23) of the anaphase promoting complex/cyclosome (APC), an E3 ubiquitin ligase that facilitates ubiquitination and degradation of target proteins (47). There were also differences in the two overexpressed G2L1 interactomes that may have resulted from steric hindrance caused by antibody binding.
Fig. 2.
The G2L1 interactomes. A, To identify new interacting partners for G2L1 in 3T3-L1 adipocytes, different G2L1 IPs were compared against negative control IPs either in the absence or presence of insulin treatment. The IPs were separated by SDS-PAGE, fractionated into gel slices, subjected to trypsin digestion, and analyzed by tandem mass spectrometry. Peptide and protein identification was performed by Mascot database searching, and the resulting spectral count data was assembled with Scaffold. The spectrum count data was then scored for enrichment using SAINT and the resulting SAINT-qualified protein spectral count data was visualized with a Spectrum Count Profile “SCP”. The SAINT-qualified proteins from the three alternative G2L1 interactomes performed were then integrated and visualized with Cytoscape. MS/MS tandem mass spectrometry. B, 150 mm plates of differentiated 3T3-L1 adipocytes infected with either mCherry or mCherry-G2L1-myc overexpressing adenovirus and were lysed in an isotonic CHAPS lysis buffer. IPs were performed as described under “Experimental procedures”. The IPs or whole cell lysates were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes. The membranes containing the immunoprecipitated proteins were subjected to Western blotting with the antibodies indicated. The labels on the right side of the blot indicate where the various proteins migrate on the gel. MK, protein ladder marker; WB, Western blotting; WCL, whole cell lysates; C, For the various G2L1 interactomes (from top to bottom, endogenous G2L1, mCherry-G2L1-myc, and mCherry-G2L1-myc), the “SAINT-qualified” proteins were ordered in a hierarchical manner, from lowest spectrum counts identified to highest, and results from two experiments were individually plotted in a SCP. Basal NIgG IPs (green), insulin NIgG IPs (magenta), basal G2L1 IPs (red), and insulin G2L1 IPs (turquoise). D, Cytoscape-based integrated visual representation of the anti-G2L1, anti-myc mCherry-G2L1-myc, and anti-mCherry mCherry-G2L1-myc SAINT-qualified proteins. The proteins listed in turquoise were identified in all three G2L1 interactomes, whereas the proteins listed in green were shared between the anti-mCherry and anti-myc antibody mCherry-G2L1-myc interactomes.
To follow up on the G2L1 interactome findings, we performed traditional co-IP and Western blotting experiments and focused on confirmatory tests for the association of G2L1 together with CLASP2, CLIP2, and EB1. We reproduced the interactome data and reciprocally confirmed the interactions between G2L1 and CLASP2, CLIP2, and EB1 (supplemental Fig. S4). We also found no significant effect of insulin on the reciprocal interactions; findings that are like those observed in the interactome studies. Collectively, we have established a new relationship between G2L1 and CLASP2, CLIP2, and EB1, in 3T3-L1 adipocytes.
Colocalization of CLASP2 and G2L1
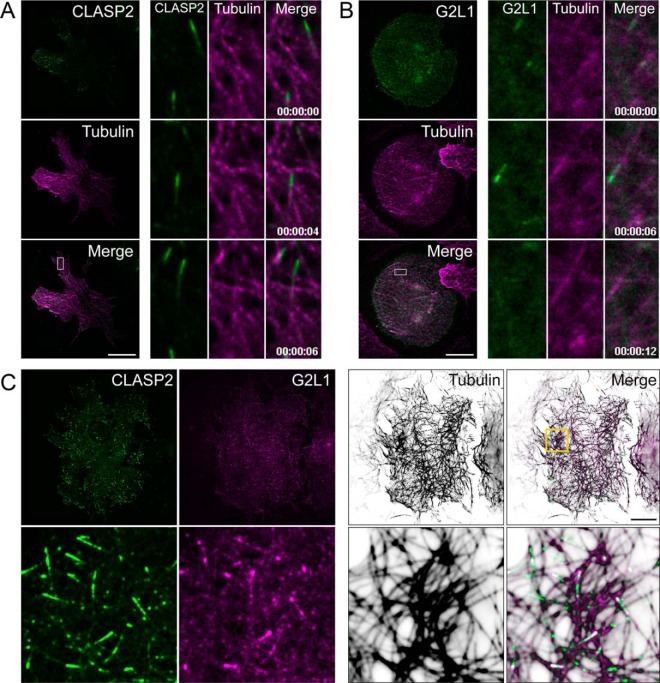
CLASP2 and G2L1 are reciprocally enriched in the interactome studies we have performed, evidence that supports potential biological cooperation between these two +TIPs. We therefore tested for colocalization of CLASP2 and G2L1 in basal state 3T3-L1 adipocytes proximal to the cell membrane using live-cell total internal reflection fluorescence microscopy (TIRFM), a powerful tool for visualizing +TIP localization and microtubule dynamics. Overexpression of either GFP-CLASP2-HA (Fig. 3A) or mCherry-G2L1-myc (Fig. 3B) in 3T3-L1 adipocytes revealed that each protein localizes to the growing plus-end of microtubules within comet-like structures, which can be observed tracking across the cell and leading a newly formed microtubule, per classic +TIP behavior. When coexpressed, CLASP2 and G2L1 are coincident on dynamic comet-like structures (supplemental Video S1). Examination of fixed samples of adipocytes coexpressing GFP-CLASP2 and mCherry-G2L1 revealed that comparatively, G2L1 occupies a smaller and more proximal area of the +TIP than CLASP2, which led a longer comet-like structure on microtubules (Fig. 3C). These additional findings validate the interactome data and solidify a potential cooperative relationship between CLASP2 and G2L1.
Fig. 3.
CLASP2 and G2L1 colocalize in 3T3-L1 adipocytes. A–B, Live-cell imaging of adipocytes cultured in complete media coexpressing mRuby2-Tubulin (magenta) and GFP-CLASP2-HA (A, green) or mCherry-G2L1-myc (B, green). Live cells were imaged using TIRFM on a 2-s acquisition interval. Time series images to the right of the whole cell image are used to highlight +TIP dynamics within the indicated ROI. C, Immunofluorescence images of adipocytes cultured in complete media cooverexpressing GFP-CLASP2-HA (green) and mCherry-G2L1-myc (magenta). Cells were fixed and immunolabeled for tubulin (inverted white). Bottom row is magnified ROI. ROI, region of interest. Scale bar = 20 μm.
The Effect of Insulin on CLASP2 and G2L1 +TIP Dynamics
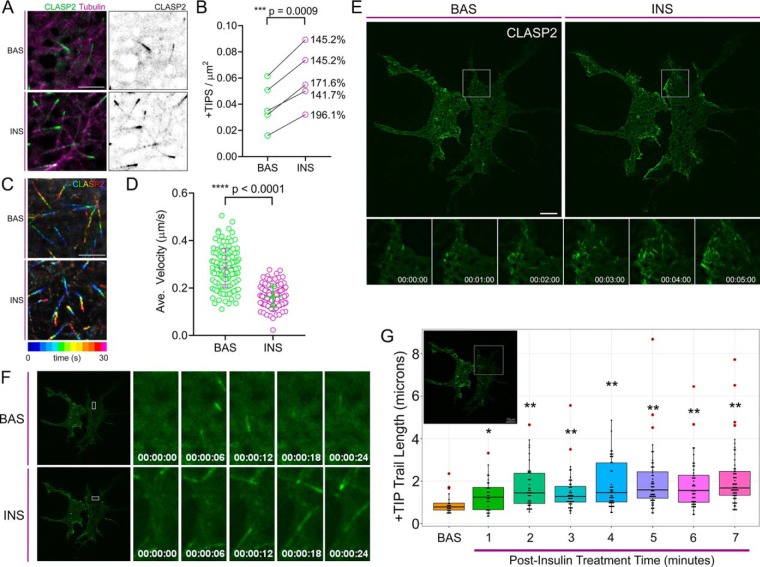
On successfully establishing colocalization of CLASP2 and G2L1 with TIRFM in 3T3-L1 adipocytes, as well as discovering that both CLASP2 (11) and G2L1 undergo insulin-regulated phosphorylation, we profiled the effect of insulin on CLASP2 and G2L1 cellular localization proximal to the inner surface of the plasma membrane in a live-cell setting. Microtubule polymerization and density have both been shown to increase after insulin treatment in 3T3-L1 adipocytes (28, 29). Comparative quantification revealed that insulin increases the number of CLASP2-containing +TIPs per unit area (Fig. 4A–4B), data that aligns with the previous findings that insulin increases microtubule density. Conversely, we discovered that insulin treatment reduces the average velocity of CLASP2-containing +TIPs (Fig. 4C–4D), suggesting that insulin slows the polymerization rate of individual microtubules with CLASP2-containing +TIPs. Taken together our data aligns with pre-existing findings that insulin increases the population of microtubules. Although this implicates insulin increases gross microtubule polymerization, we propose that whereas insulin may create more microtubules, microtubules that possess CLASP2-containing +TIPs polymerize slower in the presence of insulin.
Fig. 4.
The effect of insulin on CLASP2 +TIP dynamics. Live-cell imaging of adipocytes serum-starved for one hour and subsequently stimulated with 100 nm insulin. Live cells were imaged using TIRFM on a two-second acquisition interval. A, Single frame of live-cell imaging of adipocytes coexpressing GFP-CLASP2 (green) and mRuby2-Tubulin (magenta) at basal state (top row) or 10 mins (bottom row) following insulin stimulation. CLASP2 is displayed in inverted white to highlight +TIP density. B, Quantification of CLASP2-containing +TIP density per unit area (μm2) in adipocytes at basal state or 10 mins following insulin stimulation. Percent increase in CLASP2-containing +TIP density is indicated to the right. Statistical comparison made by paired parametric t test, n = 5 cells. C, Temporally color-coded projection of GFP-CLASP2 localization during a 30 s live-cell imaging interval. The length and extent of color overlap of time-projected CLASP2 localization indicates reduced displacement and hence lower velocity of CLASP2-containing +TIPs during the imaging interval in adipocytes after 10 mins of insulin stimulation. D, Quantification of CLASP2-containing +TIP velocity in adipocytes at basal state or 10 mins following insulin stimulation indicates reduced velocity of CLASP2-containing +TIPS after insulin treatment. Statistical comparison made by unpaired parametric t test, n = 123–125 CLASP2-containing +TIPS from five cells. Scale bars = 5 μm. E, Image of entire cells in the basal state (left panel) or 8 mins post insulin treatment (right panel) extracted from supplemental Video 2. Time series images under the whole cell image are used to highlight insulin-stimulated +TIP dynamics within the indicated ROI. F, Time series extracted from supplemental Video 2 of the indicated ROIs at either the basal state or at 4 mins post insulin treatment to present an example of changing CLASP2 microtubule plus-end dynamics with insulin stimulation. G, Live cell CLASP2-containing +TIP dynamics of the ROI extracted from supplemental Video 2 were captured in the basal state followed by stimulation with insulin. Each insulin-stimulated CLASP2-containing +TIP trail length time point was compared against the basal CLASP2-containing +TIP trail length to test for significant differences. t test; *p ≤ 0.05, **p ≤ 0.01. Red circles represent outlier data points. ROI, region of interest. BAS, basal. INS, insulin. Scale bar = 10 μm.
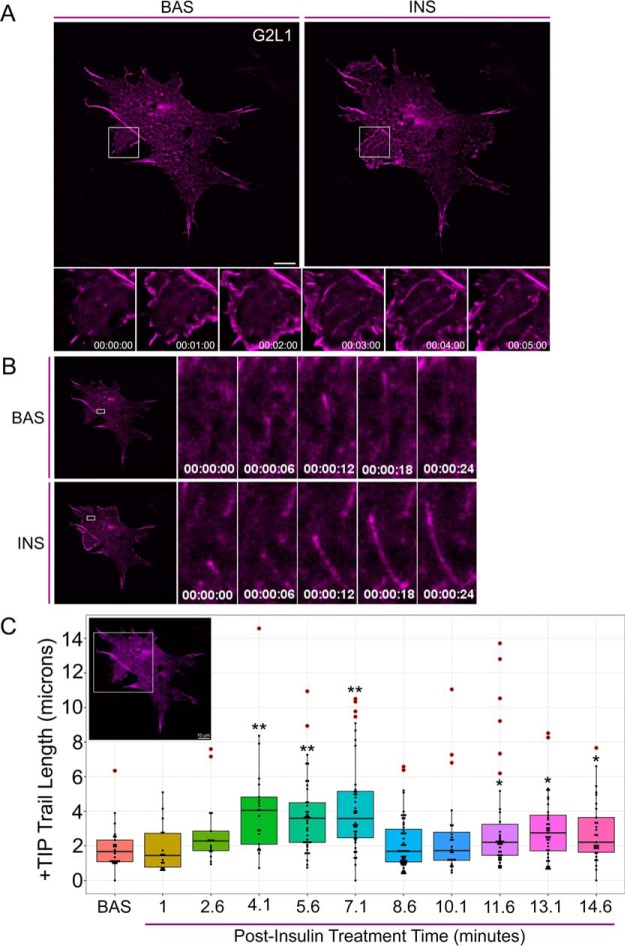
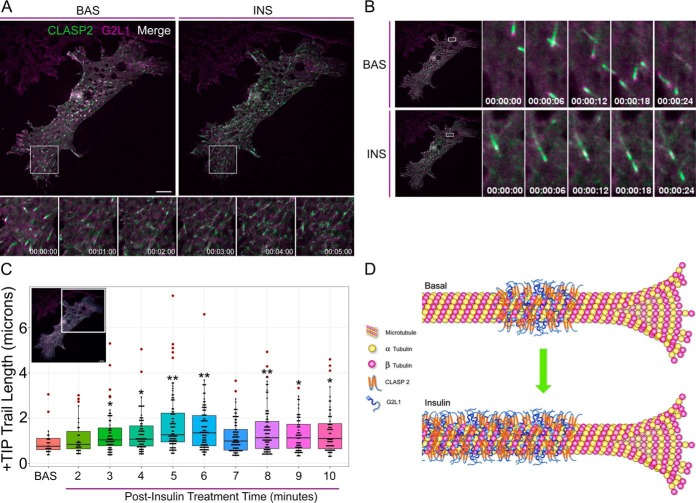
Before insulin treatment, CLASP2 moves in short, rapid bursts with a comet-like appearance per classic +TIP presentation (supplemental Video 2). After insulin stimulation, we discovered that CLASP2 shifts from being exclusively enriched at the plus-end microtubule tip to “trailing,” in which CLASP2 decorates the trailing length of the microtubule proximal to the growing plus-end (supplemental Video S2, Fig. 4E–4G, supplemental Fig. S5). Because we observed G2L1 colocalizes with CLASP2 in adipocytes, we hypothesized that G2L1 behavior might also respond to insulin stimulation. In the basal state G2L1 displays classic +TIP activity (supplemental Video S3) whereas insulin stimulates G2L1 trailing on microtubules (supplemental Video S3, Fig. 5A–5C, supplemental Fig. S5). Insulin is known to stimulate membrane ruffling in adipocytes (4). Because G2L1 is known to localize to actin in addition to microtubules, it is of interest that on insulin stimulation, G2L1 strongly enriches to lamellipodial protrusions (supplemental Video S3). On testing the effect of insulin on adipocytes coexpressing both GFP-CLASP2 and mCherry-G2L1, the insulin-stimulated trailing phenotype was definitively captured (supplemental Video S4, Fig. 6A–6C). Quantification of the +TIP trail length captured the trailing phenotype induced by insulin within each of the representative adipocytes (Fig. 4G, Fig. 5C and Fig. 6C). To rule out insulin-stimulated +TIP trailing resulting from a phototoxic effect associated with continual exposure to laser light during live cell imaging, we have reproduced the findings in cells visualized for only thirty seconds at a time at 5-minute intervals pre- and postinsulin stimulation (data not shown). The insulin-induced +TIP trailing response observed was not universal across all cells, perhaps because of the reported heterogeneous nature of adipocyte differentiation (48). We present here, for the first time, that insulin affects CLASP2 and G2L1 +TIP dynamics (modeled in Fig. 6D), direct evidence that supports the hypothesis that the +TIP network represents a new signaling system associated with insulin action.
Fig. 5.
Insulin stimulates G2L1 plus-end trailing. Live-cell imaging of adipocytes overexpressing mCherry-G2L1-myc (magenta). Adipocytes were serum starved for one hour and subsequently stimulated with 100 nm insulin. Live cells were imaged using TIRFM on a two-second acquisition interval and the displayed time series were extracted from supplemental Video 3. A, Image of entire cell in the basal state (left panel) or twelve minutes post insulin treatment (right panel). Time series images under the whole cell image are used to highlight insulin-stimulated G2L1-containing +TIP dynamics within the indicated ROI. B, Time series of the indicated ROIs at either the basal state or at 4 mins post insulin treatment to present an example of changing G2L1 plus-end dynamics with insulin stimulation. C, Live cell G2L1-containing +TIP dynamics of the ROI were captured in the basal state followed by stimulation with insulin. Each insulin-stimulated G2L1-containing +TIP trail length time point was compared against the basal G2L1-containing +TIP trail length to test for significant differences. t test; *p ≤ 0.05, **p ≤ 0.01. Red circles represent outlier data points. ROI, region of interest. BAS, basal. INS, insulin. Scale bar = 10 μm.
Fig. 6.
Insulin stimulates CLASP2 and G2L1 microtubule plus-end cotrailing. Live-cell imaging of adipocytes cooverexpressing GFP-CLASP2-HA (green) and mCherry-G2L1-myc (magenta). Adipocytes were serum starved for one hour and subsequently stimulated with 100 nm insulin. Live cells were imaged using TIRFM on a two-second acquisition interval and the displayed time series were extracted from supplemental Video 4. A, Image of entire cell in the basal state (left panel) or 8 mins post insulin treatment (right panel). Time series images under the whole cell image are used to highlight insulin-stimulated +TIP dynamics within the indicated ROI. B, Time series of the indicated ROIs at either the basal state or at 3 mins post insulin treatment to present an example of changing CLASP2 and G2L1 plus-end dynamics with insulin stimulation. C, Live cell +TIP dynamics of the ROI were captured in the basal state followed by stimulation with insulin. Each insulin-stimulated +TIP trail length time point was compared against the basal +TIP trail length to test for significant differences. t test; *p ≤ 0.05, **p ≤ 0.01. Red circles represent outlier data points. ROI, region of interest. BAS, basal. INS, insulin. Scale bar = 10 μm. D, A model depicting the insulin-stimulated shift of CLASP2 and G2L1 from growing plus-end microtubule localization to “trailing”, a behavior characterized by CLASP2 and G2L1 decorating the trailing microtubule lattice behind the growing microtubule plus-end.
Insulin Stimulates α-Tubulin Acetylation at Lysine 40 and Microtubule Stabilization
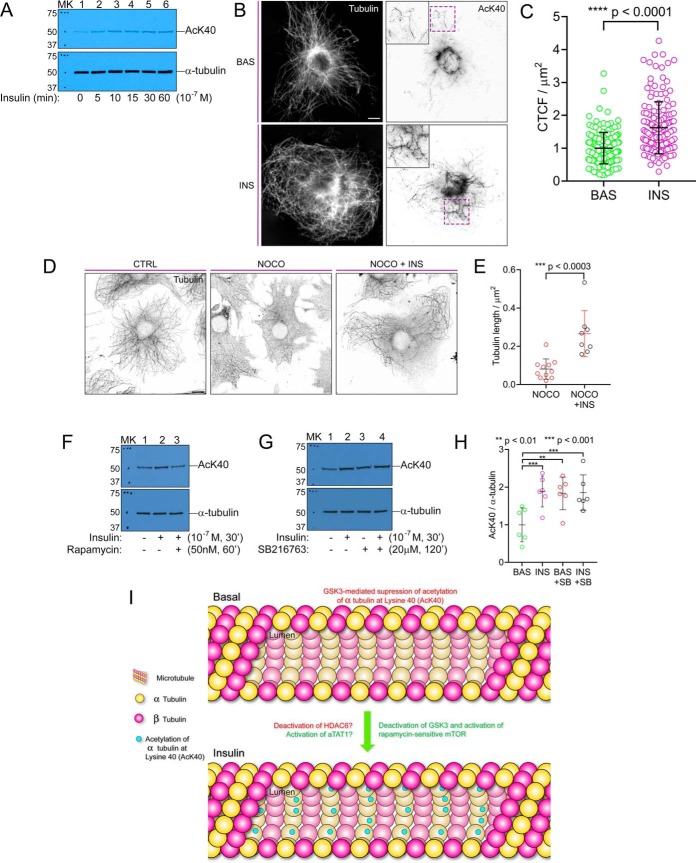
Results from the live cell experiments revealed that insulin stimulates CLASP2 and G2L1 to shift from predominant localization at the microtubule plus-end to trailing along varied lengths of the microtubule lattice proximal to the growing plus-end. Dynamic short-lived microtubules are characterized by transient periods of growth and shrinkage (49), although a subset of the microtubule population are stabilized and persist for much longer times as a means for long-range transport. These long-lived microtubules can possess α-tubulin acetylation at lysine 40 (“AcK40”), a post-translational modification that protects microtubules from mechanical stress (50, 51). Because the microtubules with insulin-stimulated CLASP2 and G2L1 trails visually presented as “brace-like” across the microtubule lattice, we first tested the hypothesis that insulin stimulates a marker for stabilized microtubules, AcK40. Western blot analysis of whole cell lysates from serum-starved 3T3-L1 adipocytes either left untreated or subjected to an insulin time course confirmed that insulin stimulates α-tubulin acetylation at lysine 40 (“AcK40”) (Fig. 7A), results that were reproduced with AcK40 immunofluorescence imaging (Fig. 7B-C). Because an insulin-stimulated increase in AcK40 is not direct evidence of microtubule stabilization, we tested whether insulin treatment results in increased resistance to the microtubule depolymerizing agent, nocodazole. Treatment of serum-starved adipocytes with nocodazole induced a loss of microtubules, whereas pretreatment of cells with insulin reduced the severity of nocodazole-stimulated microtubule depolymerization (Fig. 7D–7E), therefore, we conclude insulin increases the stability of microtubules in 3T3-L1 adipocytes.
Fig. 7.
Insulin stimulates acetylation of α-tubulin at Lysine 40 and microtubule stabilization. A, Serum-starved 3T3-L1 adipocytes were either left untreated or treated with an insulin time course for the times indicated. Adipocyte lysis was performed as described under “Experimental procedures”. The whole cell lysates were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes. The membranes were subjected to Western blotting with the antibodies indicated. Representative blot, n = 3. MK, protein ladder marker. AcK40, α-tubulin acetylation at lysine 40. B, Immunofluorescence images of the effect of insulin (30min, 100 nm) on serum-starved adipocytes. Cells were fixed and immunolabeled for α-tubulin (white) and AcK40 (inverted white). Scale bar = 10 μm. C, Quantification of AcK40 in adipocytes at basal state or 10 mins following insulin stimulation indicates increased AcK40 after insulin treatment. The images were quantified for corrected total cell fluorescence (CTCF) as described under “Experimental procedures”. Quantification was based on data collected from 25–40 cells per condition. The images are representative of five independent experiments. Error bars represent ±S.E. and statistical comparison was made by unpaired parametric t test. D, Immunofluorescence images of the effect of insulin (30min) on nocodazole-induced microtubule depolymerization in serum-starved adipocytes. Cells were left untreated or treated with either 2 μm nocodazole alone for 15min or 100 nm insulin for 30 min followed by 2 μm nocodazole for 15min, fixed and immunolabeled for α-tubulin (inverted white). CTRL, control, NOCO, nocodazole. INS, insulin. Scale bar = 10 μm. E, Quantification of microtubule density in adipocytes indicates insulin treatment results in resistance to nocodazole-induced microtubule depolymerization. Quantification was based on data collected from 8–11 cells per condition. The images are representative of two independent experiments. Error bars represent ±S.E. and statistical comparison was made by unpaired parametric t test. F, Serum-starved 3T3-L1 adipocytes were either left untreated or treated with either insulin alone or insulin together with rapamycin pretreatment to inhibit mTOR. Evaluation of AcK40 was performed as described above in A. G, Serum-starved 3T3-L1 adipocytes were either left untreated, treated with SB216763 alone (a GSK3 inhibitor), treated with insulin alone, or treated with insulin together with SB216763 pretreatment. Evaluation of AcK40 was performed as described above in A. H, Six experiments as performed in G were quantified by densitometry to evaluate the effects of SB216763-mediated inhibition of GSK3 on AcK40. The AcK40 signal was normalized to the total α-tubulin signal on the western blots. These values were then normalized by the mean value of the basal sample and then expressed as a fold change over basal ±S.E.; t test. SB, SB216763. I, In this proposed hypothetical model, GSK3 suppresses the acetylation of α-tubulin at lysine 40 (“AcK40”) in the basal state. Insulin deactivates GSK3 and stimulates AcK40 through rapamycin-sensitive mTOR.
To test for specificity of the AcK40 Western blotting signal and also to search for the pathway(s) involved in regulating insulin-controlled AcK40, we discovered we could inhibit the insulin-stimulated increase of AcK40 by treating the adipocytes with the mTOR inhibitor rapamycin (32% average decrease, n = 7, p ≤ 0.01), implicating mTOR in insulin-stimulated microtubule stabilization (Fig. 7F). The kinase GSK3 has been linked to suppressing AcK40 in the context of cell polarization (10). Because GSK3 is active in the basal state and deactivated by insulin, we hypothesized that GSK3 regulates basal levels of microtubule stabilization through suppression of AcK40. Treatment of 3T3-L1 adipocytes with the GSK3 inhibitor SB216763 reversed the decreased levels of AcK40 observed in the basal state, indicating that insulin suppresses GSK3 activity in part to encourage an increase in AcK40 (Fig. 7G and 7H). These discoveries reveal a whole new property of insulin, namely increased microtubule stabilization as well as the stimulation of α-tubulin acetylation at lysine 40, a generally accepted marker for microtubule stabilization, a mechanism that is regulated at least in part by a counterbalance between GSK3 and rapamycin-sensitive mTOR-controlled signaling elements (modeled in Fig. 7I).
DISCUSSION
We followed up on our recent CLASP2 interactome study by characterizing the G2L1 interactome within 3T3-L1 adipocytes. The +TIPs have been an encouraging fit for interactome studies, as known positive control interaction partners have been routinely detected and reciprocal interactomes have been largely confirmatory. We reproduced results from other cell lines (23, 26) that G2L1 co-IPs EB1 and discovered that G2L1 also co-IPs both CLASP2 and CLIP2 in 3T3-L1 adipocytes, data which reciprocally confirms our previous CLASP2 and CLIP2 interactome findings (24). We performed an EB1 interactome and confirmed the reciprocal presence of G2L1, although, we did not detect significant enrichment of CLASP2 in the EB1 interactome. Prior experiments on EB1 and CLASP2 have supported the concept that these two proteins may track along growing microtubule plus-ends individually from each other, as was observed in PtK1 epithelial cells (52). The lack of a detectable strong co-IP between EB1 and CLASP2 in adipocytes is consistent with our previous CLASP2 interactome work (24) although discordant from a study in COS-1 cells which proved an interaction can occur between EB1 and CLASP2 (17). There have been repeated published links between CLASP2 and the EB proteins, including a dependence of EB1 proper microtubule localization on CLASP2 within a multitude of cell types (53). All of these observed differences in the relationship between EB1 and CLASP2 could be explained by cell-line specificity, a transient nature of association as previously proposed (17), a difference in the expression levels of other +TIPs necessary for assembling specific protein complexes (9, 40, 49, 54–56), a lack of detectable interactions resulting from technical limitations, or issues with the immunoprecipitating antibodies (45, 46). Future studies will investigate whether the lack of a detectable association between CLASP2 and EB1 indicates that G2L1 and EB1 may operate independently of CLASP2 in adipocytes and whether the G2L1/EB1 relationship is affected by insulin in a spatio-temporal manner. The link between G2L1 and CLASP2 discovered during the interactome studies was solidified by the live-cell TIRFM-based discovery that G2L1 and CLASP2 colocalize at growing microtubule plus-ends. In addition to localizing to microtubule plus-ends, G2L1 also possesses binding capacity for filamentous actin (F-actin) (23, 57, 58). We have unpublished findings indicating that in addition to the insulin-regulated +TIP characteristics observed for G2L1 reported here, G2L1 also colocalizes with insulin-stimulated actin reorganization and membrane ruffling. In addition, we have now established that both CLASP2 (11) and G2L1 undergo insulin-stimulated phosphorylation. Because insulin-stimulated actin reorganization is paramount for proper insulin action (7), future studies will elucidate whether a functional association exists between insulin-stimulated actin dynamics and G2L1.
The effect of insulin on both CLASP2 and G2L1 on microtubules proximal to the interior surface of the cell (“the TIRFM zone”) represent a new mode of +TIP regulation in the context of insulin action. Live-cell TIRFM captured CLASP2 and G2L1 shifting from predominant plus-end localization to insulin-stimulated immobilization along the length of the microtubule lattice, proximal to the growing plus-end (modeled in Fig. 6D). CLASP2 has repeatedly been linked to the stabilization of microtubules through participating in microtubule rescue events and preventing microtubule depolymerization, across multiple organisms (17, 56, 59–72). These effects are imparted through the various domains in CLASP2, for example, the tumor overexpressed gene 2 (TOG2) domain was recently shown to maintain the integrity of the stabilizing cap localized at the growing microtubule tip (70). Although this is the first time CLASP2 and G2L1 have been observed to shift in real-time after insulin stimulation from predominant plus-end localization to trailing along the growing length of the microtubule, it is not the first time an alteration in the spatial relationship between CLASP2 and microtubules has been captured. In live migrating Ptk1 epithelial cells, within the cell body, CLASP2 displayed typical +TIP action, whereas lamella and lamellipodium-based CLASP2 lacked +TIP behavior and instead exhibited trailing via binding along the length of the growing microtubule lattice (52), very reminiscent of the effect of insulin we observed. As Wittmann and Waterman-Storer stated (52), CLASP2 shifts from true +TIP behavior to full lattice decoration, a style of microtubule association more reminiscent of classic MAP family members (8). Hypothetically, these newly created microtubules populated with lattice dispersion of CLASP2 and G2L1 trails may serve as a signal to assemble or initiate an as-of-yet undetermined molecular event, in this case, a signaling signature associated with insulin action. There is noticeable heterogeneity in the literature with regards to findings on CLASP2 movement, which can be explained in part by the fact that different cell types possess alternative proteomes, and these differences probably influence the cooperative nature (49) and resulting behavior of +TIPs. For example, the spatio-temporal changes in CLASP2 reported for wound healing experiments in Ptk1 epithelial cells (52) and HaCat keratinocytes (16) were not observed in motile 3T3 fibroblasts, although, when the 3T3 fibroblasts underwent serum stimulation, CLASP2 accumulated and immobilized at the leading edge of the cell in yet another pattern of localization (10, 14, 72). These different phenotypes are distinct from the population of CLASP2 that has been observed at the Golgi (73, 74). So, overall, there is support for a diverse range of CLASP2 regulation and function that can be dictated by multiple aspects, including cell-specific needs and intracellular localization.
This insulin-stimulated +TIP trailing along newly created microtubules possessed what we viewed as a “brace-like” appearance, and because prior studies support a microtubule-stabilizing role for CLASP2 (10, 69, 70), we hypothesized that insulin stimulates the stabilization of selective microtubules. Follow-up studies discovered that insulin stimulates α-tubulin Lysine 40 acetylation (“AcK40”), a post-translational modification within the 15-nm-wide lumen of the microtubule that is associated with stabilization of both long-lived and curved microtubules (50, 51, 75). AcK40 only occurs on polymerized microtubules (76–79) and is catalyzed by the tubulin acetyltransferase aTAT1 (80, 81) whereas the exclusively cytoplasmic (82) histone deacetylase 6 (HDAC6) deacetylates tubulin (83). AcK40 takes place on polymerized microtubules to reduce structural strain by increasing microtubule flexibility (50, 51). These stabilized microtubules, by becoming protected against breaking and shrinkage, exist for prolonged periods of time (77, 78, 84). Insulin-stimulated CLASP2/G2L1 trailing occurred acutely, within minutes of insulin treatment, whereas the effect of insulin on microtubule stabilization via AcK40 was gradual, indicating that trailing events precede stabilization, although whether a causal relationship exists between +TIP trailing and microtubule stabilization is unknown and whether trailing and AcK40 occur on the same microtubules will be the subject of future studies. Because the effect of insulin on AcK40 was not rapid (as seen with Akt substrates (85)) but was instead delayed and profiled more like the effects of insulin observed for mTOR substrates (85), we tested and confirmed that inhibition of mTOR with rapamycin blocks insulin-stimulated AcK40. The control of insulin-stimulated AcK40 by mTOR was found to be counterbalanced by GSK3-mediated suppression of AcK40 in the basal state. Future studies will be aimed at testing the hypothesis that, through insulin-mediated suppression of GSK3 activity and activation of mTOR, insulin influences HDAC6 and aTAT1 activity to control α-tubulin AcK40 (modeled in Fig. 7I).
Of all the microtubule-regulating protein interactome studies we have completed thus far using the label-free spectrum counting technique we have devised, none of the validated network proteins analyzed have exhibited measurable protein abundance changes after insulin-stimulation. In this report, our traditional co-IP and Western blotting confirmatory follow-up experiments led to findings that agreed with the quantitative proteomics data, the proteins did in fact co-IP although the abundance of the coimmunoprecipitated proteins was not reproducibly affected by insulin. As an alternative to changes in protein association, we hypothesized that because CLASP2 undergoes robust insulin-stimulated phosphorylation (11), perhaps the 3T3-L1 adipocyte CLASP2 protein network proteins serve as potential candidates for insulin-regulated phosphorylation. We developed a straight-forward approach to quantifying changes in protein phosphorylation that incorporates basic immunoprecipitation and in-gel protein digestion techniques together with automated label-free quantification of extracted ion abundance performed in the software program Progenesis QI for Proteomics. This protein-specific quantitative phosphoproteomics approach expanded the number of +TIPs known to undergo insulin-regulated phosphorylation from just CLASP2 to a list that now includes CLIP2, G2L1, EB1, and CKAP5, whereas we also discovered that the CLASP2 network members MARK2 and AGAP3 undergo insulin-regulated phosphorylation as well (hypothetically modeled in Fig. 1C). Of all the sites analyzed, the phosphorylation of CLIP2 (also known as CLIP-115) at Ser552 underwent the strongest increase on insulin treatment. Phosphorylation of CLIP1 (also known as CLIP-170) at Thr287 and Ser195/Ser1318 has been linked to centrosome duplication and kinetochore-microtubule attachments (86, 87), respectively, although Ser1318 of CLIP1 is not found in the shorter CLIP2 isoform, whereas Ser552, the insulin-stimulated phosphorylation site of CLIP2, is not conserved in CLIP1. Ser552 lies ∼300 amino acids past the second microtubule-binding Cytoskeleton-associated protein Gly-rich (“CAP-Gly”) domain (88) and the surrounding serine-rich basic regions within the N terminus of CLIP2 and is positioned along the extensive coiled-coil region of CLIP2 known to mediate CLIP2 dimerization (89). CLIP1 and CLIP2 association with microtubules has been shown to be regulated by phosphorylation in a negative manner (89–91). Phosphorylation also regulates CLIP1 intramolecular association and accompanying CLIP1 conformational changes (92). Each of these phospho-regulated events has been linked to phosphorylation sites near the CAP-GLY domains and the surrounding serine-rich basic regions within the N terminus of the CLIPs, rendering the insulin-stimulated Ser552 CLIP2 phosphorylation site with the potential to be functionally distinct. The CLASP2 interactome project revealed a novel association between the +TIP CLIP2 and the GTPase-activating proteins AGAP1 and AGAP3 (24). There is a key difference between the CLASP2/G2L1 interaction and the proposed complex between CLIP2 and the AGAPs, in that the AGAPs are the lone protein members of the CLASP2 network in 3T3-L1 adipocytes that act as both a GTPase (93, 94) and a GTPase activating protein (95, 96). In addition, the AGAPs also contain a Pleckstrin Homology (“PH”) domain, whose known functions include mediating protein-protein interactions as well as membrane localization through the binding of phosphoinositides (97). Of the insulin-stimulated AGAP3 phosphorylation sites we have discovered; one is within the GTPase domain (pSer300) whereas the other is located within the PH domain (pSer478). CLIP2 was the most enriched protein in the AGAP3 interactome (24), supportive of the hypothesis that functional cooperativity exists between CLIP2 and AGAP3 in the context of insulin action.
EB1, known as “the master integrator of +TIP networks” (49) as well as CKAP5 (alternatively referred to as ch-TOG) (56, 98), also undergo insulin-regulated phosphorylation. EB1, like CLASP2, G2L1, and CLIP2, tracks growing microtubule plus-ends and acts as an integrating scaffold protein that promotes a wide variety of +TIP localization to the growing microtubule plus-end (99). Phosphorylation of EB proteins is known to regulate microtubule dynamics and EB function in a variety of organisms and biological systems (100–109). The insulin-stimulated EB1 phosphorylation site we detected, Ser155, lies in the coiled-coil linker region of EB1 that bridges the N-terminal Calponin-homology (CH) domain to the C-terminal EB homology (EBH) domain and adjacent EEY/F motif. Ser155 of EB1 already has a possible connection with major players in insulin action, as AKT and GSK3 regulate levels of Ser155 phosphorylation to control EB1 localization to microtubule plus-ends (110). CKAP5 is a member of the XMAP215 family of microtubule polymerases that catalyzes the addition of tubulin dimers to elongating microtubule plus-ends. CKAP5 localizes to the extreme tip of the growing microtubule plus end whereas EB1 is further down the microtubule (111), so in an in vitro model for example, EB1 is located tens of nanometers further down the microtubule from CKAP5 (112). CKAP5 consists of five successive TOG domains that span across the majority of this 225kDa protein whereas a microtubule lattice binding domain is located between the fourth and fifth TOG domains, all of which participate in the processive addition of tubulin dimers to the growing microtubule plus-end (113, 114). Within the CKAP5 C terminus lies both a cryptic TOG domain as well as a shorter four α-helix-based domain (with no known identity) that mediates protein-protein interactions (115, 116). It is within this terminal multi-α-helix-containing domain where insulin significantly affects the phosphorylation of CKAP5 at Ser1861 in a suppressive manner. We discovered a stretch of CKAP5 phosphorylation sites spanning amino acids 1800–1870 that also trend toward insulin-mediated suppression of phosphorylation (supplemental Fig. S2F), although the four biological replicates analyzed did not achieve statistical significance. With CKAP5 playing such a vital role in cytoskeletal management, it is of future interest to elucidate the significance of insulin-mediated changes in CKAP5 phosphorylation within the context of insulin-controlled microtubule dynamics.
Another protein we discovered to undergo insulin-regulated phosphorylation was the only kinase of the group, MARK2 (also known as Par-1b). Classical MAPs bind and release along the microtubule lattice as a mode of microtubule regulation, a process that is controlled in part by transient MAP phosphorylation by the MARK family kinases (reviewed in (8)). MARK2-mediated release of the various MAPs from the microtubule lattice affects the stabilization of microtubules, microtubule polymerization, microtubule bundling, the association between microtubules and actin, and microtubule mediated motor transport of intracellular cargo (8). Insulin stimulates an increase in the phosphorylation of MARK2 at Ser40, Thr42, and Ser43, a short stretch of residues that lies adjacent to the MARK2 kinase domain, whereas phosphorylation within the spacer region of MARK2 at Ser568 undergoes insulin-mediated suppression. MARK2 makes an intriguing hypothetical candidate kinase for insulin regulation because so many MAPs are under MARK2 control and MAPs are a critical component for fine-tuning microtubule dynamics.
The novel findings we present significantly expand the number of known +TIPs affected by insulin and form a new hypothesis that a network of proteins linked to microtubule regulation synergize to coordinate insulin-regulated microtubule dynamics. On initial investigation of the effect of insulin on two of these network proteins, CLASP2 and G2L1, we discovered +TIP trailing, a new mode of insulin-regulated protein behavior. Follow-up studies determined that insulin stimulates α-tubulin Lysine 40 acetylation, a discovery that led to the finding that insulin increases microtubule stabilization. Taken together, we have expanded the protein systems and cytoskeletal elements involved in insulin action, information that is paramount for developing future studies aimed at understanding and identifying underlying mechanisms of insulin resistance, a hallmark of type 2 diabetes.
Data Availability
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (31) partner repository with the data set identifier PXD011431 and 10.6019/PXD011431.
Supplementary Material
Acknowledgments
We thank Marv Ruona and Paul Fini for their artistic renderings included in this article. We would also like to thank Mark Borgstrom for his expertise in RStudio.
Footnotes
* Imaris at the University of Arizona is supported by a TRIF Space Exploration and Optical Sciences (TRIF-SEOS) grant.
 This article contains supplemental material.
This article contains supplemental material.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
1 The abbreviations used are:
- GLUT4 solute carrier family 2
- facilitated glucose transporter member 4
- +TIP
- Plus-end tracking microtubule-associated protein
- AcK40
- α-tubulin lysine 40 acetylation
- AGAP3
- Arf-GAP with GTPase ANK repeat, and PH domain-containing protein 3
- ANK
- ankyrin
- AP-MS
- affinity purification coupled with mass spectrometry
- BAS
- basal
- CAP-Gly
- cytoskeleton-associated protein Gly-rich
- CKAP5
- cytoskeleton-associated protein 5
- CLASP1
- CLIP-associating protein 1
- CLASP2
- CLIP-associating protein 2
- CLIP1/CLIP-170
- CAP-Gly domain-containing linker protein 1
- CLIP2/CLIP-115
- CAP-Gly domain-containing linker protein 2
- co-IP(s)
- co-immunoprecipitation(s)
- CTCF
- corrected total cell fluorescence
- CTRL
- control
- EB1
- end binding protein 1
- EB3
- end binding protein 3
- EBH
- EB homology
- EEY/F
- EEY/F motif
- FA
- formic acid
- GAP
- GTPase-activating protein
- GAR
- Gas2-related
- GAS2
- growth-arrest-specific 2
- GAS2L1/G2L1
- GAS2 like protein 1
- GFP
- green fluorescent protein
- GLD
- GTP-binding protein-like domain
- GSK3
- glycogen synthase kinase 3
- HA
- hemagglutinin
- ID
- identification
- INS
- insulin
- IP(s)
- immunoprecipitation(s)
- KA1
- kinase associated domain 1
- MAP
- microtubule associated protein
- MARK2
- microtubule affinity-regulating kinase 2
- MK
- protein ladder marker
- MS/MS
- tandem mass spectrometry
- NIgG
- non-immune serum
- NOCO
- nocodazole
- PH
- pleckstrin homology
- P-Score
- probability score
- ROI
- region of interest
- SAINT
- significance analysis of interactome
- SB
- SB216763
- SC
- spectrum count
- SCP
- spectrum count profile
- SD
- standard deviation
- SEM
- standard error of the mean
- TOG
- tumor overexpressed gene
- TIRFM
- total internal reflection fluorescence microscopy
- UBA
- ubiquitin-associated
- WCL
- whole cell lysate.
REFERENCES
- 1. Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., and Hall A. (1992) The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 2. Semiz S., Park J. G., Nicoloro S. M., Furcinitti P., Zhang C., Chawla A., Leszyk J., and Czech M. P. (2003) Conventional kinesin KIF5B mediates insulin-stimulated GLUT4 movements on microtubules. EMBO J. 22, 2387–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koumanov F., Jin B., Yang J., and Holman G. D. (2005) Insulin signaling meets vesicle traffic of GLUT4 at a plasma-membrane-activated fusion step. Cell Metab. 2, 179–189 [DOI] [PubMed] [Google Scholar]
- 4. Kanzaki M., and Pessin J. E. (2001) Insulin-stimulated GLUT4 translocation in adipocytes is dependent upon cortical actin remodeling. J. Biol. Chem. 276, 42436–42444 [DOI] [PubMed] [Google Scholar]
- 5. Tong P., Khayat Z. A., Huang C., Patel N., Ueyama A., and Klip A. (2001) Insulin-induced cortical actin remodeling promotes GLUT4 insertion at muscle cell membrane ruffles. J. Clin. Invest. 108, 371–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiang Z. Y., Chawla A., Bose A., Way M., and Czech M. P. (2002) A phosphatidylinositol 3-kinase-independent insulin signaling pathway to N-WASP/Arp2/3/F-actin required for GLUT4 glucose transporter recycling. J. Biol. Chem. 277, 509–515 [DOI] [PubMed] [Google Scholar]
- 7. Chiu T. T., Jensen T. E., Sylow L., Richter E. A., and Klip A. (2011) Rac1 signalling towards GLUT4/glucose uptake in skeletal muscle. Cell Signal 23, 1546–1554 [DOI] [PubMed] [Google Scholar]
- 8. Ramkumar A., Jong B. Y., and Ori-McKenney K. M. (2018) ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule-associated proteins. Dev. Dyn. 247, 138–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galjart N. (2010) Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 20, R528–R537 [DOI] [PubMed] [Google Scholar]
- 10. Akhmanova A., Hoogenraad C. C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B. M., De Zeeuw C. I., Grosveld F., and Galjart N. (2001) Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104, 923–935 [DOI] [PubMed] [Google Scholar]
- 11. Langlais P., Dillon J. L., Mengos A., Baluch D. P., Ardebili R., Miranda D. N., Xie X., Heckmann B. L., Liu J., and Mandarino L. J. (2012) Identification of a role for CLASP2 in insulin action. J. Biol. Chem. 287, 39245–39253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Galjart N. (2005) CLIPs and CLASPs and cellular dynamics. Nat. Rev. Mol. Cell Biol. 6, 487–498 [DOI] [PubMed] [Google Scholar]
- 13. Mitchison T., Evans L., Schulze E., and Kirschner M. (1986) Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 45, 515–527 [DOI] [PubMed] [Google Scholar]
- 14. Lansbergen G., Grigoriev I., Mimori-Kiyosue Y., Ohtsuka T., Higa S., Kitajima I., Demmers J., Galjart N., Houtsmuller A. B., Grosveld F., and Akhmanova A. (2006) CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev. Cell 11, 21–32 [DOI] [PubMed] [Google Scholar]
- 15. Noordstra I., and Akhmanova A. (2017) Linking cortical microtubule attachment and exocytosis. F1000Res 6, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar P., Lyle K. S., Gierke S., Matov A., Danuser G., and Wittmann T. (2009) GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J. Cell Biol. 184, 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., and Akhmanova A. (2005) CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruiz-Saenz A., van Haren J., Laura Sayas C., Rangel L., Demmers J., Millan J., Alonso M. A., Galjart N., and Correas I. (2013) Protein 4.1R binds to CLASP2 and regulates dynamics, organization and attachment of microtubules to the cell cortex. J. Cell Sci. 126, 4589–4601 [DOI] [PubMed] [Google Scholar]
- 19. Basu S., Sladecek S., Martinez de la Pena y Valenzuela I., Akaaboune M., Smal I., Martin K., Galjart N., and Brenner H. R. (2015) CLASP2-dependent microtubule capture at the neuromuscular junction membrane requires LL5beta and actin for focal delivery of acetylcholine receptor vesicles. Mol. Biol. Cell 26, 938–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stehbens S. J., Paszek M., Pemble H., Ettinger A., Gierke S., and Wittmann T. (2014) CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16, 561–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartolini F., and Gundersen G. G. (2010) Formins and microtubules. Biochim. Biophys. Acta 1803, 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J., Yue J., and Wu X. (2017) Spectraplakin family proteins - cytoskeletal crosslinkers with versatile roles. J. Cell Sci. 130, 2447–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stroud M. J., Nazgiewicz A., McKenzie E. A., Wang Y., Kammerer R. A., and Ballestrem C. (2014) GAS2-like proteins mediate communication between microtubules and actin through interactions with end-binding proteins. J. Cell Sci. 127, 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kruse R., Krantz J., Barker N., Coletta R. L., Rafikov R., Luo M., Hojlund K., Mandarino L. J., and Langlais P. R. (2017) Characterization of the CLASP2 Protein Interaction Network Identifies SOGA1 as a Microtubule-Associated Protein. Mol. Cell Proteomics 16, 1718–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goriounov D., Leung C. L., and Liem R. K. (2003) Protein products of human Gas2-related genes on chromosomes 17 and 22 (hGAR17 and hGAR22) associate with both microfilaments and microtubules. J. Cell Sci. 116, 1045–1058 [DOI] [PubMed] [Google Scholar]
- 26. Jiang K., Toedt G., Montenegro Gouveia S., Davey N. E., Hua S., van der Vaart B., Grigoriev I., Larsen J., Pedersen L. B., Bezstarosti K., Lince-Faria M., Demmers J., Steinmetz M. O., Gibson T. J., and Akhmanova A. (2012) A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22, 1800–1807 [DOI] [PubMed] [Google Scholar]
- 27. Talior-Volodarsky I., Randhawa V. K., Zaid H., and Klip A. (2008) Alpha-actinin-4 is selectively required for insulin-induced GLUT4 translocation. J. Biol. Chem. 283, 25115–25123 [DOI] [PubMed] [Google Scholar]
- 28. Olson A. L., Eyster C. A., Duggins Q. S., and Knight J. B. (2003) Insulin promotes formation of polymerized microtubules by a phosphatidylinositol 3-kinase-independent, actin-dependent pathway in 3T3-L1 adipocytes. Endocrinology 144, 5030–5039 [DOI] [PubMed] [Google Scholar]
- 29. Dawicki-McKenna J. M., Goldman Y. E., and Ostap E. M. (2012) Sites of glucose transporter-4 vesicle fusion with the plasma membrane correlate spatially with microtubules. PLoS ONE 7, e43662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., and Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langlais P., Mandarino L. J., and Yi Z. (2010) Label-free relative quantification of co-eluting isobaric phosphopeptides of insulin receptor substrate-1 by HPLC-ESI-MS/MS. J. Am. Soc. Mass. Spectrom. 21, 1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langlais P., Yi Z., Finlayson J., Luo M., Mapes R., De Filippis E., Meyer C., Plummer E., Tongchinsub P., Mattern M., and Mandarino L. J. (2011) Global IRS-1 phosphorylation analysis in insulin resistance. Diabetologia 54, 2878–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langlais P., Yi Z., and Mandarino L. J. (2011) The identification of raptor as a substrate for p44/42 MAPK. Endocrinology 152, 1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Padilla-Rodriguez M., Parker S. S., Adams D. G., Westerling T., Puleo J. I., Watson A. W., Hill S. M., Noon M., Gaudin R., Aaron J., Tong D., Roe D. J., Knudsen B., and Mouneimne G. (2018) The actin cytoskeletal architecture of estrogen receptor positive breast cancer cells suppresses invasion. Nat. Commun. 9, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Longair M. H., Baker D. A., and Armstrong J. D. (2011) Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453–2454 [DOI] [PubMed] [Google Scholar]
- 38. Shah N., Kumar S., Zaman N., Pan C. C., Bloodworth J. C., Lei W., Streicher J. M., Hempel N., Mythreye K., and Lee N. Y. (2018) TAK1 activation of alpha-TAT1 and microtubule hyperacetylation control AKT signaling and cell growth. Nat. Commun. 9, 1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yi Z., Langlais P., De Filippis E. A., Luo M., Flynn C. R., Schroeder S., Weintraub S. T., Mapes R., and Mandarino L. J. (2007) Global assessment of regulation of phosphorylation of insulin receptor substrate-1 by insulin in vivo in human muscle. Diabetes 56, 1508–1516 [DOI] [PubMed] [Google Scholar]
- 40. Kumar P., and Wittmann T. (2012) +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 22, 418–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi H., Glatter T., Gstaiger M., and Nesvizhskii A. I. (2012) SAINT-MS1: protein-protein interaction scoring using label-free intensity data in affinity purification-mass spectrometry experiments. J. Proteome Res. 11, 2619–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Choi H., Liu G., Mellacheruvu D., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2012) Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Curr. Protoc. Bioinformatics Chapter 8, Unit8 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morris J. H., Knudsen G. M., Verschueren E., Johnson J. R., Cimermancic P., Greninger A. L., and Pico A. R. (2014) Affinity purification-mass spectrometry and network analysis to understand protein-protein interactions. Nat. Protoc. 9, 2539–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Westermarck J., Ivaska J., and Corthals G. L. (2013) Identification of protein interactions involved in cellular signaling. Mol. Cell Proteomics 12, 1752–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jin L., Williamson A., Banerjee S., Philipp I., and Rape M. (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Loo L. H., Lin H. J., Singh D. K., Lyons K. M., Altschuler S. J., and Wu L. F. (2009) Heterogeneity in the physiological states and pharmacological responses of differentiating 3T3-L1 preadipocytes. J. Cell Biol. 187, 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Akhmanova A., and Steinmetz M. O. (2015) Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16, 711–726 [DOI] [PubMed] [Google Scholar]
- 50. Portran D., Schaedel L., Xu Z., Thery M., and Nachury M. V. (2017) Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat. Cell Biol. 19, 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu Z., Schaedel L., Portran D., Aguilar A., Gaillard J., Marinkovich M. P., Thery M., and Nachury M. V. (2017) Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356, 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wittmann T., and Waterman-Storer C. M. (2005) Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J. Cell Biol. 169, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grimaldi A. D., Maki T., Fitton B. P., Roth D., Yampolsky D., Davidson M. W., Svitkina T., Straube A., Hayashi I., and Kaverina I. (2014) CLASPs are required for proper microtubule localization of end-binding proteins. Dev. Cell 30, 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gupta K. K., Alberico E. O., Nathke I. S., and Goodson H. V. (2014) Promoting microtubule assembly: A hypothesis for the functional significance of the +TIP network. Bioessays 36, 818–826 [DOI] [PubMed] [Google Scholar]
- 55. Tamura N., and Draviam V. M. (2012) Microtubule plus-ends within a mitotic cell are 'moving platforms' with anchoring, signalling and force-coupling roles. Open. Biol. 2, 120132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Al-Bassam J., and Chang F. (2011) Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21, 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gamper I., Fleck D., Barlin M., Spehr M., El Sayad S., Kleine H., Maxeiner S., Schalla C., Aydin G., Hoss M., Litchfield D. W., Luscher B., Zenke M., and Sechi A. (2016) GAR22beta regulates cell migration, sperm motility, and axoneme structure. Mol. Biol. Cell 27, 277–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gamper I., Koh K. R., Ruau D., Ullrich K., Bartunkova J., Piroth D., Hacker C., Bartunek P., and Zenke M. (2009) GAR22: a novel target gene of thyroid hormone receptor causes growth inhibition in human erythroid cells. Exp. Hematol. 37, 539–548 e534 [DOI] [PubMed] [Google Scholar]
- 59. Maiato H., Fairley E. A., Rieder C. L., Swedlow J. R., Sunkel C. E., and Earnshaw W. C. (2003) Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell 113, 891–904 [DOI] [PubMed] [Google Scholar]
- 60. Maiato H., Khodjakov A., and Rieder C. L. (2005) Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7, 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bratman S. V., and Chang F. (2007) Stabilization of overlapping microtubules by fission yeast CLASP. Dev. Cell 13, 812–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sousa A., Reis R., Sampaio P., and Sunkel C. E. (2007) The Drosophila CLASP homologue, Mast/Orbit regulates the dynamic behaviour of interphase microtubules by promoting the pause state. Cell Motil. Cytoskeleton 64, 605–620 [DOI] [PubMed] [Google Scholar]
- 63. Maton G., Edwards F., Lacroix B., Stefanutti M., Laband K., Lieury T., Kim T., Espeut J., Canman J. C., and Dumont J. (2015) Kinetochore components are required for central spindle assembly. Nat. Cell Biol. 17, 697–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lacroix B., Bourdages K. G., Dorn J. F., Ihara S., Sherwood D. R., Maddox P. S., and Maddox A. S. (2014) In situ imaging in C. elegans reveals developmental regulation of microtubule dynamics. Dev. Cell 29, 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ambrose C., Allard J. F., Cytrynbaum E. N., and Wasteneys G. O. (2011) A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bouchet B. P., Noordstra I., van Amersfoort M., Katrukha E. A., Ammon Y. C., Ter Hoeve N. D., Hodgson L., Dogterom M., Derksen P. W. B., and Akhmanova A. (2016) Mesenchymal Cell Invasion Requires Cooperative Regulation of Persistent Microtubule Growth by SLAIN2 and CLASP1. Dev. Cell 39, 708–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Moriwaki T., and Goshima G. (2016) Five factors can reconstitute all three phases of microtubule polymerization dynamics. J. Cell Biol. 215, 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu N., Signorile L., Basu S., Ottema S., Lebbink J. H. G., Leslie K., Smal I., Dekkers D., Demmers J., and Galjart N. (2016) Isolation of Functional Tubulin Dimers and of Tubulin-Associated Proteins from Mammalian Cells. Curr. Biol. 26, 1728–1736 [DOI] [PubMed] [Google Scholar]
- 69. Lawrence E. J., Arpag G., Norris S. R., and Zanic M. (2018) Human CLASP2 specifically regulates microtubule catastrophe and rescue. Mol. Biol. Cell 29, 1168–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Aher A., Kok M., Sharma A., Rai A., Olieric N., Rodriguez-Garcia R., Katrukha E. A., Weinert T., Olieric V., Kapitein L. C., Steinmetz M. O., Dogterom M., and Akhmanova A. (2018) CLASP suppresses microtubule catastrophes through a single TOG domain. Dev. Cell 46, 40–58 e48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Patel K., Nogales E., and Heald R. (2012) Multiple domains of human CLASP contribute to microtubule dynamics and organization in vitro and in Xenopus egg extracts. Cytoskeleton 69, 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Drabek K., van Ham M., Stepanova T., Draegestein K., van Horssen R., Sayas C. L., Akhmanova A., Ten Hagen T., Smits R., Fodde R., Grosveld F., and Galjart N. (2006) Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr. Biol. 16, 2259–2264 [DOI] [PubMed] [Google Scholar]
- 73. Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P. M., Andreyeva N., Gleeson P., Galjart N., Maia A. R., McLeod I. X., Yates J. R. 3rd, Maiato H., Khodjakov A., Akhmanova A., and Kaverina I. (2007) Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 12, 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Miller P. M., Folkmann A. W., Maia A. R., Efimova N., Efimov A., and Kaverina I. (2009) Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat. Cell Biol. 11, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Janke C., and Montagnac G. (2017) Causes and Consequences of Microtubule Acetylation. Curr. Biol. 27, R1287–R1292 [DOI] [PubMed] [Google Scholar]
- 76. L'Hernault S. W., and Rosenbaum J. L. (1983) Chlamydomonas alpha-tubulin is posttranslationally modified in the flagella during flagellar assembly. J. Cell Biol. 97, 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Maruta H., Greer K., and Rosenbaum J. L. (1986) The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J. Cell Biol. 103, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Piperno G., LeDizet M., and Chang X. J. (1987) Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J. Cell Biol. 104, 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bulinski J. C., Richards J. E., and Piperno G. (1988) Posttranslational modifications of alpha tubulin: detyrosination and acetylation differentiate populations of interphase microtubules in cultured cells. J. Cell Biol. 106, 1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Akella J. S., Wloga D., Kim J., Starostina N. G., Lyons-Abbott S., Morrissette N. S., Dougan S. T., Kipreos E. T., and Gaertig J. (2010) MEC-17 is an alpha-tubulin acetyltransferase. Nature 467, 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shida T., Cueva J. G., Xu Z., Goodman M. B., and Nachury M. V. (2010) The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. U.S.A. 107, 21517–21522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Verdel A., Curtet S., Brocard M. P., Rousseaux S., Lemercier C., Yoshida M., and Khochbin S. (2000) Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr. Biol. 10, 747–749 [DOI] [PubMed] [Google Scholar]
- 83. Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., and Yao T. P. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 [DOI] [PubMed] [Google Scholar]
- 84. LeDizet M., and Piperno G. (1986) Cytoplasmic microtubules containing acetylated alpha-tubulin in Chlamydomonas reinhardtii: spatial arrangement and properties. J. Cell Biol. 103, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stockli J., Yang J. Y., and James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li H. C., Liu X. S., Yang X. M., Wang Y. M., Wang Y., Turner J. R., and Liu X. Q. (2010) Phosphorylation of CLIP-170 by Plk1 and CK2 promotes timely formation of kinetochore-microtubule attachments. EMBO J. 29, 2953–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yang X., Li H., Liu X. S., Deng A., and Liu X. (2009) Cdc2-mediated phosphorylation of CLIP-170 is essential for its inhibition of centrosome reduplication. J. Biol. Chem. 284, 28775–28782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lansbergen G., Komarova Y., Modesti M., Wyman C., Hoogenraad C. C., Goodson H. V., Lemaitre R. P., Drechsel D. N., van Munster E., Gadella T. W. Jr, Grosveld F., Galjart N., Borisy G. G., and Akhmanova A. (2004) Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J. Cell Biol. 166, 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hoogenraad C. C., Akhmanova A., Grosveld F., De Zeeuw C. I., and Galjart N. (2000) Functional analysis of CLIP-115 and its binding to microtubules. J. Cell Sci. 113 (Pt 12), 2285–2297 [DOI] [PubMed] [Google Scholar]
- 90. Rickard J. E., and Kreis T. E. (1991) Binding of pp170 to microtubules is regulated by phosphorylation. J. Biol. Chem. 266, 17597–17605 [PubMed] [Google Scholar]
- 91. Choi J. H., Bertram P. G., Drenan R., Carvalho J., Zhou H. H., and Zheng X. F. (2002) The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase. EMBO Rep. 3, 988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lee H. S., Komarova Y. A., Nadezhdina E. S., Anjum R., Peloquin J. G., Schober J. M., Danciu O., van Haren J., Galjart N., Gygi S. P., Akhmanova A., and Borisy G. G. (2010) Phosphorylation controls autoinhibition of cytoplasmic linker protein-170. Mol. Biol. Cell 21, 2661–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Soundararajan M., Yang X., Elkins J. M., Sobott F., and Doyle D. A. (2007) The centaurin gamma-1 GTPase-like domain functions as an NTPase. Biochem. J. 401, 679–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Xia C., Ma W., Stafford L. J., Liu C., Gong L., Martin J. F., and Liu M. (2003) GGAPs, a new family of bifunctional GTP-binding and GTPase-activating proteins. Mol. Cell Biol. 23, 2476–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nie Z., Stanley K. T., Stauffer S., Jacques K. M., Hirsch D. S., Takei J., and Randazzo P. A. (2002) AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J. Biol. Chem. 277, 48965–48975 [DOI] [PubMed] [Google Scholar]
- 96. Luo R., Akpan I. O., Hayashi R., Sramko M., Barr V., Shiba Y., and Randazzo P. A. (2012) GTP-binding protein-like domain of AGAP1 is protein binding site that allosterically regulates ArfGAP protein catalytic activity. J. Biol. Chem. 287, 17176–17185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Halet G. (2005) Imaging phosphoinositide dynamics using GFP-tagged protein domains. Biol. Cell 97, 501–518 [DOI] [PubMed] [Google Scholar]
- 98. Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J., and Hyman A. A. (2008) XMAP215 is a processive microtubule polymerase. Cell 132, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nehlig A., Molina A., Rodrigues-Ferreira S., Honore S., and Nahmias C. (2017) Regulation of end-binding protein EB1 in the control of microtubule dynamics. Cell Mol. Life Sci. 74, 2381–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zimniak T., Stengl K., Mechtler K., and Westermann S. (2009) Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186, 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Iimori M., Ozaki K., Chikashige Y., Habu T., Hiraoka Y., Maki T., Hayashi I., Obuse C., and Matsumoto T. (2012) A mutation of the fission yeast EB1 overcomes negative regulation by phosphorylation and stabilizes microtubules. Exp. Cell Res. 318, 262–275 [DOI] [PubMed] [Google Scholar]
- 102. Ban R., Matsuzaki H., Akashi T., Sakashita G., Taniguchi H., Park S. Y., Tanaka H., Furukawa K., and Urano T. (2009) Mitotic regulation of the stability of microtubule plus-end tracking protein EB3 by ubiquitin ligase SIAH-1 and Aurora mitotic kinases. J. Biol. Chem. 284, 28367–28381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Iimori M., Watanabe S., Kiyonari S., Matsuoka K., Sakasai R., Saeki H., Oki E., Kitao H., and Maehara Y. (2016) Phosphorylation of EB2 by Aurora B and CDK1 ensures mitotic progression and genome stability. Nat. Commun. 7, 11117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Luo Y., Ran J., Xie S., Yang Y., Chen J., Li S., Shui W., Li D., Liu M., and Zhou J. (2016) ASK1 controls spindle orientation and positioning by phosphorylating EB1 and stabilizing astral microtubules. Cell Discov. 2, 16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ran J., Luo Y., Zhang Y., Yang Y., Chen M., Liu M., Li D., and Zhou J. (2017) Phosphorylation of EB1 regulates the recruitment of CLIP-170 and p150glued to the plus ends of astral microtubules. Oncotarget 8, 9858–9867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang Y., Luo Y., Lyu R., Chen J., Liu R., Li D., Liu M., and Zhou J. (2016) Proto-oncogenic Src phosphorylates EB1 to regulate the microtubule-focal adhesion crosstalk and stimulate cell migration. Theranostics 6, 2129–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Stenner F., Liewen H., Gottig S., Henschler R., Markuly N., Kleber S., Faust M., Mischo A., Bauer S., Zweifel M., Knuth A., Renner C., and Wadle A. (2013) RP1 is a phosphorylation target of CK2 and is involved in cell adhesion. PLoS ONE 8, e67595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Komarova Y. A., Huang F., Geyer M., Daneshjou N., Garcia A., Idalino L., Kreutz B., Mehta D., and Malik A. B. (2012) VE-cadherin signaling induces EB3 phosphorylation to suppress microtubule growth and assemble adherens junctions. Mol. Cell 48, 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chen J., Luo Y., Li L., Ran J., Wang X., Gao S., Liu M., Li D., Shui W., and Zhou J. (2014) Phosphoregulation of the dimerization and functions of end-binding protein 1. Protein Cell 5, 795–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Le Grand M., Rovini A., Bourgarel-Rey V., Honore S., Bastonero S., Braguer D., and Carre M. (2014) ROS-mediated EB1 phosphorylation through Akt/GSK3beta pathway: implication in cancer cell response to microtubule-targeting agents. Oncotarget 5, 3408–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nakamura S., Grigoriev I., Nogi T., Hamaji T., Cassimeris L., and Mimori-Kiyosue Y. (2012) Dissecting the nanoscale distributions and functions of microtubule-end-binding proteins EB1 and ch-TOG in interphase HeLa cells. PLoS ONE 7, e51442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Maurer S. P., Cade N. I., Bohner G., Gustafsson N., Boutant E., and Surrey T. (2014) EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr. Biol. 24, 372–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Fox J. C., Howard A. E., Currie J. D., Rogers S. L., and Slep K. C. (2014) The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Mol. Biol. Cell 25, 2375–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Widlund P. O., Stear J. H., Pozniakovsky A., Zanic M., Reber S., Brouhard G. J., Hyman A. A., and Howard J. (2011) XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc. Natl. Acad. Sci. U.S.A. 108, 2741–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Thakur H. C., Singh M., Nagel-Steger L., Kremer J., Prumbaum D., Fansa E. K., Ezzahoini H., Nouri K., Gremer L., Abts A., Schmitt L., Raunser S., Ahmadian M. R., and Piekorz R. P. (2014) The centrosomal adaptor TACC3 and the microtubule polymerase chTOG interact via defined C-terminal subdomains in an Aurora-A kinase-independent manner. J. Biol. Chem. 289, 74–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hood F. E., Williams S. J., Burgess S. G., Richards M. W., Roth D., Straube A., Pfuhl M., Bayliss R., and Royle S. J. (2013) Coordination of adjacent domains mediates TACC3-ch-TOG-clathrin assembly and mitotic spindle binding. J. Cell Biol. 202, 463–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (31) partner repository with the data set identifier PXD011431 and 10.6019/PXD011431.