Significance
Due to the steady increase of its atmospheric concentration, nitrous oxide (N2O) is among the most environmentally critical emissions of our time. It is generated both by abiotic and anthropogenic processes, but due to its chemical stability, nature has evolved only a single enzyme able to reduce the gas to uncritical N2. This nitrous oxide reductase bears a high biotechnological potential, but the complexity of the maturation pathways of its copper centers and its overall sensitivity have so far prevented its application. This work shows the recombinant production of N2O reductase in active and intact form in Escherichia coli, its structural and spectroscopic characterization, and an initial application for elucidating details of cofactor assembly.
Keywords: nitrous oxide reductase, denitrification, enzyme refactoring, cofactor biogenesis, structural biology
Abstract
The multicopper enzyme nitrous oxide reductase reduces the greenhouse gas N2O to uncritical N2 as the final step of bacterial denitrification. Its two metal centers require an elaborate assembly machinery that so far has precluded heterologous production as a prerequisite for bioremediatory applications in agriculture and wastewater treatment. Here, we report on the production of active holoenzyme in Escherichia coli using a two-plasmid system to produce the entire biosynthetic machinery as well as the structural gene for the enzyme. Using this recombinant system to probe the role of individual maturation factors, we find that the ABC transporter NosFY and the accessory NosD protein are essential for the formation of the [4Cu:2S] site CuZ, but not the electron transfer site CuA. Depending on source organism, the heterologous host E. coli can, in some cases, compensate for the lack of the Cu chaperone NosL, while in others this protein is strictly required, underlining the case for designing a recombinant system to be entirely self-contained.
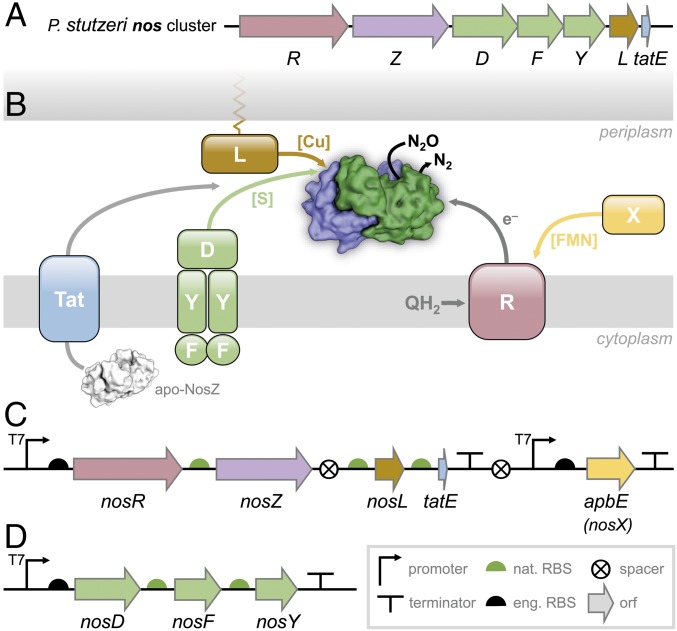
Nitrous oxide (N2O, “laughing gas”) is a potent greenhouse gas whose 100-y global warming potential exceeds the one of carbon dioxide (CO2) by a factor of 300 (1). Its atmospheric concentration increased by ∼20% since preindustrial times and is still growing at a rate of 0.2–0.3% y−1 (1, 2). This led to its designation as the most significant ozone-depleting substance of the 21st century, which it is expected to remain unless its emissions are put under tight regulation (3). More than two-thirds of N2O emissions originate from the by-products of bacterial and fungal soil nitrification and denitrification that are strongly enhanced by excessive fertilization in modern agriculture (1, 4). Although N2O reduction is thermodynamically favored (N2O + 2H+ + 2 e− → N2 + H2O; ΔG0′ = −339.5 kJ·mol−1), its activation energy barrier of 250 kJ·mol−1 leads to substantial kinetic stability and chemical inertness (4, 5). Accordingly, the biological reduction of N2O that completes denitrification is catalyzed by a specialized enzyme, nitrous oxide reductase (NosZ, the product of the nosZ gene), a periplasmic, homodimeric metalloprotein of 130 kDa that contains two copper centers, CuA and CuZ, in each monomer (6). It forms a tight head-to-tail dimer in which the CuA site of one monomer is in close proximity of about 10 Å to the CuZ site of the other, creating the composite active site of the enzyme at each dimer interface (4). CuA is a binuclear mixed-valent [Cu1.5+:Cu1.5+] site with two cysteines, two histidines, one methionine, and one tryptophan as ligands, able to accept and transfer a single electron (7), and CuZ is a tetranuclear [4Cu:2S] cluster coordinated by a unique, asymmetric histidine heptad that binds and activates N2O during catalysis (4). Initial 3D structures of NosZ from Marinobacter hydrocarbonoclasticus (8), Paracoccus denitrificans (9), and Achromobacter cycloclastes (10) consistently contained CuZ as a [4Cu:μ4-S] center, i.e., four Cu ions with a central, interstitial sulfide, while in Pseudomonas stutzeri ZoBell, the tetranuclear cluster contained an additional sulfide, revealing the complete, native [4Cu:2S] configuration (4). This form, CuZ, is readily desulfurylated during protein isolation to yield a [4Cu:S] form termed CuZ* (11). Beside its two copper centers, NosZ also binds one ion each of Ca2+, K+, and Cl− per monomer (4, 12). Currently, NosZ is the only known enzyme able to activate inert nitrous oxide. Its reaction mechanism involves both metal sites but remains to be fully elucidated. In the P. stutzeri genome, the gene nosZ forms part of the nos gene cluster (nosRZDFYL) (Fig. 1A), where the preceding NosR is a polytopic membrane protein that serves as electron donor for N2O reduction (13). Its periplasmic, N-terminal FMN-binding domain requires covalent flavinylation by an ApbE family protein forming part of the gene cluster as nosX in some organisms (14), while a C-terminal ferredoxin-like domain with two [4Fe:4S] clusters resides in the cytoplasm (5). The following ORFs, nosDFY, encode an ABC transporter (NosFY) and a periplasmic interacting protein, NosD, presumably required to shuttle a sulfur species to the periplasm for CuZ assembly (5, 15). NosL is a membrane-anchored copper chaperone that binds Cu1+ for delivery to apo-NosZ (Fig. 1B) (16, 17).
Fig. 1.
The nos cluster of P. stutzeri and its refactoring for recombinant production in E. coli. (A) In the nos cluster of P. stutzeri ZoBell, nosR encodes an integral membrane protein required for electron delivery and function, nosZ is the structural gene for N2O reductase. The ABC transporter complex NosDFY is required for sulfur delivery and NosL is a copper chaperone, and the tatE gene supports Tat-dependent export of apo-NosZ. (B) The protein products of the nos cluster comprise a functional system arranges around the cytoplasmic membrane, with N2O reductase located in the periplasm. In addition to the genes encoded in the cluster, the maturation of NosR additionally requires the flavinyltransferase AbpE that is present in some orthologous gene clusters as nosX. (C) Architecture of the construct pLZPs1containing two synthetic operons for nosRZLtatE and for abpE. The native promoter for NosR was removed for joint transcription, but the original ribosome binding sites for nosZ, nosL, and tatE were retained. (D) Construct pLZPs2 exclusively comprises the NosDFY complex required for sulfur delivery during CuZ maturation. Symbols in C and D are according to Synthetic Biology Open (http://sbolstandard.org).
Incomplete denitrification terminating with release of N2O is a major contributor to the detrimental environmental effects of excessive fertilizer use, and consequently the application of recombinant N2O reductase (rNosZ) in a suitable host is of major interest for bioremediatory applications. Besides current N2O mitigation strategies based on soil chemistry and plant community technologies (2), emerging approaches aim at harnessing protein chemistry and microbiome biotechnology (18). A prerequisite for such strategies is the availability of a recombinant, synthetically refactored enzyme produced in a heterologous host. Although homologous expression systems for N2O reductase were reported for P. stutzeri (19) and A. cycloclastes (20), and expression of the nos gene cluster of P. stutzeri in nondenitrifying Pseudomonas putida resulted in holo-NosZ (21), attempts to heterologously produce P. stutzeri (22) and Shewanella denitrificans (12) NosZ in E. coli only yielded apo-proteins. To obtain functional rNosZ, we expressed the full complement of genes required for the production, maturation, and activity maintenance of the enzyme from the model denitrifier P. stutzeri ZoBell in E. coli.
Results and Discussion
Functional Production of P. stutzeri NosZ Requires the Nos Cluster as Well as the Flavinyltransferase ApbE.
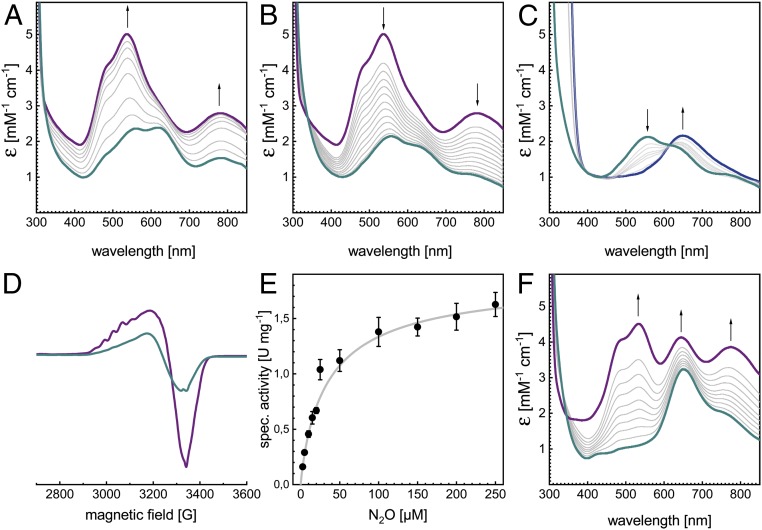
The P. stutzeri nos gene cluster contains three independent promoters for nosR, nosZ, and nosDFYL, with coupled translation of NosDFYL (15). A two-plasmid system was employed that contained nosRZL and apbE in pLZPs1, a pET22-based vector under control of two T7 promoters (Fig. 1C), and the nosDFY genes in a pET30-based vector, pLZPs2 (Fig. 1D). The ribosome binding sites (RBS) for nosR, nosD, and apbE originated from the pET vectors, while those for nosZ, nosFY, and nosL were from the native sequences and had been checked for compatibility with E. coli. The expression of all accessory proteins was verified by protein purification and mass spectrometry (SI Appendix, Fig. S1). Unsurprisingly, recombinant rNosZ protein was isolated from E. coli as a dimer of 132.5 kDa, in agreement with dimeric apo-PsNosZ and apo-SdNosZ isolated from E. coli previously (12, 22), providing further evidence that the tight dimer formation of NosZ precedes copper site maturation and likely also the Tat-dependent export to the periplasm. Oxic isolation yielded recombinant rNosZ of blue color, with an electron excitation spectrum indicative of partial oxidation of both CuA and CuZ (Fig. 2 A, blue). The exact redox state of the protein as isolated varied between preparations and was dependent on the temperature and duration of the isolation procedure. Complete oxidation with potassium ferricyanide led to a color change to purple, with absorption maxima at 538 nm and 780 nm and a shoulder at 485 nm (Fig. 2 A, purple), the defining spectral features of form I N2O reductase, with CuZ as a [4Cu:2S] cluster (4, 5, 11). From here, the addition of ascorbate resulted in the exclusive reduction of CuA, retaining a CuZ spectrum with two distinct bands at 562 nm and 625 nm (Fig. 2B), and further reduction with dithionite depleted the transition at 562 nm, leading to a single charge transfer peak at 650 nm assigned to the [3Cu+:1Cu2+] state of CuZ. This transition was direct, as evidenced by an isosbestic point at 615 nm (Fig. 2C) (11). The 7-line hyperfine pattern of the X-band electron paramagnetic resonance (EPR) spectrum (Fig. 2D) confirmed the mixed-valent [Cu1.5+:Cu1.5+] state of oxidized CuA (7, 11). These are the exact spectral features defining the native form I of the enzyme (5, 23). The molar extinction coefficient ε538 nm of 5 mM−1·cm−1 was only the half of the one reported for anoxically isolated PsNosZ (4). Nevertheless, anoxic isolation of rNosZ from E. coli yielded a similar ε538 nm of 6 mM−1·cm−1 (SI Appendix, Fig. S2). Consequently, and in line with the 3D structure, the recombinant enzyme does not contain a full complement of copper, but this partial depletion cannot be traced back to a difference between oxic and anoxic handling. We then determined the N2O-reducing activity of the oxically isolated rNosZ using reduced benzyl viologen as an electron donor (Fig. 2E). The obtained vmax of 1.73 ± 0.05 µmol N2O min−1·mg−1 and KM (N2O) of 25.6 ± 2.3 µM were comparable to those of PsNosZ isolated either oxically or anoxically from various strains (13, 21) without addition of base or reductive activation (Table 1) (24). KM values for other orthologs (A. cycloclastes NosZ: 25 µM, M. hydrocarbonoclasticus NosZ: 14 µM) were reported to be very similar (24). rNosZ from E. coli thus is intact and active, with properties very similar to the natively produced enzyme, also sharing its reduced complement of copper.
Fig. 2.
Characterization of NosZ recombinantly produced in E. coli. The protein eluted as a single, symmetric peak from size-exclusion chromatography and was characterized spectroscopically and biochemically. (A) As isolated, rNosZ was typically in a mixed oxidation state (cyan) that upon oxidation with K3Fe(CN)6 showed the properties of a form I N2O reductase (purple). (B) Reduction with ascorbate selectively removes the contribution of CuA, revealing that CuZ is in the [4Cu:2S] state, as indicated by two bands at 550 and 650 nm (cyan). (C) Upon extended reduction with Na2S2O4, the CuZ spectrum transitions to a single maximum at 650 nm, indicating the reduction from [2Cu2+:2Cu+] to [1Cu2+:3Cu+] (blue). (D) X-band EPR spectrum of rNosZ at 40 K in the as isolated (cyan) and oxidized form (purple). The seven-line hyperfine pattern originates from a mixed-valent CuA center. (E) Specific activity of isolated rNosZ with benzyl viologen as electron donor. (F) In some instances, the isolated protein (cyan) was obtained as form II, as evidenced by a single peak at 650 nm and also reflected in the 3D structure (Fig. 3). Oxidation with K3Fe(CN)6 yielded the pink/purple form II of NosZ (purple).
Table 1.
Role of accessory factors in the maturation of NosZ
| Accessory factors tested | Constructs used | Yield mg of protein/g of cells | Cu/monomer* | CuA | CuZ/CuZ* | Activity | |||
| pET22 | pET30 | vmax† | KM, μM | ||||||
| I | NosRDFYLX‡ | nosRZLX | NosDFY | 0.11 | 4.0 | + | + | 1.73 ± 0.05 | 25.56 ± 2.32 |
| II | ΔNosRX | nosZL | NosDFY | 0.80 | 3.4 | + | + | 2.08 ± 0.10 | 16.18 ± 3.01 |
| III | ΔNosDFY | nosZL | — | 0.83 | 1.2 | + | − | — | — |
| IV | NosDF*Y§ | nosZL | nosDF*Y§ | 0.83 | 0.8 | + | − | — | — |
| V | ΔNosL | nosZ | nosDFY | 0.57 | 2.4 | + | + | 1.51 ± 0.06 | 29.78 ± 4.21 |
Cu content as derived from A538nm of UV/vis spectra, w.r.t. to the Cu content of form I rNosZ as determined by ICP/MS.
Specific activity in μmol N2O·min−1·(mg protein)−1 (31).
As “NosX,” the orthologous apbE gene from P. stutzeri was used.
F* is an inactive E154Q variant of NosF.
Structural Analysis of Recombinant P. stutzeri N2O Reductase.
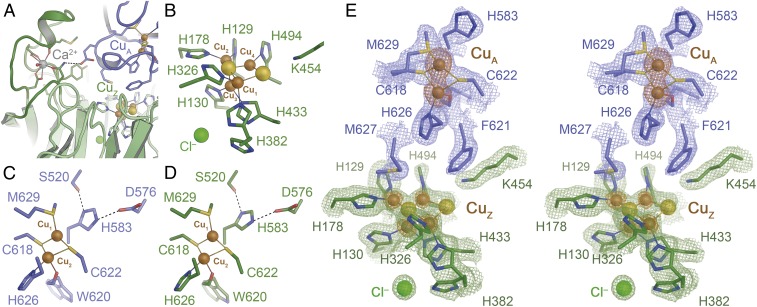
We proceeded to crystallize the protein and determined its 3D structure to 1.78 Å resolution (SI Appendix, Fig. S3 and Table S4). The fold and dimer structure of rNosZ (25) are unchanged from the native protein, with a root-mean-squared deviation of 0.2 Å for all atoms. In rNosZ, the binding of Ca2+ to a peripheral loop region in the CuZ domain (Y252–D269) stabilizes the protein as a final step of metal site assembly (12). The ion was present in the structure, and the protein backbone was entirely in the conformation observed in Ca2+-bound structures (12). The CuA ligands C618, W620, W622, H626, and M629 were in place to coordinate two copper ions, while H583, the remaining ligand to ion CuA2, was observed consistently in a conformation where it does not ligate CuA2, but rather forms a short hydrogen bond to residue D576 (Fig. 3B). This histidine flip at CuA is suggested to play a role in gating electron transfer from an external redox partner to the active site via CuA but has so far only been reported for the P. stutzeri enzyme (4).
Fig. 3.
Three-dimensional structure of rNosZ. The recombinant protein fully retained the homodimer structure of all known NosZ proteins (SI Appendix, Fig. S2). (A) The Ca2+ site fixing a loop in the β-propeller domain in mature N2O reductase following Cu insertion. (B) The CuZ site is observed in the [4Cu:2S] state indicative of form I of the enzyme. (C) The dinuclear CuA site in the A chain of the rNosZ dimer. (D) The CuA site in chain B of the dimer. In both cases, ligand His583 is rotated away from ion CuA1, a conformation that likely hinders electron transfer to the metal centers of the enzyme. (E) Stereo representation of the electron density map surrounding the CuA and CuZ sites of rNosZ form I. The blue and green maps are 2Fo − Fc difference electron density maps are contoured at the 1 σ level for the two chains of the homodimer (blue, green), and an anomalous difference Fourier map is shown at the 3.5 σ level (orange), highlighting the lower copper content of the CuZ site with respect to CuA (SI Appendix, Fig. S4).
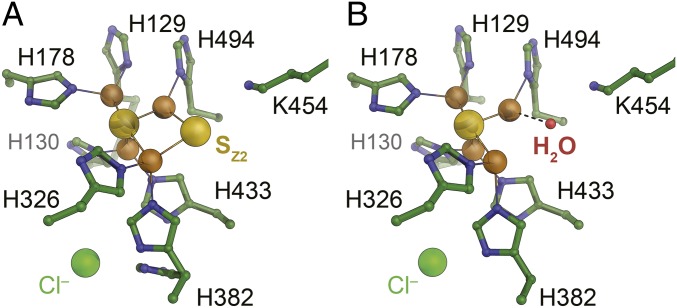
With the exception of minor dual conformations for H382 and H494, the seven histidine ligands of the CuZ site retained their orientations from the native structure, and the N2O binding position (PDB ID code 3SBR) was occupied by a water molecule. Anomalous difference Fourier maps unambiguously confirmed the correct positioning and presence of all copper ions in CuA and CuZ (Fig. 3E and SI Appendix, Fig. S4). The refined occupancies of the copper ions were 0.9–1 for CuA, but only 0.5–0.6 for CuZ, adding up to ∼8 Cu per dimer. This in in line with copper contents reported previously for native PsNosZ (23), where it has been emphasized that incomplete assembly also occurs in P. stutzeri itself (possibly only during isolation). In our preparations, copper quantification by inductively coupled plasma emission mass spectrometry yielded 7.6 Cu per dimer, so that rNosZ comprises a fully assembled CuA site and half a complement of the [4Cu:2S] CuZ cluster (SI Appendix, Fig. S4). There is currently no convincing explanation for the incomplete assembly of the CuZ site in P. stutzeri ZoBell, and the observation that this occurs in our recombinant system as much as it does in native preparations from the pseudomonad indicate that it is a feature of this particular protein. This ortholog has otherwise proven to be highly suited for refactoring in E. coli, and the knowledge gained here opens up a route toward further optimizations that include the screening for other enzymes with tighter copper binding, as well as protein design or evolutionary strategies to improve enzymatic activities. The recombinant preparation of rNosZ also showed electron density for the two labile sulfides associated with the [4Cu:2S] state that characterizes form I of the enzyme. Sulfide SZ1, located centrally in contact with all four Cu ions, was well defined in all four monomers in the asymmetric unit of the crystal structure, while the more labile SZ2 was found to bridge ions CuZ1 and CuZ4 in two monomers, but situated more closely to CuZ4 in the other two. Nevertheless, the CuZ site gave rise to the distinct absorption band at 550 nm that is associated with the presence of SZ2 (4, 6). The variable integrity of the CuZ site was further highlighted by the fact that individual preparations of the enzyme showed different spectral properties, corresponding to form II of the enzyme, in which the CuZ site has decomposed to a [4Cu:S] cluster due to the loss of SZ2 (Fig. 2F). The crystal structure of this form of recombinant NosZ was determined to 1.6 Å resolution, revealing that indeed the bridging SZ2 was absent and was replaced by a water ligand to CuZ4 (Fig. 4) (26). This variable forms of the enzyme upon isolation and the differences in occupancy of SZ2 are also observed in native purifications, emphasizing once more that the recombinant production system is a highly suitable tool to further investigate CuZ site assembly in the well-established background of the laboratory paradigm E. coli.
Fig. 4.
Forms I and II of rNosZ produced in E. coli. The observed difference in the spectroscopic signatures of individual preparations of rNosZ reflect in the composition of CuZ observed in the 3D structures. (A) Typically, isolated rNosZ contains a [4Cu:2S] center, in line with UV/vis properties of a form I enzyme (Fig. 3A). (B) In cases where spectroscopy indicated that the enzyme was obtained as form II (Fig. 2F), this was corroborated by the replacement of the bridging SZ2 sulfide for a terminal H2O molecule, leading to the [4Cu:S] center designated CuZ*.
rNosZ as a Tool for Studying Metal Site Assembly.
As a means of probing the exact role of individual maturation factors for nitrous oxide reductase, we then generated a series of knockout variants (Table 1). rNosZ isolated from a ΔnosR:ΔapbE background retained activity in vitro, although its ε538 nm was decreased with respect to that of sample I (SI Appendix, Fig. S5). This can likely be attributed to removing the effort required by the cell to produce and mature the large membrane-integral NosR protein, in line with earlier results on truncated NosR that had shown it to be dispensable for N2O reductase maturation, but had resulted in the loss of N2O-reducing activity of whole cells (13). This further supports an exclusive role for NosR as an in vivo electron donor for the reaction, making this protein the key factor to address in the next step, when rNosZ is to be coupled to the energy metabolism of E. coli to obtain in vivo activity.
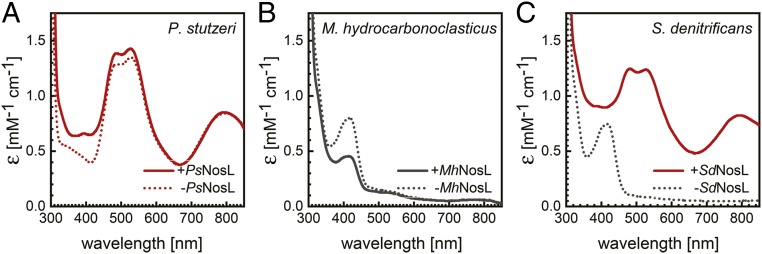
When in addition the nosDFY genes from pLZPs2 were omitted, or an inactive E154Q variant of the ATP-binding module NosF was used, the resulting rNosZ protein (samples III and IV) only contained CuA, but was fully deplete of CuZ (SI Appendix, Fig. S6). NosDFY is thus required exclusively for CuZ assembly, in line with its proposed role in the export of a sulfur species for the cluster from the cytoplasm (5, 21). Interestingly, under oxic conditions, the isolated proteins were colorless, i.e., contained reduced CuA, and were only slowly oxidized by dioxygen during several days of exposure to air. It is unclear how N2O reductase achieves this level of O2 resistance and maintains its reduced state in the oxidizing environment of the periplasm of E. coli. Unexpectedly, the deletion of the nosL gene encoding the copper chaperone still allowed for the assembly of active rNosZ in E. coli, if pLZPs1 was present for CuZ assembly (SI Appendix, Fig. S7). A similar finding was reported for the heterologous expression of nos genes from P. stutzeri in nondenitrifying P. putida (21), where copper delivery had been attributed to the presence of an intrinsic, homologous chaperone, and the same might be true in the present case. We then followed up to investigate the role of NosL by coexpression of further NosZ homologs exclusively with their respective NosL proteins in E. coli (Fig. 5). Results were diverse. While P. stutzeri NosZ contained an intact CuA site regardless of nosL coexpression, M. hydrocarbonoclasticus NosZ did not assemble the site either way (Fig. 5B). In contrast, S. denitrificans NosZ only featured an intact CuA site if its cognate SdNosL was coproduced (Fig. 5C). The intrinsic copper maturation machinery of E. coli thus seems to be compatible with PsNosZ, while MhNosL was produced (SI Appendix, Fig. S8) but could not either receive Cu or deliver it to MhNosZ. SdNosL, in contrast, worked in E. coli but could not be replaced by the host’s intrinsic system that had worked for PsNosZ. This represents a striking example for the complex problems that can be encountered during heterologous refactoring of complex proteins. Future studies will address the physiological role of NosL in detail, and in particular a crystal structure of the holoprotein should be instrumental for better understanding the mechanism of Cu site assembly in NosZ.
Fig. 5.
Maturation of N2O reductase in E. coli and the role of NosL. (A) Unexpectedly, the copper chaperone NosL was not required to assemble an intact CuA site in recombinant PsNosZ in E. coli, indicating that an intrinsic factor of the host organism can replace NosL. (B) In contrast, CuA of MhNosZ is not assembled correctly in E. coli, independent of whether NosL from the same organism is coproduced. (C) Finally, in S. denitrificans, a third denitrifier we tested, SdNosZ was produced as an apoprotein in the absence of NosL, but coexpression of the corresponding SdNosL resulted in a correctly assembled CuA center. Samples in all graphs were oxidized with K3Fe(CN)6.
Conclusions
We report on the functional maturation of a nitrous oxide reductase in E. coli and elaborate on the role of its maturation factors, revealing a strict requirement for NosL and NosDFY for the maturation of the Cu sites. In contrast, the integral membrane protein NosR and its maturation factor ApbE (NosX) were not found to be indispensable for NosZ maturation. Although NosR was previously implied to be involved in metal site biogenesis, our data speak for an exclusive role in electron transfer to NosZ in vivo. Interestingly, NosL proved to be nonessential for P. stutzeri NosZ when produced in E. coli, but the same was not true for the orthologs from M. hydrocarbonoclasticus and S. denitrificans. The two-plasmid system reported here is a functional and host-independent platform for the recombinant production of a complex metalloenzyme of high biotechnological relevance. This study is expected to facilitate future work on elucidating the mechanism of N2O reduction and on enzyme engineering through a combination of environmental chemistry and synthetic biology, aiming for the creation of biotechnological tools for the efficient bioremediation of N2O emissions.
Materials and Methods
Generation of Expression Constructs.
Bacterial strains, plasmids, and primers used in this study are listed in SI Appendix, Tables S1–S3. The work was based on plasmid pPR6hE that contains all nos genes of P. stutzeri strain ZoBell (13). Primers ZL141/ZL142 were used to add a StrepTag(II) sequence (WSHPQFEK) to the C terminus of nosZ, yielding plasmid pPR6hE-Zs. The entire nos gene cluster was then inserted into a pET30a(+) vector (Novagen) by FastCloning (27) as follows: primers ZL143/ZL144 were used to amplify the pET30a(+) backbone by PCR, and likewise primers ZL145/ZL146 were used for the nosRZDFYLtatE genes from pPR6hE-Zs. Subsequently, the two fragments were joined to generate plasmid pET30-nos(P). Primers ZL153/154 were used to excise nosDFY from pET30-nos(P) to generate pET30-nosRZL(P) by PCR. A 2.8-kb PCR fragment obtained from pET30-nosRZL(P) as a template with primers ZL161/162 was digested with NdeI and BlpI, and then ligated into pET22b(+) (Novagen) to generate pET22-nosZL(P). Primers ZL181/ZL182 were used for removing nosL from pET22-nosZL(P), generating pET22-nosZ(P) by PCR. Primers ZL143/ZL155 were used to cut nosRZ from pET30-nos(P) to form pET30-nosDFYL(P) by PCR, and primers ZL181/ZL182b were used to remove nosL from pET30-nosDFYL(P) to generate pET30-nosDFY(P), designated pLZPs2 for the functional production system. Finally, primers CM001/CM002 were used to create a defective variant of NosF, introducing an E154Q mutation in the ATP binding site by site-directed mutagenesis using pET30-nosDFY(P) as to generate pET30-nosDF*Y(E154Q). Plasmid pET22-nosRZLX(P), designated pLZPs1 for the functional production system, was constructed as follows: the attB site of the ϕBT1 integration system (28) was inserted downstream of the T7 terminator of pET30-nosRZL(P) by PCR, using primers ZL205/ZL206 to generate pET30-nosRZL(P)attB, and also upstream of the T7-promoter of pET22-apbE(P) (14) to generate pET22-apbE(P)attP. The two plasmids pET30-nosRZL(P)attB and pET22-apbE(P)attP were subsequently combined by site-specific recombination via ϕBT1 integrase (28). After digestion with ApaI and self-ligation, the final plasmid pLZPs1 was obtained.
All of the nos genes of S. denitrificans OS217 used in this study were codon-optimized for expression in E. coli (LifeTechnologies). Vector pET21a-nosZ(syn) containing a Strep-tagged nosZ gene of S. denitrificans was constructed previously (12). Plasmid pET30S-nosL(syn), containing the Strep-tagged nosL gene of S. denitrificans, was constructed as follows: a PCR fragment containing nosR of S. denitrificans was amplified using pET21a-nosR(syn) as template with primers ZL11/Strep-R and digested with NdeI/SalI, then ligated into pET30a(+) (digested with NdeI and XhoI) to generate pET30S-nosR(syn); a PCR fragment containing nosL of S. denitrificans was amplified using pET21a-nosL(syn) as a template with primers ZL29a/T7-ter and digested with NcoI/XhoI, then ligated into pET22b(+)to generate pET22b-nosL(syn) (the original signal peptide was removed and the lipid anchor was mutated in this step); then both pET30S-nosR(syn) and pET22b-nosL(syn) were digested with NdeI/XhoI, and the fragment containing nosL was ligated into a modified pET30 vector containing a Strep-tag to generate pET30S-nosL(syn).
For cloning the nosZ gene of M. hydrocarbonoclasticus DSM-8798, primers ZL197 and ZL198 were used for amplification from genomic DNA, and the PCR fragment was digested with NdeI/XhoI and ligated into pET22b(+) to generate pET22-nosZ(M). Primers ZL199 and ZL200b were used for insertion of a Strep-Tag(II) sequence into pET22-nosZ(M) to generate pET22S-nosZ(M). Primers ZL49 and ZL50 were used to amplify the M. hydrocarbonoclasticus nosL gene from genomic DNA, then the fragment was digested with MscI/XhoI and ligated into pET22b(+) to generate pET22-nosL(M); the original signal peptide sequence was replaced by a pelB leader sequence, and the lipid anchor was mutated. Both pET30S-nosL(syn) and pET22-nosL(M) were digested with NdeI/XhoI, then the fragment containing M. hydrocarbonoclasticus nosL was ligated into a pET30S vector to generate pET30S-nosL(M).
Protein Production and Isolation.
Chemically competent cells of E. coli BL21 (DE3) strain C43 (29) were transformed with the expression vectors for protein production, pLZPs1 and pLZPs2. Bacteria were grown at 37 °C in 2×YT medium supplemented with 100 µg·mL−1 ampicillin and/or 50 µg·mL−1 of kanamycin to an OD600 of 0.6. Isopropyl-β-d-1-thiogalactopyranoside was added to a final concentration of 1 mM to induce gene expression, and 0.25 mM CuSO4 or CuCl2 were supplemented for copper site maturation in overexpressed NosZ proteins. When coexpressing nosR and apbE, 50 µg·mL−1riboflavin was added as well. After further cultivation at 30 °C for 12 h with reduced agitation at 100 rpm, the cells were harvested by centrifugation for 15 min at 5,000 × g. The cells were resuspended in lysis buffer [100 mM Tris/HCl buffer at pH 8.0, 150 mM NaCl and 10% (v/v) glycerol] and disrupted by three passages through a microfluidizer (Microfluidics M-110P) at 1,000-bar pressure. The crude extract was cleared by centrifugation at 100,000 × g for 1 h, and the supernatant was loaded onto a Strep-Tactin Superflow cartridge [5 mL of bed volume, d-Desthiobiotin (IBA)]. Target proteins were eluted with lysis buffer containing 5 mM IBA and further separated by size-exclusion chromatography (Superdex S200 26/60, GE Healthcare) equilibrated in 20 mM Tris/HCl buffer at pH 8.0, 150 mM NaCl. Pure protein was concentrated by ultrafiltration, flash-frozen in liquid nitrogen and stored at −80 °C until further use. Protein was determined by the bicinchoninic acid method, using bovine serum albumin as a standard (30).
For anoxic sample preparation, oxygen was removed from all buffers. All steps were carried out under strict exclusion of dioxygen, either in an anoxic chamber containing an atmosphere of 95% N2/5% H2 at <1 ppm of O2 or using modified Schlenk techniques with nitrogen as inert gas. Cells were harvested and resuspended in anoxic lysis buffer in a glovebox and, subsequently, ruptured under exclusion of O2 in an Emulsiflex C5 homogenizer (Avestin) at 15,000 psi.
Analytical Size-Exclusion Chromatography.
Analytical size-exclusion chromatography was used to determine the oligomeric state and apparent molecular weight of PsNosZ. Triplicate chromatography runs on a Superdex 200 10/300GL column (GE Healthcare) with PsNosZ at concentrations of 1–4 mg·mL−1 were carried out on a Viscotek GPCmax VE20001 GPC solvent/sample module, integrated with a Viscotek TDA305 Triple Detector Array (refractive index, light scattering and viscosity), and a UV 2600 detector module (Malvern) (14).
Electron Excitation (UV-Visible) Spectroscopy.
UV-visible (UV/vis) absorption spectra of purified proteins (0.1 mM in 20 mM Tris/HCl buffer at pH 8.0, 150 mM NaCl) were carried out in an anoxic chamber (Coy Laboratories) and recorded over the scan range 200–900 nm using a USB 4000 Spectrometer with USB-ISS-UV/VIS Integrated Sampling System (Ocean Optics). Oxidation of protein samples was achieved by adding an increasing amount of potassium ferricyanide, stepwise reduction was done by adding sodium ascorbate for CuA, then further with sodium dithionite.
EPR Spectroscopy.
EPR spectra were recorded using a continuous-wave X-band EPR-spectrometer (Elexsys E-500, 10″ ER073 electromagnet, super high Q resonator cavity, Bruker) at a microwave frequency of 9.37 GHz, equipped with a continuous-flow liquid helium cryostat (ER 41112HV, Oxford Instruments). Three hundred microliters of as isolated or ferricyanide-oxidized rNosZ with a concentration of 47 mg·mL−1 were filled into an X-band EPR tube and flash-frozen in liquid nitrogen. The measurement was carried out at a temperature of 40 K and a power of 1 mW with a modulation amplitude of 4 G and a receiver gain of 60 dB.
Nitrous Oxide Reductase Activity Assay.
The activity of purified N2O reductase was determined spectrophotometrically with Ti(III)-reduced benzyl viologen (31) in an anoxic chamber at 25 °C. One hundred micromolar benzyl viologen was reduced with 80 µM titanium citrate in 10 mM potassium phosphate buffer at pH 8.5, and subsequently 0.5–1 µM of purified N2O reductase was added. Absorption of the reaction mixture was monitored at 578 nm for 2 min, then the reaction was initiated by the injection of dissolved N2O in a concentration range of 2.5–250 µM. For this, the solubility of N2O in water was taken as 25 mM at 1 atm, 25 °C (32). All measurements were made at least in triplicates.
Crystallization and Structure Determination.
rNosZ was crystallized by sitting drop vapor diffusion at 20 °C, using protein with a concentration of 15 mg·mL−1 in size exclusion chromatography buffer. Drops with a total volume of 0.6 µL with different protein:reservoir ratios (1:2, 1:1, 2:1) were equilibrated against 50 µL of a reservoir solution (50 µL) containing 0.1 M Bis-Tris propane buffer at pH 8.5, 0.1 M sodium formate, 0.1 M sodium chloride, and 25% (w/v) of a medium molecular weight (MMW) polyethylene glycol mixture (PEG 2K, 3350, 4K and 5K MMW) from a PEG Smears screen (33), using an automatic crystallization drop-setting system (Oryx Nano, Douglas Instruments). Single, purple crystals were obtained in combination with microseeding within 1 wk and diffracted to below 2 Å. The PEG concentration of the reservoir solution was allowed to directly mount the crystals in nylon loops for flash-cooling in liquid nitrogen before data collection. A dataset to better than 1.8 Å resolution was collected on beamline X06DA of the Swiss Light Source (Villigen, Switzerland) with a PILATUS 2M pixel detector (Dectris). Diffraction data were processed with XDS (34). Crystals of PsNosZ belonged to the monoclinic space group P21, with unit cell dimensions a = 108.9 Å, b = 73.3 Å, c = 136.3 Å, α = γ = 90.0° and β = 95.1°. The asymmetric unit contained four monomers of the protein at a solvent content of 39.5%.
The crystal structure of rNosZ was solved by molecular replacement with Phaser (35) using a monomer of PsNosZ (PDB ID code 3SBQ) as a search model. Improvement of the initial model was carried in cycles of refinement with REFMAC5 (36) and manual rebuilding in COOT (37). The final structure was refined to Rwork = 0.17 and Rfree = 0.20 at a resolution of 1.78 Å. The quality of the structure was validated by MolProbity (38). Data collection and refinement statistics are summarized in SI Appendix, Table S4.
Supplementary Material
Acknowledgments
We thank Peter Kroneck for stimulating and informative discussions; the staff of beamline X06DA at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) for providing excellent support during data collection; Dr. Anton Brausemann for assistance with X-ray dataset processing; and Dr. Julia Netzer for EPR measurements. This work was supported by the European Research Council Grant 310656.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Protein Data Bank, https://www.rcsb.org [ID codes 6RL0 (form I rNosZ) and 6RKZ (form II rNosZ)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903819116/-/DCSupplemental.
References
- 1.Butterbach-Bahl K., Baggs E. M., Dannenmann M., Kiese R., Zechmeister-Boltenstern S., Nitrous oxide emissions from soils: How well do we understand the processes and their controls? Philos Trans. R Soc. Lond. B Biol. Sci. 368, 20130122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson A. J., Giannopoulos G., Pretty J., Baggs E. M., Richardson D. J., Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1157–1168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ravishankara A. R., Daniel J. S., Portmann R. W., Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Pomowski A., Zumft W. G., Kroneck P. M. H., Einsle O., N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase. Nature 477, 234–237 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Zumft W. G., Kroneck P. M. H., Respiratory transformation of nitrous oxide (N2O) to dinitrogen by Bacteria and Archaea. Adv. Microb. Physiol. 52, 107–227 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Wüst A., et al. , Nature’s way of handling a greenhouse gas: The copper-sulfur cluster of purple nitrous oxide reductase. Biol. Chem. 393, 1067–1077 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Kroneck P. M. H., Walking the seven lines: Binuclear copper A in cytochrome c oxidase and nitrous oxide reductase. J. Biol. Inorg. Chem. 23, 27–39 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Brown K., et al. , Revisiting the catalytic CuZ cluster of nitrous oxide (N2O) reductase. Evidence of a bridging inorganic sulfur. J. Biol. Chem. 275, 41133–41136 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Haltia T., et al. , Crystal structure of nitrous oxide reductase from Paracoccus denitrificans at 1.6 A resolution. Biochem. J. 369, 77–88 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paraskevopoulos K., Antonyuk S. V., Sawers R. G., Eady R. R., Hasnain S. S., Insight into catalysis of nitrous oxide reductase from high-resolution structures of resting and inhibitor-bound enzyme from Achromobacter cycloclastes. J. Mol. Biol. 362, 55–65 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Schneider L. K., Wüst A., Pomowski A., Zhang L., Einsle O., No laughing matter: The unmaking of the greenhouse gas dinitrogen monoxide by nitrous oxide reductase. Met. Ions Life Sci. 14, 177–210 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Schneider L. K., Einsle O., Role of calcium in secondary structure stabilization during maturation of nitrous oxide reductase. Biochemistry 55, 1433–1440 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Wunsch P., Zumft W. G., Functional domains of NosR, a novel transmembrane iron-sulfur flavoprotein necessary for nitrous oxide respiration. J. Bacteriol. 187, 1992–2001 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Trncik C., Andrade S. L. A., Einsle O., The flavinyl transferase ApbE of Pseudomonas stutzeri matures the NosR protein required for nitrous oxide reduction. Biochim. Biophys. Acta Bioenerg. 1858, 95–102 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Honisch U., Zumft W. G., Operon structure and regulation of the nos gene region of Pseudomonas stutzeri, encoding an ABC-Type ATPase for maturation of nitrous oxide reductase. J. Bacteriol. 185, 1895–1902 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGuirl M. A., Bollinger J. A., Cosper N., Scott R. A., Dooley D. M., Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. J. Biol. Inorg. Chem. 6, 189–195 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Taubner L. M., McGuirl M. A., Dooley D. M., Copié V., Structural studies of Apo NosL, an accessory protein of the nitrous oxide reductase system: Insights from structural homology with MerB, a mercury resistance protein. Biochemistry 45, 12240–12252 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Hu H. W., He J. Z., Singh B. K., Harnessing microbiome-based biotechnologies for sustainable mitigation of nitrous oxide emissions. Microb. Biotechnol. 10, 1226–1231 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heikkilä M. P., Honisch U., Wunsch P., Zumft W. G., Role of the Tat ransport system in nitrous oxide reductase translocation and cytochrome cd1 biosynthesis in Pseudomonas stutzeri. J. Bacteriol. 183, 1663–1671 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita K., Chan J. M., Bollinger J. A., Alvarez M. L., Dooley D. M., Anaerobic purification, characterization and preliminary mechanistic study of recombinant nitrous oxide reductase from Achromobacter cycloclastes. J. Inorg. Biochem. 101, 1836–1844 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Wunsch P., Herb M., Wieland H., Schiek U. M., Zumft W. G., Requirements for Cu(A) and Cu-S center assembly of nitrous oxide reductase deduced from complete periplasmic enzyme maturation in the nondenitrifier Pseudomonas putida. J. Bacteriol. 185, 887–896 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viebrock A., Zumft W. G., Molecular cloning, heterologous expression, and primary structure of the structural gene for the copper enzyme nitrous oxide reductase from denitrifying Pseudomonas stutzeri. J. Bacteriol. 170, 4658–4668 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riester J., Zumft W. G., Kroneck P. M. H., Nitrous oxide reductase from Pseudomonas stutzeri. Redox properties and spectroscopic characterization of different forms of the multicopper enzyme. Eur. J. Biochem. 178, 751–762 (1989). [DOI] [PubMed] [Google Scholar]
- 24.Solomon E. I., et al. , Copper active sites in biology. Chem. Rev. 114, 3659–3853 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Wuest A., Prasser B., Mueller C., Einsle O., Recombinant Pseudomonas stutzeri nitrous oxide reductase, form I. RCSB Protein Data Bank. https://www.rcsb.org/structure/6RL0. Deposited 1 May 2019.
- 26.Zhang L., Wuest A., Prasser B., Mueller C., Einsle O., Recombinant Pseudomonas stutzeri nitrous oxide reductase, form II. RCSB Protein Data Bank. https://www.rcsb.org/structure/6RKZ. Deposited 30 April 2019.
- 27.Li C., et al. , FastCloning: A highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 11, 92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Ou X., Zhao G., Ding X., Highly efficient in vitro site-specific recombination system based on streptomyces phage phiBT1 integrase. J. Bacteriol. 190, 6392–6397 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miroux B., Walker J. E., Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Smith P. K., et al. , Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 (1985). [DOI] [PubMed] [Google Scholar]
- 31.Kristjansson J. K., Hollocher T. C., First practical assay for soluble nitrous oxide reductase of denitrifying bacteria and a partial kinetic characterization. J. Biol. Chem. 255, 704–707 (1980). [PubMed] [Google Scholar]
- 32.Weiss R. F., Price B. A., Nitrous oxide solubility in water and seawater. Mar. Chem. 8, 347–359 (1980). [Google Scholar]
- 33.Chaikuad A., Knapp S., von Delft F., Defined PEG smears as an alternative approach to enhance the search for crystallization conditions and crystal-quality improvement in reduced screens. Acta Crystallogr. D Biol. Crystallogr. 71, 1627–1639 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.