Abstract
Background and objective:
We have previously demonstrated a significant correlative relationship between PTEN deletion and ERG rearrangement both in development of clinically localized prostate cancers and metastases. Herein, we evaluate the cooperative role of ERG and PTEN in oncological outcome after radical prostatectomy for clinically localized prostate cancer.
Methods:
We evaluated ERG and PTEN status using three previously described cohorts. The first cohort included 235 clinically localized prostate cancer cases represented on tissue microarrays (TMA), evaluated using previously validated FISH assays for ERG and PTEN. The second cohort included 167 cases of clinically localized prostate cancer on TMAs evaluated for PTEN by FISH, and for PTEN and ERG by dual IHC. The third cohort comprised 59 clinically localized prostate cancer cases assessed by array comparative genomic hybridization (aCGH). Kaplan Meir plots and long rank tests were used to assess the association of ERG and PTEN status with biochemical recurrence after radical prostatectomy for clinically localized prostate cancer.
Results:
Of the 317 cases eligible for analyses with evaluable ERG and PTEN status, 88 (27.8%) patients developed biochemical recurrence over a median follow-up of 5.7 years. Overall, 45% (142/317) of cases demonstrated ERG rearrangement and 20% (62/317) of cases demonstrated PTEN loss. Hemizygous and homozygous deletion of PTEN was seen in 10% (18/175) and 3% (5/175) of ERG negative cases, respectively. In contrast, hemizygous and homozygous deletion of PTEN was seen in 11% (15/142) and 17% (24/123) of ERG positive cases, respectively. PTEN loss (heterozygous or homozygous) was significantly associated with shorter time to biochemical recurrence compared to no PTEN loss (p<0.001). However, ERG rearrangement vs. no rearrangement was not associated with time to PSA recurrence (p=0.15). Patients who exhibited ERG rearrangement and loss of PTEN had no significant difference in time to recurrence compared to patients with wild type ERG and loss of PTEN (p=0.30).
Conclusion:
Our findings confirm a mutual cooperative role of ERG and PTEN in the pathogenesis of prostate cancer, particularly for homozygous PTEN deletion. ERG did not stratify outcome either alone or in combination with PTEN in this cohort.
Keywords: ERG, PTEN, prostate cancer, fluorescent in situ hybridization (FISH), immunohistochemistry
Introduction
In a world being driven by precision medicine1, there is currently an impetus to identify biologically relevant subgroups of prostate cancer patients where therapy can be developed and targeted judiciously. Current and future studies need to identify which patients need to be treated, when the patients need the treatment, and how these patients should be treated. To address these unanswered questions, we need biomarkers that can delineate patients with prostate cancer who are at risk of disease recurrence, progression and/or metastasis (prognostic markers), and which appropriate therapy can be given to these high risk patients (predictive markers).
PTEN loss and ERG rearrangement might have a potential combined prognostic and predictive role. PTEN is a membrane-associated phosphatase encoded by a gene on chromosome 10q23, which acts as a tumor suppressor by inhibiting the pro-proliferation PI3K/AKT pathway.2 PTEN is an antagonist of PI3K (phosphatidylinositol 3–kinase) signaling and impaired PTEN function leads to phosphatidylinositol-3,4,5 trisphosphate (PIP3) accumulation in cells and to unrestrained activation of its downstream signals.3 Current estimates suggest that PI3K/AKT signaling is upregulated in 30-50% of prostate cancers, often through loss of PTEN.4 Recurrent gene fusions involving the ETS family of transcription factors (ERG, ETV1, ETV4 and ETV5) and TMPRSS2 occurs in the majority of prostate cancers.5–10 Among these aberrations, TMPRSS2:ERG fusion is the most prevalent, occurring in approximately 50% of localized prostate cancers and 30% of castration resistant metastatic cancers in PSA screened predominantly Caucasian cohorts.9,10 Emerging data suggest that TMPRSS2:ERG fusion plays an important role in carcinogenesis in vitro and in vivo.11,12 Although most post-prostatectomy studies report no or marginal association of ERG rearrangements with prostate cancer outcome, there are other cohorts where ERG deletion has been associated with a significantly poor survival. 13,14
In murine models, ERG rearrangements and PTEN deletions interact to induce prostate cancer.13–15 Several studies have confirmed a significant association between ERG gene rearrangements and PTEN genomic deletions in a subset of human prostatic adenocarcinomas, and most studies show an association of PTEN loss and poor outcome, with conflicting results on the importance of ERG status for the prognostic ability of PTEN status.16–20 Hence, in the current study, we examine the association of ERG rearrangement and PTEN loss with biochemical recurrence after radical prostatectomy in a large cohort of patients with clinically localized prostate cancer.
Materials and Methods
This study was approved by the Institutional Review Board at the University of Michigan Medical School (patient consent was deemed not necessary as part of the ethical review that was conducted). We evaluated ERG and PTEN status using three cohorts. The first cohort included 235 clinically localized prostate cancer cases represented on tissue microarrays (TMA), evaluated using previously validated FISH assays for ERG and PTEN. The second cohort included 167 cases of clinically localized prostate cancer on TMAs evaluated for PTEN by FISH, and for PTEN and ERG by dual IHC. The third cohort comprised 59 clinically localized PCa assessed by array comparative genomic hybridization (aCGH). For these analyses, we excluded any overlapping cases in the cohorts (n=25), those from patients treated with pre- or adjuvant hormone therapy or radiation (n=9), and those with no follow up information available (n=49). After excluding those without evaluable status for both ERG and PTEN, our final cohort consisted of a total of 371 unique patients.
Fluorescence In Situ Hybridization (FISH)
We performed interphase FISH as previously described by our group7,9. Briefly, bacterial artificial chromosomes (BACs) were obtained from the BACPAC Resource Center (Oakland, CA, USA), and probes were prepared as described5,9. For detection of ERG rearrangement, RP11-95I21 (5’ to ERG) and RP11-476D17 (3’ to ERG) were used with a break-apart probe strategy9. To detect PTEN deletion, a combination of PTEN gene locus-specific probe (RP11-165M8) and 10q11.1- specific probe (RP11-351D16) for chromosome 10 copy number identification were utilized. Schematic BACs for ERG and PTEN are shown in Figure 1a. The integrity and correct localization of all probes were verified by hybridization to metaphase spreads of normal peripheral lymphocytes. Slides were examined using an imaging Z1 microscope (Carl Zeiss, Oberkochen, Germany). FISH signals were scored manually (100x oil immersion) in morphologically intact and non-overlapping nuclei, and a minimum of 50 cancer cells from each site were recorded. Cancer sites with very weak or no signals were recorded as insufficiently hybridized. Cases lacking tumor tissue in all three cores were excluded. For validation of PTEN deletion, we utilized an earlier documented method with minor modification21. Briefly, based on hybridization in five control cores (data not shown), hemizygous deletion of PTEN gene was defined as >50% nuclei (mean±3 standard deviations in nonneoplastic controls) containing either one signal of locus probe and ≥2 signals of reference probe, or two signals of locus probe and ≥4 signals of reference probe. Homozygous deletion of PTEN was exhibited by the simultaneous lack of the both PTEN locus signals and the presence of control signals in >30% of cells.21–23 Representative FISH images of PTEN deletion and ERG rearrangement are shown in Figures 1b and c.
Figure 1. Fluorescent in situ hybridization (FISH) probe design and representative ERG aberrations and PTEN deletions detected in prostate cancer.
(A) Schematic of bacterial artificial chromosomes (BACs)- locus/control for PTEN and located 5’ and 3’ to ERG used as probes for interphase FISH. Chromosomal coordinates are from the March 2006 build of the human genome using the UCSC Genome Browser. BACs are indicated as numbered rectangles, with the number identifying the BAC as described below and the color indicating the probe color in the accompanying images. Genes are shown with the direction of transcription indicated by the arrowhead and exons indicated by bars. (B and C) FISH was carried out using BACs as indicated with the corresponding fluorescent label on formalin-fixed paraffin-embedded tissue sections. B. Locus/control FISH for PTEN. B1. Representative case with retained PTEN showing two red signals (10q23/PTEN locus) and two green signals (10q11.1) in tumor cells. B2. Representative case with PTEN hemizygous deletion showed one red signal and pairs of green signals in tumor cells. B3. Representative case with PTEN homozygous deletion showed absence of red signals (10q23/PTEN locus), but retained pairs of green signals (PTEN control). C. Break-apart FISH for ERG gene. Green and red colors imply individual signals, whereas yellow signals were indicated as colocalized probes. C1. C2. ERG rearrangement-positive (with deletion) case showed loss of one green labeled probe 5’ to ERG. C3. ERG rearrangement positive (translocation) case showed one pair of split 5’ and 3’ signals. For all assays, at least 50 cancer cell nuclei were evaluated.
Immunohistochemistry (IHC)
We have recently designed and validated a protocol for evaluating PTEN expression by immunohistochemistry (IHC).20 ERG–PTEN dual IHC was performed using anti-ERG (EPR3864) rabbit monoclonal primary antibody (1:100; Cat no. 790-4576, Ventana Medical Systems, Tucson, AZ, USA) and a rabbit monoclonal primary antibody against PTEN (1:25; 138G6- Cell Signaling Technology, Danvers, MA, USA). Dual IHC was performed using an automated protocol developed for the DISCOVERY XT automated slide staining system (Ventana Medical Systems) using UltraMap antirabbit HRP (Cat no. 760-4315, Ventana Medical Systems) for ERG and UltraMap anti-rabbit AP (Cat no. 760-4314, Ventana Medical Systems) for PTEN as secondary antibodies and were detected using ChromoMap DAB (Cat no. 760-159, Ventana Medical Systems) and ChromoMap Blue (Cat no. 760-161, Ventana Medical Systems) for ERG and PTEN, respectively. Nuclear Fast Red counterstain (Cat no. 780-2186 Ventana Medical Systems) was used as the counterstain. Hematoxylin II (Cat no. 790-2208 Ventana-Roche) was used as counterstain. Examples of ERG/PTEN staining in prostate cancer are shown in Figure 2.
Figure 2. Immunohistochemistry evaluation of ERG and PTEN in prostate cancer tissues.
ERG–PTEN dual immunohistochemistry was performed to evaluate ERG and PTEN status, with results shown from a representative case. Hematoxylin and eosin photomicrograph (H&E, top) showing prostatic adenocarcinoma (black arrow), benign prostatic glands (green arrow) and vessels (blue arrow). Dual ERG/PTEN staining (middle) showing positive ERG (brown chromogen) expression and loss of PTEN (purple chromogen) expression in the prostatic adenocarcinoma. Note the positive internal control for ERG antibody staining in endothelial cells and retained PTEN expression in benign prostatic glands. PTEN loss exclusively in the prostatic adenocarcinoma was confirmed by PTEN only staining (bottom). All images original magnification 20x.
Array Comparative Genomic Hybridization (aCGH)
aCGH of 59 localized prostate cancers was performed using gDNA on Agilent’s 105K or 244K aCGH microarrays (Human Genome CGH 105K or 244K Oligo Microarray) using Agilent’s standard Direct Method protocol and Wash Procedure B as reported previously.24 Briefly, 1.5 - 3 μg of gDNA from prostate specimens was restriction digested with AluI and RsaI, labeled with Cy-5 (test channel), purified using Microcon YM-30 columns and hybridized with an equal amount of Cy-3 (reference channel) labeled Human Male Genomic DNA (Promega) for 40 hours at 65 °C. Post-hybridization wash was performed with acetonitrile wash and Agilent Stabilization and Drying Solution wash according to the manufacturer’s instructions. Scanning was performed on an Agilent scanner Model G2505B (5-micron scan with software v7.0), and data was extracted using Agilent Feature Extraction software v9.5 using protocol CGH-v4_95_Feb07.
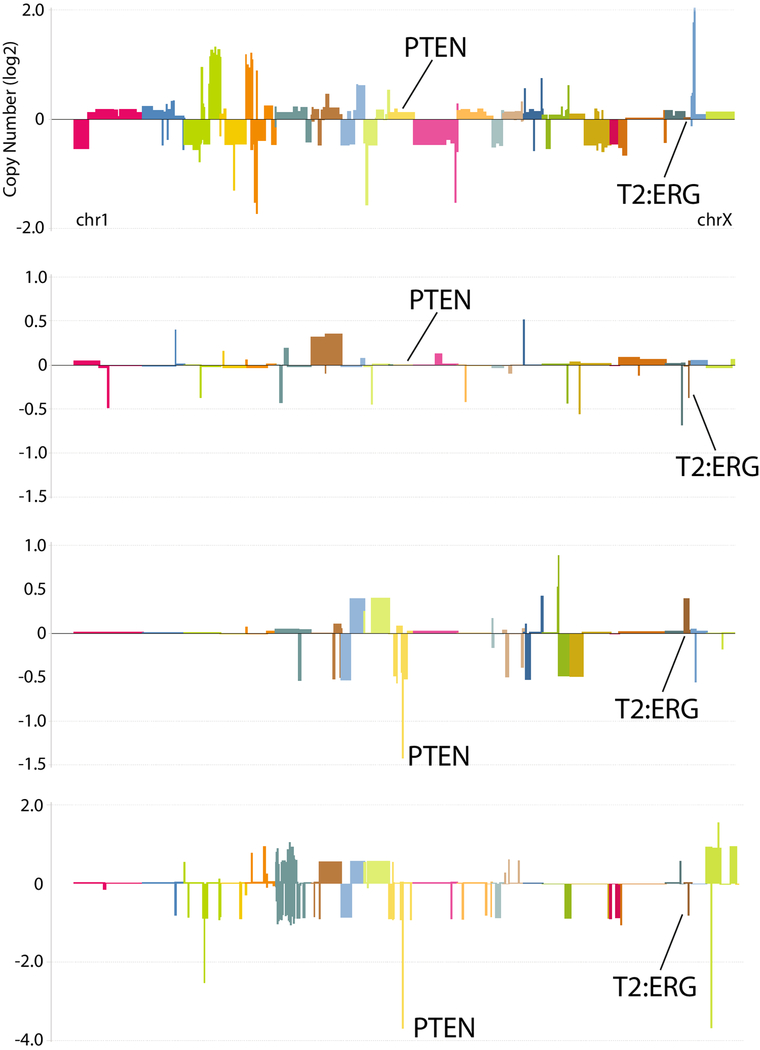
For data analysis, probes on all arrays were limited to those on the 105K array. Log(2) ratios for each probe were determined as rProcessedSignal/gProcessedSignal. The final dataset was uploaded into a custom instance of Oncomine (www.oncomine.com) for automated copy number analysis. In Oncomine, circular binary segmentation was performed on the dataset using the DNACopy package (v1.18) available via the Bioconductor package. Agilent Probe IDs are mapped to segments and reporter values are used to generate segment values (mean of reporters). Resulting segments are mapped to hg18 (NCBI 36.1) RefSeq coordinates (UCSC refGene) as provided by UCSC (UCSC refGene, July 2009, hg18, NCBI 36.1, March 2006) and segment values are assigned to each gene. Copy number profiles were visualized using Oncomine Power Tools. ERG rearrangement status was determined from qRT-PCR and DNA microarray expression profiling as reported previously. 25 Examples of cases with PTEN deletions and ERG rearrangement through deletion (assessable by aCGH) are shown in Figure 3.
Figure 3. aCGH profiles of prostate cancer with PTEN deletion and TMPRSS2:ERG gene fusions through deletion.
aCGH profiling of 59 primary prostate cancers was performed as described previously24 and uploaded into Oncomine for analysis and visualization using the Oncomine Power Tools DNA Copy Number Browser. The segmented, log2 copy number for all genes is plotted in genomic order (from chromosome 1 to X) for four representative samples showing wild type PTEN and TMPRSS2 to ERG (top panel), wild type PTEN and TMPRSS2:ERG fusion through deletion (second panel from top), homozygous PTEN deletion and wild type TMPRSS2 to ERG, and homozygous PTEN deletion and TMPRSS2:ERG fusion through deletion.
Clinical Data Collection
We reviewed patients’ medical records to abstract clinical and pathological information. Follow up data were collected including serum prostatic surface antigen (PSA). Biochemical recurrence was defined as serum PSA ≥ 0.2ng/mL following radical prostatectomy.
Statistical Analysis
Descriptive statistics were used to summarize all demographic and clinicopathologic data. Fisher’s exact test was used to test the associations between PTEN deletion and ERG rearrangement status with biochemical recurrence, with p-values 0.05 considered to be statistically significant. Statistical analyses were carried out using the R software package, version 2.7.2 (http://www.r-project.org).
Results
A total of 317 men with clinically localized prostate cancer who underwent radical prostatectomy, with long-term follow-up information, were evaluable for both ERG and PTEN evaluation. The demographic, clinical and pathological characteristics of the cohort are presented in Table 1. The median age and PSA were 61 years and 6.9 ng/mL respectively. Of the 317 eligible cases, 88 (27.8%) cases developed biochemical recurrence over a median follow-up of 5.7 years (range: 3.0 – 11.0 years). The median biochemical recurrence-free survival (RFS) was 5 years.
Table 1.
Demographics, clinical and pathologic features of the cohort
| Variable | Value* | |
|---|---|---|
| Age (years) | 61 [55 – 65] | |
| Race | White | 266 (84) |
| Black | 25 (8) | |
| Unknown/Other | 26 (8) | |
| Gland size (grams) | 45 [37 – 54] | |
| PSA (ng/mL) | 6.9 [4.9 – 10.2] | |
| Gleason score | ≤ 6 | 71 (22) |
| 7 | 209 (66) | |
| 7 with 5 | 12 (4) | |
| 8 | 11 (3) | |
| 9 | 14 (4) | |
| Maximum tumor dimension (cm) | 1.5 (1.2 – 2.0) | |
| pT stage | T2a | 33 (10) |
| T2b | 195 (62) | |
| T3a | 64 (20) | |
| T3b | 18 (6) | |
| T4 | 7 (2) | |
| pN stage | N0 | 243 (77) |
| N1 | 7 (2) | |
| Nx | 67 (21) | |
| Positive surgical margin | 101 (32) | |
| Follow up (years) | 5.7 [3.0 – 11.0] | |
Median [Interquartile range] and frequency (percentages) are presented for continuous and categorical variables respectively.
Abbreviations: PSA – Prostate specific antigen.
Overall, 44.8% (142/317) of cases demonstrated ERG rearrangement and 19.6% (62/317) of cases demonstrated PTEN loss. We observed a significant association between PTEN deletion and ERG rearrangement (p = 0.0014). Hemizygous and homozygous deletion of PTEN was seen in 10.3% (18/175) and 2.9% (5/175) of ERG negative cases, respectively. In contrast, hemizygous and homozygous deletion of PTEN was seen in 10.6% (15/142) and 19.5% (24/142) of ERG positive cases, respectively (Table 2). ERG positive cases were more likely to exhibit homozygous deletion of PTEN compared with ERG negative cases (19.5% vs. 2.9%; p < 0.0001).
Table 2.
ERG and PTEN Status of the Cohort
| PTEN Status | ERG Status |
Total | |
|---|---|---|---|
| Negative | Positive | ||
| Negative | 152 | 103 | 255 |
| Positive | 23 | 39 | 62 |
| Total | 175 | 142 | 317 |
| PTEN Status | ERG Status |
Total | |
| Negative | Positive | ||
| Negative | 152 | 103 | 255 |
| Hemizygous | 18 | 15 | 33 |
| Heterozygous | 5 | 24 | 29 |
| Total | 175 | 142 | 317 |
PTEN loss (heterozygous or homozygous) was significantly associated with time to biochemical recurrence compared to no PTEN loss (Logrank test, p= 0.0007; Figure 4a). ERG rearrangement vs. no rearrangement, on the other hand, was not associated with time to biochemical recurrence (p=0.15; Figure 4b). When PTEN deletion was stratified into homozygous versus heterozygous deletion, there appeared to be no difference in time to recurrence between the wild type PTEN and heterozygous deletion. Homozygous deletion, however, was associated with shorter time to biochemical recurrence compared with the wild type PTEN or heterozygous PTEN deletion status (p < 0.0001; Figure 4c).
Figure 4. Kaplan-Meir plots for PTEN and/or ERG status and time to biochemical recurrence.
A. Cases (n=317) were evaluated for PTEN status by FISH or aCGH, and classified as PTEN wild type or deleted. B. Cases (n = 317) were evaluated for ERG status by FISH, aCGH or IHC, and classified as ERG rearrangement negative or positive. C. As in A, except PTEN deleted cases were stratified by hemizygous or homozygous deletion status. D. Combined stratification by PTEN-ERG status as in A and C. Biochemical recurrence was censored at 10 years follow up. Log Rank p-values are indicated.
Combining the PTEN with ERG rearrangement status as depicted in Figure 4d, wild type PTEN and positive ERG rearrangement status was associated with the best biochemical RFS estimates in this cohort. Patients with prostate cancer harboring PTEN deletion and negative ERG rearrangement status suffered the worst outcome. PTEN status appears to be the driver of biochemical RFS, with wild type PTEN having an overall better biochemical RFS estimates compared with PTEN deletion, regardless of ERG rearrangement status (Figure 4d). Although the combination of PTEN and ERG status stratified patients by time to biochemical recurrence (overall 4 combinations, p= 0.001), patients who exhibited ERG rearrangement and loss of PTEN had no significant difference in time to recurrence compared to patient with wild type ERG and loss of PTEN (p=0.30; Figure 4d).
Discussion
The interaction between PTEN deletion and ERG rearrangement may play a significant role in the pathogenesis and clinical course of prostate cancer. 16–20,26,27 In our study, we report PTEN deletion in 20% of cases which was associated with worse biochemical RFS. We also found that homozygous PTEN deletion appeared to be the driver of biochemical recurrence, compared with heterozygous deletion. Although ERG rearrangement was found in 45% of cases, it was not significantly associated with recurrence. However, when stratified by PTEN status, negative ERG rearrangement was associated with worse RFS in each stratum. In addition, a significant association between PTEN deletion and ERG rearrangement was observed in our cohort.
ERG rearrangement and PTEN loss are frequent genetic alterations in prostate cancer. Bismar et al., demonstrated that ERG rearrangements and PTEN deletions were detected in 41.5% and 63.6% of castration resistant prostate cancer, with the majority (68.1%) of ERG rearrangements occurring through deletions.28 The authors also reported that, PTEN deletions were significantly associated with ERG rearrangements occurring by deletions, which is consistent with our findings. In contrast, Liu et al., detected ERG rearrangements in 13% of cases of unsuspected prostate cancers of the transition zone with 66% occurring by deletion.29 PTEN deletions were detected in 19% of cases in this cohort with only 6% having both PTEN losses and ERG rearrangements, compared with 12.3% observed in our cohort. These authors suggested that majority of transition zone tumors harbor either ERG rearrangement or PTEN deletions, compared to peripheral zone prostate cancers which are more likely to harbor both aberrations simultaneously. These differences in incidence of ERG and PTEN aberrations are likely related to different zonal origins of prostate cancer.
Genomic PTEN loss is associated with prostate cancer aggressiveness, progression or poor prognosis.8,10,18,30 Decreased PTEN expression is linked with increased risk of recurrence in patients with clinically localized prostate cancer.6–8,31 In a recent study by Lotan et al., cytoplasmic PTEN loss was identified in 84% of intraductal carcinomas of the prostate and provides a plausible molecular explanation for why intraductal carcinoma is associated with poor prognosis.32 Krohn et al., recently demonstrated that genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in both ERG fusion-positive and ERG fusion-negative prostate cancers, and characterizes a particularly aggressive subset of metastatic and castration resistant prostate cancer.16 In our study, we found that PTEN status appears to be the driver of biochemical RFS after radical prostatectomy for clinically-localized prostate cancer, with wild type PTEN having an overall better biochemical RFS estimates compared with PTEN deletion, regardless of ERG rearrangement status (Figure 4d). Our findings support the recent report that PTEN loss is associated with poor disease specific survival after radical prostatectomy.30 Although we did not evaluate PTEN levels and its correlation with genomic PTEN loss in this study, we have previously reported a significant correlation between decreased PTEN expression and genomic PTEN deletion status.17 Further, in animal and human studies, a clear dose-response relationship exists between decreased PTEN levels and prostate carcinogenesis and biological behavior, supporting our observation that only homozygous PTEN loss was associated with worse biochemical RFS. 31,33
A strong association between ERG rearrangement and PTEN deletion suggest an interactive or cooperative role in the biology of prostate cancer. 13–20,26,27 ERG rearrangement was prevalent in our cohort (45%), with a strong association with PTEN deletion, but was not associated with biochemical recurrence. Similar to our findings, Yoshimoto et al., found a strong relationship of TMPRSS2:ERG fusions with PTEN deletions, however TMPRSS2:ERG fusions did not show correlation with Gleason grade.20 Further studies are needed to characterize the cooperative role of ERG rearrangement and PTEN deletion in the pathogenesis and progression of prostate cancer, and its potential as a therapeutic target.
ETS gene fusion and members of the PI3K pathway may provide actionable targets for therapeutic intervention.34 First, the interaction between the TMPRSS2:ERG gene fusion product and PARP1 (involved in DNA repair) could be harnessed for targeted therapy. As shown in preclinical studies, the inhibition of PARP1 may result in ETS-induced DNA damage leading to cell death.35,36 In the first human study, a randomized phase II trial ( NCT01576172) of Abiraterone plus Prednisone with or without Veliparib (a PARP1 inhibitor) in treating patients with metastatic hormone-resistant prostate cancer, the final results for which are currently awaited. 37 Second, the PI3K/Akt/mTOR pathway is involved in tumorigenic processes such as cellular proliferation, cell growth, and angiogenesis. 2,4 The tumor suppressor gene, PTEN, is the primary negative regulator of this pathway and its loss results in over activation of this pathway in human cancers such as brain, breast, and prostate.38,39 Early results of a Phase I/IIa study ( NCT01458067) of GSK2636771 in PTEN deficient patients with advanced solid tumors are promising and the final report of the trial will be informative.40,41 Given the apparent cooperation or interaction between ERG rearrangement and PTEN deletion, the combined results of these two trials may provide a basis for combining these agents in a clinical trial.
Our study has several limitations. First, most of tissue-based outcome/prognostic studies for prostate cancer within the literature (including the current study) have employed TMAs where tissue cores from the index nodule have been used to construct the TMA. This is based upon the assumption that the index tumor nodule (defined as largest tumor nodule) is biologically the most representative (or most aggressive) of the disease within a single patient. Though this is true in most patients, there is a significant number of cases where the index tumor nodule indeed does not represent the highest Gleason score within a single patient. This phenomenon might explain partly why ERG fusions were not associated with poor outcome in our study, but it needs to be addressed within studies, that project use of biomarkers for clinical use. Second, we do not have data for local recurrence or metastasis in our cohort. But, PSA is a well-established marker of recurrence after radical prostatectomy for clinically localized prostate cancer. Third, there is a possibility of selection bias due to the retrospective nature of our study, however technicians who did the laboratory assessments of ERG and PTEN were blinded to the clinical or recurrence status of the patients. Nevertheless, the strength of our study lies in the exploration of ERG and PTEN in combination, and their role in long-term biochemical recurrence in a large cohort of patients who underwent radical prostatectomy for clinically localized prostate cancer.
Conclusions
Using FISH, IHC and aCGH technologies, we confirm a mutual co-operative role of ERG and PTEN in the pathogenesis of prostate cancer, particularly for homozygous PTEN deletion. Although ERG did not stratify the outcome of clinically localized prostate cancer either alone or in combination with PTEN, further studies are needed to further characterize the role of ERG in prostate cancer pathogenesis and progression. These findings improve our understanding of the underlying biology of prostate cancer and the mechanism underlying a poor outcome in clinically localized disease.
Acknowledgments
Funding: S.S.S. is supported by the Urology Care Foundation and the Prostate Cancer Foundation. S.A.T. is supported by the A. Alfred Taubman Medical Institute and the Prostate Cancer Foundation. Supported in part by the National Institutes of Health U01 CA214170 to A.M.C. and the University of Michigan Prostate Specialized Program of Research Excellence [S.P.O.R.E.] P50 CA186786-05. D.E.S. has been supported by the Department of Defense.
Footnotes
Disclosures: The University of Michigan has been issued a patent on the detection of ETS gene fusions in prostate cancer, on which R.M., S.A.T. and A.M.C. are listed as co-inventors. The University of Michigan has licensed the diagnostic field of use to Gen-Probe, Inc. (Bedford, MA), which has sublicensed some rights to Ventana Medical Systems. S.A.T. serves as a consultant to and has received honoraria from Ventana Medical Systems. A.M.C. has served as a consultant for Gen-Probe, Inc. and Ventana Medical Systems. All other authors have no disclosures.
References
- 1.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med 2011;3:111ra121–111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang B-H, Liu L-Z. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochim. Biophys. Acta 2008;1784:150–158. doi: 10.1016/j.bbapap.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan TM, Koreckij TD, Corey E. Targeted therapy for advanced prostate cancer: inhibition of the PI3K/Akt/mTOR pathway. Curr Cancer Drug Targets 2009;9:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 8.Helgeson BE, Tomlins SA, Shah N, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 9.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod. Pathol. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 10.Mehra R, Tomlins SA, Yu J, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 2008;10:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klezovitch O, Risk M, Coleman I, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc. Natl. Acad. Sci. U.S.a. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat. Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zong Y, Xin L, Goldstein AS, et al. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc. Natl. Acad. Sci. U.S.a. 2009;106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat. Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krohn A, Diedler T, Burkhardt L, et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am. J. Pathol. 2012;181:401–412. doi: 10.1016/j.ajpath.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 17.Han B, Mehra R, Lonigro RJ, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod. Pathol. 2009;22:1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshimoto M, Ding K, Sweet JM, et al. PTEN losses exhibit heterogeneity in multifocal prostatic adenocarcinoma and are associated with higher Gleason grade. Mod. Pathol. 2013;26:435–447. doi: 10.1038/modpathol.2012.162. [DOI] [PubMed] [Google Scholar]
- 19.Bismar TA, Yoshimoto M, Vollmer RT, et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107:477–485. doi: 10.1111/j.1464-410X.2010.09470.x. [DOI] [PubMed] [Google Scholar]
- 20.Bhalla R, Kunju LP, Tomlins SA, et al. Novel dual-color immunohistochemical methods for detecting ERG-PTEN and ERG-SPINK1 status in prostate carcinoma. Mod. Pathol. 2013;26:835–848. doi: 10.1038/modpathol.2012.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korshunov A, Sycheva R, Gorelyshev S, et al. Clinical utility of fluorescence in situ hybridization (FISH) in nonbrainstem glioblastomas of childhood. Mod. Pathol. 2005;18:1258–1263. doi: 10.1038/modpathol.3800415. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimoto M, Cunha IW, Coudry RA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br. J. Cancer 2007;97:678–685. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshimoto M, Joshua AM, Cunha IW, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod. Pathol. 2008;21:1451–1460. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 24.Grasso CS, Wu Y-M, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grasso CS, Cani AK, Hovelson DH, et al. Integrative molecular profiling of routine clinical prostate cancer specimens. Ann. Oncol. 2015;26:1110–1118. doi: 10.1093/annonc/mdv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell 2000;100:387–390. [DOI] [PubMed] [Google Scholar]
- 27.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat. Rev. Cancer 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bismar TA, Yoshimoto M, Duan Q, et al. Interactions and relationships of PTEN, ERG, SPINK1 and AR in castration-resistant prostate cancer. Histopathology 2012;60:645–652. doi: 10.1111/j.1365-2559.2011.04116.x. [DOI] [PubMed] [Google Scholar]
- 29.Liu S, Yoshimoto M, Trpkov K, et al. Detection of ERG gene rearrangements and PTEN deletions in unsuspected prostate cancer of the transition zone. Cancer Biol. Ther. 2011;11:562–566. [DOI] [PubMed] [Google Scholar]
- 30.Lahdensuo K, Erickson A, Saarinen I, et al. Loss of PTEN expression in ERG-negative prostate cancer predicts secondary therapies and leads to shorter disease-specific survival time after radical prostatectomy. Mod. Pathol. 2016;29:1565–1574. doi: 10.1038/modpathol.2016.154. [DOI] [PubMed] [Google Scholar]
- 31.Chaux A, Peskoe SB, Gonzalez-Roibon N, et al. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod. Pathol. 2012;25:1543–1549. doi: 10.1038/modpathol.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lotan TL, Gumuskaya B, Rahimi H, et al. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod. Pathol. 2013;26:587–603. doi: 10.1038/modpathol.2012.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trotman LC, Niki M, Dotan ZA, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Udager AM, Alva A, Mehra R. Current and proposed molecular diagnostics in a genitourinary service line laboratory at a tertiary clinical institution. Cancer J 2014;20:29–42. doi: 10.1097/PPO.0000000000000017. [DOI] [PubMed] [Google Scholar]
- 35.Brenner JC, Ateeq B, Li Y, et al. Mechanistic Rationale for Inhibition of Poly(ADP-Ribose) Polymerase in ETS Gene Fusion-Positive Prostate Cancer. Cancer Cell 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng FY, Brenner JC, Hussain M, et al. Molecular Pathways: Targeting ETS Gene Fusions in Cancer. Clin. Cancer Res. 2014;20:4442–4448. doi: 10.1158/1078-0432.CCR-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussain M, Daignault S, Twardowski P, et al. Co-targeting androgen receptor (AR) and DNA repair: A randomized ETS gene fusion-stratified trial of abiraterone + prednisone (Abi) +/− the PARP1 inhibitor veliparib for metastatic castration-resistant prostate cancer (mCRPC) patients (pts) (NCI9012)--A University of Chicago phase II consortium trial. ASCO Meeting Abstracts 2016;34:5010. [Google Scholar]
- 38.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997;275:1943–1947. [DOI] [PubMed] [Google Scholar]
- 39.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J. Clin. Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 40.Arkenau H-T, Mateo J, Lemech CR, et al. A phase I/II, first-in-human dose-escalation study of GSK2636771 in patients (pts) with PTEN-deficient advanced tumors. ASCO Meeting Abstracts 2014;32:2514. [Google Scholar]
- 41.Blackman SC, Gainer SD, Suttle BB, et al. Abstract 1752: A phase I/IIa, first time in human, open-label dose-escalation study of GSK2636771 in subjects with advanced solid tumors with PTEN deficiency. Cancer Res. 2012;72:1752–1752. doi: 10.1158/1538-7445.AM2012-1752. [DOI] [Google Scholar]