This study reports the first in-depth RNA and protein expression profiles of human sweat glands. RNA-sequencing and proteomic analysis were performed on ∼250 sweat glands collected from a healthy individual. Data revealed ∼13,300 protein-coding genes, ∼53,400 non-coding genes, and ∼6,100 proteins are expressed in eccrine sweat glands. Also, two missing and five novel proteins were identified. Integrated transcriptome and protein expression profile of eccrine sweat gland can be used to study the etiology of many diseases relevant to the sweat gland.
Keywords: Mass Spectrometry, Proteogenomics, RNA SEQ, Cystic fibrosis, Peptide Synthesis*, Proteomics, Sweat glands, Transcriptomics
Graphical Abstract

Highlights
RNA-seq and proteome analysis of human eccrine sweat glands were conducted.
Approximately 138,000 transcripts and ∼6,100 proteins were identified.
Proteins involved in secretion, reabsorption, and wound healing identified.
Two missing and five novel proteins were identified.
Abstract
The eccrine sweat gland is an exocrine gland that is involved in the secretion of sweat for control of temperature. Malfunction of the sweat glands can result in disorders such as miliaria, hyperhidrosis and bromhidrosis. Understanding the transcriptome and proteome of sweat glands is important for understanding their physiology and role in diseases. However, no systematic transcriptome or proteome analysis of sweat glands has yet been reported. Here, we isolated eccrine sweat glands from human skin by microdissection and performed RNA-seq and proteome analysis. In total, ∼138,000 transcripts and ∼6,100 proteins were identified. Comparison of the RNA-seq data of eccrine sweat glands to other human tissues revealed the closest resemblance to the cortex region of kidneys. The proteome data showed enrichment of proteins involved in secretion, reabsorption, and wound healing. Importantly, protein level identification of the calcium ion channel TRPV4 suggests the importance of eccrine sweat glands in re-epithelialization of wounds and prevention of dehydration. We also identified 2 previously missing proteins from our analysis. Using a proteogenomic approach, we identified 7 peptides from 5 novel genes, which we validated using synthetic peptides. Most of the novel proteins were from short open reading frames (sORFs) suggesting that many sORFs still remain to be annotated in the human genome. This study presents the first integrated analysis of the transcriptome and proteome of the human eccrine sweat gland and would become a valuable resource for studying sweat glands in physiology and disease.
There are ∼2–4 million sweat glands dispersed throughout the human body (1). Two distinct types of sweat glands exist in the human skin - eccrine sweat glands, which are not connected to a hair follicle; and apocrine sweat glands, which are connected to hair follicles (2). The duct of the eccrine sweat gland opens directly onto the skin surface enabling the gland to secrete water and salt-based liquid. In contrast, the apocrine sweat gland releases fluid and oily substance through the hair follicle orifice (3). Eccrine sweat glands are major sweat glands in humans and are distributed over the entire body. The physiologic role of eccrine sweat glands is to produce sweat in response to a hot environment or exercise and in response to emotional stimuli such as fear, anxiety and pain (4). The function of eccrine sweat glands is controlled by the hypothalamus through activation of post-ganglionic cholinergic branches of the sympathetic nervous system (5, 6) Beyond thermoregulation, understanding the composition and function of eccrine sweat glands can help better understand conditions such as cystic fibrosis (CF)1, hyperhidrosis, anhidrosis, and atopic dermatitis (7, 8). Furthermore, the role of eccrine sweat gland stem cells in wound healing is being increasingly recognized and regrowth of sweat glands in damaged skin such as in the case of burn victims has tremendous implications (9). One key step to the understanding the human sweat glands is to obtain an accurate catalogue of transcripts and proteins that are expressed (10). Although a tremendous effort has been put into sequencing transcripts and proteins of a number of human tissues and cell types, there are still no studies that pertain to human sweat glands despite their obvious importance in physiology and disease. Our present study concentrates on the human eccrine sweat glands. We carried out an in-depth analysis of transcriptome and proteome of eccrine sweat glands isolated by microdissections from human skin. First, we compared the RNA-seq data for eccrine sweat glands with the available transcript data of other human tissues available in the Genotype-Tissue Expression (GTEx). Next, we compared the proteome of human eccrine sweat glands to proteomes of other human tissues and cells (11). In addition, we compared the eccrine sweat gland proteome to sweat proteome. We also investigated whether we could identify missing proteins from the proteomic analysis of human eccrine sweat glands. Finally, we performed a proteogenomic analysis to identify any novel genes or protein isoforms using transcriptome and proteome data.
MATERIALS AND METHODS
Isolation of Eccrine Sweat Glands
Deidentified skin as excess material was obtained from otherwise healthy, non-CF adult patients undergoing unrelated reconstructive surgery in the Department of Plastic and Reconstructive Surgery at the Johns Hopkins Hospital. This study was exempt from IRB approval according to institutional protocol. To maintain the donor's anonymity, only the region of sampling was provided as per the approval and no clinical or demographic information was attached to the sample. 25 cm × 15 cm skin was collected from the abdominal region of a single donor. Eccrine sweat glands were dissected out from the skin dermis as previously described (12–14). Approximately 250 eccrine sweat glands were collected and pooled. Briefly, the skin was excised vertically with sharp scissors into small pieces. The cut pieces were bathed in phosphate-buffered Ringer's solution on ice. Neutral red (10 μm) was used to stain the sweat glands. After a few minutes of incubation with neutral red, the sweat glands were viewed through a Zeiss Stemi 2000 dissecting microscope. Fisher brand fine-pointed forceps were used to dissect eccrine sweat glands from the skin. Connective tissue and fat hanging to the eccrine sweat gland were removed using Fisher brand dissecting needles with attached holder. Images of the eccrine sweat glands were captured using Zeiss microscopy camera, AxioCam lCc 1, and the Axio Vision software (Release 4.8.2).
RNA Extraction
The dissected sweat glands were placed in RNAlater (Qiagen # 76104, Germany) immediately after they were dissected out from the skin. The collection tube was centrifuged to remove RNA later solution and Trizol was added to the sample and mixed. The samples were processed in a bead-beater (FastPrep-24, MP) at a setting of 4 m/s for 20 s and subsequently placed on ice for 5 mins. This was repeated three times. Chloroform was then added to the samples and incubated for 5 mins at room temperature. The samples were then centrifuged at 8000 × g for 5 min at 4 °C. The aqueous phase was separated from the organic phase containing the DNA and protein. Ethanol was added to the sample for precipitation and immediately applied to RNeasy columns (RNeasy Kit, Qiagen # 74104). The manufacturer's cleanup procedure was followed except that the RNA sample was treated with the Turbo DNA-Free kit to remove residual DNA contamination (Thermo Fisher # AM1907, Carlsbad, CA). The RNA was stored at −80 °C. The quality of RNA was measured by RNA integrity number (RIN) using Agilent 2100 Bioanalyzer.
RNA-sequencing and Data Analysis
Using 1 μg of total RNA isolated from sweat glands, library preparation and RNA-seq were performed at the Deep Sequencing and Microarray Core Facility at Johns Hopkins University. RNA-seq library was constructed using the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA), and 200 million paired-end reads were obtained on Illumina's Next Seq 500 platform. Quality assessment of raw fastq data generate from Illumina was carried out using FastQC v0.11.5. Raw read pairs were subjected to processing using fqtrim v0.9.5 by trimming low-quality bases (Q < 20 and length < 35 bp). The human genome version GRCh38 (hg38) was used as a reference with gene annotations from GENCODE. Quality processed data was then aligned using HISAT2 v2.0.1 against hg38. Default parameters of HISAT were used in “–end-to-end” mode. HISAT alignment was then converted to BAM and sorted using samtools v1.3.1 package. Transcript assembly and quantification were carried out using StringTie v1.3.3 package with default parameters (20, 21). Splice junctions from the alignment data were detected and annotated using RSeQC v2.6.4 package. Extraction of 90 bp sequence from the exon ends of novel splice junctions was carried out followed by translation into peptide sequences for database searching. GTEx pipeline was followed to align against GRCh37 (hg19) version of the human genome using GENCODE annotations. Then gene expression values were derived using RNA-SeQC v1.1.8 program (22) and compared against the median RPKM values in GTEx tissues. Eccrine sweat glands were compared with other human tissues in the GTEx using global transcriptome clustering for all genes with FPKM>1 (to remove any bias from genes with low abundance). To visualize this clustering, we generated a heat map of the RNA expression of the 1000 most abundant genes. All RNA-seq data have been deposited to the NCBI GEO with the dataset identifier GSE117885 and project name “Integrated transcriptomic and proteomic analysis of human eccrine sweat glands.”
Preparation of Sweat Glands for Proteome Analysis
To maximize the identification of proteins, the sweat grand proteins were extracted by two different methods - Filter-aided sample prep (FASP) approach and the guanidine hydrochloride (GuHCl) approach. For the FASP approach, sweat glands were sonicated in 50 mm TEAB/4% SDS/10 mm DTT with Halt protease inhibitor mixture (Thermo Scientific) for 5 min followed by heating at 95°C for 5 min. After cooling, the samples were alkylated with 30 mm iodoacetamide at room temperature for 30 min followed by centrifugation at 17,000 × g for 10 min. SDS in the sample was removed by FAST method (15). Briefly, the lysate was diluted with 50 mm TEAB/8 M urea and concentrated with centricon with 30 KDa MWCO for 40 min at RT. This buffer exchange step was repeated five times. Sweat gland proteins were digested with Lys-C at room temperature for 3 h followed by further digestion with trypsin overnight.
For the GuHCl approach, sweat glands were sonicated for 5 min with 35% amplitude in 8 m GuHCl, 50 mm HEPES, pH 7.0, 10 mm dithiothreitol in the presence of Halt protease inhibitor mixture (Thermo Scientific) followed by heating at 90°C for 3 min and centrifugation at 16,000 × g for 10 min. The pellet was resuspended in 50 mm TEAB/4% SDS/10 mm DTT and subjected to further protein extraction with barocycler for 60 cycles in which each cycle consisted of 45,000 psi at 95 °C for 50 s and atmospheric pressure at 25 °C for 10 s. The proteins alkylated with 30 mm iodoacetamide at room temperature for 15 min. The proteins were digested with trypsin at the enzyme to protein ratio of 1:50 at 37 °C overnight. Following enzyme digestion, the peptides were desalted using Sep-Pak C18 cartridge. The peptides prepared through FASP and GuHCl method were fractionated into 24 fractions by basic pH reversed phase liquid chromatography. Briefly, lyophilized samples were reconstituted in solvent A (10 mm triethylammonium bicarbonate, pH 8.5) and loaded onto XBridge C18, 5 μm 250 × 4.6 mm column (Waters, Milford, MA). Peptides were resolved using a gradient of 3 to 50% solvent B (10 mm triethylammonium bicarbonate in acetonitrile, pH 8.5) over 50 min collecting 96 fractions. The fractions were subsequently concatenated into 24 fractions followed by vacuum drying using SpeedVac. The dried peptides were resuspended in 15 μl 10% FA and the entire amount was injected. The peptides prepared through barocycler were fractionated by SCX StageTip as previously reported (16).
Mass Spectrometric Analysis
The fractionated peptides were analyzed on an LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific, Bremen, Germany) coupled to EASY-nanoLC II system or on Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific, San Jose, CA) coupled to EASY-nanoLC 1200 nanoflow liquid chromatography system (Thermo Scientific, Odense, Southern Denmark). The peptides from each fraction were reconstituted in 10% formic acid and loaded onto a trap column (100 μm × 2 cm) at a flow rate of 5 μl per minute for LTQ-Orbitrap Elite mass spectrometer or onto an Acclaim PepMap100 Nano Trap Column (100 μm × 2 cm) (Thermo Scientific) packed with 5 μm C18 particles at a flow rate of 5 μl per minute for Orbitrap Fusion Lumos Tribrid mass spectrometer. The loaded peptides were resolved at 250 nl/min flow rate using a linear gradient of 10 to 35% solvent B (0.1% formic acid in 95% acetonitrile) over 95 min on an analytical column (50 cm × 75 μm ID) packed in house for LTQ-Orbitrap Elite mass spectrometer or EASY-Spray column (50 cm × 75 μm ID, PepMap RSLC C18 and 2 μm C18 particles) (Thermo Scientific) for Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific, San Jose, CA) and was fitted on EASY-Spray ion source that was operated at 2.0 kV voltage. Mass spectrometry analysis for LTQ-Orbitrap Elite mass spectrometer was carried out in a data dependent manner with a full scan in the range of m/z 350 to 1700 in top N mode setting 8 most intense ions. Both MS and MS/MS were acquired and measured using Orbitrap mass analyzer. Full MS scans were measured at a resolution of 120,000 at m/z 400. Precursor ions were fragmented using higher-energy collisional dissociation method and detected at a mass resolution of 30,000 at m/z 400. Automatic gain control for full MS was set to 1 million ions and for MS/MS was set to 0.2 million ions with a maximum ion injection time of 200 and 300 ms, respectively. Dynamic exclusion was set to 60 s and singly charged ions were rejected. Internal calibration was carried out using lock mass option (m/z 445.1200025) from ambient air. Mass spectrometry analysis for Orbitrap Fusion Lumos Tribrid was carried out in a data dependent manner with a full scan in the range of m/z 350 to 1,550 in top speed mode setting 3 s per cycle. Both MS and MS/MS were acquired and measured using Orbitrap mass analyzer. Full MS scans were measured at a resolution of 120,000 at m/z 200. Precursor ions were fragmented using higher-energy collisional dissociation method and detected at a mass resolution of 30,000 at m/z 200. Automatic gain control for full MS was set to 1 million ions and for MS/MS was set to 0.05 million ions with a maximum ion injection time of 50 and 100 ms, respectively. Dynamic exclusion was set to 30 s and singly charged ions were rejected. Internal calibration was carried out using lock mass option (m/z 445.1200025) from ambient air.
Data Analysis
Proteome Discoverer (v 2.1; Thermo Scientific) suite was used for identification and quantitation. The tandem mass spectrometry data were searched using SEQUEST HT search algorithms against a human RefSeq database (version 70) supplemented with frequently observed contaminants (99,847 entries for the RefSeq database and 115 common contaminant proteins). The search parameters used were as follows: (1) trypsin as a proteolytic enzyme (with up to two missed cleavages) (2) peptide mass error tolerance of 10 ppm; (3) fragment mass error tolerance of 0.02 Da; (4) Carbamidomethylation of cysteine (+57.02146 Da) as fixed modification and oxidation of methionine (+15.99492 Da) as variable modifications. Number of PSMs for shared peptides was counted redundantly for all the proteins to which the share peptides were mapped. Peptides and proteins were filtered at 1% false discovery rate using Percolator node. Protein abundance values were calculated using normalized spectral abundance factor (NSAF) (17). For the spectral counting of NSAF, modified peptides and shared peptides were included. Gene ontology analysis was performed at DAVID (18, 19). The heatmap for transcriptome and proteome were generated by the pheatmap R package and Perseus software, respectively (20).
Proteogenomic Analysis
For the identification of novel genes expressed at the protein level, a custom database was generated using an in-house Python script as follows. RNA sequences from the sweat gland RNA-seq data generated in this study were translated into three frames followed by splitting the translated sequences at every stop codon into individual proteins. Protein entries that were shorter than 6 amino acids or redundant were removed leaving 4,723,976 entries in the database after filtering. To avoid forcible mapping of MS/MS spectra of peptides derived from reference protein sequences to be matched to non-reference proteins, proteins annotated in a reference database were retained. The database search against the custom database was conducted in the same way as the one against the reference database except for the enzyme non-specificity on the peptide N-terminal side. Any peptides that had identical matches to proteins in the NR human protein database were removed. For the identification of peptides mapping to novel or partially novel splice junctions, a custom database was generated using RSeQC software and an in-house Python script as follows. Splice junction sequences were extracted with RSeQC software from RNA sequences from the sweat gland RNA-seq data generated in this study followed by splitting the translated sequences at every stop codon into individual proteins. Protein entries that were shorter than 6 amino acids, redundant or not spanning a splice junction were removed leaving 130,789 entries in the database after filtering. The database search was conducted in the same way as the search against the reference database applying 1% FDR in peptide and protein levels. Any peptides that had identical matches in the NR human protein database were removed.
Validation of Peptides Mapping to Missing or Novel Proteins Using Synthetic Peptides
The identified peptides mapping to missing or novel proteins were synthesized (JPT Peptide Technologies, Berlin, Germany). The peptides were reconstituted with 0.1% formic acid and ∼1 pmol of peptides to the final concentration were mixed and analyzed on the Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific) as described in the mass spectrometric analysis section. Database search was performed in the same manner as described in the mass spectrometry data analysis section with an exception for the database that is composed of synthetic peptides and 248 common contaminant proteins. Spectra of peptides from the samples were validated by aligning those from synthetic peptides using in-house generated software written in Python programming language.
Experimental Design and Statistical Rationale
No technical and biological replicate experiments were conducted. The scarcity of the large size of human skin that was enough to provide >200 eccrine sweat glands hampered our ability to conduct replicate analysis. Although this study has been conducted with sweat glands from only one individual, this is the first study in which transcriptome and proteome of sweat gland were analyzed and, the focus of this manuscript is profiling of expressed proteins to identify missing or novel proteins.
RESULTS
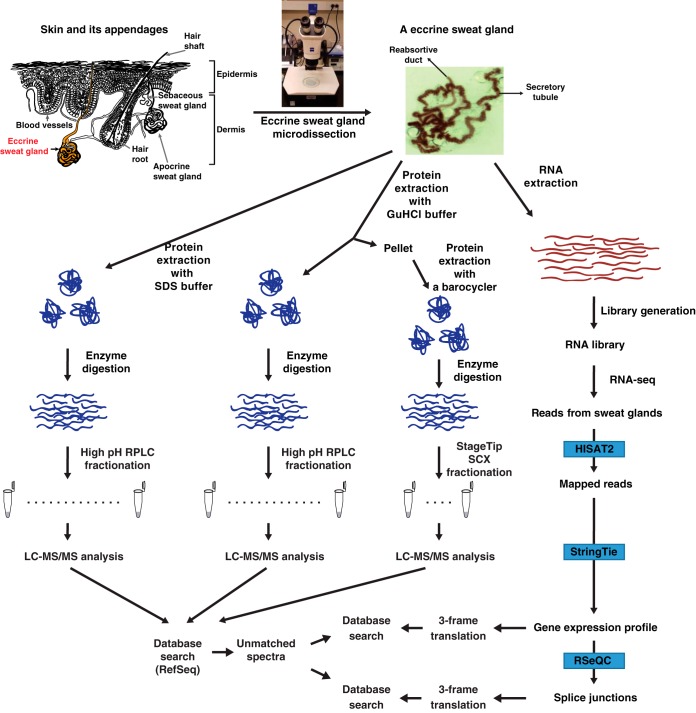
In this study, we microdissected eccrine sweat glands from healthy adult human skin to perform an integrated transcriptomic and proteomic analysis as described in the following sections and summarized in Fig. 1.
Fig. 1.
Schematic diagram for the research strategy. Human eccrine sweat glands were microdissected from human skin. Top, the left side of the figure shows a diagrammatic representation of skin and its appendages. Top, the right side of the figure shows a micrograph of an eccrine sweat gland dissected from the skin using stereomicroscope. Transcriptome and proteome analyses were conducted after the extraction of proteins and RNAs, respectively. For transcriptome analysis, the extracted mRNAs were used for the library preparation and subjected to the RNA-seq. The RNA-seq data were analyzed using HISAT2, StringTie and RSeQC. For proteome analysis, the extracted proteins were digested with Lys-C followed by trypsin. The resulting peptides were pre-fractionated by either bRPLC or an SCX StageTip followed by LC-MS/MS analyses. Acquired mass spectrometry data were subject to a reference protein database and the resulting unmatched spectra were subject to another round of a search against customized databases generated by 3-frame translation of either the RNA-seq or splice junction data.
RNA-seq Analysis of Human Eccrine Sweat Glands
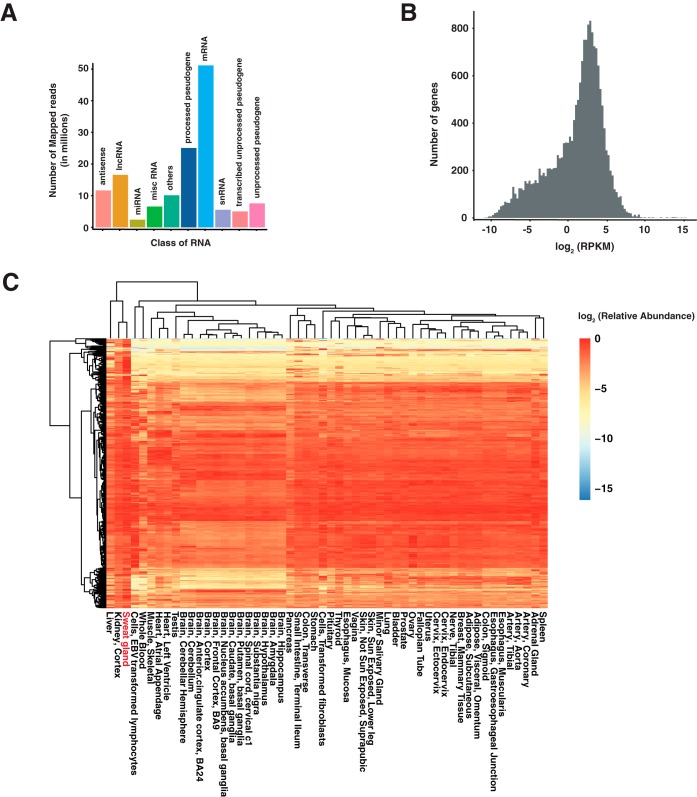
We performed RNA-seq on human eccrine sweat glands to investigate the RNA expression profile of sweat glands and to compare it to other human tissues. Two hundred twelve million paired-end reads were obtained and 193 million reads were mapped to reference human genome hg38. Of these, 50 million reads mapped to known protein-coding genes, ∼23 million reads mapped to processed pseudogenes, and about 15 million reads mapped to long non-coding RNAs (Fig. 2A). This led to the identification of 13,343 and 53,432 protein-coding genes and transcripts (RPKM≥1) respectively (Fig. 2B). Most genes followed a normal distribution, which was centered at a value of ∼2.14 log2 RPKM (∼4.39 RPKM), whereas the remaining genes formed a shoulder to the left of the normal distribution (Fig. 2B). Among the 53,432 expressed transcripts, 38,358 were annotated in GENCODE and remaining were the novel isoforms arising from alternative splicing and extension events (supplemental Table S1 and S2). Investigation of the exon-exon junctions revealed that ∼ 70% of the splice-donor and acceptor were already known, ∼ 22% of the junctions were formed by either new splice donor or acceptor, and 8% of the junctions were formed by both new splice donor and acceptor (supplemental Table S1 and S2). To determine if eccrine sweat glands exhibited a unique expression profile, we compared their RNA-seq data to that from other human tissues deposited in the GTEx repository using unsupervised hierarchical clustering of global transcriptome data (supplemental Table S3). Kidney tissue showed the closest expression profile to eccrine sweat glands (Fig. 2C).
Fig. 2.
RNA expression profile of human eccrine sweat glands. A, Classification of RNA-seq reads based on the class of RNA the reads align to. B, Density plot showing the distribution of RNA expression from genes identified in eccrine sweat glands. C, Heat map showing the relatively abundant RNAs in human eccrine sweat glands compared with other human tissues from GTEx. Top 1000 most abundant genes in eccrine sweat glands were shown on this heatmap. RPKM values of each gene were normalized by dividing by the maximum RPKM value of each gene.
Proteomic Analysis of Human Eccrine Sweat Glands
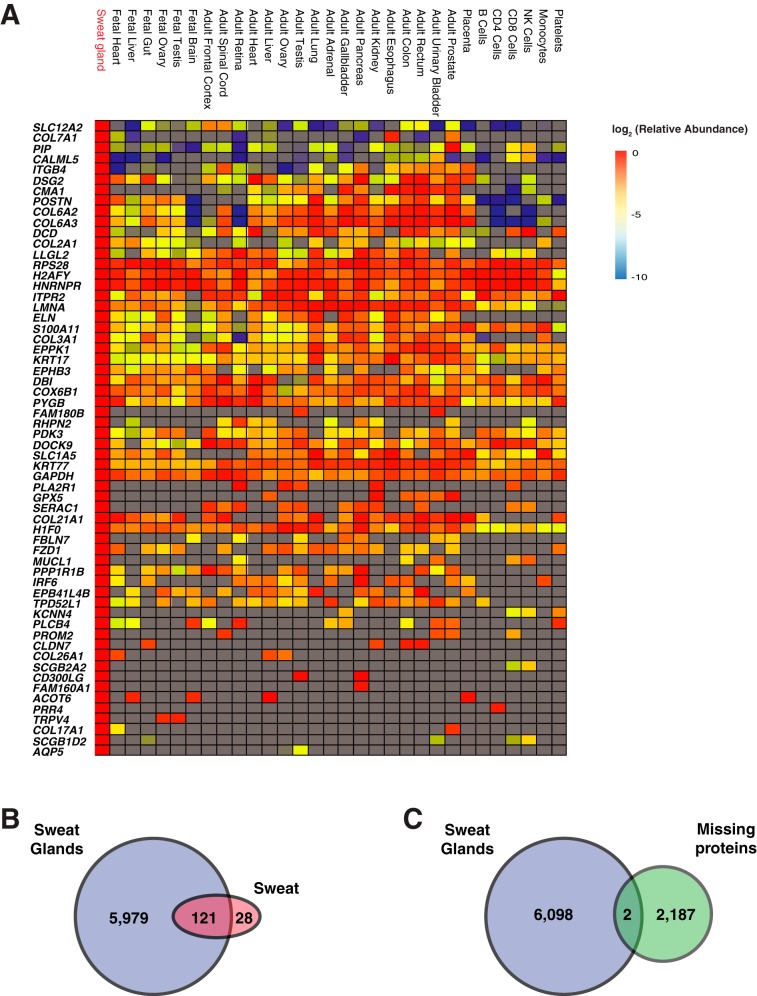
Because there has not been any published proteomic analysis of human eccrine sweat glands, we employed LC-MS/MS to investigate the proteome of eccrine sweat glands. Eccrine sweat gland proteins were extracted in two different buffers containing either SDS or guanidine hydrochloride (GuHCl). The pellet left after protein extraction with GuHCl was further subjected to further protein extraction using a barocycler to maximize protein extraction. The peptides were analyzed by LC-MS/MS, and the mass spectra were searched using Sequest HT resulting in the identification of 6100 proteins and 56,936 peptides (supplemental Table S4). This included chloride intracellular channel protein, sodium potassium-transporting ATPase subunit, sodium bicarbonate transporter, voltage-gated potassium channel, calcium-activated potassium channel, voltage-dependent calcium channels, V-H+-ATPase, AQP5, and carbonic anhydrases. The proteins extracted with three different methods were analyzed independently by LC-MS/MS. The extracted proteins were digested sequentially with Lys-C and trypsin. The proteins extracted with FASP method or GuHCl method were fractionated by basic pH RPLC whereas the proteins extracted by barocycler were fractionated by a StageTip SCX column. Next, we compared the expression profile of human eccrine sweat glands to Human Proteome Map data after normalizing all the data by NSAF quantification method (supplemental Table S5) (21). We identified proteins that showed the highest abundance in eccrine sweat glands with >2 fold expression compared with other tissue/cells with the next highest abundance. Based on this, we could classify the proteins enriched in sweat glands into three groups; proteins that are expressed in most tissues, proteins that are expressed at lower level in hematopoietic cells but at a higher level in adult tissues, and proteins that are expressed mostly in sweat glands (Fig. 3A). In the first group, proteins including LLGL2, RPS28, H2AFY, HNRNPR, ITPR2, LMNA, ELN, S100A11, COL3A1, EPPK1, KRT17, EPHB3, DBI, COX6B1, PYGB, PDK3, DOCK9, SLC1A5, KRT77, GAPDH, and H1F0 showed relatively even abundance throughout most human tissues or cells. Pathway analysis showed that these genes are enriched in protein digestion and absorption. In the second group, the proteins including SLC12A2, COL7A1, PIP, CALML5, ITGB4, DSG2, CMA1, POSTN, COL6A2, and COL6A3 showed relatively lower abundance in hematopoietic cells. Pathway analysis showed that these genes are enriched in ECM-receptor interaction, protein digestion and absorption, focal adhesion and PI3K-Akt pathway. The role of these proteins in cell-cell interactions can perhaps explain why they were in low abundance in hematopoietic cells. In the third group, proteins found to be expressed primarily in eccrine sweat glands included FAM180B, PLA2R1, MUCL, KCNN4, PLCB4, PROM2, CLDN7, COL26A1, SCGB2A2, CD300LG, FAM160A1, ACOT6, PRR4, TRPV4, COL17A1, SCGB1D2, and AQP5 (Fig. 3A). Pathway analysis suggested that these genes are enriched in salivary secretion, insulin secretion, protein digestion, reabsorption and inflammation-mediated regulation of TRP channels.
Fig. 3.
Protein expression profile of human eccrine sweat glands. A, Heat map showing the relatively abundant proteins in human eccrine sweat glands compared with other human tissues/cells. Proteins with >2-fold higher abundance in eccrine sweat glands than the one in the next most abundant tissue/cells are shown. B, Venn diagram showing overlap between the numbers of proteins identified in the sweat and the eccrine sweat gland. C, Missing proteins identified from human eccrine sweat glands.
Although this is the first study that performed human eccrine sweat gland proteome analysis, there is a study describing the expression of ∼150 proteins in human sweat (22). Thus, we were curious to examine the overlap between eccrine sweat gland and sweat proteomes. As expected, most sweat proteins (121 out of 149) were identified in eccrine sweat gland (Fig. 3B) (23). Interestingly, sixty-two of them were among the top 500 abundant proteins in eccrine sweat gland (supplemental Table S3).
Efforts are ongoing to identify all genes at the protein level, but there are still 2,187 missing proteins that have never been identified (24). This could be because of the complex architecture of human organs and the expression of proteins in specific cell types or specific locations within an organ. Because human eccrine sweat gland showed distinct patterns of protein expression, we reasoned that this unique expression profile could help us identify missing proteins. We identified 2 proteins that met the criteria of missing proteins that is the protein must be identified with 2 or more peptides, longer than 7 amino acids, and validated by synthetic peptides (Fig. 3C, Table I and supplemental Fig. S1).
Table I. Missing proteins identified from sweat glands.
| RefSeq accession | Gene symbol | Description | # Peptides | # PSMs |
|---|---|---|---|---|
| NP_001078950 | MAP1LC3B2 | Microtubule-associated proteins 1A/1B light chain 3 beta 2 | 3 | 6 |
| NP_005167 | ATP5G2 | ATP synthase F(0) complex subunit C2, mitochondrial isoform b | 2 | 10 |
These proteome analysis results for sweat glands indicate that sweat glands are quite distinct from other human organs and the proteins abundant in sweat glands were more enriched in secretion and absorption. This could stem from the fact that sweat gland is a type of secretory glands and microdissected human organ isolated from human skin. In addition, the proteome analysis of this unusual human organ enabled us to identify 2 validated missing proteins implying that more analysis on a micro scale level of a human organ such as sweat glands will lead to the identification of more missing proteins.
Proteogenomic Analysis of Human Eccrine Sweat Glands
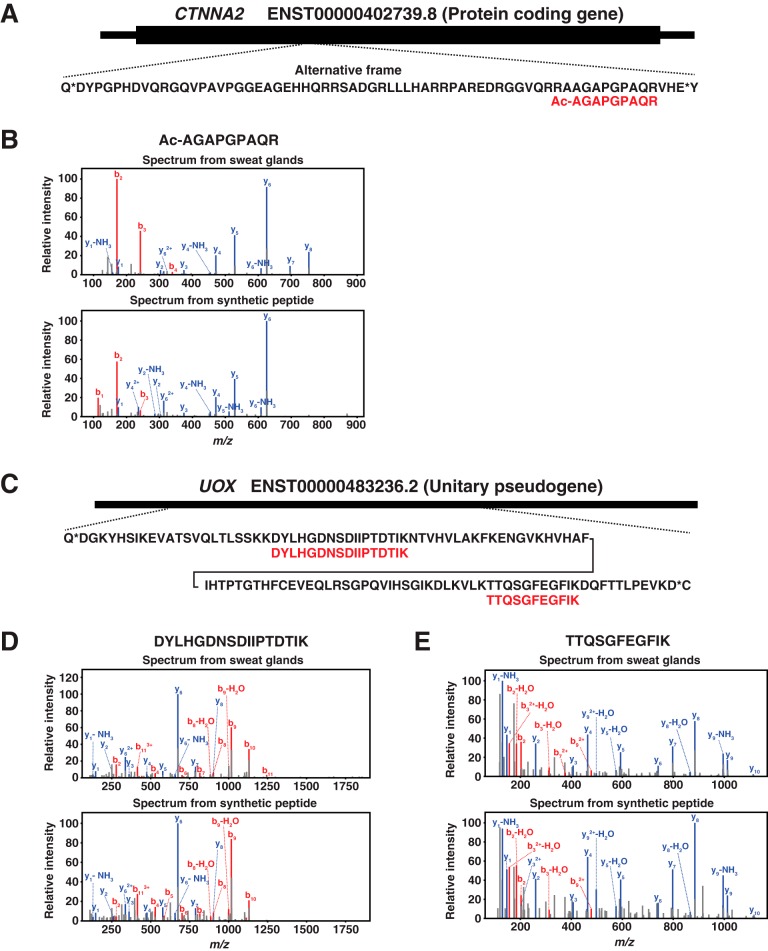
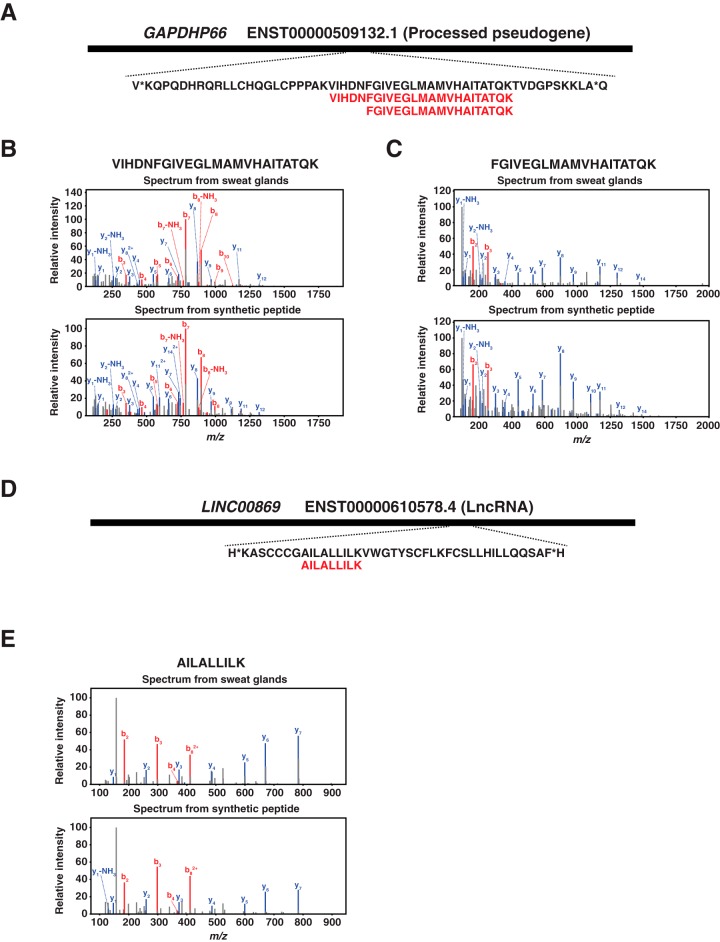
We expected that the proteome data in addition to transcriptome data will provide us the information about the expression profiles of sweat glands at two different expression levels, transcriptome and proteome. This could also lead us to identify novel genes that are expressed in sweat glands because we have expression evidence in both transcriptome and proteome levels. First, we explored the relationship between transcript and protein abundance in eccrine sweat glands by correlating the FPKM values of transcripts and NSAF values of proteins. A Spearman's correlation of 0.458 between FPKM and NSAF values was found. This was not unexpected because it has previously been shown that the integration of RNA-seq with proteomics generally has a correlation of about ∼0.40 (supplemental Fig. S2) (25). Next, we attempted to identify novel genes for which we had evidence of expression at both transcriptome and proteome levels. For this, we generated a customized protein database by 3-frame translation of the RNA sequences of eccrine sweat glands and performed a search against the newly generated database. From the database search, we identified 12 novel peptides mapping to 10 different genes composed of 5 protein-coding genes, 4 pseudogenes and 1 lncRNA. Those identified peptides were validated by comparing the MS/MS spectra of those peptides to ones of synthetic peptides. The spectra of 7 peptides mapping to 5 different genes composed of 1 protein-coding gene, 3 pseudogenes and 1 lncRNA perfectly matched to those of synthetic peptides (Table II). One out of 7 peptides, AGAPGPAQR was N-terminally acetylated one mapping to the middle part of an alternative frame of the protein-coding gene, CTNNA2 (Fig. 4A). The spectrum of the acetylated peptide showed almost similar pattern as the one of synthetic peptide (Fig. 4B). Strikingly, the 5 peptides out of 7 validated peptides were from pseudogenes. Especially, two pseudogenes, UOX and GAPDHP66, were identified by two different peptides (Table II). The two peptides of UOX was mapping to the central region of the pseudogene in the same frame with 53 amino acid distance (Fig. 4C). The spectra of two peptides from sweat glands showed perfect matches to the ones of synthetic peptides (Fig. 4D and 4E). The two peptides of GAPDHP66 mapped to almost the same region of the gene with an overlap of 19 amino acids. The spectra of the two peptides showed perfect matches to those of synthetic peptides. On the other hand, the peptide of lncRNA, LINC00869 mapped to the 3′ part of the gene (Fig. 5D). The MS/MS spectrum of the peptide, AILALLILK, showed a perfect match to the one obtained from fragmentation of the corresponding synthetic peptide (Fig. 5E).
Table II. Novel proteins identified from eccrine sweat glands.
| Peptide | Accession | Gene symbol | Gene type |
|---|---|---|---|
| Ac-AGAPGPAQR | ENST00000402739.8 | CTNNA2 | Protein coding |
| FGIVEGLMAMVHAITATQK | ENST00000509132.1 | GAPDHP66 | Processed pseudogene |
| VIHDNFGIVEGLMAMVHAITATQK | ENST00000509132.1 | GAPDHP66 | Processed pseudogene |
| AILALLILK | ENST00000610578.4 | LINC00869 | LncRNA |
| SIIIDAPGHR | ENST00000528208.2 | RP11–676F20.2 | Unprocessed pseudogene |
| DYLHGDNSDIIPTDTIK | ENST00000483236.2 | UOX | Transcribed unitary pseudogene |
| TTQSGFEGFIK | ENST00000483236.2 | UOX | Transcribed unitary pseudogene |
Fig. 4.
Novel proteins identified from human eccrine sweat glands mapping to a protein-coding gene and a unitary pseudogene. A, An N-terminally acetylated peptide, Ac-AGAPGPAQR, mapping to an alternative frame of a protein-coding gene, CTNNA2, was identified from human eccrine sweat glands. B, Ac-AGAPGPAQR was validated by a synthetic peptide with the same sequence. C, Two peptides, DYLHGDNSDIIPTDTIK and TTQSGFEGFIK, mapping to a unitary pseudogene, UOX, were identified from human eccrine sweat glands. D, DYLHGDNSDIIPTDTIK was validated by a synthetic peptide with the same sequences. E, TTQSGFEGFIK was validated by a synthetic peptide with the same sequences.
Fig. 5.
Novel proteins identified from human eccrine sweat glands that mapped to a processed pseudogene and an lncRNA. A, Two peptides, VIHDNFGIVEGLMAMVHAITATQK and FGIVEGLMAMVHAITATQK, that mapped to a processed pseudogene, GAPDHP66, were identified from human eccrine sweat glands. B, VIHDNFGIVEGLMAMVHAITATQK was validated by a synthetic peptide with the same sequences. C, FGIVEGLMAMVHAITATQK was validated by a synthetic peptide with the same sequences. D, A peptide, FGIVEGLMAMVHAITATQK, that mapped to an lncRNA, LINC00869, was identified from human eccrine sweat glands. E, AILALLILK was validated by a synthetic peptide with the same sequences.
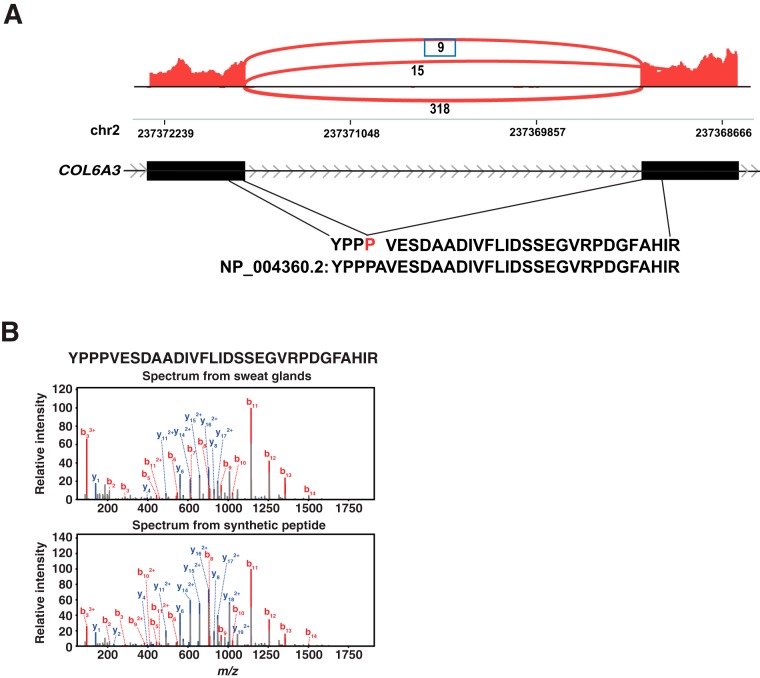
We already discovered 8% of novel and 22% of partial novel splice junctions. If we can find any splice junctions in both RNA and protein level, this could be bona fide novel junctions. Thus, we investigated whether there were any novel splice junctions that could be detected at both levels. We discovered 3 novel junctions with protein level evidence, two of them had reliable mass spectra, and one of them was validated by a synthetic peptide. A novel exon junction with three in-frame additional nucleotides was mapped to COL6A3 (Fig 6A). The spectrum of peptides from COL6A3 showed a perfect match to that of synthetic peptide (Fig 6B). These novel proteins expressed from the sweat glands suggest that there still could be many novel proteins that are expressed from sORFs or pseudogenes. UOX has been classified as a unitary pseudogene that is a pseudogene without a parent gene, but other species have a coding protein from this gene. The discovery of peptides from this unitary pseudogene suggests that this gene has been wrongly classified as a pseudogene while this gene has protein expression in human cells. Thus, these results again confirm that we need to reconsider the classification algorithm of the pseudogenes.
Fig. 6.
A novel exon junction with evidence at both RNA and protein levels in eccrine sweat glands. A, A novel exon junction mapping to COL6A3 with one peptide removed from a known protein, COL6A3, was identified. B, YPPPVESDAADIVFLIDSSEGVRPDGFAHIR was validated by a synthetic peptide with the same sequences.
DISCUSSION
In this study, we report the first in-depth RNA and protein expression profiles of human sweat glands. With the publication of human proteome (11, 26), and the availability of RNA-seq (27), there are several databases that catalogue gene and protein expression profiles from diverse tissues and cell types. However, there is no such reference available on sweat glands, despite their importance in health and disease. Our study was designed to find an answer to an open question about the diversity of RNAs (protein-coding and non-coding) and proteins that are expressed in the eccrine sweat glands. To do so, we conducted RNA-sequencing and proteomic analysis on ∼250 sweat glands collected from a healthy individual. In this pursuit, we identified 13,300 protein-coding genes that were expressed at the RNA level, 53,400 non-coding genes and detected ∼6100 proteins in eccrine sweat glands. This gene expression profile can serve as a useful resource to study the etiology of many diseases relevant to the sweat gland, such as CF, hyperhidrosis, anhidrosis and atopic dermatitis.
Comparison of eccrine sweat gland transcriptome to that of other human tissues/cells revealed its similarity to the cortex of the kidneys. One biologic explanation is that sweat glands share some of the excretory functions like kidney (28). In this regard, a striking example is the excretion of urea. The levels of urea in sweat are much higher than in serum indicating sweat glands should express urea transporters (29). A prior study has shown that urea transporter subtypes are expressed in sweat glands (30). Interestingly, urea levels in sweat are even higher in individuals with the chronic renal disease resulting in uremic frost (31). In this study, we demonstrate that the urea transporter 1 isoform 1 (SLC14A1), a urea channel known to facilitate transmembrane urea transport, is indeed detectable in both the transcriptome and proteome of eccrine sweat gland. Additionally, we demonstrate that there is extensive overlap in the gene expression between kidney and sweat glands, indicating that many other genes are also similarly expressed in the kidney and eccrine sweat glands. Together, these findings support the evidence that sweat is another way for expelling water and metabolic wastes besides urine.
Evidence from the human eccrine sweat gland proteome is consistent with previous studies that identified physiologic roles of Na+K+ATPase, K+ channels, Ca+2 channels, V-H+-ATPase, and AQP5 in secretory mechanisms (12, 32–35). Their normal activities result in sweat secretory fluid that is isotonic to plasma with the concentration of Na+ ∼145 mmol/l and Cl− ∼115 mmol/l (36, 37). Additionally, detection of sodium potassium pump, and sodium hydrogen ion exchanger 1 in the eccrine sweat gland proteome validates the concept that driving force for reabsorption is a Na+ gradient into the cell (7). The loss of Na+ from the reabsorptive duct of eccrine sweat gland creates a gradient for Cl− to pass through the cell and into the extracellular fluid via CFTR and carbonic anhydrases (7, 34, 38). As this happens, the final sweat becomes hypotonic to plasma with a concentration of Na+ and Cl- below 20 mmol/L (39). It is well established that disease-causing variants in CFTR result in the defective reabsorption and elevated sweat chloride concentration, therefore sweat chloride measurements are the gold standard method for the diagnosis of CF (40, 41). Although we have observed CFTR mRNA expression by RNA-seq, protein expression could be not be detected in the proteome of eccrine sweat gland, likely because the levels of CFTR protein were too low to detect.
Importantly, we have recently reported deleterious variants in carbonic anhydrase 12 (CA12) that resulted in dysfunctional protein in individuals exhibiting CF-like phenotypes - elevated sweat chloride concentration and lung disease (42). Our present study confirms the expression of CA12 in the proteome and transcriptome of eccrine sweat glands. This finding validates the importance of this protein in regulating sweat chloride levels. Interestingly, we were also able to detect carbonic anhydrases (2, 3, 4, 13) in the proteome and transcriptome of sweat gland, indicating that they may also alter electrolyte or pH balance in the sweat. Strikingly, proteomics data also showed enrichment for inflammation-mediated regulation of TRPV4 channels. TRPV4 is a nonselective calcium ion channel which is activated by moderate heat (25–34 °C), endogenous inflammatory metabolites (like arachidonic acid), and exogenous compounds (like phorbol esters) (43–46). TRPV4 appears to be important for skin barrier integrity and contributes to cell-cell junction development, which prevents excess skin dehydration (47). It has been shown in mouse models that sweat duct is the growth center that repairs epidermal injury (3). Interestingly, a study demonstrated that mice lacking TRPV4 have an altered temperature behavior under normal and inflammatory conditions (48). Notably, sweat glands in mice are restricted to foot pads whereas in humans there are millions of sweat glands dispersed throughout the body (4). More recent studies readdressing this issue using human skin concluded that sweat glands contribute significantly to re-epithelialization after partial-thickness wound (49). Here we provide both transcriptome and proteome level evidence that TRPV4 is indeed expressed in eccrine sweat glands. Importantly, TRPV3-related skin TRP channelopathy, defined as Olmsted syndrome, was identified in humans which is caused by gain of function missense mutations (50). Whether genetic alterations in human TRPV4 are responsible for sweat gland dysfunction that leads to skin dehydration and/or inflammation remains to be determined.
We identified two missing proteins from eccrine sweat glands. However, it is difficult to say whether these two identified missing proteins are unique to sweat glands. Because these proteins have never been identified in other proteomic studies while they were identified in eccrine sweat glands, it is reasonable to consider that these proteins are relatively more abundant in sweat glands. Lastly, we performed the proteogenomic analysis combining the transcriptome with the proteome data for the identification of novel proteins that would have never been identified if we used a reference database. We have identified 3 reliable peptides from novel splice junctions, and one of them was validated by a synthetic peptide. Strikingly, the majority of novel proteins were from pseudogenes. These results imply that our understanding about pseudogene has been incomplete and deeper proteome analyses of microscale of human tissues or individual primary cell types can potentially identify more proteins expressed from genes that have been classified as a non-coding one. In addition, most novel proteins were from sORFs. Recently there have been active researches in sORF discovering that sORFs have their own functions. However, these sORFs have been missed in considering as functional proteins. Thus, these sORFs could be one of the missing dimensions in understanding biological functions. This discovery provides us insight there are still many sORFs that remain to be discovered. We anticipate that this study will assist in a better molecular understanding of human sweat glands and their role in health and disease.
Overall, this study can be used as a starting point for future studies to assess inter-individual variability between individuals with different genetic backgrounds, sex, and age. Studies with multiple individuals should assess batch effects to correct for any technical variability (51, 52). Additionally, single-cell RNA-seq and proteome analysis of eccrine sweat gland would further help identify gene expression in specific cell types, as was observed in two recent studies that demonstrated CFTR is mostly expressed in rare cell type called an ionocyte in the airway epithelium (53, 54). Taken together, this study demonstrates the transcriptome and proteome expression profile of the eccrine sweat gland, which can be used to study the etiology of many diseases relevant to the sweat gland, such as CF, hyperhidrosis, anhidrosis and atopic dermatitis.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD010637.
Supplementary Material
Acknowledgments
We thank Prof. P.M. Quinton at the University of California-San Diego for providing training to N. Sharma on sweat gland dissection.
Footnotes
* This study was supported by a grant from Cystic Fibrosis Foundation Therapeutic (CF Foundation) and CF Research Scholar Program (Gilead Sciences) to NS; R01DK44003, CF Foundation Cuttin13A1, Cuttin16IO, to GRC. This study was supported by a grant from NCI's Clinical Proteomic Tumor Analysis Consortium initiative (U24CA210985). We acknowledge the support of the Center for Proteomics Discovery at Johns Hopkins and shared instrumentation grant S10OD021844.
 This article contains supplemental material.
This article contains supplemental material.
REFERENCES
- 1. Saga K. (2002) Structure and function of human sweat glands studied with histochemistry and cytochemistry. Prog. Histochem. Cytochem. 37, 323–386 [DOI] [PubMed] [Google Scholar]
- 2. Bovell D. L. (2018) The evolution of eccrine sweat gland research towards developing a model for human sweat gland function. Exp. Dermatol. 27, 544–550 [DOI] [PubMed] [Google Scholar]
- 3. Lu C., and Fuchs E. (2014) Sweat gland progenitors in development, homeostasis, and wound repair. Cold Spring Harb. Perspect. Med. 4, a015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fuchs E. (2016) Epithelial skin biology: three decades of developmental biology, a hundred questions answered and a thousand new ones to address. Curr. Top. Dev. Biol. 116, 357–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shibasaki M., Wilson T. E., and Crandall C. G. (2006) Neural control and mechanisms of eccrine sweating during heat stress and exercise. J. Appl. Physiol. (1985), 100, 1692–1701 [DOI] [PubMed] [Google Scholar]
- 6. Shibasaki M., and Crandall C. G. (2010) Mechanisms and controllers of eccrine sweating in humans. Front. Biosci. 2, 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quinton P. M. (2007) Cystic fibrosis: lessons from the sweat gland. Physiology 22, 212–225 [DOI] [PubMed] [Google Scholar]
- 8. Doolittle J., Walker P., Mills T., and Thurston J. (2016) Hyperhidrosis: an update on prevalence and severity in the United States. Arch. Dermatol. Res. 308, 743–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ma K., Tan Z., Zhang C., and Fu X. (2016) Mesenchymal stem cells for sweat gland regeneration after burns: From possibility to reality. Burns 42, 492–499 [DOI] [PubMed] [Google Scholar]
- 10. Pandey A., and Pevzner P. A. (2014) Proteogenomics. Proteomics 14, 2631–2632 [DOI] [PubMed] [Google Scholar]
- 11. Kim M. S., Pinto S. M., Getnet D., Nirujogi R. S., Manda S. S., Chaerkady R., Madugundu A. K., Kelkar D. S., Isserlin R., Jain S., Thomas J. K., Muthusamy B., Leal-Rojas P., Kumar P., Sahasrabuddhe N. A., Balakrishnan L., Advani J., George B., Renuse S., Selvan L. D., Patil A. H., Nanjappa V., Radhakrishnan A., Prasad S., Subbannayya T., Raju R., Kumar M., Sreenivasamurthy S. K., Marimuthu A., Sathe G. J., Chavan S., Datta K. K.2, Subbannayya Y., Sahu A., Yelamanchi S. D., Jayaram S., Rajagopalan P., Sharma J., Murthy K. R., Syed N., Goel R., Khan A. A., Ahmad S., Dey G., Mudgal K., Chatterjee A., Huang T. C., Zhong J., Wu X., Shaw P. G., Freed D., Zahari M. S., Mukherjee K. K., Shankar S., Mahadevan A., Lam H., Mitchell C. J., Shankar S. K., Satishchandra P., Schroeder J. T., Sirdeshmukh R., Maitra A., Leach S. D., Drake C. G., Halushka M. K., Prasad T. S., Hruban R. H., Kerr C. L., Bader G. D., Iacobuzio-Donahue C. A., Gowda H., and Pandey A. (2014) A draft map of the human proteome. Nature 509, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quinton P. M., and Tormey J. M. (1976) Localization of Na/K-ATPase sites in the secretory and reabsorptive epithelia of perfused eccrine sweat glands: a question to the role of the enzyme in secretion. J. Membr. Biol. 29, 383–399 [DOI] [PubMed] [Google Scholar]
- 13. Brayden D. J., and Fitzpatrick J. (1995) Cultured human sweat gland epithelia: isolation of glands using neutral red. Pharm. Res. 12, 171–175 [DOI] [PubMed] [Google Scholar]
- 14. Sharma N., LaRusch J., Sosnay P. R., Gottschalk L. B., Lopez A. P., Pellicore M. J., Evans T., Davis E., Atalar M., Na C. H., Rosson G. D., Belchis D., Milewski M., Pandey A., and Cutting G. R. (2016) A sequence upstream of canonical PDZ-binding motif within CFTR COOH-terminus enhances NHERF1 interaction. Am. J. Physiol. Lung Cell Mol. Physiol. 311, L1170–L1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wisniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 16. Kulak N. A., Pichler G., Paron I., Nagaraj N., and Mann M. (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- 17. Paoletti A. C., Parmely T. J., Tomomori-Sato C., Sato S., Zhu D., Conaway R. C., Conaway J. W., Florens L., and Washburn M. P. (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. U.S.A. 103, 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang D. W., Sherman B. T., Tan Q., Collins J. R., Alvord W. G., Roayaei J., Stephens R., Baseler M. W., Lane H. C., and Lempicki R. A. (2007) The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8, R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 20. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 21. Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., and Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 22. Csosz E., Emri G., Kalló G., Tsaprailis G., and Tõzsér J. (2015) Highly abundant defense proteins in human sweat as revealed by targeted proteomics and label-free quantification mass spectrometry. J. Eur. Acad. Dermatol. Venereol. 29, 2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raiszadeh M. M., Ross M. M., Russo P. S., Schaepper M. A., Zhou W., Deng J., Ng D., Dickson A., Dickson C., Strom M., Osorio C., Soeprono T., Wulfkuhle J. D., Petricoin E. F., Liotta L. A., and Kirsch W. M. (2012) Proteomic analysis of eccrine sweat: implications for the discovery of schizophrenia biomarker proteins. J. Proteome Res. 11, 2127–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaudet P., Michel P. A., Zahn-Zabal M., Britan A., Cusin I., Domagalski M., Duek P. D., Gateau A., Gleizes A., Hinard V., Rech de Laval V., Lin J., Nikitin F., Schaeffer M., Teixeira D., Lane L., and Bairoch A. (2017) The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 45, D177–D182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Sousa Abreu R., Penalva L. O., Marcotte E. M., and Vogel. C. (2009) Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilhelm M., Schlegl J., Hahne H., Gholami A. M., Lieberenz M., Savitski M. M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H., Mollenhauer M., Slotta-Huspenina J., Boese J. H., Bantscheff M., Gerstmair A., Faerber F., and Kuster B. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 [DOI] [PubMed] [Google Scholar]
- 27. Mortazavi A., Williams B. A., McCue K., Schaeffer L., and Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 28. Jadoon S., Karim S., Akram M. R., Kalsoom Khan A., Zia M. A., Siddiqi A. R., and Murtaza G. (2015) Recent developments in sweat analysis and its applications. Int. J. Anal. Chem. 2015, 164974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang C. T., Chen M. L., Huang L. L., and Mao I. F. (2002) Uric acid and urea in human sweat. Chin. J. Physiol. 45, 109–115 [PubMed] [Google Scholar]
- 30. Xie L., Jin L., Feng J., and Lv J. (2017) The expression of AQP5 and UTs in the sweat glands of uremic patients. Biomed. Res. Int. 2017, 8629783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saardi K. M., and Schwartz R. A. (2016) Uremic frost: a harbinger of impending renal failure. Int. J. Dermatol. 55, 17–20 [DOI] [PubMed] [Google Scholar]
- 32. Nejsum L. N., Kwon T. H., Jensen U. B., Fumagalli O., Frøkiaer J., Krane C. M., Menon A. G., King L. S., Agre P. C., and Nielsen S. (2002) Functional requirement of aquaporin-5 in plasma membranes of sweat glands. Proc. Natl. Acad. Sci. U.S.A. 99, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song Y., Sonawane N., and Verkman A. S. (2002) Localization of aquaporin-5 in sweat glands and functional analysis using knockout mice. J. Physiol. 541, 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clunes M. T., Lindsay S. L., Roussa E., Quinton P. M., and Bovell D. L. (2004) Localisation of the vacuolar proton pump (V-H+ -ATPase) and carbonic anhydrase II in the human eccrine sweat gland. J. Mol. Histol. 35, 339–345 [DOI] [PubMed] [Google Scholar]
- 35. Bovell D. L., MacDonald A., Meyer B. A., Corbett A. D., MacLaren W. M., Holmes S. L., and Harker M. (2011) The secretory clear cell of the eccrine sweat gland as the probable source of excess sweat production in hyperhidrosis. Exp. Dermatol. 20, 1017–1020 [DOI] [PubMed] [Google Scholar]
- 36. Sato K., and Sato F. (1990) Na+, K+, H+, Cl-, and Ca2+ concentrations in cystic fibrosis eccrine sweat in vivo and in vitro. J. Lab. Clin. Med. 115, 504–511 [PubMed] [Google Scholar]
- 37. Baker L. B. (2017) Sweating Rate and Sweat Sodium Concentration in Athletes: A Review of Methodology and Intra/Interindividual Variability. Sports Med. 47, 111–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quinton P. M. (1999) Physiological basis of cystic fibrosis: a historical perspective. Physiol. Rev. 79, S3–S22 [DOI] [PubMed] [Google Scholar]
- 39. Cui C. Y., and Schlessinger D. (2015) Eccrine sweat gland development and sweat secretion. Exp. Dermatol. 24, 644–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. LeGrys V. A., Yankaskas J. R., Quittell L. M., Marshall B. C., Mogayzel P. J. Jr; and Cystic Fibrosis Foundation (2007) Diagnostic sweat testing: the Cystic Fibrosis Foundation guidelines. J. Pediatr. 151, 85–89 [DOI] [PubMed] [Google Scholar]
- 41. Farrell P. M., White T. B., Ren C. L., Hempstead S. E., Accurso F., Derichs N., Howenstine M., McColley S. A., Rock M., Rosenfeld M., Sermet-Gaudelus I., Southern K. W., Marshall B. C., and Sosnay P. R. (2017) Diagnosis of Cystic Fibrosis: Consensus Guidelines from the Cystic Fibrosis Foundation. J. Pediatrics 181S, S4–S15.e1 [DOI] [PubMed] [Google Scholar]
- 42. Lee M., Vecchio-Pagán B., Sharma N., Waheed A., Li X., Raraigh K. S., Robbins S., Han S. T., Franca A. L., Pellicore M. J., Evans T. A., Arcara K. M., Nguyen H., Luan S., Belchis D., Hertecant J., Zabner J., Sly W. S., and Cutting G. R. (2016) Loss of carbonic anhydrase XII function in individuals with elevated sweat chloride concentration and pulmonary airway disease. Hum. Mol. Genet. 25, 1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watanabe H., Davis J. B., Smart D., Jerman J. C., Smith G. D., Hayes P., Vriens J., Cairns W., Wissenbach U., Prenen J., Flockerzi V., Droogmans G., Benham C. D., and Nilius B. (2002) Activation of TRPV4 channels (hVRL-2/mTRP12) by phorbol derivatives. J. Biol. Chem. 277, 13569–13577 [DOI] [PubMed] [Google Scholar]
- 44. Chung M. K., Lee H., and Caterina M. J. (2003) Warm temperatures activate TRPV4 in mouse 308 keratinocytes. J. Biol. Chem. 278, 32037–32046 [DOI] [PubMed] [Google Scholar]
- 45. Kida N., Sokabe T., Kashio M., Haruna K., Mizuno Y., Suga Y., Nishikawa K., Kanamaru A., Hongo M., Oba A., and Tominaga M. (2012) Importance of transient receptor potential vanilloid 4 (TRPV4) in epidermal barrier function in human skin keratinocytes. Pflugers Arch. 463, 715–725 [DOI] [PubMed] [Google Scholar]
- 46. Rajasekhar P., Poole D. P., and Veldhuis N. A. (2017) Role of Nonneuronal TRPV4 Signaling in Inflammatory Processes. Adv. Pharmacol. 79, 117–139 [DOI] [PubMed] [Google Scholar]
- 47. Caterina M. J., and Pang Z. (2016) TRP Channels in Skin Biology and Pathophysiology. Pharmaceuticals 9, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee H., Iida T., Mizuno A., Suzuki M., and Caterina M. J. (2005) Altered thermal selection behavior in mice lacking transient receptor potential vanilloid 4. J. Neurosci. 25, 1304–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rittie L., Sachs D. L., Orringer J. S., Voorhees J. J., and Fisher G. J. (2013) Eccrine sweat glands are major contributors to reepithelialization of human wounds. Am. J. Pathol. 182, 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Duchatelet S., Pruvost S., de Veer S., Fraitag S., Nitschké P., Bole-Feysot C., Bodemer C., and Hovnanian A. (2014) A new TRPV3 missense mutation in a patient with Olmsted syndrome and erythromelalgia. JAMA Dermatol. 150, 303–306 [DOI] [PubMed] [Google Scholar]
- 51. Leek J. T., Scharpf R. B., Bravo H. C., Simcha D., Langmead B., Johnson W. E., Geman D., Baggerly K., and Irizarry R. A. (2010) Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11, 733–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nygaard V., Rodland E. A., and Hovig E. (2016) Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics 17, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Plasschaert L. W., Žilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A. M., and Jaffe A. B. (2018) A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Montoro D. T., Haber A. L., Biton M., Vinarsky V., Lin B., Birket S. E., Yuan F., Chen S., Leung H. M., Villoria J., Rogel N., Burgin G., Tsankov A. M., Waghray A., Slyper M., Waldman J., Nguyen L., Dionne D., Rozenblatt-Rosen O., Tata P. R., Mou H., Shivaraju M., Bihler H., Mense M., Tearney G. J., Rowe S. M., Engelhardt J. F., Regev A., and Rajagopal J. (2018) A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD010637.