Significance
Despite being a leading cause of foodborne illnesses, accounting for 58% of all outbreaks and over 96% of nonbacterial outbreaks, there are no approved treatments available for norovirus infections. Assembled shells of the viruses without genetic materials enclosed are currently being used as candidates for vaccine trials. Although the virus shells have been thought to exist in a single-sized assembly, our structures in near-atomic detail reveal clear variations in size between different outbreak strains, and in spatial and angular arrangements of the antigenic surface spikes. The structures we present serve as valuable templates for facilitating vaccine formulations.
Keywords: norovirus, cryo-EM, foodborne illnesses
Abstract
Noroviruses are a leading cause of foodborne illnesses worldwide. Although GII.4 strains have been responsible for most norovirus outbreaks, the assembled virus shell structures have been available in detail for only a single strain (GI.1). We present high-resolution (2.6- to 4.1-Å) cryoelectron microscopy (cryo-EM) structures of GII.4, GII.2, GI.7, and GI.1 human norovirus outbreak strain virus-like particles (VLPs). Although norovirus VLPs have been thought to exist in a single-sized assembly, our structures reveal polymorphism between and within genogroups, with small, medium, and large particle sizes observed. Using asymmetric reconstruction, we were able to resolve a Zn2+ metal ion adjacent to the coreceptor binding site, which affected the structural stability of the shell. Our structures serve as valuable templates for facilitating vaccine formulations.
Noroviruses are a leading cause of foodborne illnesses, accounting for 58% of all outbreaks and over 96% of nonbacterial outbreaks, causing ∼21 million cases in the United States and 685 million cases worldwide each year (1–3). Also commonly referred to as stomach flu or winter vomiting bug, the viruses cause frequent outbreaks of acute gastroenteritis in hospitals, nursing homes, day care centers, schools, and restaurants, and over 90% of diarrheal outbreaks on cruise ships. Norovirus illnesses are estimated at a cost of $2 billion for treatment and lost productivity in the United States and $60 billion worldwide annually (1–3). The viruses are highly contagious, with as few as 18 virus particles needed to cause infection (4), and spread by the fecal–oral route through direct contact with patients and aerosolized viruses from vomiting and contaminated surfaces, foods, and water supplies (5, 6).
Noroviruses are round, nonenveloped viruses with (+)ssRNA genomes that belong to the Caliciviridae family and are divided into at least 6 genogroups that are subdivided into 30 or more genotypes, of which genogroups I, II, and IV cause illnesses in humans (5, 6). The norovirus genomes encode two structural proteins, one major capsid protein (VP1) that forms the icosahedral shell enclosing the genome and a minor structural protein (VP2) that is positively charged and may interact with and stabilize its genome (7, 8). More recently, the VP2 of feline vesivirus was shown to form a portal structure upon binding of the feline receptor (9).
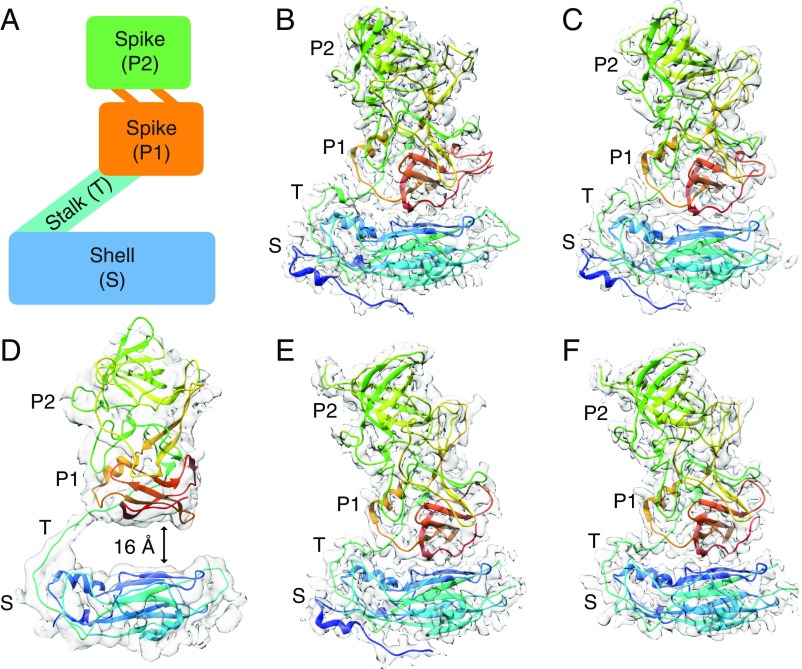
The major capsid protein consists of a shell (S) domain that forms the icosahedral enclosure and a protruding (P) domain that forms dimeric spikes on the virus surface and bears the antigenic features involved in host interactions (Fig. 1). The P domain consists of two subdomains: P1 emerging from the S domain and P2, which is an insertion within P1 and positioned at the outermost surface of the virus (SI Appendix, Fig. S1). The P2 subdomains have been shown to bind human histo-blood group antigens (HBGAs) as attachment factors, although some strains including GII.2 Snow Mountain virus (SMV) show only weak or no binding to HBGAs (5, 10, 11).
Fig. 1.
Modular organization of norovirus capsid proteins. Side views of human norovirus major capsid protein A subunits, showing the cryo-EM maps in gray and the fitted atomic models colored in rainbow representation from N to C termini. (A) A schematic diagram showing the modular organization of capsid subunits, consisting of trapezoid-shaped shell, long and flexible stalk, and protruding spike domains. The P domain consists of a P1 subdomain emerging from the S domain and P2, which is an insertion in P1 and positioned at the outermost surface of the virus. (B) GI.1 subunit, showing the P domain placed immediately above and forming contacts with the S domain. (C) GI.7 subunit, with the P domain placed close to the S domain. (D) GII.4 subunit, with the P domain lifted significantly (∼16 Å) above the shell domain through the long and flexible stalk region. (E) GII.2 T = 3 particle subunit, with the P domain making contacts with the S domain. (F) GII.2 T = 1 particle subunit, also with the P domain placed close to the S domain.
Recombinant major capsid proteins assemble into virus-like particles (VLPs) that preserve the structure and antigenicity of infectious virions (7, 12). VLPs and animal noroviruses have been used as model systems, due to difficulties in propagating human virions in cell-culture systems (7, 13). There are no approved treatments available for norovirus infections, and VLPs are currently being used as candidates for vaccine trials; one candidate has reached a phase II clinical trial (12). Although crystal structures of P domains are available (10, 14), currently there is only one high-resolution VLP crystal structure of one norovirus strain (GI.1) available, which was solved at 3.4-Å resolution (7). The GI.1 VLP exhibits T = 3 icosahedral symmetry, an assembly of 180 subunits, with 90 dimeric spikes on its surface.
To further our understanding of human norovirus capsid architectures and the relationship to disease, we report high-resolution cryo-EM VLP structures of four outbreak strains in the frozen hydrated state: GII.2 SMV (2.7 and 3.1 Å), GII.4 Minerva (4.1 Å), GI.7 Houston (2.9 Å), and GI.1 Norwalk (2.6 Å) strains (Fig. 2 and SI Appendix, Fig. S2 and Table S1). Notably, GII.4 strains have been responsible for 70–80% of all norovirus outbreaks, representing the most epidemiologically prevalent strain (5, 6). The VLP structures reveal polymorphism with T = 1, T = 3, and T = 4 assemblies observed. These structures serve as valuable templates for facilitating VLP vaccine formulations.
Fig. 2.
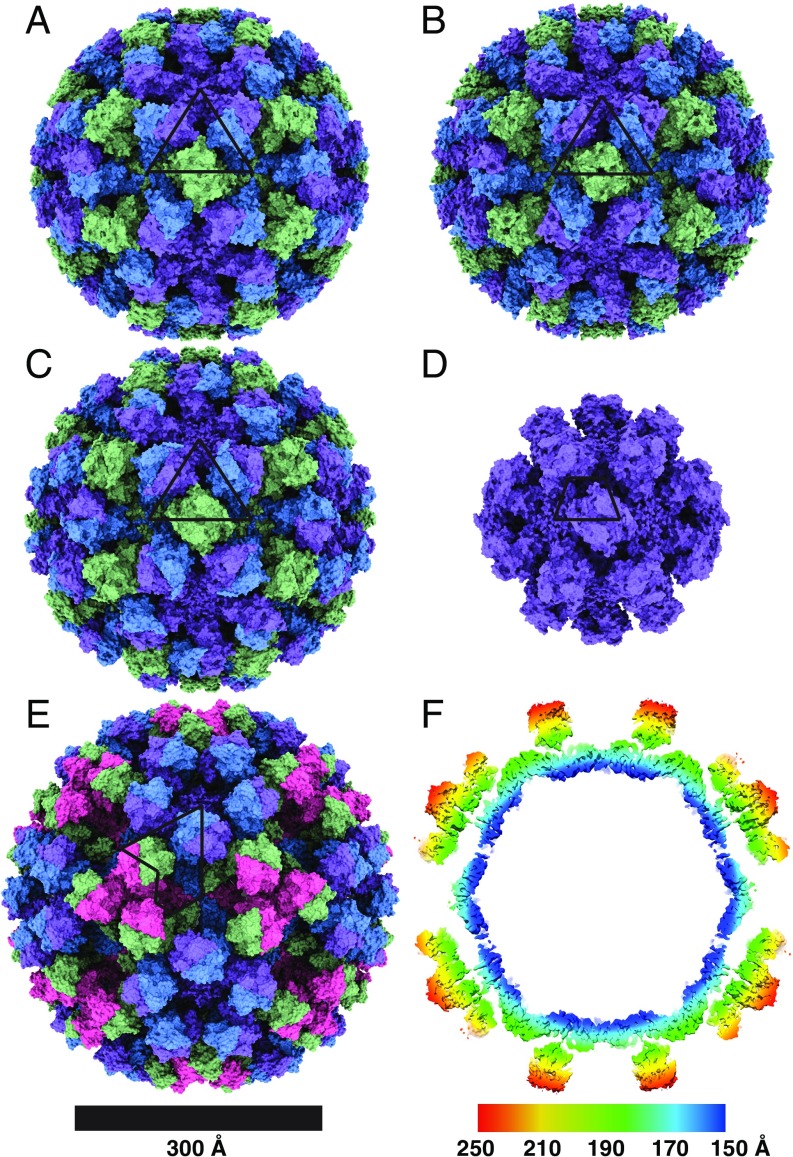
Cryo-EM structures of human norovirus outbreak strain capsids. Shaded depth-cue representations of norovirus VLP structures viewed along the icosahedral twofold axis, colored purple (subunit A), blue (subunit B), green (subunit C), and pink (subunit D). The positions of asymmetric units are identified by black lines. (A) GI.1 Norwalk strain at 2.9-Å resolution in T = 3 icosahedral symmetry with 90 dimeric P-domain spikes assembled from 180 subunits and 410 Å in diameter. (B) GI.7 Houston strain at 2.9-Å resolution in T = 3 symmetry and 420 Å in diameter. (C) GII.2 Snow Mountain virus strain at 3.1-Å resolution in T = 3 symmetry and 430 Å in diameter. The spikes of GII.2 SMV are twisted ∼15° counterclockwise relative to GI.1 and GI.7. (D) GII.2 SMV at 2.7-Å resolution in T = 1 symmetry with 30 spikes and 60 subunits and 310 Å in diameter. The spikes are placed significantly farther apart (∼10–25 Å) in T = 1 symmetry. (E) GII.4 Minerva strain at 4.1-Å resolution in T = 4 symmetry with 120 spikes and 240 subunits and 490 Å in diameter. The spikes of GII.4 are twisted ∼50° clockwise relative to the orientation of GI.1 and GI.7. (F) A central slice view of a GII.4 cryo-EM map colored by radial distance from the center (blue to red), showing a two-layered architecture with a secondary layer of spikes suspended ∼16 Å above the primary layer of the icosahedral shell domain.
Results and Discussion
Shell Domain and Size Variation.
Movies of VLPs embedded in vitreous ice on holey carbon grids were collected using a Titan Krios G3 transmission electron microscope (ThermoFisher) equipped with a K2 Summit direct electron detector (Gatan) (Materials and Methods and SI Appendix, Fig. S3). To improve the maps, we used an asymmetric focused reconstruction method that resolved features lost with symmetry imposition, using the program symmetry_expand_stack_and_par included with cisTEM).
Although norovirus VLPs have been thought to exist only in T = 3 assemblies, our cryo-EM VLP structures revealed structural polymorphism between and within genogroups in identical buffer conditions (15, 16). Our GI.1 structure is in good agreement with the previous GI.1 crystal structure (0.8-Å rmsd). The GI.7 and GI.1 strains are also both in T = 3 assemblies (Fig. 2 and SI Appendix, Fig. S2). The external diameters of GI.7 and GI.1 particles are 420 and 410 Å, respectively, and the internal diameter of both strains is 240 Å. The GII.4 VLPs, on the other hand, are exclusively in T = 4 icosahedral symmetry consisting of 120 spikes and 240 subunits. Accordingly, the larger particles are 490 Å in external diameter and 280 Å in internal diameter. The GII.2 strain particles are observed in a T = 3 form as well as in T = 1 symmetry, the latter of which is likely to exist as an empty subviral form, with 30 spikes and 60 subunits. The external diameters of GII.2 T = 3 and T = 1 particles are 430 and 310 Å, respectively, and the internal diameters are 240 and 120 Å, respectively. We note that the T = 3 and T = 1 structures were observed together and solved from micrographs of the same sample.
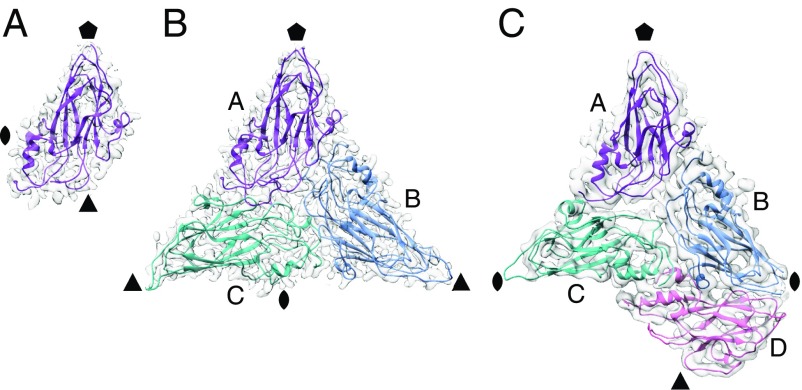
The observed variances in the T numbers of norovirus VLPs are examples of quasi-equivalence, with identical capsid subunits undergoing conformational adjustments to fit multiple symmetry arrangements (17). Each T = 1 particle icosahedral asymmetric unit (ASU) consists of one subunit (A), whereas the T = 3 particle ASU consists of A, B, and C subunits in a quasi-threefold symmetry arrangement (Fig. 3). The T = 4 particle ASU consists of four subunits, A, B, C, and D, with the C–D dimer taking up a position similar to the C–C dimer of T = 3 particles. In the T = 4 particles, the icosahedral threefold axes are formed between three D subunits (Fig. 2 and SI Appendix, Fig. S2). In the T = 3 particles, B and C subunits interdigitate around the icosahedral threefold axes in quasi-sixfold symmetry. The T = 4 particle icosahedral twofold symmetry axes are formed between two sets of B, C, and D subunits in quasi-sixfold arrangements. The C–C dimer interfaces form the icosahedral twofold axes in T = 3 particles.
Fig. 3.
Asymmetric units of GII.2 T = 1, GII.2 T = 3, and GII.4 T = 4 particles. The icosahedral symmetry axes positions are indicated by a pentagon (fivefold), triangle (threefold), and oval (twofold). (A) Shell domain of a single subunit in each ASU of GII.2 T = 1 particles. (B) Shell domains of three subunits in quasi-equivalent positions, A, B, and C, that constitute each ASU of GII.2 T = 3 particles. (C) GII.4 shell domains in the T = 4 particle ASU with four subunits, A, B, C, and D. The C–D dimer takes up a position similar to the C–C dimer in T = 3 particles.
In the T = 3 particles of GII.2, GI.7, and GI.1, the two S domains of A–B dimers are in a bent arrangement and the C–C dimers are in plane (SI Appendix, Fig. S4). The N-terminal arm of GI.1 VP1 on the inner surface of the S domain has been suggested as a potential molecular switch that transitions between ordered and disordered states, where the ordered arm from the bent A–B dimer interacts with and stabilizes the neighboring flat C–C dimer conformation during T = 3 particle assembly (7, 17, 18). In the morphologically closely related tomato bushy stunt virus capsid with T = 3 symmetry, the arm of the C subunit wedges the two S domains apart in the C–C dimer interface to control the bent-to-flat conformation switching (17, 19). In our cryo-EM structures of GII.2, GI.7, and GI.1, the arm is ordered only on the B subunit that latches onto the neighboring C subunit.
In the large, T = 4, GII.4 particles, the two S domains of A–B dimers are bent as in the A–B dimers of T = 3 particles, and the C–D dimers are flat like the C–C dimers of those particles (SI Appendix, Fig. S4). However, the arm is disordered in all four subunits. It appears, therefore, that an ordered arm is not required to stabilize the flat dimer conformation in large norovirus particles. Indeed, truncation of the arm still resulted in self-assembly of GI.1 T = 3 particles, and the arm was dispensable for stabilization of the flat dimers in a previous study (18). In GII.2 small, T = 1, particles, all VP1 dimers are bent and the arms are disordered.
Protruding Spike Domain.
Variations in the spatial and angular orientations of subunits and their domains would have consequences in host interactions, such as bivalent Ig binding. The P domain of GII.4 is lifted significantly (∼4–28 Å) from the S domain through a long and flexible stalk region (Fig. 1). The P-domain densities were not as well resolved as those of the S domain. This may be due to the greater flexibility between the P domains and the shell in this assembly. Interactions between the elevated P domains form a double-layered structure with a distinct layer of spikes suspended above the icosahedral S-domain layer (Fig. 2). The dimeric P-domain spikes of A–B and C–D dimers are tilted toward one side (SI Appendix, Fig. S4). The distance between P and S domains is 16 and 24 Å on the A and B subunits, respectively, and 4 and 28 Å on the C and D subunits, respectively. On the other hand, the P domains of GII.2, GI.7, and GI.1 are placed close to the S domains, and the P1 subdomain makes contacts with the S domains of its own and of a neighboring subunit (Fig. 1). The lifted P-domain positioning and secondary-layer formation do not appear to be universally shared features among genogroup II strains. The P-domain spikes of GII.2 are twisted ∼25° counterclockwise relative to the orientation of GI.1 and GI.7 (Fig. 2). The spikes of GII.4 are twisted ∼40° clockwise relative to GI.1 and GI.7. Similarly lifted, twisted, and tilted P-domain positioning and orientations were previously reported in an 8-Å cryo-EM reconstruction of murine norovirus 1 native infectious particles in T = 3 form (13, 20).
Recently reported cocrystal structures of murine norovirus P domains in complex with their protein receptor, CD300lf, revealed the receptors bind at the interfaces between three P-domain spikes when superimposed on the map of the assembled virus (21–23). In GII.4 T = 4 particles, the interface between dimeric P-domain spikes forms around the 20 icosahedral threefold axes between D subunits, and the 60 quasi-threefold axes between A, B, and C subunits (Fig. 2). In the T = 3 particles of GII.2, GI.7, and GI.1, the equivalent interface surrounds the 60 quasi-threefold axes. Thus, a consequence of the larger, T = 4, form is the presence of a larger number of potential receptor attachment sites at the interfaces between three spikes, compared with those in T = 3 particles. On the other extreme, the spikes are placed significantly farther apart (∼10–25 Å) in the GII.2 T = 1 particle architecture, and no contacts are made.
Asymmetric Reconstruction Reveals a Zn2+ Ion That Affects Shell Stability.
Positioned at the outermost surface of the virus, the P2 subdomain bears many of the antigenic features involved in host interactions. Two hypervariable loops (loop A residues 378–381 and loop B residues 295–298) on the outermost apex of P2 in GII.2 are not observed in the existing P-domain crystal structure (11). These loops are only partially resolved in our reconstructions with symmetry imposed. However, subsequent symmetry expansion assigning and extracting 60 icosahedrally related views of the ASU from each whole-particle image, and signal subtraction outside of each ASU, followed by asymmetric focused reconstruction of the ASU enabled near-complete observation of the two missing loops (Materials and Methods and SI Appendix, Fig. S5).
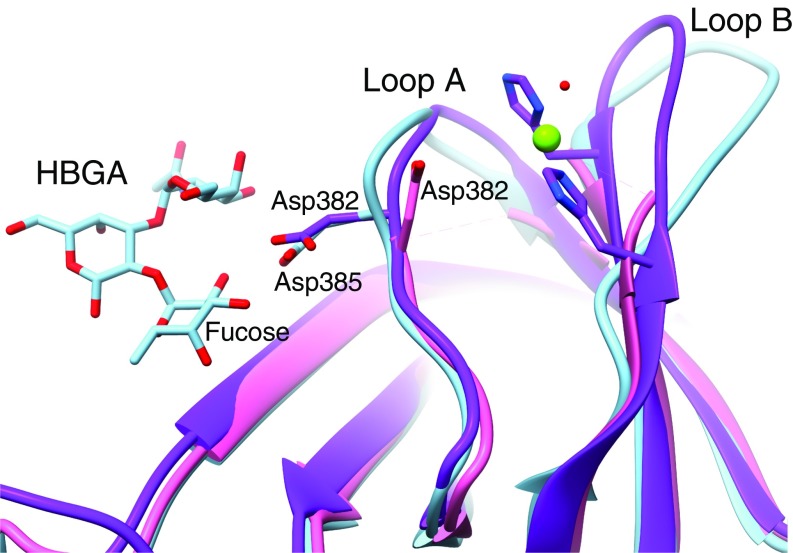
In HBGA-binding genogroup II strains, the conserved Asp382 (loop A) binds the fucose moiety of human HBGAs (14). In the GII.2 P-domain crystal structure, Asp382 was the last residue observed and the side chain was turned away ∼180° from the fucose binding site, which was proposed along with the dynamic loop movement as the possible molecular basis of weak or no HBGA binding by the GII.2 SMV strain. In our cryo-EM structures in the vitrified hydrated state, however, the Asp382 side chain is correctly oriented toward the fucose binding site on all subunits observed in a self-consistent manner (Fig. 4 and SI Appendix, Fig. S6).
Fig. 4.
Histo-blood group antigen binding site in GII.2. The P-domain crystal structure of GII.10 with HBGA bound (PDB ID code 3PA1) was superimposed on the partial P-domain crystal structure of GII.2 (PDB ID code 4RPB) and our GII.2 full-subunit cryo-EM structure without HBGA bound to show the expected positioning of the HBGA binding site of GII.2. In HBGA-binding strains such as GII.10 (light blue), a conserved residue (Asp385) binds the fucose moiety of HBGAs. In the P-domain crystal structure of GII.2 (pink), the side chain of the equivalent residue (Asp382) was turned ∼180° away from the fucose binding site, and the hypervariable loop A bearing Asp382 and loop B were not observed. In our cryo-EM structures (purple), the hypervariable loops A and B are resolved with the Asp382 side chain pointed correctly toward the fucose binding site. A Zn2+ metal ion (green sphere) is bound between loops A and B immediately adjacent to the fucose binding site that may play a role as a coligand that enhances HBGA binding of the GII.2 strain.
The two flexible loops appear to be held by a metal-ion density on each subunit. Although partial disorder around the dynamic loop regions limits precise positioning, the metal appears to be bound between the His293 and His299 side chains and a water molecule. The metal-binding site is immediately adjacent to the fucose-binding Asp382 residue. To confirm the identity and significance of this observed metal binding, we added EDTA, EGTA, or 1,10-phenanthroline to GII.2 VLP samples at the same concentration. The addition of either EDTA or 1,10-phenanthroline that preferentially chelates Zn2+ caused an over threefold decrease in the number of intact VLPs observed, and the metal density disappeared in the asymmetric and symmetric reconstructions from the remaining intact particles, whereas EGTA, which preferentially chelates Ca2+, did not cause such changes. The infectivities of murine norovirus and feline caliciviruses that do not bind HBGAs have been reported to be metal dependent (21, 22, 24). Moreover, GII.2 VLPs were reported to bind HBGAs when mixed with GII.2-positive patient stool, underscoring the involvement of coligands (25). This opens a possibility that metal ions may be required for the infectivity and stability of GII.2 SMV and possibly other strains by coordination between the flexible loops as an allosteric coligand, enhancing the human HBGA recognition and virus attachment.
The high-resolution structures of the human outbreak norovirus strain VLPs we have presented here serve as valuable templates for vaccine formulations and development of antivirals that inhibit host attachment and encapsidation. The structural polymorphism between and within genogroups should be valuable in guiding vaccine formulations that cover the broad spectrum of structural differences observed. Careful isolation of specific symmetry forms or a mixed sampling of all forms for vaccine manufacture may help to ensure maximum immunogenicity. The metal ions we identified present the surface antigen loops in the HBGA-binding conformations and allow raising neutralizing immunoglobulins. The addition of metal ions in the vaccine formulations would also be important in ensuring long-term structural integrity of the VLPs during storage of vaccines.
Materials and Methods
VLP Preparation.
The GI.1 Norwalk, GII.4 Minerva, and GII.2 Snow Mountain virus strain VLPs were expressed in tobacco leaves. The GI.1 and GII.4 VLPs were prepared commercially by Kentucky Bioprocessing, and the GII.2 VLPs were produced by Hugh S. Mason and Andrew G. Diamos, Arizona State University, Tempe, AZ (26). The GI.7 VLPs were a kind gift from Robert L. Atmar, Baylor College of Medicine, Houston, TX. The VLPs were purified further by size-exclusion chromatography using a Superose 6 Increase 10/300 column (GE) in 20 mM MES⋅OH (pH 5.75) and 50 mM NaCl, and the VLPs were collected from the void-volume fractions. The samples were concentrated to 3–5 mg/mL using centrifugal concentrators (Amicon).
Cryo-EM Sample Preparation.
Lacey carbon grids (300 mesh; Electron Microscopy Sciences) were glow discharged using an easiGlow glow discharger (Pelco). Aliquots (4 μL) of VLP samples were applied to the grids and blotted for 1.7 s using an EM GP2 blotting system (Leica) at 22 °C and 95% humidity level with 10-s preblot incubation time before plunging in liquid ethane for vitrification. For metal-free experiments, the blotting papers were pretreated with 5 mM EDTA and then thoroughly washed in ultrapure water and dried before blotting.
Data Collection.
Movies of VLPs embedded in vitreous solution were collected at liquid-nitrogen temperature using a Titan Krios G3 transmission electron microscope (ThermoFisher) equipped with a K2 Summit direct electron detector (Gatan) and a GIF Quantum LS imaging filter (Gatan). The movies were recorded in superresolution mode using EPU acquisition software (ThermoFisher) at 130,000× magnification with a pixel size of 0.535 Å per pixel, later resampled twofold to 1.07 Å per pixel, and nominal defocus range of 1.0–2.8 μm. The total electron exposure of each movie was ∼70–85 e/Å2 with a nominal exposure rate of 2.0 electrons⋅Å−2⋅s−1 per frame, fractionated into 35 movie frames with 200 ms per frame exposure time.
Data Processing.
Many of the software packages used for data processing were available through SBGrid (27). Beam-induced motion of particles was corrected using UNBLUR (cisTEM) on whole frames (28, 29). Contrast transfer function (CTF) parameters were estimated from sums of three movie frames using CTFFIND4 (cisTEM) (29, 30). Particles were automatically picked ab initio using soft-edged disk templates internally generated in cisTEM (29, 31). The picked particle images were boxed, extracted, and 2D-classified ab initio into 64 classes using cisTEM (SI Appendix, Fig. S7) (29). The initial 3D reconstructions were carried out ab initio, followed by 3D refinement using FrealignX (cisTEM) with refinement of CTF estimations (SI Appendix, Fig. S8) (29, 32). The numbers of movies and particles used toward final reconstructions are in SI Appendix, Table S1.
For asymmetric focused reconstruction, a 3D binary mask of an ASU was generated using Chimera and IMAGIC (33, 34). An inverse version of this mask was then applied to the symmetrical 3D reconstruction using apply_mask within cisTEM. This masked reconstruction was used to subtract out all but one ASU from the raw images in combination with symmetry expansion. For symmetry expansion, each image was subtracted 60 times using each of the 60 icosahedral ASUs to create a dataset of isolated asymmetric units, which were automatically centered. This dataset of centered isolated asymmetric units was further refined in cisTEM without applying symmetry. Symmetry expansion, signal subtraction, and ASU particle image cropping were carried out using the program symmetry_expand_stack_and_par, developed for this project and included with cisTEM.
The crystal structure of GI.1 (PDB ID code 1IHM) was docked into the GI.1 cryo-EM densities using MOLREP (CCP-EM) (35, 36) and rebuilt where needed. Water molecules were added using Coot to fit the cryo-EM densities (37). The S-domain structure of GI.1 was initially fitted into the GI.7 EM densities using MOLREP, and manually rebuilt with the correct amino acid sequence for GI.7 using Coot. A P-domain crystal structure of GI.7 (PDB ID code 4P26) was fitted into the EM densities using MOLREP, and modified using Coot to fit the EM densities.
The GII.2 S-domain atomic models were built manually into the EM densities using Coot. The partial crystal structure of GII.2 (PDB ID code 4RPB) was fitted using MOLREP, modified to fit the EM densities, and manually built in previously missing regions using Coot. The manually built GII.2 S-domain model was fitted into the GII.4 EM densities using MOLREP, and manually rebuilt with the correct amino acid sequence for GII.4 using Coot. A crystal structure of the GII.4 P domain (PDB ID code 5IYN) was fitted and modified into the P-domain EM densities using MOLREP and Coot.
All protein models were real space-refined using PHENIX (38), and evaluated using Coot and the MolProbity server (39). Water molecules were added based on identifying EM density peaks with appropriate shape and hydrogen-bond interactions. The symmetric and asymmetric reconstruction cryo-EM maps were deposited in the Electron Microscopy Databank, and the coordinates of the atomic models were deposited in the Protein Data Bank (40, 41). The figures were generated using UCSF Chimera and ChimeraX (33).
Further details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Kentucky BioProcessing (GI.1 Norwalk and GII.4 Minerva strains); Hugh S. Mason and Andrew G. Diamos (Biodesign Institute, Arizona State University; GII.2 SMV strain); and Robert L. Atmar (Baylor College of Medicine; GI.7 Houston strain) for producing the norovirus VLP samples. We thank Stephen C. Harrison, Stephen A. Johnston, and members of the L.J. lab for discussion and advice. D.R.T. and the cryo-EM facility are supported by the Cold Spring Harbor Laboratory. N.G. and L.J. are Investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and cryo-EM maps reported in this paper have been deposited in the Protein Data Bank (PDB) and Electron Microscopy Databank (EMDB) under the following ID codes: EMD-20199 for the GI.1 T = 3 symmetric reconstruction map, EMD-20205 for the GI.1 T = 3 asymmetric reconstruction map, and PDB 6OUT for the GI.1 T = 3 atomic coordinates; EMD-20197 for the GI.7 T = 3 symmetric reconstruction map, EMD-20198 for the GI.7 T = 3 asymmetric reconstruction map, and PDB 6OU9 for the GI.7 T = 3 atomic coordinates; EMD-20195 for the GII.2 T = 3 symmetric reconstruction map and PDB 6OTF for the GII.2 T = 3 atomic coordinates; EMD-20201 for the GII.2 T = 1 symmetric reconstruction map, EMD-20202 for the GII.2 T = 1 asymmetric reconstruction map, and PDB 6OUC for the GII.2 T = 1 atomic coordinates; and EMD-20206 for the GII.4 T = 4 symmetric reconstruction map and PDB 6OUU for the GII.4 T = 4 atomic coordinates.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903562116/-/DCSupplemental.
References
- 1.Centers for Disease Control and Prevention (CDC) , Norovirus worldwide. https://www.cdc.gov/norovirus/trends-outbreaks/worldwide.html. Accessed 22 May 2019.
- 2.Hall A. J., et al. ; Division of Viral Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention , Updated norovirus outbreak management and disease prevention guidelines. MMWR Recomm. Rep. 60, 1–18 (2011). [PubMed] [Google Scholar]
- 3.Kirk M. D., et al. , World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 12, e1001921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teunis P. F. M., et al. , Norwalk virus: How infectious is it? J. Med. Virol. 80, 1468–1476 (2008). [DOI] [PubMed] [Google Scholar]
- 5.de Graaf M., van Beek J., Koopmans M. P. G., Human norovirus transmission and evolution in a changing world. Nat. Rev. Microbiol. 14, 421–433 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Donaldson E. F., Lindesmith L. C., Lobue A. D., Baric R. S., Viral shape-shifting: Norovirus evasion of the human immune system. Nat. Rev. Microbiol. 8, 231–241 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad B. V. V., et al. , X-ray crystallographic structure of the Norwalk virus capsid. Science 286, 287–290 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Vongpunsawad S., Venkataram Prasad B. V., Estes M. K., Norwalk virus minor capsid protein VP2 associates within the VP1 shell domain. J. Virol. 87, 4818–4825 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conley M. J., et al. , Calicivirus VP2 forms a portal-like assembly following receptor engagement. Nature 565, 377–381 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Choi J. M., Hutson A. M., Estes M. K., Prasad B. V., Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U.S.A. 105, 9175–9180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh B. K., Leuthold M. M., Hansman G. S., Structural constraints on human norovirus binding to histo-blood group antigens. mSphere 1, e00049-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leroux-Roels G., et al. , Safety and immunogenicity of different formulations of norovirus vaccine candidate in healthy adults: A randomized, controlled, double-blind clinical trial. J. Infect. Dis. 217, 597–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katpally U., Wobus C. E., Dryden K., Virgin H. W. IV, Smith T. J., Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. J. Virol. 82, 2079–2088 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh B. K., Leuthold M. M., Hansman G. S., Human noroviruses’ fondness for histo-blood group antigens. J. Virol. 89, 2024–2040 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pogan R., Dülfer J., Uetrecht C., Norovirus assembly and stability. Curr. Opin. Virol. 31, 59–65 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Shoemaker G. K., et al. , Norwalk virus assembly and stability monitored by mass spectrometry. Mol. Cell. Proteomics 9, 1742–1751 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison S. C., The familiar and the unexpected in structures of icosahedral viruses. Curr. Opin. Struct. Biol. 11, 195–199 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Bertolotti-Ciarlet A., White L. J., Chen R., Prasad B. V. V., Estes M. K., Structural requirements for the assembly of Norwalk virus-like particles. J. Virol. 76, 4044–4055 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison S. C., Olson A. J., Schutt C. E., Winkler F. K., Bricogne G., Tomato bushy stunt virus at 2.9 Å resolution. Nature 276, 368–373 (1978). [DOI] [PubMed] [Google Scholar]
- 20.Hansman G. S., et al. , Structural basis for broad detection of genogroup II noroviruses by a monoclonal antibody that binds to a site occluded in the viral particle. J. Virol. 86, 3635–3646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilic T., Koromyslova A., Malak V., Hansman G. S., Atomic structure of the murine norovirus protruding domain and soluble CD300lf receptor complex. J. Virol. 92, e00413-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson C. A., et al. , Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor. Proc. Natl. Acad. Sci. U.S.A. 115, E9201–E9210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orchard R. C., et al. , Discovery of a proteinaceous cellular receptor for a norovirus. Science 353, 933–936 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Z., Ledgerwood E. D., Hinchman M. M., Dick R., Parker J. S. L., Conserved surface residues on the feline calicivirus (FCV) capsid are essential for interaction with its receptor feline junctional adhesion molecule A (fJAM-A). J. Virol. 92, e00035-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrington P. R., Vinjé J., Moe C. L., Baric R. S., Norovirus capture with histo-blood group antigens reveals novel virus-ligand interactions. J. Virol. 78, 3035–3045 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diamos A. G., Mason H. S., High-level expression and enrichment of norovirus virus-like particles in plants using modified geminiviral vectors. Protein Expr. Purif. 151, 86–92 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Morin A., et al. , Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell M. G., et al. , Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure 20, 1823–1828 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sigworth F. J., Classical detection theory and the cryo-EM particle selection problem. J. Struct. Biol. 145, 111–122 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Lyumkis D., Brilot A. F., Theobald D. L., Grigorieff N., Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 183, 377–388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 34.van Heel M., et al. , “Four-dimensional cryo-electron microscopy at quasi-atomic resolution: IMAGIC 4D” in International Tables for Crystallography, E. Arnold, D. M. Himmel, M. G. Rossmann, Eds. (John Wiley & Sons, 2012), vol. F, Crystallography of Biological Macromolecules, pp. 624–628.
- 35.Burnley T., Palmer C. M., Winn M., Recent developments in the CCP-EM software suite. Acta Crystallogr. D Struct. Biol. 73, 469–477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagin A., Teplyakov A., Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berman H. M., et al. , The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 58, 899–907 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Lawson C. L., et al. , EMDataBank unified data resource for 3DEM. Nucleic Acids Res. 44, D396–D403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.