SUMMARY
Circular RNAs (circRNAs) are an intriguing class of RNA due to their covalently closed structure, high stability, and implicated roles in gene regulation. Here, we used an exome capture RNA sequencing protocol to detect and characterize circRNAs across >2,000 cancer samples. When compared against Ribo-Zero and RNase R, capture sequencing significantly enhanced the enrichment of circRNAs and preserved accurate circular-to-linear ratios. Using capture sequencing, we built the most comprehensive catalogue of circRNA species to date: MiOncoCirc, the first database to be composed primarily of circRNAs directly detected in tumor tissues. Using MiOncoCirc, we identified candidate circRNAs to serve as biomarkers for prostate cancer and were able to detect circRNAs in urine. We further detected a novel class of circular transcripts, termed read-through circRNAs, that involved exons originating from different genes. MiOncoCirc will serve as a valuable resource for the development of circRNAs as diagnostic or therapeutic targets across cancer types.
Keywords: circRNA, exome capture sequencing, biomarkers, cancer
INTRODUCTION
Circular RNAs (circRNAs) are single-stranded, covalently closed RNA molecules that are produced from pre-mRNAs through a process called backsplicing and were initially proposed to be splicing-associated noise (Capel et al., 1993). However, recent studies have shown that circRNAs may be involved in miRNA inhibition (Hansen et al., 2013), epithelial-mesenchymal transition (Conn et al., 2015), and tumorigenesis (Guarnerio et al., 2016). Further, circRNA expression can be tissue-specific (Conn et al., 2015) and some evidence support the translation of some circRNAs (Pamudurti et al., 2017; Legnini et al., 2017). When compared to linear counterparts, circRNAs are highly stable and can be found in exosomes, cell-free saliva, and plasma (Li et al., 2015; Bahn et al., 2016). Therefore, with improved detection and characterization methodologies, circRNAs may be potential biomarkers or therapeutic targets.
Advances in high-throughput sequencing technology and novel bioinformatics algorithms have facilitated the systematic detection of circRNAs (Salzman et al., 2013). Although short-read paired-end RNA-seq technology does not fully unveil the whole body of circRNAs, it can reliably identify backspliced junctions and thus allows for the robust identification and quantification of several circRNA species per sample. Detection of circRNAs through high-throughput RNA-sequencing (RNA-seq) technologies requires a protocol that can profile non-polyadenylated (non-poly(A)) transcripts (Hansen et al., 2013; Jeck et al., 2013). Currently, RNase R and Ribo-Zero are the gold standard methods for detecting circRNAs. With exoribonuclease RNase R, enrichment of circRNAs within samples is achieved at the expense of degrading linear RNAs (Jeck et al., 2013). This method, therefore, proves impractical for further downstream analysis since it interferes with the quantification of mRNA. On the other hand, Ribo-Zero enriches circRNAs while depleting rRNAs and preserving linear transcripts but requires at least 5 μg of total RNA to yield reliable results (Giannoukos et al., 2012).
Here, we present a novel use of our exome capture RNA-seq protocol (Cieslik et al., 2015) to profile circRNAs across more than 800 human cancer samples. In exome capture sequencing, RNA probes that target gene bodies hybridize with cDNA fragments and enrich exonic circRNA. Upon validation against Ribo-Zero and RNase R, our method consistently achieved significantly better enrichment for circRNAs than Ribo-Zero, and, unlike RNase R treatment, preserved accurate circular-to-linear ratios. We not only detected more circRNA transcripts per gene, but also uncovered circularized read-through transcripts. Furthermore, we used less than 5 μg of total RNA to achieve our results, thus demonstrating our protocol as a preferable alternative to Ribo-Zero when using samples with limited total RNA, an important advantage in clinical settings for analysis of biospecimens. To extend the catalog of reported circRNAs, we developed a compendium, MiOncoCirc, that will be an open, comprehensive resource for facilitating the exploration of circRNA as a new type of cancer biomarker and for aiding in the elucidation of circRNA function. Of note, MiOncoCirc is the first cancer-focused circRNA resource to be generated from an extensive array of tumor tissues.
RESULTS
Exome capture RNA-seq is an effective method to profile circRNAs
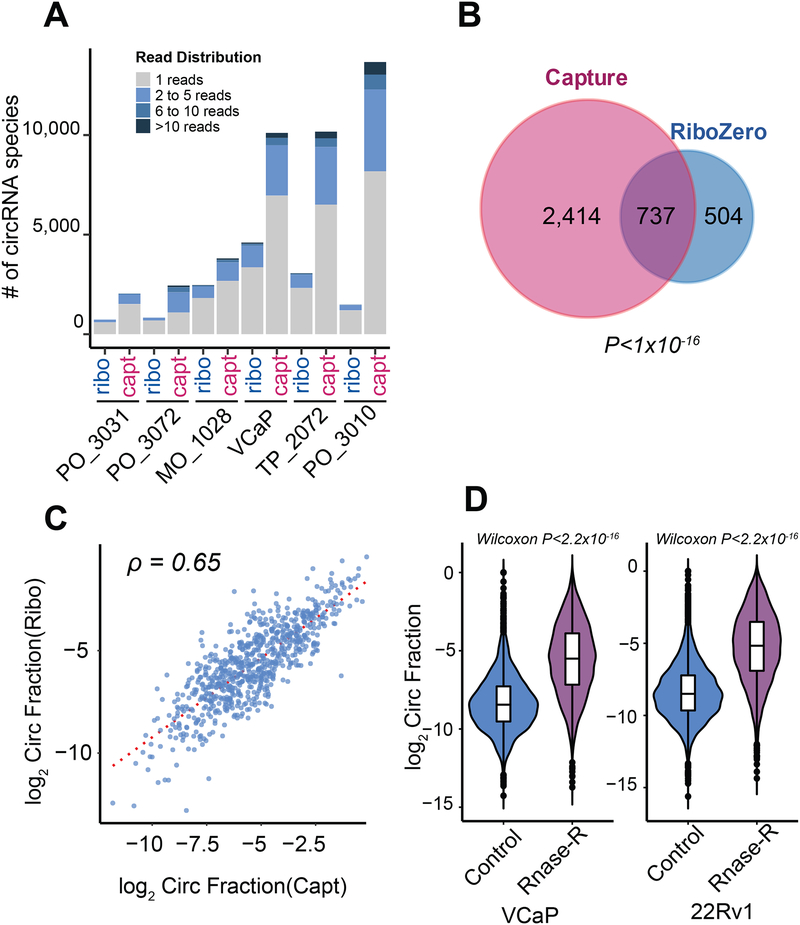
Our previously described exome capture transcriptome protocol, exome capture RNA-seq, is a poly(A)-independent RNA sequencing method that targets the gene body, thus rendering it suitable for the study of circRNAs (Cieslik et al., 2015). We validated the performance of exome capture RNA-seq against Ribo-Zero sequencing, the most frequently used high-throughput method for detecting circRNAs. Relative to Ribo-Zero, capture sequencing consistently detected more circRNA species per library in a panel of cell lines and frozen tissues (Figure 1A, Table S1). Furthermore, using VCaP prostate cancer cells, we validated that the majority of circRNA species present in the Ribo-Zero library were also present in the matched capture library from this cell line (Figure 1B). The circular to linear fractions were retained in capture as in Ribo-Zero (Figure 1C). In addition, in post RNase R treatment, capture sequencing yielded an overall elevated circular-to-linear ratio of all circRNA species (Figure 1D), thus confirming that our capture sequencing method detected true circular molecules and not potential ligation artifacts inherent to the protocol.
Figure 1. Validation of the exome capture RNA-seq method for circRNA detection.
A) When compared to matched Ribo-Zero samples, capture transcriptome sequencing consistently detected more circular RNA (circRNA) in six paired libraries from clinical samples and cell lines. Capt: exome capture RNA-seq; Ribo: Ribo-Zero. Details about sequencing depths and the number of circRNAs stratified by detected number of backspliced reads can be found in Table S1. To validate that the higher numbers of detected circRNAs in capture sequencing libraries were not due to the differences in sequencing depth, some libraries were down-sampled to make sure that the sequencing depth of any capture library was no more than the depth of its matched Ribo-Zero library.
B) In a VCaP cell line library, the overlap of detected circRNA species in exome capture and Ribo-Zero sequencing platforms was significant (Fisher exact test, P-value < 1×10−16). A threshold of two backspliced reads was applied.
C) In VCaP, capture sequencing retained the relative abundance of circRNAs to their linear counterparts, comparable to Ribo-Zero library (Spearman Rank Correlation ρ = 0.65) (see STAR Methods). A threshold of two backspliced reads was applied.
D) Backspliced events called from our MiOncoCirc pipeline were elevated (Mann-Whitney-Wilcoxon < 2.2×10−16) post RNase R treatment in VCaP and 22RV1 cell lines, further confirming that they were true circRNAs and not ligation artifacts.
See also Table S1.
Building the MiOncoCirc compendium with exome capture RNA-seq
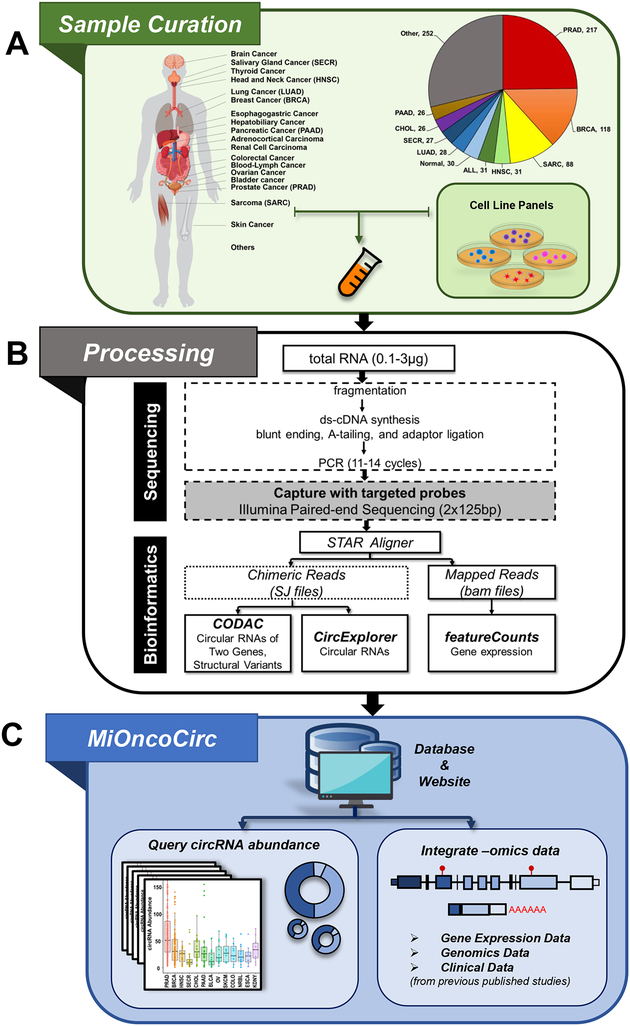
Using data generated with the exome capture RNA-seq protocol, we developed MiOncoCirc, an accessible compendium of cancer-focused circRNAs for the scientific community. The version of MiOncoCirc reported here was comprised of 868 samples obtained from previously published data sets of clinical samples (Mody et al., 2015, Robinson et al., 2015, Robinson et al., 2017) and cancer cell lines as well as pooled normal tissues (Figure 2A, Table S1). The protocol and bioinformatics pipeline used to create MiOncoCirc are detailed in Figure 2B and STAR Methods. Briefly, the transcriptome for each sample was profiled by paired-end, strand specific capture RNA-seq with moderate depth (median 49M ± 14M paired-reads). To detect backspliced (circular) reads from RNA-seq libraries, we used the pipeline CIRCexplorer (Zhang et al., 2014). CIRCexplorer has been shown to achieve high sensitivity and specificity among current circRNA bioinformatics tools (Hansen at al., 2015). In addition, we employed our in-house computational pipeline CODAC to discover read-through circRNAs (rt-circRNA), a novel class of circRNAs only recently discovered (Liang et al., 2017). CODAC was initially developed to call structural rearrangements in paired-end RNA-seq but has been extended to the annotation of circRNAs, especially to those involving more than one gene. Information about circRNAs and rt-circRNAs can be found on our MiOncoCirc website, which also enables the querying and downloading of circRNA abundances across different cancer types, as well as the expression of their parent genes (Figure 2C, STAR Methods).
Figure 2. Construction and overview of the MiOncoCirc compendium.
A) 868 high-depth, paired-end RNA-seq samples from previously published data sets as well as cell line panels and normal tissues were included. Additional details and abbreviations can be found in Table S1.
B) Exome capture RNA-seq protocol and the bioinformatics pipeline for creation of MiOncoCirc. The unmapped reads from chimeric aligner (STAR) were annotated against the exon junctions. CIRCexplorer was used to call circRNA transcripts, and CODAC was used to annotate circRNAs involving two genes. FeatureCounts was used to quantify gene expression.
C) MiOncoCirc is an online database that enables querying and downloading of circRNAs abundance across different tissues. Additional genomic data can be retrieved from previous studies (STAR Methods).
See also Table S1.
The characteristics of circRNAs in MiOncoCirc
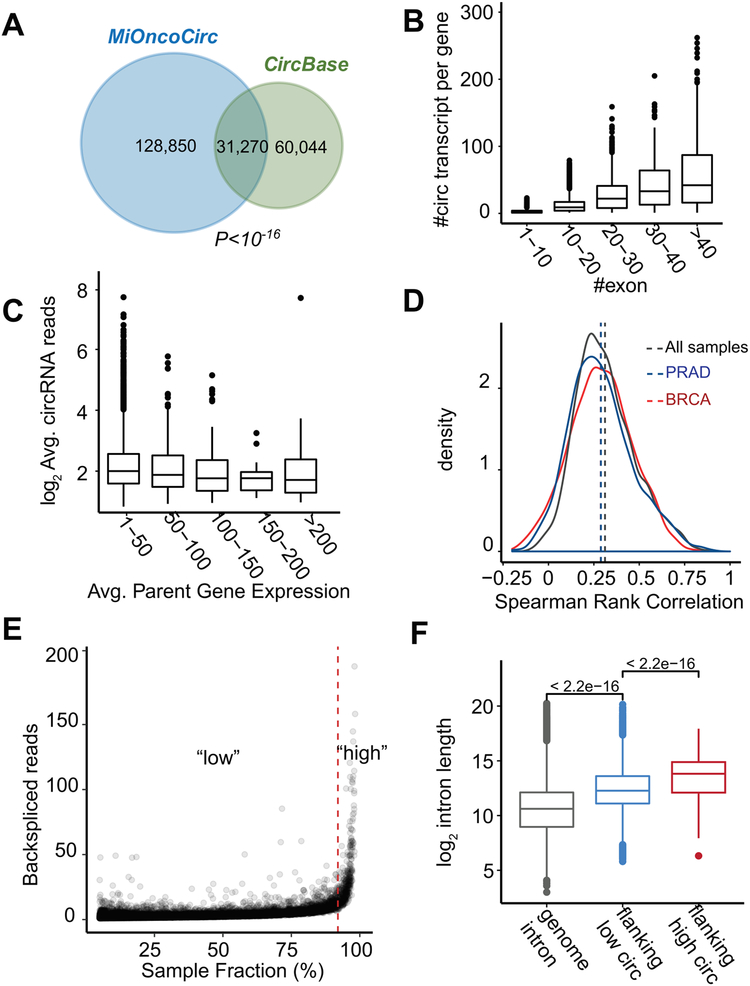
MiOncoCirc uncovered a significant number of circRNA species in addition to the species provided by CircBase, a compendium of circRNA compiled from different sources (Glažar et al., 2014). To analyze the overlap, we first used a stringent cut-off that included only circRNAs that appeared in at least five different samples from the data sets (Table S2). Using this criterion, MiOncoCirc and CircBase significantly overlapped in terms of circular transcripts (Figure 3A, Fisher Exact Test P < 1 × 10−16) and parent genes (Figure S1A, Fisher Exact Test P < 2.2 × 10−16). The overlap was even more significant if the criteria were relaxed to include circRNAs detected in any one sample (Figure S1B). Among genes that were found in both databases to produce circRNAs, MiOncoCirc detected twice the number of circular isoforms (Figure S1C). The non-overlapping sets of genes and circular transcripts may have resulted from differences in tissue types included in the two compendia. For instance, the cell lines included in the CircBase compendium are heavily endometrial, fibroblastic, and myoblastic while the tissues in MiOncoCirc are primarily epithelial (carcinoma) and mesenchymal (sarcoma). Indeed, when we compared the circRNAs in MiOncoCirc with CircBase from the same cell/tissue type (lung and breast, both epithelial), we found significant overlap (Figure S1D).
Figure 3. Features of circRNAs in MiOncoCirc and properties associated with expression.
A) The overlap of circRNA species in MiOncoCirc and CircBase was significant (Fisher Exact Test P-Value < 1×10−16). Only high confidence circRNAs which appeared in five or more samples were included in this comparison.
B) Genes can form multiple circRNA transcripts. The number of circular transcripts increased proportionally with the number of exons per gene (binned to 10).
C) Average expression of circRNA abundance (in normalized backspliced reads) vs. average expression of parental expression (in FPKM). Parent gene expression was grouped into bins of 50. Overall, there was no different in the mean of the bins (ANOVA P-Value = 0.12). This result agreed with Figure S2A in that the correlation was weak (Spearman’s ρ = 0.12).
D) The distribution of Spearman’s rank correlation between circRNA abundance and their cognate parent gene expression, across all samples (gray), in prostate adenocarcinoma (PRAD, blue), and breast cancer (BRCA, red). Overall, the correlations were low (all medians < 0.28).
E) Circular RNA abundance (in normalized backspliced reads) vs. sample fraction (%). There was a small portion of circRNAs (<2% of all circRNAs, generated from 589 genes, marked as “high”) that were detected in more than 90% of all samples. They also had higher expression compared to the median of all circRNAs (marked as “high” in the density plot of Figure S2A).
F) These “high” circRNAs were flanked by significantly longer introns (Mann-Whitney-Wilcoxon P < 2.2×10−16) than the remaining 98% of circRNAs. Genome-wide introns were included as the control.
See also Table S2 and Figures S1–S2.
By further examining the circRNA species in MiOncoCirc, we found that the genes that generated circRNAs tended to form multiple circular isoforms. The number of circular isoforms increased proportionally with the number of exons per gene (Figure 3B). One extreme example from our compilation, the gene BIRC6, could generate more than 500 different circular isoforms. Motif analysis of the exon-intron boundaries revealed that the majority (>99.2%) of circRNAs were flanked by the canonical splicing motif, AG-GT (Figure S1E). Among the non-canonical splicing signals, the most commonly observed were GC-AG (0.7%) or AT-AC (0.05%), while the rest were from other combinations of non-canonical motifs.
We further determined that the average abundance of the parent genes (as quantified by FPKM) only weakly correlated with the average abundance of their associated circRNAs (in normalized backspliced reads, see STAR Methods; Spearman’s ρ = 0.12, Figures S2A and 3C). These results indicate that the baseline expression of the parent gene is not a reliable predictor of the corresponding abundance of its circRNA. In addition, the Spearman’s rank correlations of the circRNAs and their parental expression across all 868 samples and within two sample cohorts (prostate and breast cancers) were low (all medians ρ < 0.3, Figure 3D). These findings suggest that the variability of circRNA abundances cannot be directly explained by the variability of their parental expression, but rather, may involve regulatory splicing mechanisms or varying rates of circRNA turnover in tumors.
We also discovered a subset of circRNAs in MiOncoCirc that could be consistently detected in ≥ 90% of our samples and with higher abundances (≥ 5×) than the median abundance of all circRNAs (marked as “high” in Figures 3E and S2A). This subset of highly abundant circRNAs was generated from 589 genes with low and modest average expression (≤ 50 FPKM) (Figure 3C) and enriched for functional categories fundamental to cells (Figure S2B). While these genes were not more highly expressed than the parental genes of other circRNAs (Figure S2C), they possessed interesting genomic features. First, the circRNAs generated from this subset of genes were characterized by significantly longer flanking introns (Wilcoxon rank-sum test P< 2.2 × 10−16, Figure 3F) that harbored more repetitive elements (Wilcoxon rank-sum test P< 4.5 × 10−8, Figure S2D) than the introns flanking circRNAs with lower abundance (marked as “low”). Long flanking introns, which may harbor more repetitive and reverse complement elements that promote circularization, were found to be the hallmark of circular RNA biogenesis (Ivanov et al., 2015). Indeed, even the class of circRNAs with “low” abundance in our compendium exhibited significantly longer flanking introns with more repetitive elements than all introns genome-wide (Wilcoxon rank-sum test P< 2.2 × 10−16, Figures 3F and S2D). Additionally, genes generating circRNAs with outlier expression were significantly longer and contained more exons (Figures S2E–F). Together, these results suggest that this “high” class of circRNAs is more consistently detected and presented at higher abundance because their unique genomic structures support circularization.
The prevalence and characteristics of read-through circRNAs
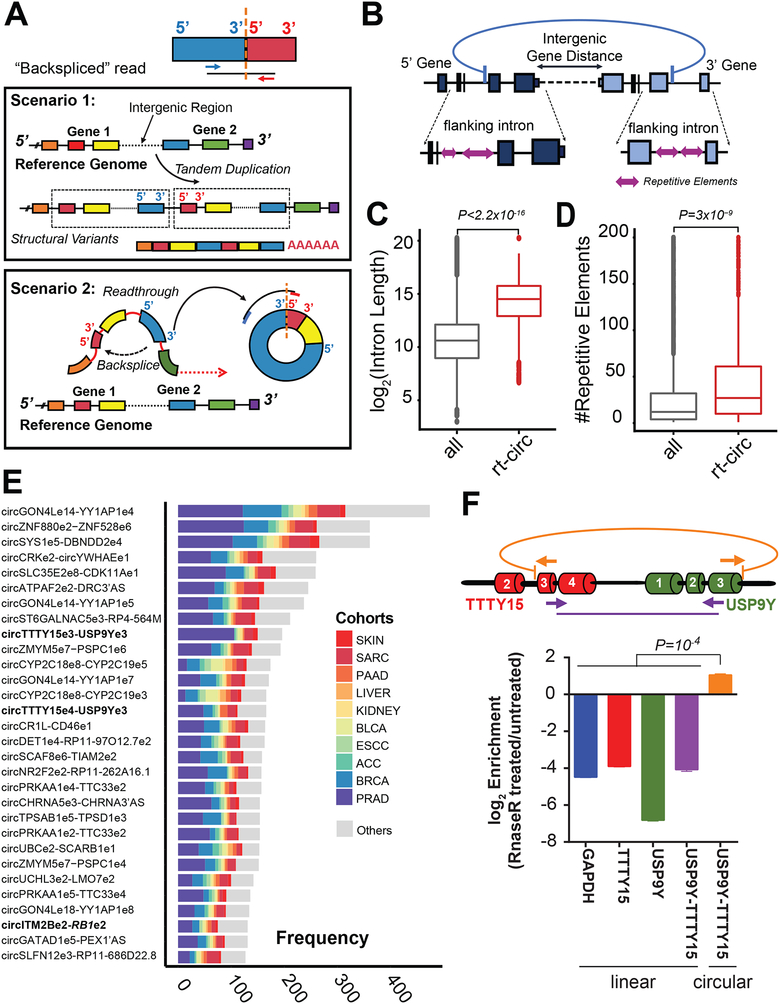
Circular RNAs produced from exons originating from different genes were previously reported to be products of gene fusions, in which each fusion partner donated their exons for backsplicing, and were named f-circRNAs (Guarnerio et al., 2016). We recently developed a novel annotation pipeline, CODAC, that could annotate backsplicing events involving two genes (see STAR Methods). Since pairs of homologous/paralogous genes can give rise to mapping ambiguities and false positives, we performed preliminary filtering (see STAR Methods) as well as indicated pairs with high degrees of similarity in Table S3. Although we did not detect any f-circRNAs in MiOncoCirc resulting from chromosomal translocations and deletions, we discovered a novel class of circular transcripts that involved exons originating from two adjacent genes on the same strand: the read-through circRNA (rt-circRNA). Without the genomic information from matched whole-genome sequencing (WGS) and whole-exome sequencing (WES) acquired through our integrative clinical sequencing approach (Robinson et al., 2017), rt-circRNAs would have appeared deceptively similar to linear transcripts resulting from tandem duplications in RNA-seq (Figures 4A–B). In general, rt-circRNAs comprised a small portion of all circRNAs in each sample (average 2.5%, Figure S3A) and were detected at lower abundance (average 3.1× lower, Wilcoxon rank-sum test P-Value = 1 × 10−12, Figure S3B) than most other circRNAs from a single gene.
Figure 4. Identification of novel read-through (rt) circRNA species.
A) Schematic showing that genomic tandem duplications and circRNAs involving two genes can appear similar in paired-end RNA-seq. Specifically, when mates of a paired-end read were aligned in divergent orientation to exons of two adjacent genes, the result could be interpreted as either a duplication of a group of exons from two genes (Scenario 1), or a circularization from the downstream gene back to the upstream gene (Scenario 2).
B) Schematic depicting the circular read-through event that can be generated from two adjacent genes, and their genomic features.
C) The introns flanking rt-circRNAs were longer than genome-wide introns (Mann-Whitney-Wilcoxon P < 2.2×10−16).
D) The introns flanking rt-circRNAs harbored more repetitive elements than genome-wide introns (Mann-Whitney-Wilcoxon P < 3×10−9).
E) The frequency and distribution of the top 30 most abundant backspliced events involved neighboring genes in our compendium.
F) The circular read-through event involving exon 3 of TTTY15 and exon 3 of USP9Y was chosen for validation in LNCaP cells. Post RNase R treatment, only the RT-qPCR product of outward facing primers was resistant to exoribonuclease degradation (see STAR Methods).
See also Tables S3 and S7 and Figures S3–S5.
Similar to circRNAs generated from single genes, the expression of the respective parental genes upstream and downstream of rt-circRNAs varied greatly (Figures S3C–D). The rt-circRNAs also demonstrated the genomic trademarks of typical circRNAs in that they were flanked by introns that were longer (Wilcoxon rank-sum test P-Value = 2.2 × 10−16, Figure 4C) and harbored more repetitive elements (Wilcoxon rank-sum test P-Value = 3 × 10−9, Figure 4D). In addition, pairs of genes that generated rt-circRNAs were slightly shorter than any random pairs of adjacent genes on the same strand (median 37kb vs. median 48.8kb apart, Wilcoxon rank-sum Test P-Value = 1 × 10−4, Figure S3E).
Some of these backspliced reads involving two genes were commonly found across different cancer types (Figure 4E) and were even detected in normal tissues or in samples with normal copy-number (diploid) of the parent genes (Figure S4A and Table S3), which further suggested that they were true common transcriptomic processes rather than rare genomic events. To further experimentally validate by RT-qPCR that this class of readthrough transcript was circularized, we searched for transcripts that were expressed in cell lines and selected a backspliced event spanning two adjacent genes, TTTY15 and USP9Y, which were less than 9 kb apart on chromosome Y and detected in several prostate cancer tissue samples (Figure 4F). The product of outward facing primers involving exon 3 of TTTY15 and exon 3 of USP9Y were detected via RT-qPCR in LNCaP prostate cancer cells, as well as the product of inward facing primers involving the same pair of exons (see STAR Methods). However, only the product of outward facing amplification was resistant to RNase R degradation (Figure 4F), and the backspliced exon-exon junctions of USP9Y and TTTY15 were validated by Sanger sequencing (Figure S4B), confirming that the target was a circular molecule.
Among the most commonly found rt-circRNAs (Figure 4E), we also identified an event that involved exon 2 of the well-known tumor suppressor RB1 with an upstream gene, ITM2B (Figure S4C). An RB1-ITM2B backspliced transcript involving exon 2 of RB1 and exon 3 of ITM2B (ITM2Be3-RB1e2) was reported in a few melanoma cases (Berger et al., 2010) and was confirmed to be the result of focal amplification via SNP array. In contrast, the ITM2Be2-RB1e2 rt-circRNA from the MiOncoCirc compilation was commonly found across cancer types and even detected in normal samples (Table S3). Copy-number analysis from one case of non-small cell lung cancer (NSCLC) and bladder cancer (BLCA) inferred from target capture panels (Robinson et al., 2017) did not show any copy-number alterations, further confirming that ITM2Be2-RB1e2 was a rt-circRNA (Figure S4C).
Finally, even though read-through circularization was largely widespread across cancer types (Figure 4E), we were able to nominate a small set of select rt-circRNAs that were tissue-specific in the MiOncoCirc compendium (Figure S5A). Their tissue specificity could be explained by the tissue-specific expression of the genes involved in the generation of the corresponding rt-circRNAs (Figure S5B). The overlap between all tissue-specific genes and rt-circRNA parental genes was minimal, which was consistent with our ability to detect a limited number of tissue-specific rt-circRNAs (Figure S5C).
The properties of circRNAs in cancer
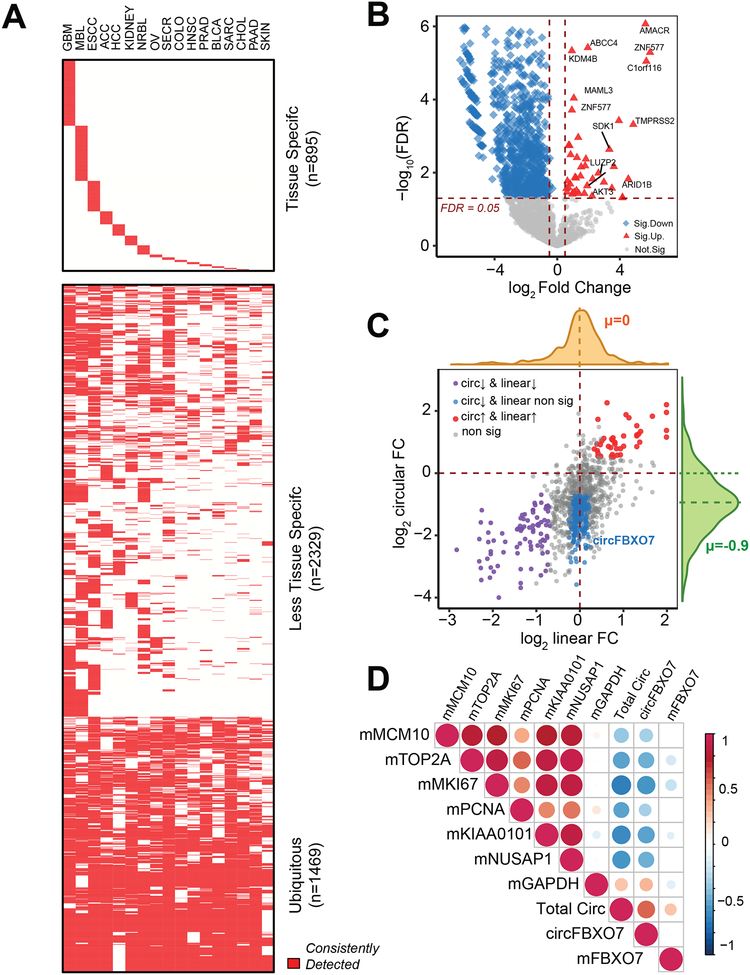
Non-coding RNAs, such as lncRNAs (long non-coding RNAs), pseudogenes, and miRNAs, have demonstrated lineage-specific patterns (Kalyana-Sundaram et al., 2012; Guo et al., 2014; Iyer et al., 2015). We investigated whether the overall set of genes that could form circRNAs exhibited lineage specificity in cancer samples. To test this, we analyzed 17 different cancer cohorts from the MiOncoCirc compendium consisting of a range of lineages. Since many samples from these cohorts were obtained from metastatic sites, contamination from tissue at the biopsy site was a possible caveat. To enrich for signals specific to the tissue of origin, we considered a gene to be positive for circRNA production if it formed at least one circRNA that could be consistently detected in at least 30% of a cancer lineage (STAR Methods). Based on these analyses, we determined that genes that formed circRNAs in our compendium could be classified into three categories: tissue-specific (n=895), less tissue-specific (n=2,329), and ubiquitous (n=1,469) (Figure 5A and Table S3). The tissue specificity of the first group could be explained by the tissue specificity of the parental genes (Figure S6A). These data suggest that there are indeed lineage-specific genes that form circRNAs in the MiOncoCirc resource that could be used as biomarkers to discriminate between different tissue types.
Figure 5. Expression patterns and characteristics of circRNAs in cancer.
A) Tissue-specific heatmap of genes that can generate circRNAs, as demonstrated in 17 cancer cohorts from the MiOncoCirc compendium. A gene was considered to be consistently detected if it generated at least one high-confidence circRNA in more than 30% of samples of any given lineage (see STAR Methods).
B) Volcano plot of circRNA abundances in 25 matched pairs of normal/localized prostate adenocarcinoma. Horizontal dash-line corresponded to FDR = 0.05. Vertical dash-line corresponded to fold-change > 1.5× (up-regulation) and fold-change < −1.5× (down-regulation).
C) The correlation of log fold-change (FC) of circular RNA vs. log FC of linear expression. Again, circRNA abundances were downregulated overall (mean circular logFC = −0.9). We further stratified genes into groups based on the relationship between the linear and circular fold change. Group 1 (red) were circRNAs that were upregulated in cancer because their parent genes were upregulated. Group 2 (purple) were those circRNAs that were downregulated in cancer because their parent genes were also downregulated. However, there was a subset of circRNAs (Group 3, blue) downregulated in cancer with no corresponding change in parent gene expression.
D) Total circRNA correlated with prostate cancer mRNA (“m”) proliferation markers calculated in FPKM (MCM10, TOP2A, MKI67, PCNA, KIAA0101, and NUSAP1). The size and the color scale of the dots indicate the values of pair-wise Spearman Rank Correlation. GAPDH mRNA expression was included as a negative control. Circular FBXO7, a highly abundant circRNA also showed remarkable negative correlation with proliferation index, even though its parental gene expression did not correlate.
See also Tables S3–S4 and Figures S6–S7.
To characterize whether the expression patterns of circRNAs varied between tumor and normal cells, we performed differential abundance analysis for circRNAs in 25 pairs of matched tumor/normal for localized primary prostate adenocarcinoma (PRAD) samples (Figures 5B and S6B). Among circular transcripts that showed differential abundance (n=652) in cancer compared to normal (FDR < 0.01), a majority (n=629) were downregulated in cancer (average log-fold change of circRNAs was −0.9, Figures 5C and S6B, Table S4). While the downregulation of some circRNAs could be explained by the downregulation of the parent genes (Figure 5C, purple data points), there were also circRNAs with relatively low abundance in prostate cancer (< −1.5× fold change compared to matched normal, FDR < 0.01) without significant associated changes in the parental gene expression (Figure 5C, blue data points, circ-FSX07 as an example).
In tumor tissues, we demonstrated that cellular proliferation may lead to the downregulation of circRNAs, a mechanism also proposed by Bachmayr-Heyda et al. (2015). In our 25 pairs of matched normal/PRAD, total circRNA abundance showed a consistent negative correlation with expressions of MKI67and PCNA, and with a panel of cell cycle progression genes (e.g., MCM10, T0P2A, KIAA0101, and NUSAP1), whose mRNA levels have been used as proliferation markers for prostate cancer (Cuzick et al., 2011, Figures 5D and S6C). A similar pattern was seen when individual circRNAs, such as FBXO7, were analyzed. Furthermore, we compared the global circRNA profile in non-matched tumor/normal across six other tissue types: bone-osteosarcoma (n=8), colon-colorectal adenocarcinoma (n=13), kidney-renal cell carcinoma (n=12), liver-hepatocellular carcinoma (n=10), lung-lung adenocarcinoma (n=10), and stomach-gastric adenocarcinoma (n=9). The normal samples of each tissue were pooled from healthy donors, and the tumor samples were from the MiOncoCirc cohort. Across diverse lineages, total circRNA abundance was lower in cancer compared to normal (Figure S7A), suggesting that the downregulation of circRNA was a universal observation regardless of cell lineage. To further confirm the negative correlation between total circRNA abundance and proliferation, we treated LNCaP cells with dinaciclib, a potent kinase inhibitor that inhibits a wide range of kinases (Parry et al., 2010) and thus decreases cellular proliferation. Capture sequencing was then performed on control (N=3) and dinaciclib-treated (N=3) samples. After 24 hours of treatment, we observed an overall increase in total circRNA abundance independent of any change in parent gene expression (Figures S7B–C, Table S4).
Interestingly, despite a general decrease in total circRNA abundance in cancer samples, we observed a small subset (n=23) of circRNAs to be expressed more highly in tumor samples compared to normal (Figures 5B and S6B). This subset included the circular isoforms of AKT3, SDK1, LUZP2, ABCC4, and AMACR, a gene whose mRNA is currently used as a biomarker of prostate cancer and was characterized previously by our group (Rubin et al., 2002). The upregulation of these circRNAs could be directly explained by the elevated expression of the parent genes (Figure 5C, red data points). This finding implicates a potential association between the upregulation of circRNAs and genomic amplification, a common mechanism for gene overexpression in cancer. For instance, AR, the androgen receptor gene, is frequently amplified in metastatic castration-resistant prostate cancer (mCRPC) (Robinson et al., 2015). We compared 70 cases of mCRPC with amplified AR (> than 5 copies) against 50 cases of hormone-naive primary prostate cancer without amplified AR and found that circ-AR (backspliced from exon 4 to exon 3) was detected in mCRPC (54/70 cases) but not in primary cancers at the current sequencing depth. This interesting result could be explained by the massive upregulation of AR via focal amplification in mCRPC samples (Figure S7D).
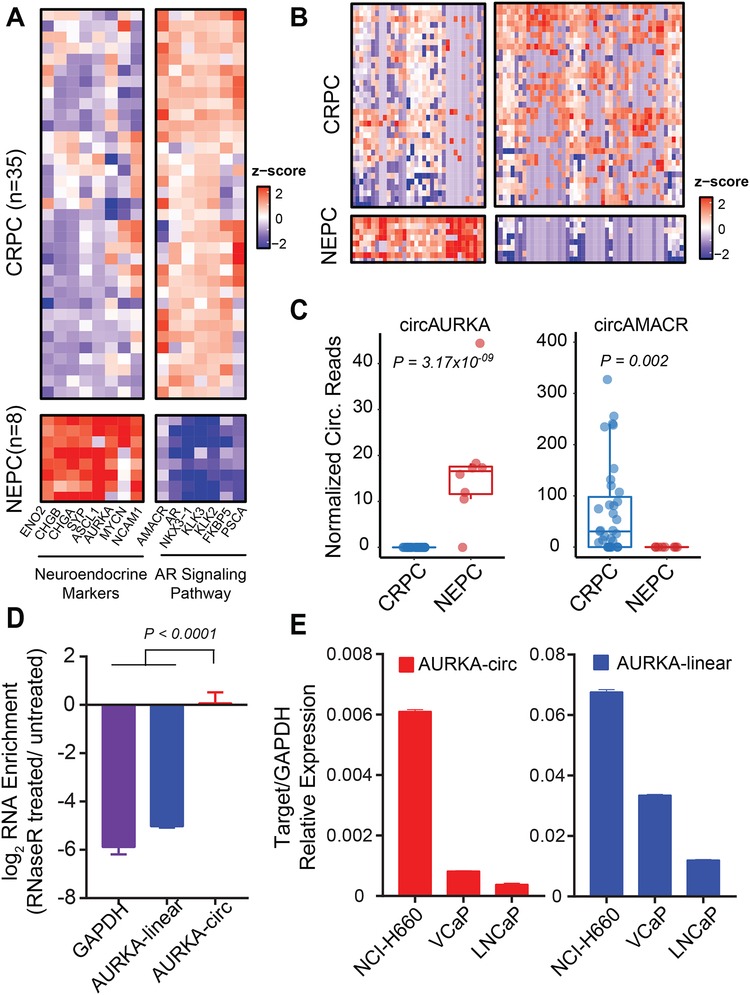
Because circRNAs are “spliced out” from mRNAs, any cellular process or transformation that has a profound impact on the transcriptome should likewise alter the circRNA landscape of the cell. Thus, we characterized the circRNA landscape of prostate cancers undergoing neuroendocrine differentiation. Neuroendocrine prostate cancer (NEPC) is a rare, aggressive subtype of prostate cancer that can arise post-hormonal therapy for PRAD (Beltran et al., 2011) and has a poor prognosis (Conteduca et al., 2014). In our cohort, pathologists diagnosed eight NEPC cases based on cell morphology. To further validate neuroendocrine differentiation at the transcriptomic level, we performed differential gene (mRNA) analysis of the eight NEPC cases versus 35 CRPC cases with the highest tumor purity. Our eight NEPC cases were all characterized by the upregulation of neuroendocrine markers (e.g., SYP, CHGA, CHGB, NCAM1, ENO2, ASCL1, MYCN, and AURKA), and downregulation of genes in the AR signaling pathway (e.g., AR, AMACR, KLK3, KLK2, FKBP5, and PSCA) (Figure 6A). This gene expression signature agrees with the well-established literature on NEPC (Beltran et al., 2011). We then performed differential circRNA expression analyses (Figure 6B and Table S5) and uncovered 34 upregulated and 48 downregulated circRNAs with statistical significance (P-Value < 0.01). In NEPC, the most significantly upregulated and downregulated circRNAs were circ-AURKA (Mann-Whitney U test P=3.17× 10−9) and circ-AMACR (Mann-Whitney U test P= 0.002), respectively (Figure 6C). This finding is consistent with the change in parental gene expression of AURKA and AMACR (Figure 6A). We carried out RT-qPCR in RNase R treated NCI-H660, a neuroendocrine cell line, and confirmed that circ-AURKA was generated from exon 6 backspliced to exon 3 (Figure 6D). Finally, we confirmed that circ-AURKA was expressed more highly in the NEPC cell line NCI-H660 than in the non-NEPC prostate cell lines, LNCaP and VCaP, a result that was consistent with the expression of its parent gene (Figure 6E).
Figure 6. Differential circRNAs in neuroendocrine prostate.
A) NEPC cases from our cohort, classified by cell morphology from pathology assessments, were all characterized by the upregulation of neuroendocrine markers (SYP, CHGA, CHGB, NCAM1, ENO2, ASCL1, MYCN, and AURKA) and downregulation of genes in the AR signaling pathway (AR, AMACR, KLK3, KLK2, FKBP5, and PSCA) compared to castration-resistant prostate cancer (CRPC) cases.
B) The heatmap of 34 upregulated and 48 downregulated circRNAs with statistical significance (P < 0.01) in NEPC compared to CRPC cases.
C) Comparing NEPC to CRPC, the most significantly upregulated circRNA was circ-AURKA (Mann-Whitney U test P = 3.17×10−9); the most significantly downregulated circRNA was circ-AMACR (Mann-Whitney U test P = 0.002).
D) RT-qPCR of outward-facing primers of AURKA (backspliced from exon 6 to exon 3) in RNase R treated NCI-H660, a NEPC cell line, confirmed the circular structure of this molecule. P < 0.0001 calculated from one-way ANOVA.
E) RT-qPCR of circular and linear AURKA in prostate cancer cell lines. Both circ-AURKA and linear-AURKA were expressed higher in NCI-H660 than in two non-NEPC cell lines, LNCaP and VCaP.
The stability of circRNAs in prostate cancer cells and detection in urine of prostate cancer patients
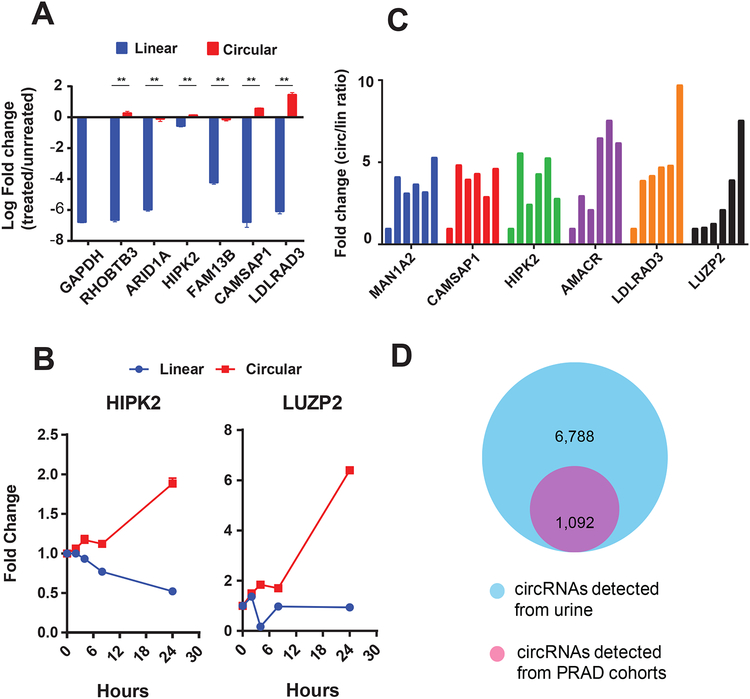
Due to their lack of open ends, circRNAs are resistant to exoribonuclease (RNase R treatment) and are potentially more stable than their cognate linear transcripts, thus making them ideal candidates for biomarker development. To evaluate the stability of circRNAs identified in MiOncoCirc, total RNA was first isolated from LNCaP prostate cancer cells and incubated with RNase R for 30 minutes. The ratio of circular-to-linear RNA species was then quantified by RT-qPCR. All circRNA species tested showed resistance to exoribonuclease and were thus significantly more stable than their linear counterparts (Figure 7A). In an orthogonal method to assess the stability of circRNAs, we compared concentrations of circular and linear transcripts in LNCaP cells that were treated with actinomycin D, a transcription inhibitor, over time. In LNCaP cells harvested at 0, 2, 4, 8, and 24 hours after actinomycin D treatment, circRNA levels increased while mRNA levels decreased (Figure 7B), thus demonstrating the relatively higher stability of circular transcripts.
Figure 7. Circular RNAs are more stable than cognate linear transcripts and can be detected in urine samples from prostate cancer patients.
A) Compared to their linear counterparts, circRNAs were resistant to RNase R degradation. Linear transcripts were detected by inward-facing RT-qPCR primers, while circular transcripts were detected by outward-facing RT-qPCR primers (**P < 0.0001, calculated from Student’s t test).
B) After transcription inhibition by actinomycin D in LNCaP cells, linear transcripts (Linear) degraded faster than their corresponding circular transcripts (Circular). Samples were harvested at 0, 2, 4, 8, and 24 hours post-treatment. GAPDH was used as the control. The fold changes were calculated relative to the starting time point. circHIPK2 was selected to represent “high” class circRNAs. circLUZP2 represented “low” class circRNAs but with elevated expression in prostate cancer compared to normal.
C) After incubating VCaP RNAs in plasma, the circular-to-linear ratio of circRNAs increased over time. Samples were harvested at 0, 15, 30, 45, 60, and 75 minutes.
D) Circular RNAs were detected by exome capture RNA-seq of 3 urine samples from prostate cancer patients. These circRNAs greatly overlapped with circRNAs identified in prostate cancer tissues from the MiOncoCirc cohorts.
See also Tables S6–S7 and Figure S7E.
Identification of biomarker species that are resistant to degradation is desirable for clinical settings. To determine whether circRNAs are more stable than their cognate linear RNAs in biospecimens, we analyzed the stability of circRNA in human blood plasma. VCaP RNAs were incubated in the plasma to simulate an environment of circulating RNAs. Indeed, as assessed by the ratio of circular-to-linear transcripts of select candidates, circRNAs were more stable than linear RNA in plasma after incubation (Figure 7C). Because noninvasive methods of detection are more ideal for screening assays in the clinic, we assessed whether circRNAs could be reliably detected in urine samples. Analysis of circRNAs by RT-qPCR showed that circRNA species could be detected in urine from prostate cancer patients (Figure S7E). Furthermore, we generated three libraries with exome capture RNA-seq and detected 1092 circRNAs in urine samples from prostate cancer patients that completely overlapped with circRNAs identified in PRAD tissue samples from the MiOncoCirc compendium (Figure 7D and Table S6). These data demonstrate that, even with low starting amounts of RNA (50 ng), exome capture RNA-seq of urine samples is a promising assay for profiling circRNAs of prostate cancer patients in a noninvasive manner.
DISCUSSION
For the research community, we have developed MiOncoCirc, an openly available circRNA compendium with a focus on clinical cancer samples. MiOncoCirc provides circRNAs characterized by our exome capture RNA-seq protocol, a poly(A)-independent RNA sequencing method that outperforms Ribo-Zero (Zhang et al., 2012) in terms of sensitivity and, unlike RNase R (Jeck et al., 2013), preserves linear transcripts. By using capture RNA-seq and including data from clinical samples, MiOncoCirc differs from most of the currently available large-scale consortiums (e.g., TCGA and Genotype-Tissue Expression [GTEX]), which only provide data generated from poly(A)-selected RNA-seq methods.
Emerging interests in identifying and developing circRNAs for diagnostic and therapeutic purposes have produced several circRNA databases, including CircBase (Glazar et al., 2014), CIRCpedia (Zhang et al., 2016), circRNAdb (Chen et al., 2016), and CSCD (Xia et al., 2017) (Table S1). However, MiOncoCirc is the first extensive clinical, cancer-centric resource of circRNAs. Importantly, our database has been largely constructed from clinical tumor samples (2,000+) across a plethora of disease sites, while other resources have characterized circRNAs from cell lines (Table S1). The transcriptional processes, and resulting circRNA formation, that occur in a native tumor microenvironment undoubtedly differ from those that occur in vitro, making MiOncoCirc a better representation of the true circRNA profile associated with cancer. Furthermore, MiOncoCirc presents a novel, rich resource containing circRNAs from primary tumors, metastases, and very rare cancer types. Researchers who are interested can also query mutations and copy-number since MiOncoCirc samples are collected from previously published genomic papers (STAR Methods).
Additionally, all MiOncoCirc samples are presented in capture-sequencing RNA-seq libraries. The data were, therefore, uniformly processed, normalized, and ready to query for meta-analysis and downstream analysis. By contrast, data from circBase and circRNAdb are not quantitative in nature. While CIRCpedia V2 has provided a “FPM” value to quantify circRNA abundances and CSCD has provided the circular junction counts, data in both databases are presented as a mixture of Ribo-Minus, poly(A)-Minus, RNase R RNA-seq, and total RNA libraries (Zhang et al., 2016, Dong et al., 2018, and Xia et al., 2017). This mixture of multiple sequencing protocols can obstruct analysis, as performing meta-analysis across different sequencing protocols requires additional cross-platform statistical methods for normalization and the careful removal of platform-specific biases. Thus, MiOncoCirc is a uniformly curated and processed quantitative database constructed from the most extensive clinical cancer sources to-date, thus uniquely positioning it for candidate biomarker nomination. Indeed, studies have shown that circular RNAs are an evolving class of promising cancer biomarkers (Kristensen et al., 2017; Zhang et al., 2018). As we confirm here, circRNAs exhibit increased stability over their corresponding linear transcripts (Figures 7A–C), an advantage for biomarker development (Li et al., 2015, and Bahn et al., 2016). Certain circRNAs identified in the MiOncoCirc compendium also displayed lineage- and cancer-specific expression. Importantly, we were able to demonstrate that our methods could detect prostate cancer tissue-associated circRNAs, such as circ-CPNE4 and circ-ACPP (Table S6 and S3), in a noninvasive urine assay for prostate cancer patients, starting with a low amount of RNA (50 ng).
We observed an interesting downregulation of circRNAs in proliferative cells across different tumor types (Figures 5B–D, S6B–C, and S7A), which could indicate that some circRNAs may have tumor suppressive roles but could also be explained as the dilution of the circRNA concentration upon cell division (Bachmayr-Heyda et al., 2015). Interestingly, the accumulation of circRNAs in non-proliferative cells, such as in aging nervous tissue (Gruner et al., 2016), supports the latter explanation. Similarly, we demonstrated that by slowing down cell growth with a kinase inhibitor, we could elevate the global abundance of circRNAs (Figure S7B). The universal downregulation of circRNAs should not discourage future studies exploring the use of circRNAs in translational cancer research and biomarker development. For example, the negative correlation we observed between total circRNA abundance with proliferation could help position some circRNAs as proliferation markers (Figure S6C). Additionally, our analysis still revealed several interesting genes that were upregulated in cancer, which resulted in elevated circRNAs compared to normal tissues (Figures 5B–C and S6B). Furthermore, some upregulated circRNAs could also be used as potential surrogate markers to distinguish subtypes of cancer, such as circ-AURKA in NEPC (Figures 6B–E), or as a potential indicator of genomic amplification, such as circ-AR in CRPC (Figure S7D).
MiOncoCirc allowed for a large-scale genomic analysis that characterized features associated with the formation and abundance of circRNAs. In Figures 3C–D and S2A, we have confirmed that the baseline expression of the parent gene does not serve as an effective predictor of the abundance of the circRNA originating from that gene (Salzman et al., 2013). Furthermore, we also confirmed that the most abundant and consistently detected subset of circRNAs were flanked by longer introns and harbored more repetitive elements than genome-wide introns (Figures 3F and S2D). Long flanking introns were previously found to harbor more repetitive and reverse complement elements that pull two splicing sites into proximity and facilitate the process of backsplicing (Ivanov et al., 2015). These data highlight the importance of intronic architecture as the predominant contributor of circRNA formation rather than expression of the parental gene.
Read-through chimeric transcripts are widespread phenomena and may represent a mechanism for the evolution of protein complexes (Akiva et al., 2006), which may explain the detection of some rt-circRNA at high frequency in our consortium (Figure 4E and Table S3). Circularized read-through events may have several important implications. First, whether the process of circularization (“backsplicing”) is a co-transcriptional or post-transcriptional process has remained an ongoing debate (Ebbesen et al., 2017; Ashwal-Fluss et al.,2014; Liang and Wilusz.,2014; Kramer et al., 2015; Zhang et al., 2016). However, recent evidence provided by Liang et al., 2017 shows that depleting CPSF3, a 3′ end processing endonuclease, could increase of the intergenic readthrough as well as circularization at one specific locus. Further, the formation of rt-circRNAs, as reported in-depth by our study, confirms that in some pairs of genes, circularization must occur prior to cleavage/polyadenylation. Our data, therefore, contribute to clarifying the timing of the circularization process and provides evidence for the co-transcriptional model. Second, the backspliced reads from those resulting from RNA circularization, genomic tandem duplications, or some structural rearrangements in RNA-seq appear identical (Figure 4A). Indeed, one of our most commonly detected “backspliced” events, from exon 3 of USP9Y to exon 3 of TTTY15, was previously proposed to be a “fusion” or “translocation” in prostate cancer (see the Supplementary Figure S2 from Ren et al., 2013). Our validation via RNase R treatment and Sanger sequencing (Figure 4F and S4B), however, proved that it is a rt-circRNA. The MiOncoCirc resource will thus serve as a highly valuable tool for cancer genomic researchers who wish to “filter” out rt-circRNA transcripts from a list of potential structural rearrangement candidates.
In conclusion, MiOncoCirc will serve as an important resource for scientists who wish to explore the lineage-specific and expression patterns of circRNAs in cancer, as well as the intriguing mechanism of read-through splicing. Such studies may shed light into the function of circRNAs and help develop the use of circRNAs in diagnostic medicine.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Arul M. Chinnaiyan (arul@med.umich.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
LNCaP, VCaP, 22Rv1 (male, prostate adenocarcinoma), and NCI-H660 (male, prostate epithelial neuroendocrine) cell lines were obtained from the American Type Culture Collection (ATCC). LNCaP and 22Rv1 cells were cultured in ATCC-formulated RPMI1640 medium supplemented with 10% fetal bovine serum (FBS; Invitrogen). VCaP cells were maintained in ATCC-formulated Dulbecco’s Modified Eagle’s Medium supplemented with 1% penicillin/streptomycin (Invitrogen) and 10% FBS (Invitrogen). NCI-H660 cells were maintained in ATCC-formulated RPMI-1640 medium, supplemented with 0.005 mg/mL insulin, 0.01 mg/mL transferrin, 30 nM sodium selenite, 10 nM hydrocortisone, 10 nM beta-estradiol, 2 mM L-glutamine, and 5% FBS. All cell lines were genotyped to confirm their identity at the University of Michigan Sequencing Core. We maintained cell lines at 37°C in a 5% CO2 cell culture incubator and tested all cell lines routinely for Mycoplasma contamination.
Human subjects and patient inclusion
Sequencing of clinical samples was approved by the Institutional Review Board of the University of Michigan (Michigan Oncology Sequencing Protocol, MI-ONCOSEQ, IRB # HUM00046018, HUM00067928, and HUM00056496). Detailed information about patient selection and sample collection were described in previous studies (Mody et al., 2015, Robinson et al., 2015, and Robinson et al., 2017). Information about additional cases in MiOncoCirc can be download directly from the website (https://mioncocirc.github.io/download/). All patients provided written informed consent to obtain fresh tumor biopsies and to perform comprehensive molecular profiling of tumor and germline exomes and tumor transcriptomes. Total RNA from normal tissues were purchased from two different commercial sources, Takara and Origene. The normal RNA samples profiled included bone marrow, colon, liver, spinal cord, stomach, small intestine, heart, placenta, spleen, brain, kidney, testis, and uterus.
METHOD DETAILS
Exome capture mRNA and Ribo-Zero sequencing
Exome capture RNA-seq was performed as previously described (Cieslik et al, 2015). We started with 0.1–3 μg of total RNA and proceeded through fragmentation, first-strand synthesis, second-strand synthesis, end repair, A-tailing, adapter ligation, size selection on a 3% agarose gel, and uridine digestion, according to Illumina’s TruSeq RNA protocol. Agilent SureSelect Human All Exon v4 probes, designed to target 20,965 genes and 334,378 exons, were then used to capture cDNA. Ribo-Zero RNA-seq followed a modified protocol described by Zhang et al., 2012. Briefly, beginning with at least 5 μg of total RNA, we first applied the Ribo-Zero rRNA Removal Kit (Illumina) to remove ribosomal RNA, and then proceeded with fragmentation, first- and second-strand synthesis, end repair, A-tailing, adapter ligation, size selection, and uridine digestion. RNA integrity was measured on an Agilent 2100 Bioanalyzer using RNA Nano reagents (Agilent Technologies). For both capture and Ribo-Zero sequencing, the stranded RNA materials were sequenced by Illumina HiSeq 2000 or HiSeq 2500 with median coverage of 49 million paired reads. Illumina BaseCall software was used to assess the quality of reads and filter the reads before processing.
RNase R treatment
Total RNA was isolated by TRIZOL lysis followed by purification using the miRNeasy Mini Kit (QIAGEN) with DNase digestion step. 2 μg of total RNA was either treated with 0 units (control) or 20 units of RNase R (Lucigen) in reaction buffer consisting 20 mM Tris-HCl (pH 8.0), 100 mM KCl, and 0.1 mM MgCl2, respectively. Treatment was conducted at 37°C for 1 hour, followed by RNase R inactivation at 65°C for 20 minutes. RNA was then extracted using miRNeasy Mini Kit (QIAGEN) and eluted in 15 μl of water. Reverse transcription was performed using SuperScript III Reverse Transcriptase (Invitrogen) and random primers (Invitrogen) following manufacturer’s standard protocol.
Dinaciclib treatment and sequencing
LNCaP cells were treated with DMSO (control) or 10 nM dinaciclib (Selleckchem) for 24 hours in triplicate for each data point. Total RNAs were extracted using the AllPrep DNA/RNA/miRNA kit (Qiagen), and capture transcriptome libraries were generated and sequenced following the protocol described above.
RT-qPCR and validation of circRNA
To assess relative expression of circRNA candidates, quantitative Real-time PCR (qRT-PCR) assays were performed using Power SYBR Green Master Mix (Applied Biosystems) and were carried out with the StepOne Real-Time PCR System (Applied Biosystems). Sequences of oligonucleotide primers were included in Table S7, with the following abbreviations used- li: linear RNAs; circ: circular RNAs; in: inward facing direction; out: outward facing direction; F: forward; R: reverse. Linear version of a housekeeping gene, GAPDH, were amplified as control. Expression of targets were calculated relative to the housekeeping gene. Fold changes following Rnase R treatment were calculated relative to the control untreated samples. The genomic sequence of qPCR products from the circRNA backspliced junction (Figure S4B) was further validated with Sanger Sequencing at the University of Michigan Sequencing Core.
Actinomycin D treatment
To validate the stability of RNAs, LNCaP cells were plated in 6-well plates and incubated for 12 hours. After incubation, cells were treated with 2.5 μg/mL of actinomycin D (Sigma) for 0–24 hours. Cells were harvested in Qiazol at 0, 2, 4, 8, and 24 hours post-treatment. RNA was isolated using the miRNeasy mini kit (Qiagen). RNA was quantified, and 1 μg of RNA was used to make cDNA using SuperScript® III First-Strand Synthesis System for RT-qPCR (Invitrogen) using random primers. We then performed RT-qPCR and analyzed data with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as a normalization control.
RNA stability in blood plasma
To check the stability of various linear and circular transcripts in plasma, we first isolated the blood plasma from fresh blood taken from healthy individual (male, age 30). In short, a total of 15 ml blood was collected in a vacutainer tube containing EDTA as the anticoagulant and mixed well before centrifugation at 2,000 RCF for 20 minutes at room temperature. The plasma layer was then carefully aspirated and stored at −80°C in cryovials. Next, we incubated 1 μg of VCaP RNA with 100 μl of plasma for 0, 15, 30, 45, 60, and 75 minutes. After incubation, total RNA was isolated and various linear and circular transcripts were quantified using qRT-PCR. The expression of transcripts at the zero-minute time point was considered as the control. Relative levels of circular and linear transcripts were calculated and shown.
Urine RNA extraction for RNA-seq and qRT-PCR
Post-digital rectal examination (Post-DRE) urine was collected from 13 prostate cancer patients presenting for diagnostic prostate biopsy using standardized protocols at University of Michigan Rogel Cancer Center. Urine was collected in an equal volume of RNA Protection Reagent and then frozen at −80°C until extraction of RNA was performed. Urine RNA was isolated by MagMAX™ mirVana™ Total RNA Isolation Kit (Invitrogen), which allows for recovery of total RNA (both intra- and extracellular) in urine. Capture sequencing was performed on three urine samples (Figure 7D), and qRT-PCR was performed on another 10 urine samples (Figure S7E).
QUANTIFICATION AND STATISTICAL ANALYSIS
Chimeric alignment and circRNA quantification
Reads that passed queue thresholds were trimmed of adaptor sequences and aligned to the GRCh38 reference genome. The aligner STAR 2.4 (Dobin et al., 2013) was used for alignment with the following settings (customized for chimeric alignment, “--outFilterType BySJout”): alignIntronMax: 400000; alignMatesGapMax: 400000; chimSegmentMin: 10; chimJunctionOverhangMin: 1; chimScoreSeparation: 0; chimScoreJunctionNonGTAG: 0; chimScoreDropMax: 1,000; chimScoreMin: 1.
To annotate circRNAs to genes, we employed the CIRCexplorer pipeline (Zhang et al., 2014) on the junction files generated by the above chimeric aligning step. For the number of circRNA isoforms reported in Figure 3A and Table S2, we only included circRNAs called from backsplicing events that appeared in at least five samples. Circular RNA abundances were normalized by the median mapped read of all libraries in our cohort. One “normalized backspliced read” is thus equivalent to one backspliced read discovered per 49 million mapped reads of linear gene. To compare and intersect our compendium with CircBase, we used the tools in CrossMap (Zhao et al., 2013) to lift over the coordinates from hg19 to hg38. In addition, the circular-to-linear fraction (Figures 1C–D) was calculated as the ratio between backspliced reads and all spliced reads (linear plus circular) involving the same junction.
To discover circRNAs involving two genes, including rt-circRNAs, we used the pipeline CODAC (M.C., Y.M.W., D.R.R., and A.M.C., manuscript in preparation). CODAC was developed as a pipeline to call all classes of chimeric RNAs, including circRNAs and structural rearrangements from paired-end (PE) RNA-seq. Briefly, PE sequencing reads are aligned to the reference genome in two independent runs, following a step of read-merging. In the read-merging, PE reads are merged into a synthetic single-end (SE) read if the insert size of a fragment is smaller than double the sequencing length. This results in two sets of FASTQ files (PE and SE) that are independently aligned to the reference genome using STAR as described above. The resulting chimeric alignments from STAR were filtered for recurrent sequencing artifacts, breakpoints within repetitive regions, segmental duplications, and possible alignment errors. Depending on the breakpoint position, customized thresholds of supporting reads were required. A comprehensive list of black-list regions, recurrent false-positive junctions, and problematic regions that have required increased filtering thresholds is included in the CODAC software. All events involving backspliced reads of two genes with the orientation of interest (Figures 4A–B) nominated by CODAC were collected as the candidate rt-circRNAs (Table S3). We further filtered out difficult-to-interpret partners, i.e., the pair of parental genes with high degree of homology, especially the human leukocyte antigen (HLA) genes. Pairs of homologous/paralogous genes with lesser degrees of similarity were also indicated in Table S3 to inform the users about the possibilities of mapping artifact and false positives.
All backspliced junctions of circRNAs and rt-circRNAs, as well as normalized abundances of circRNAs and parental gene expression of all samples are provided on the “Download Data” page of the MiOncoCirc website (https://mioncocirc.github.io/). The MiOncoCirc “Query Data” page is a user-friendly and interactive interface built with R shiny (http://shiny.rstudio.com/) that allows the user to browse all circular isoforms per gene and click on each isoform to browse the abundance (normalized circular reads) in all tissues.
Differential mRNA/circRNA analysis/clustering
All fragment quantifications were computed using featureCounts (Liao et al., 2013). Gene expression was measured in fragments per kilobase per million (FPKM). Differential mRNA and circRNA abundance analysis were carried out with edgeR (Robinson et al., 2010). Independent filtering was carried out before differential analysis to increase detection power of moderate to high mRNAs (or circRNAs) (Bourgon et al., 2010). For mRNA, genes with a count per million (CPM) <1 in more samples than the sample size of one of the groups being compared were filtered from the analysis. For circRNA, a detection threshold of 5-reads was applied to avoid “shot noises” for low read counts (Anders et al., 2010). All clustering was performed with a hierarchical clustering method using Manhattan Distance. For the analysis of NEPC vs. CRPC, the eight NEPC cases were diagnosed by the pathologists based on cell morphology, and the 35 cases of metastasis CRPC with the highest tumor content (> 40%) were selected from data provided by Robinson et al., 2015 to avoid contamination signals from biopsy sites.
RNA-seq reads sampling
Seqtk was used to perform down-sampling of paired-end RNA-seq samples (Figure 1A) using the command:
seqtk sample [seed] read_1.fq [depth] > sub_1.fq; seqtk sample [seed] read_2.fq [depth] > sub_2.fq.
Repetitive elements analysis
All annotated repeat sites were retrieved from the UCSC Genome Browser’s RepeatMasker track as a bed file, June 2018. BEDtools (Quinlan et al., 2010) were used to intersect the RepeatMasker bed file with all introns flanking circRNAs and rt-circRNAs from MiOncoCirc.
Tissue specificity analysis
Since the majority of our clinical samples were collected from advanced metastasized tumors (Mody et al., 2015, Robinson et al, 2015, Robinson et al., 2017), we “binarized” the frequency per tissue/cancer type of a circRNA (Figure 5A) and rt-circRNA (Figure S5A) to enrich for tissue specificity and to avoid picking up contaminating signals. A circular transcript was labeled as “consistently detected” if it appeared in at least 30% of a cancer cohort. Similarly, a gene was labeled as “consistently detected” (in terms of the ability to generate circRNA) if it could generate at least one circRNA that could be detected in at least 30% of a cancer cohort. To calculate the tissue specificity of a gene (Figure S5C) or a circRNA (Figure S6A), we used the Shannon Entropy score as proposed by Kryuchkova-Mostacci et al. (2016). The distribution of the Shannon score of genes in our cohort also follows a bimodal distribution. Tissue-specfic genes (Figure S5C) were defined as those genes with Shannon scores >= 0.75.
Motif analysis
All introns flanking circRNAs were collected, and the 13-mers spanning the splicing donor and acceptor were retrieved according to hg38. The position weight matrix (PWM) in Figure S1E was plotted using R package seqLogo.
Proliferation analysis
The expressions of the 31 cell cycle progression (CCP) genes (Cuzick et al., 2011) (calculated in FPKM) were used as the proliferation markers of the 25 matched pairs of prostate cancer. These 31 CCP genes include: FOXM1, ASPM, TK1, PRC1, CDC20, BUB1B, PBK, DTL, CDKN3, RRM2, ASF1B, CEP55, CDC2, DLGAP5, C18orf24, RAD51, KIF11, BIRC5, RAD54L, CENPM, KIAA0101, KIF20A, PTTG1, CDCA8, NUSAP1, PLK1, CDCA3, ORC6L, CENPF, TOP2A, and MCM10. The total circular RNA abundance was calculated using the sum of all detected backspliced reads, normalized by the total mapped read in each library.
Exome-sequencing and copy-number analysis
An in-house pipeline constructed for analysis of paired tumor/normal data was used to process FASTQ sequence files from whole exome libraries. Using Novoalign (version 3.02.08) (Novocraft) and SAMtools (version 0.1.19), respectively, the sequencing reads were aligned to the GRCh37 reference genome and converted into BAM files. Novosort (version 1.03.02) was used for the sorting, indexing, and duplicate marking of BAM files. Freebayes (version 1.0.1) and pindel (version 0.2.5b9) were used to perform mutational analysis. Variants were annotated to RefSeq (via the UCSC genome browser) COSMIC v79, dbSNP v146, ExAC v0.3, and 1000 Genomes phase 3 databases using snpEff and snpSift (version 4.1g).
Using the DNAcopy (version 1.48.0) implementation of the Circular Binary Segmentation algorithm, exome data were analyzed for copy-number aberrations by jointly segmenting B-allele frequencies and log2-transformed tumor/normal coverage ratios across targeted regions. The Expectation-Maximization Algorithm was used to jointly estimate tumor purity and classify regions by copy-number status. To allow for the possibility of non-diploid tumor genomes, additive adjustments were made to the log2-transformed coverage ratios. The adjustment resulting in the best fit to the data using minimum mean-squared error was chosen automatically and, if necessary, manually overridden.
DATA AND SOFTWARE AVAILABILITY
All circRNAs and rt-circRNAs, as well as their expression patterns, can be found in our MiOncoCirc website, currently hosted at: https://mioncocirc.github.io/. The pipeline CODAC can be found at https://github.com/mcieslik-mctp/codac.
Supplementary Material
Related to Figure 3. A) The overlap of genes that could form circRNAs in MiOncoCirc and CircBase was significant (Fisher Exact test P < 2.2×10−16).
B) The overlap of circRNA transcripts detected in MiOncoCirc and CircBase was even more significant without the stringent cut-off used in Figure 3A.
C) Among overlapping genes, MiOncoCirc could detect twice the number of circular isoforms compared to CircBase (paired t-test P < 1×10−10).
D) The overlaps of breast and lung circRNAs found in MiOncoCirc and CircBase were significant (Fisher Exact Test P < 2.2×10−16).
E) Left: Circular RNA was predominantly flanked by a canonical splicing motif, AG-GT (99.2%). Right: Among the non-canonical splicing signals, the most commonly observed were GC-AG (0.7%) and AT-AC (0.05%).
Related to Figure 6.
Related to Figure 3. A) The correlation of average circRNA abundance (in normalized backspliced reads) and parental expression (in FPKM) was low (Spearman’s ρ=0.12).
B) Gene set enrichment analysis for the genes that could form circRNAs with outlier expression (“high” group). The Reactome pathways were ranked by the P-values.
C) Parental genes that produced highly abundant (outlier) circRNAs were not more highly expressed than other circRNA parental genes (Mann-Whitney-Wilcoxon P = 0.15).
D) The “high” circRNAs were flanked by introns that harbored more repetitive elements (Mann-Whitney-Wilcoxon P < 4.5×10−8) than the remaining 98% of circRNAs. The introns flanking “low” circRNAs also contained more repetitive elements than all introns genome-wide (“background”) (Mann-Whitney-Wilcoxon P < 2.2×10−16).
E) The genes that could form circRNAs with outlier expression (“high” group) had more exons (Mann-Whitney-Wilcoxon P < 1×10−09) than the parental gene of the remaining 98% of circRNAs. The parent genes of low circRNAs, in turn, had more exons than all genes (“background”) (Mann-Whitney-Wilcoxon P < 2.2×10−16).
F) The genes that could form the “high” group of outlier circRNAs were significantly longer (Mann-Whitney-Wilcoxon P < 2.2×10−16) than the parental gene of the remaining circRNAs. Similarly, the parent genes of low circRNAs were longer than all genes (“background”) (Mann-Whitney-Wilcoxon P < 2.2×10−16).
Related to Figure 4 and Table S3. A) Density plot demonstrated that rt-circRNAs comprised a small portion of total circRNAs in each sample (average 2.5% per sample).
B) The rt-circRNAs were detected at lower abundance (average 3.1-fold lower, Wilcoxon rank-sum Test P-Value = 1×10−12) than other circRNAs. The vertical dashed-lines corresponded to the medians of rt-circRNAs and circRNAs abundances, log2 transformed.
C) The expressions of parental genes could range from as low as 0.05 FPKM up to more than 2,000 FPKM.
D) The ratio of gene expression of upstream to downstream genes involved in circRNA generation varied widely (from 0.001 to 1000).
E) The distances between two genes that can generate rt circRNAs were shorter than the entire population (median 37 kb vs. median 48.8 kb apart, Wilcoxon rank-sum Test P-Value = 10−4).
Related to Figure 4 and Table S3. A) 542 out of 1359 (39.88%) putative rt-circRNAs can be detected in at least one normal (healthy) tissue. In addition, 1129 out of 1359 (83%) putative rt-circRNAs have parent genes (upstream and downstream) with normal karyotype (two copies). Copy-number data of cases with matched RNA-seq and whole exome/capture gene panel were retrieved from Mody et al., (2015), Robinson et al., (2015), and Robinson et al., (2017). Thus, a majority of these events were generated from genes with no evidence of alteration in copy-numbers.
B) Schematic (top) depicting the circular read-through event that generated circTTTY15e3-USP9Ye3 transcript detected in the MiOncoCirc compendium. The Sanger sequencing result of circTTTY15-USP9Y RT-qPCR product showed the correct sequence spanning the backsplice exon-exon junctions of USP9Y and TTTY15.
C) Schematic (top) depicting the circular read-through event that generated the ITM2Be2-RB1e2 transcript detected in the MiOncoCirc compendium. Copy-number data (bottom) from capture genome sequencing of a case of non-small cell lung cancer (NSCLC) and a case of bladder cancer (BLCA) did not show any copy-number alterations in ITM2B and RB1 (STAR Methods).
Related to Figure 4 and Table S3. A) The tissue specificity heatmap of rt-circRNAs (see STAR Methods). The rt-circRNAs specific to liver and prostate cancer are labeled.
B) The gene expression, in transcript per million (TPM), of select parental genes of tissue-specific rt-circRNAs associated with prostate and liver cancer shown in A. These parent genes also showed tissue-specific patterns.
C) The intersection of tissue-specific genes with the list of rt-circRNA parental genes. Tissue-specific genes were defined as genes with Shannon Scores >= 0.75 (STAR Methods). The overlap was minimal (n=22), which suggested that few parental genes of rt-circRNA were tissue-specific. This result agrees with our initial analyses in Figure S5A, that only a few rt-circRNAs are tissue-specific (36 out of 1,359).
Related to Figure 5 and Table S3–4. A) The correlation of Shannon Scores of tissue-specific circRNAs (>0.5) against the Shannon Scores of their parent genes. When multiple circular isoforms of a gene were available, the circular isoform with the highest score was chosen. The Spearman rank correlation was ρ=0.62.
B) Heatmap of circRNA abundance in 25 pairs of matched tumor and normal of localized PRAD (prostate adenocarcinoma). The circRNAs whose FDR < 0.01 were included. The majority (n=629) of these circRNAs were downregulated in cancer compared to normal. However, there was a small subset of circRNAs upregulated in cancer (n=23).
C) The Spearman rank correlation heatmap in Figure 5D was extended to a panel of 31 cell cycle progression (CCP) genes associated with proliferation in localized PRAD, as curated by a meta-analysis by Cuzick et al. (2011). The normalized total circRNA abundance, as well as some of the most abundant circRNAs (circ-FBXO7, circ-ELK4, and circ-FNDC3B), negatively correlated with the expressions (in FPKM) of the whole CCP gene panel. The housekeeping genes GAPDH, HSPA4, TBP, B2M, and HPRT1 were used as the negative control, which showed no changes in expression between cancer and normal.
Related to Figures 5 and 7 and Tables S4 and S6–S7. A) The global circRNA profile in non-matched tumor/normal across six other tissue types: bone-osteosarcoma (n=8), colon-colorectal adenocarcinoma (n=13), kidney-renal cell carcinoma (n=12), liver-hepatocellular carcinoma (n=10), lung-lung adenocarcinoma (n=10), and stomach-gastric adenocarcinoma (n=9). The normal samples of each tissue were pooled from healthy donors (STAR Methods). The tumor samples were from the MiOncoCirc cohorts. The total circRNA abundance (total backspliced reads normalized by sequencing depth) are downregulated in cancer compared to normal across these six lineages.
B) Twenty-four hours post-treatment with the cell cycle inhibitor dinaciclib, there was an overall increase of total circRNA abundance, calculated by the total circular reads normalized by sequencing depth.
C) The correlation of log fold-change (FC) of circular RNA vs. log FC of linear expression in dinaciclib-treated samples compared to controls. The upregulation of total circRNAs was not due to the change in expression of parental genes: in general, the circRNA abundances were upregulated (mean circular logFC = 0.3), while the mean linear logFC = 0.
D) Circular AR (backsplice from exon 4 to exon 3) was detected in mCRPC with amplified AR (more than 5 copies, 54/70 cases), but not in primary tumors. This result could be directly explained by the higher expression of AR in mCRPC (Wilcoxon rank-sum test P < 2.2×10−16).
E) The relative expression of circRNAs detected in urine from 10 prostate cancer patients. Linear GAPDH expression was used for normalization, and KLK3 was included as a positive control.
Related to Figure 3.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| N/A | ||
| Bacterial and Virus Strains | ||
| N/A | ||
| Biological Samples | ||
| Tumor/normal tissues from various cancer patients | University of Michigan MI-ONCOSEQ collection | See STAR Methods Table S1, and Meta data file (meta.xlsx) at https://mioncocirc.github.io/download/ |
| Bone tissues from healthy individuals | Origene | CR559716, CR559724, CR561975 |
| Colon tissues from healthy individuals | Takara | 636553 |
| Kidney tissues from healthy individuals | Takara | 636529 |
| Liver tissues from healthy individuals | Origene | CR560592, CR560901, CR560916 |
| Lung tissues from healthy individuals | Takara | 636524 |
| Stomach tissues from healthy individuals | Takara | 636578 |
| Other normal tissues | Takara | 636591, 636554, 636539, 636532, 636527, 636585 636535, 636533, 636551 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Actinomycin D | Sigma-Aldrich | A1410–10MG |
| Dinaciclib | Selleckchem | S2768 |
| RNase R | Lucigen | RNR07250 |
| RQ1 RNase-Free DNase | Promega | M6101 |
| Superscript II Reverse Transcriptase | Invitrogen | 18064–071 |
| RNase H | Invitrogen | 18021–071 |
| Fetal Bovine Serum | Invitrogen | A3160701 |
| Dulbecco’s Modified Eagle’s Medium | Invitrogen | 10569010 |
| RPMI-1640 Medium | ATCC | ATCC® 30–2001™ |
| Penicillin-Streptomycin | Invitrogen | 15140122 |
| Power SYBR Green Master Mix | Applied Biosystems | 4367659 |
| DNA Polymerase I | New England Biolabs | M0209L |
| USER Enzyme | New England Biolabs | M5505L |
| Critical Commercial Assays | ||
| AllPrep DNA/RNA/miRNA Universal Kit | Qiagen | 80224 |
| KAPA Hyper Prep Kit for Illumina | Kapa Biosystems | KK8504 |
| SureSelect XT Human All Exon V4 library | Agilent Technologies | 5190–4632 |
| SureSelectXT Reagent kit | Agilent Technologies | G9611B |
| RNA 6000 Nano kit | Agilent Technologies | 5067–1511 |
| DNA 1000 kit | Agilent Technologies | 5067–1504 |
| QIAGEN Multiplex PCR Kit | Qiagen | 206143 |
| Ribo-Zero rRNA Removal Kit | Illumina | MRZH11124 |
| MagMAX mirVana Total RNA Isolation Kit | ThermoFisher | A27828 |
| Deposited Data | ||
| FASTQ files of mCRPC in Mi-Oncoseq program, University of Michigan Clinical Sequencing Exploratory Research (CSER) | Robinson et al., 2017 | dbGaP (phs000673.v2.p1) |
| BAM files of the SU2C-PCF CRPC150 cohort | Robinson et al., 2015 | dbGaP (phs000915.v1.p1) |
| FASTQ files of pediatric tumors in Mi-Oncoseq program, University of Michigan Clinical Sequencing Exploratory Research (CSER) | Mody et al., 2015 | dbGaP (phs000673.v1.p1) |
| Tab-delimited files for all circular RNAs, their genomics information and abundances in samples included in this study | MiOncoCirc | https://nguyenjoshvo.github.io/ |
| Experimental Models: Cell Lines | ||
| LNCaP | ATCC | CRL-1740 |
| VCaP | ATCC | CRL-2876 |
| 22Rv1 | ATCC | CRL-2505D |
| NCI-H660 | ATCC | CRL-5813 |
| Experimental Models: Organisms/Strains | ||
| N/A | ||
| Oligonucleotides | ||
| NEBNext Multiplex Oligos for Illumina | New England Biolabs | E7535L |
| NEBNext Multiplex Oligos for Illumina Index Set 2 | New England Biolabs | E7500L |
| Random Primers | Invitrogen | 48190–011 |
| Recombinant DNA | ||
| N/A | ||
| Software and Algorithms | ||
| NCBI Multiple Sequence Alignment Viewer | NCBI | https://www.ncbi.nlm.nih.gov/projects/msaviewer/# |
| STAR 2.4 | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| CIRCexplorer | Zhang et al., 2014 | https://github.com/YangLab/CIRCexplorer |
| CrossMap | Zhao et al., 2013 | https://github.com/gantzgraf/CrossMap |
| featureCounts | Liao et al., 2013 | http://bioinf.wehi.edu.au/featureCounts/ |
| seqtk | https://github.com/lh3/seqtk | https://github.com/lh3/seqtk |
| BEDtools | Quinlan et al., 2010 | https://github.com/arq5x/bedtools |
| seqLogo | Bembom O (2018) | https://bioconductor.org/packages/release/bioc/html/seqLogo.html |
| Comprehensive Detection and Analysis of Chimeras (CODAC) | This paper and Robinson et al., 2017 | https://github.com/mcieslik-mctp/codac |
| ggplot2 | http://ggplot2.org/book/ | https://cran.r-project.org/web/packages/ggplot2/index.html |
| Shiny Server | https://www.rstudio.com/products/shiny/shiny-server/ | https://www.rstudio.com/products/shiny/shiny-server/ |
| DNACopy | Olshen et al., 2004 | http://bioconductor.org/packages/release/bioc/html/DNAcopy.html |
| edgeR | Robinson et al., 2010 | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Novoalign | Novocraft | http://www.novocraft.com/products/novoalign |
| Freebayes | https://github.com/ekg/freebayes | https://github.com/ekg/freebayes |
| Pindel | https://github.com/genome/pindel | https://github.com/genome/pindel |
| SnpEff | http://snpeff.sourceforge.net | http://snpeff.sourceforge.net |
| SnpSift | http://snpeff.sourceforge.net/SnpSift.html | http://snpeff.sourceforge.net/SnpSift.html |
| Other | ||
| SeqCap EZ HE-Oligo Kit A | Roche | 06777287001 |
| SeqCap EZ HE-Oligo Kit B | Roche | 06777317001 |
| Agencourt RNAClean XP | Beckman Coulter | A63987 |
| AMPURE XP beads | Beckman Coulter | A63882 |
| Dynabeads MyOne Streptavidin T1 | Invitrogen | 65602 |
HIGHLIGHTS.
Use of exome capture transcriptome sequencing to compile a cancer circRNA landscape
MiOncoCirc is the most comprehensive catalogue of cancer-based circRNA species
MiOncoCirc contains circRNA from cancer cell lines as well as tumor samples
Novel biomarkers can be nominated through MiOncoCirc
ACKNOWLEDGMENTS
We gratefully acknowledge all patients who participated in these studies. We also thank all members of the Chinnaiyan Lab and the Michigan Center for Translational Pathology (MCTP) as well as the MI-OncoSeq team. We greatly thank Sisi Gao, Ph.D., and Stephanie Ellison, Ph.D., for their efforts in preparing this manuscript, and Jin Chen for assistance with computational resources. We also acknowledge the efforts of the MI-OncoSeq team. This work was supported by the Early Detection Research Network Grant U01 CA214170, Prostate SPORE Grants P50 CA186786 and P50 CA097186, and the Prostate Cancer Foundation (PCF). J.N.V., M.C., A.N. and A.M.C. are supported by funding from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC, Grant U24CA210967). M.C. is supported by a PCF Young Investigator Grant and a Department of Defense (DOD) Prostate Cancer Research Program Idea Development Award (PC160429). Yajia.Z. is supported by a DOD Early Investigator Research Award (W81XWH-17-1-0134). L.X. is supported by a DOD Postdoctoral Fellowship (W81XWH-16-1-0195). A.M.C. is a Howard Hughes Medical Institute Investigator, Taubman Scholar, and American Cancer Society Professor.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, and Sorek R (2005). Transcription-mediated gene fusion in the human genome. Genome Research 16, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biology 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti N, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, and Kadener S (2014). circRNA Biogenesis Competes with Pre-mRNA Splicing. Molecular Cell 56, 55–66. [DOI] [PubMed] [Google Scholar]
- Bachmayr-Heyda A, Reiner A, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt T, Zeillinger R, and Pils D (2015). Correlation of circular RNA abundance with proliferation - exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis and normal human tissues. Scientific Reports 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J, Zhang Q, Li F, Chan T, Lin X, Kim Y, Wong D, and Xiao X (2014). The Landscape of MicroRNA, Piwi-Interacting RNA, and Circular RNA in Human Saliva. Clinical Chemistry 61, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Rickman D, Park K, Chae S, Sboner A, MacDonald T, Wang Y, Sheikh K, Terry S, and Tagawa S et al. (2011). Molecular Characterization of Neuroendocrine Prostate Cancer and Identification of New Drug Targets. Cancer Discovery 1, 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M, Levin J, Vijayendran K, Sivachenko A, Adiconis X, Maguire J, Johnson L, Robinson J, Verhaak R, and Sougnez C et al. (2010). Integrative analysis of the melanoma transcriptome. Genome Research 20, 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgon R, Gentleman R, and Huber W (2010). Independent filtering increases detection power for high-throughput experiments. Proceedings of the National Academy of Sciences 107, 9546–9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, and Lovell-Badge R (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73, 1019–1030. [DOI] [PubMed] [Google Scholar]
- Chen X, Han P, Zhou T, Guo X, Song X, and Li Y (2016). circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Scientific Reports 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslik M, Chugh R, Wu Y, Wu M, Brennan C, Lonigro R, Su F, Wang R, Siddiqui J, and Mehra R et al. (2015). The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Research 25, 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn S, Pillman K, Toubia J, Conn V, Salmanidis M, Phillips C, Roslan S, Schreiber A, Gregory P, and Goodall G (2015). The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 160, 1125–1134. [DOI] [PubMed] [Google Scholar]
- Conteduca V, Aieta M, Amadori D, and De Giorgi U (2014). Neuroendocrine differentiation in prostate cancer: Current and emerging therapy strategies. Critical Reviews in Oncology/Hematology 92, 11–24. [DOI] [PubMed] [Google Scholar]
- Cuzick J, Swanson G, Fisher G, Brothman A, Berney D, Reid J, Mesher D, Speights V, Stankiewicz E, and Foster C et al. (2011). Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. The Lancet Oncology 12, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis C, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras T (2012). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Ma X, Li G, and Yang L (2018). CIRCpedia v2: An Updated Database for Comprehensive Circular RNA Annotation and Expression Comparison. Genomics, Proteomics & Bioinformatics (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbesen K, Hansen T, and Kjems J (2016). Insights into circular RNA biology. RNA Biology 14, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoukos G, Ciulla D, Huang K, Haas B, Izard J, Levin J, Livny J, Earl A, Gevers D, and Ward D et al. (2012). Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biology 13, r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glažar P, Papavasileiou P, and Rajewsky N (2014). circBase: a database for circular RNAs. RNA 20, 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner H, Cortés-López M, Cooper D, Bauer M, and Miura P (2016). CircRNA accumulation in the aging mouse brain. Scientific Reports 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnerio J, Bezzi M, Jeong J, Paffenholz S, Berry K, Naldini M, Lo-Coco F, Tay Y, Beck A, and Pandolfi P (2016). Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 165, 289–302. [DOI] [PubMed] [Google Scholar]
- Guo Z, Maki M, Ding R, Yang Y, zhang B, and Xiong L (2014). Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Scientific Reports 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T, Jensen T, Clausen B, Bramsen J, Finsen B, Damgaard C, and Kjems J (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Hansen T, Venø M, Damgaard C, and Kjems J (2015). Comparison of circular RNA prediction tools. Nucleic Acids Research 44, e58–e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath H, Orejuela M, Piechotta M, Levanon E, Landthaler M, and Dieterich C et al. (2015). Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Reports 10, 170–177. [DOI] [PubMed] [Google Scholar]
- Iyer M, Niknafs Y, Malik R, Singhal U, Sahu A, Hosono Y, Barrette T, Prensner J, Evans J, and Zhao S et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nature Genetics 47, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W, Sorrentino J, Wang K, Slevin M, Burd C, Liu J, Marzluff W, and Sharpless N (2012). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson D, Wu Y, Cao X, Asangani I, Kothari V, Prensner J, and Lonigro R et al. (2012). Expressed Pseudogenes in the Transcriptional Landscape of Human Cancers. Cell 149, 1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M, Liang D, Tatomer D, Gold B, March Z, Cherry S, and Wilusz J (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes & Development 29, 2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen L, Hansen T, Venø M, and Kjems J (2017). Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryuchkova-Mostacci N, and Robinson-Rechavi M (2016). A benchmark of gene expression tissue-specificity metrics. Briefings in Bioinformatics 18, 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, and Wade M et al. (2017). Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Molecular Cell 66, 22–37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, and Huang S (2015). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Research 25, 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, and Wilusz J (2014). Short intronic repeat sequences facilitate circular RNA production. Genes & Development 28, 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Tatomer D, Luo Z, Wu H, Yang L, Chen L, Cherry S, and Wilusz J (2017). The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Molecular Cell 68, 940–954.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth G, and Shi W (2013). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Mody R, Wu Y, Lonigro R, Cao X, Roychowdhury S, Vats P, Frank K, Prensner J, Asangani I, and Palanisamy N et al. (2015). Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA 314, 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, and Wigler M (2004). Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5, 557–572. [DOI] [PubMed] [Google Scholar]
- Pamudurti N, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, and Ramberger E et al. (2017). Translation of CircRNAs. Molecular Cell 66, 9–21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, Seghezzi W, Paruch K, Dwyer M, and Doll R et al. (2010). Dinaciclib (SCH 727965), a Novel and Potent Cyclin-Dependent Kinase Inhibitor. Molecular Cancer Therapeutics 9, 2344–2353. [DOI] [PubMed] [Google Scholar]
- Quinlan A, and Hall I (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Peng Z, Mao J, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, and Xu W et al. (2012). RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Research 22, 806–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, McCarthy D, and Smyth G (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen E, Wu Y, Schultz N, Lonigro R, Mosquera J, Montgomery B, Taplin M, Pritchard C, and Attard G et al. (2015). Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 161, 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Wu Y, Lonigro R, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, and Raymond V et al. (2017). Integrative clinical genomics of metastatic cancer. Nature 548, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin M, Zhou M, Dhanasekaran S, Varambally S, Barrette T, Sanda M, Pienta K, Ghosh D, and Chinnaiyan A (2002). α-Methylacyl Coenzyme A Racemase as a Tissue Biomarker for Prostate Cancer. JAMA 287, 1662. [DOI] [PubMed] [Google Scholar]
- Salzman J, Chen R, Olsen M, Wang P, and Brown P (2013). Cell-Type Specific Features of Circular RNA Expression. Plos Genetics 9, e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, Jin Y, Gao Y, Xia L, and Chang H et al. (2017). CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Research 46, D925–D929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, and Yuan H (2018). Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Scientific Reports 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang H, Zhang Y, Lu X, Chen L, and Yang L (2014). Complementary Sequence-Mediated Exon Circularization. Cell 159, 134–147. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dong R, Zhang Y, Zhang J, Luo Z, Zhang J, Chen L, and Yang L (2016). Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Research 26, 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang J, Yang L, and Chen L (2016). The Biogenesis of Nascent Circular RNAs. Cell Reports 15, 611–624. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Theurkauf W, Weng Z, and Zamore P (2012). Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection. Silence 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sun Z, Wang J, Huang H, Kocher J, and Wang L (2013). CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 3. A) The overlap of genes that could form circRNAs in MiOncoCirc and CircBase was significant (Fisher Exact test P < 2.2×10−16).
B) The overlap of circRNA transcripts detected in MiOncoCirc and CircBase was even more significant without the stringent cut-off used in Figure 3A.
C) Among overlapping genes, MiOncoCirc could detect twice the number of circular isoforms compared to CircBase (paired t-test P < 1×10−10).
D) The overlaps of breast and lung circRNAs found in MiOncoCirc and CircBase were significant (Fisher Exact Test P < 2.2×10−16).
E) Left: Circular RNA was predominantly flanked by a canonical splicing motif, AG-GT (99.2%). Right: Among the non-canonical splicing signals, the most commonly observed were GC-AG (0.7%) and AT-AC (0.05%).
Related to Figure 6.
Related to Figure 3. A) The correlation of average circRNA abundance (in normalized backspliced reads) and parental expression (in FPKM) was low (Spearman’s ρ=0.12).
B) Gene set enrichment analysis for the genes that could form circRNAs with outlier expression (“high” group). The Reactome pathways were ranked by the P-values.
C) Parental genes that produced highly abundant (outlier) circRNAs were not more highly expressed than other circRNA parental genes (Mann-Whitney-Wilcoxon P = 0.15).
D) The “high” circRNAs were flanked by introns that harbored more repetitive elements (Mann-Whitney-Wilcoxon P < 4.5×10−8) than the remaining 98% of circRNAs. The introns flanking “low” circRNAs also contained more repetitive elements than all introns genome-wide (“background”) (Mann-Whitney-Wilcoxon P < 2.2×10−16).
E) The genes that could form circRNAs with outlier expression (“high” group) had more exons (Mann-Whitney-Wilcoxon P < 1×10−09) than the parental gene of the remaining 98% of circRNAs. The parent genes of low circRNAs, in turn, had more exons than all genes (“background”) (Mann-Whitney-Wilcoxon P < 2.2×10−16).
F) The genes that could form the “high” group of outlier circRNAs were significantly longer (Mann-Whitney-Wilcoxon P < 2.2×10−16) than the parental gene of the remaining circRNAs. Similarly, the parent genes of low circRNAs were longer than all genes (“background”) (Mann-Whitney-Wilcoxon P < 2.2×10−16).
Related to Figure 4 and Table S3. A) Density plot demonstrated that rt-circRNAs comprised a small portion of total circRNAs in each sample (average 2.5% per sample).
B) The rt-circRNAs were detected at lower abundance (average 3.1-fold lower, Wilcoxon rank-sum Test P-Value = 1×10−12) than other circRNAs. The vertical dashed-lines corresponded to the medians of rt-circRNAs and circRNAs abundances, log2 transformed.
C) The expressions of parental genes could range from as low as 0.05 FPKM up to more than 2,000 FPKM.
D) The ratio of gene expression of upstream to downstream genes involved in circRNA generation varied widely (from 0.001 to 1000).
E) The distances between two genes that can generate rt circRNAs were shorter than the entire population (median 37 kb vs. median 48.8 kb apart, Wilcoxon rank-sum Test P-Value = 10−4).
Related to Figure 4 and Table S3. A) 542 out of 1359 (39.88%) putative rt-circRNAs can be detected in at least one normal (healthy) tissue. In addition, 1129 out of 1359 (83%) putative rt-circRNAs have parent genes (upstream and downstream) with normal karyotype (two copies). Copy-number data of cases with matched RNA-seq and whole exome/capture gene panel were retrieved from Mody et al., (2015), Robinson et al., (2015), and Robinson et al., (2017). Thus, a majority of these events were generated from genes with no evidence of alteration in copy-numbers.
B) Schematic (top) depicting the circular read-through event that generated circTTTY15e3-USP9Ye3 transcript detected in the MiOncoCirc compendium. The Sanger sequencing result of circTTTY15-USP9Y RT-qPCR product showed the correct sequence spanning the backsplice exon-exon junctions of USP9Y and TTTY15.
C) Schematic (top) depicting the circular read-through event that generated the ITM2Be2-RB1e2 transcript detected in the MiOncoCirc compendium. Copy-number data (bottom) from capture genome sequencing of a case of non-small cell lung cancer (NSCLC) and a case of bladder cancer (BLCA) did not show any copy-number alterations in ITM2B and RB1 (STAR Methods).
Related to Figure 4 and Table S3. A) The tissue specificity heatmap of rt-circRNAs (see STAR Methods). The rt-circRNAs specific to liver and prostate cancer are labeled.
B) The gene expression, in transcript per million (TPM), of select parental genes of tissue-specific rt-circRNAs associated with prostate and liver cancer shown in A. These parent genes also showed tissue-specific patterns.
C) The intersection of tissue-specific genes with the list of rt-circRNA parental genes. Tissue-specific genes were defined as genes with Shannon Scores >= 0.75 (STAR Methods). The overlap was minimal (n=22), which suggested that few parental genes of rt-circRNA were tissue-specific. This result agrees with our initial analyses in Figure S5A, that only a few rt-circRNAs are tissue-specific (36 out of 1,359).
Related to Figure 5 and Table S3–4. A) The correlation of Shannon Scores of tissue-specific circRNAs (>0.5) against the Shannon Scores of their parent genes. When multiple circular isoforms of a gene were available, the circular isoform with the highest score was chosen. The Spearman rank correlation was ρ=0.62.
B) Heatmap of circRNA abundance in 25 pairs of matched tumor and normal of localized PRAD (prostate adenocarcinoma). The circRNAs whose FDR < 0.01 were included. The majority (n=629) of these circRNAs were downregulated in cancer compared to normal. However, there was a small subset of circRNAs upregulated in cancer (n=23).
C) The Spearman rank correlation heatmap in Figure 5D was extended to a panel of 31 cell cycle progression (CCP) genes associated with proliferation in localized PRAD, as curated by a meta-analysis by Cuzick et al. (2011). The normalized total circRNA abundance, as well as some of the most abundant circRNAs (circ-FBXO7, circ-ELK4, and circ-FNDC3B), negatively correlated with the expressions (in FPKM) of the whole CCP gene panel. The housekeeping genes GAPDH, HSPA4, TBP, B2M, and HPRT1 were used as the negative control, which showed no changes in expression between cancer and normal.
Related to Figures 5 and 7 and Tables S4 and S6–S7. A) The global circRNA profile in non-matched tumor/normal across six other tissue types: bone-osteosarcoma (n=8), colon-colorectal adenocarcinoma (n=13), kidney-renal cell carcinoma (n=12), liver-hepatocellular carcinoma (n=10), lung-lung adenocarcinoma (n=10), and stomach-gastric adenocarcinoma (n=9). The normal samples of each tissue were pooled from healthy donors (STAR Methods). The tumor samples were from the MiOncoCirc cohorts. The total circRNA abundance (total backspliced reads normalized by sequencing depth) are downregulated in cancer compared to normal across these six lineages.
B) Twenty-four hours post-treatment with the cell cycle inhibitor dinaciclib, there was an overall increase of total circRNA abundance, calculated by the total circular reads normalized by sequencing depth.
C) The correlation of log fold-change (FC) of circular RNA vs. log FC of linear expression in dinaciclib-treated samples compared to controls. The upregulation of total circRNAs was not due to the change in expression of parental genes: in general, the circRNA abundances were upregulated (mean circular logFC = 0.3), while the mean linear logFC = 0.
D) Circular AR (backsplice from exon 4 to exon 3) was detected in mCRPC with amplified AR (more than 5 copies, 54/70 cases), but not in primary tumors. This result could be directly explained by the higher expression of AR in mCRPC (Wilcoxon rank-sum test P < 2.2×10−16).
E) The relative expression of circRNAs detected in urine from 10 prostate cancer patients. Linear GAPDH expression was used for normalization, and KLK3 was included as a positive control.
Related to Figure 3.
Data Availability Statement
All circRNAs and rt-circRNAs, as well as their expression patterns, can be found in our MiOncoCirc website, currently hosted at: https://mioncocirc.github.io/. The pipeline CODAC can be found at https://github.com/mcieslik-mctp/codac.