Abstract
HIV-1 protease inhibitors are effective in HIV/AIDS therapy, although drug resistance is a severe problem. This study examines the effects of four investigational inhibitors against HIV-1 protease with drug resistant mutations of V32I, I47V and V82I (PRTri) that model the inhibitor-binding site of HIV-2 protease. These inhibitors contain diverse chemical modifications on the darunavir scaffold and form new interactions with wild type protease, however, the measured inhibition constants for PRTri mutant range from 17 to 40 nM or significantly worse than picomolar values reported for wild type enzyme. The X-ray crystal structure of PRTri mutant in complex with inhibitor 1 at 1.5 Å resolution shows minor changes in interactions with inhibitor compared with the corresponding wild type PR complex. Instead, the basic amine at P2 of inhibitor together with mutation V82I induces two alternate conformations for the side chain of Arg8 with new interactions with inhibitor and Leu10. Hence, inhibition is influenced by small coordinated changes in hydrophobic interactions.
Keywords: HIV-1 protease, Antiviral inhibitors, Drug resistance, X-ray crystallography
1. Introduction
HIV/AIDS is a pandemic disease with about 37 million people infected worldwide [1]. HIV infection can be controlled by antiviral drugs targeting different stages of viral replication, however, the genetic diversity of the virus and rapid selection of drug resistant strains pose a severe challenge [2,3]. The retrovirus HIV includes two types, HIV-1 and HIV-2, and HIV-1 is subdivided into three groups (M, O, and N) and subtypes with different geographical distribution. HIV-2 infections are common in West Africa, however, some drugs designed for HIV-1 are not effective for HIV-2 infections [4].
One important class of antiviral drugs targets the viral protease (PR), which is crucial for production of infectious virus. PR processes cleavage sites in Gag-Pol region during viral maturation to produce individual structural proteins [5]. This aspartic protease is catalytically active as a dimer of 99-residue monomers [6]. Clinical inhibitors bind in the active site cavity of the enzyme and block binding of substrates. Some HIV-1 PR inhibitors, including amprenavir (APV), are significantly less potent against HIV-2 infections probably because the amino acid sequences of HIV-1 and HIV-2 PRs show only about 40% sequence identity [7]. In addition to its natural genetic diversity, the virus has evolved resistance mutations for all clinical protease inhibitors (PIs) [8]. Single, major mutations decrease binding of inhibitor, however, they can be deleterious for viral replication. Resistance mutations can alter the catalytic activity, binding affinity and stability of PR [9,10]. The virus evolves additional mutations that compensate by restoring effective viral replication in the presence of inhibitor [11].
HIV-1 PR with drug resistant mutations V32I, I47V and V82I (PRTri) has been evaluated as a model for inhibition of HIV-2 PR to overcome the problem of autoproteolysis of HIV-2 PR. The three mutations in PRTri alter residues in the inhibitor-binding cavity (Fig. 1A) and represent the changes in the inhibitor binding site of HIV-2 protease. APV showed poor inhibition of both PRTri and HIV-2 PR at 15- and 19-fold worse than for HIV-1 PR, while darunavir (DRV) and saquinavir (SQV) were effective inhibitors of all three enzymes [12]. Moreover, the individual mutations of V32I, I47V and V82I are associated with resistance to one or more clinical inhibitors [8]. Hence, PRTri was chosen to evaluate the ef-ficacy of investigational inhibitors for drug-resistant mutants and HIV-2 PR.
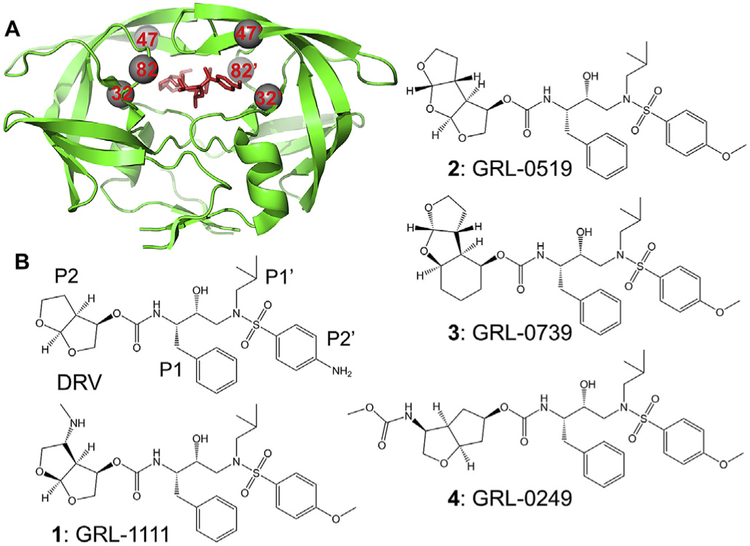
Fig. 1. HIV PR dimer with sites of mutations and selected inhibitors.
A) Sites of mutation in HIV-1 PR dimer with inhibitor GRL-1111. PR is shown as green ribbons with sites of the three mutations, V32I, I47V and V82I, indicated as grey spheres. Inhibitor is shown in red sticks.
B) Structures of darunavir and investigational inhibitors. The P2-P2’ groups are indicated for DRV. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The four investigational antiviral inhibitors were designed on the DRV scaffold (Fig. 1B) with the aim of introducing new interactions of the P2 group with PR. They exhibited potent antiviral activity on wild type and drug resistant strains of HIV-1. Crystal structures confirmed the presence of new interactions of wild type PR with the P2 groups of these inhibitors compared to DRV. Inhibitor 1 has a novel P2 bis-THF group with a basic amine that forms direct and water-mediated hydrogen bond interactions with the main chain carbonyl oxygen and amide of Gly48 in the PR flap [13]. Inhibitor 2 contains tris-tetrahydrofuranylurethane (tris-THF) as the P2 ligand instead of bis-THF in DRV [14–16]. The third THF group introduces new water-mediated hydrogen bond interactions with Gly27 and Arg87 in the PR dimer interface. Inhibitor 3 has a novel tricycle cyclohexyl-bis-tetrahydrofuranylurethane at P2, and resembles inhibitor 2 in its new water-mediated interactions [17]. This inhibitor showed favorable penetration of the central nervous system (CNS) in an in vitro model [18]. Inhibitor 4 has a 3-(S)eN-methoxycarbonyl amino substituted cyclopentyltetrahydrofuranyl (Cp-THF) at the P2 group [19]. The carbamate NH of the ligand forms hydrogen bonds with the main chain of Gly48 similar to those of inhibitor 1 and the carbamate carbonyl has a water-mediated interaction with the guanidinium group of Arg8. Therefore, we hypothesized they would show good inhibition of PRTri. Inhibition constants were measured for the four compounds, and a high resolution X-ray structure was determined for PRTri in complex with inhibitor 1.
2. Materials and methods
2.1. Inhibitors
Amprenavir (APV) (HPLC purity of 99.7%) was obtained from the AIDS Reagent Program, Division of AIDS, NIAID, NIH. Compounds 1–4 (>95.0% purity by HPLC) were provided by Dr. Arun Ghosh at Purdue University.
2.2. Protein purification
The clone for triple mutant PRTri (V32I, I47V and V82I) includes optimizing mutations of Q7K, L33I, and L63I to decrease autoproteolysis, and C67A, C95A to eliminate cysteine-thiol oxidation [20]. Protein was expressed in E. coli BL21 and purified from inclusion bodies as described previously [21,22] using gel filtration followed by reverse phase chromatography and refolding. Samples were concentrated to 5.0 mg/mL for crystallization or diluted for kinetic assays.
2.3. Protein crystallography
PRTri was mixed with inhibitor (dissolved in dimethylsulfoxide) at a molar ratio of 1:5. Crystals were grown at room temperature by hanging drop vapor diffusion. Each drop contained 1 mL protein and 1 mL reservoir solution. Crystals of PRTri in complex with compound 1 were grown from 0.1 M sodium acetate, pH 4.6, and 2 M NaCl. Crystals were cryo-protected in 30% glycerol and flash frozen in liquid nitrogen. X-ray diffraction data were collected on the SERCAT 22BM beamline, Advanced Photon Source, Argonne National Laboratory (Argonne, IL), and processed using HKL-2000 [23]. The structure was solved by molecular replacement with PR complex with APV (3NU3) [24] using CCP4i Phaser [25,26]. The structure was refined with SHELX-2014 [27], followed by REFMAC5 [28]. COOT [29] was used for visualization and refitting. Alternate conformations were modeled according to the electron density maps. Anisotropic B factors were applied in the refinement. Structural figures were made using PyMOL [30]. Atomic coordinates and structure factors for PRTri/1 have been deposited in the PDB [31] with ID: 6OTG.
2.4. Enzyme kinetic assays
Kinetic parameters of PRTri were determined in 3e5 replicate runs by monitoring hydrolysis of fluorescence substrate derived from the HIV-1 p2/NC cleavage site: Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 (BACHEM H-2992) (where Abz is anthranilic acid, Nle is norleucine, and p-nitro-Phe is p-nitrophenylalanine). Samples were equilibrated at 37 °C for 5 min prior to initiating the reactions. Enzyme activity was measured at 37 °C using a PolarStar Optima microplate reader (BMG Labtech) with excitation wavelength at 340 nm and emission wavelength at 420 nm, as described previously [24,32]. To determine catalytic efficiency, 10 μL of PRTri (final well concentration of 40–100 nM determined by active site titration with APV) was mixed with 100 μL reaction buffer (100 mMMES pH 5.6, 400 mM NaCl, 1 mM EDTA, and 5% glycerol). Reaction was initiated by adding 100 μL substrate (12–96 μM final concentration). Initial velocities (V0) were determined using MARS software (BMG Labtech). The Km and kcat were determined by fitting data to the Michaelis-Menten plot of V0 vs [substrate].
For inhibition studies, 10 μL PRTri was mixed with 98 μL reaction buffer and 2 μL inhibitor in DMSO (final well concentration 0–40 mM). Reaction was initiated with 90 μL substrate (final well concentration 60 mM). IC50 values were determined using Sigma-Plot (Systat Software) by non-linear regression curve fitting to a dose-response plot of V0 vs [inhibitor]. Ki values were calculated using the equation for tight-binding inhibitor of Ki= (IC50 - 0.5[E])/ (1 þ [S]/Km) [33].
For the urea denaturation assay, 10 mL PRTri was mixed with 100 mL reaction buffer containing 8 different urea concentrations (0–4 μM). Reaction was initiated with 90 μL substrate (final well concentration 60–72 μM) in 0–4 M urea. The urea concentration in each well remained the same throughout the experiment. A plot of V0 vs [urea] was constructed using SigmaPlot (Systat Software) and sigmoidal curve fitting used to determine the urea concentration for 50% maximum velocity (UC50).
3. Results
3.1. Kinetic parameters, inhibition and stability of PRTri mutant
Kinetic parameters were determined for PRTri hydrolysis of a fluorescent substrate analog at 37 °C prior to assaying the effects of compounds 1–4 under the same conditions. PRTri mutant had kcat of 299 ± 48 μM/min and Km of 72 ± 17.7 μM. The catalytic efficiency (kcat /Km) of 4.2 μM−1 min 1 is similar to that value of 6.5 μM 1 min 1 for wild type enzyme at 26° C [34].
The measured Ki values were 31.4 ± 3.9 nM for compound 1, 38.4 ± 1.2 nM for compound 2, 16.7 ± 0.8 nM for compound 3, and 39.5 ± 6.1 nM for compound 4. These Ki values were significantly worse for the mutant compared to Ki values of 2 to 10 pM reported for the wild type enzyme [13,15,17,19]. Therefore, these antiviral inhibitors are unlikely to be effective on HIV-1 with these mutations or on HIV-2.
The stability of the mutant and wild type proteases were assessed by measuring enzyme activity under urea denaturation. A UC50 value of 0.97 ± 0.05 M was obtained for PRTri mutant. In comparison, the wild type enzyme gave a UC50 value of 0.70 ± 0.07 M under the same conditions. These values suggest the mutant dimer is somewhat more stable than the wild type protease.
3.2. Crystallographic analysis of PRTri complex with inhibitor 1
Co-crystallization experiments were performed for the four compounds, however, crystals grew only for PRTri with compound 1. The crystal structure of PRTri/1 was solved at 1.50 Å resolution in the P21212 space group, and refined to R/Rfree values of 13.4/17.2%. Crystallographic statistics are listed in Table 1. The asymmetric unit of the crystal structure contained a dimer of PRTri, and the inhibitor was bound at the active site in two alternate orientations with an occupancy ratio of 0.5/0.5. The solvent was modeled with 133 water, 1 glycerol and 4 formic acid molecules from the crystallization and cryo-protectant solutions. The crystal structure of PRTri/1 was compared with the corresponding complex of wild type PR (PDB ID: 5BRY), which was determined at 1.34 Å resolution in the same space group with isomorphous unit cell dimensions and contained PR dimer with inhibitor bound in two orientations with 0.6/0.4 relative occupancy [13]. The two dimer structures superimposed with a low RMSD value of 0.23 Å on Cα atoms.
Table 1. Crystallographic Data Collection and Refinement Statistics.
(Values in parentheses are given for the highest resolution shell.).
| PRTri/GRL-1111 | |
|---|---|
| Space group | P21212 |
| Unit cell dimensions: (Å) | |
| A | 58.57 |
| B | 86.52 |
| C | 45.32 |
| Resolution range (Final Shell)(Å) | 50–1.50 (1.55–1.50) |
| Unique reflections | 37,188 (2,865) |
| Rmerge (%) overall (final shell) | 5.9 (49.2) |
| I/σ(I) overall (final shell) | 26.6 (3.1) |
| Completeness (%) overall (final shell) | 96.8 (76.4) |
| Redundancy (final shell) | 6.7 (4.1) |
| R (%) | 13.4 |
| Rfree (%) | 17.2 |
| No. of solvent atoms | 151 |
| RMS deviation from ideality | |
| Bonds (Å) | 0.020 |
| Angle distance (degree) | 2.29 |
| Average B-factors (A2) | |
| Wilson B-factor | 17.7 |
| Main-chain atoms | 17.0 |
| Side-chain atoms | 23.6 |
| Inhibitor | 15.7 |
| Solvent | 28.3 |
The interactions of PRTri with inhibitor were analyzed in comparison to those in wild type PR complex. Inhibitor 1 forms six direct hydrogen bonds with PRTri, excluding the interactions of the central hydroxyl with the catalytic Asp25 and 25’ (Fig. 2). Additionally, it forms water-mediated interactions with flap residues Gly48, Ile50 and Ile50’, while one orientation of inhibitor (designated as the “major” conformation) has a water-mediated interaction with the carboxylate side chain of Asp29. PR-inhibitor hydrogen bond interactions are conserved in the mutant and wild type complexes with differences of no greater than 0.2 Å in length for the major inhibitor conformations. This inhibitor was designed to incorporate an amine on bis-THF at P2 that forms a new direct hydrogen bond interaction with the carbonyl oxygen of Gly48, a water-mediated interaction with the amide of Gly48 in the flap region, and a second water-mediated interaction with the side chain of Asp29. These interactions of compound 1 cannot occur with clinical inhibitor DRV, which lacks the basic amine. However, the minor conformation of 1 in PRTri complex has lost the second water and its interaction with Asp29.
Fig. 2. Hydrogen bond interactions of PRTri with inhibitor 1.
Protein is shown as grey sticks and inhibitor in green sticks. Dotted lines indicate hydrogen bond interactions with interatomic distances in Å. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The hydrophobic side chains of residues 32, 47 and 82 are important components of the PR binding site for substrates or inhibitors. In wild type PR/1, Val82 interacts with the P1 and P1’ groups of inhibitors, while Val32 and Ile47 contribute to the binding site for P2 and P2’ groups. Ile82 in the mutant has similar interactions with inhibitor P1 and P1’ as seen for Val82 of wild type enzyme. Val32 and Ile32 show similar van der Waals contacts with the P2 group of 1, while Val47 in the mutant has shifted to form new interactions with P2 compared to Ile47 in the flap. The P2’ group has gained van der Waals interactions with Ile32’ and lost contacts with Val47’ relative to those in the wild type structure.
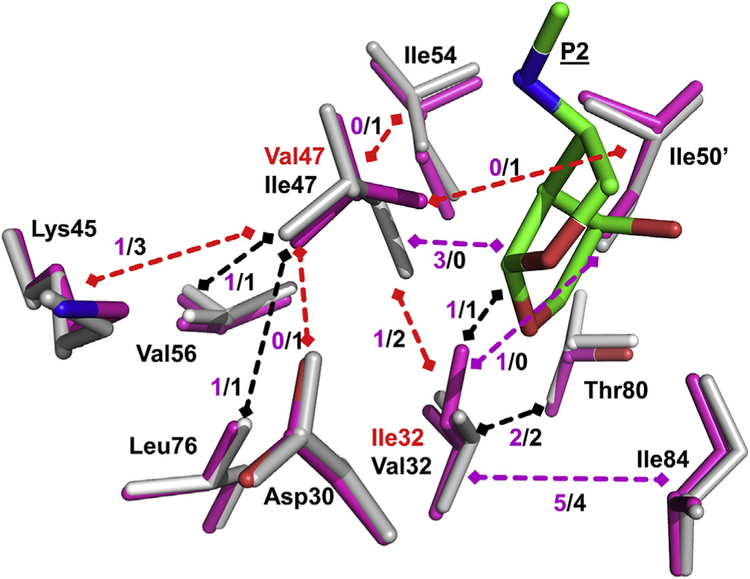
In addition to the contacts with inhibitors, these residues interact with adjacent PR side chains. The side chains of residue 32 and 47 form hydrophobic contacts with each other and with neighboring side chains of Asp30, Lys45, Ile54, Val56, Leu76, Thr80, Ile84 and Ile50’ (Fig. 3). The majority of these internal hydrophobic contacts are retained in the PRTri mutant. The most significant change is a shift of about 0.5 Å in the Ca atoms of both Val47 and 47’ relative to their locations in wild type PR. This change is illustrated in Fig. 3 for Val47/Ile47. This shift enables Val47 to form 3 new hydrophobic contacts with the P2 group of inhibitor, while losing contacts with neighboring side chains of Asp30, Lys45, Ile54 and Ile50’.
Fig. 3. Hydrophobic interactions of residues 32 and 47.
Residues in PRTri are shown in magenta sticks superimposed on corresponding residues of wild type PR in grey. Mutations Ile32 and Val47 are labeled in red, and P2 group of inhibitor is green. Hydrophobic interactions between residues are shown as dashed arrows with the number of contacts indicated. Black arrows indicate identical contacts in both structures, red arrows mark fewer contacts, and magenta arrows show more contacts in mutant than in wild type PR. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Mutation of Val82 to the longer Ile introduces new van der Waals interactions and significant changes in the side chain conformation of Arg8. In the wild type PR complex, the side chain of Arg8 has a single conformation that forms a key intersubunit ion pair with Asp29’ and no contacts with inhibitor. In the PRTri/1 structure, the longer side chain of Ile82/82’ appears to induce two alternate conformations of Arg8/8’ with equivalent changes in both subunits (Fig. 4A). The guanidinium group of one conformation of Arg8 retains the ion pair with Asp29’ and also forms a new van der Waals contact with the P2 group of inhibitor. The second conformation of Arg8 is rotated away to form van der Waals contacts with Leu10 instead of the intersubunit ionic interaction with Asp29’. The second conformation of Arg8/8’ is not observed in the corresponding complex of PRTri/DRV, probably because DRV lacks the extra amine of compound 1 (Fig. 4B). These changes may be due to the local environment, since a multiple mutant PR20 has similar alternative conformations of Arg8/8’ related to the substitution of the larger Phe side chain instead of Leu10 [35].
Fig. 4. Interactions of alternative conformations of Arg8′ with neighboring side chains and inhibitor.
A) Mutant PRTri is shown in magenta sticks with alternate conformation of Arg8’ in cyan and inhibitor 1 in green. Wild type PR and inhibitor are grey. Dotted lines indicate ionic interaction, and dashed lines indicate hydrophobic interactions. CH-p interactions with the aromatic P1 group of inhibitor are indicated by a single dashed line. B) Mutant PRTri in peach bonds with DRV in yellow sticks. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
The four tested investigational antiviral compounds were poor inhibitors (Ki values of 16.7–39.5 nM) of HIV-1 PRTri despite their excellent pM inhibition of wild type enzyme [13,15,17,19]. Analysis of protease-inhibitor interactions in the crystal structure of PRTri/1 revealed the loss of one water-mediated polar interaction in one orientation of inhibitor and small changes in hydrophobic contacts. In particular, Val47 loses hydrophobic contacts with flap residues Lys45, Ile50’ and Ile54 compared to those of wild type complex. The most substantial change compared to the wild type complex occurred for mutated residue Ile82/82’, where the larger side chain produces two alternate conformations for the side chain of Arg8/8’. One conformation of the guanidinium group of Arg8/8’ forms new van der Waals contacts with Leu10 and the P2 group of inhibitor 1 instead of its typical intersubunit ion pair with Asp29’/29. We previously reported the crystal structures of PRTri complexes with APV, DRV and SQV [12]. These structures exhibited a single conformation for Arg8/8’, except for the APV complex which had two alternate conformations for Arg8’ side chain in one subunit and new van der Waals contacts with Ile82’ and Leu10’ similar to those seen in the PRTri/1 structure. Moreover, only a single conformation was observed for Arg8/8’ in our structures of HIV-2 PR with different inhibitors [12,36]. This change in conformation seen for Arg8/8’ in PRTri/1, comprising partial loss of its intersubunit ion pair and new intra-subunit interactions, might be expected to alter the stability of the mutant, however, the UC50 for urea denaturation remains close to the value for wild type PR. In contrast, PR mutant with the single substitution of R8Q, which completely eliminated the ion pair with Asp29’, showed decreased stability with UC50 of 0.7 relative to wild type enzyme [37]. Overall, the loss of internal contacts among flap residues and disruption of the ion pair between Arg8 and Asp29’ are consistent with molecular dynamics simulations suggesting substitutions V32I, I47V and V82I in HIV-2 PR decrease the hydrophobic interactions with APV and DRV [38].
This new structure of PRTri/1 suggests how drug resistant mutations of V32I and I47V on opposite sides of the S2 and S2’ subsites can partially compensate for altered hydrophobic interactions with inhibitors and other protease residues, while mutation V82I induces alternate conformations of Arg8/8’ and introduces new interactions with Leu10 and the P2 group of 1. Structural analysis suggests these changes in hydrophobic interactions are specific for this combination of compound 1 and mutant PRTri and act to decrease the inhibition.
Supplementary Material
Acknowledgements
We thank the staff at the Southeast Regional-Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory, for assistance during X-ray data collection. Supporting institutions may be found at http://www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31–109-Eng-38.
Funding
This research was supported by the National Institutes of Health grant GM062920 and a Georgia State University Molecular Basis of Disease fellowship. Design and synthesis of the compounds was supported by National Institutes of Health grant GM53386.
Footnotes
Conflicts of interest
None declared.
Transparency document
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrc.2019.05.064.
References
- [1].World Health Organization. http://www.who.int/mediacentre/factsheets/fs360/en/,, 2016 April 2019.
- [2].Hemelaar J, Implications of HIV diversity for the HIV-1 pandemic, J. Infect 66 (2013) 391–400. 10.1016/j.jinf.2012.10.026. [DOI] [PubMed] [Google Scholar]
- [3].Clutter DS, Jordan MR, Bertagnolio S, Shafer RW, HIV-1 drug resistance and resistance testing, Infect. Genet. Evol 46 (2016) 292–307. 10.1016/j.meegid.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Menendez-Arias L, Alvarez M, Antiretroviral therapy and drug resistance in human immunodeficiency virus type 2 infection, Antivir. Res 102 (2014) 70–86. 10.1016/j.antiviral.2013.12.001. [DOI] [PubMed] [Google Scholar]
- [5].Konvalinka J, Krausslich HG, Muller B, Retroviral proteases and their roles in virion maturation, Virology 479–480 (2015) 403–417. 10.1016/j.virol.2015.03.021. [DOI] [PubMed] [Google Scholar]
- [6].Tang C, Louis JM, Aniana A, Suh JY, Clore GM, Visualizing transient events in amino-terminal autoprocessing of HIV-1 protease, Nature 455 (2008) 693–696. 10.1038/nature07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Menendez-Arias L, Tozser J, HIV-1 protease inhibitors: effects on HIV-2 replication and resistance, Trends Pharmacol. Sci 29 (2008) 42–49. 10.1016/j.tips.2007.10.013. [DOI] [PubMed] [Google Scholar]
- [8].Wensing AM, Calvez V, Gunthard HF, et al. , Update of the drug resistance mutations in HIV-1, Top Antivir. Med 24 (2017) 132–133, 2017, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5677049/pdf/tam-24-132.pdf. [PMC free article] [PubMed] [Google Scholar]
- [9].Menendez-Arias L, Molecular basis of human immunodeficiency virus type 1 drug resistance: overview and recent developments, Antivir. Res 98 (2013) 93–120. 10.1016/j.antiviral.2013.01.007. [DOI] [PubMed] [Google Scholar]
- [10].Weber IT, Agniswamy J, HIV-1 protease: structural perspectives on drug resistance, Viruses 1 (2009) 1110–1136. 10.3390/v1031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nijhuis M, Schuurman R, de Jong D, et al. , Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy, AIDS 13 (1999) 2349–2359. [DOI] [PubMed] [Google Scholar]
- [12].Tie Y, Wang YF, Boross PI, et al. , Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors, Protein Sci. 21 (2012) 339–350. 10.1002/pro.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ghosh AK, Martyr CD, Osswald HL, et al. , Design of HIV-1 protease inhibitors with amino-bis-tetrahydrofuran derivatives as P2-ligands to enhance backbone-binding interactions: synthesis, biological evaluation, and protein-ligand X-ray studies, J. Med. Chem 58 (2015) 6994–7006. 10.1021/acs.jmedchem.5b00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang H, Wang YF, Shen CH, et al. , Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease, J. Med. Chem 56 (2013) 1074–1083. 10.1021/jm301519z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ghosh AK, Xu CX, Rao KV, et al. , Probing multidrug-resistance and protein-ligand interactions with oxatricyclic designed ligands in HIV-1 protease inhibitors, ChemMedChem 5 (2010) 1850–1854. 10.1002/cmdc.201000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Amano M, Tojo Y, Salcedo-Gomez PM, et al. , GRL-0519, a novel oxatricyclic ligand-containing nonpeptidic HIV-1 protease inhibitor (PI), potently suppresses replication of a wide spectrum of multi-PI-resistant HIV-1 variants in vitro, Antimicrob. Agents Chemother 57 (2013) 2036–2046. 10.1128/AAC.02189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ghosh AK, Parham GL, Martyr CD, et al. , Highly potent HIV-1 protease inhibitors with novel tricyclic P2 ligands: design, synthesis, and protein-ligand X-ray studies, J. Med. Chem 56 (2013) 6792–6802. 10.1021/jm400768f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Amano M, Tojo Y, Salcedo-Gomez PM, et al. , A novel tricyclic ligand-containing nonpeptidic HIV-1 protease inhibitor, GRL-0739, effectively inhibits the replication of multidrug-resistant HIV-1 variants and has a desirable central nervous system penetration property in vitro, Antimicrob. Agents Chemother 59 (2015) 2625–2635. 10.1128/AAC.04757-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ghosh AK, Chapsal BD, Steffey M, et al. , Substituent effects on P2-cyclopentyltetrahydrofuranyl urethanes: design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors, Bioorg. Med. Chem. Lett 22 (2012) 2308–2311. 10.1016/j.bmcl.2012.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wondrak EM, Louis JM, Influence of flanking sequences on the dimer stability of human immunodeficiency virus type 1 protease, Biochemistry 35 (1996) 12957–12962. 10.1021/bi960984y. [DOI] [PubMed] [Google Scholar]
- [21].Sayer JM, Agniswamy J, Weber IT, Louis JM, Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance, Protein Sci 19 (2010) 2055–2072. 10.1002/pro.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Louis JM, Ishima R, Aniana A, Sayer JM, Revealing the dimer dissociation and existence of a folded monomer of the mature HIV-2 protease, Protein Sci 18 (2009) 2442–2453. 10.1002/pro.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Otwinowski Z, Minor W, Processing of X-ray diffraction data collected in oscillation mode, Methods Enzymol. 276 (1997) 307–326. [DOI] [PubMed] [Google Scholar]
- [24].Shen CH, Wang YF, Kovalevsky AY, et al. , Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters, FEBS J. 277 (2010) 3699–3714. 10.1111/j.1742-4658.2010.07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McCoy AJ, Grosse-Kunstleve RW, Adams PD, et al. , Phaser crystallographic software, J. Appl. Crystallogr 40 (2007) 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Winn MD, Ballard CC, Cowtan KD, et al. , Overview of the CCP4 suite and current developments, Acta Crystallogr. Sect. D Biol. Crystallogr 67 (2011) 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sheldrick GM, Crystal structure refinement with SHELXL, Acta Crystallogr. C Struct. Chem 71 (2015) 3–8. 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Murshudov GN, Vagin AA, Dodson EJ, Refinement of macromolecular structures by the maximum-likelihood method, Acta Crystallogr. Sect. D Biol. Crystallogr 53 (1997) 240–255. 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- [29].Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of coot, Acta Crystallogr. Sect. D Biol. Crystallogr 66 (2010) 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].DeLano WL, Pymol: an Open-Source Molecular Graphics Tool, CCP4 Newsletter on Protein Crystallography, 2002, pp. 82–92. https://www.ccp4.ac.uk/newsletters/newsletter40/11_pymol.pdf.
- [31].Berman HM, Westbrook J, Feng Z, et al. , The protein data bank, Nucleic Acids Res. 28 (2000) 235–242. 10.1107/S0907444902003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wong-Sam A, Wang YF, Zhang Y, et al. , Drug resistance mutation L76V alters nonpolar interactions at the flap-core interface of HIV-1 protease, ACS Omega 3 (2018) 12132–12140. 10.1021/acsomega.8b01683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Copeland RA, Lombardo D, Giannaras J, Decicco CP, Estimating K-I values for tight-binding inhibitors from dose-response plots, Bioorg. Med. Chem. Lett 5 (1995) 1947–1952. 10.1016/0960-894x(95)00330-V. [DOI] [Google Scholar]
- [34].Liu F, Kovalevsky AY, Tie Y, et al. , Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir, J. Mol. Biol 381 (2008) 102–115. 10.1016/j.jmb.2008.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Agniswamy J, Shen CH, Aniana A, et al. , HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements, Biochemistry 51 (2012) 2819–2828. 10.1021/bi2018317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kovalevsky AY, Louis JM, Aniana A, et al. , Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 pro-tease, J. Mol. Biol 384 (2008) 178–192. 10.1016/j.jmb.2008.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mahalingam B, Louis JM, Reed CC, et al. , Structural and kinetic analysis of drug resistant mutants of HIV-1 protease, Eur. J. Biochem 263 (1999) 238–245. 10.1046/j.1432-1327.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- [38].Chen J, Drug resistance mechanisms of three mutations V32I, I47V and V82I in HIV-1 protease toward inhibitors probed by molecular dynamics simulations and binding free energy predictions, RSC Adv. (2016) 58573–58585. 10.1039/C6RA09201B. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.