Abstract
Bacteria use a cell-cell communication process called quorum sensing to coordinate collective behaviors. Quorum sensing relies on production and group-wide detection of extracellular signal molecules called autoinducers. Here, we probe the activity of the Pseudomonas aeruginosa LasR quorum-sensing receptor using synthetic agonists based on the structure of the native homoserine lactone autoinducer. The synthetic compounds range from low to high potency, and agonist activity tracks with the ability of the agonist to stabilize the LasR protein. Structural analyses of the LasR ligand binding domain complexed with representative synthetic agonists reveal two modes of ligand binding, one mimicking the canonical autoinducer binding arrangement and the other with the lactone head group rotated approximately 150°. Iterative mutagenesis combined with chemical synthesis reveals the amino acid residues and the chemical moieties, respectively, that are key to enabling each mode of binding. Simultaneous alteration of LasR residues Thr75, Tyr93, and Ala127 converts low-potency compounds into high-potency compounds and converts ligands that are nearly inactive into low-potency compounds. These results show that the LasR binding pocket displays significant flexibility in accommodating different ligands. The ability of LasR to bind ligands in different conformations, and in so doing, alter their potency as agonists, could explain the difficulties that have been encountered in the development of competitive LasR inhibitors.
Graphical Abstract
Quorum sensing (QS) is a process of cell-cell communication used by bacteria to orchestrate collective behaviors. QS involves the production, detection, and group-wide response to extracellular signal molecules called autoinducers (Al)1–5. QS controls genes involved in pathogenesis and biofilm formation in many bacteria6–8. As a result, disrupting QS is viewed as a possible therapeutic alternative to traditional antibiotics9–11. The QS bacterium Pseudomonas aeruginosa is an opportunistic pathogen that affects patients suffering from cystic fibrosis, patients with implanted medical devices, and those with suppressed immune systems, such as cancer sufferers12. P. aeruginosa has developed resistance to many commonly used antibiotics and represents a significant burden with respect to nosocomial infections, making the development of new treatments for P. aeruginosa infections vital13–17.
QS controls P. aeruginosa pathogenesis18. The QS system employs two LuxI/R homoserine lactone (HSL) AI synthase/receptor pairs, Lasl/R and Rhll/R, which produce and detect N-3-oxo-dodecanoyl-L-homoserine lactone (3OC12HSL) and N-butyryl-L-homoserine lactone (C4HSL), respectively7, 19–24. These native Als are structurally characterized by a polar 3-amino-γ-butyrolactone “head group” attached, via an amide bond, to a nonpolar hydrocarbon-based “tail group” (Figure 1A). LuxR-type QS receptors are transcription factors that possess variable N-terminal ligand-binding domains (LBD) and well-conserved C-terminal helix-turn-helix DNA-binding domains25–30. Like other LuxR-type QS receptors, LasR and RhlR require their cognate Als to be bound in order to properly fold, dimerize, bind to target promoter DNA, and initiate transcription28. The LasR-AI complex, in addition to modulating genes required for pathogenesis, activates expression of rhlR and rhll, thus launching the second QS system. This regulatory arrangement enables the two QS systems to function in tandem7, 20. Due to its location at the top of the QS hierarchy, its connection to pathogenesis, and its reliance on a small molecule to function, LasR has garnered interest as a target for medicinal chemistry approaches aimed at limiting virulence31. Efforts to identify potent inhibitors of LasR have been described, but no compound has yet been moved forward to the clinic32–38. Understanding the structural basis underlying LasR activation by different agonists could be informative for addressing challenges in LasR inhibitor design.
Figure 1. Structure activity relationship for AI analogs and LasR.
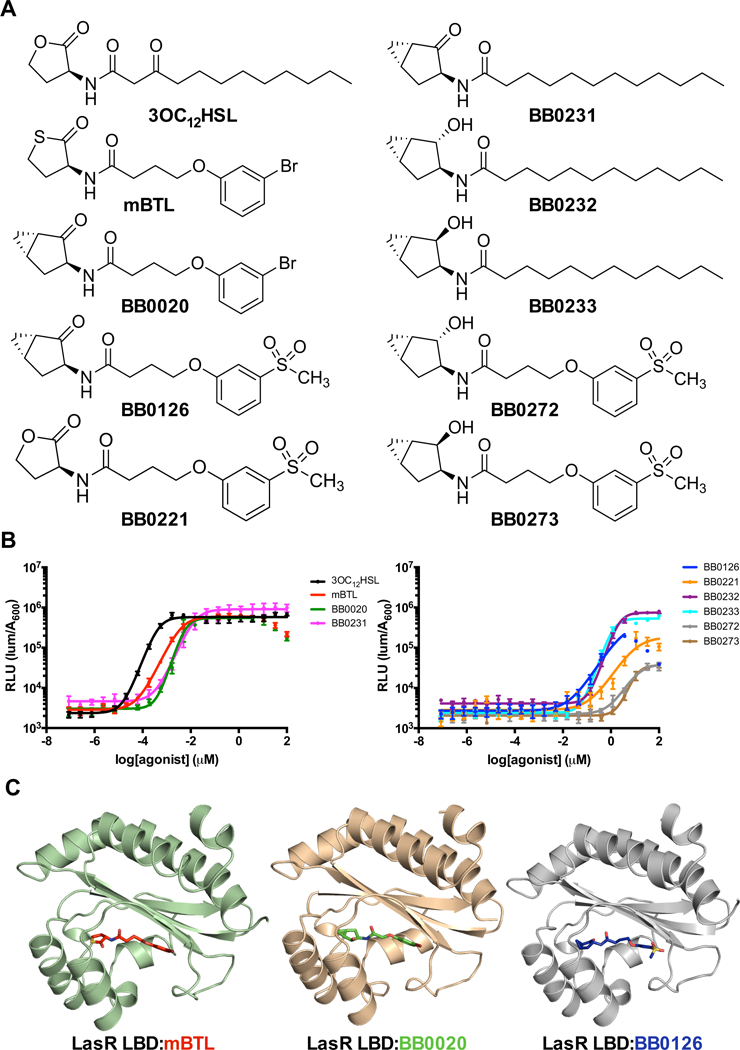
A) Structure of the LasR cognate AI, 3OC12HSL, a previously reported agonist, mBTL, and the AI analogs developed and studied here. B) Dose response analyses for the test compounds using the E. coli lasB-lux reporter strain that contains LasR. RLU is defined as light production per OD (see Methods). Dose response data are depicted as curve fits with the raw data plotted as individual points. Error bars represent SEM, n=3. At high concentrations of mBTL, BB0020, and BB0126, reductions in light levels occur due to off-target (i.e., non LasR-directed) inhibition of luciferase. In these instances, the data points from those concentrations were omitted from the curve fit analyses. C) Crystal structures of LasR LBDs bound to two Class I molecules, mBTL (left) and BB0020 (middle), and a Class II molecule, BB0126 (right).
Analyses of the features driving ligand accommodation by LasR were accelerated when crystal structures were solved of the LasR LBD bound to its cognate HSL, noncognate HSLs, and other agonists39–41. Subsequent mutational analyses revealed that modification of a single LasR residue, Trp60, that hydrogen bonds with the carbonyl moiety of the ligand lactone ring can convert an agonist into an antagonist and vice versa42. Consistent with these findings, substituting the lactone head group on the native AI with a phenyl head group converts the ligand into an antagonist42, 43.
We previously reported that the compound meta-bromo-thiolactone (mBTL) (Figure 1A) was a QS antagonist based on measurements of QS-controlled pyocyanin production. We suggested that mBTL could function by inhibition of one or both QS receptors44. Subsequently, it was shown that mBTL, while indeed inhibiting pyocyanin, is a LasR agonist45. We agree with the validity of these authors’ findings. Here, we report the agonist activity of a set of compounds that combine features from 3OC12HSL and mBTL with novel head groups designed as metabolically stable bioisosteres of the native AI homoserine lactone head group. Our characterization shows that the compounds vary from low to high potency. Crystal structures of the LasR LBD bound to mBTL and to two of these analogs, 4-(3-bromophenoxy)-N-(rel-(1S,3S,5S)-2-oxobicyclo[3.1.0]hexan-3-yl)butanamide (BB0020) and 4-(3-(methylsulfonyl)phenoxy)-N-(rel-(1S,3S,5S)-2-oxobicyclo[3.1.0]hexan-3-yl)butanamide (BB0126), reveal two distinct modes of ligand binding. Strikingly, in the case of BB0126, the head group lies in the opposite orientation relative to that of 3OC12HSL, mBTL, and BB0020. Mutagenesis reveals that LasR residues Thr75, Tyr93, and Ala127 stabilize the alternative BB0126-mode of binding. Indeed, substitutions at these three residues cause BB0126 to reorient back into the canonical binding conformation, as revealed by the crystal structure of the LasR triple mutant LBD complexed with BB0126. Collectively, our findings show that LasR possesses a flexible ligand binding pocket that accommodates multiple orientations of the ligand head and tail groups. The results also reveal the crucial structural elements in LasR that bind to the head and tail groups to confer ligand potency.
RESULTS AND DISCUSSION
Design and analysis of synthetic LasR agonists.
A number of replacements for the hydrolytically sensitive lactone moiety in 3OC12HSL have been reported35, 46–51, however, most of these compounds showed a considerable loss of functional potency. To identify new ligands that modulate QS receptors, we sought to develop structurally novel head groups that would maintain potency comparable to that of the native HSL for LasR while providing improved chemical and metabolic stability. Suga and coworkers52 have reported that replacement of the 3OC12HSL head group with an unsubstituted cyclopentanone led to poor activity for LasR. Nonetheless, our inspection of ligand-bound LasR crystal structures indicated that there exists additional space for placement of small hydrophobic groups around the 5-membered lactone ring system. We reasoned that placement of a fused cyclopropane ring such as that in a bicyclo [3.1.0]hexanone system might provide additional hydrophobic interactions necessary to maintain LasR potency while also providing robust chemical and metabolic stability. We synthesized the requisite (1S,3S,5S)-3-aminobicyclo[3.1.0]hexan-2-one and corresponding 3-aminobicyclo [3.1.0]hexan-2-ols according to a published procedure53 and attached these new head groups to three different tail groups: the dodecanoyl side chain of 3OC12HSL (BB0231, BB0232, and BB0233), the 3-bromophenoxybutanoyl tail group found in mBTL (BB0020), and a novel 3-(methylsulfonyl)phenoxybutanoyl tail group in which the bromine of mBTL was replaced with a methylsulfone group (BB0126, BB0272, BB0273). We also prepared the analog with the lactone head group found in 3OC12HSL and the 3-(methylsulfonyl)phenoxybutanoyl tail piece (BB0221). The structures of the compounds are shown in Figure 1A, the synthetic schemes are in Supplemental Figure 1, and synthetic procedures are in the Supporting Information.
We assessed the activities of our compounds in dose response assays (Figure 1B) using a recombinant E. coli strain that produces LasR and harbors the LasR-activated lasB promoter fused to luxCDABE. Briefly, in this assay, arabinose is used to induce LasR production and LasR is activated by exogenously-supplied ligand. LasR:ligand complexes bind the lasB promoter and activate transcription of the fused lux operon resulting in light production (for more information, see Materials and Methods). High concentrations of some compounds (e.g., mBTL, BB0020, and BB0126) caused off-target inhibition of luciferase; we included these data in the figures but excluded them from the curve fitting used to estimate EC50 values (Table 1).
Table 1.
EC50 values (nM) for LasR agonists in the plasB-lux assay.
| Compound Class | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| I | I | I | II | II | I | II | II | III | III | |
| LasR | 3OC12HSL | mBTL | BB0020 | BB0126 | BB0221 | BB0231 | BB0232 | BB0233 | BB0272 | BB0273 |
| WT | 1 | 4 | 6 | >3100 | 10000 | 20 | 2300 | 1200 | >20000 | >20000 |
| T75V | 1 | 6 | 15 | 1500 | 5600 | 8 | 50 | 390 | NR | NR |
| Y93F | 1 | 10 | 15 | 500 | 7000 | 15 | 1600 | 220 | NR | NR |
| A127W | 5200 | 60 | 200 | 500 | 1200 | 4000 | NR | NR | NR | NR |
| T75V/Y93F | 2 | 10 | 20 | 600 | 3500 | 7 | 100 | 90 | >25000 | >25000 |
| T75V/A127W | 600 | 7 | 20 | 50 | 170 | 330 | NR | NR | 3400 | 11000 |
| Y93F/A127W | 8200 | 9 | 100 | 350 | 400 | 1700 | NR | NR | NR | 11000 |
| T75V/Y93F/A127W | 900 | 10 | 20 | 50 | 200 | 230 | NR | NR | 3700 | 2700 |
| R61A | 140 | 40 | 130 | >10000 | >50000 | 90 | 4800 | 2200 | NR | NR |
| NR = non-responsive | ||||||||||
The compound activities fell into three classes: I) strong agonists with EC50 values comparable to the cognate AI, II) weak agonists with ~1000-fold higher EC50 values than the cognate AI, and III) nearly inactive compounds with ~20000-fold higher EC50 values than the cognate AI (Figure 1B and Table 1). Class I molecules included mBTL, BB0020, and BB0231. Class II molecules included BB0126, BB0221, BB0232, and BB0233. Class III molecules included BB0272 and B0273. Replacement of the lactone/thiolactone head group with our novel bicyclo[3.1.0]hexan-2-one head group (BB0020, BB0231) maintained agonist potency for LasR (Figure 1B). Replacing the bromine in the tail group of BB0020 or mBTL with a methylsulfonyl group dramatically decreased potency for LasR by 780-fold and 2500-fold for BB0126 and BB0221, respectively (Figure 1B and Table 1). Reduction of the ketone moiety to provide the corresponding α- or β-bicyclo [3.1.0]hexan-2-ol derivatives also led to a 120-fold and 60-fold loss in potency (compare BB0231 to BB0232 and BB0233, respectively; Table 1). The loss of potency in going from the ketone to the α- or β-hydroxyl head group was unexpected, as several active HSL head group replacements containing hydroxyl moieties have been reported37, 46, 54. Indeed, it has been shown that both the 2-cyclohexanol and 2-cyclopentanol head groups are more potent LasR agonists than their corresponding 2-ketone derivatives52. Not surprisingly, analogs BB0272 and BB0273, which contain both the alcohol head groups and sulfone tail group, showed almost no activity (Figure 1B and Table 1).
Crystal structures of the LasR LBD bound to mBTL, BB0020, and BB0126 reveal an alternative binding mode.
To understand how our active analogs are accommodated in the LasR ligand binding pocket, we obtained X-ray co-crystal structures of LasR LBD bound to mBTL, BB0020, and BB0126. We chose these three complexes because each could be stably expressed and purified to homogeneity from Escherichia coli. Our crystals diffracted to 1.5 Å (mBTL), 2.2 Å (BB0020), and 2.8 Å (BB0126) (Figure 1C, Table 2, and Supplemental Figure 2A–C). None of the other complexes, surprisingly including LasR bound to BB0231, yielded diffracting crystals.
Table 2.
Crystallographic Statistics
| Dataset | LasR LBD:mBTL | LasR LBD:BB0020 |
LasR LBD:BB0126 |
LasR LBD*:BB0126 |
|---|---|---|---|---|
| Space Group | P 1 21 1 | P 1 21 1 | P 1 21 1 | P 1 21 1 |
| Unit Cell Dimensions (Å) | a=52.1, b=67.3, c=53.8; α=90.0°, β=117°, γ=90.0° | a=42.7, b=62.1, c=50.6; α=90.0°, β=91.8°, γ=90.0° | a=50.9, b=63.6, c=51.8; α=90.0°, b=112°, γ=90.0° | a=45.0, b=72.4, c=51.9, α=90.0°, β=92.6°, γ=90.0° |
| Resolution (Å) | 20.2–1.50 (1.55–1.50) | 29.2–2.19 (2.27–2.19) | 23.78–2.76 (2.86–2.76) | 29.7–1.7 (1.69–1.66) |
| Unique Reflections | 49779 | 12675 | 7739 | 39323 |
| Completeness (%) | 99.1 (92.8) | 92.7 (80.0) | 97.2 (91.6) | 99.67 (97.0) |
| <Redundancy> | 3.4 | 2.0 | 3.2 | 6.8 |
| Rmeas (%) | 4.5 (65.7) | 5.3 (65.9) | 21.1 (92.3) | 4.2 (60.2) |
| < I > / < sigma > | 18.5 (1.8) | 16.5 (1.9) | 7.6 (1.5) | 22.4 (2.7) |
| Total atoms | 2880 | 2651 | 2624 | 2832 |
| Rwork (%) | 19.3 (36.2) | 19.7 (29.3) | 21.8 (29.9) | 20.7 (26.3) |
| Rfree (%) | 22.2 (37.4) | 27.5 (47.0) | 27.7 (30.5) | 23.7 (27.9) |
| Average B-factor | 18.4 | 33.2 | 37.8 | 29.4 |
| R.m.s. deviation from ideality | ||||
| Bond lengths (Å) | 0.007 | 0.011 | 0.010 | 0.006 |
| Bond angles (°) | 1.09 | 1.27 | 1.15 | 0.914 |
| Dihedral angles (°) | 11.2 | 4.2 | 7.4 | 16.7 |
| Phi-Psi values (Ramachandran) | ||||
| Most favored | 98.1 | 96.8 | 96.9 | 98.7 |
| Additionally allowed | 0.9 | 3.2 | 3.1 | 1.0 |
| Outliers | 0.9 | 0 | 0 | 0.3 |
denotes the LasR LBD T75V/Y93F/A127W mutatiion
In the previously reported LasR:3OC12HSL structure39, the ligand acyl tail forms extensive hydrophobic interactions with the hydrophobic walls of the binding pocket, with the terminal carbon adjacent to residue Ala127 in β4 (Figure 2A). The phenyl rings of mBTL, BB0020, and BB0126, bind in a different conformation, with the meta-positioned bromine atoms of mBTL and BB0020 and the sulfonyl group of BB0126 residing near the backbone of Leu39 in β1 (Figure 2A). In all cases, the ligands fill similar volumes in the LBD (Figure 2A). This finding is consistent with recent evidence suggesting that the LasR ligand binding pocket is malleable and can accommodate a variety of ligands41, 55. As mentioned above, LasR Trp60 forms a hydrogen bond with the ketone in the lactone head group of 3OC12HSL. mBTL and BB0020 use this identical interaction for head group binding (Figure 2A). However, the head group of BB0126 is rotated approximately 150° into an opposite orientation, causing the head group ketone to point toward residues Thr75 and Tyr93, at distances of 3.3 Å and 4.9 Å, respectively. We call these two orientations the “canonical binding mode” and the “alternative binding mode”, respectively.
Figure 2. Crystal structure of LasR LBD bound to BB0126 reveals an alternative mode of binding.
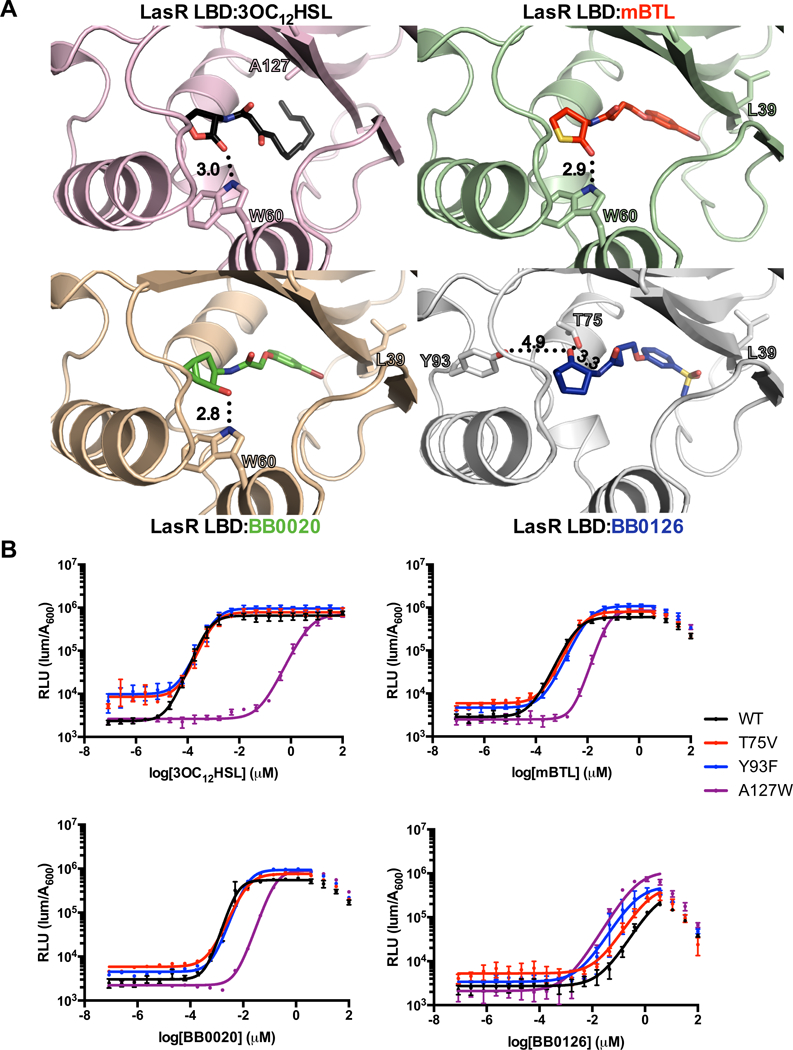
A) Crystal structures of the LasR LBD bound to 3OC12HSL (top left; black; PDB: 2UV039), mBTL (top right; red), BB0020 (bottom left; green), and BB0126 (bottom right; blue). Note that the orientation of the ketone in the lactone head group of BB0126 points in the opposite direction to those in the other three structures. In the structures with 3OC12HSL, mBTL, and BB0020, the W60 residue interacts with the head group ketone whereas in the structure with BB0126, T75 and Y93 are highlighted because of their putative roles in accommodating the alternative mode of binding. Residue A127 is highlighted in the structure with 3OC12HSL and L39 is highlighted in the other three structures for their roles in tail interactions. Important hydrogen bonds are depicted as black dotted lines and the calculated bond distances (Å) are shown. B) Dose response analyses using the E. coli lasB-lux reporter strain that produces WT LasR (black), LasR T75V (red), LasR Y93F (blue), and LasR A127W (purple) with 3OC12HSL (top left), mBTL (top right), BB0020 (bottom left), and BB0126 (bottom right). Dose response data are depicted as curve fits with the raw data plotted as individual points. Error bars represent SEM, n=3. At high concentrations of mBTL, BB0020, and BB0126, reductions in light levels occur due to off-target (i.e., non LasR-directed) inhibition of luciferase. In these instances, data points from those concentrations were omitted from the curve fit analyses.
The surprising finding of the alternative binding mode for BB0126, which differs from BB0020 only by replacement of a bromine atom with a methylsulfonyl group in the tail, indicates that the tail sulfonyl moiety can interact with LasR so as to cause the amide bond connecting the head and tail to rotate and, thus, reorient the head group (Figure 2A). Consequently, the carbonyl oxygen in the BB0126 head group hydrogen bonds with Thr75 (Figure 2A). The rotation of the head group eliminates the key stabilizing hydrogen-bonding interaction with Trp60. The alternative hydrogen bond with Thr75 is longer (3.0 vs 3.3 Å) and therefore, presumably, weaker. We hypothesize that the methylsulfonyl moiety of BB0126, being considerably more polar than the bromine moiety of BB0020, makes fewer favorable stereoelectronic interactions within the hydrophobic tail binding pocket of LasR39–41. This change causes the ligand to shift within the pocket resulting in the reorientation of the head group and, consequently, contributes both directly and indirectly to the overall reduction in agonist potency (Figure 1B, 2A, and Table 1).
Mutational analysis of LasR to probe ligand interactions.
To explore the ramifications of the different ligand head group orientations on LasR activity, we used our crystal structures to guide mutagenesis of the LasR ligand binding pocket followed by analysis of mBTL, BB0020, and BB0126 agonism. We focused on residues Thr75 and Tyr93 due to their potential roles in conferring the alternative binding mode to the BB0126 ligand and on residue A127 for its role in accommodating the AI acyl tail (Figure 2A). We mutated Thr75 to Val and Tyr93 to Phe to eliminate the ability of these side chains to form hydrogen bonds with the ligands, while conserving their space filling roles. We mutated Ala127 to Trp to introduce a larger hydrophobic element into the binding pocket, potentially enabling stabilization of the tails of the synthetic compounds. The LasR T75V and Y93F mutations caused, at most, a modest decrease in the potencies of 3OC12HSL, mBTL, and BB0020, with the EC50 values increasing less than three-fold (Figure 2B and Table 1). The LasR A127W mutation caused much larger (15- to 1000-fold) reductions in the potencies of 3OC12HSL, mBTL, and BB0020 (Figure 2B and Table 1). Unexpectedly, the potency of BB0126 was increased by all three mutations, with the EC50 values at least 2-to 6-fold lower (Figure 2B and Table 1). Similar reductions in EC50 values occurred for BB0221 (Supplemental Figure 3A and Table 1), which also contains a sulfonyl tail group, indicating that this moiety drives this structure-activity relationship. These results bolster our structure-based hypothesis that the sulfonyl tail group is instrumental for the BB0126 alternative binding mode.
To explore the mechanism underlying the increased potency of BB0126 on the mutant LasR proteins relative to that with WT LasR, we combined the LasR T75V, Y93F, and A127W mutations and examined the consequences on the activities of the test compounds. EC50 values for the test molecules with all of the combinations of mutations are shown in Table 1. As expected based on our data for the single point mutations in LasR, the double and triple mutations reduced, sometimes drastically, the potencies of 3OC12HSL, mBTL, and BB0020 (Figure 3A). Strikingly however, combining mutations further increased the potency of BB0126 (Figure 3A), with the triple mutation converting a Class II molecule (EC50 = >3.1 μM) into a Class I molecule (EC50 = 50 nM) (Figure 3A and Table 1). In the case of BB0221, both the potency and the efficacy increased in the LasR T75V/Y93F/A127W mutant relative to WT (Table 1 and Supplemental Figure 3B). Presumably, these mutations enable LasR to place BB0126 (and likely BB0221) into a highly stable conformation, potentially a canonical binding conformation.
Figure 3. LasR T75V/Y93F/A127W enhances BB0126, BB0272, and BB0273 potency.
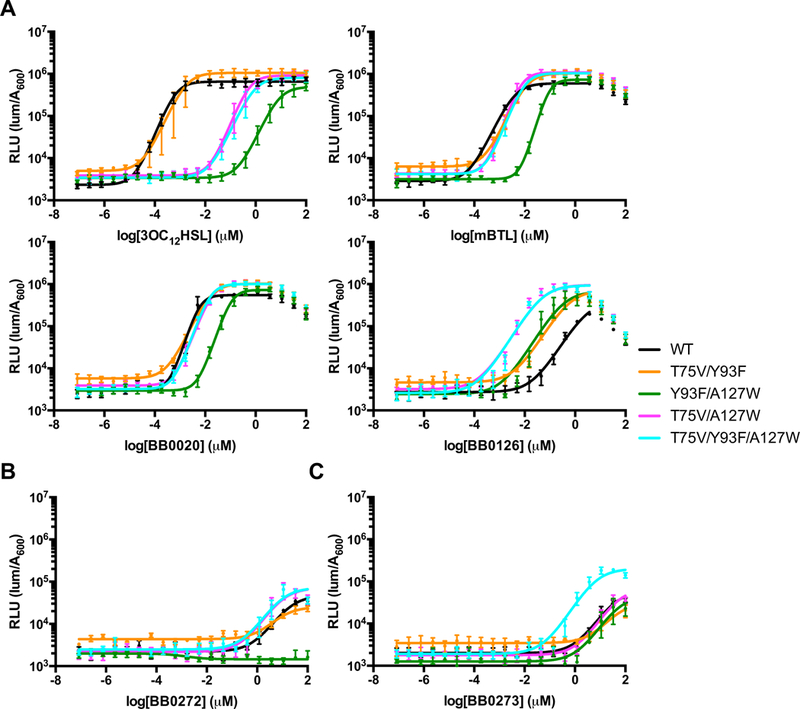
A) Dose response analyses using the E. coli lasB-lux reporter strain that contains WT LasR (black), LasR T75V/Y93F (orange), LasR Y93F/A127W (green), LasR T75V/A127W (magenta), and LasR T75V/Y93F/A127W (cyan) to 3OC12HSL (top left), mBTL (top right), BB0020 (bottom left), and BB0126 (bottom right). B) As in panel A for the BB0272 ligand. C) As in panel A for the BB0273 ligand. Dose response data are depicted as curve fits with the raw data plotted as individual points. Error bars represent SEM, n=3. At high concentrations of mBTL, BB0020, and BB0126, reductions in light levels occur due to off-target (i.e., non LasR-directed) inhibition of luciferase. In these instances, data points from those concentrations were omitted from the curve fit analyses.
To determine whether triple mutation of LasR exclusively enhanced BB0126 and BB0221 activity, or whether the changes had broader consequences – for example, on Class III ligands – we tested the response of LasR T75V/Y93F/A127W to the Class III molecules BB0272 and BB0273. Indeed, BB0272 and BB0273 had at least 5-fold and 7-fold lower EC50 values for LasR T75V/Y93F/A127W than for WT LasR (Figure 3B and 3C, respectively). Below, we focus on the LasR T75V/Y93F/A127W mutant protein because it had the most pronounced effects on ligand potency.
The LasR T75V/Y93F/A127W mutation enhances BB0126 recognition in P. aeruginosa.
To examine the consequences that mutations altering LasR ligand preference have on QS-controlled gene expression in vivo, we engineered P. aeruginosa PA14 to express lasR T75V/Y93F/A127W from the native lasR chromosomal locus and assayed rhlA expression using a rhlA promoter fusion to mNeonGreen (prhlA-mNG). We performed this analysis in a Δlasl parent strain to eliminate endogenously-produced AI. We tested 3OC12HSL, BB0020, and BB0126. Figure 4A shows that WT LasR activated prhlA-mNG in response to 3OC12HSL and BB0020, but not in response to BB0126. By contrast, LasR T75V/Y93F/A127W activated the reporter in response to BB0020 and BB0126, but not in response to 3OC12HSL.
Figure 4. LasR T75V/Y93F/A127W enhances BB0126 efficacy in P. aeruginosa.
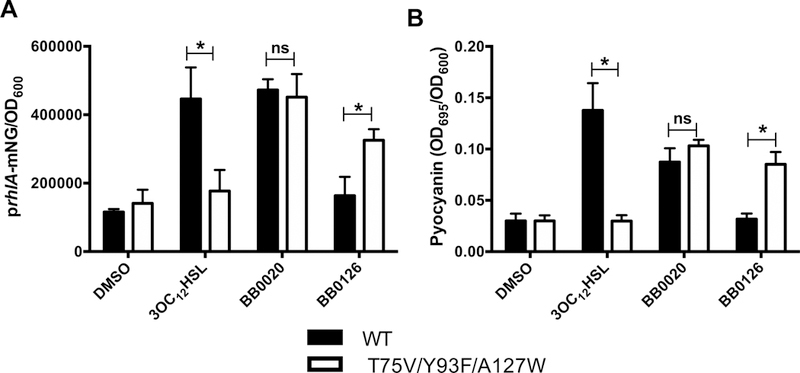
A) The rhlA promoter was fused to mNeonGreen (mNG) ad introduced into the endogenous rhlA locus in the chromosome of ΔlasI P. aeruginosa PA14. Either WT lasR (black) or lasR T75V/Y93F/A127W (white) was engineered into the lasR chromosomal locus. Strains were grown to OD600 = 2.0 and the responses to 10 μM 3OC12HSL, BB0020, or BB0126 were monitored by measuring fluorescence. B) As in panel A except pyocyanin production was measured by A695. Error bars represent SEM, n=3.Statistical significance was determined by multiple t-tests comparing grouped columns. * P < 0.005.
To assess how alterations in ligand preference influence an endogenous LasR-controlled behavior, we monitored the production of pyocyanin in response to 3OC12HSL, BB0020, and BB0126 in the Δlasl strain containing WT LasR or LasR T75V/Y93F/A127W. WT LasR induced pyocyanin production in response to 3OC12HSL, whereas LasR T75V/Y93F/A127W did not (Figure 4B). Conversely, LasR T75V/Y93F/A127W responded to BB0126 while WT LasR did not (Figure 4B). As was the case for the prhlA-mNG reporter, BB0020 activated both LasR alleles equally, resulting in nearly identical pyocyanin production levels (Figure 4B).
LasR:ligand interactions drive stability.
Our previous work showed that ligand potency for LasR tracked with the ability of the ligand to stabilize the LasR protein, thus enhancing the solubility of the complex41. To determine whether this mechanism underpinned the differential activation of WT LasR by the synthetic molecules, we assessed overall soluble protein levels as a measure of LasR-ligand complex stability. Following growth at 37°C, we induced LasR LBD production in E. coli at either 25°C or 37°C in the presence of 10 μM of our test compounds. At 25°C, 3OC12HSL, mBTL, BB0020, and BB0126 yielded soluble WT LasR LBD (Figure 5A and 5B). However, at 37°C, the WT LasR LBD was significantly less soluble in the presence of BB0126 (Figure 5A and 5B). Thus, the LasR LBD:BB0126 complex is less stable than the LasR LBD bound to 3OC12HSL, mBTL, or BB0020.
Figure 5. Ligand potency drives LasR stability.
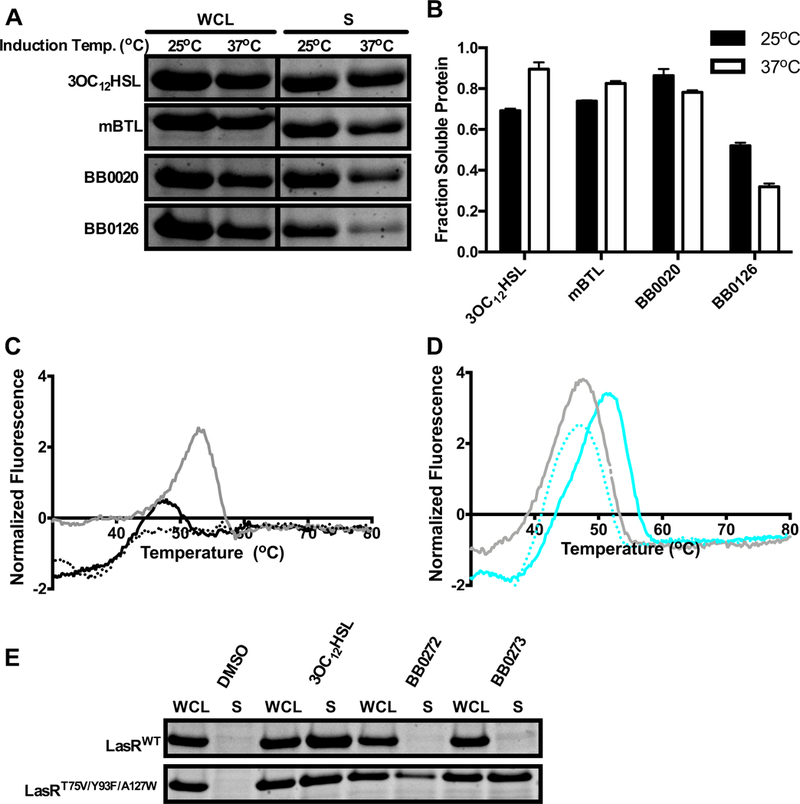
A) Comparison of whole cell lysates (WCL) and the soluble fractions (S) from E. coli overexpressing the LasR LBD after 4 h of protein induction at 25°C or 37°C in the presence of 10 μM 3OC12HSL, mBTL, BB0020, or BB0126. B) Quantification of the data in panel A as the fraction of soluble protein at induction temperatures of 25°C (black) and 37°C (white). The fraction of soluble protein was calculated by dividing the quantified band intensity of the soluble fraction (S) by the quantified band intensity of the whole cell lysate (WCL). C) Thermal shift analyses of LasR LBD:BB0126 without additional BB0126 (dotted black line), with additional BB0126 (solid black line), or with additional 3OC12HSL provided (solid gray line). All molecules were supplied at 10 μM. D) Thermal shift analyses of LasR LBD T75V/Y93F/A127W:BB0126 without additional BB0126 (dotted cyan line), with additional BB0126 (solid cyan line), or with additional 3OC12HSL (solid gray line). All molecules were supplied at 10 μM. E) Comparison of whole cell lysates (WCL) and the soluble fractions (S) from E. coli overexpressing LasR LBD T75V/Y93F/A127W in the absence of ligand (DMSO) or in the presence of 3OC12HSL, BB0272, or BB0273.
To explore the mechanism underlying the inability of BB0126 to stabilize LasR at high temperature, we purified the LasR LBD bound to BB0126 and subjected it to thermal shift analysis. LasR LBD:BB0126 (Figure 5C, black dotted line) was unstable above room temperature, and therefore we could not calculate an accurate melting temperature. This result is consistent with the findings from the solubility assay (Figure 5A and 5B). In the presence of excess BB0126, however, LasR exhibited a melting temperature peak of 49.9°C (Figure 5C, black solid line), indicating that as pre-bound ligand is released, exogenously supplied ligand can bind and stabilize the protein. Exogenous 3OC12HSL (gray solid line) was highly effective at binding to and stabilizing the LasR LBD as the BB0126 ligand was released (Figure 5C). These results indicate that WT LasR is more stable when bound to 3OC12HSL than BB0126, explaining the relative potencies of these molecules in activating WT LasR.
To assess whether the increased potency of BB0126 for LasR T75V/Y93F/A127W could be due to enhanced stability of the LasR T75V/Y93F/A127W:BB0126 complex compared to the WT LasR:BB0126 complex, we purified the LasR LBD T75V/Y93F/A127W bound to BB0126 for thermal shift analysis. This complex was stable and, unlike the WT complex, displayed a well-defined melting transition (Tm = 46.8°C; cyan dotted line in Figure 5D). Excess BB0126 further enhanced the Tm (solid cyan line). In contrast, excess 3OC12HSL had no effect (solid gray line), consistent with its inability to activate LasR T75V/Y93F/A127W (Figure 3A). Notably, the finding that LasR T75V/Y93F/A127W:BB0126 is more stable than WT LasR:BB0126 strongly supports the notion that the stability of the LasR:ligand complex plays a major role in determining the potency of a ligand.
To further explore the relationship between ligand potency and the ability of ligands to stabilize soluble LasR, we examined representative Class III molecules. Neither BB0272 nor BB0273 could solubilize the WT LasR LBD (Figure 5E), likely leading to the low potency observed with these ligands. Conversely, the LasR T75V/Y93F/A127W LBD was solubilized by BB0272 and even more so by BB0273 (Figure 5E), in good agreement with the improvements in potency conferred to these two molecules by the triple mutation (Figure 3B, 3C and Table 1). These findings are not the result of LasR T75V/Y93F/A127W being inherently more stable than WT LasR, as both LasR proteins were insoluble in the absence of ligand (Figure 5E).
Crystal structure of LasR T75V/Y93F/A127W bound to BB0126 reveals a new mode of binding for high-potency ligands.
To understand how the LasR low-potency ligand BB0126 becomes a high-potency ligand for LasR T75V/Y93F/A127W, we determined the crystal structure of the LasR LBD T75V/Y93F/A127W:BB0126 complex. Unlike WT LasR, the triple mutant binds the BB0126 head group in the canonical orientation (Figure 6A, Table 2, and Supplemental Figure 2D). Reorientation of the head group back to the canonical state is presumably caused by the loss of hydrogen bonds between the head group carbonyl and Thr75 and/or Tyr93. Combined, the mutations allow the ligand to adopt the high-potency head group conformation in the ligand binding pocket. Additionally, the LasR A127W mutation shifts the sulfonyl group in the meta-position of the tail phenyl ring by ~8 Å compared to the LasR LBD:BB0126 structure (Figure 6A). This change enables the sulfonyl-containing tail group to reside in a conformation similar to those shown for the natural HSL AI tails in the CviR, TraR, and SdiA structures, all of which are high potency agonists41. We note that the observed shift in the sulfonyl group is accommodated by the displacement of LasR residue Arg61, which normally forms a hydrogen bond with the ketone group at carbon 3 of the acyl tail of 3OC12HSL (Figure 6B)39. A similar shift is observed in the structure of the LasR LBD bound to TP-1, a previously reported high-potency LasR agonist (Figure 6B and 6C)40. A more modest shift occurs in LasR LBD:mBTL (Figure 6B), likely due to the absence of hydrogen bonding between LasR R61 and the mBTL tail. The R61A mutant exhibits reduced potency for all ligands tested here (Supplemental Figure 4A-J and Table 1), suggesting a general role for the R61 side chain in stabilizing LasR LBD:ligand complexes.
Figure 6. Crystal structure of LasR LBD T75V/Y93F/A127W bound to BB0126 reveals the canonical binding mode for the ligand head group and a new binding mode for the ligand tail group.
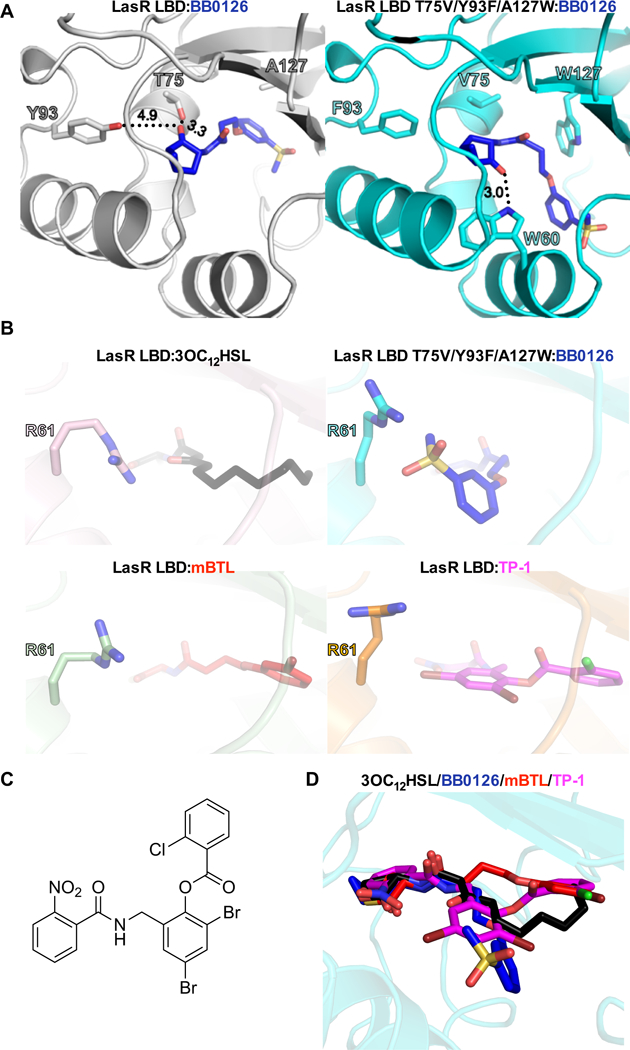
A) Crystal structures of the LasR LBD (left; silver) and LasR LBD T75V/Y93F/A127W (right; cyan), both bound to BB0126 (blue). LasR residues 75, 93, and 127 are highlighted in both structures. LasR residue W60 is highlighted in the LasR LBD T75V/Y93F/A127W structure because, due to the reorientation of the BB126 head group, W60 forms a hydrogen bond with the ligand. Important hydrogen bonds are depicted as black dotted lines and the calculated bond distances (Å) are shown. B) Crystal structures of LasR LBD (top left, bottom left, and bottom right) and LasR LBD T75V/Y93F/A127W (top right) bound to 3OC12HSL (black), BB0126 (blue), mBTL (red), and TP-1 (magenta; PDB: 3IX4). Note the orientation of LasR R61 relative to its respective ligand across all four structures. C) Structure of the LasR agonist called TP-167. D) Composite model of LasR LBD bound to 3OC12HSL (black), BB0126 (blue), mBTL (red), and TP-1 (magenta; PDB: 3IX4).
In conclusion, we note that every reported structure of a LasR LBD complexed with a high-potency ligand shows the head group bound in the canonical mode. We interpret our results with LasR LBD:BB0126 and LasR LBD T75V/Y93F/A127W:BB0126 to mean that having the ligand head group in the canonical position could be a requirement for high potency.
Our structure of LasR LBD T75V/Y93F/A127W:BB0126 represents a new mode of binding for the ligand tail group compared to 3OC12HSL, mBTL, and TP-1 (Figure 6D) as well as all other published TP-1 derivatives40, 55. Indeed, each of the four ligands shown in Figure 6D adopts a distinct tail group conformation. It is now clear that LasR can accommodate a diversity of ligands, especially in terms of the tail group, while maintaining high-potency activation. This broad specificity is presumably enabled by a LasR binding pocket that is remarkably plastic, with a strong preference to bind ligands as agonists. This feature might explain why inhibitor campaigns targeting the LasR ligand binding pocket have, to date, been unsuccessful in identifying high potency inhibitors.
Theoretical models of LasR bound to different compounds have been put forward based on the crystal structure of the LasR LBD:3OC12HSL complex39. One goal of such modeling efforts has been to infer the kinds of compounds that would act as LasR inhibitors. For example, molecular docking of inhibitory compounds in the LasR LBD suggested that ligands could bind in an alternative conformation, with head group carbonyl or hydroxyl moieties interacting with LasR residues Asn73, Thr75, and Tyr9339, in an orientation similar to what we observe for BB0126 in our structural analysis. It was suggested that compounds containing acyl tails and modified head groups would adopt unfavorable (i.e., inhibitory) conformations due to steric hindrance between residues in the LasR ligand binding site and the modified head groups39. Indeed, such steric hindrance likely drives head group reorientation for BB0126, as discussed above. The authors proposed that such compounds should be explored as LasR inhibitors56–61. These theoretical predictions are borne out by the crystal structure of LasR LBD:BB0126, with the crucial difference that reorientation does not yield inhibition. Rather, BB0126 is a weak agonist. Our findings suggest that because LasR can bind molecules in markedly different orientations, and moreover can structurally rearrange the ligands into conformations that stabilize and activate the protein, it will be challenging to design an inhibitory ligand that competes effectively with 3OC12HSL and destabilizes LasR in vivo.
The mechanism of inhibition proposed from these earlier theoretical studies differs from the one we reported for CviR inhibition by the small molecule antagonist chlorolactone (CL)62. CL and mBTL differ in that the former has a lactone and the latter has a thiolactone head group. Also, on the phenyl rings, chlorine is in the para-position in CL and bromine is in the meta-position in mBTL (Supplemental Figure 5A). CL functions as a CviR inhibitor by stabilizing a “closed” configuration in which the DNA-binding domains pack against the LBDs in a configuration that is incompatible with DNA binding62. The CviR:CL contacts differ from the LasR:mBTL contacts we observe (Supplemental Figure 5B). When the two complexes are aligned, the substituted phenyl rings of CL and mBTL are nearly 9 A apart (Supplemental Figure 5B). Docking studies suggest that, when bound to the LBD of LasR, CL would adopt a conformation similar to that of mBTL (Supplemental Figure 5C). This notion, coupled with our structure-function studies of the LasR LBD:BB0126 interaction, suggest that the mechanism underlying antagonism in one LuxR-type family member might not be applicable to another family member even if the receptors naturally accommodate nearly identical HSL ligands.
LasR inhibition via ligand-induced destabilization of LasR continues to be a challenging goal, and alternative approaches that interrogate different features required for activity such as DNA binding or RNA polymerase engagement should be explored. To successfully do so will likely require structures of full-length LasR bound to DNA and/or to RNA polymerase. Toward that end, the work presented here should prove informative for establishing highly stable LasR:ligand complexes amenable to crystallography.
METHODS
Strain construction.
lasR mutations were constructed on the pBAD-lasR plasmid. Primers were designed using the Quikchange primer design tool and PCR was carried out with the pFUltra polymerase kit (Agilent). PCR reactions were incubated with DpnI for 30 min to digest parental plasmid DNA. Plasmids with mutant lasR genes were transformed into One Shot T0P10 chemically competent E. coli cells (Invitrogen). Transformants were plated on LB agar plates containing ampicillin (50 μg/mL), individual colonies were selected, and they were screened for mutations via sequencing with primers flanking the lasR gene (ARM203 and ARM204). In-frame, marker-less mutations were made as previously described41. Primers and strains used in this study are listed in Supplemental Table 1 and Supplemental Table 2, respectively.
E. coli reporter assay and data processing.
TOP10 E. coli cells (Invitrogen) containing pBAD-lasR and plasB-luxCDABE were grown overnight in 3 mL of LB medium containing ampicillin (100 μg/mL) and kanamycin (50 μg/mL) at 37°C. Cultures were back-diluted 1:1000 and grown in fresh medium containing antibiotics at 37°C until A600 = 0.5 or for ~4 h. For dose response assays with different compounds, 100× stocks were generated in DMSO. 10 mM stocks of all compounds were serially diluted in DMSO and 1 μL of ligands were added to 100 μL of cell culture. Assays were carried out in 96-well plates (Corning). Plates were incubated at 30°C for 4 h and bioluminescence was measured on an Envision 2103 Multilabel Reader (PerkinElmer Life Sciences) with a measurement time of 0.1s. A600 was measured using a photometric 600-nm filter at 100% light emission. Relative light units (RLU) were calculated by dividing the bioluminescence measurement by the A600 measurement. To determine EC50 values, RLU versus agonist concentration data were fit using a variable slope model (four-parameter dose-response curve) using GraphPad Prism. Data for the highest concentrations (>10 μM) of mBTL, BB0020, and BB0126 were excluded, since they were influenced by non-LasR-dependent inhibition of luciferase. Nonetheless, all data points are shown in the graphs for completeness. The R2 values for each fit are displayed in Supplemental Table 3. When a lack of data at sufficiently high ligand concentration precluded accurate EC50 determination, the provided values are denoted in Table 1 as “>X”. When molecules did not activate LasR at even the highest concentrations tested, EC50 values are denoted in Table 1 as non-responsive (NR).
Protein production, purification, and crystallography.
Recombinant 6xHis-LasR LBD and 6xHis-LasR LBD T75V/Y93F/A127W proteins bound to 3OC12HSL, mBTL, BB0020, and BB0126 were expressed in E. coli BL21 (DE3) cells (Invitrogen). The strains were supplied with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and 100 μM of test compound and grown for 4 h at 25°C. The cells were harvested via centrifugation at 16,100 × g for 15 min. LasR complexes were purified as previously described for LasR LBD:3OC12HSL using Ni-NTA affinity columns (Qiagen) followed by size exclusion chromatography (GE Healthcare)38. 6xHis-LasR LBD and 6xHis-LasR LBD T75V/Y93F/A127W proteins complexed with mBTL, BB0020, and BB0126 were crystallized by the hanging drop diffusion method. Diffraction data were processed using the HKL-300 software package63. The structures were solved using Phaser in Phenix by molecular replacement, with the structure of LasR LBD:3OC12HSL used as the search model39, 64, 65. Model building was performed using Coot66 and further refinement was accomplished using Phenix64.
Thermal shift assay.
Thermal shift analyses of 6xHis-LasR LBD and 6xHisLasR LBD T75V/Y93F/A127W bound to 3OC12HSL and BB0126 were performed as previously described38. Briefly, ligand-bound LasR protein was diluted to 5 μM in reaction buffer (20 mM Tris-HCL pH 8, 200 mM NaCl, and 1 mM DTT (dithiothreitol)) containing either DMSO, 10 μM 3OC12HSL, or 10 μM BB0126 in 18 μL total volume. The mixtures were incubated at room temperature for 15 min. 2 μL of 200x SYPRO Orange (Thermo-Fisher) was added to the 18 μL sample immediately prior to the assay. Samples were subjected to a linear heat gradient of 0.05°C/s, from 25°C to 99°C in a Quant Studio 6 Flex System (Applied Biosystems) using the melting curve setting. Fluorescence was measured using the ROX reporter setting.
Protein solubility assay.
E. coli BL21 DE3 (Invitrogen) containing plasmid-borne 6xHis-LasR LBD or 6xHis-LasR LBD T75V/Y93F/A127W were grown overnight and back diluted 1:500 in 20 mL of LB medium containing ampicillin (100 μg/mL). Cultures were grown to OD600 of 0.5 and protein production was induced with 1 mM IPTG. Upon addition of IPTG, the desired test compound was also added at a final concentration of 10 μM, and the cultures were incubated at 25°C or 37°C with shaking for 4 h. Cells were harvested and soluble fractions were obtained as previously described38.
P. aeruginosa PA14 rhlA-mNG reporter assay.
P. aeruginosa PA14 strains carrying WT and mutant lasR genes and the prhlA-mNG reporter fusion on the chromosome were grown overnight and, subsequently, diluted 1:1000 in 2 mL of LB medium. 3OC12HSL, BB0020, and BB0126 were added at 10 μM and cultures were grown at 37°C for 8 h. The cultures were pelleted via centrifugation at 845 × g and cells were resuspended in 2 mL of PBS. 200 μL of cells were transferred to a 96-well plate (Corning) and fluorescence was measured using an Envision 2103 Multilabel Reader (Perkin Elmer) using the FITC filter with an excitation of 485 nm and emission of 535 nm. In parallel, cell density was measured by A600 on a UV/vis spectrophotometer (Beckman Coulter).
Pyocyanin assay.
Overnight cultures of the P. aeruginosa PA14 ΔlasI and ΔlasI lasR T75V Y93F A127W strains were grown in LB medium with shaking at 37°C. 100 μL of each culture was diluted into 3 mL of fresh LB medium and shaken at 37°C for 8 h. Subsequently, 2 μL of each culture was diluted into 2 mL of fresh LB medium, and DMSO or appropriate compounds were added at the concentrations described in the figures. Cultures were shaken at 37°C for 17 h. 1 mL aliquots were removed and cell density (OD600 nm) was measured using a Beckman Coulter DU730 spectrophotometer. The aliquots were subjected to centrifugation at 16,100 × g for 2 min and the clarified supernatants were removed. The OD695 nm of the supernatants were measured. Pyocyanin activity was determined as the OD695 nm/OD600 nm for each strain.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Bassler group for insightful ideas. We thank A. Miller of ChemEnvision LLC for computational chemistry guidance concerning compound design and mutagenesis as well as analysis of crystal structures. The research used the FMX beamline of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health Grant 5R37GM065859, and National Science Foundation Grant MCB-1713731 (B.L.B.), National Institutes of Health Grant R56AI091681 (F.M.H.), a Jane Coffin Childs Memorial Fund for Biomedical Research Postdoctoral Fellowship (J.E.P.), and NIGMS T32GM007388 (A.R.M.).
Footnotes
ACCESSION CODES
The coordinates and structure factors for LasR LBD bound to mBTL, BB0020, and BB0126 as well as LasR LBD T75V/Y93F/A127W bound to BB0126 have been deposited in the Protein Data Bank with accession codes 6MWL, 6MWH, 6MWW, and 6MWZ, respectively.
Supporting Information Available: This material is available free of charge via the internet.
REFERENCES
- [1].Albus AM, Pesci EC, Runyen-Janecky LJ, West SE, and Iglewski BH (1997) Vfr controls quorum sensing in Pseudomonas aeruginosa, J Bacteriol 179, 3928–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bassler BL, Wright M, Showalter RE, and Silverman MR (1993) Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence, Mol Microbiol 9, 773–786. [DOI] [PubMed] [Google Scholar]
- [3].Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, and Oppenheimer NJ (1981) Structural identification of autoinducer of Photobacterium fischeri luciferase, Biochemistry 20, 2444–2449. [DOI] [PubMed] [Google Scholar]
- [4].Engebrecht J, Nealson K, and Silverman M (1983) Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri, Cell 32, 773–781. [DOI] [PubMed] [Google Scholar]
- [5].Papenfort K, and Bassler BL (2016) Quorum sensing signal-response systems in Gram-negative bacteria, Nat Rev Microbiol 14, 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, and Greenberg EP (1994) Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes, Proc Natl Acad Sci U S A 91, 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pearson JP, Pesci EC, and Iglewski BH (1997) Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes, J Bacteriol 179, 5756–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, and Greenberg EP (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm, Science 280, 295–298. [DOI] [PubMed] [Google Scholar]
- [9].LaSarre B, and Federle MJ (2013) Exploiting quorum sensing to confuse bacterial pathogens, Microbiol Mol Biol Rev 77, 73–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bhardwaj AK, Vinothkumar K, and Rajpara N (2013) Bacterial quorum sensing inhibitors: attractive alternatives for control of infectious pathogens showing multiple drug resistance, Recent Pat Antiinfect Drug Discov 8, 68–83. [DOI] [PubMed] [Google Scholar]
- [11].Davies J, and Davies D (2010) Origins and evolution of antibiotic resistance, Microbiol Mol Biol Rev 74, 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tang HB, DiMango E, Bryan R, Gambello M, Iglewski BH, Goldberg JB, and Prince A (1996) Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection, Infect Immun 64, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lister PD, Wolter DJ, and Hanson ND (2009) Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms, Clin Microbiol Rev 22, 582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cabot G, Zamorano L, Moya B, Juan C, Navas A, Blazquez J, and Oliver A (2016) Evolution of Pseudomonas aeruginosa Antimicrobial Resistance and Fitness under Low and High Mutation Rates, Antimicrob Agents Chemother 60, 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vincent JL, Bihari DJ, Suter PM, Bruining HA, White J, Nicolas-Chanoin MH, Wolff M, Spencer RC, and Hemmer M (1995) The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) Study. EPIC International Advisory Committee, JAMA 274, 639–644. [PubMed] [Google Scholar]
- [16].Peleg AY, and Hooper DC (2010) Hospital-acquired infections due to gram-negative bacteria, N Engl J Med 362, 1804–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Storey DG, Ujack EE, Rabin HR, and Mitchell I (1998) Pseudomonas aeruginosa lasR transcription correlates with the transcription of lasA, lasB, and toxA in chronic lung infections associated with cystic fibrosis, Infect Immun 66, 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mukherjee S, Moustafa D, Smith CD, Goldberg JB, and Bassler BL (2017) The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer, PLoS Pathog 13, e1006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pearson JP, Passador L, Iglewski BH, and Greenberg EP (1995) A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa, Proc Natl Acad Sci U S A 92, 1490–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pesci EC, Pearson JP, Seed PC, and Iglewski BH (1997) Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa, J Bacteriol 179, 3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gambello MJ, and Iglewski BH (1991) Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression, J Bacteriol 173, 3000–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gambello MJ, Kaye S, and Iglewski BH (1993) LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression, Infect Immun 61, 1180–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Toder DS, Gambello MJ, and Iglewski BH (1991) Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR, Mol Microbiol 5, 2003–2010. [DOI] [PubMed] [Google Scholar]
- [24].Brint JM, and Ohman DE (1995) Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family, J Bacteriol 177, 7155–7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Urbanowski ML, Lostroh CP, and Greenberg EP (2004) Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein, J Bacteriol 186, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chen J, and Xie J (2011) Role and regulation of bacterial LuxR-like regulators, J Cell Biochem 112, 2694–2702. [DOI] [PubMed] [Google Scholar]
- [27].Qin Y, Luo ZQ, Smyth AJ, Gao P, Beck von Bodman S, and Farrand SK (2000) Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm, EMBO J 19, 5212–5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schuster M, Urbanowski ML, and Greenberg EP (2004) Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR, Proc Natl Acad Sci U S A 101, 15833–15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tsai CS, and Winans SC (2010) LuxR-type quorum-sensing regulators that are detached from common scents, Mol Microbiol 77, 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nasser W, and Reverchon S (2007) New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators, Anal Bioanal Chem 387, 381–390. [DOI] [PubMed] [Google Scholar]
- [31].Smith RS, and Iglewski BH (2003) Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target, J Clin Invest 112, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Choi H, Ham SY, Cha E, Shin Y, Kim HS, Bang JK, Son SH, Park HD , and Byun Y (2017) Structure-Activity Relationships of 6- and 8-Gingerol Analogs as Anti-Biofilm Agents, J Med Chem 60, 9821–9837. [DOI] [PubMed] [Google Scholar]
- [33].Amara N, Gregor R, Rayo J, Dandela R, Daniel E, Liubin N, Willems HM, Ben-Zvi A, Krom BP, and Meijler MM (2016) Fine-Tuning Covalent Inhibition of Bacterial Quorum Sensing, Chembiochem 17, 825–835. [DOI] [PubMed] [Google Scholar]
- [34].Goh WK, Gardner CR, Chandra Sekhar KV, Biswas NN, Nizalapur S, Rice SA, Willcox M, Black DS, and Kumar N (2015) Synthesis, quorum sensing inhibition and docking studies of 1,5-dihydropyrrol-2-ones, Bioorg Med Chem 23, 7366–7377. [DOI] [PubMed] [Google Scholar]
- [35].O’Reilly MC, and Blackwell HE (2016) Structure-Based Design and Biological Evaluation of Triphenyl Scaffold-Based Hybrid Compounds as Hydrolytically Stable Modulators of a LuxR-Type Quorum Sensing Receptor, ACS Infect Dis 2, 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Geske GD, O’Neill JC, Miller DM, Mattmann ME, and Blackwell HE (2007) Modulation of bacterial quorum sensing with synthetic ligands: systematic evaluation of N-acylated homoserine lactones in multiple species and new insights into their mechanisms of action, J Am Chem Soc 129, 13613–13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Smith KM, Bu Y, and Suga H (2003) Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs, Chem Biol 10, 81–89. [DOI] [PubMed] [Google Scholar]
- [38].Paczkowski JE, Mukherjee S, McCready AR, Cong JP, Aquino CJ, Kim H, Henke BR, Smith CD, and Bassler BL (2017) Flavonoids Suppress Pseudomonas aeruginosa Virulence through Allosteric Inhibition of Quorum-sensing Receptors, J Biol Chem 292, 4064–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bottomley MJ, Muraglia E, Bazzo R, and Carfi A (2007) Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer, J Biol Chem 282, 13592–13600. [DOI] [PubMed] [Google Scholar]
- [40].Zou Y, and Nair SK (2009) Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor, Chem Biol 16, 961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].McCready AR, Paczkowski JE, Henke BR, and Bassler BL (2018) Structural determinants driving homoserine lactone ligand selection in the Pseudomonas aeruginosa LasR quorum-sensing receptor, Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gerdt JP, McInnis CE, Schell TL, Rossi FM, and Blackwell HE (2014) Mutational analysis of the quorum-sensing receptor LasR reveals interactions that govern activation and inhibition by nonlactone ligands, Chem Biol 21, 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gerdt JP, McInnis CE, Schell TL, and Blackwell HE (2015) Unraveling the contributions of hydrogen-bonding interactions to the activity of native and non-native ligands in the quorum-sensing receptor LasR, Org Biomol Chem 13, 1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, and Bassler BL (2013) A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation, Proc Natl Acad Sci U S A 110, 17981–17986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Welsh MA, Eibergen NR, Moore JD, and Blackwell HE (2015) Small molecule disruption of quorum sensing cross-regulation in pseudomonas aeruginosa causes major and unexpected alterations to virulence phenotypes, J Am Chem Soc 137, 1510–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Glansdorp FG, Thomas GL, Lee JK, Dutton JM, Salmond GP, Welch M, and Spring DR (2004) Synthesis and stability of small molecule probes for Pseudomonas aeruginosa quorum sensing modulation, Org Biomol Chem 2, 3329–3336. [DOI] [PubMed] [Google Scholar]
- [47].Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, Goldner M, Dessaux Y, Camara M, Smith H, and Williams P (2002) N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa, Infect Immun 70, 5635–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Galloway WR, Hodgkinson JT, Bowden SD, Welch M, and Spring DR (2011) Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways, Chem Rev 111, 28–67. [DOI] [PubMed] [Google Scholar]
- [49].Hodgkinson JT, Galloway WR, Wright M, Mati IK, Nicholson RL, Welch M, and Spring DR (2012) Design, synthesis and biological evaluation of non-natural modulators of quorum sensing in Pseudomonas aeruginosa, Org Biomol Chem 10, 6032–6044. [DOI] [PubMed] [Google Scholar]
- [50].Park S, Kim HS, Ok K, Kim Y, Park HD, and Byun Y (2015) Design, synthesis and biological evaluation of 4-(alkyloxy)-6-methyl-2H-pyran-2-one derivatives as quorum sensing inhibitors, Bioorg Med Chem Lett 25, 2913–2917. [DOI] [PubMed] [Google Scholar]
- [51].Nizalapur S, Kimyon O, Biswas NN, Gardner CR, Griffith R, Rice SA, Manefield M, Willcox M, Black DS, and Kumar N (2016) Design, synthesis and evaluation of N-aryl-glyoxamide derivatives as structurally novel bacterial quorum sensing inhibitors, Org Biomol Chem 14, 680–693. [DOI] [PubMed] [Google Scholar]
- [52].Jog GJ, Igarashi J, and Suga H (2006) Stereoisomers of P. aeruginosa autoinducer analog to probe the regulator binding site, Chem Biol 13, 123–128. [DOI] [PubMed] [Google Scholar]
- [53].Bullock K, Chong P, Davis R, Elitzin V, Hatcher M, Jackson M, Liu B, Patterson D, Powers J, Salmons M, Tabet E, and Toczko M (2012) Ir C-H Activation and Other Catalysis Applied to a Complex Drug Candidate, Top Catal 55, 446–452. [Google Scholar]
- [54].Smith KM, Bu Y, and Suga H (2003) Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer, Chem Biol 10, 563–571. [DOI] [PubMed] [Google Scholar]
- [55].O’Reilly MC, Dong SH, Rossi FM, Karlen KM, Kumar RS, Nair SK, and Blackwell HE (2018) Structural and Biochemical Studies of Non-native Agonists of the LasR Quorum-Sensing Receptor Reveal an L3 Loop “Out” Conformation for LasR, Cell Chem Biol 25, 1128–1139 e1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hentzer M, and Givskov M (2003) Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections, J Clin Invest 112, 1300–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Hoiby N, Kjelleberg S, and Givskov M (2002) Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound, Microbiology 148, 87–102. [DOI] [PubMed] [Google Scholar]
- [58].Hjelmgaard T, Persson T, Rasmussen TB, Givskov M, and Nielsen J (2003) Synthesis of furanone-based natural product analogues with quorum sensing antagonist activity, Bioorg Med Chem 11, 3261–3271. [DOI] [PubMed] [Google Scholar]
- [59].Manefield M, de Nys R, Kumar N, Read R, Givskov M, Steinberg P, and Kjelleberg S (1999) Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein, Microbiology 145 ( Pt 2), 283–291. [DOI] [PubMed] [Google Scholar]
- [60].Manefield M, Rasmussen TB, Henzter M, Andersen JB, Steinberg P, Kjelleberg S, and Givskov M (2002) Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover, Microbiology 148, 1119–1127. [DOI] [PubMed] [Google Scholar]
- [61].Wu H, Song Z, Hentzer M, Andersen JB, Molin S, Givskov M, and Hoiby N (2004) Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice, J Antimicrob Chemother 53, 1054–1061. [DOI] [PubMed] [Google Scholar]
- [62].Chen G, Swem LR, Swem DL, Stauff DL, O’Loughlin CT, Jeffrey PD, Bassler BL, and Hughson FM (2011) A strategy for antagonizing quorum sensing, Mol Cell 42, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Minor W, Cymborowski M, Otwinowski Z, and Chruszcz M (2006) HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes, Acta Crystallogr D Biol Crystallogr 62, 859–866. [DOI] [PubMed] [Google Scholar]
- [64].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2011) The Phenix software for automated determination of macromolecular structures, Methods 55, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine, Acta Crystallogr D Biol Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot, Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Muh U, Hare BJ, Duerkop BA, Schuster M, Hanzelka BL, Heim R, Olson ER, and Greenberg EP (2006) A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal, Proc Natl Acad Sci U S A 103, 16948–16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


