Abstract
Olfactomedin 4 (OLFM4) induces signal transducer and activator of transcription 3 (STAT3) activation by inhibiting gene associated with retinoid‐interferon‐induced mortality 19 (GRIM19), a strong STAT3 suppressor gene; however, the mechanisms of OLFM4 for regulating GRIM19‐STAT3 cascade in hepatocellular carcinoma (HCC) remain unclear. The functions and regulations of OLFM4, GRIM19, and STAT3 activation in HCC progression were evaluated using surgical specimens collected from 111 HCC patients or 2 HCC cell lines in vitro. Moreover, the cancer stem cell–like property of OLFM4 mediated by leucine‐rich repeat‐containing G protein‐coupled receptor 5 (LGR5), known as an intestinal stem cell marker, was investigated. OLFM4 was increased in HCC compared with adjacent liver tissue. The multivariate analysis revealed that high OLFM4 expression was an independent factor for poor prognosis. OLFM4 expression was negatively correlated with GRIM19 expression and positively correlated with STAT3 activation in HCC, thereby increasing cell cycle progression. OLFM4 knockdown in HCC cells increased GRIM19 expression and inhibited STAT3 activation; however, after double knockdown of GRIM19 and OLFM4, STAT3 activation decreased by OLFM4 knockdown was increased again. OLFM4 knockdown increased cell apoptosis, inhibited cell proliferation, and suppressed cancer stem cell–like property in HCC cells. The incidence of hematogenous recurrence was higher in HCC patients with high OLFM4 expression, suggesting that anoikis resistance of HCC was enhanced by OLFM4. In clinical cases, LGR5 expression and CD133 expression was correlated with OLFM4 expression in HCC, leading to poor patient prognosis. In vitro, LGR5 enhanced cancer stem cell–like property by up‐regulating OLFM4 through the Wnt signaling pathway. Conclusion: OLFM4 is induced by the LGR5‐Wnt signaling pathway and is strongly associated with aggressive tumor progression and poor prognosis in HCC by regulating STAT3‐induced tumor cell proliferation and cancer stem cell–like property. Therefore, OLFM4 is a novel prognostic predictor and a potential therapeutic target for patients with HCC.
Abbreviations
- AUC
area under the curve
- BrdU
5‐bromo‐2′‐deoxyuridine
- EMSA
electrophoretic mobility‐shift assay
- EMT
epithelial‐mesenchymal transition
- GRIM19
gene associated with retinoid‐interferon‐induced mortality 19
- HCC
hepatocellular carcinoma
- LGR5
leucine‐rich repeat‐containing G protein‐coupled receptor 5
- NF‐κB
nuclear factor kappa B
- NL
normal liver
- OLFM4
olfactomedin 4
- p‐
phosphorylated
- RIU
relative intensity unit
- ROC
receiver operating characteristic
- siRNA
small interfering RNA
- STAT3
signal transducer and activator of transcription 3
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer‐related death worldwide, particularly in Asian countries.1, 2 Despite improvements in multidisciplinary therapy, the prognosis of advanced HCC is still poor because of the lack of effective treatments. Therefore, it is essential to elucidate the mechanisms of HCC progression, discover new prognostic factors, and identify useful therapeutic targets.
Signal transducer and activator of transcription 3 (STAT3) is a transcriptional factor that is known to be activated in many malignancies, including HCC, and to promote tumor progression. Activation of STAT3 induces tumor cell proliferation by promoting cell‐cycle progression,3, 4 enhances tumor cell survival by inhibiting apoptosis,5 and accelerates tumor invasion and migration by promoting epithelial‐mesenchymal transition (EMT).6, 7 In addition, STAT3 is reported to mediate cancer stem cell–like property by enhancing anoikis resistance, which is related to chemoresistance and poor prognosis.8, 9 However, in HCC, little is known about the clinical significance of STAT3 function, and the precise mechanisms of its activation are still unclear. Moreover, because STAT3 is essential for cell survival in normal organs, therapies targeting STAT3 are not common.
Several factors regulating STAT3 activation have been reported. Because STAT3 is strongly related to inflammatory responses, its activation is induced by inflammatory cytokines and chemokines. In addition, activation of nuclear factor kappa B (NF‐κB), a potent inflammation‐related transcriptional factor, is known to accelerate STAT3 activation.9 In contrast, gene associated with retinoid‐interferon‐induced mortality 19 (GRIM19) is known to be a strong suppressor for STAT3 activation.10, 11 It has been reported that GRIM19 expression is decreased in many malignancies, such as breast cancer,12 colon cancer,13 ovarian cancer,14 and HCC,15 which conversely induces STAT3 activation and promotes cancer progression. However, the mechanisms of decreased GRIM19 expression in malignancies are still unclear. In addition, therapies up‐regulating GRIM19 expression in tumor are not yet established.
Olfactomedin 4 (OLFM4), also known as GW112, is a member of the OLFM family, which has been identified from myeloid cells.16 Initially, OLFM4 is recognized to regulate inflammatory response and innate immunity17, 18; however, recent studies have demonstrated that OLFM4 is associated with cancer progression in several malignancies. OLFM4 is reported to increase in cancers, including gastric cancer,19, 20, 21, 22 colon cancer,23, 24 and pancreatic cancer,25, 26 and to promote tumor progression by inducing cell‐cycle progression and enhancing tumor invasion and metastasis, which resulted in poor prognosis. Moreover, recent studies have suggested the usefulness of OLFM4 as a stem cell marker of the intestine, because OLFM4 expression in the epithelium of the intestine is significantly correlated with the expression levels of leucine‐rich repeat‐containing G protein‐coupled receptor 5 (LGR5), which is known to enhance stem cell property in the intestine through the Wnt signaling pathway.27, 28, 29 In contrast, several studies have demonstrated that OLFM4 suppressed tumor proliferation, invasion, and metastasis in prostate cancer30, 31 and triple‐negative breast cancer.32 Therefore, the role of OLFM4 in cancer progression remains uncertain. In relation to STAT3 activation, only a few experiments have shown that OLFM4 attenuates the ability of GRIM19 and enhances STAT3 activation in Hela cells33 and gastric cancer cells34 in vitro. Based on these results, controlling STAT3 activation through OLFM4‐GRIM19 cascade is a potentially useful therapeutic target for HCC patients. However, no reports have evaluated OLFM4 expression in HCC in any experimental models.
The aim of this study was to evaluate the expression of OLFM4 in patients with HCC in order to investigate the correlations with clinicopathological variables, including patient survival, and to determine whether STAT3 activation mediated by OLFM4‐GRIM19 cascade is a relevant factor in the regulation of HCC progression.
Materials and Methods
Tissue Samples
We studied 111 HCC patients who underwent primary surgical resection between 2009 and 2012 at our institution. None of the patients received preoperative treatments. Fresh surgical specimens were obtained from these patients during operation. Normal liver tissues were obtained from patients with liver metastases of colorectal cancer as controls. Portions of the samples were fixed in formalin, embedded in paraffin, and stained with hematoxylin and eosin. The pathological features of HCC were evaluated based on the classification proposed by the Liver Cancer Study Group of Japan (6th ed., 2015). Whole‐tissue lysates and nuclear extracts were collected from 35 HCC samples. Fully informed consent was obtained from all patients. The study was performed in accordance with the guidelines of the Helsinki Declaration and approved by the Chiba University Human Research Committee.
Immunohistochemical and Immunofluorescence Staining
Immunohistochemical staining was performed with anti‐OLFM4, anti‐GRIM19, anti‐Ki‐67, anti‐CD133, anti‐LGR5 (Abcam, Cambridge, UK), or anti‐phosphorylated (p‐)STAT3 (Ser727) antibody (Santa Cruz Biotechnology, Santa Cruz, CA) using the EnVision Kit or Universal LSAB Kit (Dako, Copenhagen, Denmark) and was counterstained with hematoxylin. A negative control obtained by omitting the secondary antibody was included in each target protein. Isotype‐matched controls were used as negative controls. The expression levels of OLFM4 or GRIM19 were evaluated based on the staining intensity and percentage of positive cells. The intensity grade was evaluated by two clinical pathologists based on the grading scale and ranged from 0 (no signal) to 1 (very weak signal), 2 (weak signal), 3 (strong signal), and 4 (the strongest signal), and the average score was recognized as the intensity score. The percentage score for the positive cells was determined from 1 (0%‐5%), 2 (6%‐25%), 3 (26%‐50%), and 4 (51%‐100%) using the ImageJ software. The product of the intensity score and the percentage score was defined as the labeling index (0‐16). The cell proliferation index was evaluated by Ki‐67 immunohistochemistry based on the percentage of positive nuclei in 400 to 600 tumor cells. Determination of p‐STAT3 (Ser727) labeling index was evaluated by immunohistochemistry based on the percentage of positive nuclei and was expressed as the labeling index of HCC. Overexpression of CD133 was defined as 30% or more of cancer cells expressing CD133 compared with adjacent liver tissue. For LGR5 expression, HCC with more than 30% of cancer cells losing LGR5 expression compared with adjacent liver tissue was defined as HCC with low LGR5 expression. Immunofluorescence staining was done as described previously. Anti‐GRIM19, anti‐Ki‐67, and anti‐green fluorescent protein antibody (Abcam) were used for immunofluorescence staining.
Western Blotting
Western blotting was performed using whole cell lysates as described.35 PhosphoBLOCKER (Cell Biolabs, Inc., San Diego, CA) was used for blocking nonspecific binding sites. Antibodies against OLFM4, GRIM19, LGR5, CD133 (Abcam), p‐STAT3 (Ser727), β‐actin, or caspase‐3 (Cell Signaling Technology, Danvers, MA) were used for primary antibody. Immunoreactive proteins were detected by enhanced chemiluminescence and quantified by image analysis.
Electrophoretic Mobility‐Shift Assay
Nuclear extracts were prepared by the method of Deryckere and Gannon, and the DNA‐binding activity was evaluated by electrophoretic mobility‐shift assay (EMSA). Double‐stranded consensus oligonucleotides to STAT3 (Santa Cruz Biotechnology) was end‐labeled with γ[32P] adenosine triphosphate (Perkin Elmer, Inc., Boston, MA). Binding reaction products were separated in a polyacrylamide gel and analyzed by autoradiography.
Cell Culture
Human HCC cell lines, Huh‐7 cells and HepG2 cells, purchased from the JCRB Cell Bank (Osaka, Japan), were distributed onto a 6‐cm dish at a concentration of 3.5 × 105 cells/5 mL. After overnight incubation, small interfering RNA (siRNA) specific for OLFM4, GRIM19, or LGR5, or negative control siRNA (QIAGEN, Inc., Valencia, CA), was transfected into HCC cells using RNAiMAX Transfection Reagent (Thermo Fisher Scientific, New Hampshire). In some experiments, additional knockdown of OLFM4 was performed in GRIM19‐knockdown HCC cells to achieve double knockdown of OLFM4 and GRIM19. The Biotrak cell proliferation enzyme‐linked immunosorbent assay system (GE Healthcare, Buckingham, United Kingdom) was used for evaluating DNA incorporation of 5‐bromo‐2′‐deoxyuridine (BrdU), and a cell counting kit (Dojindo Laboratories, Kumamoto, Japan) was used for counting total cell number after 24, 48, or 72 hours of treatment as an index of HCC cell proliferation. Cell lysates and nuclear extracts were prepared for western blotting or EMSA. For the apoptosis assay, cells were stained with propidium ionide and Annexin V‐FITC 72 hours after treatment. Apoptotic cells were identified using a FACS CANTO II flow cytometer (BD Biosciences, Franklin Lakes, NJ). In other experiments, cells were incubated for 72 hours in medium containing 0 or 50 μmol/L IWP‐2, a Wnt signaling inhibitor (Sigma‐Aldrich, St. Louis, MO).
Anoikis Assay
To evaluate the resistance for apoptosis after losing contact with the extracellular matrix (cancer stem cell–like property), anoikis assay was performed using OLFM4‐depleted HCC cells. After incubating these cells with medium without growth factor with rotation for 24 hours, colony formation assay was performed as previously described. The number of colonies was determined at 14 days after cell seeding.
Statistical Analysis
Data are expressed as means ± SEM. The Kaplan‐Meier method was used for estimating overall survival and relapse‐free survival and analyzed by the log‐rank test. Significant prognostic factors by univariate analysis were included in a multivariable analysis to identify independent factors for poor prognosis. Statistical comparisons for significance were performed using the Mann‐Whitney U test, Student t test, Fisher's exact test, or regression analysis. Probability (P) values of 0.05 or less were considered significant. Statistical analyses were performed using the commercially available software JMP 11 (SAS Institute, Inc., Cary, NC).
Results
OLFM4 Expression in Normal Liver Tissue, Adjacent Liver Tissue, and HCC
When OLFM4 expression was evaluated by immunohistochemistry (Fig. 1A), it was weak in normal liver tissue and adjacent liver tissue. Increased OLFM4 expression was seen in some cases of HCC compared with adjacent liver tissue, but not in other cases of HCC. Based on receiver operating characteristic (ROC) analysis in accordance with the 3‐year survival (Supporting Fig. S1A), 111 HCC patients were divided into two groups (cutoff value = 6; P = 0.014; AUC [area under the curve] = 0.636). When the relationships between OLFM4 expression in HCC and several clinicopathological variables were evaluated, high OLFM4 expression was significantly correlated with younger age (P = 0.047), larger tumor size (P < 0.001), positive portal vein invasion (P = 0.005), and the incidence of hematogenous recurrence (P = 0.030) (Table 1). Overall survival time and relapse‐free survival time were significantly poorer in HCC patients with high OLFM4 expression (P = 0.010 and P < 0.001, respectively) (Fig. 1B,C). The univariate and multivariate analyses revealed that the presence of intrahepatic metastasis (P = 0.045), positive portal vein invasion (P = 0.003), positive venous invasion (P = 0.042), and high OLFM4 expression in HCC (P = 0.002) were independent poor prognostic factors associated with relapse‐free survival (Table 2).
Figure 1.

(A) Immunohistochemical staining for OLFM4 in normal liver (NL) tissues from patients with liver metastasis and HCC with high and low OLFM4 expression. Results are representative of 111 HCC sections. Original magnification was ×100. Overall survival (B) and relapse‐free survival (C) of 111 HCC patients in relation to OLFM4 expression in HCC were analyzed by the Kaplan‐Meier methods (P = 0.010 and P < 0.001, respectively). (D) Immunohistochemical staining for GRIM19 in NL and HCC with high and low GRIM19 expression. Results are representative of 111 HCC sections. Original magnification was ×100. Overall survival (E) and relapse‐free survival (F) of 111 HCC patients in relation to GRIM19 expression in HCC were analyzed by the Kaplan‐Meier methods (P = 0.002 and P = 0.004, respectively).
Table 1.
Relationship Between OLFM4 Expression and Clinicopathological Characteristics in HCC
| OLFM4 Expression in HCC | P Value | |||
|---|---|---|---|---|
| High | Low | |||
| Patients (n) | 53 | 58 | ||
| Age (years; means ± SEM) | 63.9 ± 1.5 | 67.4 ± 1.4 | 0.047 | |
| Gender | Male | 44 | 44 | 0.351 |
| Female | 9 | 14 | ||
| Virus infection | HBV+ or HCV+ | 37 | 39 | 0.771 |
| NBNC | 16 | 19 | ||
| AST levels (U/L) (mean ± SEM) | 48.4 ± 4.1 | 43.2 ± 3.9 | 0.818 | |
| Tumor capsule | + | 43 | 46 | 0.652 |
| − | 9 | 12 | ||
| Capsule infiltration | + | 33 | 30 | 0.170 |
| − | 18 | 28 | ||
| Septum formation | + | 44 | 44 | 0.351 |
| − | 9 | 14 | ||
| Differentiation | Well or moderately | 47 | 55 | 0.233 |
| Poorly | 6 | 3 | ||
| Tumor size (mm) (means ± SEM) | 64.2 ± 5.0 | 38.4 ± 4.8 | <0.001 | |
| Number of tumors | Solitary | 42 | 39 | 0.153 |
| Multiple | 11 | 19 | ||
| Intrahepatic metastasis | + | 19 | 21 | 0.969 |
| − | 34 | 37 | ||
| Portal vein invasion | + | 23 | 11 | 0.005 |
| − | 30 | 47 | ||
| Venous invasion | + | 12 | 9 | 0.338 |
| − | 41 | 49 | ||
| Serosal infiltration | + | 2 | 3 | 0.882 |
| − | 50 | 55 | ||
| PIVKA‐II levels (mAU/mL) (mean ± SEM) | 3,901 ± 960.5 | 6,019 ± 3,425 | 0.277 | |
| AFP levels (ng/mL) (mean ± SEM) | 3,366 ± 1,800 | 5,254 ± 3,754 | 0.326 | |
| Hematogenous recurrence | + | 5 | 1 | 0.030 |
| − | 47 | 57 | ||
Abbreviations: AFP, alpha fetoprotein; AST, aspartate aminotransferase; HBV, hepatitis B virus; HCV, hepatitis C virus; NBNC, non‐B and non‐C; PIVKA‐II, protein induced by vitamin K absence or antagonist II. The bold‐faced values were statistically significant by the student t test.
Table 2.
Univariate and Multivariate Analyses of Relapse‐Free Survival in 111 Patients With HCC
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| Hazard Ratio | P Value | Hazard Ratio | P Value | |
| Age (≥70, n = 45 vs. <70, n = 66) | 1.341 (0.812‐2.191) | 0.248 | ||
| Gender (male, n = 88 vs. female, n = 23) | 1.221 (0.674‐2.399) | 0.526 | ||
| Virus infection (HBV/HCV, n = 76 vs. NBNC, n = 35) | 1.799 (1.074‐2.577) | 0.026 | 1.499 (0.850‐2.597) | 0.158 |
| Tumor size (≥50 mm, n = 38 vs. <50 mm, n = 73) | 3.145 (1.901‐5.177) | <0.001 | 1.350 (0.695‐2.577) | 0.371 |
| Tumor capsule (absent, n = 21 vs. present, n = 89) | 1.709 (0.878‐ 3.759) | 0.120 | ||
| Capsule infiltration (present, n = 63 vs. absent, n = 46) | 1.812 (1.082‐3.118) | 0.024 | 1.068 (0.577‐1.964) | >0.999 |
| Septum formation (present, n = 88 vs. absent, n = 23) | 1.655 (0.881‐3.455) | 0.122 | ||
| Number of tumors (multiple, n = 30 vs. solitary, n = 81) | 1.232 (0.712‐2.060) | 0.444 | ||
| Intrahepatic metastasis (present, n = 40 vs. absent, n = 71) | 2.462 (1.485‐4.039) | <0.001 | 1.845 (1.015‐3.310) | 0.045 |
| Portal vein invasion (positive, n = 34 vs. negative, n = 77) | 5.579 (3.243‐9.509) | <0.001 | 3.177 (1.481‐6.820) | 0.003 |
| Venous invasion (positive, n = 21 vs. negative, n = 90) | 5.518 (2.989‐9.754) | <0.001 | 2.226 (1.031‐4.747) | 0.042 |
| Serosal infiltration (present, n = 5 vs. absent, n = 105) | 1.938 (0.599‐11.905) | 0.310 | ||
| Differentiation (poorly, n = 9 vs. moderately and well, n = 102) | 1.854 (0.645‐4.214) | 0.226 | ||
| PIVKA‐Ⅱ (≥40 mAU/mL, n = 70 vs. <40 mAU/mL, n = 41) | 2.260 (1.340‐3.965) | 0.002 | 1.016 (0.508‐2.033) | 0.934 |
| AFP (≥40 ng/mL, n = 52 vs. <40 ng/mL, n = 59) | 1.549 (0.950‐2.535) | 0.079 | ||
| OLFM4 expression (high, n = 53 vs. low, n = 58) | 2.646 (1.603‐4.429) | <0.001 | 2.555 (1.423‐4.655) | 0.002 |
Abbreviations: AFP, alpha fetoprotein; HBV, hepatitis B virus; HCV, hepatitis C virus; NBNC, non‐B and non‐C; PIVKA‐II, protein induced by vitamin K absence or antagonist‐II. The bold‐faced values were statistically significant by the regression analysis.
GRIM19 Expression in Normal Liver Tissue, Adjacent Liver Tissue, and HCC
Immunohistochemical staining revealed abundant GRIM19 expression in normal liver tissue and adjacent liver tissue (Fig. 1D). GRIM19 expression was decreased in some cases of HCC. When 111 HCC patients were divided into two groups by ROC analysis in accordance with the 3‐year survival (Supporting Fig. S1B) (cutoff value = 6; P = 0.040; AUC = 0.623), low GRIM19 expression was significantly correlated with larger tumor size (P = 0.019), positive septum formation (P = 0.018), poor differentiation (P = 0.014), and positive portal vein invasion (P = 0.030) (Supporting Table S1). Kaplan‐Meier analyses revealed that overall survival time and relapse‐free survival time for patients with low GRIM19 expression were significantly poorer when compared with those with high GRIM19 expression (P = 0.002 and P = 0.004, respectively) (Fig. 1E,F).
Clinical Relevance Between OLFM4 and GRIM19
Immunohistochemical staining revealed that OLFM4 expression was negatively correlated with GRIM19 expression in clinical cases of HCC (P = 0.002) (Fig. 2A). These data were confirmed by western blotting using whole‐tissue lysates from HCC (Fig. 2B). Consistent with the results of immunohistochemistry, western blotting results showed the negative correlation between OLFM4 expression and GRIM19 expression (P = 0.043). When the overall survival time and relapse‐free survival time in patients with HCC in relation to the expression levels of OLFM4 and GRIM19 were evaluated, low OLFM4 expression with high GRIM19 expression in HCC was a significant predictor for better prognosis (P < 0.05, compared with other groups) (Supporting Fig. S1C,D). In most cases, the labeling index of GRIM19 in tumor‐adjacent liver tissues was very high (mean + SEM of the GRIM19 labeling index = 12.4 + 0.3). In contrast, the OLFM4 labeling index was very low in tumor‐adjacent liver tissues in most cases (mean + SEM of the OLFM4 labeling index = 3.4 + 0.2). These data were consistent with the correlations in HCC.
Figure 2.
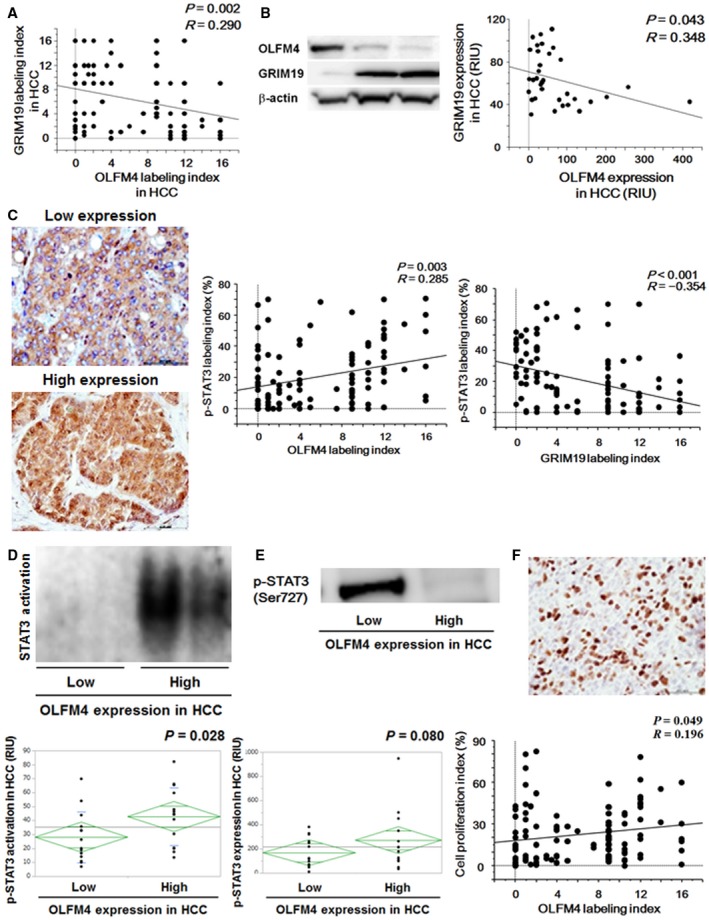
(A) Correlation between OLFM4 labeling index and GRIM19 labeling index in HCC (n = 111). A significant correlation was seen by regression analysis (P = 0.002). (B) OLFM4 and GRIM19 expression evaluated by western blotting (n = 35). A significant correlation was seen by regression analysis (P = 0.043). Data are shown by relative intensity unit (RIU). (C) Nuclear accumulation of p‐STAT3 (Ser727) was evaluated by immunohistochemical staining. Results are representative of 111 HCC sections. Original magnification was ×400. The p‐STAT3 (Ser727) labeling index was positively correlated with the OLFM4 labeling index (P = 0.003) and negatively correlated with the GRIM19 labeling index (P < 0.001). (D) STAT3 activation in HCC with low and high OLFM4 expression, analyzed by EMSA (n = 16‐19 per group). *P = 0.028 between these groups. (E) Expression levels of p‐STAT3 (Ser727) evaluated by western blotting (n = 16‐19 per group). P = 0.080 between these groups. (F) Cell proliferation index evaluated by Ki‐67 immunohistochemistry. Results are representative of 111 HCC sections. Original magnification was ×100. The cell proliferation index was significantly correlated with the OLFM4 labeling index (P = 0.049).
STAT3 Activation in Relation to OLFM4 and GRIM19 Expression in HCC
Following activation, Ser727 residue of STAT3 is phosphorylated and translocated into the nucleus. Therefore, the activation of STAT3 was evaluated by nuclear accumulation of p‐STAT3 (Ser727) by immunohistochemistry (Fig. 2C). Interestingly, the p‐STAT3 (Ser727) labeling index was positively correlated with OLFM4 expression and negatively correlated with GRIM19 expression in HCC (P = 0.003 and P < 0.001, respectively). In accordance with STAT3 activation in tumor‐adjacent liver tissues, its p‐STAT3 (Ser727) labeling index was extremely low in most cases (mean + SEM of the pSTAT3 [Ser727] labeling index = 4.2 + 0.9).
STAT3 activation in relation to OLFM4 expression in HCC was confirmed by EMSA and western blotting. EMSA results revealed that STAT3 activation was significantly increased in HCC with high OLFM4 expression (P = 0.028; Fig. 2D). Similar to the results from EMSA, p‐STAT3 (Ser727) expression tended to be higher in HCC with high OLFM4 expression than that with low OLFM4 expression (P = 0.080; Fig. 2E). When the cell proliferation index was determined by Ki‐67 immunohistochemistry, it was significantly correlated with STAT3 activity (P = 0.002; Supporting Fig. S1E). Consistent with the results of STAT3 activation, the cell proliferation index by Ki‐67 was significantly correlated with the OLFM4 labeling index (P = 0.049) (Fig. 2F). When 111 HCC patients were divided into two groups, high and low p‐STAT3 (Ser727) labeling index, based on ROC analysis in accordance with the 3‐year survival (cutoff value = 19.1; P = 0.002; AUC = 0.600) (Supporting Fig. S2A), overall survival time and relapse‐free survival time were significantly poorer in HCC patients with high p‐STAT3 (Ser727) labeling index (Supporting Fig. S2B,C).
Effects of OLFM4 and GRIM19 Knockdown on HCC Cells In Vitro
Expression of OLFM4 and GRIM19 in Huh‐7 cells and HepG2 cells was confirmed by western blotting (Supporting Fig. S2D). In Huh‐7 cells, knockdown of OLFM4 by its siRNAs significantly increased GRIM19 expression (P < 0.01 for each), thereby reducing p‐STAT3 (Ser727) expression (P < 0.01 for each) (Fig. 3A). Increased expression of GRIM19 in OLFM4‐depleted Huh‐7 cells was confirmed by immunofluorescence staining (Fig. 3B). Strong OLFM4 expression was seen in negative control cells, which expressed low GRIM19. After OLFM4 knockdown, GRIM19 expression was significantly increased in these OLFM4‐depleted Huh‐7 cells. When double knockdown of OLFM4 and GRIM19 was further performed in Huh‐7 cells, cell proliferation suppressed by OLFM4 knockdown was enhanced again after additional knocking down of GRIM19 (P < 0.001 for each) (Fig. 3C). Consistent, decreased STAT3 activation by OLFM4 knockdown was recovered to the basal line in Huh‐7 cells with OLFM4 knockdown followed by GRIM19 knockdown (P < 0.001 for each) (Fig. 3D). In contrast, only GRIM19 knockdown had no effects on OLFM4 expression in Huh‐7 cells (Fig. 3E), suggesting that OLFM4 is the upstream factor in the OLFM4‐GRIM19 cascade. These results in Huh‐7 cells were confirmed using another HCC cell line, HepG2 cells. Consistent with the results of Huh‐7 cells, OLFM4 knockdown in HepG2 cells significantly increased GRIM19 expression (P < 0.01) and decreased p‐STAT3 (Ser727) expression (P < 0.01) evaluated by western blotting and immunofluorescence staining (Supporting Fig. S3A,B), whereas cell proliferation suppressed by OLFM4 knockdown was enhanced again and STAT3 activation was recovered to the basal line in OLFM4 and GRIM19 double‐knockdown HepG2 cells (P < 0.001 for each) (Supporting Fig. S3C,D). Similar to Huh‐7 cells, GRIM19 knockdown alone had no effects on OLFM4 expression in HepG2 cells (Supporting Fig. S3E).
Figure 3.
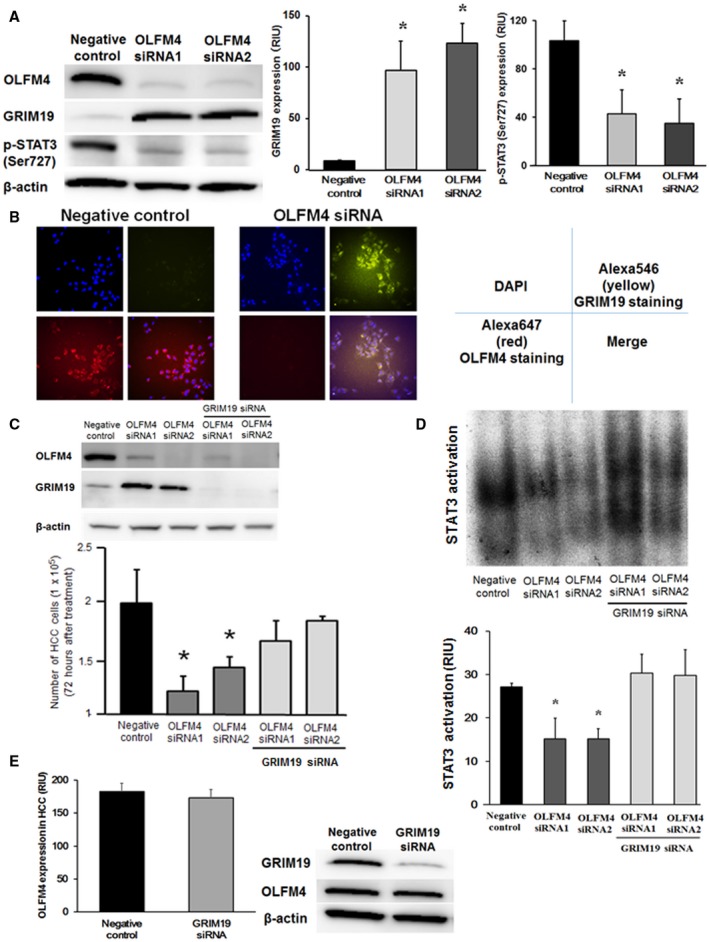
(A) Effects of OLFM4 knockdown by siRNA in Huh‐7 cells evaluated by western blotting (n = 4‐6 per group). *P < 0.01 compared with negative controls. (B) Effects of OLFM4 knockdown in Huh‐7 cells evaluated by immunofluorescence staining. 4′6‐diamidino‐2‐phenylindole (DAPI) was used for nuclear staining. Results are representative of n = 6 per group. Original magnification was ×400. (C) Double knockdown of OLFM4 and GRIM19 in Huh‐7 cells. GRIM19 knockdown was further performed in addition to knocking down of OLFM4. Cell proliferation was evaluated by a cell counting kit (n = 4‐6 per group). *P < 0.001 compared with negative controls and double‐knockdown cells. (D) Effects of OLFM4 and GRIM19 double knockdown on STAT3 activation assessed by EMSA (n = 4‐6 per group). *P < 0.001 compared with negative controls and double‐knockdown cells. (E) Effects of GRIM19 knockdown in Huh‐7 cells evaluated by western blotting (n = 4‐6 per group). No differences were seen between the two groups.
Effects of OLFM4 Knockdown on HCC Cell Apoptosis, Proliferation, and Cancer Stem Cell–Like Property In Vitro
In Huh‐7 cells, OLFM4 knockdown significantly increased the expression levels of caspase‐3, the indicators of cell apoptosis (Fig. 4A). When flow cytometry was performed to evaluate the effects of OLFM4 knockdown on Huh‐7 cell apoptosis, OLFM4 knockdown significantly increased the population of apoptotic cells at 72 hours after treatment (P < 0.001; Fig. 4B). DNA incorporation of BrdU was reduced in OLFM4‐depleted Huh‐7 cells, when compared with negative controls (P < 0.001 for each; Fig. 4C). Consistent with these results, cell proliferation was inhibited in Huh‐7 cells with OLFM4 knockdown (Fig. 4D). The results of apoptosis and proliferation in Huh‐7 cells were confirmed in HepG2 cells. OLFM4 knockdown in HepG2 cells significantly increased caspase‐3 expression by western blotting (Supporting Fig. S4A), thereby inducing apoptosis evaluated by flow cytometry (P < 0.001; Supporting Fig. S4B). In addition, OLFM4 knockdown significantly inhibiting DNA incorporation of BrdU and cell proliferation in HepG2 cells (P < 0.001 for each; Supporting Fig. S4C,D). Anoikis resistance is known as one of the potent functions for cancer stem cell–like property; therefore, cancer stem cell–like property in OLFM4‐depleted HCC cells was evaluated by anoikis assay. Interestingly, the number of colony formation was significantly reduced after OLFM4 knockdown in both Huh‐7 cells and HepG2 cells, suggesting that OLFM4 enhances cancer stem cell–like property of HCC cells (P < 0.001 for each) (Fig. 4E and Supporting Fig. S4E).
Figure 4.

(A) Effects of OLFM4 knockdown on caspase‐3 expression in Huh‐7 cells evaluated by western blotting (n = 3 per group). *P < 0.001 compared with negative controls. (B) Huh‐7 cell apoptosis at 72 hours after OLFM4 knockdown was evaluated by apoptosis assay using flow cytometry (n = 3‐4 per group). *P < 0.001 compared with negative controls. Effects of OLFM4 knockdown on DNA incorporation of BrdU by absorptiometry (optimal density = 450 nm) (C) and cell proliferation in Huh‐7 cells (n = 4‐6 per group) (D). *P < 0.001 compared with negative controls at 72 hours after incubation. (E) Effects of OLFM4 knockdown on cancer stem cell–like property in Huh‐7 cells evaluated by anoikis assay (n = 4‐6 per group). Original magnification was ×400. *P < 0.001 compared with negative controls. Abbreviation: O.D., optimal density.
Clinical Relevance of OLFM4, LGR5, and CD133 in HCC
The upstream factors regulating OLFM4 expression in malignancies are not yet fully understood. Recent studies have suggested that LGR5, a receptor of the Wnt signaling pathway that is known as a stem cell marker of the intestine, mediates OLFM4 expression through the Wnt signaling pathway. Our findings showed that OLFM4 enhanced cancer stem cell–like property in HCC; therefore, the expression levels of LGR5 and CD133, a well‐known cancer stem cell marker for HCC, in 111 HCC patients were evaluated by immunohistochemistry. LGR5 expression was abundantly seen in normal liver tissue and adjacent liver tissue, whereas its expression was weak in some cases of HCC compared with adjacent liver tissue. OLFM4 labeling index was significantly higher in HCC with high LGR5 expression compared with HCC with low LGR5 expression (P < 0.001) (Fig. 5A). Instead, CD133 was weak in normal liver tissue and adjacent liver tissue; however, it was strongly expressed in some cases of HCC. Interestingly, CD133 expression in HCC was significantly correlated with OLFM4 expression in HCC (P = 0.003) (Fig. 5B). Moreover, overall survival time and relapse‐free survival time were significantly poorer in HCC patients with high LGR5 expression (P = 0.018 and P = 0.002, respectively; Fig. 5C,D).
Figure 5.
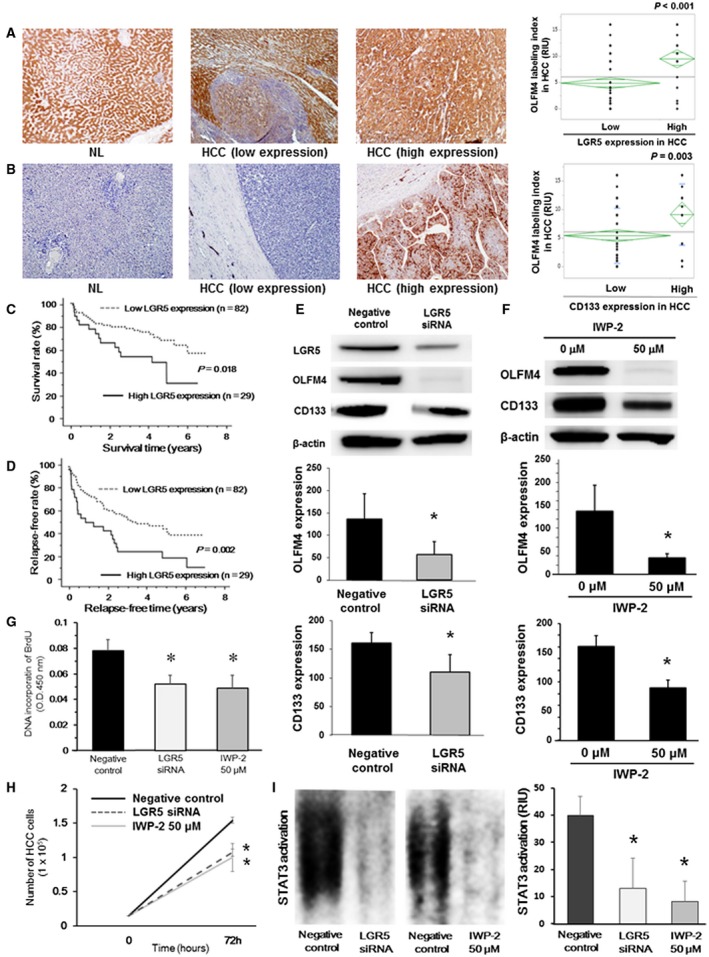
(A) Immunohistochemical staining for LGR5 in NL tissues from patients with liver metastasis and HCC with high and low LGR5 expression. Results are representative of 111 HCC sections. Original magnification was ×100. The OLFM4 labeling index was significantly higher in HCC with high LGR5 expression (*P < 0.001 between these groups). (B) Immunohistochemical staining for CD133 in NL tissues from patients with liver metastasis and HCC with high and low CD133 expression. Results are representative of 111 HCC sections. Original magnification was ×100. OLFM4 labeling index was significantly higher in HCC with high CD133 expression. *P = 0.003 between these groups. Overall survival (C) and relapse‐free survival (D) of 111 HCC patients in relation to LGR5 expression in HCC were analyzed by the Kaplan‐Meier methods (P = 0.018 and P = 0.002, respectively). (E) Effects of LGR5 knockdown in Huh‐7 cells evaluated by western blotting in vitro (n = 4‐6 per group). *P < 0.001 compared with negative controls. (F) Effects of treatment with the Wnt signaling inhibitor on OLFM4 expression in Huh‐7 cells in vitro (n = 4‐6 per group). Huh‐7 cells were stimulated with 50 μM IWP‐2, a Wnt signaling inhibitor, for 72 hours. *P < 0.001 compared with 0 μM. Effects of LGR5 knockdown or treatment with Wnt signaling inhibitor on DNA incorporation of BrdU (G) and cell proliferation in Huh‐7 cells (H) (n = 4‐6 per group). *P < 0.001 compared with negative controls at 72 hours after incubation. (I) Effects of LGR5 knockdown or treatment with the Wnt signaling inhibitor on STAT3 activation in Huh‐7 cells evaluated by EMSA (n = 4‐6 per group). *P < 0.001 compared with negative controls at 72 hours after incubation. Abbreviation: O.D., optimal density.
Cancer Stem Cell–Like Property Mediated by LGR5‐Wnt Signaling‐OLFM4 Pathway In Vitro
When LGR5 knockdown was performed in Huh‐7 cells in vitro, expression levels of OLFM4 and CD133 were significantly decreased after knocking down of LGR5 (P < 0.001 for each) (Fig. 5E). In addition, stimulation with a Wnt signaling inhibitor, IWP‐2, significantly decreased OLFM4 expression in Huh‐7 cells (P < 0.001), thereby reducing CD133 expression (P < 0.001) (Fig. 5F). These findings were confirmed using HepG2. Consistently, LGR5 knockdown and IWP‐2 stimulation significantly inhibited OLFM4 expression, leading to decreased expression of CD133 in HepG2 cells (P < 0.001 for each) (Supporting Fig. S5A,B). In addition, DNA incorporation of BrdU was reduced in Huh‐7 cells and HepG2 cells after LGR5 knockdown or Wnt signaling inhibition by IWP‐2, when compared with negative controls (P < 0.001 for each) (Fig. 5G and Supporting Fig. S5C). Consistent with these results, cell proliferation was inhibited in Huh‐7 cells and HepG2 cells after LGR5 knockdown or Wnt signaling inhibition by IWP‐2 (P < 0.001 for each, compared with negative controls) (Fig. 5H and Supporting Fig. S5D). Moreover, STAT3 activation evaluated by EMSA was significantly decreased after LGR5 knockdown or IWP‐2‐treatment in Huh‐7 cells or HepG2 cells compared with negative controls (Fig. 5I and Supporting Fig. S5E). Furthermore, GRIM19 expression evaluated by western blotting was increased in LGR5‐depleted or LGR5‐inhibited Huh‐7 cells or HepG2 cells in vitro (Supporting Fig. S5F). These results suggest that the LGR5‐Wnt signaling pathway is the upstream cascade for increasing OLFM4 expression in HCC, which enhances cancer stem cell–like property and tumor cell proliferation in HCC by up‐regulating OLFM4‐mediated STAT3 activation.
Discussion
OLFM4 has been reported to increase in cancers and promote tumor progression by several mechanisms. OLFM4 promotes tumor cell proliferation by activating STAT3 and accelerating cell‐cycle progression in gastric cancer22 and pancreatic cancer.25 Moreover, OLFM4 inhibits tumor cell apoptosis through NF‐κB pathway in gastric cancer.20 Regarding tumor invasiveness and metastasis, OLFM4 is reported to increase liver metastasis in colon cancer24 and promote lymph node invasion and metastasis in gastric cancer by enhancing EMT signaling.20 As these results show, increased OLFM4 expression is associated with poor survival in patients with pancreatic cancer, colon cancer, and gastric cancer. In contrast, OLFM4 has been reported to inhibit tumor proliferation and metastasis in prostate cancer by inhibiting cathepsin and stromal cell–derived factor 1.30, 31 Moreover, lymph node metastasis and distant metastasis are significantly increased in triple‐negative breast cancer patients with low OLFM4 expression, which is associated with poor prognosis.32 Therefore, the roles of OLFM4 on cancer progression might be different among different types of cancer. However, the function of OLFM4 in HCC has not been reported in any experimental model. In the present study, we propose precise mechanisms of OLFM4‐mediated HCC progression (Fig. 6) and reveal that OLFM4 expression is increased in HCC, and increased OLFM4 expression is associated with poor prognosis in patients with HCC.
Figure 6.
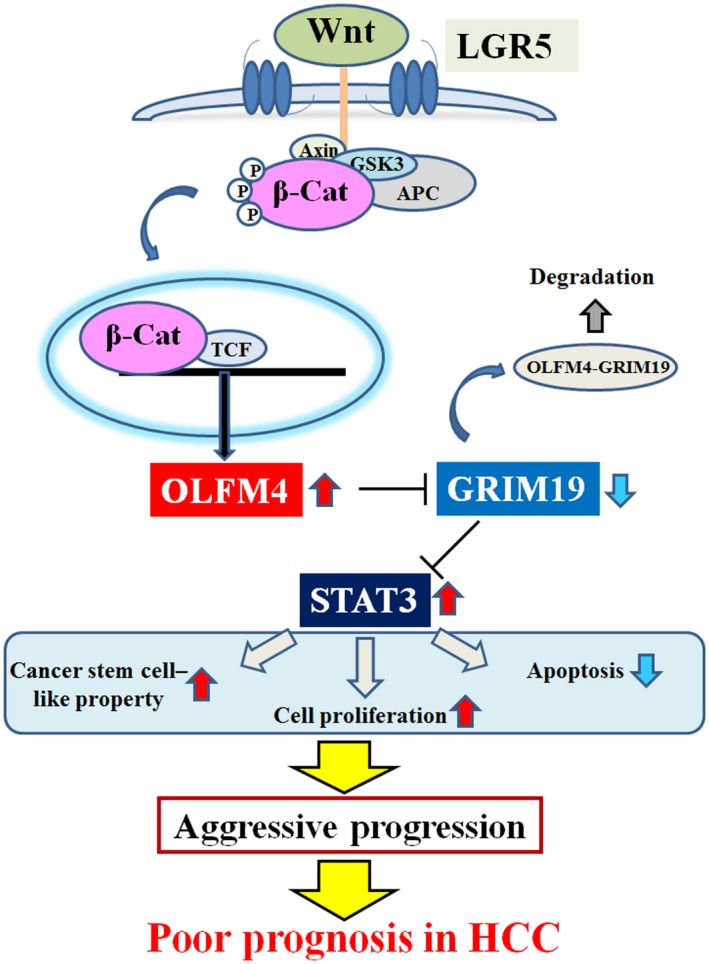
Proposed model of OLFM4‐induced STAT3 activation mediated by the LGR5‐Wnt signaling pathway in HCC. Abbreviations: APC, adenomatous polyposis coli protein; β‐Cat, β‐catenin; GSK3, glycogen synthase kinase 3; TCF, T cell factor.
To clarify the precise mechanisms of OLFM4‐mediated HCC progression, we focus on the GRIM19‐STAT3 cascade. GRIM19 is known as a strong tumor suppressor gene, and the effects were induced by STAT3 inhibition. As STAT3 promotes tumor progression by inducing tumor cell proliferation, inhibiting apoptosis and increasing tumor invasion and metastasis by enhancing EMT,3, 4, 5, 6, 7 GRIM19‐mediated STAT3 inhibition is also reported to be associated with tumor suppression in many cancers. In brief, GRIM19 expression is decreased and negatively correlates with STAT3 activation in gastric cancer,36 colon cancer,13 breast cancer,12 ovarian cancer, and HCC,15 leading to poor patient prognosis. As we show that GRIM19 expression is negatively correlated with STAT3 activation, low GRIM19 expression is the significant factor for poor prognosis in patients with HCC. In the present study, we evaluated p‐STAT3 (Ser727) expression in HCC, as GRIM19 is known to recognize and bind to p‐STAT3 (Ser727) and induce dephosphorylation of STAT3 (Ser727), resulting in suppressed STAT3 activation.37 STAT3 (Tyr705) phosphorylation was the classical pattern for STAT3 activation, and many studies have demonstrated that STAT3 (Tyr705) phosphorylation promotes cancer progression. However, the roles of STAT3 (Ser727) phosphorylation on cancer progression are not well defined. A recent study demonstrated that STAT3 (Ser727) phosphorylation suppressed HCC progression38; however, several other papers have shown that STAT3 (Ser727) phosphorylation promotes HCC progression by activating STAT3.39, 40 In addition, we have recently reported that STAT3 (Ser727) phosphorylation by Pin1, a peptidyl‐prolyl isomerase, activates STAT3 and enhances tumor progression in gallbladder cancer.41 In the present study, overall survival time and relapse‐free survival time were significantly poorer in HCC patients with high p‐STAT3 (Ser727) labeling index. Moreover, significant correlations were found between p‐STAT3 (Ser727) expression and STAT3 activation in HCC. These results in the present study clearly showed that STAT3 (Ser727) phosphorylation promoted HCC progression by activating STAT3. Therefore, STAT3 (Ser727) phosphorylation is a useful target for evaluating STAT3 activity mediated by OLFM4‐GRIM19 cascade in the present study.
The interaction between OLFM4 and GRIM19 is not fully understood. Only a few reports have suggested that OLFM4 negatively regulates GRIM19 expression in Hela cells and gastric cancer cells in vitro.33 However, no reports have shown the correlation in clinical patients. In the present study, we demonstrate that OLFM4 expression is negatively correlated with GRIM19 expression and positively correlated with STAT3 activation. Because STAT3 activation is correlated with cell proliferation in clinical cases of HCC, these data suggest that OLFM4 promotes STAT3 activation by inhibiting GRIM19‐mediated STAT3 inactivation, leading to poor prognosis in patients with HCC. The results that low OLFM4 expression with high GRIM19 expression in HCC was a significant predictor for better prognosis support our hypothesis.
The precise mechanisms of HCC progression by OLFM4‐GRIM19‐STAT3 cascade are confirmed in vitro using two HCC cell lines. We reveal that OLFM4 is the upstream factor in the OLFM4‐GRIM19‐STAT3 cascade, because OLFM4 knockdown significantly increases GRIM19 expression; however, GRIM19 knockdown shows no effects on OLFM4 expression in HCC cells. Moreover, we clarify that STAT3 activation by OLFM4 is mediated through inhibition of GRIM19 function. In brief, OLFM4 knockdown reduces STAT3 activation; however, the effects of OLFM4 knockdown on STAT3 activation are recovered by double knockdown of OLFM4 and GRIM19. OLFM4‐induced STAT3 activation inhibits apoptosis and induces HCC cell proliferation by enhancing cell‐cycle progression. Based on these results, OLFM4‐GRIM19‐STAT3 signaling is a potentially excellent therapeutic target for controlling tumor progression in patients with HCC. Direct inhibition of STAT3 is critical for the survival of normal organs; therefore, STAT3 inhibitor is difficult for clinical use. Up‐regulation of GRIM19 expression in HCC might be another chance for molecular targeting therapy; however, no effective drugs exist for up‐regulating GRIM19 expression. Moreover, the development of drugs up‐regulating the target factor is sometimes difficult. We believe that OLFM4 is the favorable target for controlling OLFM4‐GRIM19‐STAT3 cascade. At present, OLFM4 inhibitor has not developed yet; therefore, we focus on the upstream pathway of this cascade.
Inflammation‐related factors, such as activator protein 1, NF‐κB, retinoic acid, and granulocyte colony‐stimulating factor, have been reported to regulate OLFM4 expression.20, 42, 43, 44 In addition, recent studies have demonstrated that LGR5 increases OLFM4 expression through the Wnt signaling pathway and promotes stem cell property in the intestine.27, 28, 29 LGR5 is reported to be expressed in HCC45, 46; however, no reports have focused on the interaction between LGR5 and OLFM4. Moreover, no previous studies have shown the effects of LGR5 or OLFM4 on cancer stem cell–like property in any malignancies. In the present study, we clearly demonstrate that LGR5 induces OLFM4 expression through the Wnt signaling pathway and up‐regulates STAT3 activation, leading to enhanced cancer stem cell–like property and accelerated cell‐cycle progression in HCC. As these results show, increased LGR5 expression is associated with poor prognosis in patients with HCC. Cancer stem cell–like property, such as anoikis resistance, is known to be strongly correlated with hematogenous distant metastasis, because anoikis resistance enhances the resistance to apoptosis after losing contact with the extracellular matrix and accelerates colonization at the distant site. In the present study, we reveal that the incidence of hematogenous recurrence is higher in HCC patients with high OLFM4 expression. Therefore, therapies controlling the LGR5‐Wnt signaling pathway are assumed to suppress STAT3 activation by down‐regulating OLFM4‐GRIM19 cascade and inhibit HCC metastasis by reducing cancer stem cell–like property. Further in vivo and clinical studies using LGR5 inhibitor or Wnt inhibitor are needed for clinical use.
In conclusion, OLFM4 expression is induced by the LGR5‐Wnt signaling pathway and is strongly associated with aggressive tumor progression and poor prognosis in patients with HCC by accelerating cell‐cycle progression and enhancing cancer stem cell–like property through STAT3 activation. Therefore, OLFM4 is a novel prognostic predictor and a potential therapeutic target for patients with HCC.
Supporting information
Supported by Japan Society for the Promotion of Science (Grant/Award Number: 26462036).
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. Hashem B, El‐Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365:1118‐1127. [DOI] [PubMed] [Google Scholar]
- 2. Rawla P, Sunkara T, Muralidharan P, Raj JP. Update in global trends and etiology of hepatocellular carcinoma. Contemp Oncol (Pozn) 2018;22:141‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C, et al. STAT3 is necessary for proliferation and survival in colon cancer–initiating cells. Cancer Res 2011;71:7226‐7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu WY, Li J, Wu ZS, Zhang CL, Meng XL. STAT3 activation in monocytes accelerates liver cancer progression. BMC Cancer 2011;11:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deng JY, Sun D, Liu XY, Pan Y, Liang H. STAT‐3 correlates with lymph node metastasis and cell survival in gastric cancer. World J Gastroenterol 2010;16:5380‐5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y, Wu C, Zhang C, Li Z, Zhu T, Chen J, et al. TGF‐β‐induced STAT3 overexpression promotes human head and neck squamous cell carcinoma invasion and metastasis through malat1/miR‐30a interactions. Cancer Lett 2018;436:52‐62. [DOI] [PubMed] [Google Scholar]
- 7. Zhang X, Hu F, Li G, Li G, Yang X, Liu L, et al. Human colorectal cancer‐derived mesenchymal stem cells promote colorectal cancer progression through IL‐6/JAK2/STAT3 signaling. Cell Death Dis 2018;9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He W, Wu J, Shi J, Huo YM, Dai W, Geng J, et al. IL22RA1/STAT3 signaling promotes stemness and tumorigenicity in pancreatic cancer. Cancer Res 2018;78:3293‐3305. [DOI] [PubMed] [Google Scholar]
- 9. Alduais S, Alduais Y, Wu X, Li H, Mao J. HMGB1 knock‐down promoting tumor cells viability and arrest pro‐apoptotic proteins via Stat3/NFκB in HepG2 cells. BioFactors 2018;44:570‐576. [DOI] [PubMed] [Google Scholar]
- 10. Lufei C, Ma J, Huang G, Zhang T, Novotny DV, Ong CT, et al. GRIM‐19, a death‐regulatory gene product, suppresses Stat3 activity via functional interaction. EMBO J 2003;22:1325‐1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Okamoto T, Inozume T, Mitsui H, Kanzaki M, Harada K, Shibagaki N, et al. Overexpression of GRIM‐19 in cancer cells suppresses STAT3‐mediated signal transduction and cancer growth. Mol Cancer Ther 2010;9:2333‐2343. [DOI] [PubMed] [Google Scholar]
- 12. Zhou T, Chao L, Rong G, Wang C, Ma R, Wang X. Down‐regulation of GRIM‐19 is associated with STAT3 overexpression in breast carcinomas. Hum Pathol 2013;44:1773‐1779. [DOI] [PubMed] [Google Scholar]
- 13. Hao M, Shu Z, Sun H, Sun R, Wang Y, Liu T, et al. GRIM‐19 expression is a potent prognostic marker in colorectal cancer. Hum Pathol 2015;46:1815‐1820. [DOI] [PubMed] [Google Scholar]
- 14. Ilelisa F, Amarala NS, Alvesa MR, Costab AA, Calsavarac VF, Lordellod L, et al. Prognostic value of GRIM‐19, NF‐κB and IKK2 in patients with high‐grade serous ovarian cancer. Pathol Res Pract 2018;214:187‐194. [DOI] [PubMed] [Google Scholar]
- 15. Li F, Ren W, Zhao Y, Fu Z, Ji Y, Zhu Y, et al. Downregulation of GRIM‐19 is associated with hyperactivation of p‐STAT3 in hepatocellular carcinoma. Med Oncol 2012;29:3046‐3054. [DOI] [PubMed] [Google Scholar]
- 16. Snyder DA, Rivers AM, Yokoe H, Menco BP, Anholt RR. Olfactomedin: purification, characterization, and localization of a novel olfactory glycoprotein. Biochemistry 1991;30:9143‐9153. [DOI] [PubMed] [Google Scholar]
- 17. Shinozaki S, Nakamura T, Iimura M, Kato Y, Iizuka B, Kobayashi M, et al. Upregulation of Reg 1alpha and GW112 in the epithelium of inflamed colonic mucosa. Gut 2001;48:623‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu W, Yan M, Liu Y, Wang R, Li C, Deng C, et al. Olfactomedin 4 down‐regulates innate immunity against Helicobacter pylori infection. Proc Natl Acad Sci USA 2010;107:11056‐11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oue N, Sentani K, Noguchi T, Ohara S, Sakamoto N, Hayashi T, et al. Serum olfactomedin 4 (GW112, hGC) in combination with Reg IV is a highly sensitive biomarker for gastric cancer patients. Int J Cancer 2009;125:2383‐2392. [DOI] [PubMed] [Google Scholar]
- 20. Kim KK, Park KS, Song SB, Kim KE. Up regulation of GW112 gene by NF kappaB promotes an antiapoptotic property in gastric cancer cells. Mol Carcinog 2010;49:259‐270. [DOI] [PubMed] [Google Scholar]
- 21. Liu RH, Yang MH, Xiang H, Bao LM, Yang HA, Yue LW, et al. Depletion of OLFM4 gene inhibits cell growth and increases sensitization to hydrogen peroxide and tumor necrosis factor‐alpha induced‐apoptosis in gastric cancer cells. J Biomed Sci 2012;19:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ran X, Xu X, Yang Y, She S, Yang M, Li S, et al. A quantitative proteomics study on olfactomedin 4 in the development of gastric cancer. Int J Oncol 2015;47:1932‐1944. [DOI] [PubMed] [Google Scholar]
- 23. Liu W, Liu Y, Zhu J, Wright E, Ding I, Rodgers GP. Reduced hGC‐1 protein expression is associated with malignant progression of colon carcinoma. Clin Cancer Res 2008;14:1041‐1049. [DOI] [PubMed] [Google Scholar]
- 24. Huang MY, Wang HM, Chang HJ, Hsiao CP, Wang JY, Lin SR. Overexpression of S100B, TM4SF4, and OLFM4 genes is correlated with liver metastasis in Taiwanese colorectal cancer patients. DNA Cell Biol 2012;31:43‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kobayashi D, Koshida S, Moriai R, Tsuji N, Watanabe N. Olfactomedin 4 promotes S‐phase transition in proliferation of pancreatic cancer cells. Cancer Sci 2007;98:334‐340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takadate T, Onogawa T, Fukuda T, Motoi F, Suzuki T, Fujii K, et al. Novel prognostic protein markers of resectable pancreatic cancer identified by coupled shotgun and targeted proteomics using formalin‐fixed paraffin‐embedded tissues. Int J Cancer 2013;132:1368‐1382. [DOI] [PubMed] [Google Scholar]
- 27. Jang BG, Lee BL, Kim WH. Intestinal stem cell markers in the intestinal metaplasia of stomach and Barrett's esophagus. PLoS One 2015;21:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu W, Li H, Hong SH, Piszczek GP, Chen W, Rodgers GP. Olfactomedin 4 deletion induces colon adenocarcinoma in ApcMin/+ mice. Oncogene 2016;35:5237‐5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol 2009;10:468‐477. [DOI] [PubMed] [Google Scholar]
- 30. Chen L, Li H, Liu W, Zhu J, Zhao X, Wright E, et al. Olfactomedin 4 suppresses prostate cancer cell growth and metastasis via negative interaction with cathepsin D and SDF‐1. Carcinogenesis 2011;32:986‐994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Rodriguez‐Canales J, Liu W, Zhu J, Hanson JC, Pack S, et al. Deletion of the olfactomedin 4 gene is associated with progression of human prostate cancer. Am J Pathol 2013;183:1329‐1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiong B, Lei X, Zhang L, Fu J. The clinical significance and biological function of olfactomedin 4 in triple negative breast cancer. Biomed Pharmacother 2017;86:67‐73. [DOI] [PubMed] [Google Scholar]
- 33. Zhang X, Huang Q, Yang Z, Li Y, Li CY. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res 2004;64:2474‐2481. [DOI] [PubMed] [Google Scholar]
- 34. Huang Y, Yang M, Yang H, Zeng ZC. Upregulation of the GRIM‐19 gene suppresses invasion and metastasis of human gastric cancer SGC‐7901 cell line. Exp Cell Res 2010;316:2061‐2070. [DOI] [PubMed] [Google Scholar]
- 35. Shinoda K, Kuboki S, Shimizu H, Ohtsuka M, Kato A, Yoshitomi H, et al. Pin1 facilitates NF‐κB activation and promotes tumour progression in human hepatocellular carcinoma. Br J Cancer 2015;113:1323‐1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang Y, Yang M, Hu H, Zhao X, Bao L, Huang D, et al. Mitochondrial GRIM‐19 as a potential therapeutic target for STAT3‐dependent carcinogenesis of gastric cancer. Oncotarget 2016;7:41404‐41420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang J, Yang J, Roy SK, Tininini S, Hu J, Bromberg JF, et al. The cell death regulator GRIM‐19 is an inhibitor of signal transducer and activator of transcription 3. Proc Nat Acad Sci USA 2003;100:9342‐9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D, et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin‐6/STAT3 signaling pathway. Hepatology 2018;67:171‐187. [DOI] [PubMed] [Google Scholar]
- 39. Kong J, Kong F, Gao J, Zhang Q, Dong S, Gu F, et al. YC‐1 enhances the anti‐tumor activity of sorafenib through inhibition of signal transducer and activator of transcription 3 (STAT3) in hepatocellular carcinoma. Mol Cancer 2014;13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miyakoshi M, Yamamoto M, Tanaka H, Ogawa K. Serine 727 phosphorylation of STAT3: an early change in mouse hepatocarcinogenesis induced by neonatal treatment with diehtylnitrosamine. Mol Carcinog 2014;53:67‐76. [DOI] [PubMed] [Google Scholar]
- 41. Nakada S, Kuboki S, Nojima H, Yoshitomi H, Furukawa K, Takayashiki T, et al. Roles of Pin1 as a key molecule for EMT induction by activation of STAT3 and NF‐κB in human gallbladder cancer. Ann Surg Oncol 2019;26:907‐917. [DOI] [PubMed] [Google Scholar]
- 42. Chin KL, Aerbajinai W, Zhu J, Drew L, Chen L, Liu W, et al. The regulation of OLFM4 expression in myeloid precursor cells relies on NF‐kappaB transcription factor. Br J Haematol 2008;143:421‐432. [DOI] [PubMed] [Google Scholar]
- 43. Liu W, Lee HW, Liu Y, Wang R, Rodgers GP. Olfactomedin 4 is a novel target gene of retinoic acids and 5‐aza‐2′‐deoxycytidine involved in human myeloid leukemia cell growth, differentiation, and apoptosis. Blood 2010;116:4938‐4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang J, Liu WL, Tang DC, Chen L, Wang M, Pack SD, et al. Identification and characterization of a novel member of olfactomedin‐related protein family, hGC‐1, expressed during myeloid lineage development. Gene 2002;283:83‐93. [DOI] [PubMed] [Google Scholar]
- 45. Chen W, Fu Q, Fang F, Fang J, Zhang Q, Hong Y. Overexpression of leucine‐rich repeat‐containing G protein‐coupled receptor 5 predicts poor prognosis in hepatocellular carcinoma. Saudi J Biol Sci 2018;25:904‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Effendi K, Yamazaki K, Fukuma M, Sakamoto M. Overexpression of leucine‐rich repeat‐containing G protein‐coupled receptor 5 (LGR5) represents a typical Wnt/β‐catenin pathway‐activated hepatocellular carcinoma. Liver Cancer 2014;3:451‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


